Abstract
To clarify difference in the responses on the reprogramming of metabolism toward carcinogenesis between genotoxic and non-genotoxic hepatocarcinogens in the liver, rats were repeatedly administered genotoxic hepatocarcinogens (N-nitrosodiethylamine, aflatoxin B1, N-nitrosopyrrolidine, or carbadox) or non-genotoxic hepatocarcinogens (carbon tetrachloride, thioacetamide, or methapyrilene hydrochloride) for 28, 84, or 90 days. Non-genotoxic hepatocarcinogens revealed transcript expression changes suggestive of suppressed mitochondrial oxidative phosphorylation (OXPHOS) after 28 days and increased glutathione S-transferase placental form-positive (GST-P+) foci downregulating adenosine triphosphate (ATP) synthase subunit beta, mitochondrial precursor (ATPB), compared with genotoxic hepatocarcinogens after 84 or 90 days, suggesting that non-genotoxic hepatocarcinogens are prone to suppress OXPHOS from the early stage of treatment, which is in contrast to genotoxic hepatocarcinogens. Both genotoxic and non-genotoxic hepatocarcinogens upregulated glycolytic enzyme genes and increased cellular membrane solute carrier family 2, facilitated glucose transporter member 1 (GLUT1) expression in GST-P+ foci for up to 90 days, suggesting induction of a metabolic shift from OXPHOS to glycolysis at early hepatocarcinogenesis by hepatocarcinogens unrelated to genotoxic potential. Non-genotoxic hepatocarcinogens increased c-MYC+ cells after 28 days and downregulated Tp53 after 84 or 90 days, suggesting a commitment to enhanced metabolic shift and cell proliferation. Genotoxic hepatocarcinogens also enhanced c-MYC activation-related metabolic shift until 84 or 90 days. In addition, both genotoxic and non-genotoxic hepatocarcinogens upregulated glutaminolysis-related Slc1a5 or Gls, or both, after 28 days and induced liver cell foci immunoreactive for neutral amino acid transporter B(0) (SLC1A5) in the subpopulation of GST-P+ foci after 84 or 90 days, suggesting glutaminolysis-mediated facilitation of cell proliferation toward hepatocarcinogenesis. These results suggest differential responses between genotoxic and non-genotoxic hepatocarcinogens on reprogramming of energy metabolic pathways toward carcinogenesis in liver cells from the early stage of hepatocarcinogen treatment.
Keywords: hepatocarcinogenesis, genotoxicity, rat, liver, oxidative phosphorylation, glycolysis
Introduction
Evaluation of chemical carcinogenicity is crucial for the assessment of chemical safety. However, administering test compounds to hundreds of rodents over a prolonged period in standard carcinogenicity bioassays is time-consuming and costly. In previous studies to identify early prediction marker molecules of hepatocarcinogenesis in rats, we reported that administration of carcinogens for 28 days induces expression changes in cell cycle-related molecules resulting in cell cycle arrest in many target organs1, 2, 3. Considering that cell cycle arrest is a typical feature of cellular senescence4, our previous study results suggest an increased number of liver cells undergoing cellular senescence after repeated carcinogenic stimuli.
Carcinogens are currently categorized into two classes, genotoxic and non-genotoxic carcinogens, which are subject to different regulatory policies5. Genotoxic carcinogens exert carcinogenicity through induction of mutations6, and there is thought to be no safe exposure threshold or dose because of their DNA interaction properties. Genotoxic carcinogens are regulated under the assumption that they pose a cancer risk for humans, even at very low doses7. In contrast, non-genotoxic carcinogens, which induce cancer through mechanisms other than mutations, such as cytotoxicity, cell proliferation, hormonal influence, or epigenetic alterations, are thought to have a safe exposure threshold or dose. Thus, use of non-genotoxic carcinogens is permitted unless the exposure or intake level exceeds the threshold7. Therefore, understanding the mode of action of carcinogens in relation to carcinogenic potential, whether through a genotoxic or non-genotoxic mechanism, is important for risk assessment of chemical carcinogens.
We have previously reported that thioacetamide (TAA) and methapyrilene hydrochloride (MP), non-genotoxic hepatocarcinogens that facilitate target cellular proliferation with repeated administration in rats for up to 90 days, clearly facilitate cell cycle arrest during both the G1/S and G2/M phases through the mechanism involving upregulation of Tp53 and p21WAF1/CIP1 activation in liver cells8. In contrast, carbadox (CRB), a genotoxic hepatocarcinogen, slightly induces p21WAF1/CIP1 activation alone even after administration for up to 90 days8. These results indicate that the responses of cellular senescence-related molecules may differ between genotoxic and non-genotoxic hepatocarcinogens.
Normal mammalian cells generate adenosine triphosphate (ATP) by mitochondrial oxidative phosphorylation (OXPHOS) through the tricarboxylic acid (TCA) cycle, which utilizes oxygen. On the other hand, cancer cells alter their metabolism in order to support the increased energy requirement due to continuous growth, rapid proliferation, and other characteristics typical of neoplastic cells9. This phenomenon of changes of tumor cellular bioenergetics, called “metabolic reprogramming”, has been recognized as one of the hallmarks of cancer10. The “Warburg effect”, which refers to active utilization of a glycolytic system with low efficiency of ATP production, represents one of the metabolic reprogrammings found in cancer cells11. In fact, the enhanced tumor uptake of 2-deoxy-2(18F)-fluoro-D-glucose in positron emission tomography scans is now exploited in clinics for diagnostic purposes12. In addition to glycolysis, it has been reported that cancer cells also utilize glutaminolysis to increase ATP production13. On the other hand, ATP synthase of OXPHOS is downregulated in many types of carcinoma14. Furthermore, mitochondrial dysfunction promotes secondary glycolysis in RasV12-transformed cells surrounded by normal cells15, suggesting that suppression of OXPHOS induces activation of the other energy metabolic pathways. We have previously found downregulation of a mitochondrial OXPHOS-related protein, transmembrane protein 70 (TMEM70), which is suggestive of disrupted cellular senescence, in glutathione S-transferase placental form (GST-P)-expressing (+) proliferative lesions in rat hepatocarcinogenesis using an initiation promotion model and non-genotoxic hepatocarcinogens as tumor promoters16. In that study, GST-P+ preneoplastic lesions showing TMEM70 downregulation also downregulated the ATP synthase subunit beta, mitochondrial precursor (ATPB), but upregulated solute carrier family 2, facilitated glucose transporter member 1 (GLUT1) and glucose-6-phosphate 1-dehydrogenase (G6PD), suggesting a metabolic shift via the Warburg effect.
We hypothesize that the responses on reprogramming of energy metabolic pathways toward carcinogenesis may differ between genotoxic and non-genotoxic hepatocarcinogens from the early stage of hepatocarcinogen treatment. It is important to elucidate the carcinogenic pathways of cellular metabolism that can distinguish the respective types of hepatocarcinogens. To identify the difference in pattern of cellular metabolism between genotoxic and non-genotoxic hepatocarcinogens, the present study examined the transcript levels of cellular metabolism-related genes in the liver of rats treated with genotoxic or non-genotoxic hepatocarcinogens for 28 and 84 or 90 days, as well as the immunohistochemical cellular distribution of the cellular metabolism-related molecules in the liver of rats treated with these hepatocarcinogens for up to 90 days.
Materials and Methods
Chemicals
Carbon tetrachloride (CCl4; CAS No. 56-23-5, purity ≥ 99.5%), thioacetamide (TAA; CAS No. 62-55-5, purity ≥ 98%), carbadox (CRB; CAS No. 6804-07-5, purity ≥99%), dimethyl sulfoxide (DMSO; purity ≥99.5%), and corn oil were obtained from Fujifilm Wako Pure Chemical Corporation (Osaka, Japan). Aflatoxin B1 (AFB1; CAS No. 1162-65-8) was extracted from medial and mycelial fractions of cultivated A. flavus in M1 medium and purified by high-performance liquid chromatography as previously described17. According to AOAC official method 970.44, the purity of aflatoxin B1 was calculated to be approximately 90% based on the absorption peak ratios of ultraviolet measurements on methanol18. N-nitrosodiethylamine (DEN; CAS No. 55-18-5, purity ≥99%) was purchased from Tokyo Chemical Industry Co. Ltd. (Tokyo, Japan), and N-nitrosopyrrolidine (NPYR; CAS No. 930-55-2, purity ≥99%) and methapyrilene hydrochloride (MP; CAS No. 135-23-9, purity ≥98%) were purchased from MilliporeSigma (St. Louis, MO, USA).
Animal experiments
Five-week-old male F344/NSlc rats were purchased from Japan SLC, Inc. (Hamamatsu, Japan) and acclimatized to a basal diet (CRF-1; Oriental Yeast Co., Ltd., Tokyo, Japan) and tap water ad libitum. They were housed in plastic cages with paper chip bedding in a barrier-maintained animal room under standard conditions (room temperature, 23 ± 2°C; relative humidity, 55 ± 15%; 12-h light-dark cycle). After a one-week acclimatization period, animals were randomized into three groups (Experiment 1) or six groups (Experiment 2) of 10 animals per group. In Experiment 1, animals were provided a basal diet (untreated controls) or treated with DEN (4 mg/5 mL/kg body weight, dissolved in saline) or CCl4 (100 mg/5 mL/kg body weight, dissolved in corn oil) daily by gavage for 28 days or 90 days. In Experiment 2, animals were provided a basal diet (untreated controls) or treated with AFB1 (15 µg/0.5 mL/kg body weight, dissolved in DMSO) daily by gavage, NPYR (13 mg/5 mL/kg body weight, dissolved in saline) daily by gavage, CRB (300 ppm) in diet, TAA (400 ppm) in diet, or MP (1,000 ppm) in diet for 28 or 90 days. In the CCl4 group in Experiment 1, the initial dose was set at 100 mg/kg body weight daily by gavage. However, as two animals died and the general conditions of the remaining animals worsened at day 80, the dose was reduced to 50 mg/kg body weight after 80 days from the start of administration of CCl4. At day 84, another animal died, and the general conditions of the other animals worsened in CCl4 group; therefore, it was decided to terminate Experiment 1 at this time point. DEN, AFB1, NPYR, and CRB were selected as genotoxic hepatocarcinogens in rats; the dose level used for each of these compounds has been shown to induce liver tumors or preneoplastic liver lesions after 5 or 13 weeks of treatment or neoplastic lesions after 10 months of treatment, respectively19, 20, 21, 22, 23. CCl4, TAA, and MP were selected as non-genotoxic hepatocarcinogens; the dose level of each of these compounds, even after the dose change of CCl4, has been shown to induce neoplastic liver lesions in carcinogenicity bioassays24, 25, 26, 27. The animals of all groups were euthanized by exsanguination from the posterior vena cava and abdominal aorta under CO2/O2 anesthesia at the next day of the 28 days (Experiments 1 and 2), or 84 days (Experiment 1), or 90 days (Experiment 2) of treatment. At necropsy, livers were removed, weighed, and then cut into small pieces (approximately 30 mg/sample). All samples were immediately frozen in liquid nitrogen and stored at −80°C until total RNA extraction. In addition, liver slices (2 slices per animal, one from the median lobe and another from the left lateral lobe) were fixed in 4% (w/v) paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) overnight and processed for histopathological examinations. Animal samples were identical to those previously reported28.
All animal experiments of this study were conducted in compliance with the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan, 1 June 2006), and the protocols were approved by the Animal Care and Use Committee of Tokyo University of Agriculture and Technology. All efforts were made to minimize animal suffering.
Transcript expression analysis
Real-time reverse transcription-polymerase chain reaction (RT-PCR) quantification of mRNA was performed for cellular metabolism-related genes and transcription factor genes on RNA samples (n=6/group) isolated from the untreated controls and each treatment groups in Experiments 1 and 2. Total RNA was extracted with an RNeasy Mini Kit (Qiagen, Hilden, Germany), and first-strand complementary DNA was synthesized from 2 µg of total RNA using SuperScript® III Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). Real-time PCR was performed using Power SYBR® Green PCR Master Mix and an Applied Biosystems StepOnePlusTM Real-Time PCR System (Thermo Fisher Scientific). The PCR primers listed in Supplementary Table 1 (online only) for target genes were designed using Primer Express version 3.0 (Thermo Fisher Scientific). Using the threshold cycle (CT) values of actin, beta (Actb), or hypoxanthine phosphoribosyltransferase 1 (Hprt1) in the same sample as the endogenous control, the relative differences in gene expression were calculated using the 2-ΔΔCT method29.
Histopathology and immunohistochemistry
Liver slices in Experiments 1 and 2 (n=10/group) were processed using a standard protocol for paraffin embedding and were serially sectioned in 3-µm thick sections. Immunohistochemistry was performed by incubating liver tissue sections overnight at 4°C with primary antibodies against GST-P, a preneoplastic liver cell lesion marker in rats30, 31; ATPB, which catalyzes ATP synthesis and utilizes an electrochemical gradient of protons across the inner membrane during oxidative phosphorylation32; GLUT1, which facilitates the glucose transport across the plasma membranes of mammalian cells33; G6PD, an enzyme which is responsible for the first step in the pentose phosphate pathway (PPP) to participate in nucleotide synthesis33; pyruvate kinase L/R (PKLR), a major isoform that plays a part in the glycolysis of the normal liver34; pyruvate kinase isozyme M2 (PKM2), which promotes aerobic glycolysis in cancer cells33; neutral amino acid transporter B(0) (SLC1A5), also known as ASCT2, which facilitates the glutamine transport across the plasma membranes of mammalian cells35; and c-MYC, a regulator of glycolysis and glutaminolysis36. Antigen retrieval conditions and the concentration of each antibody are shown in Supplementary Table 2 (online only). To inhibit endogenous peroxidase, deparaffinized sections were incubated in 0.3% H2O2 solution in absolute methanol for 30 min. Immunodetection was performed using a Vectastain® Elite ABC Kit (PK6101, PK6102, PK6105, Vector Laboratories Inc., Burlingame, CA, USA) with the primary antibodies and 3,3’-diaminobenzidine/H2O2 as the chromogen. All immunostained slides were counterstained with hematoxylin and coverslipped for microscopic examination.
Analysis of immunolocalization
The number and area of GST-P+ liver cell foci larger than 200 μm in diameter in liver sections from Experiments 1 and 2 (n=10/group) were determined as described previously37. In the DEN and CCl4 groups after 84 days of treatment in Experiment 1 and the AFB1, NPYR, TAA, and MP groups after 90 days of treatment in Experiment 2 (n=10/group), the immunoreactivity of ATPB, GLUT1, G6PD, PKLR, PKM2, and SLC1A5 was classified as increased (+) or decreased (−) in the GST-P+ foci compared with the surrounding hepatocytes, and the incidences of ATPB−, GLUT1+, G6PD+, PKLR−, PKM2+, and SLC1A5+ expression in total GST-P+ foci that appeared in liver sections per animal were estimated. In the DEN and CCl4 groups after 28 days of treatment in Experiment 1 and NPYR and TAA groups after 28 days of treatment in Experiment 2 (n=10/group), the ratio of nuclear c-MYC+ cells to total liver cells was calculated in 10 randomly selected areas at a magnification of 400×. In the DEN and CCl4 groups after 84 days of treatment in Experiment 1 and the NPYR and TAA groups after 90 days of treatment in Experiment 2 (n=10/group), the ratio of nuclear c-MYC+ cells to total liver cells was also calculated for each of the inside and outside regions of GST-P+ foci in 10 randomly selected areas at a magnification of 400×.
Statistical analysis
Numerical data are presented as the mean ± SD. For comparison of the numerical data between multiple groups, values were analyzed by Bartlett’s test for homogeneity of variance. If there was no significant difference in variance, Dunnett’s test was performed for comparison between the untreated controls and each treatment group. If a significant difference was found in variance, Steel’s test was performed. In case of comparison of data among all pairs, values were analyzed by Bartlett’s test for homogeneity of variance. If there was no significant difference in variance, Tukey’s test was performed for comparison among the groups. If a significant difference was found in variance, Steel-Dwass test was performed. With regard to categorical data, Fisher’s exact test was performed. All analyses were performed using Excel Statistics 2013 software package version 2.02 (Social Survey Research Information Co., Ltd., Tokyo, Japan), and P<0.05 was considered statistically significant.
Results
Transcript expression changes
Table 1 and 2 summarizes the data regarding the transcript levels of the genes determined by real-time RT-PCR in groups of genotoxic hepatocarcinogens (DEN, AFB1, NPYR, or CRB) or non-genotoxic hepatocarcinogens (CCl4, TAA, or MP) and comparisons with the levels in untreated controls after 28 days and 84 or 90 days of treatment in Experiments 1 and 2 (Table 1 and 2, Supplementary Table 3–6: online only).
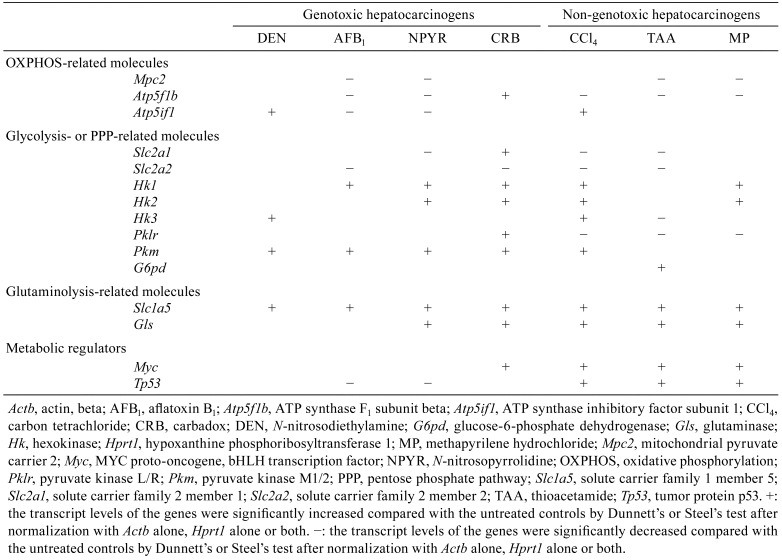
Table 1. Summary of Transcript Expression Levels in the Liver of Rats after Treatment with Genotoxic or Non-genotoxic Hepatocarcinogen for 28 Days.
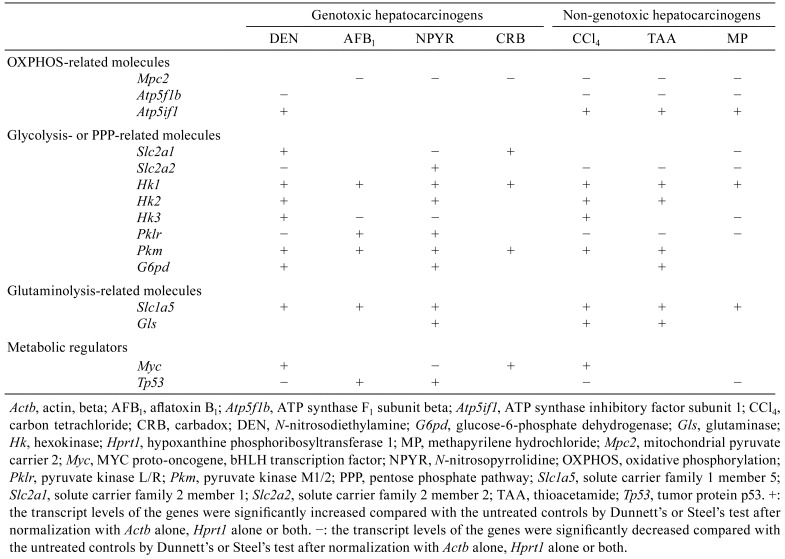
Table 2. Summary of Transcript Expression Levels in the Liver of Rats after Treatment with Genotoxic or Non-genotoxic Hepatocarcinogen for 84 or 90 Days.
After 28 days of treatment, the transcript level of Mpc2, which encodes a mitochondrial pyruvate transporter playing a role for OXPHOS, was significantly decreased in the AFB1, NPYR, TAA, and MP groups compared with untreated controls. The transcript level of Atp5f1b (also known as Atp5b), which encodes an ATP synthase, was significantly decreased in AFB1 group, NPYR group, and all non-genotoxic hepatocarcinogen groups and significantly increased in the CRB group compared with untreated controls. The transcript level of Atp5if1, which encodes a mitochondrial ATPase inhibitor, was significantly increased in the DEN and CCl4 groups and significantly decreased in the AFB1 and NPYR groups compared with untreated controls. With regard to genes related to glycolysis, the transcript level of Slc2a1, which encodes a glucose transporter, was significantly decreased in the NPYR, CCl4, and TAA groups and significantly increased in the CRB group compared with untreated controls. The transcript level of Slc2a2 was significantly decreased in the AFB1, CRB, CCl4, and TAA groups compared with untreated controls. With regard to genes encoding glycolytic enzymes, the transcript level of Hk1 was significantly increased in the AFB1, NPYR, CRB, CCl4, and MP groups compared with untreated controls. The transcript level of Hk2 was significantly increased in the NPYR, CRB, CCl4, and MP groups compared with untreated controls. The transcript level of Hk3 was significantly increased in the DEN and CCl4 groups and significantly decreased in the TAA group compared with untreated controls. The transcript level of Pklr was significantly increased in the CRB group and significantly decreased in all non-genotoxic hepatocarcinogen groups compared with untreated controls. The transcript level of Pkm was significantly increased in all genotoxic hepatocarcinogen groups and the CCl4 group compared with untreated controls. With regard to genes related to PPP, the transcript level of G6pd was significantly increased in the TAA group compared with untreated controls. With regard to genes related to glutaminolysis, the transcript level of Slc1a5, which encodes a glutamine transporter, was significantly increased in all hepatocarcinogen groups compared with untreated controls. The transcript level of Gls, which encodes a glutaminase, was significantly increased in the NPYR group, CRB group, and all non-genotoxic hepatocarcinogen groups compared with untreated controls. With regard to genes related to metabolic regulators, the transcript level of Myc was significantly increased in the CRB group and all non-genotoxic hepatocarcinogen groups compared with untreated controls. The transcript level of Tp53 was significantly decreased in the AFB1 and NPYR groups and significantly increased in all non-genotoxic hepatocarcinogen groups compared with untreated controls.
After 84 or 90 days of treatment, the transcript level of Mpc2, one of the genes related to OXPHOS, was significantly decreased in the AFB1 group, NPYR group, CRB group, and all non-genotoxic hepatocarcinogen groups compared with untreated controls. The transcript level of Atp5f1b was significantly decreased in the DEN and all non-genotoxic hepatocarcinogen groups compared with untreated controls. The transcript level of Atp5if1 was significantly increased in the DEN and all non-genotoxic hepatocarcinogen groups compared with untreated controls. With regard to genes related to glycolysis, the transcript level of Slc2a1 was significantly increased in the DEN and CRB groups and significantly decreased in the NPYR and MP groups compared with untreated controls. The transcript level of Slc2a2 was significantly decreased in the DEN and all non-genotoxic hepatocarcinogen groups and significantly increased in the NPYR group compared with untreated controls. With regard to genes encoding glycolytic enzymes, the transcript level of Hk1 was significantly increased in all hepatocarcinogen groups compared with untreated controls. The transcript level of Hk2 was significantly increased in the DEN, NPYR, CCl4, and TAA groups compared with untreated controls. The transcript level of Hk3 was significantly increased in the DEN and CCl4 groups and significantly decreased in the AFB1, NPYR, and MP groups compared with untreated controls. The transcript level of Pklr was significantly decreased in the DEN group and all non-genotoxic hepatocarcinogen groups and significantly increased in the AFB1 and NPYR groups compared with untreated controls. The transcript level of Pkm was significantly increased in all genotoxic hepatocarcinogen groups and the CCl4 and TAA groups compared with untreated controls. With regard to genes related to PPP, the transcript level of G6pd was significantly increased in the DEN, NPYR, and TAA groups compared with untreated controls. With regard to genes related to glutaminolysis, the transcript level of Slc1a5 was significantly increased in the DEN, AFB1, NPYR groups, and all non-genotoxic hepatocarcinogen groups compared with untreated controls. The transcript level of Gls was significantly increased in the NPYR, CCl4, and TAA groups compared with untreated controls. With regard to genes related to metabolic regulators, the transcript level of Myc was significantly increased in the DEN, CRB, and CCl4 groups and significantly decreased in the NPYR group compared with untreated controls. The transcript level of Tp53 was significantly decreased in the DEN, CCl4, and MP groups and significantly increased in the AFB1 and NPYR groups compared with untreated controls.
Measurement of proliferative lesions
In Experiments 1 and 2, there were no significant changes in the number and area of GST-P+ foci in any of the treatment groups compared with untreated controls after 28 days of treatment (Supplementary Tables 7 and 8: online only). There were significantly more and larger GST-P+ foci compared with untreated controls after 84 or 90 days of treatment with DEN, CCl4, AFB1, NPYR, TAA, or MP.
Distribution of immunolocalized cells
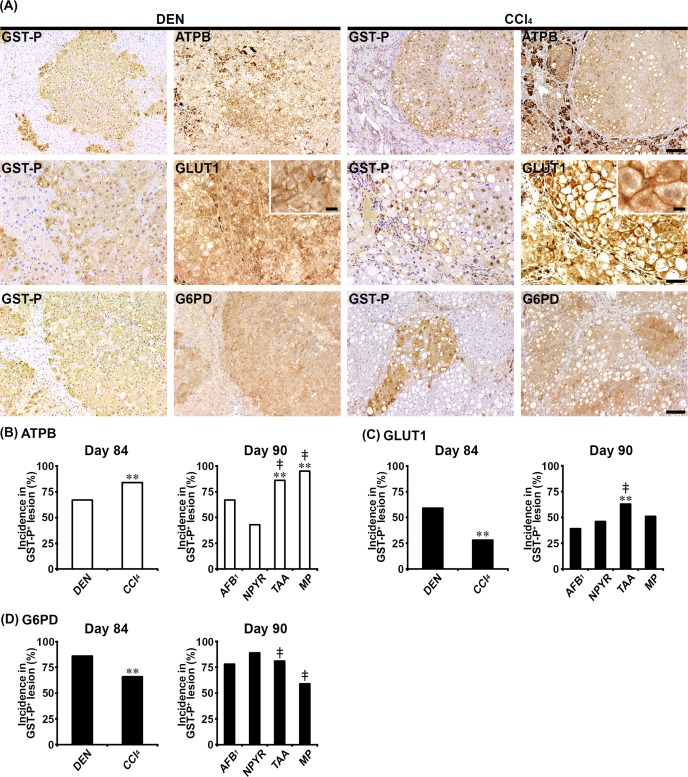
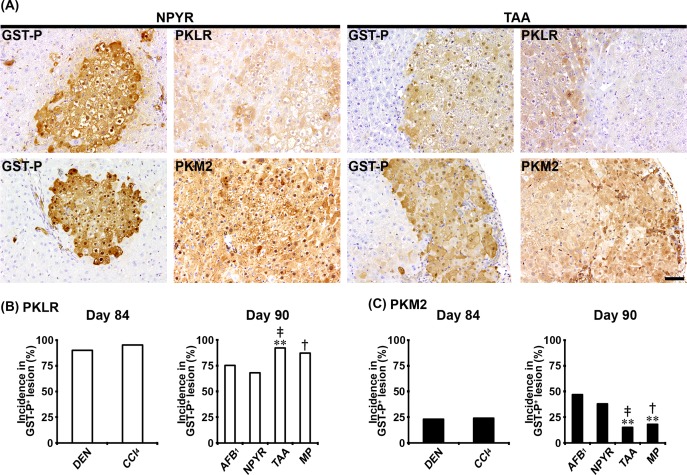
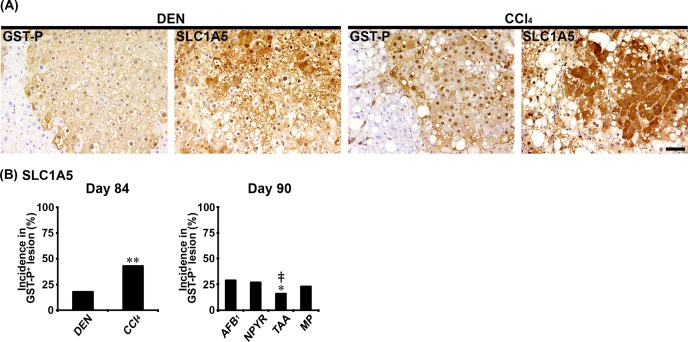
In Experiments 1 and 2, ATPB, GLUT1, G6PD, PKLR, PKM2, and SLC1A5 showed cytoplasmic expression, and GLUT1 also showed cell membrane expression in non-proliferative and proliferative liver cells. In both genotoxic (DEN, AFB1, and NPYR) and non-genotoxic hepatocarcinogens (CCl4, TAA, and MP), GST-P+ foci showed either increased or decreased expression of these molecules. With regard to ATPB, the population of GST-P+ foci downregulating expression of ATPB in non-genotoxic hepatocarcinogens was increased, and the incidences of ATPB− foci in GST-P+ foci were significantly increased compared with genotoxic hepatocarcinogens in Experiments 1 and 2 (Fig. 1A and B). With regard to GLUT1, the population of GST-P+ foci upregulating membranous GLUT1 expression was observed in both genotoxic and non-genotoxic hepatocarcinogens (Fig. 1A). In the CCl4 group, the incidence of GLUT1+ foci in GST-P+ foci was significantly decreased compared with the DEN group in Experiment 1 (Fig. 1C). In contrast, the incidence of GLUT1+ foci in GST-P+ foci in the TAA group was significantly increased compared with the AFB1 and NPYR groups in Experiment 2 (Fig. 1C). With regard to G6PD, the population of GST-P+ foci upregulating expression of G6PD in the DEN and NPYR groups was increased compared with non-genotoxic hepatocarcinogens (Fig. 1A). In non-genotoxic hepatocarcinogens, the incidences of G6PD+ foci in GST-P+ foci were significantly decreased compared with the DEN or NPYR groups in Experiments 1 and 2 (Fig. 1D). With regard to PKLR, the population of GST-P+ foci downregulating expression of PKLR was increased in the TAA and MP groups. The incidence of PKLR− foci in GST-P+ foci in the TAA group was significantly increased compared with the AFB1 group, and the incidences of PKLR− foci in GST-P+ foci in the TAA and MP groups were significantly increased compared with the NPYR group in Experiment 2 (Fig. 2A and B). With regard to PKM2, the population of GST-P+ foci upregulating PKM2 expression was observed in both genotoxic and non-genotoxic hepatocarcinogens (Fig. 2A). In the TAA and MP groups, the incidences of PKM2+ foci in GST-P+ foci were significantly decreased compared with the AFB1 and NPYR groups in Experiment 2 (Fig. 2C). With regard to SLC1A5, the population of GST-P+ foci upregulating SLC1A5 expression was observed in both genotoxic and non-genotoxic hepatocarcinogens (Fig. 3A). In the CCl4 group, the incidence of SLC1A5+ foci in GST-P+ foci was significantly increased compared with the DEN group in Experiment 1 (Fig. 3B). In contrast, the incidence of SLC1A5+ foci in GST-P+ foci in the TAA group was significantly decreased compared with the AFB1 and NPYR groups in Experiment 2 (Fig. 3B).
Fig. 1.
Immunohistochemical cellular distribution of adenosine triphosphate (ATP) synthase subunit beta, mitochondrial precursor (ATPB), solute carrier family 2, facilitated glucose transporter member 1 (GLUT1), and glucose-6-phosphate 1-dehydrogenase (G6PD) in association with glutathione S-transferase placental form-positive (GST-P+) liver cell foci after treatment with genotoxic [N-nitrosodiethylamine (DEN), aflatoxin B1 (AFB1), or N-nitrosopyrrolidine (NPYR)] or non-genotoxic hepatocarcinogens [carbon tetrachloride (CCl4), thioacetamide (TAA), or methapyrilene hydrochloride (MP)] for 84 or 90 days. (A) Representative images of the expression of ATPB, GLUT1, and G6PD in GST-P+ foci in the DEN and CCl4 groups (×10 objective; GLUT1 ×20 objective; inset ×60 objective). Bar = 100 µm, 50 µm, or 10 µm (inset). (B) Incidences of ATPB− foci in GST-P+ foci in genotoxic and non-genotoxic hepatocarcinogens. (C) Incidences of GLUT1+ foci in GST-P+ foci in genotoxic and non-genotoxic hepatocarcinogens. (D) Incidences of G6PD+ foci in GST-P+ foci in genotoxic and non-genotoxic hepatocarcinogens. Graphs in (B), (C), and (D) show incidences (% value, n=10) of GST-P+ foci showing altered expression of each molecule (open column, decreased; filled column, increased) in each group. **P<0.01, significantly different from the DEN or AFB1 group by Fisher’s exact test. ‡P<0.01, significantly different from the NPYR group by Fisher’s exact test.
Fig. 2.
Immunohistochemical cellular distribution of pyruvate kinase L/R (PKLR) and pyruvate kinase isozyme M2 (PKM2) in association with glutathione S-transferase placental form-positive (GST-P+) liver cell foci after treatment with genotoxic [N-nitrosodiethylamine (DEN), aflatoxin B1 (AFB1), or N-nitrosopyrrolidine (NPYR)] or non-genotoxic hepatocarcinogens [carbon tetrachloride (CCl4), thioacetamide (TAA), or methapyrilene hydrochloride (MP)] for 84 or 90 days. (A) Representative images of the expression of PKLR and PKM2 in GST-P+ foci in the NPYR and TAA groups (×20 objective). Bar = 50 µm. (B) Incidences of PKLR− foci in GST-P+ foci in genotoxic and non-genotoxic hepatocarcinogens. (C) Incidences of PKM2+ foci in GST-P+ foci in genotoxic and non-genotoxic hepatocarcinogens. Graphs in (B) and (C) show incidences (% value, n=10) of GST-P+ foci showing altered expression of each molecule (open column, decreased; filled column, increased) in each group. **P<0.01, significantly different from the AFB1 group by Fisher’s exact test. †P<0.05, significantly different from the NPYR group by Fisher’s exact test. ‡P<0.01, significantly different from the NPYR group by Fisher’s exact test.
Fig. 3.
Immunohistochemical cellular distribution of neutral amino acid transporter B(0) (SLC1A5) in association with glutathione S-transferase placental form-positive (GST-P+) liver cell foci after treatment with genotoxic [N-nitrosodiethylamine (DEN), aflatoxin B1 (AFB1), or N-nitrosopyrrolidine (NPYR)] or non-genotoxic hepatocarcinogens [carbon tetrachloride (CCl4), thioacetamide (TAA), or methapyrilene hydrochloride (MP)] for 84 or 90 days. (A) Representative images of the expression of SLC1A5 in GST-P+ foci in the DEN and CCl4 groups (×20 objective). Bar = 50 µm. (B) Incidences of SLC1A5+ foci in GST-P+ foci in genotoxic and non-genotoxic hepatocarcinogens. Graphs in (B) show incidences (% value, n=10) of GST-P+ foci showing altered expression of each molecule (filled column, increased) in each group. *P<0.05, significantly different from the DEN or AFB1 group by Fisher’s exact test. **P<0.01, significantly different from the DEN or AFB1 group by Fisher’s exact test. ‡P<0.01, significantly different from the NPYR group by Fisher’s exact test.
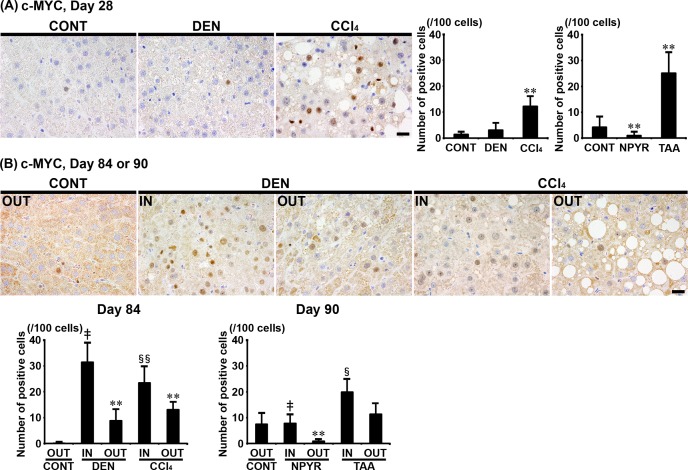
In Experiments 1 and 2, c-MYC showed immunolocalization in the nucleus of liver cells. Furthermore, the numbers of c-MYC+ cells were significantly increased in the CCl4 and TAA groups and significantly decreased in the NPYR group compared with untreated controls after 28 days of treatment (Fig. 4A). The numbers of c-MYC+ cells were significantly increased in liver cells distributed outside of GST-P+ foci in both the DEN and CCl4 groups after 84 days of treatment compared with untreated controls in Experiment 1 and significantly decreased in the liver cells distributed outside of GST-P+ foci in the NPYR group after 90 days of treatment compared with untreated controls in Experiment 2 (Fig. 4B). The numbers of c-MYC+ cells were significantly increased in liver cells inside of GST-P+ foci in both genotoxic and non-genotoxic hepatocarcinogens compared with those distributed outside of GST-P+ foci in each group after 84 or 90 days of treatment in Experiments 1 and 2 (Fig. 4B).
Fig. 4.
Distribution of c-MYC+ cells in the liver of rats after treatment with genotoxic [N-nitrosodiethylamine (DEN) or N-nitrosopyrrolidine (NPYR)] or non-genotoxic hepatocarcinogens [carbon tetrachloride (CCl4) or thioacetamide (TAA)] for 28 days and distribution of c-MYC+ cells in association with glutathione S-transferase placental form-positive (GST-P+) liver cell foci after treatment with genotoxic (DEN or NPYR) or non-genotoxic hepatocarcinogens (CCl4 or TAA) for 84 or 90 days. (A) Representative images of the expression of c-MYC in the liver in the DEN and CCl4 groups (×40 objective). Bar = 20 µm. (B) Representative images of the expression of c-MYC of inside (IN) or outside (OUT) of GST-P+ foci in the DEN and CCl4 groups (×40 objective). Bar = 20 µm. Graphs in (A) and (B) show the number of c-MYC+ cells (/100 cells; value, mean + SD) IN or OUT of GST-P+ foci in each group. **P<0.01, significantly different from OUT of untreated controls by Tukey’s or Steel-Dwass test. ‡P<0.01, significantly different from OUT of GST-P+ foci in the DEN or NPYR group by Tukey’s or Steel-Dwass test. §P<0.05, significantly different from OUT of GST-P+ foci in the CCl4 or TAA group by Tukey’s or Steel-Dwass test. §§P<0.01, significantly different from OUT of GST-P+ foci in the CCl4 or TAA group by Tukey’s or Steel-Dwass test.
Discussion
In the present study, non-genotoxic hepatocarcinogens started to downregulate OXPHOS-related genes, Mpc2 and Atp5f1b, after 28 days of treatment, and after 84 or 90 days of treatment, all non-genotoxic hepatocarcinogens downregulated these genes. In contrast, non-genotoxic hepatocarcinogens upregulated Atp5if1, which encodes mitochondrial ATPase inhibitor, after 84 or 90 days of treatment. While genotoxic hepatocarcinogens did not specifically change the expression of these genes after 28 days of treatment, most genotoxic hepatocarcinogens later decreased the Mpc2 transcript level. Immunohistochemically, the incidences of ATPB− foci in GST-P+ foci induced by treatment with non-genotoxic hepatocarcinogens after 84 or 90 days of treatment were higher than those with genotoxic hepatocarcinogens. ATP synthase is downregulated in many types of cancer14, and we have previously found an increase in the number of GST-P+ liver foci reducing mitochondrial ATP synthase in the early stage of tumor promotion by non-genotoxic hepatocarcinogens16. Therefore, our results suggest the onset of suppressed OXPHOS toward carcinogenesis from as early as 28 days of non-genotoxic hepatocarcinogen treatment before the formation of GST-P+ foci.
With regard to glycolysis-related cellular events, we found that treatment with non-genotoxic hepatocarcinogens for 28 days and for 84 or 90 days downregulated genes encoding glucose transporters. On the other hand, genotoxic hepatocarcinogens did not specifically change the expression of these genes at any time point. Immunohistochemically, a subpopulation of GLUT1+ foci appeared in GST-P+ foci with all hepatocarcinogens after 84 or 90 days of treatment without relation to genotoxic potential. Moreover, transcript upregulation was observed in genes encoding glycolytic enzymes with both genotoxic and non-genotoxic hepatocarcinogens after treatment for 28 days and for 84 or 90 days. Glycolysis produces ATP with lower efficiency but at a faster rate than OXPHOS, suggesting that this faster rate of ATP production aids the rapid proliferation of cancer cells38, 39. Moreover, we have previously found a catastrophic cellular senescence-related metabolic shift from OXPHOS to glycolysis in GST-P+ proliferative lesions from the early stage of tumor promotion by non-genotoxic hepatocarcinogens16. Therefore, it could be suggested that the metabolic shift from OXPHOS to glycolysis is the initial cellular event that activates cell proliferation beneficial for hepatocarcinogenesis without relation to the genotoxic potential of hepatocarcinogens.
With regard to expression changes of metabolic regulators, we found that the transcript level of Myc and nuclear c-MYC+ cells were increased by all non-genotoxic hepatocarcinogens after 28 days of treatment. In addition, we observed that the number of c-MYC+ cells inside of GST-P+ foci was increased compared with that of those distributed outside of GST-P+ foci after 84 or 90 days of treatment with any of the genotoxic and non-genotoxic hepatocarcinogens. c-MYC, a well-known driver of cell proliferation, stimulates glycolysis by activation of glucose transporters and glycolytic enzymes40. On the other hand, it has been reported that suppression of c-MYC induces cellular senescence41. Therefore, it can be postulated that activation of c-MYC-mediated gene transcription occurs from as early as 28 days of treatment with non-genotoxic hepatocarcinogens before the formation of GST-P+ foci. While we could not identify the candidate molecules, target genes of c-MYC-mediated transcription may be those contributing to facilitation of glycolysis and cell proliferation, which promoted escape from cellular senescence and advancement to carcinogenesis in the present study. In this study, we found that the transcript level of Tp53 was increased by all non-genotoxic hepatocarcinogens after 28 days of treatment. In contrast, expression of Tp53 was decreased after treatment with non-genotoxic hepatocarcinogen for 84 or 90 days, except for TAA. In contrast, genotoxic hepatocarcinogens did not consistently change the transcript level of Tp53 at any time point. It has been reported that p53 promotes mitochondrial OXPHOS by activation of Sco2, which encodes cytochrome c oxidase 242. Moreover, p53 regulates glucose transporters and glycolytic enzymes13, 33. These findings suggest that a metabolic shift from mitochondrial OXPHOS to glycolysis in liver cells is promoted by downregulation of Tp53 after repeated treatment with non-genotoxic hepatocarcinogens. Moreover, a combined effect of c-MYC upregulation and p53 downregulation facilitates glucose consumption and utilization in tumor cells, suggesting reprogramming of cellular metabolism for acquiring the hallmark capabilities of cell proliferation, avoidance of cytostatic controls, and attenuation of apoptosis43. Therefore, our results indicate that non-genotoxic hepatocarcinogens enhance a metabolic shift by inducing a combined effect of c-MYC upregulation and p53 downregulation, resulting in the facilitation of carcinogenic steps.
Among genes encoding glycolytic enzymes, we observed transcript upregulation of Pkm, which encodes pyruvate kinase isozymes M1/M2 (PKM1/PKM2), after 28 days of treatment with any of the genotoxic hepatocarcinogens and after 84 or 90 days of treatment with non-genotoxic hepatocarcinogens, except for MP. Immunohistochemically, the incidences of PKM2+ foci in GST-P+ foci induced by treatment with genotoxic hepatocarcinogens, AFB1 and NPYR, for 90 days were higher than those with non-genotoxic hepatocarcinogens, whereas no changes in the incidence were observed with genotoxic DEN and non-genotoxic CCl4 after 84 days of treatment. PKM2, the major isozyme in the fetus, is expressed in the majority of proliferating cells and essentially all cancer cells44. PKM2 not only plays a role in glycolysis to achieve the nutrient demands of cancer cell proliferation but also contributes to carcinogenesis as a coactivator and protein kinase45. These findings may suggest an onset of disruptive activation of PKM2 toward carcinogenesis from as early as 28 days of genotoxic hepatocarcinogen treatment before the formation of GST-P+ foci. On the other hand, we found downregulation of Pklr, which encodes pyruvate kinase isozymes L/R (PKL/PKR), after treatment with any of the non-genotoxic hepatocarcinogens for 28 days and for 84 or 90 days. In addition, the incidences of PKLR− foci in GST-P+ foci induced by treatment with non-genotoxic hepatocarcinogens, TAA and MP, for 90 days were higher than those with genotoxic hepatocarcinogens, whereas no changes in the incidence were observed with genotoxic DEN and non-genotoxic CCl4 after 84 days of treatment. PKL is the major isoform in the normal liver34. We have also previously reported that non-genotoxic hepatocarcinogen treatment for up to 90 days induces a molecular shift from Pklr to Pkm16. Therefore, it could be suggested that the molecular shift from PKLR to PKM2 is the initial cellular event that activates cell proliferation beneficial for hepatocarcinogenesis by repeated treatment with non-genotoxic hepatocarcinogens. These findings suggest that these cellular metabolism-related pyruvate kinase genes and molecules provide early detection markers of non-genotoxic hepatocarcinogens in a scheme of 28- or 90-day repeated administration studies in rats.
In the present study, we found that genotoxic hepatocarcinogen treatment for 84 or 90 days increased the incidences of G6PD+ foci in GST-P+ foci compared with those with non-genotoxic hepatocarcinogen treatment. G6PD catalyzes the conversion of glucose-6-phosphate to 6-phosphogluconate to facilitate the process of PPP, which participates in nucleotide synthesis and produces nicotinamide adenine dinucleotide phosphate (NADPH) to reduce DNA damage caused by oxidative stress33. G6PD activity is increased in tumor cells, and overexpression of G6PD stimulates cell growth in tumor cells by providing ribose-5-phosphate for nucleic acid synthesis46. Therefore, an increase of G6PD in GST-P+ foci by repeated treatment with genotoxic hepatocarcinogens suggests an enhanced synthesis of ribose-5-phosphate and NADPH by activation of G6PD-mediated PPP to facilitate carcinogenesis steps.
With regard to glutaminolysis, we found upregulation of Slc1a5, which encodes a glutamine transporter, after treatment with both genotoxic and non-genotoxic hepatocarcinogens for 28 days and for 84 or 90 days. Immunohistochemically, SLC1A5+ foci in GST-P+ foci were observed with all hepatocarcinogens after treatment for 84 or 90 days without relation to genotoxic potential in this study. SLC1A5 is reported to be overexpressed in cancer cells and is transcriptionally upregulated by c-MYC13, 47. Moreover, glutamine, which is transported into cells through SLC1A5, is not only used as a major substrate for OXPHOS but also used for the synthesis of other macromolecules, such as nucleotides, proteins, and hexosamines for cell growth and survival47. These findings suggest that the disruptive activation of c-MYC-mediated Slc1a5 facilitates cell proliferation toward carcinogenesis from as early as 28 days of non-genotoxic hepatocarcinogen treatment and at 84 or 90 days of genotoxic hepatocarcinogen treatment. In this study, we also found upregulation of Gls, which encodes glutaminase, after treatment with any of the non-genotoxic hepatocarcinogens for 28 days and after treatment with 2 of 3 non-genotoxic hepatocarcinogens for 84 or 90 days. On the other hand, genotoxic hepatocarcinogens did not consistently change the transcript level of Gls at any time point. GLS catalyzes the conversion of glutamine to glutamate in mitochondria and is expressed in a wide variety of tumors, and its upregulation correlates with tumor growth47. Interestingly, Gls expression is also regulated by c-MYC, resulting in the promotion of tumor development13. Therefore, it could be suggested that activation of c-MYC induces glutaminolysis-related genes in liver cells to facilitate carcinogenesis from as early as 28 days of non-genotoxic hepatocarcinogen treatment before the formation of GST-P+ foci.
Conclusion
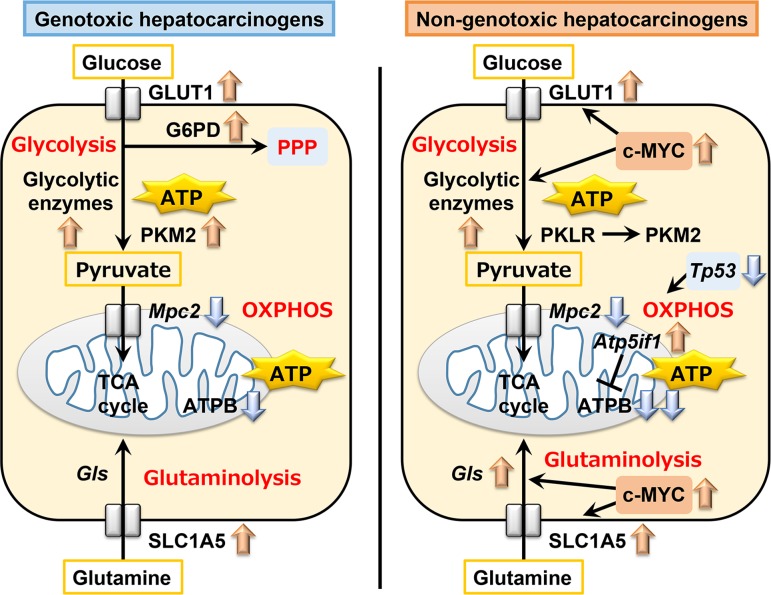
Both genotoxic and non-genotoxic hepatocarcinogens facilitated glycolysis after 28 days of repeated treatment in rats. Non-genotoxic hepatocarcinogens suppressed mitochondrial OXPHOS and activated c-MYC, suggesting enhancement of a metabolic shift from OXPHOS to glycolysis to cause disruptive cellular senescence and facilitation of cell proliferation from as early as 28 days of treatment before the formation of GST-P+ foci. Later, non-genotoxic hepatocarcinogens caused Tp53 downregulation in addition to c-MYC activation, contributing to further facilitation of the metabolic shift and cell proliferation. This resulted in promoting the escape from cellular senescence and advancement to carcinogenesis. Until 84 or 90 days of treatment, genotoxic hepatocarcinogens also enhanced a metabolic shift via c-MYC activation. However, Tp53 downregulation was not essential in this case. In addition, both genotoxic and non-genotoxic hepatocarcinogens upregulated either or both of glutaminolysis-related Slc1a5 and Gls after 28 days of treatment, and induced liver cell foci immunoreactive for SLC1A5 in a subpopulation of GST-P+ foci after 84 or 90 days of treatment, suggesting that glutaminolysis-mediated cell proliferation undergoes a hepatocarcinogenesis step. These results suggest differential responses between genotoxic and non-genotoxic hepatocarcinogens on the reprogramming of energy metabolic pathways toward carcinogenesis in liver cells from the early stage of hepatocarcinogen treatment (Fig. 5). Further study may be necessary to address the underlying mechanism for producing these differences between genotoxic and non-genotoxic hepatocarcinogens to clarify the respective carcinogenic mechanisms.
Fig. 5.
Schematic summary of the responses on energy metabolic pathway reprogramming by genotoxic or non-genotoxic hepatocarcinogens in rat liver cells.
Disclosure of Potential Conflicts of Interest
All authors declare that there are no conflicts of interest that influenced the outcome of the present study.
Supplemental Table
Sequence of primers used for real-time RT-PCR analysis
Acknowledgments
The authors thank Shigeko Suzuki for her technical assistance in preparing the histological specimens. All authors declare that there are no conflicts of interest that influenced the outcome of the present study. This work was supported by research funds from the Chemicals Evaluation and Research Institute, Japan, and the Institute of Global Innovation Research, Tokyo University of Agriculture and Technology.
References
- 1.Kimura M, Abe H, Mizukami S, Tanaka T, Itahashi M, Onda N, Yoshida T, and Shibutani M. Onset of hepatocarcinogen-specific cell proliferation and cell cycle aberration during the early stage of repeated hepatocarcinogen administration in rats. J Appl Toxicol. 36: 223–237. 2016. [DOI] [PubMed] [Google Scholar]
- 2.Taniai E, Yafune A, Hayashi H, Itahashi M, Hara-Kudo Y, Suzuki K, Mitsumori K, and Shibutani M. Aberrant activation of ubiquitin D at G2 phase and apoptosis by carcinogens that evoke cell proliferation after 28-day administration in rats. J Toxicol Sci. 37: 1093–1111. 2012. [DOI] [PubMed] [Google Scholar]
- 3.Yafune A, Taniai E, Morita R, Hayashi H, Suzuki K, Mitsumori K, and Shibutani M. Aberrant activation of M phase proteins by cell proliferation-evoking carcinogens after 28-day administration in rats. Toxicol Lett. 219: 203–210. 2013. [DOI] [PubMed] [Google Scholar]
- 4.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 75: 685–705. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi Y. Overview of genotoxic carcinogens and non-genotoxic carcinogens. Exp Toxicol Pathol. 44: 465–471. 1992. [DOI] [PubMed] [Google Scholar]
- 6.MacGregor JT, Frötschl R, White PA, Crump KS, Eastmond DA, Fukushima S, Guérard M, Hayashi M, Soeteman-Hernández LG, Johnson GE, Kasamatsu T, Levy DD, Morita T, Müller L, Schoeny R, Schuler MJ, and Thybaud V. IWGT report on quantitative approaches to genotoxicity risk assessment II. Use of point-of-departure (PoD) metrics in defining acceptable exposure limits and assessing human risk. Mutat Res Genet Toxicol Environ Mutagen. 783: 66–78. 2015. [DOI] [PubMed] [Google Scholar]
- 7.Nohmi T. Thresholds of genotoxic and non-genotoxic carcinogens. Toxicol Res. 34: 281–290. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura M, Mizukami S, Watanabe Y, Hasegawa-Baba Y, Onda N, Yoshida T, and Shibutani M. Disruption of spindle checkpoint function ahead of facilitation of cell proliferation by repeated administration of hepatocarcinogens in rats. J Toxicol Sci. 40: 855–871. 2015. [DOI] [PubMed] [Google Scholar]
- 9.Cazzaniga M, and Bonanni B. Understanding the anticancer effect of metformin and its clinical implications. relationship between metabolic reprogramming and mitochondrial activity in cancer cells. Anticancer Res. 35: 5789–5796. 2015. [PubMed] [Google Scholar]
- 10.Yoshida GJ. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J Exp Clin Cancer Res. 34: 111 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warburg O. On the origin of cancer cells. Science. 123: 309–314. 1956. [DOI] [PubMed] [Google Scholar]
- 12.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat Rev Cancer. 2: 683–693. 2002. [DOI] [PubMed] [Google Scholar]
- 13.Amoêdo ND, Valencia JP, Rodrigues MF, Galina A, and Rumjanek FD. How does the metabolism of tumour cells differ from that of normal cells. Biosci Rep. 33: e00080 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isidoro A, Martínez M, Fernández PL, Ortega AD, Santamaría G, Chamorro M, Reed JC, and Cuezva JM. Alteration of the bioenergetic phenotype of mitochondria is a hallmark of breast, gastric, lung and oesophageal cancer. Biochem J. 378: 17–20. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kon S, Ishibashi K, Katoh H, Kitamoto S, Shirai T, Tanaka S, Kajita M, Ishikawa S, Yamauchi H, Yako Y, Kamasaki T, Matsumoto T, Watanabe H, Egami R, Sasaki A, Nishikawa A, Kameda I, Maruyama T, Narumi R, Morita T, Sasaki Y, Enoki R, Honma S, Imamura H, Oshima M, Soga T, Miyazaki JI, Duchen MR, Nam JM, Onodera Y, Yoshioka S, Kikuta J, Ishii M, Imajo M, Nishida E, Fujioka Y, Ohba Y, Sato T, and Fujita Y. Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes. Nat Cell Biol. 19: 530–541. 2017. [DOI] [PubMed] [Google Scholar]
- 16.Mizukami S, Watanabe Y, Nakajima K, Hasegawa-Baba Y, Jin M, Yoshida T, and Shibutani M. Downregulation of TMEM70 in rat liver cells after hepatocarcinogen treatment related to the Warburg effect in hepatocarcinogenesis producing GST-P-expressing proliferative lesions. Toxicol Sci. 159: 211–223. 2017. [DOI] [PubMed] [Google Scholar]
- 17.De Jesus AE, Gorst-Allman CP, Horak RM, and Vleggaar R. Large-scale purification of the mycotoxins aflatoxin B1, B2 and G1. J Chromatogr A. 450: 101–104. 1988. [DOI] [PubMed] [Google Scholar]
- 18.AOAC. Chapter 49 Official Method 970.44, Preparation of standards for aflatoxins. In: Official Methods of Analysis of AOAC International. 18th ed. W Horwitz., G.W Latimer Jr. (eds.). AOAC International Press, Gaithersburg. 47–53, 2005. [Google Scholar]
- 19.Verna L, Whysner J, and Williams GM. N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol Ther. 71: 57–81. 1996. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki S, Takeshita K, Asamoto M, Takahashi S, Kandori H, Tsujimura K, Saito F, Masuko K, and Shirai T. High mobility group box associated with cell proliferation appears to play an important role in hepatocellular carcinogenesis in rats and humans. Toxicology. 255: 160–170. 2009. [DOI] [PubMed] [Google Scholar]
- 21.Qian G, Wang F, Tang L, Massey ME, Mitchell NJ, Su J, Williams JH, Phillips TD, and Wang JS. Integrative toxicopathological evaluation of aflatoxin B1 exposure in F344 rats. Toxicol Pathol. 41: 1093–1105. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanki K, Nishikawa A, Masumura K, Umemura T, Imazawa T, Kitamura Y, Nohmi T, and Hirose M. In vivo mutational analysis of liver DNA in gpt delta transgenic rats treated with the hepatocarcinogens N-nitrosopyrrolidine, 2-amino-3-methylimidazo[4,5-f]quinoline, and di(2-ethylhexyl)phthalate. Mol Carcinog. 42: 9–17. 2005. [DOI] [PubMed] [Google Scholar]
- 23.King TO. Target organ toxicity of GS-6244 (carbadox) and CP-17,056 (desoxycarbadox) with chronic administration in rats. Submitted to WHO by Pfizer Central Research, Groton, CT, USA. 1976. [Google Scholar]
- 24.Weisburger EK. Carcinogenicity studies on halogenated hydrocarbons. Environ Health Perspect. 21: 7–16. 1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barber ED, Donish WH, and Mueller KR. A procedure for the quantitative measurement of the mutagenicity of volatile liquids in the Ames Salmonella/microsome assay. Mutat Res. 90: 31–48. 1981. [DOI] [PubMed] [Google Scholar]
- 26.Becker FF. Thioacetamide hepatocarcinogenesis. J Natl Cancer Inst. 71: 553–558. 1983. [PubMed] [Google Scholar]
- 27.National Toxicology Program. NTP hepatotoxicity studies of the liver carcinogen methapyrilene hydrochloride (CAS no. 135–23–9) administered in feed to male F344/N rats. Toxic Rep Ser. 46: 1–C7. 2000. [PubMed] [Google Scholar]
- 28.Ito Y, Nakajima K, Masubuchi Y, Kikuchi S, Saito F, Akahori Y, Jin M, Yoshida T, and Shibutani M. Expression characteristics of genes hypermethylated and downregulated in rat liver specific to non-genotoxic hepatocarcinogens. Toxicol Sci. 169: 122–136. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, and Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 25: 402–408. 2001. [DOI] [PubMed] [Google Scholar]
- 30.Ito N, Imaida K, Tamano S, Hagiwara A, and Shirai T. Medium-term bioassays as alternative carcinogenicity test. J Toxicol Sci. 23(Suppl 2): 103–106. 1998. [DOI] [PubMed] [Google Scholar]
- 31.Shirai T. A medium-term rat liver bioassay as a rapid in vivo test for carcinogenic potential: a historical review of model development and summary of results from 291 tests. Toxicol Pathol. 25: 453–460. 1997. [DOI] [PubMed] [Google Scholar]
- 32.Luciaková K, and Kužela S. Increased steady-state levels of several mitochondrial and nuclear gene transcripts in rat hepatoma with a low content of mitochondria. Eur J Biochem. 205: 1187–1193. 1992. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, and Yang JM. Altered energy metabolism in cancer: a unique opportunity for therapeutic intervention. Cancer Biol Ther. 14: 81–89. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imamura K, and Tanaka T. Multimolecular forms of pyruvate kinase from rat and other mammalian tissues. I. Electrophoretic studies. J Biochem. 71: 1043–1051. 1972. [DOI] [PubMed] [Google Scholar]
- 35.Pavlova NN, and Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 23: 27–47. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller DM, Thomas SD, Islam A, Muench D, and Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 18: 5546–5553. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniai E, Kawai M, Dewa Y, Nishimura J, Harada T, Saegusa Y, Matsumoto S, Takahashi M, Mitsumori K, and Shibutani M. Crosstalk between PTEN/Akt2 and TGFβ signaling involving EGF receptor down-regulation during the tumor promotion process from the early stage in a rat two-stage hepatocarcinogenesis model. Cancer Sci. 100: 813–820. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeiffer T, Schuster S, and Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 292: 504–507. 2001. [DOI] [PubMed] [Google Scholar]
- 39.Lunt SY, and Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 27: 441–464. 2011. [DOI] [PubMed] [Google Scholar]
- 40.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, and Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 275: 21797–21800. 2000. [DOI] [PubMed] [Google Scholar]
- 41.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, and Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci USA. 104: 13028–13033. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, and Hwang PM. p53 regulates mitochondrial respiration. Science. 312: 1650–1653. 2006. [DOI] [PubMed] [Google Scholar]
- 43.Hanahan D, and Weinberg RA. Hallmarks of cancer: the next generation. Cell. 144: 646–674. 2011. [DOI] [PubMed] [Google Scholar]
- 44.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, and Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 452: 230–233. 2008. [DOI] [PubMed] [Google Scholar]
- 45.Dong G, Mao Q, Xia W, Xu Y, Wang J, Xu L, and Jiang F. PKM2 and cancer: The function of PKM2 beyond glycolysis. Oncol Lett. 11: 1980–1986. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanton RC. Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life. 64: 362–369. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin L, Alesi GN, and Kang S. Glutaminolysis as a target for cancer therapy. Oncogene. 35: 3619–3625. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence of primers used for real-time RT-PCR analysis