Abstract
Papillary renal cell carcinoma (PRCC) accounts for about 10 percent of all renal cell carcinomas, and the prognosis is poor for people with advanced disease. Interleukin-20 receptor subunit beta (IL20RB) is a single-pass type I membrane protein of the type II cytokine receptor family and is related to the pathogenesis of chronic inflammation and autoimmune diseases, including psoriasis, glaucoma, vitiligo, rheumatoid arthritis, and inflammatory bowel disease. However, little has been reported on IL20RB with respect to cancer, especially in PRCC. Thus, we performed this study to explore its biological characteristics in PRCC. Data from the TCGA database were used to analyze the expression and prognosis of IL20RB. qRT-PCR was used to detect the expression of IL20RB in PRCC cells in vitro. After knockdown of IL20RB with small interfering RNA (siRNA) technology, the proliferation, migration, and invasion of Ketr-3 cells and the expression of related proteins in the epithelial-mesenchymal transition (EMT) pathway were measured with Cell Counting Kit-8 (CCK-8), transwell, and western blot assays. The findings demonstrated that the expression of IL20RB was upregulated in both PRCC tissues and cells and that the high expression of IL20RB led to low overall survival (OS). Furthermore, after knockdown of IL20RB in vitro, the proliferation, migration, and invasion of Ketr-3 cells were reduced, and the expression of related proteins in the EMT pathway declined, suggesting that IL20RB plays a vital role in PRCC through the EMT pathway. These results reveal the biological significance of IL20RB in PRCC and provide new insight for future targeted drugs.
Keywords: papillary renal cell carcinoma (PRCC), interleukin-20 receptor subunit beta (IL20RB), invasion, migration, epithelial-mesenchymal transition (EMT)
Introduction
Renal cell carcinoma (RCC) is the most common tumor to affect the adult kidney, accounting for 80–90% of primary malignant renal neoplasms in adults1. The pathogenesis of RCC is still unclear, which makes it difficult to treat. Papillary renal cell carcinoma (PRCC) is the second most common histologic subtype, accounting for approximately 10% of all renal cell cancers2. Papillary tumors are subdivided into type I tumors, which occur sporadically and metastasize somewhat late, and type II papillary RCC, which is more likely inherited and has a poorer prognosis3. Considering the above, it is necessary to identify therapeutic targets and diagnostic biomarkers for the treatment of PRCC.
The interleukin-20-receptor I complex (IL-20-RI) is composed of two chains, interleukin-20 receptor subunit alpha (IL20RA) and interleukin-20 receptor subunit beta (IL20RB). IL-20-RI is a receptor for IL-19, IL-20, and IL-24. IL20RB can also form a heterodimer with IL22RA1. The IL22RA1/IL20RB dimer is a receptor for IL20 and IL24. IL-20R cytokines can be expressed by both immune cells and epithelial cells and are important for their interaction4. IL20RB may affect the IL20 signaling pathway and may be involved in the pathogenesis of glaucoma5, vitiligo6, and psoriasis7. Meanwhile, multiple studies have also reported that IL20RB carries a big weight in various cancers, for instance non-small-cell lung cancer (NSCLC)8, nasopharyngeal carcinoma (NPC)9, breast cancer (BC)10, and pancreatic ductal adenocarcinoma (PDAC)11. Based on the above research, IL20RB plays an important role in many cancers, but the function of IL20RB in PRCC has not been described. The present study was designed to explore its role in PRCC. Currently, there is no report on IL20RB in PRCC. Therefore, much work remains to be done to explore the function of IL20RB in PRCC.
In the present study, the expression pattern, correlation to clinical characteristics, and prognostic significance of IL20RB; the capabilities of proliferation, invasion and migration of PRCC cells; and the underlying mechanism were investigated for the first time, to the best of our knowledge. Thus, our results provide new insights into possible therapeutic interventions for further preclinical or clinical studies.
Materials and Methods
Data collection
Data concerning the expression of IL20RB in tumor and normal tissues were downloaded from The Cancer Genome Atlas (TCGA) database ( https://cancergenome.nih.gov/), including 32 normal cases and 289 tumor cases, and 68 cases with complete clinical data were used to explore the correlation of IL20RB expression and clinical characteristics (TCGA Project ID: TCGA-KIRP). The Kaplan-Meier method was used for analysis of the prognostic survival curve.
Cell culture
We obtained human PRCC cell lines A498, 786-0, ACHN, and Ketr-3 and normal control cells, Human Renal Cortical Epithelial (HRCE) cells from the Shanghai cell bank of the Chinese Academy of Medical Sciences (Shanghai, China). PRCC cells were cultured in RPMI-1640 with a 10% serum concentration, 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Gibco, Invitrogen, Carlsbad, CA, USA) at 37°C with 5% CO2 in air. HRCE cells were cultured in MEM with a 10% serum concentration, 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Gibco, Invitrogen) under the same conditions. The cells were washed three times in PBS and then digested with trypsin (Gibco, Invitrogen) when they under the logarithmic growth stage. When the cells became round, culture medium was added to stop digestion, the cells were repeatedly blown into single-cell suspension, and they were then placed into a six-well plate for subsequent experiments.
Transfection
Transfection was performed according to the instructions of Lipofectamine 2000 transfection kits (Invitrogen). The transfection efficiency was detected by western blot and quantitative real-time PCR (qRT-PCR) 24 h after transfection. The siRNA sequences were synthesized by GenePharma (Shanghai, China): for si-IL20RB, 5’-GTTCCAAGGAGAAGCCCACA-3’, forward, 5’-CCGGTC CACATTCTCTGGAA-3’, reverse; for Si-con, 5’-GGAGCG AGATCCCTCCAAAAT-3’, forward, 5’-GGCTGTTGTCAT ACTTCTCATGG-3’, reverse.
RNA extraction and qRT-PCR
Total RNA was extracted using an RNA extraction kit (Invitrogen). RNA was reverse transcribed into cDNA with Superscript III reverse transcriptase (Invitrogen), and qRT-PCR was used to explore the expression of IL20RB. qRT-PCR was executed with 40 circles consisting of 95°C for 5 min, 95°C for 30 s, 60°C for 45 s, and then 72°C for 30 min. The primers used were 5’-GTTCCAAGGAGAAGCCCACA-3’, forward, and 5’-CCGGTCCACATTCTCTGGAA-3’, reverse, for IL20RB and 5’-GGAGCGAGATCCCTCCAAAAT-3’, forward, and 5’-GGCTGTTGTCATACTTCTCATGG-3’, reverse, for GAPDH. Three plates were used for each group, and experiments were repeated three times independently. The 2−ΔΔCT method was used to analyze the relative expression of IL20RB.
Western blot
Total protein was extracted from the cells 24 h after transfection with si-IL20RB, and the concentration was measured by the BCA method. Twenty micrograms of protein from each sample was subjected to SDS-PAGE and electrotransferred onto PVDF membranes. The membrane was incubated with 5% skim milk powder for 1 h at room temperature and then incubated with the primary antibodies, GAPDH (1:1,000, Cell Signaling Technology Inc., Beverly, MA, USA), IL20RB, E-cadherin, N-cadherin, Vimentin, Snail1, and Snail2 (1:1,000, Abcam, Cambridge, MA,USA) at 4°C overnight. The membrane was washed with PBS three times and then incubated with the secondary antibodies (1:1,000, Cell Signaling Technology Inc.) for 1 h at room temperature. The signal of the protein was visualized by ECL. Quantity One software was used to measure the relative expression of the protein, and GAPDH was used as the internal control.
CCK-8 assay
To digest and count the cells 24 h after transfection with siRNA and prepare cell suspensions, cell suspensions were placed into 96-well plates at a density of 1000 cells/well and cultured at 37°C in a 5% CO2 incubator. Cell activity was measured by CCK-8 every 24 h, and the cells were incubated at 37°C in the incubator for 1.5 h after adding CCK-8 reagent (Bestbio, Shanghai, China) to each well. A microplate reader was used to analyze the optical density (OD) at a wavelength of 450 nm and to plot the proliferation curve.
Transwell
After thawing 100 μL of matrigel overnight, it was added to a 24-well plate in the upper chamber of a transwell chamber, shaken gently, and placed into a carbon dioxide incubator for 4-6 h to form a gel. Next, 500 μL serum-free medium was added into the lower chamber after the culture medium dried, and the substrate membrane was hydrated for half an hour. Cells were cultured in serum-free culture for 24 h after transfection with siRNA, 100 μL suspension was then put into the upper chamber at a density of 1×103 cells/μL, and 500 μL complete culture medium was added into the lower chamber. After overnight culture, the lower chamber was removed, the cells remaining in the upper chamber were cleaned, washed with PBS, fixed with 4% paraformaldehyde for 30 min, and stained with 0.1% crystal violet for 20 min. After cleaning with PBS, 5 visual fields were chosen at random under a microscope to observe and count.
Compared with cell invasion assays, the transwell chamber does not need to be coated with matrigel in the migration procedure.
Colony-forming assay
Cell exhibiting logarithmic growth were digested with trypsin (Gibco, Invitrogen) and blown into a single-cell suspension. The cell suspension was prepared at a density of 400 cells per dish and inoculated into a petri dish. It was then cultured at 37°C in a 5% CO2 incubator for approximately 1–2 weeks. When visible clones appeared in the petri dish, the culture was terminated. The cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Colonies were visualized and subjected to statistical analysis. The experiment was repeated three times independently.
Statistical analysis
The IBM SPSS22.0 statistical analysis software and GraphPad Prism version 5.0 were used to analyze the experimental data. The chi-square test was used to analyze the correlation between genes and clinical characteristics. Survival data were evaluated using univariate and multivariate Cox proportional hazards models. Student’s test was performed to examine the difference in two groups. One-way ANOVA analysis with Dunnett’s post hoc test was used to compare the mean values of multiple samples. P<0.05 was considered significantly.
Results
Overexpression IL20RB in PRCC patients is related to poor overall survival
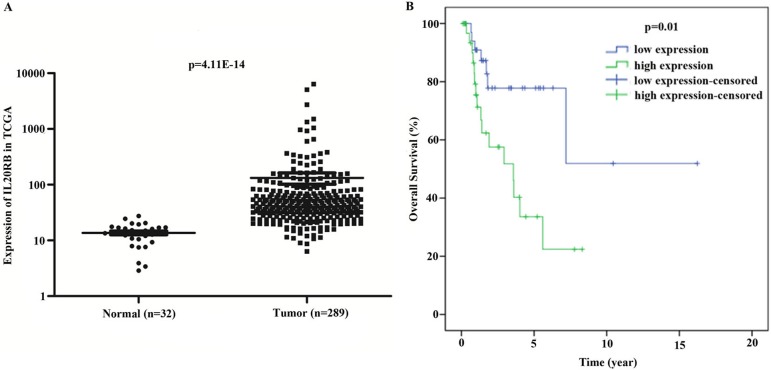
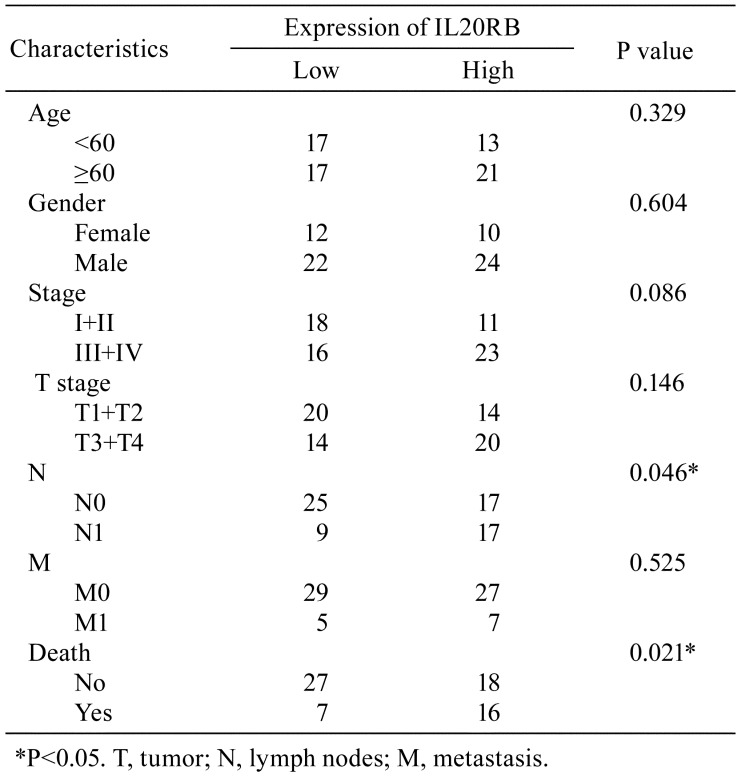
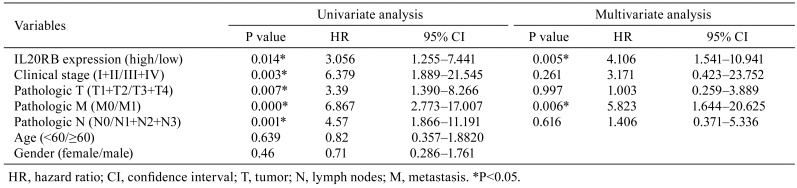
Data concerning IL20RB expression in PRCC tissue were extracted from the TCGA database, and it was revealed that IL20RB was highly expressed in PRCC tissue compared with the normal tissues (Fig. 1A, P<0.01). We further studied the association of IL20RB expression and clinical features (Table 1). The data indicated that the expression of IL20RB was related to lymph nodes (P=0.046) and death (P=0.021) in patients with PRCC. Furthermore, COX regression analysis suggested that IL20RB may be an independent predictor of prognosis in PRCC patients (Table 2). In univariate analysis, factors such as IL20RB expression, stage (I+II/III+IV), and TNM stage were connected with PRCC. In multivariate analysis, IL20RB expression and pathologic metastasis were correlated with PRCC, suggesting that IL20RB may be used as an independent predictor of prognosis in patients with PRCC. Moreover, the overall survival rate of PRCC patients with high IL20RB expression was lower than that of patients with low IL20RB expression (Fig. 1B, P<0.01). The results indicated that IL20RB was overexpressed in PRCC tissues and resulted in a poor prognosis.
Fig. 1.
Interleukin-20 receptor subunit beta (IL20RB) was highly expressed, and its overexpression in papillary renal cell carcinoma (PRCC) led to a poor prognosis. A: The expression of IL20RB was higher in tumor tissue than in normal tissue. B: The overall survival of patients with high expression of IL20RB was decreased compared with those with low expression.
Table 1. Relationship Between Interleukin-20 Receptor Subunit Beta (IL20RB) Expression and Clinicopathologic Features of Patients with Papillary Renal Cell Carcinoma.
Table 2. Cox Univariate and Multivariate Analysis of Interleukin-20 Receptor Subunit Beta (IL20RB) in Papillary Renal Cell Carcinoma Patients.
IL20RB was highly expressed in the Ketr-3 cell line
To further investigate the role of IL20RB in PRCC, the expression levels of IL20RB in different PRCC cell lines, including A498, ACHN, 786-0, and Ketr-3, and normal control HRCE cells were measured by qPCR. The data showed that IL20RB was highly expressed in PRCC cell lines, especially in Ketr-3, compared with the normal control HRCE cells (Fig. 2, P<0.01). Thus, in the following experiment, Ketr-3 cells were used to explore the biological function of IL20RB in PRCC. The experimental results in vitro were consistent with the results of analysis at the tissue level, indicating that IL20RB was highly expressed in PRCC.
Fig. 2.
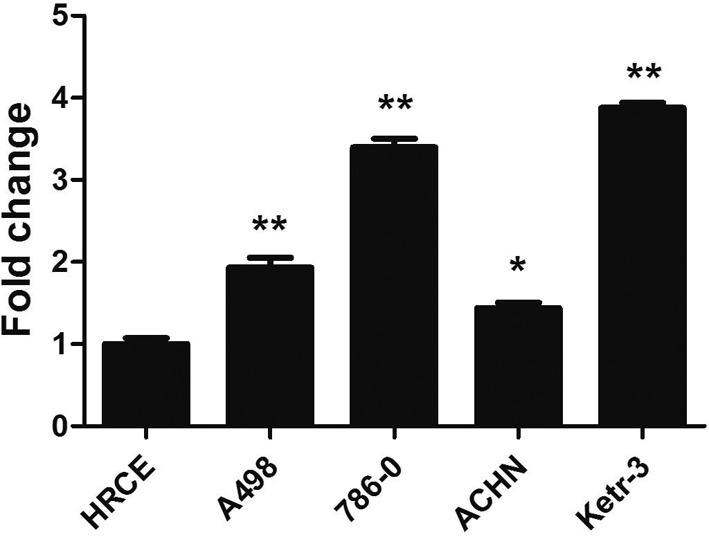
The overexpression of interleukin-20 receptor subunit beta (IL20RB) in papillary renal cell carcinoma (PRCC) cell lines. The expression levels of IL20RB in different cell lines, including A498, ACHN, 786-0, Ketr-3, and human renal cortical epithelial (HRCE) cells, were analyzed by quantitative real-time PCR (qRT-PCR). *P<0.05; **P<0.01.
Proliferation of PRCC cells was significantly reduced after knockdown of IL20RB
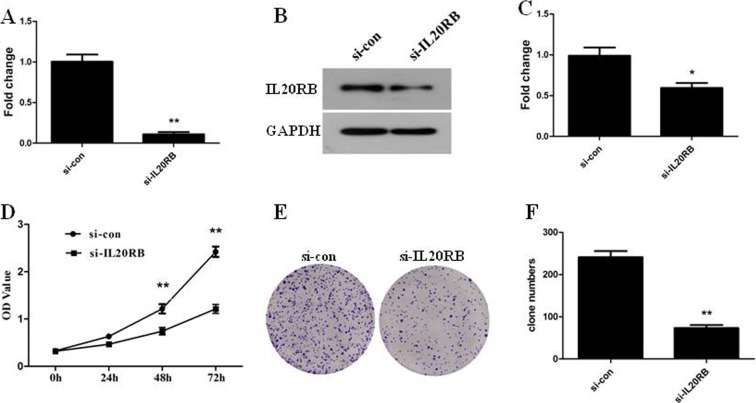
Next, based on the importance of IL20RB in PRCC and the discovery that IL20RB was overexpressed in PRCC and resulted in a poor prognosis, we investigated IL20RB effects on PRCC cells. We began by silencing IL20RB using siRNA technology in Ketr-3 cells. Obvious reductions in both mRNA and protein levels were observed (Fig. 3A, P<0.01, Fig. 3B and 3C, P<0.05). Then, in order to observe the functions of IL20RB in terms of PRCC cell characteristics after knockdown of IL20RB, we used CCK-8 and colony formation methods to investigate the proliferation activity of Ketr-3 cells. The results showed that the OD value in Ketr-3 cells was remarkably decreased after knockdown for 48 h and 72 h (Fig. 3D, P<0.01). Furthermore, the colony formation efficiency of Ketr-3 cells treated with si-IL20RB was measured. The number of colonies formed was obviously decreased compared with the control group (Fig. 3E and 3F, P<0.01). These results demonstrated that IL20RB silencing can reduce the proliferation of PRCC cells.
Fig. 3.
Knockdown of interleukin-20 receptor subunit beta (IL20RB) by small interfering RNA (siRNA) reduced the proliferation of Ketr-3 cells. A: Detection of interference efficiency by quantitative real-time PCR (qRT-PCR) after knockdown of IL20RB. **P<0.01. B: The protein levels of IL20RB were decreased. C: Quantification of the protein levels of IL20RB. *P<0.05. D: The results of CCK-8 indicated that knockdown of IL20RB restrained Ketr-3 cell proliferation. **P<0.01. E: The number of colonies formed was decreased. F: Quantification of the number of colonies formed. **P<0.01.
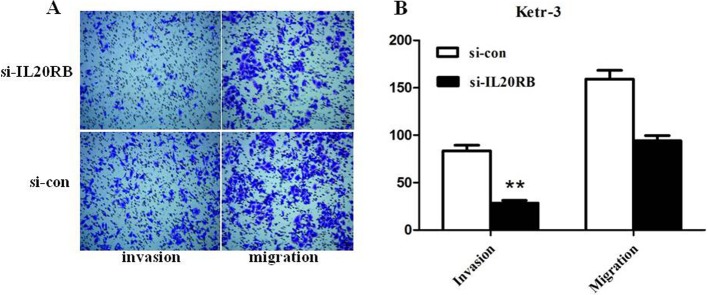
Silencing of IL20RB reduced Ketr-3 cells migration and invasion
Subsequently, a transwell assay was used to measure migration and invasion abilities in Ketr-3 cells after knockdown of IL20RB. The number of invading Ketr-3 cells was obviously reduced compared with the control group (Fig. 4A and 4B, P<0.01), indicating that their invasion ability was obviously inhibited after infection with si-IL20RB. Meanwhile, the number of migrating Ketr-3 was relatively decreased compared with the control group (Fig. 4A and 4B). The data indicated that knockdown of IL20RB impaired the invasion and migration abilities of PRCC cells.
Fig. 4.
A transwell assay was used to assess the effect of interleukin-20 receptor subunit beta (IL20RB) on cell invasion and migration. A: Compared with the Si-con group, invasion and migration of Ketr-3 cells were decreased. B: Quantification of invasion and migration of Ketr-3 cells. **P<0.01.
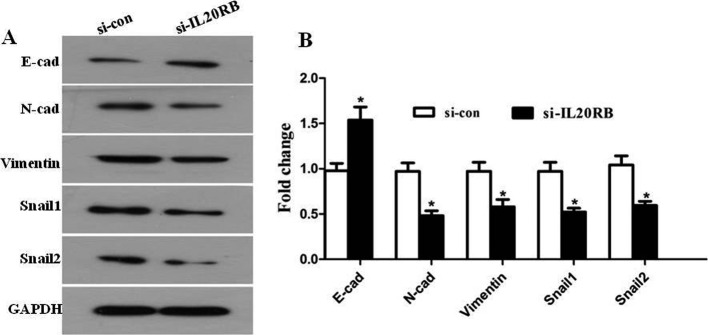
Knockdown of IL20RB affected epithelial-mesenchymal transition in Ketr-3 cells
Epithelial-mesenchymal transition (EMT) is recognized to be a process in which epithelial cells transdifferentiate into motile mesenchymal cells, and it contributes pathologically to fibrosis and cancer progression12. Hallmarks of EMT include the loss of expression or function of E-cadherin, reduced abundance of tight junction proteins, and high expression of N-cadherin, Snail1, Snail2, and Vimentin13. In our study, we found that after knockdown of IL20RB in Ketr-3 cells, the expression of E-cadherin was obviously increased; however, the N-cadherin, Snail1, Snail2, and Vimentin were reduced by varying degrees compared with the control group (Fig. 5A and 5B, P<0.05). The findings indicated that IL20RB may function via the EMT to influence the invasion and migration of PRCC cells.
Fig. 5.
Silencing of interleukin-20 receptor subunit beta (IL20RB) affected the epithelial-mesenchymal transition (EMT) in papillary renal cell carcinoma (PRCC) cells. A: Western blot was performed to evaluate the expression of EMT-related proteins including E-cadherin, N-cadherin, Snail1, Snail2, and Vimentin. B: Quantification of the expression levels of EMT-related proteins, *P<0.05.
Discussion
Renal cell carcinoma, which accounts for 90% of renal malignancies, is the most lethal tumor of the urinary system14. In 2004, the World Health Organization classification of adult renal tumors stratified renal cell carcinoma into several subtypes, of which clear cell, papillary, and chromophobe tumors accounted for 70%, 10–15% and 5% of the tumors, respectively15. Different types of RCC have different biological functions, prognoses, and treatment options. PRCC, as the second largest type of RCC, is difficult to diagnose and has a poor prognosis and limited therapeutic options16. Therefore, surgical treatment is the first choice, and interferon and interleukin-2 are the main immunotherapy methods at present. For metastatic RCC, targeted therapy is becoming the standard adjuvant therapy to improve overall survival. Thus, it is extremely important to discover the target genes to cure PRCC.
The IL-20 subfamily is involved both in amplified inflammatory responses, particularly during autoimmunity and chronic inflammation, and in anti-inflammatory responses, such as tissue protection and regeneration. This subfamily includes the cytokines IL-19, IL-20, IL-22, IL-24, and IL-26 and the receptors IL-20RA, IL-20RB, IL-10RB, and IL-22RA117. Different ligands have unique biological activities; for instance, IL-19 has been reported to directly affect immune cells18, IL-20 has shown activity on skin biology19, and IL24 plays a role in promoting apoptosis in tumors20. IL20RB, as an IL20 subfamily receptor, can be combined with these ligands to perform a variety of functions, such as activation of the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway (JAK-STAT)21 and stimulation of stable proliferation of BaF3 cells. However, the function of IL20R in PRCC has not yet been reported. Hence, we analyzed the expression of IL20RA and IL20RB in PRCC data from the TCGA database. The data showed no significant difference in the expression level of IL20RA in PRCC tissues; however, the abnormal expression of IL20RB in PRCC tissues and the characteristics significantly related to prognosis led us to focus of IL20RB. To the best of our knowledge, this is the first report on the expression and prognosis of IL20RB in PRCC, as well as the effect on the proliferation, invasion, and migration of PRCC cells after knockdown of IL20RB. IL20RB expression is related to the poor prognosis of patients with PRCC, suggesting that IL20RB, as a tumor-promoting gene, may have a positive regulatory role in PRCC. The findings of the present study showed that IL20RB has a positive effect on the proliferation, invasion, and migration abilities of PRCC cells and that silencing of IL20RB had an effect on the expression of EMT pathway-related proteins. Therefore, we hypothesized that IL20RB regulated the occurrence of PRCC through the EMT pathway, but the specific mechanism remains unclear.
The epithelial-mesenchymal transition is a key regulator of metastasis through promotion of tumor cell invasion and metastasis to distant organs22. Recent studies have reported that the EMT plays vital role in the occurrence and development of various tumors, including RCC23. During the EMT, E-cadherin is one of the strongest markers for cancer diagnosis and progression routinely used in the clinic24. Furthermore, abnormal expression of Snail was related to poor survival in breast25, hepatocellular26, and ovarian27 cancers. The overexpression of Vimentin in cancer and its relationship with growth and metastasis suggested that it may be an indicator of poor prognosis in many cancers28. In the present study, we detected the key proteins, including E-cadherin, N-cadherin, Snail1, Snail2, and Vimentin in the EMT pathway after knockdown of IL20RB. Compared with the control, the expression of E-cadherin was obviously upregulated, while the other proteins exhibiting positive correlation, including N-cadherin, Snail1, Snail2, and Vimentin, were significantly downregulated. The results showed that silencing of IL20RB limited the invasion and migration of PRCC cells probably through the EMT. However, the detailed molecular mechanism has not been well defined, and this will require further research.
Overall, all the data in the present study demonstrated that IL20RB was highly expressed in both PRCC tissues and cells and that overexpression of IL20RB in PRCC led to poor overall survival. Based on the findings of the knockdown experiment, IL20RB repressed the proliferation, invasion, and migration capacities of PRCC cells by regulating the EMT pathway. Therefore, we surmised that IL20RB exists as a key molecule in the occurrence and development of PRCC. We predicted that IL20RB could possibly be used for diagnosis of PRCC and a novel targeted therapy.
Disclosure of Potential Conflicts of Interest
None
References
- 1.Ng CS, Wood CG, Silverman PM, Tannir NM, Tamboli P, and Sandler CM. Renal cell carcinoma: diagnosis, staging, and surveillance. AJR Am J Roentgenol. 191: 1220–1232. 2008. [DOI] [PubMed] [Google Scholar]
- 2.Liu K, Ren Y, Pang L, Qi Y, Jia W, Tao L, Hu Z, Zhao J, Zhang H, Li L, Yue H, Han J, Liang W, Hu J, Zou H, Yuan X, and Li F. Papillary renal cell carcinoma: a clinicopathological and whole-genome exon sequencing study. Int J Clin Exp Pathol. 8: 8311–8335. 2015. [PMC free article] [PubMed] [Google Scholar]
- 3.Koul H, Huh JS, Rove KO, Crompton L, Koul S, Meacham RB, and Kim FJ. Molecular aspects of renal cell carcinoma: a review. Am J Cancer Res. 1: 240–254. 2011. [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Caspi RR, and Chong WP. IL-20 receptor cytokines in autoimmune diseases. J Leukoc Biol. 104: 953–959. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wirtz MK, and Keller KE. The role of the IL-20 subfamily in glaucoma. Mediators Inflamm. 2016: 4083735 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reimann E, Kingo K, Karelson M, Reemann P, Loite U, Sulakatko H, Keermann M, Raud K, Abram K, Vasar E, Silm H, and Kõks S. The mRNA expression profile of cytokines connected to the regulation of melanocyte functioning in vitiligo skin biopsy samples and peripheral blood mononuclear cells. Hum Immunol. 73: 393–398. 2012. [DOI] [PubMed] [Google Scholar]
- 7.Kingo K, Mössner R, Rätsep R, Raud K, Krüger U, Silm H, Vasar E, Reich K, and Kõks S. Association analysis of IL20RA and IL20RB genes in psoriasis. Genes Immun. 9: 445–451. 2008. [DOI] [PubMed] [Google Scholar]
- 8.Baird AM, Gray SG, and O’Byrne KJ. IL-20 is epigenetically regulated in NSCLC and down regulates the expression of VEGF. Eur J Cancer. 47: 1908–1918. 2011. [DOI] [PubMed] [Google Scholar]
- 9.Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC, Li J, Zhang YQ, Shi JW, Lin XL, Yang S, Xie RY, Liu W, Zhang TT, Sun YL, Xu K, Yao KT, and Xiao D. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. 431: 610–616. 2013. [DOI] [PubMed] [Google Scholar]
- 10.Omarini C, Bettelli S, Caprera C, Manfredini S, Caggia F, Guaitoli G, Moscetti L, Toss A, Cortesi L, Kaleci S, Maiorana A, Cascinu S, Conte PF, and Piacentini F. Clinical and molecular predictors of long-term response in HER2 positive metastatic breast cancer patients. Cancer Biol Ther. 19: 879–886. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haider S, Wang J, Nagano A, Desai A, Arumugam P, Dumartin L, Fitzgibbon J, Hagemann T, Marshall JF, Kocher HM, Crnogorac-Jurcevic T, Scarpa A, Lemoine NR, and Chelala C. A multi-gene signature predicts outcome in patients with pancreatic ductal adenocarcinoma. Genome Med. 6: 105 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamouille S, Xu J, and Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 15: 178–196. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalluri R, and Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 119: 1420–1428. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel RL, Miller KD, and Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 65: 5–29. 2015. [DOI] [PubMed] [Google Scholar]
- 15.Low G, Huang G, Fu W, Moloo Z, and Girgis S. Review of renal cell carcinoma and its common subtypes in radiology. World J Radiol. 8: 484–500. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choueiri TK, Plimack E, Arkenau HT, Jonasch E, Heng DYC, Powles T, Frigault MM, Clark EA, Handzel AA, Gardner H, Morgan S, Albiges L, and Pal SK. Biomarker-Based Phase II Trial of Savolitinib in Patients With Advanced Papillary Renal Cell Cancer. J Clin Oncol. 35: 2993–3001. 2017. [DOI] [PubMed] [Google Scholar]
- 17.Rutz S, Wang X, and Ouyang W. The IL-20 subfamily of cytokines--from host defence to tissue homeostasis. Nat Rev Immunol. 14: 783–795. 2014. [DOI] [PubMed] [Google Scholar]
- 18.Gallagher G, Dickensheets H, Eskdale J, Izotova LS, Mirochnitchenko OV, Peat JD, Vazquez N, Pestka S, Donnelly RP, and Kotenko SV. Cloning, expression and initial characterization of interleukin-19 (IL-19), a novel homologue of human interleukin-10 (IL-10). Genes Immun. 1: 442–450. 2000. [DOI] [PubMed] [Google Scholar]
- 19.Blumberg H, Conklin D, Xu WF, Grossmann A, Brender T, Carollo S, Eagan M, Foster D, Haldeman BA, Hammond A, Haugen H, Jelinek L, Kelly JD, Madden K, Maurer MF, Parrish-Novak J, Prunkard D, Sexson S, Sprecher C, Waggie K, West J, Whitmore TE, Yao L, Kuechle MK, Dale BA, and Chandrasekher YA. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell. 104: 9–19. 2001. [DOI] [PubMed] [Google Scholar]
- 20.Lebedeva IV, Su ZZ, Chang Y, Kitada S, Reed JC, and Fisher PB. The cancer growth suppressing gene mda-7 induces apoptosis selectively in human melanoma cells. Oncogene. 21: 708–718. 2002. [DOI] [PubMed] [Google Scholar]
- 21.Parrish-Novak J, Xu W, Brender T, Yao L, Jones C, West J, Brandt C, Jelinek L, Madden K, McKernan PA, Foster DC, Jaspers S, and Chandrasekher YA. Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions. J Biol Chem. 277: 47517–47523. 2002. [DOI] [PubMed] [Google Scholar]
- 22.Santamaria PG, Moreno-Bueno G, Portillo F, and Cano A. EMT: Present and future in clinical oncology. Mol Oncol. 11: 718–738. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasti A, Madjd Z, Abolhasani M, Mehrazma M, Janani L, Saeednejad Zanjani L, and Asgari M. Cytoplasmic expression of Twist1, an EMT-related transcription factor, is associated with higher grades renal cell carcinomas and worse progression-free survival in clear cell renal cell carcinoma. Clin Exp Med. 18: 177–190. 2018. [DOI] [PubMed] [Google Scholar]
- 24.Pasquier J, Abu-Kaoud N, Al Thani H, and Rafii A. Epithelial to mesenchymal transition in a clinical perspective. J Oncol. 2015: 792182 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanco MJ, Moreno-Bueno G, Sarrio D, Locascio A, Cano A, Palacios J, and Nieto MA. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene. 21: 3241–3246. 2002. [DOI] [PubMed] [Google Scholar]
- 26.Miyoshi A, Kitajima Y, Kido S, Shimonishi T, Matsuyama S, Kitahara K, and Miyazaki K. Snail accelerates cancer invasion by upregulating MMP expression and is associated with poor prognosis of hepatocellular carcinoma. Br J Cancer. 92: 252–258. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai T, Horiuchi A, Wang C, Oka K, Ohira S, Nikaido T, and Konishi I. Hypoxia attenuates the expression of E-cadherin via up-regulation of SNAIL in ovarian carcinoma cells. Am J Pathol. 163: 1437–1447. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satelli A, and Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 68: 3033–3046. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]