Abstract
Several members of the short-chain dehydrogenase/reductase (SDR) enzyme family play fundamental roles in adrenal and gonadal steroidogenesis as well as in the metabolism of steroids, oxysterols, bile acids, and retinoids in peripheral tissues, thereby controlling the local activation of their cognate receptors. Some of these SDRs are considered as promising therapeutic targets, for example to treat estrogen-/androgen-dependent and corticosteroid-related diseases, whereas others are considered as anti-targets as their inhibition may lead to disturbances of endocrine functions, thereby contributing to the development and progression of diseases. Nevertheless, the physiological functions of about half of all SDR members are still unknown. In this respect, in silico tools are highly valuable in drug discovery for lead molecule identification, in toxicology screenings to facilitate the identification of hazardous chemicals, and in fundamental research for substrate identification and enzyme characterization. Regarding SDRs, computational methods have been employed for a variety of applications including drug discovery, enzyme characterization and substrate identification, as well as identification of potential endocrine disrupting chemicals (EDC). This review provides an overview of the efforts undertaken in the field of virtual screening supported identification of bioactive molecules in SDR research. In addition, it presents an outlook and addresses the opportunities and limitations of computational modeling and in vitro validation methods.
Keywords: Short-chain dehydrogenase/reductase, hydroxysteroid dehydrogenase, drug development, endocrine disrupting chemicals, in silico, virtual screening
1. Introduction
1.1. The short-chain dehydrogenase/reductase (SDR) superfamily
The SDR enzyme family consists of over 47,000 members found in archaea, bacteria, and eukaryota, with more than 80 members identified in the human genome [1]. They share a common core structure, the so-called Rossmann-fold, consisting of up to seven stranded parallel β–sheets flanked by three α–helices on each side. This core structure is crucial for NAD(P)(H) binding and includes a Tyr-(Xaa)3-Lys motif essential for the catalytic center. The conserved Tyr residue acts as a catalytic amino acid promoting the proton transfer by the support of a hydrogen bond between Lys and nicotinamide ribose, which lowers the pKa of the Tyr [2]. The catalytic center is frequently occurring with a conserved Ser residue, stabilizing the bound substrate. Despite this substantial structural similarity, SDRs generally share low sequence identity of 20-30%. In addition to the classic SDRs, consisting of one globular structure, there are extended forms with additional domains fused to the N- or C-terminus [3].
SDRs are catalyzing carbonyl-alcohol oxidoreduction, isomerization, decarboxylation, epimerization, C=N reduction, enoyl-CoA reduction, dehydration, and dehalogenation reactions. They are involved in the metabolism of a wide range of molecules, including steroid hormones, oxysterols, bile acids, prostaglandins, retinoids, fatty acids, amino acids, sugars, and various xenobiotics [3]. Probably the most extensively studied SDRs are hydroxysteroid dehydrogenases (HSDs) with key roles in adrenal and gonadal steroidogenesis, including 3β-HSDs and 17β-HSDs, as well as enzymes with 3α-HSD, 11β-HSD and 17β-HSD activities catalyzing the metabolism of steroids in peripheral tissues and thereby controlling local steroid hormone action [4]. Generally, 3α-HSDs are assigned to the family of aldo-keto reductases (AKR); however, several SDRs are reported to have 3α-HSD activity such as 17β-HSD6, 17β-HSD10 or members of the retinol dehydrogenase (RODH) subfamily [5, 6].
Some of these HSDs are investigated as potential therapeutic targets for estrogen- and androgen-dependent diseases such as osteoporosis, endometriosis, and breast and prostate cancer or corticosteroid-related diseases such as dyslipidemia, visceral obesity and diabetes, wound healing, atherosclerosis, osteoporosis, glaucoma, neurodegenerative disease, and cognitive impairment [7–13].
The similarity of the core structure of various SDRs needs to be taken into account when developing specific inhibitors to avoid the inhibition of other members causing off-target effects. In this respect, a major challenge remains the identification of the substrates and functions of “orphan” enzymes with yet unknown substrates and physiological functions. Approximately 50% of the SDRs have been poorly or not investigated so far, although some of these orphan enzymes have been associated with diseases [14–17]. Molecular modeling and virtual screening (VS) approaches can not only facilitate the identification of selective inhibitors by excluding molecules that bind to off-targets, but they may also support the identification of substrates for orphan enzymes [18]. Another application of the modeling approach includes the identification of toxic industrial and environmentally relevant chemicals [19–21]. Due to their involvement in steroid biosynthesis and metabolism, SDRs represent potential sites for molecular initiating events of endocrine disrupting chemicals (EDCs) [22–27].
1.2. Computer-aided drug design
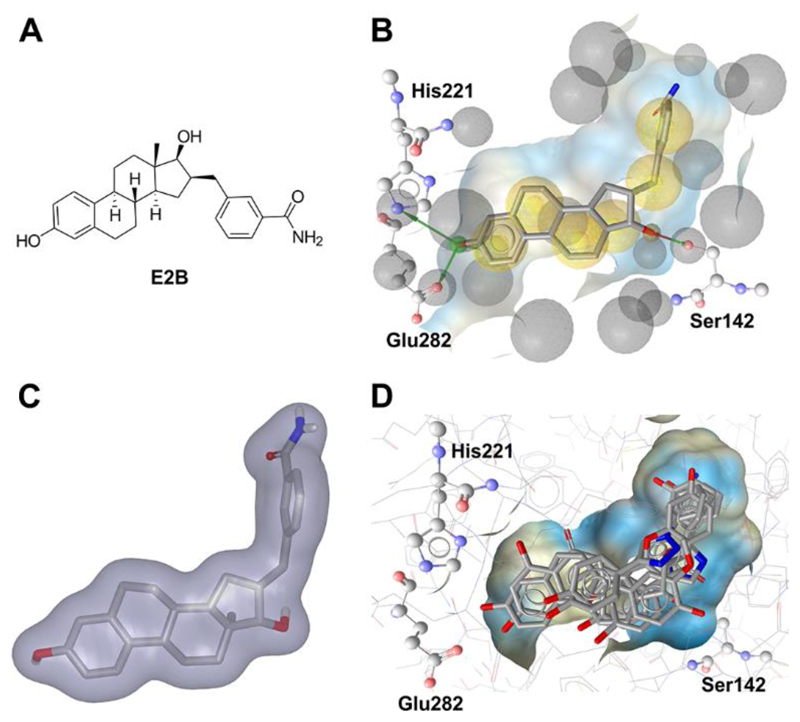
Besides experimental methods, a plethora of computational techniques is available to support the identification of novel bioactive molecules in the context of both drug discovery and toxicology. The majority of these techniques rely on the concept of similarity introduced by Johnson and Maggiora, based on the assumption that similar compounds exert similar bioactivities [28]. Computational models can be generated based on the properties of known active compounds (preferably in comparison to known inactive molecules) to search for similar compounds in large chemical databases in the course of a VS. For example, 2D similarity-based methods (Fig. 1A) can employ molecular fingerprints to represent the 2D structure of molecules. The degree of similarity among the molecules is then determined with similarity coefficients, most prominently among them the Tanimoto coefficient (Tc) [29, 30].
Figure 1.
Principles of commonly applied virtual screening tools exemplified on the crystal structure of 17β-HSD1 in complex with a steroidal inhibitor. (A) Based on the 2D structure of the inhibitor (the estradiol analogue E2B, 3-[3′,17′β-dihydroxyestra-1′,3′,5′(10′)-trien-16′β-methyl]benzamide, PDB code 3HB5 [211]), structurally similar compounds can be retrieved from a compound database in the course of a 2D similarity-based search. (B) A pharmacophore model can be created based on the ligand-target interactions patterns in the crystal complex. Exclusion volumes (Xvols, gray spheres) can be added on residues lining the binding site, thereby mimicking the steric constraints of the pocket. Red arrows: HBA, green arrows: HBD, yellow spheres: H features. (C) The shape of the inhibitor defines the 3D space in which other active chemicals may fit. (D) Diverse compounds from a chemical database docked into the binding pocket of 17β-HSD1.
Structure-based pharmacophore models (Fig. 1B) use a higher degree of abstraction, as they solely represent the interaction patterns between a ligand and its macromolecular target. According to IUPAC, pharmacophore models are defined as “the ensemble of steric and electronic features that is necessary to ensure the optimal supra-molecular interactions with a specific biological target and to trigger (or block) its biological response” [31]. These features do not describe specific functional groups, but the type of interactions these chemical functionalities can be involved in. For example, many pharmacophore modeling tools include hydrogen bond donor (HBD) and acceptor (HBA), hydrophobic (H), positively (PI) or negatively (NI) ionizable, and aromatic (Aro) features in their default settings [32–35]. In addition, some types of steric constraints, either shape or exclusion volumes (XVols) (or both) are commonly available. XVols for example can be added to mimic the binding site and to prevent the mapping of compounds that would clash with the binding site and therefore be inactive. The shape of a known active molecule can also be added to a model to restrict the virtual hits to those with similar volumes and geometries compared to the initial training compound. A scoring function is then employed to calculate how well a compound geometrically fits a pharmacophore model. Widely used pharmacophore modeling programs include Phase (Schrödinger Inc.), DS Catalyst (Biovia), LigandScout (Inte:Ligand GmbH), Molecular Operating Environment (MOE), and others (as reviewed in [36]).
Shape-based methods (Fig. 1C) in principle rely on the shape similarity between a query compound and the molecules under investigation to prioritize compounds for biological testing. Additionally, many shape-based modeling tools provide the option to include chemical information such as pharmacophore features [37, 38] (also referred to as color features in ROCS [39, 40]), atom types as in Phase Shape [41], or electrostatic potentials in ShaEP [42]) to improve the performance of the shape model. Similar to pharmacophore modeling, scoring functions are employed to determine the degree of shape overlay and, if applicable, the extent to which a compound fulfills additional requirements of the model.
Other than the methods mentioned so far, docking does not rely on the concept of similarity, but rather aims to calculate the free binding energy between a macromolecular target and potential ligands. For this purpose, the molecules under investigation are placed within the empty binding pocket of the target, which needs to be defined by the user prior to docking. Each created docking pose is then evaluated with a score estimating the binding energy [43], thus predicting the likelihood of binding (Fig. 1D). For a comprehensive description of docking and scoring as well as frequently used programs, a recent work by Sotriffer is recommended [44]. In some docking programs, also the flexibility of amino acids in the binding site is considered, e.g. in GOLD [45]. The in silico methods described in this section have also been employed for the investigation of SDRs. Successful application examples for selected SDRs are described in detail in the following sections.
2. Examples from the SDR family
2.1. Drug development
2.1.1. 11β-hydroxysteroid dehydrogenase type 1
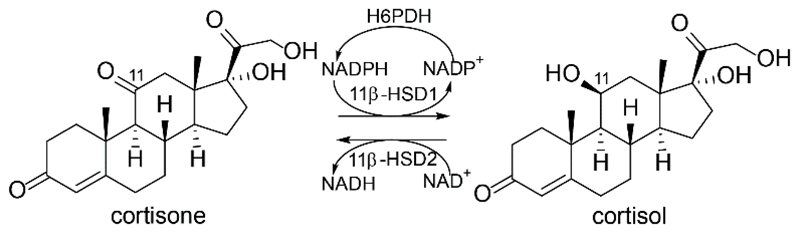
The inactive glucocorticoid cortisone is converted to the biologically active cortisol by 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) using NADPH as cofactor (Fig. 2). 11β-HSD1 is expressed in tissues such as liver, adipose tissue, adrenals, skeletal muscle, skin, pancreas, hippocampus, as well as in macrophages [46, 47]. The reverse reaction is catalyzed by 11β-HSD2, thereby ensuring mineralocorticoid receptor (MR) activation by aldosterone in kidney and colon as well as fetal protection in the placenta from excess amounts of maternal glucocorticoids [48]. Transgenic mice selectively overexpressing 11β-HSD1 in the adipose tissue develop metabolic syndrome including insulin-resistant diabetes, hyperlipidemia, hypertension, and visceral obesity [49, 50], whereas hepatic overexpression of 11β-HSD1 caused metabolic syndrome without obesity [51]. Enhanced 11β-HSD1 expression can also be detected in adipose tissue of obese patients and in skeletal muscles of diabetic patients [52–56]. In contrast, 11β-HSD1 knock-out mice were found to be resistant against the development of diet- or stress-induced diabetes [57], suggesting pharmacological inhibition of this enzyme as a therapeutic option for metabolic diseases. Furthermore, inhibition of 11β-HSD1 showed favorable therapeutic effects in wound healing [58, 59], skin aging [60], osteoporosis [8, 61], atherosclerosis [62–65], glaucoma [66–68], and cognitive functions [69–73]. Although various 11β-HSD1 inhibitors have been reported and a few also have entered clinical trials, no 11β-HSD1 inhibitor has reached the market so far [74]. Structural variety is prevailing between the different 11β-HSD1 inhibitors; nevertheless, the crystalized protein structures are comparable [75]. However, for selection of a protein structure for in silico evaluations, the detected dissimilarities upon ligand binding should be considered. The protein data bank (PDB) currently contains 29 human, 5 mouse, and 3 guinea-pig 11β-HSD1 crystal structures. To date, only X-ray crystal structures in complex with inhibitors but not with a substrate are accessible for the human isoenzyme. In contrast, no crystal structure of 11β-HSD2 is available to date.
Figure 2.
Conversion of cortisone to cortisol by 11β-HSD1 using NADPH, supplied by hexose-6-phosphate dehydrogenase (H6PDH), and oxidation of cortisol to cortisone catalyzed by 11β-HSD2 using NAD+ as cofactor.
Schuster et al. first reported the use of pharmacophore modeling to identify structurally new classes of 11β-HSD1 inhibitors [76]. To perform VS of different databases, they designed two ligand-based multi-feature pharmacophore models as the 11β-HSD1 protein structure was not experimentally resolved at the beginning of their study. They classified their pharmacophore models according to the 11β-HSD activity of the training compounds employed for the model generation as 11β-HSD1-selective and 11β-HSD non-selective. The virtual hits found by the pharmacophore models contained steroid-like compounds, the known unselective 11β-HSD inhibitor glycyrrhetinic acid (GA), related triterpenoids, and novel structural classes. The in vitro 11β-HSD1 and 11β-HSD2 inhibition profile of the active VS hits showed similar selectivities as the training set compounds used for the 11β-HSD1-selective and the 11β-HSD-unselective pharmacophore model construction.
Suitable enzyme activity assays are fundamental for selectivity testing of potential inhibitors. Regarding 11β-HSD1 inhibitors, 11β-HSD2 is usually chosen as counter screen because cross-inhibition of 11β-HSD2 would cause cortisol-induced MR activation in the kidney, resulting in hypertension. Taking into account that the different SDR family members share considerable structural similarity, but rather low primary sequence similarity, other enzymes such as 3β-HSDs and 17β-HSDs should be included in the selectivity assessment of 11β-HSD1 inhibitors.
By determining the biological activities in HEK-293 cell lysates or intact cells expressing human recombinant 11β-HSD1, 11β-HSD2, 17β-HSD1 or 17β-HSD2, Schuster et al. tested the potency and selectivity of their VS hits [76]. 11β-HSD1 activity was inhibited by more than 70% at 10 μM by 7 out of 30 tested compounds, but only 3 of them displayed reasonable selectivity over the other tested SDRs. This is not surprising as some of the unselective hits belonged to the triterpenoids resembling GA. The authors observed similar kinetic parameters for the three 11β-HSD1-selective chemicals in differentiated mouse adipocytes and myotubes, metabolically relevant tissues endogenously expressing 11β-HSD1. Other studies showed significant species-specific variability in the potency of various 11β-HSD1 inhibitors [77–79], indicating significant differences in the 3D organization of the hydrophobic substrate-binding pocket of human and mouse 11β-HSD1. Thus, species-specific variability must be considered and the use of suitable human cell lines endogenously expressing 11β-HSD1 is indicated.
Glucocorticoids are recognized by several different proteins during synthesis (CYP11B1), distribution (cortisol-binding globulin, transport proteins such as P-glycoprotein), peripheral metabolism (11β-HSDs), receptor action (MR, glucocorticoid receptor (GR)), and degradation (5β-reductase, CYP3A4). These proteins recognize some common structural features and inhibitors of 11β-HSD1 might therefore bind to other glucocorticoid recognizing proteins as well. To address this, Schuster et al. tested their most active hits in GR- and MR-dependent reporter gene assays. Reduced tissue-specific glucocorticoid reactivation and therefore blockade of the GR-mediated gene expression are responsible for the therapeutic effects of an 11β-HSD1 inhibitor. The ability of an 11β-HSD1 inhibitor to also act as a GR or MR antagonist would rather enhance its therapeutic benefit by reducing GR-dependent stimulation of hepatic gluconeogenesis and decreasing cortisol-mediated MR activation in macrophages. Their most active compound showed only weak GR and MR antagonistic effects, with a 6-10-fold preference for 11β-HSD1 inhibition and can therefore be used as starting point for further investigations.
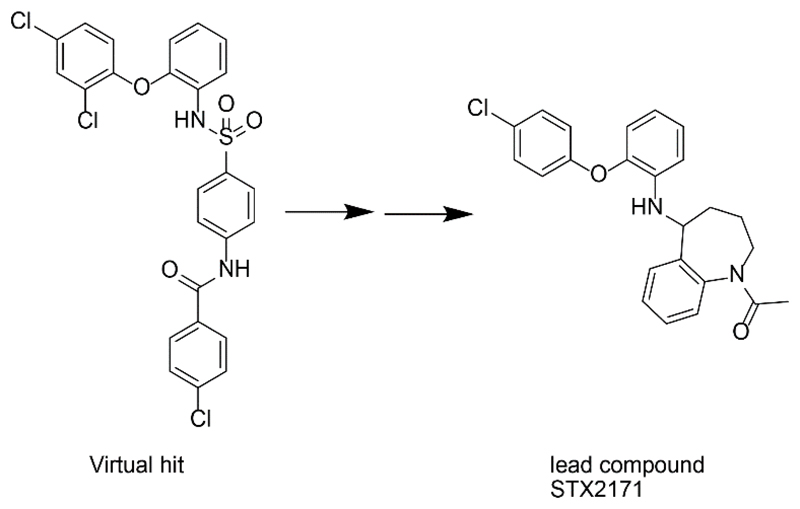
The selective 11β-HSD1 pharmacophore model generated by Schuster et al. [76] was further used to assess its potential to identify new lead structures for 11β-HSD1 inhibitor development [80]. Enzymatic testing of the virtual hits led to the discovery of an 11β-HSD1 inhibitor with an IC50 of 4.8 μM. Lead optimization studies revealed arylsulfonylpiperazine scaffolds as a new class of selective 11β-HSD1 inhibitors.
This pharmacophore model was additionally employed to search for selective 11β-HSD1 inhibitors derived from constituents of medicinal plants [81]. The virtual hit list contained to a large extent scaffolds from the chemical class of triterpenoids such as corosolic acid. This is a known constituent of Eriobotrya japonica, which is used in the traditional Chinese medicine as antidiabetic treatment. An earlier study, investigating the potential of extracts from traditionally used antidiabetic medical plants to inhibit 11β-HSD1 activity, found leave extracts of E. japonica preferentially inhibiting 11β-HSD1 over 11β-HSD2 [82]. Thus, the VS hit corosolic acid was tested for inhibition of the human 11β-HSD enzymes in a lysate-based assay and revealed selective inhibition of 11β-HSD1 with an IC50 of 810 nM. In order to discover additional secondary metabolites inhibiting 11β-HSD1, bioassay-guided phytochemical analyses were implemented. These investigations led to the identification of several molecules from the triterpenoid ursane type with IC50 between 1.9 and 17.4 μM. However, an enhanced 11β-HSD1 inhibitory activity could be detected by mixtures of these moderately active compounds. Additive effects of constituent mixtures are a common finding in phytotherapy and often explain their therapeutic effect. Binding mode prediction performed by docking studies indicated a flipped interaction pattern of the triterpenoids with interactions to Thr124 and Tyr177 instead of the catalytic residues. The identification of 11β-HSD1 inhibitors in traditionally used antidiabetic medical plants indicated a possible mode of action – a further application field for pharmacophore modeling.
Further studies using the 11β-HSD pharmacophore models from Schuster et al. [76] for model refinement [83] and subsequently as screening tool for 11β-HSD1 inhibitor identification among constituents of the traditionally used Greek medical plant Pistacia lenticus [84], are described in the supplementary information.
For the identification of new 11β-HSD1 inhibitors Miguet et al. [85] developed a homology model to predict the 3D structure of 11β-HSD1. Structure-based VS of a reference database composed of molecules with known activities towards 11β-HSD1 was used to validate the model showing its ability to discriminate between 11β-HSD1 inhibitors. The reference database included 19 11β-HSD1 inhibitors, 3 weak inhibitors, 3 non-inhibitors, and 2 substrates. To distinguish between virtual hits based on activity data of a reference database, it would be advantageous if the different activity categories would be more equal. Scoring calculations were further used as numerical cut-offs, filtering the virtual hit list after VS of a natural molecules database. As in the meantime, the first experimentally derived 11β-HSD1 X-ray structure became available, the results derived from the homology model were confirmed by molecular modeling based on the crystallographic structure. Several hits of the VS belonged to the flavonoids, with 2 hits already known as 11β-HSD1 inhibitors. The remaining candidates were not enzymatically tested, and, unfortunately, no follow up evaluation of these hits was reported.
Yang et al. combined ligand-based pharmacophore modeling and molecular docking for the identification of synthetic 11β-HSD1 inhibitors [86]. In a virtual docking approach, the SPECS database was screened and the 3000 compounds with the highest docking score were selected for a second, more computationally expensive docking calculation. Furthermore, a ligand-based pharmacophore model on the basis of three selective 11β-HSD1 inhibitors was generated and used as a query to additionally filter the 3000 selected compounds. High fit and docking scores, as well as drug likeness were selection criterions for compounds to be further biologically tested for their activity on human and mouse 11β-HSDs. Therefore, a scintillation proximity assay (SPA) was performed using microsomes prepared from HEK-293 cells stably expressing human and mouse 11β-HSD1 or 11β-HSD2, respectively. Significant differences in the inhibitory potential of the compounds were observed when comparing their activities against human and mouse 11β-HSD1. Whereas 11 out of 121 tested compounds revealed IC50 values of 0.26 – 14.6 μM against the human enzyme, 6 substances showed IC50 values between 0.48 – 12.49 μM against the mouse enzyme. Among these inhibitors, only two displayed overlapping activity for human and mouse 11β-HSD1 with IC50 values of 0.69 μM and 3.57 μM, and 0.48 μM and 2.09 μM, respectively. In regard to subsequent animal studies, selectivity over 11β-HSD2 was tested only for the mouse isoenzyme and just for compounds inhibiting mouse 11β-HSD1. Selectivity was ensured; however, appropriate selectivity determination requires at least the inclusion of human 11β-HSD2, and ideally also other SDRs. The ideal case for preclinical assessments in drug development would include cross-species activity. Importantly, significant species-specific differences regarding the potency of diverse 11β-HSD1 inhibitors have previously been reported, implying critical variability in the 3D conformation of the active site of human and mouse 11β-HSD1 [77–79].
In a consecutive study, Yang et al. successfully used 11β-HSD1 structure-based pharmacophore models as initial screening tools, followed by a docking approach for hit selection [87]. Only compounds interacting with the catalytic residues Tyr183 and Ser170 were chosen after the docking evaluation for further biological assessment. In contrast to their earlier study where they found 11 out of 121 hits as inhibitors for the human 11β-HSD1 [86], 9 out of 56 tested compounds displayed selective and dose-dependent 11β-HSD1 inhibition with IC50 values of 0.85 - 7.98 μM. The mouse enzyme was inhibited by 6 compounds with IC50 values between 0.44 μM and 8.48 μM, of which 4 inhibited human and mouse 11β-HSD1 with comparable IC50 values.
Shape-based screening combined with fast rigid docking was applied by Xia et al. to find 11β-HSD1 inhibitors [88]. The 1000 best ranked compounds of each screening were combined for further ligand-flexible docking calculations. By manual inspection of the top 200 molecules in the final hit list, 70 structurally diverse molecules were chosen for biological testing by SPA, of which 14 compounds inhibited 11β-HSD1 by more than 50% at 1 μM, 8 of them had IC50 values ≤100 nM, and 3 inhibitors already being reported [89–91]. In addition, by analyzing the binding mode conformations, a new hydrophobic sub-pocket was discovered. However, the interacting residues of this sub-pocket were not described, although this would further support inhibitor development. During the validation of this finding with a molecule fitting into this pocket, a novel scaffold was identified that inhibited 11β-HSD1 with an IC50 of 45 nM. Selectivity over 11β-HSD2 was only verified for the two compounds with the most favorable ADME prediction profiles. However, since pharmacokinetics can be improved after lead identification, it would be of interest to obtain selectivity information for all 11β-HSD1 inhibitors. Moreover, to improve further study designs, it would be important to know the difference between the hit rates of shape-based screening and rigid docking. In an independent, subsequently performed study, Xia et al. designed a new class of derivatives of 1-arylsulfonyl piperidine-3-carboxamides using medicinal chemistry tools [92]. For lead structure selection and following animal studies, the compounds were tested against mouse and human 11β-HSD1 and 11β-HSD2. They found a large lipophilic group at the amino moiety as favorable for cross-species potency. Unfortunately, they did not mention if this bulky lipophilic group also targets their previously identified hydrophobic sub-pocket, which could be interesting for further inhibitor development studies.
A similar approach was performed by Lagos et al. who used shape-based query hypotheses as a filter during a structure-based VS process [93]. Steroidal compounds were excluded from the query in order to avoid similarity of the virtual hits with this scaffold type. Top scored compounds were visually analyzed for their binding features and selected for testing in cell-based assays. For this purpose, they used the liposarcoma-derived adipose cell line LS14, differentiated into adipocytes and endogenously expressing 11β-HSD1 [94]. Of 39 compounds tested, two selectively inhibited 11β-HSD1 over 11β-HSD2 with IC50 values around 5 μM. Selectivity over 11β-HSD2 was also tested in differentiated LS14 cells, although its expression was marginal. However, the use of an intact cell-based testing system as an initial biological assessment tool has the disadvantage that the compounds do not have direct access to their target and the ranking of the obtained biological activities cannot be used to draw conclusions on the performance of the VS approach.
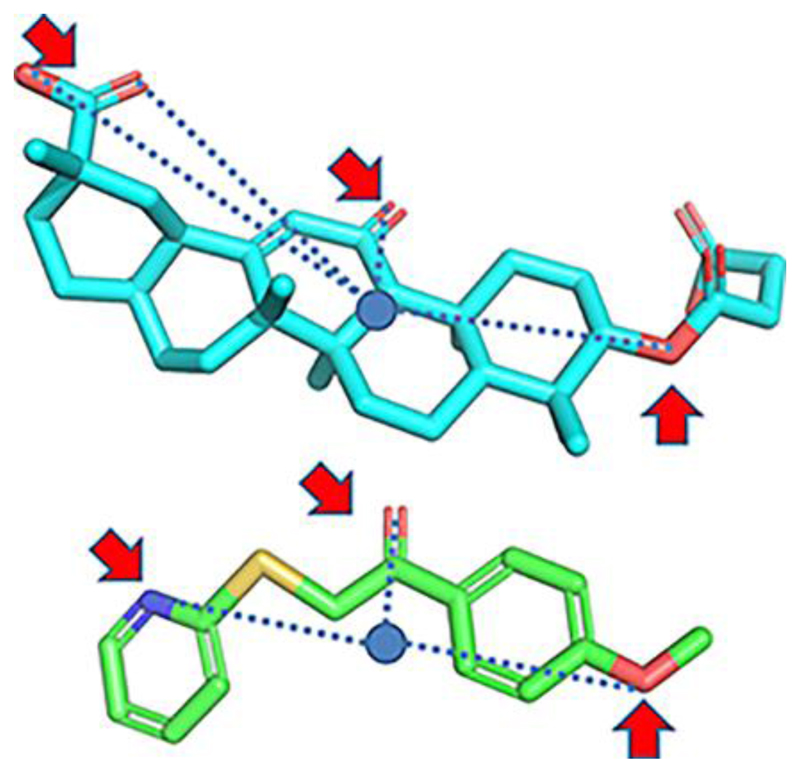
Shave et al. employed another method containing shape-based calculations but without the need for any detailed structural information of the target [95]. They used Ultra-fast Recognition with Atom Types (UFSRAT), an algorithm that considers the shape and the electrostatics of atoms to score and retrieve candidate molecules capable to make similar interactions to those of the supplied query. The non-selective 11β-HSD1 inhibitor carbenoxolone was used to generate the query. VS of a database against the query resulted in a hit list of the most similar compounds. Biological testing was implemented against 11β-HSD1 reductase and dehydrogenase activity. Out of 26 tested compounds, 4 inhibited the reductase activity in a SPA cell-based assay with IC50 values between 0.067 and 11.3 μM and the dehydrogenase activity of recombinant human 11β-HSD1 protein with Kiapp of 26 - 248 μM. Interestingly, the top virtual hits displayed totally different scaffolds compared to the query molecule but showed similarity to already known 11β-HSD1 inhibitors (Fig. 3). Thus, UFSRAT has demonstrated its ability for scaffold hopping during the VS screening process. However, the query molecule carbenoxolone is an unselective 11β-HSD inhibitor, exhibiting activity against 11β-HSD1 and 11β-HSD2. For this reason, biological testing of the selected virtual hits against 11β-HSD2 would be required for further development of these compounds. Moreover, choosing a selective 11β-HSD1 inhibitor as a query molecule for the UFSRAT algorithm may even improve the success of this approach.
Figure 3.
Carbenoxolone (top) and hit molecule with a preserved key pattern of atoms involved in hydrogen bonding [95].
A number of adamantine-containing selective 11β-HSD1 inhibitors have been reported [96]. Tice et al. used them to generate models of the 11β-HSD1 binding site with the help of a proprietary structure-based drug design program of Vitae Pharmaceuticals called Contour [97]. The most satisfactory poses were selected, and based on them a medicinal chemistry program was initiated to improve potency, selectivity, and physical properties, supported by additional modeling. This led to the identification of a class of spirocyclic ureas selectively inhibiting 11β-HSD1 with IC50 values in the lower nanomolar range.
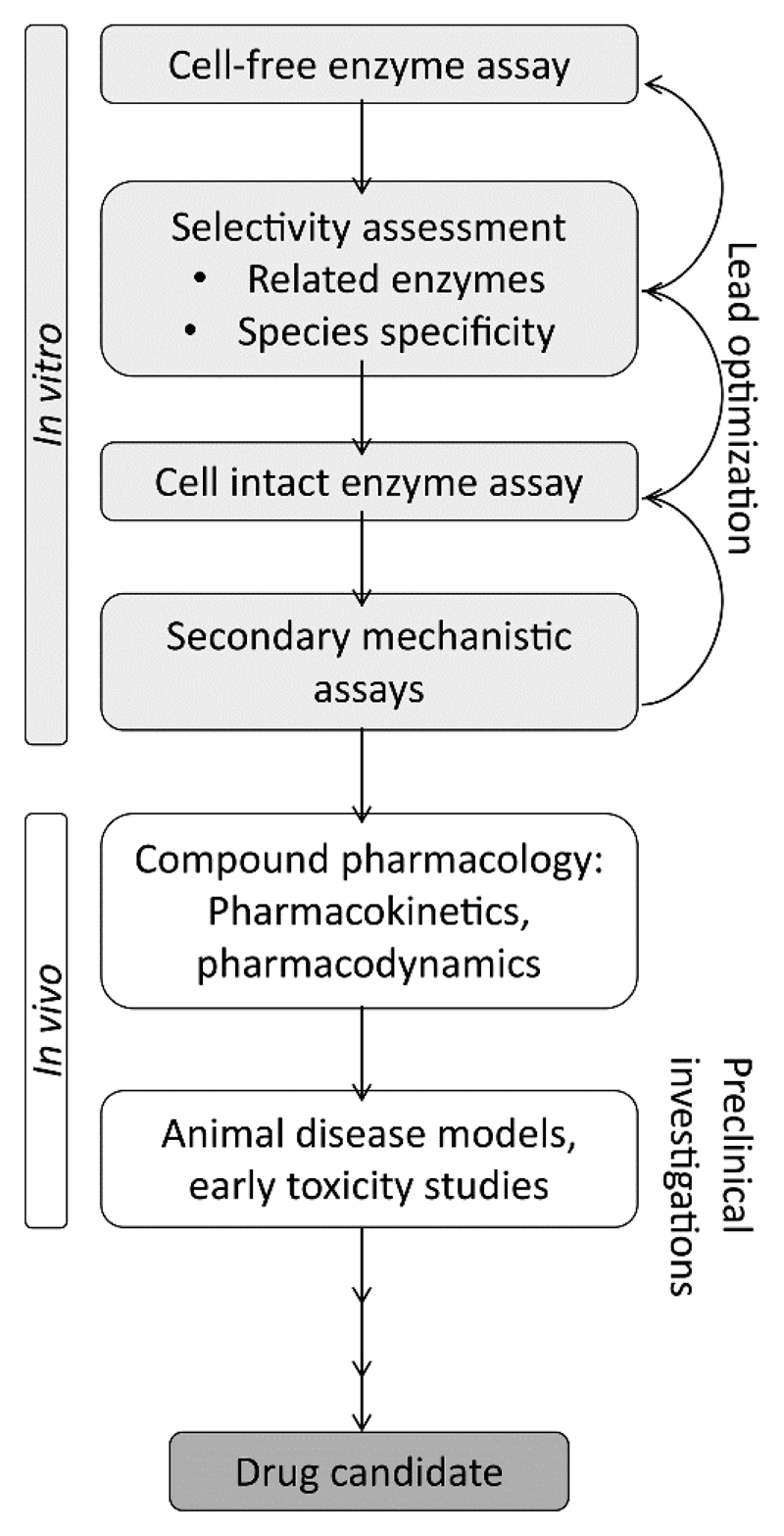
Using the same drug design platform Contour, Xu et al. developed a novel class of 11β-HSD1 inhibitors by incorporating a 1,3-oxazinan-2-one ring system [98]. Prior to the medicinal chemistry program, the available 11β-HSD1 X-ray structures were examined and a template compound bearing a 1,3-oxazinan-2-one ring docked into the 2BEL structure of 11β-HSD1. Subsequently, more than 5000 molecules were designed in silico by adding fragments directly to the template compound. Structure-based drug design and lead compound optimization studies revealed a highly potent 11β-HSD1 inhibitor with IC50 values of 0.8 nM using recombinant human 11β-HSD1 in a microsomal preparation of CHO cells and 2.5 nM in differentiated human adipocytes. Testing inhibitory activity against 11β-HSD2, 17β-HSD1, 3β-HSD2, and three CYP isoenzymes showed >1000-fold selectivity for 11β-HSD1. The same results were observed when examining its potential to bind to GR, MR, FXR or hERG. In regard to subsequently performed animal studies, pharmacokinetic parameters were measured in several species. In addition, distribution into mouse adipose tissue could be observed with proportional plasma concentrations levels and three times higher concentrations in the liver. However, due to the poor potency against mouse 11β-HSD1, but comparable activity towards human and cynomolgus monkey 11β-HSD1, the latter species was selected as in vivo model for 11β-HSD1 inhibition. Oral administration, after suppression of endogenous plasma glucocorticoid levels with dexamethasone and challenge with cortisone 21-acetate after 5 h of compound administration, revealed reduced cortisol production by 85% compared to the vehicle control. However, the authors described no further details how the animal study was conducted as for instance the number of animals used. As they aimed to specifically target the adipose tissue, this measurement only provides data about the overall 11β-HSD1 activity. Therefore, they determined in a consecutive preclinical characterization study the inhibitory activity of their lead compound ex vivo in cynomolgus monkey and human adipose tissue [99]. Remarkably, the enzyme inhibition was minor in cynomolgus monkey tissue and 30-fold less pronounced in human adipose tissue compared to cultured differentiated preadipocytes. They proposed the high lipophilic nature of the compound and therefore its uptake and sequestration into lipid droplets as a possible reason for this observation. Based on these investigations, they established a modified assay strategy for lead compound identification, newly including analysis in human and non-human primate adipose tissue. This approach led to the identification of a new 11β-HSD1 inhibitor candidate, of which toxicological assessment was introduced in regard to Phase I clinical studies. An adapted, more general version of this testing cascade is shown in the biological limitation section.
Several different computational methods have been applied to discover new selective 11β-HSD1 inhibitors and successfully identified structurally diverse virtual hits in biological assays as potential lead compounds for further drug development. However, computer-aided drug design is also advantageous for lead optimization. In order to improve selectivity, potency and pharmacokinetic parameters of initially discovered 11β-HSD1 inhibitors, several groups implemented scaffold hopping and structure-activity relationship (SAR) studies on the basis of docking studies [100–105]. Following chemical synthesis the same approach is often also used for binding mode explanations [106–108].
2.1.2. 17β-hydroxysteroid dehydrogenase type 1
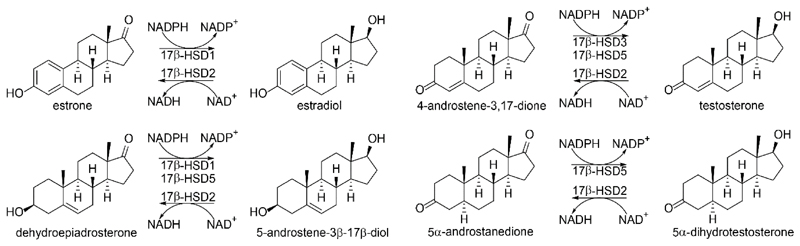
To date, 14 different human 17β-hydroxysteroid dehydrogenase (17β-HSD) enzymes, belonging to the SDR family with the exception of the aldo-keto reductase AKR1C3 (17β-HSD5), have been described [109]. Several of the 17β-HSDs are essentially involved in the local metabolism of estrogens and androgens, thereby controlling ER and AR signaling in a tissue- and cell-dependent manner (Fig. 4).
Figure 4.
Selected 17β-HSDs involved in estrogen and androgen steroid metabolism.
17β-HSD1 reduces in an NADPH-dependent reaction the weak estrogen estrone to the potent estradiol. It also catalyzes the conversion of dehydroepiandrosterone (DHEA) to 5-androstene-3β,17β-diol [110]. The human placenta, ovaries, and mammary gland are the predominant expression sites of 17β-HSD1 and therefore of considerable relevance for the gonadal and peripheral synthesis of estradiol [111]. Studies demonstrating a correlation of 17β-HSD1 mRNA expression levels and poor breast cancer prognosis [112–114] suggest the local inhibition of estradiol biosynthesis by targeting 17β-HSD1 as a promising therapeutic strategy against breast cancer, especially in postmenopausal women, where estradiol originates mainly from extragonadal sites. Importantly, in vivo studies found a reduction in tumor size in mice stimulated with exogenous estrone after co-treatment with specific 17β-HSD1 inhibitors [115, 116]. In addition, high 17β-HSD1 expression levels were shown to be associated with endometriosis [117, 118], endometrial cancer [119], and uterine leiomyoma [120], offering additional therapeutic opportunities.
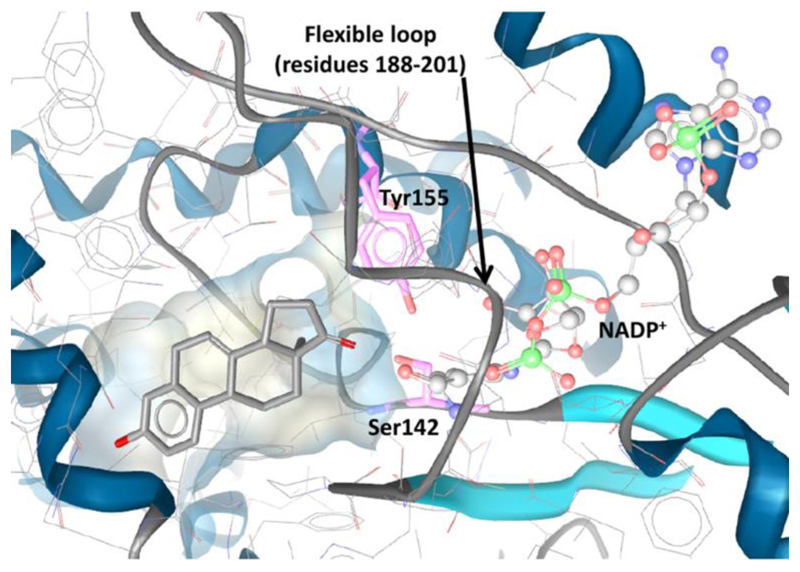
Although the number of reported 17β-HSD1 inhibitors is increasing, to date no compound made it into clinical trials. Currently, over 20 crystal structures of the 17β-HSD1 protein have been published. The binding pocket consists of an elongated hydrophobic channel, formed by Leu149, Val225, Phe226, and Phe259, and hydrophilic residues at each end allowing interactions with the catalytic essential residues Ser142 and Tyr155 on one side and His221 and Glu282 on the other side. A flexible loop (amino acids 188-201) in the crystal structure, which compromises the exact definition of the substrate binding pocket and therefore influences the predictivity of VS studies, is not well resolved [121] (Fig. 5).
Figure 5.
Substrate binding pocket of 17β-HSD1 with the co-crystallized ligand equilin (PDB 1EQU), catalytic key residues (Ser142 and Tyr155), a flexible loop (residues 188-201) and NADP+.
Hoffrén et al. were the first to describe structure-based pharmacophore models for the discovery of 17β-HSD1 inhibitors [122]. They validated their pharmacophore models with molecules bearing the structural and chemical features of steroids and flavonoids. The most potent training compound applied in the model validation was coumestrol. However, coumestrol also displays inhibitory activity against 17β-HSD5 and is not selective for 17β-HSD1, which needs to be taken into account in model and hit validation [123]. The selectivity and sensitivity of a pharmacophore model strongly depend on the compounds selected for its generation and validation. Ideally, if available, selective inhibitors are chosen for model development. Thus, because phytoestrogens and steroidal scaffolds frequently display cross-reactivity against other enzymes and receptors involved in steroid action, non-steroidal structures are preferred, not only for the modeling and validation, but also as lead structures to increase selectivity. Since the VS hits from Hoffrén et al. were not validated by biological testing, no further information is available on the selectivity of the pharmacophore model as well as on the identified hits.
Even though selective inhibitors are usually chosen for therapeutic applications, polyvalent inhibitors with synergistic beneficial effects may be advantageous in some circumstances. Chanplakorn et al. reported a significant increase of estrogen sulfatase and 17β-HSD1 expression after neoadjuvant therapy with aromatase inhibitors (AIs) in postmenopausal women suffering from estrogen receptor-α (ERα)-positive breast cancer [124]. They proposed that the observed expression changes are a result of compensatory responses to estrogen depletion in breast carcinoma tissue. To prevent the compensatory estradiol production triggered by chronic treatment with AIs, Dual Aromatase-Sulfatase Inhibitors (DASI) have been developed as an alternative to administration of a combination of drugs for each target [125–127]. Aromatase catalyzes the conversion of 4-androstene-3,7-dione to estrone, which is then conjugated by estrogen sulfotransferase to estrone sulfate that can serve as a storage upon hydrolysis in breast cancer tissue by steroid sulfatase (STS), and further reduction by 17β-HSD1 leads to estradiol production. Thus, hormone-dependent breast cancer might be more effectively treated using a polyvalent drug. Designing these DASIs, Woo et al. integrated the inhibitory STS pharmacophore into the scaffold search for AIs, allowing minimal structural changes to preserve aromatase inhibition [128]. Thus, the use of specific inhibitors for each relevant target as well as inhibitors with activities against synergistic targets represents a promising approach to prevent the development of resistance.
Focusing exclusively on 17β-HSD1, inhibitors can target several sites including reversible and irreversible inhibition of the binding of the substrate, of the cofactor NADPH at the Rossmann-fold, or both by so-called hybrid compounds (Fig. 6) consisting of a steroidal core and extended side chains to occupy the cofactor binding site [129, 130].
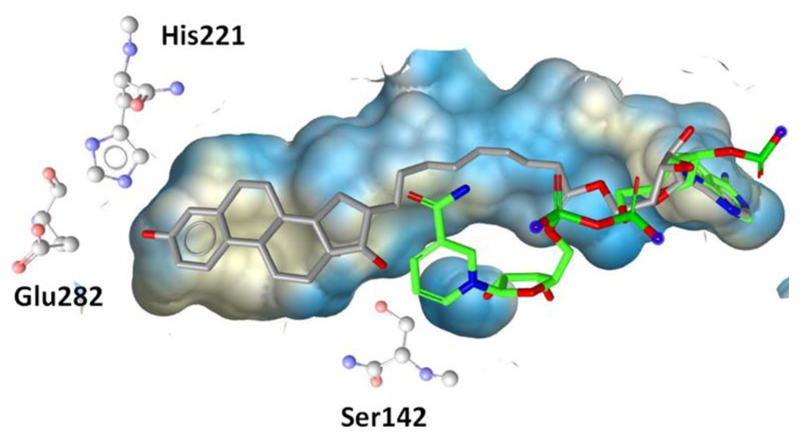
Figure 6.
The hybrid inhibitor EM1745 (gray) occupies both the steroid and the co-factor (green) binding site in 17β-HSD1 (PDB entry 1I5R). The co-factor conformation was taken from the PDB entry 3HB5.
Due to the lack of 17β-HSD1 X-ray structures co-crystallized with nonsteroidal inhibitors, Schuster et al. constructed two structure-based pharmacophore models based on crystal structures containing steroidal inhibitors in order to find new nonsteroidal 17β-HSD1 inhibitor scaffolds [131]. Whereas one model was developed based on the steroidal scaffold equilin and expected to be an appropriate general screening tool, yielding a higher number of false positive and unselective hits, the second model was constructed based on a hybrid inhibitor, suggested to be more restrictive because of the underlying unique scaffold. In vitro testing of 14 selected virtual hits led to the identification of two nonsteroidal, selective 17β-HSD1 inhibitors with moderate activities (IC50 5.7 μM and 19 μM). Selectivity was tested against 17β-HSD2, 17β-HSD3, the AKR 17β-HSD5, 11β-HSD1, and 11β-HSD2, additionally revealing a nonsteroidal and a steroidal 11β-HSD1 inhibitor (IC50 6.2 μM and 3.8 μM, respectively) and a nonsteroidal 17β-HSD3 inhibitor (IC50 19 μM). These results emphasize the relevance of including several structurally related enzymes for selectivity evaluation. To increase the exclusion rate of compounds potentially causing off-target effects, Schuster et al. tested an alternative approach using pharmacophore models of structurally related enzymes [131]. Using these models as additional filters to exclude compounds with a low degree of selectivity enriched the virtual hit list with more selective compounds and therefore reduced the efforts for laborious biological testing. They applied their previously constructed selective 11β-HSD1 pharmacophore model for a VS of the scaffolds identified as 11β-HSD1 inhibitors [76]. The nonsteroidal 11β-HSD1 inhibitor mentioned above was identified as hit, and it was also found when removing the shape restriction. Therefore, applying nonrestrictive pharmacophore models of related enzymes as additional filter tools can assist the selection of virtual hits for biological testing by eliminating promiscuous inhibitors.
Regarding the structural similarity of different proteins, Brown et al. demonstrated inhibition of human lactate dehydrogenase (LDH) and of 17β-HSD1 by binding of gossypol derivatives to the Rossmann fold [132]. Thus, on one hand structural conservation provides a basis for lead compounds targeting several related proteins but on the other hand it raises concerns about their selectivity. Brown et al. did not test their gossypol derivatives against other SDRs. Since gossypol was also found to inhibit 3β-HSD1, 17β-HSD3 [133], and 11β-HSD2 [134], testing of the gossypol derivatives against other structurally related enzymes will be crucial.
To create a pharmacophore model and define the ligand-protein interactions for both, the ligand and the active site of the protein, Sparado et al. [135] superimposed five 17β-HSD1 X-ray structures. VS of a small in-house compound library and experimental validation of the hits in a cell-free assay led to the identification of a moderately active 17β-HSD1 inhibitor. Further SAR analysis, including scaffold hopping and rigidification, resulted in two benzothiazole-scaffold bearing 17β-HSD1 inhibitors with IC50 values of 44 and 243 nM, respectively. The subsequent selectivity testing not only consisted of an assay for the related enzyme 17β-HSD2, but also for the ERα and ERβ. Depending on whether a compound also binds to ERα and/or ERβ and acts as agonist or antagonist, different effects on ER-mediated signaling can be expected. Both compounds showed selectivity over 17β-HSD2 but differences regarding their binding affinities against both ERs. The more potent compound displayed considerable affinity to bind to ERα and ERβ, whereas the less active compound was marginally active against both ERs. Further biological evaluation in a human cell system endogenously expressing 17β-HSD1 (T47-D cells) revealed potent inhibition of estrogen formation with an IC50 of 245 nM for the less active compound. Moreover, docking investigations revealed a 180° flipped orientation of the two compounds, although they differ only in a carbonyl and an amide-bridge, respectively. As described earlier, a flipped binding orientation was also observed for corosolic acid and other triterpenes inhibiting 11β-HSD1 [81]. To improve the activity and selectivity for in vivo use of their two 17β-HSD1 inhibitors, Sparado et al. conducted a follow-up optimization study [136]. SAR experiments led to the identification of two new lead compounds, which were highly active against 17β-HSD1 with IC50 values in a cell-free assay of 27 nM and 13 nM and in T47-D cells of 258 nM and 37 nM, respectively. Both inhibitors were selective over 17β-HSD2 and ERα/β. Furthermore, the potency of the inhibitors was tested against marmoset 17β-HSD1 and 17β-HSD2 as the marmoset monkey can be used as an animal model for endometriosis. The lead compounds almost completely inhibited 17β-HSD1 at 50 nM when tested in marmoset placenta microsomes. However, the compounds were less selective towards marmoset 17β-HSD2 compared to the human enzymes (50 nM of the compounds inhibited marmoset 17β-HSD2 activity by 51% and 40%, respectively).
Pharmacophore modeling using structure-based and ligand-based concepts were also applied by Karkola et al. [137]. Four different approaches using docking, alignment of known inhibitors, molecular dynamics (MD) simulation, and automated model generation on the basis of a 17β-HSD1 crystal structure were implemented. VS led to the discovery of several potential 17β-HSD1 inhibitors; however, their biological activities were not determined. Biological testing of these hits would allow validating them as 17β-HSD1 inhibitors and could provide information on the selectivity and sensitivity of the different pharmacophore models.
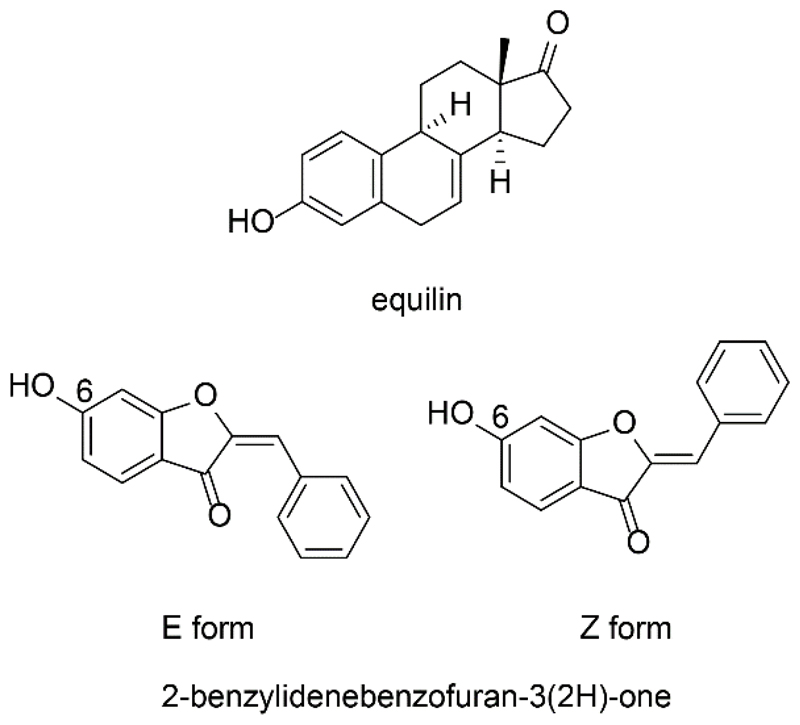
Starčević et al. [138] performed a virtual high-throughput screening based on the 3D structure of 17β-HSD1 in complex with equilin. The database was pre-filtered to reduce it to compounds with similar size and shape as estrone. Concerning the large scaffold of hybrid inhibitors, this approach may bias the VS and its hit list, and potentially active hits may be missed. During the visual inspection of the virtual hit list, compounds with potential estrogenic effects such as steroids, flavonoids, or other phytoestrogens were eliminated. Of 18 enzymatically tested substances, three compounds bearing the central scaffold of aurones, a 2-benzylidenebenzofuan-3(2H)-one structure, showed potent inhibition with IC50 values in the lower nanomolar range (Fig. 7). Additionally, one hit was an already known 17β-HSD1 inhibitor, thus validating the approach. However, a 2D similarity search with the aurone derivatives as queries identified no new 17β-HSD1 inhibitors. A SAR analysis revealed the presence of a 6-OH group as essential for potent 17β-HSD1 inhibition by 2-benzylidenebenzofuan-3(2H)-ones. Docking studies suggested that these inhibitors occupy an ideal orientation in the active site by forming triple hydrogen bonds with the catalytic residues Ser142 and Tyr155 and the cofactor.
Figure 7.
Equilin and E respectively Z form of 2-benzylidenebenzofuran-3(2H)-one structure.
A docking approach was also applied by Frotscher et al. [139], who studied the 3D architecture of 17β-HSD1 in complex with estradiol for important chemical features involved in the protein-ligand interactions to design steroid mimetics with nonsteroidal scaffolds. These inhibitors should contain two polar groups with 11 Å distance in between to imitate the A-ring and the D-ring and a flat conformation similar to the steroids. In addition, they discovered two residues in the active site, Tyr218 and Ser222, which are not directly involved in steroid binding but may display promising new interaction partners for the development of new inhibitors. Accordingly, phenyltetralone, phenylnaphthalene, phenylquinoline, and phenylindole scaffolds were chosen for a medicinal chemistry program with scaffold hopping and SAR analysis based on biological analysis with 17β-HSD1 and 17β-HSD2. Docking and MD simulations thereby helped to reveal the molecular interactions of the synthesized compounds with the protein. This led to the discovery of a (hydroxyphenyl)naphthalene derivative as potent 17β-HSD1 inhibitor with selectivity towards 17β-HSD2, ERα, and ERβ. Moreover, pharmacokinetic studies demonstrated Caco-2 penetration, low inhibitory effects on the most important hepatic CYP enzymes, and moderate metabolic stability in rat liver microsomes. For optimization of inhibitory activity, selectivity and pharmacokinetic properties, novel substituted 6-phenyl-2-naphtols were synthesized [140]. Molecular modeling supported SAR and binding mode explanation. The new lead compound showed improved overall properties and can be further tested in vivo. The inhibitory activity was tested only against human 17β-HSD1. Thus, prior to animal experiments, possible species-specific differences should be considered.
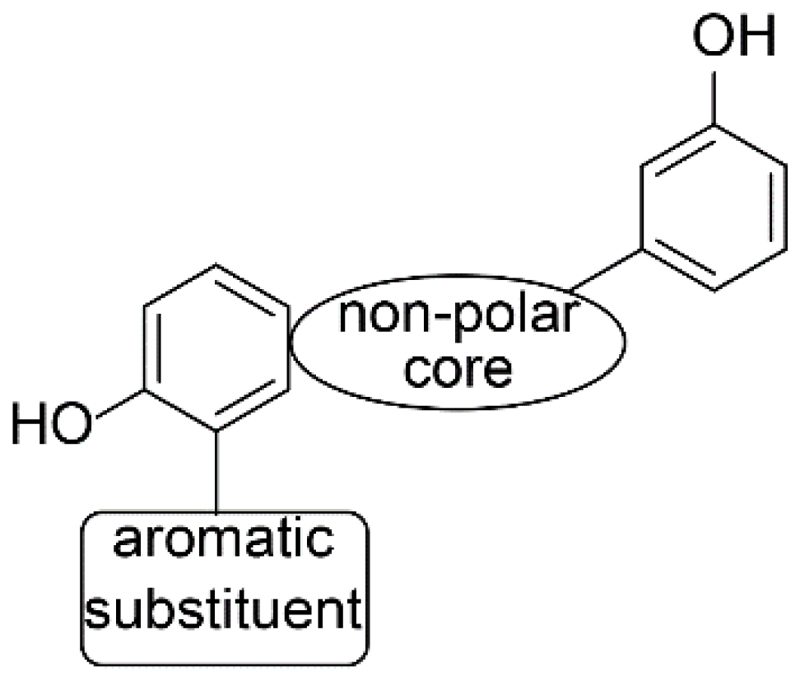
In a follow-up project, Marchais-Oberwinkler et al. [141] performed a SAR study to optimize the 17β-HSD1 pharmacophore of Frotscher et al. [139] (Fig. 8). The study highlights the restricted flexibility of the active site of 17β-HSD1. Therefore, the enzyme might not be able to adjust its geometry upon inhibitor binding. Targeting the polar ends of the active site, the positions of the hydroxyl groups of the (hydroxphenyl)naphthalene derivatives were optimized in order to allow formation of hydrogen bonds. By the introduction of a hydrophobic core, the inhibitors are stabilized in the hydrophobic tunnel of the enzyme. Furthermore, biological results and modeling studies indicated that the amino acids Tyr218 and Ser222, characterized by Frotscher et al. as potential interaction partners [139], are unlikely to form hydrogen bonds with this class of inhibitors. Available space with potential π-π interactions close to position 1 of the naphthalene ring may be exploited by introducing an aromatic substituent.
Figure 8.
Revised pharmacophore for 17β-HSD1 inhibitors (adapted from Marchais-Oberwinkler et al. [141]).
Applying a similar strategy of a ligand- and structure-based drug design mimicking steroids, Bey et al. identified bis(hydroxyphenyl) azoles as potent 17β-HSD1 inhibitors [121]. Different azoles and hydroxyl substitutions were synthesized and evaluated for activity and selectivity. Thereby, a 2,5-disubstituted oxazole displayed the most potent inhibitory effects with good selectivity and pharmacokinetic properties. Further structural optimization resulted in enhanced IC50 values in the low nanomolar range, high selectivity profiles, and good pharmacokinetic properties [142]. Interestingly, compared to the (hydroxyphenyl)naphthalene class of 17β-HSD1 inhibitors, the active bis(hydroxyphenyl) azoles were predicted to form hydrogen bonds with Tyr218.
The approach of identifying new 17β-HSD1 inhibitors by structure-based drug design was applied by several groups and led to the discovery of 17β-HSD1 inhibitors based on 2-substitutions of estrone and D-homo-estrone [143] and on pyrimidinones [144]. This strategy is commonly used in medicinal chemistry for optimization of known inhibitors and lead compounds in a rational manner. Thus, VS offers an inexpensive and rapid approach to identify novel compounds and helps to enrich compounds within a set of similar compounds from the same scaffold [145].
2.1.3. 17β-hydroxysteroid dehydrogenase type 2
17β-HSD2 catalyzes the NAD+-dependent conversion of the active estradiol into the weak estrogen estrone. Moreover, 17β-HSD2 is able to convert testosterone to 4-androstene-3,17-dione (androstenedione), 5α-dihydrotestosterone (DHT) to 5α-androstanedione, 5-androstene-3β,17β-diol to DHEA, and 20α-dihydroprogesterone to progesterone (Fig. 4) [146, 147]. Several tissues such as bone, endometrium, uterus, breast, placenta, stomach, small intestine, and colon epithelium express 17β-HSD2 [148, 149]. In the placenta, 17β-HSD2 protects the fetus from maternal androgens and estrogens. Decreased levels of estrogens in postmenopausal women and androgens in elderly men result in an imbalance between bone formation and bone loss, ultimately causing osteoporosis [150]. Among the most frequent pharmacological interventions in Europe against osteoporosis is the administration of bisphosphonates and selective estrogen-receptor modulators (SERMs) [151]. However, both treatment options have limitations and there is a great demand for novel therapies. Especially SERMs are associated with an increased risk for cardiovascular complications. Since 17β-HSD2 is expressed in osteoblasts, its inhibition may provide a new approach to treat osteoporosis by increasing local estradiol availability. This strategy is supported by an in vivo study in ovariectomized cynomolgus monkeys, where oral administration of a 17β-HSD2 inhibitor led to maintenance of bone formation and strength [152]. Nevertheless, as increased estradiol concentrations can also be related to severe disorders such as endometriosis or breast cancer, it may be important to design 17β-HSD2 inhibitors that exclusively act in bone tissue. Thus, the application route of these inhibitors might be a major challenge to overcome.
Because of the lack of 17β-HSD2 crystal structures, Vuorinen et al. applied ligand-based pharmacophore modeling to find novel inhibitors [153]. Biological testing of the VS hits in a cell-free assay revealed 7 out of 29 tested compounds (of initially 202,906 compounds subjected to in silico screening) with IC50 values between 0.24 μM and 33 μM. Among the active compounds, phenylbenzene-sulfonamides and –sulfonates displayed the main class of structural scaffolds. In addition, a search for structurally similar molecules was performed using this new class of 17β-HSD2 inhibitors. With a simple 2D similarity search, one (IC50 of 3.3 μM) out of 16 compounds was found to inhibit 17β-HSD2. In parallel, the pharmacophore model was refined, used for VS, and 14 derivatives were biologically tested. Among them, 5 hits revealed IC50 values between 1-15 μM. Although the compounds used for pharmacophore model generation were selective against 17β-HSD1, the selectivity of the newly identified 17β-HSD2 inhibitors remains to be determined. The active inhibitor-derivatives were tested for their selectivity against 17β-HSD1, 17β-HSD3, 11β-HSD1, and 11β-HSD2. Only one of the overall 13 discovered inhibitors showed activity against 17β-HSD1 (IC50 18 μM), being still 18 times more active against 17β-HSD2. Unfortunately, the most potent 17β-HSD2 inhibitor (IC50 0.24 μM) was also active against 17β-HSD3 (IC50 8.5 μM) and 11β-HSD1 (IC50 2.1 μM). Two compounds were equipotent against 17β-HSD2 and 11β-HSD1 and two substances showed even higher inhibitory activity against 17β-HSD3 than 17β-HSD2. However, all tested compounds were selective against 11β-HSD2. In addition, the specificity of the initial pharmacophore model was improved by creating a refinement database including the original training compounds, the newly discovered active compounds, as well as the inactive substances. Thus, the ability of the model was enhanced to find active hits from a database.
Wetzel et al. [154] used the findings from their previous 17β-HSD1 inhibitor study [155] for the development of a new class of 17β-HSD2 inhibitors. Their former project was based on a structure- and ligand-based design strategy including docking of two potent heterocyclic substituted biphenylol 17β-HSD1 inhibitors and evaluation of their interaction pattern in the active site of the enzyme. A three-point pharmacophore model built the basis for a medicinal chemistry inhibitor design concept. One of the most promising 17β-HSD1 inhibitors (IC50 8 nM) showed 48-fold selectivity against 17β-HSD2 (IC50 382 nM) in cell-free lysate assays. Nevertheless, this compound was selected by Wetzel et al. as starting point for 17β-HSD2 activity optimization experiments in order to gain selectivity against 17β-HSD1. However, considerably more potent 17β-HSD2 inhibitors with lower selectivity factors toward 17β-HSD1 were obtained as well. Unfortunately, the authors did not provide a reason for their particular selection of the starting compound. Structural optimization led to the discovery of bicyclic substituted hydroxyphenylmethanone derivatives as a new class of 17β-HSD2 inhibitors. The most promising compound displayed 13-fold selectivity over 17β-HSD1 with an IC50 value of 101 nM.
2.1.4. 17β-hydroxysteroid dehydrogenase type 3
17β-HSD3 reduces androstenedione to testosterone using cofactor NADPH. It is almost exclusively expressed in the testis [156]. The rare autosomal recessive disorder 46, XY disorder of sex development (also known as male pseudohermadophroditism) emphasizes the importance of 17β-HSD3 for testosterone production [157, 158]. 17β-HSD3 deficiency leads to impaired masculinization of male external genitalia and the affected individuals are born with female or ambiguous external genitalia [159]. However, even though 17β-HSD3 is predominantly expressed in testis, it was reported that 17β-HSD3 mRNA was upregulated in prostate cancer [160]. Importantly, Day et al. recently reported that treatment with specific 17β-HSD3 inhibitors significantly decreased androgen-dependent growth of xenografts expressing 17β-HSD3 in castrated mice [161]. The therapeutic efficacy of 17β-HSD3 inhibitors might be limited by the coexpression of 17β-HSD5 (AKR1C3), which catalyzes the same reaction and is expressed in prostate cancer [162, 163]. Therefore, a combined treatment targeting both enzymes should be envisaged. Like 17β-HSD2, 17β-HSD3 is anchored to the endoplasmic reticulum membrane by an N-terminal transmembrane domain, and its catalytic domain faces the cytoplasmic compartment [164, 165]. For both membrane proteins experimentally derived 3D structures are still not available. Thus, Vicker et al. built a homology model, validated by known 17β-HSD3 inhibitors, to support structure-based drug design [166]. Homology models depict the active site not as accurate as crystal structure-derived models. However, used in a docking approach, it allows for a prediction of an inhibitor’s interactions with the active site and helps to uncover the chemical features that are important for its activity. The established 17β-HSD3 homology model revealed the highly hydrophobic nature of the active site. Potential interactions include π-π interactions of aromatic rings with Phe205 and other hydrophobic interactions with residues such as Val213, Ile148, Phe151, Trp153, and Leu252. The homology model was then used for docking-based VS. Biological testing in a cell-based assay revealed a novel lead compound with an IC50 of 770 nM for 17β-HSD3, with selectivity over 17β-HSD1 and 17β-HSD2. Docking into the homology model and subsequent scaffold hopping, considering potential interactions with the active site, led to the discovery of new compounds, with the most potent inhibitor showing an IC50 of 200 nM in intact cells of human origin (Fig. 9). The activity of this 17β-HSD3 inhibitor was further confirmed in an in vivo hormone-dependent prostate cancer model, where castrated mice showed significantly decreased androgen-dependent growth of tumor xenografts expressing human 17β-HSD3 [161]. Moreover, significantly lowered plasma testosterone levels were observed in treated mice.
Figure 9.
Virtual screening hit compared to 17β-HSD3 inhibitor STX2171 found by Vicker et al. [166].
The hydrophobic character of the active site of 17β-HSD3 described by Vicker et al. was further supported by two ligand-based pharmacophore models generated by Schuster et al [167]. One model was based on steroidal and another one on nonsteroidal 17β-HSD3 inhibitors as different interaction patterns might be established within the binding pocket. After the screening of several databases and an additional in silico filtering approach, the selected virtual hits were tested in a cell-based assay for 17β-HSD3 inhibition. In addition, specificity was tested against 17β-HSD1, 17β-HSD2, 17β-HSD4, 17β-HSD5, 17β-HSD7, 11β-HSD1, and 11β-HSD2. Two out of 15 tested compounds found by the steroid-based pharmacophore model and 2 out of 16 hits form the non-steroidal model showed > 40% inhibition of 17β-HSD3 at 2 μM of test substance. Of the tested compounds, 3 derived from the steroidal and 5 from the non-steroidal model revealed 2-4 fold more potent inhibition against 17β-HSD5 (AKR1C3) than 17β-HSD3. Furthermore, one 17β-HSD3 inhibitor displayed equipotent activity to 17β-HSD5 and even stronger inhibition against 11β-HSD1, whereas as a second 17β-HSD3 inhibitor was capable to inhibit 17β-HSD1. These investigations again emphasize the importance of an expanded selectivity profiling including structurally and functionally related enzymes.
2.2. Enzyme characterization and substrate identification
Although the sequence of the human genome has been solved and all genes are accessible, to date the physiological role of more than half of all SDR members still remains unknown or poorly examined. In silico approaches assist not only in the process of lead compound identification during drug development for well-characterized enzymes, but can also support the deorphanization and characterization of enzymes in order to explore their physiological functions and to identify additional drug targets.
2.2.1. 17β-hydroxysteroid dehydrogenase type 10
The homotetrameric single domain multifunctional enzyme 17β-HSD10 has diverse substrate specificity and, in healthy tissues, is located in mitochondria [168, 169]. 17β-HSD10 is expressed in various regions of the brain, the liver, heart, kidney, and gonads. In addition to its promiscuous substrate spectrum, it was shown that proteins and peptides such as the Alzheimer’s disease (AD) related amyloid-β (Aβ) peptide or ERα can bind to 17β-HSD10, thereby inhibiting its activity [170]. 17β-HSD10 contains a unique β-hairpin structure at residues 102-107, which is distinct from all other NAD+-dependent SDRs and thought to be the recognition site for Aβ [171, 172]. Thus, 17β-HSD10 is also known as Aβ binding alcohol dehydrogenase (ABAD). The inhibition of 17β-HSD10 by ERα binding seems to be estrogen-dependent [173], and high levels of intracellular estradiol disrupt the described interaction and the released unbound 17β-HSD10 then was suggested to convert estradiol to estrone. However, regarding its mitochondrial localization it remains to be demonstrated that interaction of 17β-HSD10 with Aβ and ERα indeed occurs under patho-physiological conditions. Furthermore, 17β-HSD10 has been proposed to catalyze the oxidation of steroid modulators of GABA(A) receptors [174]. Thus, clearly further research is needed to elucidate the physiological substrates and interactions of this enzyme. Nevertheless, altered 17β-HSD10 function can be found in patients suffering from AD, certain cognitive disabilities, multiple sclerosis, and in chemotherapy-resistant osteosarcoma patients showing an overexpression of 17β-HSD10 [170].
In order to understand the molecular basis of the substrate promiscuity of human 17β-HSD10, Nordling et al. [175] generated a homology model and performed docking experiments with known substrates to examine structure-function relationships. The active site was found to be a wide cleft, consisting of mainly hydrophobic residues and containing at the bottom of the pocket a polar region with the highly conserved catalytic triad Ser155, Tyr168, and Lys172. For the docking calculations different steroids were selected to simulate the following site- and stereo-specific enzyme activities: 3α-OH to 3-oxo conversion and 17β-OH dehydrogenase activity. 17β-HSD activity was mimicked with optimal distances and geometry for estradiol, testosterone, DHT, and 3β-androstanediol. Five-α reduced steroids such as 3α,5α-androstane,17-one revealed ideal hydrogen bond distances to Tyr168, Ser155, and NAD+ for 3α-HSD activity. In contrast, 3β-hydroxylated compounds or 5β-reduced steroids such as the bile acid ursodeoxycholic acid (UDCA) showed inappropriate distances to Ser155 (>4 Å), therefore restricting the oxidation reaction. However, in an expanded follow-up study including kinetic measurements, they found 17β-HSD10 acting as 7β-hydroxysteroid dehydrogenase for the bile acids UDCA and isoUDCA, respectively [176]. To explain this novel substrate specificity on both equatorial and axial positions of the steroidal compounds, they generated again a homology model, this time based on the orthologous rat crystal structure (PDB entry 1E6W [177]), whereas the model of the former study was obtained using the related 7α-HSD (PDB entry 1FMC [178]). Interestingly, the observed molecular distances for isoUDCA to the catalytic triad were identical for the hydroxyl group of Tyr and the C4 atom of NAD+, but decreased in this docking application from 4.26 Å to 2.1 Å. Unfortunately, the exact pose of isoUDCA and therefore the parts of the substrate involved in the interaction, was not displayed for both approaches. This might clarify structural differences of the homology models and may explain the observed distance difference. In addition to the 7β-hydroxysteroid dehydrogenase activity of 17β-HSD10, they detected novel activities, namely the oxidation of 20β- and 21-hydroxyl groups in C21 steroids such as glucocorticoids.
Although homology models are not as precise as high-resolution X-ray structures, they represent a valuable basis for the analysis of substrate recognition and specificity. Furthermore, the 17β-HSD10 crystal structure resolved by Kissinger et al. [172] almost entirely confirmed the hydrophobic residues predicted by the homology model generated by Nordling et al. to be involved in substrate binding [175].
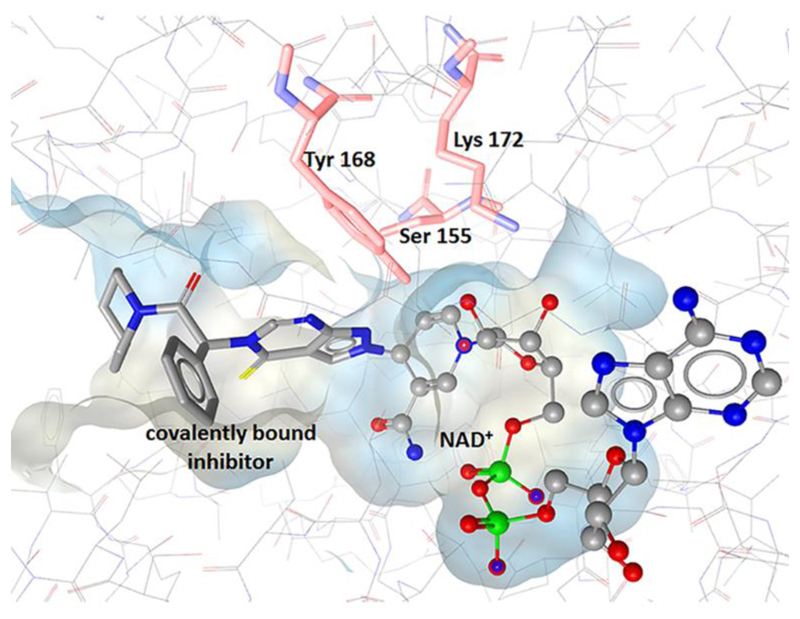
To analyze the role of 17β-HSD10 in AD pathogenesis and as part of a structure-based drug design process, Kissinger et al. crystallized the human 17β-HSD10 in complex with NAD+ and a small molecule inhibitor (PDB 1U7T) (Fig. 10). In line with Nordling et al., they described the substrate binding pocket as flexible and highly hydrophobic cleft, supporting the multi-substrate specificity of the enzyme. Superimposition with the rat enzyme, for which already three resolved protein structures were available, showed very similar overall chain fold [177]. Interestingly, the inhibitor was found to form covalent adducts with the bound cofactor NAD+. Although the human protein structure was successfully resolved, no molecular modeling studies for substrate or inhibitor identification were implemented so far.
Figure 10.
Binding pocket of 17β-HSD10 with residues of the catalytic triad and co-crystallized ligand covalently bound to NAD+ (PDB 1U7T).
2.2.2. 17β-hydroxysteroid dehydrogenase type 14
The physiological functions of 17β-HSD14 still remain unclear. Expression analysis revealed a cytoplasmic localization of 17β-HSD14 with high expression in liver, placenta, and brain, but absence in steroidogenic tissues such as testis and ovary [179]. In contrast, Sivik et al. [180] reported intense 17β-HSD14 immunohistochemical staining patterns in breast, ovary, and testis. The observed discrepancy may be due to differences in antibody specificity; thus, further confirmatory expression studies are necessary.
Lukacik et al. provided structural and functional information building a basis for the deorphanization of this enzyme and leading to the renaming of the DHRS10 gene to HSD17B14 [179]. The resolution of the 17β-HSD14 holo enzyme crystal structure represented a major achievement to deduce functional consequences. Besides the typically conserved regions of SDRs, such as the Rossmann-fold or the catalytic triad consisting of Ser141, Tyr154, and Lys158, the active site of 17β-HSD14 displayed a deep and broad hydrophobic cleft. An in vitro substrate screening using purified recombinant 17β-HSD14 and including 50 different steroids comprising androgens, estrogens, progestin, glucocorticoids, bile acids and oxysterols revealed rather low catalytic turnover of estradiol and 5-androstene-3β,17β-diol using NAD+ as cofactor. Non-saturable kinetics were found for testosterone. Docking calculations for structural comparison of estradiol binding within 17β-HSD1 and other 17β-HSDs showed a comparatively loose binding of estradiol in the active site cleft of 17β-HSD14. However, this extensive and open active cleft may also be due to lack of a co-crystallized substrate in the X-ray structure, and substrate binding might result in an induced fit. Evidence for an induced fit upon cofactor binding was recently provided by Bertoletti and Braun et al., who resolved the crystal structure of 17β-HSD14 as the holo form with NAD+ and as ternary complexes with estrone and the first nonsteroidal inhibitor of 17β-HSD14 [181]. The residues 189-212 were found to form a flexible loop adopting a closed conformation in the presence of cofactor and reducing the size of the active site. The geometry of the active site in the ternary complexes with the inhibitor or estrone bound showed the same closed state delimiting an elongated, conical shaped binding site with the catalytic triade at the apex of the cone and a solvent-exposed opening site. However, estrone was found in an atypical binding pose with the A-ring next to the nicotinamide structure of the cofactor and forming an H-bond with Tyr154 of the catalytic triade instead of the actual reaction position 17. Furthermore, the position 17 itself was not observed to introduce any other interaction. Chemical modification of this inhibitor scaffold applying a ligand-based approach led to the identification of five new 17β-HSD14 inhibitors with Ki values in the lower nanomolar range [182]. The 17β-HSD14 crystal structure determination in complex with these new inhibitors revealed a highly similar binding mode compared to previously reported non-steroidal 17β-HSD14 inhibitor.
2.2.3. Application of structural modeling for substrate identification
Favia et al. [183] introduced a molecular docking protocol to identify candidate substrates for 27 SDR members with resolved X-ray structures and known catalytic function. The enzymes included oxidoreductases, lyases, and isomerases, whereby half of the proteins were from bacterial organisms and half from eukaryotes. They docked the known substrates and products together with > 900 human metabolites from the KEGG pathway metabolite database to each protein. In two thirds of all cases, the actual known substrate or product was found within the top 5% of all docked compounds. For the remaining third, allowing full flexibility of the side chains enhanced the rate of recognition of their natural substrates. However, increasing the degrees of freedom is not a practicable solution for docking of large libraries as it also increases the required computational time and capacity. Nevertheless, rigidity of a docking calculation can result in an increased rate of false negative and false positive hits. A closer look at the 2D structural similarity of the top-ranking compounds to the substrate revealed rather weak correlation. Metabolites resembling the natural substrate were indeed identified; however, only some of the top-ranking compounds showed reasonable correlation of the similarity with the docking rank. This may be explained by the large hydrophobic binding pockets and induced fit, allowing the enzyme to bind various molecules, an essential property of multi-functional SDR enzymes. In addition, Favia et al. clustered the substrates into steroids, small polar molecules, coenzyme A derivatives, nucleotide sugars, and others, and investigated whether the use of representatives of each structural class might provide similar information on substrate preferences of an enzyme as the whole dataset. Even though they found that the rank of a group representative correlated well with the mean rank of the corresponding cluster, this approach seemed to be too abstract for substrate identification calculations. The diversity within a cluster is important as it covers structurally related compounds with great affinity variations; thus defining a class representative will decrease the ability to identify potential candidate substrates.
In this respect, Hermann et al. conducted a structure-based docking study on a selection of high-energy intermediate forms of potential substrates for the amidohydrolase superfamily member Tm0936, and they were able to predict three substrates with substantial catalytic rate constants [184]. Their study was based on two important facts: first the X-ray structure was already resolved, and second the number of possible catalytic reactions could be reduced to a limited set of mechanistically associated conversions. For this approach to be successful, it seems crucial to already have some information on the mechanistic details of the enzyme. Regarding SDRs, they show very broad substrate diversity, making it difficult to restrict possible substrates to a specific subclass of molecules. A pharmacophore-based VS approach for substrate identification of enzymes not belonging to the SDR family where no prior knowledge about the binding site of a protein is necessary was employed by Mallipeddi and Joshi et al. [185]. Detailed description of this study can be found in the supplementary information. However, all of the above mentioned studies validated their approaches by modeling known substrates and they did not include a subsequent in vitro evaluation of the substrate-like compounds.
Reinhardt et al. examined tropinone reductase-like SDRs (TLRs) of the Brassicaceae Cochlearia officinalis and the closely related Arabidopsis thaliana in silico and in vitro for their catalytic capacities due to the uncertainty of their denomination and biological function [186]. Two TLRs sharing 79% sequence identity and one sharing 61% identity with a known tropinone reductase were chosen for homology modeling, substrate docking and in vitro validation. Although tropinone was successfully docked in a favorable position into the binding pockets of all three enzymes, none of them was able to reduce tropinone or nitrogen-containing analogues in vitro. A more detailed investigation of the substrate binding sites revealed a small and hydrophobic pocket, where compounds with a charged nitrogen atom can hardly be accepted as substrates. Therefore, small lipophilic carbonyl compounds were used for further docking applications, in which they were predicted as substrates. Biological testing confirmed these in silico predictions, even though highly different kinetic characteristics were obtained. These results enabled them to generate pharmacophore models to identify further substrates by VS of small compound libraries. The resulting hit list was again docked into the binding sites to evaluate for keto or aldehyde functions in the reactive position. The four scaffolds recognized by all of the pharmacophore models were selected for biological assays. All three TRLs reduced the four compounds in vitro, however, with different kinetics. Thus, this pharmacophore-based approach succeeded in the identification of new substrates. This study also emphasizes the importance of in vitro testing of hits from VS when using models based on reference proteins sharing high sequence identity. Especially small scaffolds may easily adopt a favorable docking pose without acting as a substrate. Nevertheless, VS techniques for substrate identification are always dependent on the available database. A potential substrate can only be found if contained in the database. Another limitation of pharmacophore models is that only compounds matching the defined chemical features are retrieved. If a model contains one feature more or less than a potential substrate, it will not be found in the virtual hit list. Hence, applying this approach for substrate identification should rather include open models, retrieving a higher number of hits.
Another example of rather unusually high sequence identity (71%) among SDRs can be found for carbonyl reductases (CBR) 1 and CBR3. CBR1 has a role in phase I metabolism of a wide range of carbonyl containing xenobiotics, and it also catalyzes the conversion of some endogenous substrates including prostaglandins, steroids, and lipid aldehyde, although the physiological role in converting these endogenous substrates requires further research. In contrast, the function of CBR3 has not yet been extensively characterized [187]. Pilka et al. established an in vitro substrate profile for CBR3 that they then used for an in silico structure-activity relationship comparison with CBR1 [188]. The results revealed a much narrower substrate spectrum for CBR3 compared with CBR1. Orthoquinones, isatin-like compounds, and oracin were the only tested substrates shared between the two enzymes. None of the endogenous substrates of CBR1 served as substrate of CBR3. The resolution of a CBR3 crystal structure, substrate docking calculations, and site-directed mutagenesis studies allowed them to identify residues that are critical for substrate recognition and enzyme conformation. Although CBR1 and CBR3 share high sequence similarity, their active sites differ in regard to shape, size, and surface properties. A major difference lies in a short segment of the substrate binding loop containing Trp229 and Ala235 in CBR1 and Pro230 and Asp236 at analogous positions in CBR3, respectively. Thus, a large hydrophobic barrier is formed in CBR1 by Trp229, in contrast to a rather open binding pocket with an additional charge contributed by Asp236 in CBR3. Furthermore, Met141 and Gln142 in the active site of CBR1 and CBR3, respectively, had significantly different effects on the catalytic activity, although the residues are of similar size. The observations of this study were further supported by El-Hawari et al. [189]. Thus, CBR3 clearly has a substrate spectrum distinct form that of CBR1.
CBR1 shares 27% sequence identity with the fruit-fly Drosophila melanogaster carbonyl reductase sniffer, which was reported to prevent age-related neurodegeneration [190]. The mechanism underlying the role of sniffer in neurodegeneration remains incompletely understood. To study the substrate binding site of sniffer and obtain hints towards potential physiological substrates, Sgraja et al. resolved the crystal structure of sniffer in complex with NAD+ [191]. The structure revealed that the dinucleotide-binding site and the substrate-binding loop adopt similar conformations compared with porcine and human CBR1. Compared to other SDRs, the substrate-binding loop is shorter in all three enzymes. For most SDRs, this loop remains disordered until the substrate has bound. However, in the sniffer protein this loop adopts a well-defined conformation even in the absence of a bound substrate. Crystallization of the sniffer protein in complex with an artificial substrate such as 9,10-phenanthrenequinone, p-nitrobenzaldehyde, or menadione, which were described earlier as sniffer substrates, was not successful. Thus, these compounds were computationally docked into the binding site after crystallization. Plausible docking poses could be found for 9,10-phenanthrenequinone and menadione. The previously mentioned tryptophane residue corresponding to position 229 in the human enzyme seems to be highly conserved and was also found to be involved in substrate binding in the sniffer protein. Furthermore, the observed binding modes led the authors to suggest that the unoccupied space in the binding pocket should allow binding of larger substrate molecules such as steroids or prostaglandins. In this respect, Martin et al. recently showed that sniffer is able to catalyze the NADPH-dependent reduction of the lipid peroxidation product 4-oxonon-2-enal into the less reactive 4-hydroxynon-2-enal, thereby providing a possible explanation for the mechanism of protection from oxidative stress in Drosophila melanogaster [192].
2.3. Virtual screening applications in toxicology focusing on SDRs
Molecular modeling applications are not only useful for substrate and lead compound identification, but they can also facilitate the identification of hazardous chemicals in predicting interactions of compounds with so-called anti-targets that may lead to severe adverse health effects. Although the different applications pursue distinct goals, the actual computational algorithm distinguishes not between screening of a drug candidate, potential substrate or hazardous compound. There are only few VS reports focusing on SDRs and toxicological questions so far, and they will be introduced briefly.
A large number of exogenous substances such as dyes, food additives, body care products and cosmetics, as well as chemicals used for industrial production or agriculture are produced and placed on the market every year, often with insufficient safety assessment. Hazard characterization and risk evaluation of synthetic chemicals on human health and the environment are important for safety management strategies; and in this respect the interference of xenobiotics with the endocrine system may cause harmful effects and is a topic of high actual interest [19, 193]. These so-called endocrine disrupting chemicals (EDCs) can disturb hormone synthesis, metabolism, and hormonal signaling, thereby potentially contributing to major diseases.
Nuclear steroid receptors such as ER, AR, MR, GR, and progesterone receptor (PR) are among the most extensively investigated targets for EDC action. However, biosynthesis and peripheral metabolism essentially impact on tissue- and cell-specific regulation of steroid levels and action. Several SDRs are involved in the local interconversion of inactive and active steroid metabolites, and it is important to include them in the evaluation of potential EDCs. Covering all these potential enzymes and receptors for biological testing represents a major challenge. Computational approaches can be highly useful in a first step to identify new EDCs and to gain insights into the mechanism of action, thus helping to prioritize the chemicals for further biological evaluation [19]. In such a proof of concept study, Vuorinen et al. aimed at identifying potential EDCs inhibiting 11β-HSD2, which converts the potent glucocorticoid cortisol into the inactive cortisone, thereby protecting MR from excessive cortisol [21]. Elevated MR activation by glucocorticoids due to 11β-HSD2 disruption or congenital deficiency causes the syndrome of apparent mineralocorticoid excess (AME), characterized by severe hypertension, hypokalemia, and hypernatremia [194, 195]. 11β-HSD2 is also expressed in non-mineralocorticoid target tissues such as the placenta, where it ensures fetal protection from maternal glucocorticoids [196, 197]. Impaired 11β-HSD2 function has been associated with altered fetal growth, impaired angiogenesis, and a higher risk for cardio-metabolic and neuropsychiatric disorders in later life [198, 199]. Thus, Nashev et al. applied a ligand-based 11β-HSD pharmacophore model for VS of a database containing putative EDCs [21]. In total, 29 hit compounds virtually matched with the chemical features of the model, of which 5 substances were tested in a cell-free assay for 11β-HSD1 and 11β-HSD2 inhibition. AB110873, a silane-coupling agent used as rubber additive, and the antibiotic lasalocid, applied in chicken and turkey breeding, were thereby found to inhibit 11βHSD2 activity (IC50 6.1 μM and 14 μM, respectively). Besides 11β-HSD2 inhibition, the silane AB110873 showed concentration-dependent MR activation, which resulted in enhanced interleukin-6 (IL-6) expression and mitochondrial reactive oxygen species (ROS) production in macrophage exposed to this compound. Docking studies were implemented to understand the binding mode of AB110873. Interestingly, AB110873 adopted the same hydrophobic interactions as aldosterone and in addition occupied a hydrophobic side pocket of the receptor. A newly generated structure-based MR pharmacophore recognized AB110873 as a hit, demonstrating the ability to find compounds with quite different, non-steroidal scaffolds. However, regarding risk assessment, it remains to be shown whether relevant concentrations of these substances can be observed in the human body, especially in industry workers producing and processing them.
The importance of 17β-HSD3 during male sexual development is highlighted by specific defects in HSD17B3, leading to 46, XY disorder of sex development [143–145]. Inhibition of 17β-HSD3 activity during early development until puberty by EDCs might lower plasma testosterone levels, thereby disturbing the masculinization process and contributing to male reproductive disorders. For this purpose, Nashev et al. developed a ligand-based pharmacophore model to search for 17β-HSD3 inhibitors from a database containing putative EDCs [19]. VS revealed several benzophenones (Fig. 11) used as UV-filters in cosmetics, sunscreens, and as plastic additives. Upon biological testing of selected virtual hits and structurally similar derivatives that are environmentally relevant in intact cells, benzophenone-1 (BP-1) was found to be the most potent 17β-HSD3 inhibitor with an IC50 value of 1.05 μM. Moderate inhibition with IC50 between 5.9 μM and 10.3 μM were found for BP-2, 3-benzylidene camphor (3-BC) and 4-methylbenzylidene camphor (4-MBC). Notably, BP-1, 3-BC and 4-MBC were not found as virtual hits but were included in the in vitro testing because of their structural similarity to the initially identified UV-filter hits. In addition to 17β-HSD3, the related SDRs 17β-HSD2 and 11β-HSD1 were also inhibited by these UV-filter chemicals (17β-HSD2: IC50 for BP-2, 3-BC and 4-MBC of 10.0 μM, 6.3 μM and 5.8 μM; 11β-HSD1: IC50 for BP-2, 3-BC and 4-MBC of 12.2 μM, 2.3 μM and 2.9 μM, respectively), emphasizing the need to include several related SDRs for the safety assessment of these chemicals.
Figure 11.
Benzophenones (BP-1, BP-2, BP-3), 3-benzylidene camphor (3-BC), and 4-methylbenzylidene camphor (4-MBC).
As benzophenones may be regarded as steroid mimetics, they may directly bind to nuclear steroid receptors. Therefore, the investigated UV-filter chemicals were also tested for direct effects on the androgen receptor (AR). Several UV-filters antagonized the AR, with BP-1, BP-2, and BP-3 displaying the most potent activities with IC50 values between 2.2 μM and 5.7 μM. These results emphasize that VS as initial filtering tool for the discovery of active compound classes can aid the identification of EDCs. Additionally, as metabolites can be equally or even more active than their parent substances, major metabolites should be included in safety analyses. For example, whilst benzophenone-3 (BP-3) displayed no inhibitory activity against 17β-HSD3, it is rapidly demethylated in vivo to the potent 17β-HSD3 inhibitor BP-1 [200]. Of note, pharmacophore-based SAR studies, implemented to analyze the differences in the activities of the tested UV-filters on 17β-HSD3, indicated that the loss of activity of BP-3 was due to the ether group. Nevertheless, for the risk assessment, the concentrations of BP-1 reached in vivo in the testes need to be determined in order to assess the relevance of 17β-HSD3 inhibition.
2.4. Limitations
2.4.1. Computational applications
Despite the many reported success stories, in silico approaches for the identification of chemicals binding to a cognate protein are facing several challenges. The predictive power of VS workflows is determined by the quality of the data underlying the model or workflow. Thus, the biological data of the training and test compounds used for model generation and validation must be critically evaluated [201]. The following aspects should therefore be considered: the biological data should be based on suitable in vitro testing systems employed for activity determination (i.e. purified protein or cell lysates allowing for a direct access of a given chemical to the corresponding protein), activity cut-offs to exclude unspecific effects, use of suitable positive and negative controls, consideration of species differences (i.e. data and model must be for the same species), and verification of the original references when using compound databases. The majority of these issues is equally important for the biological validation of virtual hits, to guarantee a correct subsequent model refinement. Hence, they are discussed in more detail below. These issues refer not only to compounds determined as active, but also to confirm inactive molecules. These data are scarce because proven inactive compounds for a specific target, i.e. negative results, are rarely reported. Compounds confirmed as inactive can provide valuable insights into unwanted properties that cannot be tolerated for a compound to interact with a macromolecular target. In addition, they can help estimating and adjusting the selectivity and sensitivity of an in silico workflow. Rather restrictive models are often employed in the drug development process to find few potential drug candidates and keep the number of false positive virtual hits low. In contrast, toxicological applications aim to identify all conceivably harmful substances and can accept lower VS hit rates. Additionally, most in silico workflows were generated with drug-like compounds where a lot of data may already be available. Toxicologically relevant molecules such as environmental chemicals occupy a different chemical space, for which a workflow was not optimized. Nevertheless, the identification of structural compound classes acting on a given target, which are then further biologically evaluated, can yield important information for the prioritization of follow-on experiments.
For many members of the SDR family, structural information is currently not or only partially available. For example, many available crystal structures are not including the cofactor, and several SDRs bear a transmembrane binding anchor that is usually not covered in the crystal structures. The experimental 3D resolution of these membrane proteins still remains challenging as is the alternative approach of homology modeling due to the low sequence similarity among SDRs. The lack of suitable x-ray crystallographic data thus limits structure-guided design strategies for several SDR family members. Alternatively, ligand-based methods can be applied to target these molecules.
Different computational programs can generate different VS results even when the same chemical library and protein structural information are used, and each approach by its own yields valuable results [202, 203]. Thus, combining different computational programs may be crucial for substrate identification or toxicological approaches, where a more complete recovery of the active compounds is needed [204]. Another drawback in the VS process, especially for substrate identification purposes, represents the fact that the available chemical databases are not fully representative. While a lot of databases containing small drug-like compounds are available (e. g. ChEMBL [205]), less data is easily accessible for endogenous substrates or environmental chemicals. The gaining knowledge of existing metabolites and extending existing databases such as the human metabolome database [206], will improve future VS applications aiming at substrate identification.
Regarding successful VS applications, in silico tools should not be seen as isolated approaches, but rather as complementary to experimental techniques. Thus, the biological validation of virtual hits using appropriate in vitro testing systems is of utmost importance and will therefore be discussed in detail in the following section.
2.4.2. Biological validation
As mentioned above, biological assays are essential to validate virtual hits. To avoid biased results, the limitations of the in vitro testing systems should be known and considered for data interpretation. The enzyme activity analysis of selected virtual hits has to be conducted under cell-free conditions (i.e. purified protein or lysates expressing the protein in an environment with very low background). Equally important is the appropriate selection of the assay detection technology. Compounds can be regarded as false positive due to interference issues with the assay signaling (for example autofluorescence of certain compounds for fluorescence detectors). Data obtained from intact cells can only be used to verify active hits but do not allow appropriate ranking of hits with different activities, nor does it allow exclusion of inactive molecules. Whereas a test compound has direct access to its target in cell-free systems, a compound’s concentration in intact cells depends on the presence of transport proteins, intracellular binding proteins, as well as metabolizing enzymes. Thus, negative results in cell-based assays do not allow drawing unambiguous conclusions about the biological activity of a virtual hit. Moreover, results from cell-based assays can be influenced by parameters such as passage number of a cell line and therefore different expression levels, conditions of cell handling, and medium composition. Similarly, problems may be evident in preparation of cell lysates and protein purification, where different handling can lead to different activity of an enzyme preparation. Thus, ideally data on a series of compounds should be obtained with the same material. Using the same procedure and inclusion of appropriate positive and negative controls facilitates comparison with results obtained other studies. Nevertheless, upon initial confirmation, active hits need further comprehensive biological characterization including selectivity assessment, analysis of species-specific differences and in vivo pharmacodynamics and pharmacokinetics.
Although the members of the SDR family share considerable structural similarity, the primary sequence similarity is rather low. Koch et al. [207] proposed that structural similarity rather than primary sequence similarity should be chosen as criterion for whether a certain chemical affects the activity of a related enzyme. Therefore, the closest structurally related enzymes should be included for selectivity testing. Because of the high number of orphan SDRs, the function and substrate specificity of the closest relatives of a given enzyme are often unknown and no suitable assay read-outs for selectivity assessment are available. Thus, deorphanization of SDRs is important to improve the physiological understanding of these enzymes and to discover potentially novel drug or anti-drug targets.
The lack of assay read-outs for testing potential substrates to deorphanize enzymes hampers the attempt of substrate identifications. Unlike other protein families, SDRs do not share a common activity element that can be used as read-out. For instance, the activation of G protein coupled-receptors (GPCRs) involves well-described steps that can be monitored, such as the accumulation of second messengers. This allows for the detection of specific changes upon receptor activation [208]. SDRs, however, are enzymes with a remarkably broad substrate specificity, which impedes the monitoring of activation-specific changes.
Species-specific variabilities have been reported for inhibitors and substrates of different HSDs [77, 79, 209]. These observations suggested significant differences in the 3D conformations of the enzymes from different species. During the drug development process, a lead compound ideally inhibits the human enzyme as well as the orthologue of the species (usually rodents) in which the preclinical or safety studies are conducted. However, Möller et al. compared the activities of 17β-HSD1 inhibitors in different species and found that the most potent inhibitors of the human enzyme lacked inhibition of the rodent enzymes [209]. Moreover, they were not able to predict the species-specific effects by molecular docking calculations. In this regard, an initial enzymatic assay including only the orthologue of the subsequently used animal model species would miss potential inhibitors for human applications. Abdelsamie et al. performed structural optimization studies to enhance the potency of a 17β-HSD1 inhibitor against the rodent enzyme and performed docking and homology modeling applications to elucidate the interspecies differences [210]. The observed species-specific protein-ligand interactions might offer valuable information for the design of new inhibitors.
Achieving tissue-specific enzyme inhibition can be important for therapeutic applications in order to reduce potential side effects. Therefore, a compound should be tested in suitable in vitro and in vivo testing systems to elucidate its tissue distribution. Moreover, regarding toxicological studies, tissue distribution and accumulation studies could help to assess the potential of compounds acting as EDCs. In this context, Hamilton et al. developed a test cascade for the development of 11β-HSD1 inhibitors as described above [99]. To summarize the limitations involved in the biological validation of virtual hits described in this review and to provide a potential solution approach, we adapted the former cascade to a more general screening strategy (Fig. 12).
Figure 12.
Potential screening strategy for biological validation of in silico derived hits.
3. Conclusion
Computational tools offer a rational, cost-effective addition to large high-throughput screening (HTS) studies by enriching virtual hits from large databases applying target-specific filters. They can be employed in various application fields such as lead compound identification during drug development, toxicological screenings including EDC assessment, but also for substrate identification and enzyme characterization. This review highlights the efforts made in these areas for the family of SDRs, particularly focusing on HSDs. Several SDRs are involved in steroid synthesis and metabolism as well as in the metabolism of xenobiotics, suggesting that these enzymes may be susceptible to endocrine disruption. In silico design for therapeutic 11β-HSD1 and 17β-HSD1 inhibitors comprise the majority of studies conducted. Only few studies investigated HSDs as EDC targets and applied VS tools as pre-filter to identify compound classes rather than specific substances. Regarding the large number of orphan SDRs, new methods are required for substrate identification, where computational approaches could offer valuable support. However, these investigations into SDRs are rather at the beginning, and need to be extended.
For the appropriate application of in silico tools and subsequent biological validation of virtual hits, careful consideration of the limitations of the individual approaches is crucial. The reported successful studies including computational and biological analyses of SDRs raise the expectations of increasing numbers of studies performed in this area.
Supplementary Material
Highlights.
Review of virtual screening approaches in SDR research including drug development, enzyme characterization and toxicological applications
In silico design for therapeutic 11β-HSD1 and 17β-HSD1 inhibitors comprise the majority of studies
Few studies investigated SDRs as EDC targets
Computational approaches could offer valuable support for substrate identification
Discussion of limitations of computational modeling and in vitro validation methods
Acknowledgements
We are grateful for financial support by the Swiss National Science Foundation (No 31003A-159454 to A.O.), the Austrian Science Fund (FWF, project P26782 “Safety of environmental chemicals” to D.S.), and the Swiss Center for Applied Human Toxicology and the Swiss Federal Office of Public Health (to A.O.). A.O. was supported as Chair for Molecular and Systems Toxicology by the Novartis Research Foundation. D.S. holds an Ingeborg Hochmair Professorship of the University of Innsbruck.
Abbreviations
- 3-BC
3-benzylidene camphor
- 4-MBC
4-methylbenzylidene camphor
- Aβ
amyloid-β
- AD
Alzheimer’s disease
- AI
aromatase inhibitor
- AKR
aldo-keto reductase
- AME
apparent mineralocorticoid excess
- AR
androgen receptor
- Aro
aromatic feature
- BP
benzophenone
- CBR
carbonyl reductase
- CYP
cytochrome P450
- DASI
dual aromatase-sulfatase inhibitor
- DHEA
dehydroepiandrosterone
- DHT
5α-dihydrotestosterone
- EDCs
endocrine disrupting chemicals
- ER
estrogen receptor
- FXR
farnesoid X receptor
- GA
glycyrrhetinic acid
- GPCR
G-protein-coupled receptor
- GR
glucocorticoid receptor
- H6PDH
Hexose-6-phosphate dehydrogenase
- H
hydrophobic feature
- HBA
hydrogen bond acceptor
- HBD
hydrogen bond donor
- hERG
human ether-a-go-go related gene
- HSD
hydroxysteroid dehydrogenase
- HTS
high-throughput screening
- LDH
lactate dehydrogenase
- MR
mineralocorticoid receptor
- MD
molecular dynamics
- MOE
Molecular Operating Environment
- NI
negatively ionizable
- PAINS
Pan-Assay Interference Compounds
- PI
positively ionizable
- PDB
Protein Data Bank
- PR
progesterone receptor
- ROS
reactive oxygen species
- SAR
structure-activity relationship
- SDR
short-chain dehydrogenase/reductase
- SERM
selective estrogen receptor modulator
- SPA
scintillation proximity assay
- STS
steroid sulfatase
- Tc
Tanimoto coefficient
- TRL
tropinone reductase-like
- UDCA
ursodeoxycholic acid
- UFSRAT
Ultra-fast recognition with atom types
- VS
virtual screening
- XVols
exclusion volumes
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kallberg Y, Oppermann U, Persson B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models. FEBS J. 2010;277:2375–2386. doi: 10.1111/j.1742-4658.2010.07656.x. [DOI] [PubMed] [Google Scholar]
- [2].Hosfield DJ, Wu Y, Skene RJ, Hilgers M, Jennings A, Snell GP, Aertgeerts K. Conformational flexibility in crystal structures of human 11β-hydroxysteroid dehydrogenase type I provide insights into glucocorticoid interconversion and enzyme regulation. J Biol Chem. 2005;280:4639–4648. doi: 10.1074/jbc.M411104200. [DOI] [PubMed] [Google Scholar]
- [3].Kavanagh KL, Jornvall H, Persson B, Oppermann U. Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65:3895–3906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Penning TM, Bauman DR, Jin Y, Rizner TL. Identification of the molecular switch that regulates access of 5alpha-DHT to the androgen receptor. Mol Cell Endocrinol. 2007;265–266:77–82. doi: 10.1016/j.mce.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mohler JL, Titus MA, Bai S, Kennerley BJ, Lih FB, Tomer KB, Wilson EM. Activation of the androgen receptor by intratumoral bioconversion of androstanediol to dihydrotestosterone in prostate cancer. Cancer Res. 2011;71:1486–1496. doi: 10.1158/0008-5472.CAN-10-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang SY, He XY, Isaacs C, Dobkin C, Miller D, Philipp M. Roles of 17β-hydroxysteroid dehydrogenase type 10 in neurodegenerative disorders. J Steroid Biochem Mol Biol. 2014;143:460–472. doi: 10.1016/j.jsbmb.2014.07.001. [DOI] [PubMed] [Google Scholar]
- [8].Gathercole LL, Lavery GG, Morgan SA, Cooper MS, Sinclair AJ, Tomlinson JW, Stewart PM. 11β-hydroxysteroid dehydrogenase 1: Translational and therapeutic aspects. Endocr Rev. 2013;34:525–555. doi: 10.1210/er.2012-1050. [DOI] [PubMed] [Google Scholar]
- [9].Luu-The V. Assessment of steroidogenesis and steroidogenic enzyme functions. J Steroid Biochem Mol Biol. 2013;137:176–182. doi: 10.1016/j.jsbmb.2013.05.017. [DOI] [PubMed] [Google Scholar]
- [10].Marchais-Oberwinkler S, Henn C, Moller G, Klein T, Negri M, Oster A, Spadaro A, Werth R, Wetzel M, Xu K, Frotscher M, et al. 17β-Hydroxysteroid dehydrogenases (17β-HSDs) as therapeutic targets: protein structures, functions, and recent progress in inhibitor development. J Steroid Biochem Mol Biol. 2011;125:66–82. doi: 10.1016/j.jsbmb.2010.12.013. [DOI] [PubMed] [Google Scholar]
- [11].Hadoke PW, Kipari T, Seckl JR, Chapman KE. Modulation of 11β-hydroxysteroid dehydrogenase as a strategy to reduce vascular inflammation. Curr Atheroscler Rep. 2013;15:320. doi: 10.1007/s11883-013-0320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Penning TM. Human hydroxysteroid dehydrogenases and pre-receptor regulation: Insights into inhibitor design and evaluation. J Steroid Biochem Mol Biol. 2011;125:46–56. doi: 10.1016/j.jsbmb.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sun D, Wang M, Wang Z. Small molecule 11β-hydroxysteroid dehydrogenase type 1 inhibitors. Curr Top Med Chem. 2011;11:1464–1475. doi: 10.2174/156802611795860988. [DOI] [PubMed] [Google Scholar]
- [14].Seibert JK, Quagliata L, Quintavalle C, Hammond TG, Terracciano L, Odermatt A. A role for the dehydrogenase DHRS7 (SDR34C1) in prostate cancer. Cancer medicine. 2015;4:1717–1729. doi: 10.1002/cam4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kowalik D, Haller F, Adamski J, Moeller G. In search for function of two human orphan SDR enzymes: Hydroxysteroid dehydrogenase like 2 (HSDL2) and short-chain dehydrogenase/reductase-orphan (SDR-O) J Steroid Biochem Mol Biol. 2009;117:117–124. doi: 10.1016/j.jsbmb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- [16].Kitamoto A, Kitamoto T, Nakamura T, Ogawa Y, Yoneda M, Hyogo H, Ochi H, Mizusawa S, Ueno T, Nakao K, Sekine A, et al. Association of polymorphisms in GCKR and TRIB1 with nonalcoholic fatty liver disease and metabolic syndrome traits. Endocr J. 2014;61:683–689. doi: 10.1507/endocrj.ej14-0052. [DOI] [PubMed] [Google Scholar]
- [17].Chen X, Yamamoto M, Fujii K, Nagahama Y, Ooshio T, Xin B, Okada Y, Furukawa H, Nishikawa Y. Differential reactivation of fetal/neonatal genes in mouse liver tumors induced in cirrhotic and non-cirrhotic conditions. Cancer Sci. 2015;106:972–981. doi: 10.1111/cas.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dhoke GV, Ensari Y, Davari MD, Ruff AJ, Schwaneberg U, Bocola M. What's My Substrate? Computational Function Assignment of Candida parapsilosis ADH5 by Genome Database Search, Virtual Screening, and QM/MM Calculations. J Chem Inf Model. 2016;56:1313–1323. doi: 10.1021/acs.jcim.6b00076. [DOI] [PubMed] [Google Scholar]
- [19].Vuorinen A, Odermatt A, Schuster D. In silico methods in the discovery of endocrine disrupting chemicals. J Steroid Biochem Mol Biol. 2013;137:18–26. doi: 10.1016/j.jsbmb.2013.04.009. [DOI] [PubMed] [Google Scholar]
- [20].Vitku J, Starka L, Bicikova M, Hill M, Heracek J, Sosvorova L, Hampl R. Endocrine disruptors and other inhibitors of 11β-hydroxysteroid dehydrogenase 1 and 2: Tissue-specific consequences of enzyme inhibition. J Steroid Biochem Mol Biol. 2016;155:207–216. doi: 10.1016/j.jsbmb.2014.07.007. [DOI] [PubMed] [Google Scholar]
- [21].Nashev LG, Vuorinen A, Praxmarer L, Chantong B, Cereghetti D, Winiger R, Schuster D, Odermatt A. Virtual screening as a strategy for the identification of xenobiotics disrupting corticosteroid action. PLoS One. 2012;7:e46958. doi: 10.1371/journal.pone.0046958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Odermatt A, Nashev LG. The glucocorticoid-activating enzyme 11β-hydroxysteroid dehydrogenase type 1 has broad substrate specificity: Physiological and toxicological considerations. J Steroid Biochem Mol Biol. 2010;119:1–13. doi: 10.1016/j.jsbmb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- [23].Maser E, Oppermann UC. Role of type-1 11β-hydroxysteroid dehydrogenase in detoxification processes. Eur J Biochem. 1997;249:365–369. doi: 10.1111/j.1432-1033.1997.00365.x. [DOI] [PubMed] [Google Scholar]
- [24].Maser E. Xenobiotic carbonyl reduction and physiological steroid oxidoreduction. The pluripotency of several hydroxysteroid dehydrogenases. Biochem Pharmacol. 1995;49:421–440. doi: 10.1016/0006-2952(94)00330-o. [DOI] [PubMed] [Google Scholar]
- [25].Nashev LG, Schuster D, Laggner C, Sodha S, Langer T, Wolber G, Odermatt A. The UV-filter benzophenone-1 inhibits 17β-hydroxysteroid dehydrogenase type 3: Virtual screening as a strategy to identify potential endocrine disrupting chemicals. Biochem Pharmacol. 2010;79:1189–1199. doi: 10.1016/j.bcp.2009.12.005. [DOI] [PubMed] [Google Scholar]
- [26].Yuan K, Zhao B, Li XW, Hu GX, Su Y, Chu Y, Akingbemi BT, Lian QQ, Ge RS. Effects of phthalates on 3beta-hydroxysteroid dehydrogenase and 17β-hydroxysteroid dehydrogenase 3 activities in human and rat testes. Chem-Biol Interact. 2012;195:180–188. doi: 10.1016/j.cbi.2011.12.008. [DOI] [PubMed] [Google Scholar]
- [27].Zhao B, Chu Y, Hardy DO, Li XK, Ge RS. Inhibition of 3β- and 17β-hydroxysteroid dehydrogenase activities in rat Leydig cells by perfluorooctane acid. J Steroid Biochem Mol Biol. 2009;118:13–17. doi: 10.1016/j.jsbmb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- [28].Johnson MA, Maggiora GM. Concepts and Applications of Molecular Similarity. Wiley; New York: 1990. [Google Scholar]
- [29].Tanimoto TT. IBM Internal Report. 1957. Nov 17, [Google Scholar]
- [30].Jaccard P. Distribution de la flore alpine dans le bassin des Dranses et dans quelques régions voisines. Bull Soc Vaud sci nat. 1901;37:241–272. [Google Scholar]
- [31].Wermuth G, Ganellin CR, Lindberg P, Mitscher LA. Glossary of terms used in medicinal chemistry (IUPAC Recommendations 1998) Pure Appl Chem. 1998;70:1129–1143. [Google Scholar]
- [32].Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model. 2005;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- [33].Discovery Studio Modeling Environment, Dassault Systèmes BIOVIA. San Diego: Dassault Systèmes; [Google Scholar]
- [34].Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE, Friesner RA. PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des. 2006;20:647–671. doi: 10.1007/s10822-006-9087-6. [DOI] [PubMed] [Google Scholar]
- [35].C.C.G. Inc. Molecular Operating Environment (MOE) 2015 1010 Sherbooke St. West, Suite #910, Montreal, QC Canada, H3A 2R7.
- [36].Akram MK, T, Schuster D. Pharmacophore modeling and pharmacophore-based virtual screening. In: Cavasotto CN, editor. In silico drug discovery and design. Theory, Methods, Challenges, and Applications. CRC Press; Boca Raton, FL: 2016. pp. 123–154. [Google Scholar]
- [37].Liu X, Jiang H, Li H. SHAFTS: A Hybrid Approach for 3D Molecular Similarity Calculation. 1. Method and Assessment of Virtual Screening. J Chem Inf Model. 2011;51:2372–2385. doi: 10.1021/ci200060s. [DOI] [PubMed] [Google Scholar]
- [38].Lu W, Liu X, Cao X, Xue M, Liu K, Zhao Z, Shen X, Jiang H, Xu Y, Huang J, Li H. SHAFTS: A Hybrid Approach for 3D Molecular Similarity Calculation. 2. Prospective Case Study in the Discovery of Diverse p90 Ribosomal S6 Protein Kinase 2 Inhibitors To Suppress Cell Migration. J Med Chem. 2011;54:3564–3574. doi: 10.1021/jm200139j. [DOI] [PubMed] [Google Scholar]
- [39].vROCS. Santa FE, NM: OpenEye Scientific Software. http://www.eyesopen.com. [Google Scholar]
- [40].Hawkins PC, Skillman AG, Nicholls A. Comparison of Shape-Matching and Docking as Virtual Screening Tools. J Med Chem. 2007;50:74–82. doi: 10.1021/jm0603365. [DOI] [PubMed] [Google Scholar]
- [41].Sastry GM, Dixon SL, Sherman W. Rapid shape-based ligand alignment and virtual screening method based on atom/feature-pair similarities and volume overlap scoring. J Chem Inf Model. 2011;51:2455–2466. doi: 10.1021/ci2002704. [DOI] [PubMed] [Google Scholar]
- [42].Vainio MJ, Puranen JS, Johnson MS. ShaEP: Molecular Overlay Based on Shape and Electrostatic Potential. J Chem Inf Model. 2009;49:492–502. doi: 10.1021/ci800315d. [DOI] [PubMed] [Google Scholar]
- [43].Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discovery. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- [44].A SC. Protein-Ligand Docking: From Basic Principles to Advanced Applications. In: Cavasotto CN, editor. In silico drug discovery and design. Theory, Methods, Challenges, and Applications. CRC Press; Boca Raton, FL: 2016. pp. 155–188. [Google Scholar]
- [45].Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- [46].Chapman K, Holmes M, Seckl J. 11β-hydroxysteroid dehydrogenases: intracellular gate-keepers of tissue glucocorticoid action. Physiol Rev. 2013;93:1139–1206. doi: 10.1152/physrev.00020.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Odermatt A, Kratschmar DV. Tissue-specific modulation of mineralocorticoid receptor function by 11β-hydroxysteroid dehydrogenases: An overview. Mol Cell Endocrinol. 2012;350:168–186. doi: 10.1016/j.mce.2011.07.020. [DOI] [PubMed] [Google Scholar]
- [48].White PC, Mune T, Agarwal AK. 11β-Hydroxysteroid dehydrogenase and the syndrome of apparent mineralocorticoid excess. Endocr Rev. 1997;18:135–156. doi: 10.1210/edrv.18.1.0288. [DOI] [PubMed] [Google Scholar]
- [49].Masuzaki H, Paterson J, Shinyama H, Morton NM, Mullins JJ, Seckl JR, Flier JS. A transgenic model of visceral obesity and the metabolic syndrome. Science. 2001;294:2166–2170. doi: 10.1126/science.1066285. [DOI] [PubMed] [Google Scholar]
- [50].Masuzaki H, Yamamoto H, Kenyon CJ, Elmquist JK, Morton NM, Paterson JM, Shinyama H, Sharp MG, Fleming S, Mullins JJ, Seckl JR, et al. Transgenic amplification of glucocorticoid action in adipose tissue causes high blood pressure in mice. J Clin Invest. 2003;112:83–90. doi: 10.1172/JCI17845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Paterson JM, Morton NM, Fievet C, Kenyon CJ, Holmes MC, Staels B, Seckl JR, Mullins JJ. Metabolic syndrome without obesity: Hepatic overexpression of 11β-hydroxysteroid dehydrogenase type 1 in transgenic mice. Proc Natl Acad Sci USA. 2004;101:7088–7093. doi: 10.1073/pnas.0305524101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lindsay RS, Wake DJ, Nair S, Bunt J, Livingstone DE, Permana PA, Tataranni PA, Walker BR. Subcutaneous adipose 11β-hydroxysteroid dehydrogenase type 1 activity and messenger ribonucleic acid levels are associated with adiposity and insulinemia in Pima Indians and Caucasians. J Clin Endocrinol Metab. 2003;88:2738–2744. doi: 10.1210/jc.2002-030017. [DOI] [PubMed] [Google Scholar]
- [53].Valsamakis G, Anwar A, Tomlinson JW, Shackleton CH, McTernan PG, Chetty R, Wood PJ, Banerjee AK, Holder G, Barnett AH, Stewart PM, et al. 11β-hydroxysteroid dehydrogenase type 1 activity in lean and obese males with type 2 diabetes mellitus. J Clin Endocrinol Metab. 2004;89:4755–4761. doi: 10.1210/jc.2003-032240. [DOI] [PubMed] [Google Scholar]
- [54].Rask E, Walker BR, Soderberg S, Livingstone DE, Eliasson M, Johnson O, Andrew R, Olsson T. Tissue-specific changes in peripheral cortisol metabolism in obese women: increased adipose 11β-hydroxysteroid dehydrogenase type 1 activity. J Clin Endocrinol Metab. 2002;87:3330–3336. doi: 10.1210/jcem.87.7.8661. [DOI] [PubMed] [Google Scholar]
- [55].Paulmyer-Lacroix O, Boullu S, Oliver C, Alessi MC, Grino M. Expression of the mRNA coding for 11β-hydroxysteroid dehydrogenase type 1 in adipose tissue from obese patients: An in situ hybridization study. J Clin Endocrinol Metab. 2002;87:2701–2705. doi: 10.1210/jcem.87.6.8614. [DOI] [PubMed] [Google Scholar]
- [56].Kannisto K, Pietilainen KH, Ehrenborg E, Rissanen A, Kaprio J, Hamsten A, Yki-Jarvinen H. Overexpression of 11β-hydroxysteroid dehydrogenase-1 in adipose tissue is associated with acquired obesity and features of insulin resistance: studies in young adult monozygotic twins. J Clin Endocrinol Metab. 2004;89:4414–4421. doi: 10.1210/jc.2004-0153. [DOI] [PubMed] [Google Scholar]
- [57].Kotelevtsev Y, Holmes MC, Burchell A, Houston PM, Schmoll D, Jamieson P, Best R, Brown R, Edwards CR, Seckl JR, Mullins JJ. 11β-hydroxysteroid dehydrogenase type 1 knockout mice show attenuated glucocorticoid-inducible responses and resist hyperglycemia on obesity or stress. Proc Natl Acad Sci USA. 1997;94:14924–14929. doi: 10.1073/pnas.94.26.14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tiganescu A, Hupe M, Uchida Y, Mauro T, Elias PM, Holleran WM. Increased glucocorticoid activation during mouse skin wound healing. J Endocrinol. 2014;221:51–61. doi: 10.1530/JOE-13-0420. [DOI] [PubMed] [Google Scholar]
- [59].Youm JK, Park K, Uchida Y, Chan A, Mauro TM, Holleran WM, Elias PM. Local blockade of glucocorticoid activation reverses stress- and glucocorticoid-induced delays in cutaneous wound healing. Wound Repair Regen. 2013;21:715–722. doi: 10.1111/wrr.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tiganescu A, Tahrani AA, Morgan SA, Otranto M, Desmouliere A, Abrahams L, Hassan-Smith Z, Walker EA, Rabbitt EH, Cooper MS, Amrein K, et al. 11β-Hydroxysteroid dehydrogenase blockade prevents age-induced skin structure and function defects. J Clin Invest. 2013;123:3051–3060. doi: 10.1172/JCI64162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wu L, Qi H, Zhong Y, Lv S, Yu J, Liu J, Wang L, Bi J, Kong X, Di W, Zha J, et al. 11β-Hydroxysteroid dehydrogenase type 1 selective inhibitor BVT.2733 protects osteoblasts against endogenous glucocorticoid induced dysfunction. Endocr J. 2013;60:1047–1058. doi: 10.1507/endocrj.ej12-0376. [DOI] [PubMed] [Google Scholar]
- [62].Kipari T, Hadoke PW, Iqbal J, Man TY, Miller E, Coutinho AE, Zhang Z, Sullivan KM, Mitic T, Livingstone DE, Schrecker C, et al. 11β-hydroxysteroid dehydrogenase type 1 deficiency in bone marrow-derived cells reduces atherosclerosis. FASEB J. 2013;27:1519–1531. doi: 10.1096/fj.12-219105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hermanowski-Vosatka A, Balkovec JM, Cheng K, Chen HY, Hernandez M, Koo GC, Le Grand CB, Li Z, Metzger JM, Mundt SS, Noonan H, et al. 11β-HSD1 inhibition ameliorates metabolic syndrome and prevents progression of atherosclerosis in mice. J Exp Med. 2005;202:517–527. doi: 10.1084/jem.20050119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Garcia RA, Search DJ, Lupisella JA, Ostrowski J, Guan B, Chen J, Yang WP, Truong A, He A, Zhang R, Yan M, et al. 11β-hydroxysteroid dehydrogenase type 1 gene knockout attenuates atherosclerosis and in vivo foam cell formation in hyperlipidemic apoE(-)/(-) mice. PLoS One. 2013;8:e53192. doi: 10.1371/journal.pone.0053192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Luo MJ, Thieringer R, Springer MS, Wright SD, Hermanowski-Vosatka A, Plump A, Balkovec JM, Cheng K, Ding GJ, Kawka DW, Koo GC, et al. 11β-HSD1 inhibition reduces atherosclerosis in mice by altering proinflammatory gene expression in the vasculature. Physiol Genomics. 2013;45:47–57. doi: 10.1152/physiolgenomics.00109.2012. [DOI] [PubMed] [Google Scholar]
- [66].Rauz S, Cheung CM, Wood PJ, Coca-Prados M, Walker EA, Murray PI, Stewart PM. Inhibition of 11β-hydroxysteroid dehydrogenase type 1 lowers intraocular pressure in patients with ocular hypertension. QJM. 2003;96:481–490. doi: 10.1093/qjmed/hcg085. [DOI] [PubMed] [Google Scholar]
- [67].Rauz S, Walker EA, Shackleton CH, Hewison M, Murray PI, Stewart PM. Expression and putative role of 11β-hydroxysteroid dehydrogenase isozymes within the human eye. Invest Ophthalmol Visual Sci. 2001;42:2037–2042. [PubMed] [Google Scholar]
- [68].Anderson S, Carreiro S, Quenzer T, Gale D, Xiang C, Gukasyan H, Lafontaine J, Cheng H, Krauss A, Prasanna G. In vivo evaluation of 11β-hydroxysteroid dehydrogenase activity in the rabbit eye. J Ocul Pharmacol Ther. 2009;25:215–222. doi: 10.1089/jop.2008.0120. [DOI] [PubMed] [Google Scholar]
- [69].Sooy K, Webster SP, Noble J, Binnie M, Walker BR, Seckl JR, Yau JL. Partial deficiency or short-term inhibition of 11β-hydroxysteroid dehydrogenase type 1 improves cognitive function in aging mice. J Neurosci. 2010;30:13867–13872. doi: 10.1523/JNEUROSCI.2783-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yau JL, McNair KM, Noble J, Brownstein D, Hibberd C, Morton N, Mullins JJ, Morris RG, Cobb S, Seckl JR. Enhanced hippocampal long-term potentiation and spatial learning in aged 11β-hydroxysteroid dehydrogenase type 1 knock-out mice. J Neurosci. 2007;27:10487–10496. doi: 10.1523/JNEUROSCI.2190-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yau JL, Noble J, Seckl JR. 11β-hydroxysteroid dehydrogenase type 1 deficiency prevents memory deficits with aging by switching from glucocorticoid receptor to mineralocorticoid receptor-mediated cognitive control. J Neurosci. 2011;31:4188–4193. doi: 10.1523/JNEUROSCI.6145-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mohler EG, Browman KE, Roderwald VA, Cronin EA, Markosyan S, Scott Bitner R, Strakhova MI, Drescher KU, Hornberger W, Rohde JJ, Brune ME, et al. Acute inhibition of 11β-hydroxysteroid dehydrogenase type-1 improves memory in rodent models of cognition. J Neurosci. 2011;31:5406–5413. doi: 10.1523/JNEUROSCI.4046-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sooy K, Noble J, McBride A, Binnie M, Yau JL, Seckl JR, Walker BR, Webster SP. Cognitive and Disease-Modifying Effects of 11β-Hydroxysteroid Dehydrogenase Type 1 Inhibition in Male Tg2576 Mice, a Model of Alzheimer's Disease. Endocrinology. 2015;156:4592–4603. doi: 10.1210/en.2015-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Scott JS, Goldberg FW, Turnbull AV. Medicinal chemistry of inhibitors of 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) J Med Chem. 2014;57:4466–4486. doi: 10.1021/jm4014746. [DOI] [PubMed] [Google Scholar]
- [75].Thomas MP, Potter BV. Crystal structures of 11β-hydroxysteroid dehydrogenase type 1 and their use in drug discovery. Future Med Chem. 2011;3:367–390. doi: 10.4155/fmc.10.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Schuster D, Maurer EM, Laggner C, Nashev LG, Wilckens T, Langer T, Odermatt A. The discovery of new 11β-hydroxysteroid dehydrogenase type 1 inhibitors by common feature pharmacophore modeling and virtual screening. J Med Chem. 2006;49:3454–3466. doi: 10.1021/jm0600794. [DOI] [PubMed] [Google Scholar]
- [77].Arampatzis S, Kadereit B, Schuster D, Balazs Z, Schweizer RA, Frey FJ, Langer T, Odermatt A. Comparative enzymology of 11β-hydroxysteroid dehydrogenase type 1 from six species. J Mol Endocrinol. 2005;35:89–101. doi: 10.1677/jme.1.01736. [DOI] [PubMed] [Google Scholar]
- [78].Barf T, Vallgarda J, Emond R, Haggstrom C, Kurz G, Nygren A, Larwood V, Mosialou E, Axelsson K, Olsson R, Engblom L, et al. Arylsulfonamidothiazoles as a new class of potential antidiabetic drugs. Discovery of potent and selective inhibitors of the 11β-hydroxysteroid dehydrogenase type 1. J Med Chem. 2002;45:3813–3815. doi: 10.1021/jm025530f. [DOI] [PubMed] [Google Scholar]
- [79].Hult M, Shafqat N, Elleby B, Mitschke D, Svensson S, Forsgren M, Barf T, Vallgarda J, Abrahmsen L, Oppermann U. Active site variability of type 1 11β-hydroxysteroid dehydrogenase revealed by selective inhibitors and cross-species comparisons. Mol Cell Endocrinol. 2006;248:26–33. doi: 10.1016/j.mce.2005.11.043. [DOI] [PubMed] [Google Scholar]
- [80].Hofer S, Kratschmar DV, Schernthanner B, Vuorinen A, Schuster D, Odermatt A, Easmon J. Synthesis and biological analysis of benzazol-2-yl piperazine sulfonamides as 11beta-hydroxysteroid dehydrogenase 1 inhibitors. Bioorg Med Chem Lett. 2013;23:5397–5400. doi: 10.1016/j.bmcl.2013.07.047. [DOI] [PubMed] [Google Scholar]
- [81].Rollinger JM, Kratschmar DV, Schuster D, Pfisterer PH, Gumy C, Aubry EM, Brandstötter S, Stuppner H, Wolber G, Odermatt A. 11β-Hydroxysteroid dehydrogenase 1 inhibiting constituents from Eriobotrya japonica revealed by bioactivity-guided isolation and computational approaches. Bioorg Med Chem. 2010;18:1507–1515. doi: 10.1016/j.bmc.2010.01.010. [DOI] [PubMed] [Google Scholar]
- [82].Gumy C, Thurnbichler C, Aubry EM, Balazs Z, Pfisterer P, Baumgartner L, Stuppner H, Odermatt A, Rollinger JM. Inhibition of 11β-hydroxysteroid dehydrogenase type 1 by plant extracts used as traditional antidiabetic medicines. Fitoterapia. 2009;80:200–205. doi: 10.1016/j.fitote.2009.01.009. [DOI] [PubMed] [Google Scholar]
- [83].Vuorinen A, Nashev LG, Odermatt A, Rollinger JM, Schuster D. Pharmacophore model refinement for 11β-hydroxysteroid dehydrogenase inhibitors: Search for modulators of intracellular glucocorticoid concentrations. Mol Inf. 2014;33:15–25. doi: 10.1002/minf.201300063. [DOI] [PubMed] [Google Scholar]
- [84].Vuorinen A, Seibert J, Papageorgiou VP, Rollinger JM, Odermatt A, Schuster D, Assimopoulou AN. Pistacia lentiscus oleoresin: Virtual screening and identification of masticadienonic and isomasticadienonic acids as inhibitors of 11β-hydroxysteroid dehydrogenase 1. Planta Med. 2015;81:525–532. doi: 10.1055/s-0035-1545720. [DOI] [PubMed] [Google Scholar]
- [85].Miguet L, Zhang Z, Barbier M, Grigorov MG. Comparison of a homology model and the crystallographic structure of human 11β-hydroxysteroid dehydrogenase type 1 (11βHSD1) in a structure-based identification of inhibitors. J Comput-Aided Mol Des. 2006;20:67–81. doi: 10.1007/s10822-006-9037-3. [DOI] [PubMed] [Google Scholar]
- [86].Yang H, Dou W, Lou J, Leng Y, Shen J. Discovery of novel inhibitors of 11β-hydroxysteroid dehydrogenase type 1 by docking and pharmacophore modeling. Bioorg Med Chem Lett. 2008;18:1340–1345. doi: 10.1016/j.bmcl.2008.01.020. [DOI] [PubMed] [Google Scholar]
- [87].Yang H, Shen Y, Chen J, Jiang Q, Leng Y, Shen J. Structure-based virtual screening for identification of novel 11β-HSD1 inhibitors. Eur J Med Chem. 2009;44:1167–1171. doi: 10.1016/j.ejmech.2008.06.005. [DOI] [PubMed] [Google Scholar]
- [88].Xia G, Xue M, Liu L, Yu J, Liu H, Li P, Wang J, Li Y, Xiong B, Shen J. Potent and novel 11β-HSD1 inhibitors identified from shape and docking based virtual screening. Bioorg Med Chem Lett. 2011;21:5739–5744. doi: 10.1016/j.bmcl.2011.08.019. [DOI] [PubMed] [Google Scholar]
- [89].Aertgeerts K, Brennan N, Cao SX, Chang E, Kiryanov AA, Liu Y. Arylsulfonylpiperazines and related compounds as hydroxysteroid dehydrogenase inhibitors and their preparation and pharmaceutical compositions. Takeda San Diego, Inc. USA: 2006. [Google Scholar]
- [90].Eddine JSSXiang, Tam Steve Y, Mckew John C, Chen Lihren, Ipek Manus, Lee Katherine, Li Huan-Qui, Li Jianchang, Li Wei, Mansour Tarek Suhayl, Suri Vipin, et al. Sulfonylated piperidine and piperazine derivatives as 11β-HSD1 inhibitors and their preparation, pharmaceutical compositions and use in the treatment of diseases. Wyeth, John, and Brother Ltd; USA: 2007. [Google Scholar]
- [91].William PWRMCaulkett, Packer Martin, Whittamore Paul Robert Owen. Preparation of 1,4-disubstituted piperazine or diazepane derivatives as 11β-hydroxysteroid dehydrogenase 1 inhibitors, AstraZeneca AB. Swed: AstraZeneca UK Limited; 2007. [Google Scholar]
- [92].Xia G, Liu L, Xue M, Liu H, Yu J, Li P, Chen Q, Xiong B, Liu X, Shen J. Discovery of novel sulfonamides as potent and selective inhibitors against human and mouse 11β-hydroxysteroid dehydrogenase type 1. Mol Cell Endocrinol. 2012;358:46–52. doi: 10.1016/j.mce.2012.02.017. [DOI] [PubMed] [Google Scholar]
- [93].Lagos CF, Vecchiola A, Allende F, Fuentes CA, Tichauer JE, Valdivia C, Solari S, Campino C, Tapia-Castillo A, Baudrand R, Villarroel P, et al. Identification of novel 11β-HSD1 inhibitors by combined ligand- and structure-based virtual screening. Mol Cell Endocrinol. 2014;384:71–82. doi: 10.1016/j.mce.2014.01.011. [DOI] [PubMed] [Google Scholar]
- [94].Hugo ER, Brandebourg TD, Comstock CE, Gersin KS, Sussman JJ, Ben-Jonathan N. LS14: A novel human adipocyte cell line that produces prolactin. Endocrinology. 2006;147:306–313. doi: 10.1210/en.2005-0989. [DOI] [PubMed] [Google Scholar]
- [95].Shave S, Blackburn EA, Adie J, Houston DR, Auer M, Webster SP, Taylor P, Walkinshaw MD. UFSRAT: Ultra-fast Shape Recognition with Atom Types--the discovery of novel bioactive small molecular scaffolds for FKBP12 and 11βHSD1. PLoS One. 2015;10:e0116570. doi: 10.1371/journal.pone.0116570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Boyle CD, Kowalski TJ. 11β-hydroxysteroid dehydrogenase type 1 inhibitors: A review of recent patents. Expert Opin Ther Pat. 2009;19:801–825. doi: 10.1517/13543770902967658. [DOI] [PubMed] [Google Scholar]
- [97].Tice CM, Zhao W, Xu Z, Cacatian ST, Simpson RD, Ye YJ, Singh SB, McKeever BM, Lindblom P, Guo J, Krosky PM, et al. Spirocyclic ureas: orally bioavailable 11β-HSD1 inhibitors identified by computer-aided drug design. Bioorg Med Chem Lett. 2010;20:881–886. doi: 10.1016/j.bmcl.2009.12.082. [DOI] [PubMed] [Google Scholar]
- [98].Xu Z, Tice CM, Zhao W, Cacatian S, Ye YJ, Singh SB, Lindblom P, McKeever BM, Krosky PM, Kruk BA, Berbaum J, et al. Structure-based design and synthesis of 1,3-oxazinan-2-one inhibitors of 11β-hydroxysteroid dehydrogenase type 1. J Med Chem. 2011;54:6050–6062. doi: 10.1021/jm2005354. [DOI] [PubMed] [Google Scholar]
- [99].Hamilton BS, Himmelsbach F, Nar H, Schuler-Metz A, Krosky P, Guo J, Guo R, Meng S, Zhao Y, Lala DS, Zhuang L, et al. Pharmacological characterization of the selective 11β-hydroxysteroid dehydrogenase 1 inhibitor, BI 135585, a clinical candidate for the treatment of type 2 diabetes. Eur J Pharmacol. 2015;746:50–55. doi: 10.1016/j.ejphar.2014.10.053. [DOI] [PubMed] [Google Scholar]
- [100].Lepifre F, Christmann-Franck S, Roche D, Leriche C, Carniato D, Charon C, Bozec S, Doare L, Schmidlin F, Lecomte M, Valeur E. Discovery and structure-guided drug design of inhibitors of 11β-hydroxysteroid-dehydrogenase type I based on a spiro-carboxamide scaffold. Bioorg Med Chem Lett. 2009;19:3682–3685. doi: 10.1016/j.bmcl.2009.02.123. [DOI] [PubMed] [Google Scholar]
- [101].Valeur E, Christmann-Franck S, Lepifre F, Carniato D, Cravo D, Charon C, Bozec S, Musil D, Hillertz P, Doare L, Schmidlin F, et al. Structure-based design of 7-azaindole-pyrrolidine amides as inhibitors of 11β-hydroxysteroid dehydrogenase type I. Bioorg Med Chem Lett. 2012;22:5909–5914. doi: 10.1016/j.bmcl.2012.07.070. [DOI] [PubMed] [Google Scholar]
- [102].Xia G, You X, Liu L, Liu H, Wang J, Shi Y, Li P, Xiong B, Liu X, Shen J. Design, synthesis and SAR of piperidyl-oxadiazoles as 11β-hydroxysteroid dehydrogenase 1 inhibitors. Eur J Med Chem. 2013;62:1–10. doi: 10.1016/j.ejmech.2012.12.059. [DOI] [PubMed] [Google Scholar]
- [103].Hong SP, Nam KY, Shin YJ, Kim KW, Ahn SK. Discovery of 11beta-hydroxysteroid dehydrogenase type 1 inhibitor. Bioorg Med Chem Lett. 2015;25:3501–3506. doi: 10.1016/j.bmcl.2015.06.099. [DOI] [PubMed] [Google Scholar]
- [104].Lee Y, Shin YJ, Ahn SK. 3-Amino-N-adamantyl-3-methylbutanamide derivatives as 11β-hydroxysteroid dehydrogenase 1 inhibitor. Bioorg Med Chem Lett. 2014;24:1421–1425. doi: 10.1016/j.bmcl.2014.01.017. [DOI] [PubMed] [Google Scholar]
- [105].Vicker N, Su X, Ganeshapillai D, Smith A, Purohit A, Reed MJ, Potter BV. Novel non-steroidal inhibitors of human 11β-hydroxysteroid dehydrogenase type 1. J Steroid Biochem Mol Biol. 2007;104:123–129. doi: 10.1016/j.jsbmb.2007.03.023. [DOI] [PubMed] [Google Scholar]
- [106].Ye XY, Yoon D, Chen SY, Nayeem A, Golla R, Seethala R, Wang M, Harper T, Sleczka BG, Apedo A, Li YX, et al. Synthesis and structure-activity relationship of 2-adamantylmethyl tetrazoles as potent and selective inhibitors of human 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) Bioorg Med Chem Lett. 2014;24:654–660. doi: 10.1016/j.bmcl.2013.11.066. [DOI] [PubMed] [Google Scholar]
- [107].Blum A, Favia AD, Maser E. 11β-Hydroxysteroid dehydrogenase type 1 inhibitors with oleanan and ursan scaffolds. Mol Cell Endocrinol. 2009;301:132–136. doi: 10.1016/j.mce.2008.08.028. [DOI] [PubMed] [Google Scholar]
- [108].Su X, Halem HA, Thomas MP, Moutrille C, Culler MD, Vicker N, Potter BV. Adamantyl carboxamides and acetamides as potent human 11β-hydroxysteroid dehydrogenase type 1 inhibitors. Bioorg Med Chem. 2012;20:6394–6402. doi: 10.1016/j.bmc.2012.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Moeller G, Adamski J. Integrated view on 17β-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2009;301:7–19. doi: 10.1016/j.mce.2008.10.040. [DOI] [PubMed] [Google Scholar]
- [110].Poirier D. Inhibitors of 17β-hydroxysteroid dehydrogenases. Curr Med Chem. 2003;10:453–477. doi: 10.2174/0929867033368222. [DOI] [PubMed] [Google Scholar]
- [111].Lukacik P, Kavanagh KL, Oppermann U. Structure and function of human 17β-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2006;248:61–71. doi: 10.1016/j.mce.2005.12.007. [DOI] [PubMed] [Google Scholar]
- [112].Jansson A. 17β-hydroxysteroid dehydrogenase enzymes and breast cancer. J Steroid Biochem Mol Biol. 2009;114:64–67. doi: 10.1016/j.jsbmb.2008.12.012. [DOI] [PubMed] [Google Scholar]
- [113].Oduwole OO, Li Y, Isomaa VV, Mantyniemi A, Pulkka AE, Soini Y, Vihko PT. 17β-hydroxysteroid dehydrogenase type 1 is an independent prognostic marker in breast cancer. Cancer Res. 2004;64:7604–7609. doi: 10.1158/0008-5472.CAN-04-0446. [DOI] [PubMed] [Google Scholar]
- [114].Miyoshi Y, Ando A, Shiba E, Taguchi T, Tamaki Y, Noguchi S. Involvement of up-regulation of 17β-hydroxysteroid dehydrogenase type 1 in maintenance of intratumoral high estradiol levels in postmenopausal breast cancers. Int Cancer J. 2001;94:685–689. doi: 10.1002/ijc.1525. [DOI] [PubMed] [Google Scholar]
- [115].Husen B, Huhtinen K, Poutanen M, Kangas L, Messinger J, Thole H. Evaluation of inhibitors for 17β-hydroxysteroid dehydrogenase type 1 in vivo in immunodeficient mice inoculated with MCF-7 cells stably expressing the recombinant human enzyme. Mol Cell Endocrinol. 2006;248:109–113. doi: 10.1016/j.mce.2005.11.042. [DOI] [PubMed] [Google Scholar]
- [116].Ayan D, Maltais R, Roy J, Poirier D. A new nonestrogenic steroidal inhibitor of 17β-hydroxysteroid dehydrogenase type I blocks the estrogen-dependent breast cancer tumor growth induced by estrone. Mol Cancer Ther. 2012;11:2096–2104. doi: 10.1158/1535-7163.MCT-12-0299. [DOI] [PubMed] [Google Scholar]
- [117].Smuc T, Pucelj MR, Sinkovec J, Husen B, Thole H, Rizner TL. Expression analysis of the genes involved in estradiol and progesterone action in human ovarian endometriosis. Gynecol Endocrinol. 2007;23:105–111. doi: 10.1080/09513590601152219. [DOI] [PubMed] [Google Scholar]
- [118].Delvoux B, D'Hooghe T, Kyama C, Koskimies P, Hermans RJ, Dunselman GA, Romano A. Inhibition of type 1 17β-hydroxysteroid dehydrogenase impairs the synthesis of 17β-estradiol in endometriosis lesions. J Clin Endocrinol Metab. 2014;99:276–284. doi: 10.1210/jc.2013-2503. [DOI] [PubMed] [Google Scholar]
- [119].Cornel KM, Kruitwagen RF, Delvoux B, Visconti L, Van de Vijver KK, Day JM, Van Gorp T, Hermans RJ, Dunselman GA, Romano A. Overexpression of 17β-hydroxysteroid dehydrogenase type 1 increases the exposure of endometrial cancer to 17β-estradiol. J Clin Endocrinol Metab. 2012;97:E591–601. doi: 10.1210/jc.2011-2994. [DOI] [PubMed] [Google Scholar]
- [120].Kasai T, Shozu M, Murakami K, Segawa T, Shinohara K, Nomura K, Inoue M. Increased expression of type I 17β-hydroxysteroid dehydrogenase enhances in situ production of estradiol in uterine leiomyoma. J Clin Endocrinol Metab. 2004;89:5661–5668. doi: 10.1210/jc.2003-032085. [DOI] [PubMed] [Google Scholar]
- [121].Bey E, Marchais-Oberwinkler S, Kruchten P, Frotscher M, Werth R, Oster A, Algül O, Neugebauer A, Hartmann RW. Design, synthesis and biological evaluation of bis(hydroxyphenyl) azoles as potent and selective non-steroidal inhibitors of 17β-hydroxysteroid dehydrogenase type 1 (17β-HSD1) for the treatment of estrogen-dependent diseases. Bioorg Med Chem. 2008;16:6423–6435. doi: 10.1016/j.bmc.2008.04.073. [DOI] [PubMed] [Google Scholar]
- [122].Hoffren AM, Murray CM, Hoffmann RD. Structure-based focusing using pharmacophores derived from the active site of 17β-hydroxysteroid dehydrogenase. Curr Pharm Des. 2001;7:547–566. doi: 10.2174/1381612013397870. [DOI] [PubMed] [Google Scholar]
- [123].Krazeisen A, Breitling R, Moller G, Adamski J. Phytoestrogens inhibit human 17β-hydroxysteroid dehydrogenase type 5. Mol Cell Endocrinol. 2001;171:151–162. doi: 10.1016/s0303-7207(00)00422-6. [DOI] [PubMed] [Google Scholar]
- [124].Chanplakorn N, Chanplakorn P, Suzuki T, Ono K, Chan MSM, Miki Y, Saji S, Ueno T, Toi M, Sasano H. Increased estrogen sulfatase (STS) and 17β-hydroxysteroid dehydrogenase type 1(17 beta-HSD1) following neoadjuvant aromatase inhibitor therapy in breast cancer patients. Breast Cancer Res Treat. 2010;120:639–648. doi: 10.1007/s10549-010-0785-3. [DOI] [PubMed] [Google Scholar]
- [125].Woo LW, Sutcliffe OB, Bubert C, Grasso A, Chander SK, Purohit A, Reed MJ, Potter BV. First dual aromatase-steroid sulfatase inhibitors. J Med Chem. 2003;46:3193–3196. doi: 10.1021/jm034033b. [DOI] [PubMed] [Google Scholar]
- [126].Woo LW, Bubert C, Sutcliffe OB, Smith A, Chander SK, Mahon MF, Purohit A, Reed MJ, Potter BV. Dual aromatase-steroid sulfatase inhibitors. J Med Chem. 2007;50:3540–3560. doi: 10.1021/jm061462b. [DOI] [PubMed] [Google Scholar]
- [127].Thomas MP, Potter BV. Discovery and Development of the Aryl O-Sulfamate Pharmacophore for Oncology and Women's Health. J Med Chem. 2015;58:7634–7658. doi: 10.1021/acs.jmedchem.5b00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Woo LW, Jackson T, Putey A, Cozier G, Leonard P, Acharya KR, Chander SK, Purohit A, Reed MJ, Potter BV. Highly potent first examples of dual aromatase-steroid sulfatase inhibitors based on a biphenyl template. J Med Chem. 2010;53:2155–2170. doi: 10.1021/jm901705h. [DOI] [PubMed] [Google Scholar]
- [129].Berube M, Poirier D. Synthesis of simplified hybrid inhibitors of type 1 17β-hydroxysteroid dehydrogenase via cross-metathesis and sonogashira coupling reactions. Org Lett. 2004;6:3127–3130. doi: 10.1021/ol048820u. [DOI] [PubMed] [Google Scholar]
- [130].Fournier D, Poirier D, Mazumdar M, Lin SX. Design and synthesis of bisubstrate inhibitors of type 1 17β-hydroxysteroid dehydrogenase: Overview and perspectives. Eur J Med Chem. 2008;43:2298–2306. doi: 10.1016/j.ejmech.2008.01.044. [DOI] [PubMed] [Google Scholar]
- [131].Schuster D, Nashev LG, Kirchmair J, Laggner C, Wolber G, Langer T, Odermatt A. Discovery of nonsteroidal 17β-hydroxysteroid dehydrogenase 1 inhibitors by pharmacophore-based screening of virtual compound libraries. J Med Chem. 2008;51:4188–4199. doi: 10.1021/jm800054h. [DOI] [PubMed] [Google Scholar]
- [132].Brown WM, Metzger LE, Barlow JP, Hunsaker LA, Deck LM, Royer RE, Vander Jagt DL. 17β-Hydroxysteroid dehydrogenase type 1: Computational design of active site inhibitors targeted to the Rossmann fold. Chem-Biol Interact. 2003;143–144:481–491. doi: 10.1016/s0009-2797(02)00184-9. [DOI] [PubMed] [Google Scholar]
- [133].Hu GX, Zhou HY, Li XW, Chen BB, Xiao YC, Lian QQ, Liang G, Kim HH, Zheng ZQ, Hardy DO, Ge RS. The (+)- and (-)-gossypols potently inhibit both 3β-hydroxysteroid dehydrogenase and 17β-hydroxysteroid dehydrogenase 3 in human and rat testes. J Steroid Biochem Mol Biol. 2009;115:14–19. doi: 10.1016/j.jsbmb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- [134].Chen BB, Lin H, Hu GX, Su Y, Zhou HY, Lian QQ, Cai H, Hardy DO, Gu DY, Ge RS. The (+)- and (-)-gossypols potently inhibit human and rat 11β-hydroxysteroid dehydrogenase type 2. J Steroid Biochem Mol Biol. 2009;113:177–181. doi: 10.1016/j.jsbmb.2008.12.006. [DOI] [PubMed] [Google Scholar]
- [135].Spadaro A, Negri M, Marchais-Oberwinkler S, Bey E, Frotscher M. Hydroxybenzothiazoles as new nonsteroidal inhibitors of 17β-hydroxysteroid dehydrogenase type 1 (17beta-HSD1) PLoS One. 2012;7:e29252. doi: 10.1371/journal.pone.0029252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Spadaro A, Frotscher M, Hartmann RW. Optimization of hydroxybenzothiazoles as novel potent and selective inhibitors of 17β-HSD1. J Med Chem. 2012;55:2469–2473. doi: 10.1021/jm201711b. [DOI] [PubMed] [Google Scholar]
- [137].Karkola S, Alho-Richmond S, Wahala K. Pharmacophore modelling of 17β-HSD1 enzyme based on active inhibitors and enzyme structure. Mol Cell Endocrinol. 2009;301:225–228. doi: 10.1016/j.mce.2008.08.030. [DOI] [PubMed] [Google Scholar]
- [138].Starcevic S, Turk S, Brus B, Cesar J, Lanisnik Rizner T, Gobec S. Discovery of highly potent, nonsteroidal 17β-hydroxysteroid dehydrogenase type 1 inhibitors by virtual high-throughput screening. J Steroid Biochem Mol Biol. 2011;127:255–261. doi: 10.1016/j.jsbmb.2011.08.013. [DOI] [PubMed] [Google Scholar]
- [139].Frotscher M, Ziegler E, Marchais-Oberwinkler S, Kruchten P, Neugebauer A, Fetzer L, Scherer C, Muller-Vieira U, Messinger J, Thole H, Hartmann RW. Design, synthesis, and biological evaluation of (hydroxyphenyl)naphthalene and -quinoline derivatives: potent and selective nonsteroidal inhibitors of 17β-hydroxysteroid dehydrogenase type 1 (17beta-HSD1) for the treatment of estrogen-dependent diseases. J Med Chem. 2008;51:2158–2169. doi: 10.1021/jm701447v. [DOI] [PubMed] [Google Scholar]
- [140].Marchais-Oberwinkler S, Kruchten P, Frotscher M, Ziegler E, Neugebauer A, Bhoga U, Bey E, Muller-Vieira U, Messinger J, Thole H, Hartmann RW. Substituted 6-phenyl-2-naphthols. Potent and selective nonsteroidal inhibitors of 17β-hydroxysteroid dehydrogenase type 1 (17β-HSD1): Design, synthesis, biological evaluation, and pharmacokinetics. J Med Chem. 2008;51:4685–4698. doi: 10.1021/jm800367k. [DOI] [PubMed] [Google Scholar]
- [141].Marchais-Oberwinkler S, Frotscher M, Ziegler E, Werth R, Kruchten P, Messinger J, Thole H, Hartmann RW. Structure-activity study in the class of 6-(3'-hydroxyphenyl)naphthalenes leading to an optimization of a pharmacophore model for 17β-hydroxysteroid dehydrogenase type 1 (17β-HSD1) inhibitors. Mol Cell Endocrinol. 2009;301:205–211. doi: 10.1016/j.mce.2008.09.024. [DOI] [PubMed] [Google Scholar]
- [142].Bey E, Marchais-Oberwinkler S, Werth R, Negri M, Al-Soud YA, Kruchten P, Oster A, Frotscher M, Birk B, Hartmann RW. Design, synthesis, biological evaluation and pharmacokinetics of bis(hydroxyphenyl) substituted azoles, thiophenes, benzenes, and azabenzenes as potent and selective nonsteroidal inhibitors of 17β-hydroxysteroid dehydrogenase type 1 (17β-HSD1) J Med Chem. 2008;51:6725–6739. doi: 10.1021/jm8006917. [DOI] [PubMed] [Google Scholar]
- [143].Moller G, Deluca D, Gege C, Rosinus A, Kowalik D, Peters O, Droescher P, Elger W, Adamski J, Hillisch A. Structure-based design, synthesis and in vitro characterization of potent 17β-hydroxysteroid dehydrogenase type 1 inhibitors based on 2-substitutions of estrone and D-homo-estrone. Bioorg Med Chem Lett. 2009;19:6740–6744. doi: 10.1016/j.bmcl.2009.09.113. [DOI] [PubMed] [Google Scholar]
- [144].Messinger J, Hirvela L, Husen B, Kangas L, Koskimies P, Pentikainen O, Saarenketo P, Thole H. New inhibitors of 17β-hydroxysteroid dehydrogenase type 1. Mol Cell Endocrinol. 2006;248:192–198. doi: 10.1016/j.mce.2005.11.044. [DOI] [PubMed] [Google Scholar]
- [145].Schuster D, Spetea M, Music M, Rief S, Fink M, Kirchmair J, Schütz J, Wolber G, Langer T, Stuppner H, Schmidhammer H, et al. Morphinans and isoquinolines: Acetylcholinesterase inhibition, pharmacophore modeling, and interaction with opioid receptors. Bioorg Med Chem. 2010;18:5071–5080. doi: 10.1016/j.bmc.2010.05.071. [DOI] [PubMed] [Google Scholar]
- [146].Wu L, Einstein M, Geissler WM, Chan HK, Elliston KO, Andersson S. Expression cloning and characterization of human 17β-hydroxysteroid dehydrogenase type 2, a microsomal enzyme possessing 20α-hydroxysteroid dehydrogenase activity. J Biol Chem. 1993;268:12964–12969. [PubMed] [Google Scholar]
- [147].Puranen TJ, Kurkela RM, Lakkakorpi JT, Poutanen MH, Itaranta PV, Melis JP, Ghosh D, Vihko RK, Vihko PT. Characterization of molecular and catalytic properties of intact and truncated human 17β-hydroxysteroid dehydrogenase type 2 enzymes: Intracellular localization of the wild-type enzyme in the endoplasmic reticulum. Endocrinology. 1999;140:3334–3341. doi: 10.1210/endo.140.7.6861. [DOI] [PubMed] [Google Scholar]
- [148].Dong Y, Qiu QQ, Debear J, Lathrop WF, Bertolini DR, Tamburini PP. 17β-hydroxysteroid dehydrogenases in human bone cells. J Bone Miner Res. 1998;13:1539–1546. doi: 10.1359/jbmr.1998.13.10.1539. [DOI] [PubMed] [Google Scholar]
- [149].Vihko P, Isomaa V, Ghosh D. Structure and function of 17β-hydroxysteroid dehydrogenase type 1 and type 2. Mol Cell Endocrinol. 2001;171:71–76. doi: 10.1016/s0303-7207(00)00389-0. [DOI] [PubMed] [Google Scholar]
- [150].Michael H, Harkonen PL, Vaananen HK, Hentunen TA. Estrogen and testosterone use different cellular pathways to inhibit osteoclastogenesis and bone resorption. J Bone Miner Res. 2005;20:2224–2232. doi: 10.1359/JBMR.050803. [DOI] [PubMed] [Google Scholar]
- [151].Kanis JA, McCloskey EV, Johansson H, Cooper C, Rizzoli R, Reginster JY. European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporosis Int. 2013;24:23–57. doi: 10.1007/s00198-012-2074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Bagi CM, Wood J, Wilkie D, Dixon B. Effect of 17β-hydroxysteroid dehydrogenase type 2 inhibitor on bone strength in ovariectomized cynomolgus monkeys. J Musculoskeletal Neuronal Interact. 2008;8:267–280. [PubMed] [Google Scholar]
- [153].Vuorinen A, Engeli R, Meyer A, Bachmann F, Griesser UJ, Schuster D, Odermatt A. Ligand-based pharmacophore modeling and virtual screening for the discovery of novel 17β-hydroxysteroid dehydrogenase 2 inhibitors. J Med Chem. 2014;57:5995–6007. doi: 10.1021/jm5004914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Wetzel M, Gargano EM, Hinsberger S, Marchais-Oberwinkler S, Hartmann RW. Discovery of a new class of bicyclic substituted hydroxyphenylmethanones as 17β-hydroxysteroid dehydrogenase type 2 (17β-HSD2) inhibitors for the treatment of osteoporosis. Eur J Med Chem. 2012;47:1–17. doi: 10.1016/j.ejmech.2011.09.004. [DOI] [PubMed] [Google Scholar]
- [155].Oster A, Hinsberger S, Werth R, Marchais-Oberwinkler S, Frotscher M, Hartmann RW. Bicyclic substituted hydroxyphenylmethanones as novel inhibitors of 17β-hydroxysteroid dehydrogenase type 1 (17β-HSD1) for the treatment of estrogen-dependent diseases. J Med Chem. 2010;53:8176–8186. doi: 10.1021/jm101073q. [DOI] [PubMed] [Google Scholar]
- [156].Geissler WM, Davis DL, Wu L, Bradshaw KD, Patel S, Mendonca BB, Elliston KO, Wilson JD, Russell DW, Andersson S. Male pseudohermaphroditism caused by mutations of testicular 17β-hydroxysteroid dehydrogenase 3. Nat Genet. 1994;7:34–39. doi: 10.1038/ng0594-34. [DOI] [PubMed] [Google Scholar]
- [157].Boehmer AL, Brinkmann AO, Sandkuijl LA, Halley DJ, Niermeijer MF, Andersson S, de Jong FH, Kayserili H, de Vroede MA, Otten BJ, Rouwe CW, et al. 17β-hydroxysteroid dehydrogenase-3 deficiency: diagnosis, phenotypic variability, population genetics, and worldwide distribution of ancient and de novo mutations. J Clin Endocrinol Metab. 1999;84:4713–4721. doi: 10.1210/jcem.84.12.6174. [DOI] [PubMed] [Google Scholar]
- [158].Phelan N, Williams EL, Cardamone S, Lee M, Creighton SM, Rumsby G, Conway GS. Screening for mutations in 17β-hydroxysteroid dehydrogenase and androgen receptor in women presenting with partially virilised 46,XY disorders of sex development. Eur J Endocrinol. 2015;172:745–751. doi: 10.1530/EJE-14-0994. [DOI] [PubMed] [Google Scholar]
- [159].Chuang J, Vallerie A, Breech L, Saal HM, Alam S, Crawford P, Rutter MM. Complexities of gender assignment in 17β-hydroxysteroid dehydrogenase type 3 deficiency: Is there a role for early orchiectomy? Int J Pediatr Endocrinol. 2013;2013:15. doi: 10.1186/1687-9856-2013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Koh E, Noda T, Kanaya J, Namiki M. Differential expression of 17β-hydroxysteroid dehydrogenase isozyme genes in prostate cancer and noncancer tissues. Prostate. 2002;53:154–159. doi: 10.1002/pros.10139. [DOI] [PubMed] [Google Scholar]
- [161].Day JM, Foster PA, Tutill HJ, Schmidlin F, Sharland CM, Hargrave JD, Vicker N, Potter BV, Reed MJ, Purohit A. STX2171, a 17β-hydroxysteroid dehydrogenase type 3 inhibitor, is efficacious in vivo in a novel hormone-dependent prostate cancer model. Endocr-Rela Cancer. 2013;20:53–64. doi: 10.1530/ERC-12-0231. [DOI] [PubMed] [Google Scholar]
- [162].Hamid AR, Pfeiffer MJ, Verhaegh GW, Schaafsma E, Brandt A, Sweep FC, Sedelaar JP, Schalken JA. Aldo-keto reductase family 1 member C3 (AKR1C3) is a biomarker and therapeutic target for castration-resistant prostate cancer. Mol Med. 2012;18:1449–1455. doi: 10.2119/molmed.2012.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Penning TM, Burczynski ME, Jez JM, Lin HK, Ma H, Moore M, Ratnam K, Palackal N. Structure-function aspects and inhibitor design of type 5 17β-hydroxysteroid dehydrogenase (AKR1C3) Mol Cell Endocrinol. 2001;171:137–149. doi: 10.1016/s0303-7207(00)00426-3. [DOI] [PubMed] [Google Scholar]
- [164].Legeza B, Balazs Z, Nashev LG, Odermatt A. The microsomal enzyme 17β-hydroxysteroid dehydrogenase 3 faces the cytoplasm and uses NADPH generated by glucose-6-phosphate dehydrogenase. Endocrinology. 2013;154:205–213. doi: 10.1210/en.2012-1778. [DOI] [PubMed] [Google Scholar]
- [165].Tsachaki M, Birk J, Egert A, Odermatt A. Determination of the topology of endoplasmic reticulum membrane proteins using redox-sensitive green-fluorescence protein fusions. Biochim Biophys Acta. 2015;1853:1672–1682. doi: 10.1016/j.bbamcr.2015.04.002. [DOI] [PubMed] [Google Scholar]
- [166].Vicker N, Sharland CM, Heaton WB, Gonzalez AM, Bailey HV, Smith A, Springall JS, Day JM, Tutill HJ, Reed MJ, Purohit A, et al. The design of novel 17β-hydroxysteroid dehydrogenase type 3 inhibitors. Mol Cell Endocrinol. 2009;301:259–265. doi: 10.1016/j.mce.2008.08.005. [DOI] [PubMed] [Google Scholar]
- [167].Schuster D, Kowalik D, Kirchmair J, Laggner C, Markt P, Aebischer-Gumy C, Strohle F, Moller G, Wolber G, Wilckens T, Langer T, et al. Identification of chemically diverse, novel inhibitors of 17β-hydroxysteroid dehydrogenase type 3 and 5 by pharmacophore-based virtual screening. J Steroid Biochem Mol Biol. 2011;125:148–161. doi: 10.1016/j.jsbmb.2011.01.016. [DOI] [PubMed] [Google Scholar]
- [168].He XY, Merz G, Yang YZ, Mehta P, Schulz H, Yang SY. Characterization and localization of human type10 17β-hydroxysteroid dehydrogenase. Eur J Biochem. 2001;268:4899–4907. doi: 10.1046/j.0014-2956.2001.02421.2421.x. [DOI] [PubMed] [Google Scholar]
- [169].He XY, Merz G, Yang YZ, Pullakart R, Mehta P, Schulz H, Yang SY. Function of human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase in androgen metabolism. Biochim Biophys Acta. 2000;1484:267–277. doi: 10.1016/s1388-1981(00)00014-7. [DOI] [PubMed] [Google Scholar]
- [170].Yang SY, He XY, Miller D. Hydroxysteroid (17β) dehydrogenase X in human health and disease. Mol Cell Endocrinol. 2011;343:1–6. doi: 10.1016/j.mce.2011.06.011. [DOI] [PubMed] [Google Scholar]
- [171].Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, Caspersen C, Chen X, Pollak S, Chaney M, Trinchese F, et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- [172].Kissinger CR, Rejto PA, Pelletier LA, Thomson JA, Showalter RE, Abreo MA, Agree CS, Margosiak S, Meng JJ, Aust RM, Vanderpool D, et al. Crystal structure of human ABAD/HSD10 with a bound inhibitor: implications for design of Alzheimer's disease therapeutics. J Mol Biol. 2004;342:943–952. doi: 10.1016/j.jmb.2004.07.071. [DOI] [PubMed] [Google Scholar]
- [173].Jazbutyte V, Kehl F, Neyses L, Pelzer T. Estrogen receptor alpha interacts with 17β-hydroxysteroid dehydrogenase type 10 in mitochondria. Biochem Biophys Res Commun. 2009;384:450–454. doi: 10.1016/j.bbrc.2009.04.139. [DOI] [PubMed] [Google Scholar]
- [174].He XY, Wegiel J, Yang YZ, Pullarkat R, Schulz H, Yang SY. Type 10 17β-hydroxysteroid dehydrogenase catalyzing the oxidation of steroid modulators of gamma-aminobutyric acid type A receptors. Mol Cell Endocrinol. 2005;229:111–117. doi: 10.1016/j.mce.2004.08.011. [DOI] [PubMed] [Google Scholar]
- [175].Nordling E, Oppermann UC, Jornvall H, Persson B. Human type 10 17β-hydroxysteroid dehydrogenase: molecular modelling and substrate docking. J Mol Graph Model. 2001;19:514–520. doi: 10.1016/s1093-3263(00)00098-x. 591-513. [DOI] [PubMed] [Google Scholar]
- [176].Shafqat N, Marschall HU, Filling C, Nordling E, Wu XQ, Bjork L, Thyberg J, Martensson E, Salim S, Jornvall H, Oppermann U. Expanded substrate screenings of human and Drosophila type 10 17β-hydroxysteroid dehydrogenases (HSDs) reveal multiple specificities in bile acid and steroid hormone metabolism: characterization of multifunctional 3alpha/7alpha/7beta/17beta/20beta/21-HSD. Biochem J. 2003;376:49–60. doi: 10.1042/BJ20030877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Powell AJ, Read JA, Banfield MJ, Gunn-Moore F, Yan SD, Lustbader J, Stern AR, Stern DM, Brady RL. Recognition of structurally diverse substrates by type II 3-hydroxyacyl-CoA dehydrogenase (HADH II)/amyloid-beta binding alcohol dehydrogenase (ABAD) J Mol Biol. 2000;303:311–327. doi: 10.1006/jmbi.2000.4139. [DOI] [PubMed] [Google Scholar]
- [178].Tanaka N, Nonaka T, Tanabe T, Yoshimoto T, Tsuru D, Mitsui Y. Crystal structures of the binary and ternary complexes of 7α-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry. 1996;35:7715–7730. doi: 10.1021/bi951904d. [DOI] [PubMed] [Google Scholar]
- [179].Lukacik P, Keller B, Bunkoczi G, Kavanagh KL, Lee WH, Adamski J, Oppermann U. Structural and biochemical characterization of human orphan DHRS10 reveals a novel cytosolic enzyme with steroid dehydrogenase activity. Biochem J. 2007;402:419–427. doi: 10.1042/BJ20061319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Sivik T, Vikingsson S, Green H, Jansson A. Expression patterns of 17β-hydroxysteroid dehydrogenase 14 in human tissues. Horm Metab Res. 2012;44:949–956. doi: 10.1055/s-0032-1321815. [DOI] [PubMed] [Google Scholar]
- [181].Bertoletti N, Braun F, Lepage M, Moller G, Adamski J, Heine A, Klebe G, Marchais-Oberwinkler S. New Insights into Human 17β-Hydroxysteroid Dehydrogenase Type 14: First Crystal Structures in Complex with a Steroidal Ligand and with a Potent Nonsteroidal Inhibitor. J Med Chem. 2016;59:6961–6967. doi: 10.1021/acs.jmedchem.6b00293. [DOI] [PubMed] [Google Scholar]
- [182].Braun F, Bertoletti N, Moller G, Adamski J, Steinmetzer T, Salah M, Abdelsamie AS, van Koppen CJ, Heine A, Klebe G, Marchais-Oberwinkler S. First Structure-Activity Relationship of 17β-Hydroxysteroid Dehydrogenase Type 14 Nonsteroidal Inhibitors and Crystal Structures in Complex with the Enzyme. J Med Chem. 2016;59:10719–10737. doi: 10.1021/acs.jmedchem.6b01436. [DOI] [PubMed] [Google Scholar]
- [183].Favia AD, Nobeli I, Glaser F, Thornton JM. Molecular docking for substrate identification: the short-chain dehydrogenases/reductases. J Mol Biol. 2008;375:855–874. doi: 10.1016/j.jmb.2007.10.065. [DOI] [PubMed] [Google Scholar]
- [184].Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Mallipeddi PL, Joshi M, Briggs JM. Pharmacophore-based virtual screening to aid in the identification of unknown protein function. Chem Biol Drug Des. 2012;80:828–842. doi: 10.1111/j.1747-0285.2012.01408.x. [DOI] [PubMed] [Google Scholar]
- [186].Reinhardt N, Fischer J, Coppi R, Blum E, Brandt W, Drager B. Substrate flexibility and reaction specificity of tropinone reductase-like short-chain dehydrogenases. Bioorg Chem. 2014;53:37–49. doi: 10.1016/j.bioorg.2014.01.004. [DOI] [PubMed] [Google Scholar]
- [187].Oppermann U. Carbonyl reductases: the complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol. 2007;47:293–322. doi: 10.1146/annurev.pharmtox.47.120505.105316. [DOI] [PubMed] [Google Scholar]
- [188].Pilka ES, Niesen FH, Lee WH, El-Hawari Y, Dunford JE, Kochan G, Wsol V, Martin HJ, Maser E, Oppermann U. Structural basis for substrate specificity in human monomeric carbonyl reductases. PLoS One. 2009;4:e7113. doi: 10.1371/journal.pone.0007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].El-Hawari Y, Favia AD, Pilka ES, Kisiela M, Oppermann U, Martin HJ, Maser E. Analysis of the substrate-binding site of human carbonyl reductases CBR1 and CBR3 by site-directed mutagenesis. Chem Biol Interact. 2009;178:234–241. doi: 10.1016/j.cbi.2008.11.004. [DOI] [PubMed] [Google Scholar]
- [190].Botella JA, Ulschmid JK, Gruenewald C, Moehle C, Kretzschmar D, Becker K, Schneuwly S. The Drosophila carbonyl reductase sniffer prevents oxidative stress-induced neurodegeneration. Curr Biol. 2004;14:782–786. doi: 10.1016/j.cub.2004.04.036. [DOI] [PubMed] [Google Scholar]
- [191].Sgraja T, Ulschmid J, Becker K, Schneuwly S, Klebe G, Reuter K, Heine A. Structural insights into the neuroprotective-acting carbonyl reductase Sniffer of Drosophila melanogaster. J Mol Biol. 2004;342:1613–1624. doi: 10.1016/j.jmb.2004.08.020. [DOI] [PubMed] [Google Scholar]
- [192].Martin HJ, Ziemba M, Kisiela M, Botella JA, Schneuwly S, Maser E. The Drosophila carbonyl reductase sniffer is an efficient 4-oxonon-2-enal (4ONE) reductase. Chem Biol Interact. 2011;191:48–54. doi: 10.1016/j.cbi.2010.12.006. [DOI] [PubMed] [Google Scholar]
- [193].Blumberg B, Iguchi T, Odermatt A. Endocrine disrupting chemicals. J Steroid Biochem Mol Biol. 2011;127:1–3. doi: 10.1016/j.jsbmb.2011.07.012. [DOI] [PubMed] [Google Scholar]
- [194].Mune T, Rogerson FM, Nikkila H, Agarwal AK, White PC. Human hypertension caused by mutations in the kidney isozyme of 11β-hydroxysteroid dehydrogenase. Nat Genet. 1995;10:394–399. doi: 10.1038/ng0895-394. [DOI] [PubMed] [Google Scholar]
- [195].Wilson RC, Harbison MD, Krozowski ZS, Funder JW, Shackleton CHL, Hanauskeabel HM, Wei JQ, Hertecant J, Moran A, Neiberger RE, Balfe JW, et al. Several homozygous mutations in the gene for 11β-hydroxysteroid dehydrogenase type-2 in patients with apparent mineralocorticoid excess. J Clin Endocrinol Metab. 1995;80:3145–3150. doi: 10.1210/jcem.80.11.7593417. [DOI] [PubMed] [Google Scholar]
- [196].Lindsay RS, Lindsay RM, Edwards CR, Seckl JR. Inhibition of 11β-hydroxysteroid dehydrogenase in pregnant rats and the programming of blood pressure in the offspring. Hypertension. 1996;27:1200–1204. doi: 10.1161/01.hyp.27.6.1200. [DOI] [PubMed] [Google Scholar]
- [197].Nyirenda MJ, Lindsay RS, Kenyon CJ, Burchell A, Seckl JR. Glucocorticoid exposure in late gestation permanently programs rat hepatic phosphoenolpyruvate carboxykinase and glucocorticoid receptor expression and causes glucose intolerance in adult offspring. J Clin Invest. 1998;101:2174–2181. doi: 10.1172/JCI1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Cottrell EC, Seckl JR, Holmes MC, Wyrwoll CS. Foetal and placental 11β-HSD2: a hub for developmental programming. Acta Physiol (Oxf) 2014;210:288–295. doi: 10.1111/apha.12187. [DOI] [PubMed] [Google Scholar]
- [199].Togher KL, O'Keeffe MM, Khashan AS, Gutierrez H, Kenny LC, O'Keeffe GW. Epigenetic regulation of the placental HSD11B2 barrier and its role as a critical regulator of fetal development. Epigenetics. 2014;9:816–822. doi: 10.4161/epi.28703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Wang L, Kannan K. Characteristic profiles of benzonphenone-3 and its derivatives in urine of children and adults from the United States and China. Environ Sci Technol. 2013;47:12532–12538. doi: 10.1021/es4032908. [DOI] [PubMed] [Google Scholar]
- [201].Fourches D, Muratov E, Tropsha A. Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J Chem Inf Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Temml V, Voss CV, Dirsch VM, Schuster D. Discovery of new liver X receptor agonists by pharmacophore modeling and shape-based virtual screening. J Chem Inf Model. 2014;54:367–371. doi: 10.1021/ci400682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Cross JB, Thompson DC, Rai BK, Baber JC, Fan KY, Hu Y, Humblet C. Comparison of several molecular docking programs: pose prediction and virtual screening accuracy. J Chem Inf Model. 2009;49:1455–1474. doi: 10.1021/ci900056c. [DOI] [PubMed] [Google Scholar]
- [204].Kaserer T, Höferl M, Müller K, Elmer S, Ganzera M, Jäger W, Schuster D. In silico predictions of drug-drug interactions caused by CYP1A2, 2C9, and 3A4 inhibition - a comparative study of virtual screening performance. Mol Inf. 2015;34:431–457. doi: 10.1002/minf.201400192. [DOI] [PubMed] [Google Scholar]
- [205].Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrian-Uhalte E, Davies M, et al. The ChEMBL database in 2017. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Koch MA, Wittenberg LO, Basu S, Jeyaraj DA, Gourzoulidou E, Reinecke K, Odermatt A, Waldmann H. Compound library development guided by protein structure similarity clustering and natural product structure. Proc Natl Acad Sci U S A. 2004;101:16721–16726. doi: 10.1073/pnas.0404719101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Beets I, Lindemans M, Janssen T, Verleyen P. Deorphanizing g protein-coupled receptors by a calcium mobilization assay. Methods Mol Biol. 2011;789:377–391. doi: 10.1007/978-1-61779-310-3_25. [DOI] [PubMed] [Google Scholar]
- [209].Moller G, Husen B, Kowalik D, Hirvela L, Plewczynski D, Rychlewski L, Messinger J, Thole H, Adamski J. Species used for drug testing reveal different inhibition susceptibility for 17β-hydroxysteroid dehydrogenase type 1. PLoS One. 2010;5:e10969. doi: 10.1371/journal.pone.0010969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Abdelsamie AS, Bey E, Gargano EM, van Koppen CJ, Empting M, Frotscher M. Towards the evaluation in an animal disease model: Fluorinated 17β-HSD1 inhibitors showing strong activity towards both the human and the rat enzyme. Eur J Med Chem. 2015;103:56–68. doi: 10.1016/j.ejmech.2015.08.030. [DOI] [PubMed] [Google Scholar]
- [211].Mazumdar M, Fournier D, Zhu D-W, Cadot C, Poirier D, Lin S-X. Binary and ternary crystal structure analyses of a novel inhibitor with 17β-HSD type 1: a lead compound for breast cancer therapy. Biochem J. 2009;424:357–366. doi: 10.1042/BJ20091020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.