Abstract
Early intervention for newborns who are deaf or hard-of-hearing leads to improved language, communication, and social–emotional outcomes. Universal physiologic newborn hearing screening has been widely implemented across the United States with the goal of identifying newborns who are deaf or hard-of-hearing, thereby reducing time to diagnosis and intervention. The current physiologic newborn hearing screen is generally successful in accomplishing its goals but improvements could be made. In the past ten years, genetic testing has emerged as the most important etiological diagnostic test for evaluation of children with deafness and congenital cytomegalovirus has been recognized as a major cause of childhood deafness that may be treatable. A comprehensive newborn hearing screen that includes physiologic, genetic, and cytomegalovirus testing would have multiple benefits, including (1) identifying newborns with deafness missed by the current physiologic screen, (2) providing etiologic information, and (3) possibly decreasing the number of children lost to follow up. We present a framework for integrating limited genetic testing and cytomegalovirus screening into the current physiologic newborn hearing screening. We identify needed areas of research and include an overview of genome sequencing, which we believe will become available over the next decade as a complement to universal physiologic newborn hearing screening.
Keywords: newborn hearing screening, deafness, genetics, genomics, cytomegalovirus
INTRODUCTION
The impact of permanent deafness on a child’s development is profound. It affects not only language acquisition but also social development and quality of life.1 Early detection of congenital deafness with targeted intervention significantly reduces negative impacts in these areas.2–4 In 1994, the Joint Committee on Infant Hearing (JCIH) published a position statement that endorsed the goal of universal detection of infants with deafness and encouraged continuing research and development to improve techniques for detection of and intervention for deafness as early as possible.5,6 Today, the crucial role of newborn hearing screening (NBHS) is emphasized by the fact that 43 states and territories of the United States have passed laws mandating NBHS, with the remainder of states having implemented universal NBHS without legislation. Currently, the JCIH recommends universal NBHS by 1 month of age, diagnosis by 3 months of age, and early intervention by 6 months of age to allow optimal intervention for children with deafness, if warranted and if desired by the family.7
The most recent data show that 98.2% of newborns in the United States receive NBHS.8 Universal screening has led to a significant reduction in the average age at which newborns with congenital deafness are identified in this country.9 Although the current universal NBHS has been remarkably successful in improving the early diagnosis and intervention in newborns who are deaf or hard-of-hearing (DHH), as will be detailed below, knowledge gained from the universal NBHS, an improved understanding of the genetics of hearing loss, as well as an increased recognition of the contribution of congenital cytomegalovirus (cCMV) to childhood deafness have provided an opportunity to improve the current NBHS.
Over the past 20 years, our understanding of genetic deafness has greatly improved. Along with diagnostic audiologic evaluation, diagnostic genetic testing platforms now form a cornerstone for evaluation of DHH newborns and children.10–12 An etiological diagnosis is provided in 49% of DHH newborns who undergo genetic testing.13 Genetic testing can also identify mild deafness, later-onset childhood deafness, syndromic forms of deafness, risk factors for aminoglycoside-induced deafness, and auditory neuropathy that may not be detected by the current physiologic NBHS. A genetic test for deafness that could be translated into a universal genetic screening test would form a powerful complement to the current NBHS; however, to date no genetic screening has been incorporated as part of a NBHS program in the United States.
An additional important contributor to childhood deafness is congenital cytomegalovirus infection (cCMV), estimated to be a cause of ~10% of congenital deafness and 15–20% of childhood deafness.14,15 Because cCMV often presents as mild, fluctuating, and progressive deafness, detection with a physiologic screen can be challenging.16 The incorporation of cCMV screening in a NBHS program would provide etiological information, improve ascertainment, and further complement the physiologic NBHS.
Creating a comprehensive newborn hearing screen that includes physiologic, genetic, and cytomegalovirus screening would have multiple benefits including (1) identifying newborns at risk for deafness and who could benefit from early intervention but are missed by the current physiologic screen, (2) providing etiologic information as part of the screen, (3) possibly decreasing the number of children who are lost to follow up, and (4) potentially saving costs by reducing the frequency of later testing. In this paper, we aim to provide a conceptual framework for a comprehensive NBHS program that incorporates the current physiologic screening as well as molecular screening that includes genetic screening and cCMV screening (Fig. 1). A comprehensive NBHS of this nature would require three concurrent screening methods, with results that inform the final screening result. Its implementation will likely require incremental steps and goals, particularly with respect to the incorporation of a form of nucleotide-level genetic screening, as there is no precedent in the United States. Targeted cCMV screening for those newborns who fail the physiologic screen has been incorporated in several states and therefore there is precedent for this form of molecular screening.
Fig. 1.
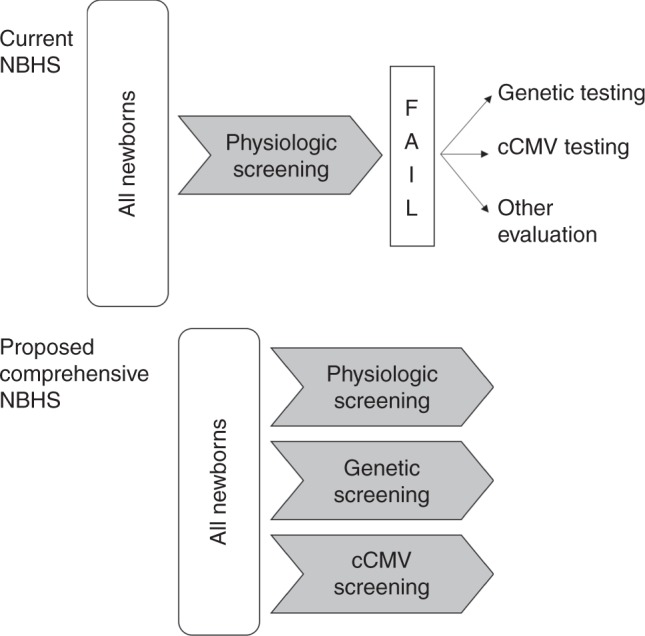
Comparison schematic of current and proposed newborn hearing screening (NBHS). cCMV congenital cytomegalovirus.
We first provide an overview of the current physiologic NBHS, including examination of areas of needed improvement. We next examine the available evidence that genetic and cCMV testing could improve the NBHS. Finally, we address challenges and barriers to accomplishing the goal of an improved NBHS, including patient and family perspectives. We believe that a comprehensive NBHS would significantly improve detection of newborns with deafness, advance our understanding of deafness in children, improve time to diagnosis and time-to-intervention, provide more intervention options for families across the childhood age span, and lay the foundation for improved treatment and support for children who are DHH.
THE CURRENT NBHS: STRENGTHS AND WEAKNESSES
The prevalence of permanent deafness identified by state-based Early Hearing Detection and Intervention (EHDI) programs and reported to the Centers for Disease Control and Prevention (CDC) is 1.7 per 1000 births.8 This prevalence makes deafness the most common sensory deficit in humans. Universal NBHS is widely implemented in the United States, with 98.2% of newborns undergoing screening. From this perspective, the physiologic NBHS has been tremendously successful in regard to universal implementation and in its primary goal of decreasing the average age of identification of children with deafness.9
The goal of universal NBHS is to identify all newborns with permanent deafness. Both detecting and establishing an etiologic diagnosis for deafness in children are challenging because of the large number of possible causes and the wide clinical variability in presentation (Table 1). Hearing loss is described as slight (16–25 decibel hearing level, dB HL), mild (26–40 dB HL), moderate (41–55 dB HL), moderately severe (56–70 dB HL), severe (71–90 dB HL), or profound (>90 dB HL); can be unilateral or bilateral; and is either asymmetric or symmetric (Fig. 2). Select frequencies can be affected that can give an audiogram a specific shape or profile associated with specific genetic forms of deafness (for example, downsloping if high frequencies are impacted more than low frequencies, or upsloping if the reverse is true; Fig. 2). Deafness can also be defined by the site of impairment in the auditory system. A conductive hearing loss (CHL, see a list of definitions in Box 1) implies that transmission of sound through the external ear canal or middle ear is impaired, as in the case of a middle ear effusion, stenosis of the ear canal, or fixation of the ossicular chain. A sensorineural hearing loss (SNHL), in comparison, reflects compromised transmission of the neural signal along the auditory pathway, be it in the cochlea, the auditory nerve, or more proximally in the brainstem and cortex. In the United States and other developed countries, most permanent congenital deafness is SNHL and the majority of SNHL (~70%) is due to a genetic cause (Table S1).14,17 Environmental causes of SNHL, such as infections, hypoxia, and trauma, are a significant but smaller contributor to congenital deafness compared with genetic causes.
Table 1.
Classification of permanent childhood deafness
| Classification | Category |
|---|---|
| Mechanism | Conductive |
| Sensorineural | |
| Mixed (conductive and sensorineural components) | |
| Auditory neuropathy | |
| Degree | Slight (16–25 dB HL) |
| Mild (26–40 dB HL) | |
| Moderate (41–55 dB HL) | |
| Moderately severe (56–70 dB HL) | |
| Severe (71–90 dB HL) | |
| Profound (>90 dB HL) | |
| Symmetry | Bilateral–symmetric |
| Bilateral–asymmetric | |
| Unilateral | |
| Progression | Stable |
| Progressive | |
| Fluctuating | |
| Onset | Congenital |
| Prelingual | |
| Infantile | |
| Postlingual | |
| Childhood | |
| Adult | |
| Frequency pattern of deafness | High frequency (downsloping) |
| Low frequency (upsloping) | |
| Mid-frequency (cookie bite) | |
| All frequencies (flat) | |
| Associated clinical findings | Nonsyndromic (no other clinical findings) |
| Syndromic (other clinical findings) | |
| Nonsyndromic mimics (syndromic HL diagnosed when other clinical findings are not yet apparent) |
HL hearing level.
Fig. 2.
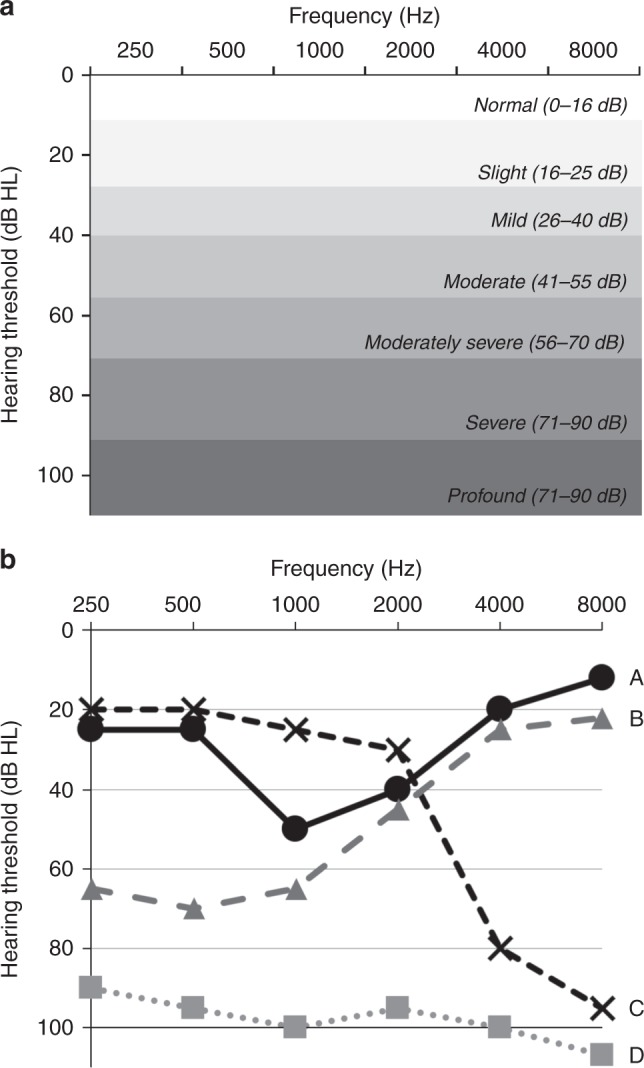
Evaluation of hearing with audiometry. (a) Audiogram showing hearing threshold in decibels (dB) of hearing level (HL) on the y-axis and frequency in hertz (Hz) on the x-axis. The degrees of hearing loss from slight to profound are labeled. (b) Characteristic forms of deafness including: A—mid-frequency (associated with variants in TECTA), B—low frequency (associated with variants in WFS1), C—high frequency (associated with variants in KCNQ4), and D—severe to profound (associated with variants in GJB2).
The ideal NBHS, as is the case for any screening test, must be inexpensive, easy to learn and to administer by screeners, acceptable to the screened individuals, and able to identify the presence or absence of deafness with few false positives (high specificity) and few false negatives (high sensitivity). Nearly all current NBHS programs in the United States use a physiologic evaluation of auditory function in response to sound. Methods used include an otoacoustic emission (OAE) screening test, which measures responses from the outer hair cells of the cochlea; an automated auditory brainstem response (AABR) screening test, which records response to sound based on the neural transmission of a signal from the cochlea to the brainstem; or a combination of both. These screening tests are low cost and can be administered within minutes by a trained screener. As highly validated testing methods, they have been widely adopted. The NBHS results in a “pass” (screened negative) or a “refer/fail/did not pass” (screened positive) indicating possible deafness and the need for further evaluation.
For newborns who do not pass the NBHS, diagnostic/confirmatory audiological testing is required to determine the type, degree, and configuration of hearing loss. Further clinical evaluation focuses on determining the etiological cause of deafness and is performed in concert between the primary care provider and otolaryngologist. This evaluation varies based on type of deafness but by current guidelines should include clinical examination and genetic testing, followed by imaging, other laboratory tests, and referrals to specialists as indicated.11 Further referrals and follow ups in the health-care system are guided by findings determined by evaluation and frequently include referral to a geneticist and other relevant medical specialists, as determined by clinical findings. Typically, the finding of deafness is also reported to the state EHDI program. A treatment or habilitation plan is then developed that can range from preferential seating in school and speech therapy, to sign language, assistive devices, and/or surgical intervention such as cochlear implantation. Although it has been remarkably successful in improving identification of children with hearing loss, there are several ways in which the current physiologic NBHS could be improved, and these are detailed below.
The current physiologic NBHS has a low positive predictive value
Any screening test must weigh false negatives versus false positives to best serve the population screened. “False positive” may refer to the actual screening device used but here we refer to the overall result given to parents—“pass” or “refer”—versus the subsequent diagnostic confirmatory physiologic testing, which serves as the “gold standard.” The positive predictive value (PPV) is the probability that subjects who screen positive truly have the disease. An ideal screening test would have a high PPV and a high sensitivity. While negative predictive value and specificity are also considered in design of screening tests, a confirmatory or diagnostic test should aim for a high negative predictive value and a high specificity as false negatives are reduced.
Data show that of the 3,866,820 newborns who underwent NBHS in 2015, 64,978 (1.7%) did not pass. Of these children, 39,468 (60.7%) went on to have confirmatory audiologic testing.8 Of these, only 6442 (16.3%) were found to have deafness on confirmatory diagnostic audiometric testing. Therefore, 83.7% of children who screened positive did not have deafness, equating to a PPV of 16.3% and a specificity of 98.6% for the NBHS. The true sensitivity of this screening program is not known, as there are no large studies that have performed diagnostic testing on all newborns to identify the number of false negative instances (i.e., newborns who passed screening but were found to have deafness). From a practical sense, this means that the vast majority of newborns who refer on the physiologic NBHS do not have permanent deafness.
While the false positive rate is a concern for the physiologic NBHS, other newborn screening tests have similar performance. For example, a study in California showed that of 755,673 newborns screened for a panel of inborn errors of metabolism using tandem mass spectrometry, 461 newborns screened positive (0.13%) (ref. 18). Of the 386 children who underwent confirmatory testing, 335 (86.7%) were found to be false positives and have no inborn errors of metabolism. For this panel, the overall PPV is 13.2%. Other newborn screenings for inborn errors of metabolism have similar false positives rates and positive predictive values.19,20
As a universal screening test performed on a vulnerable population, reducing the false positive rate for the NBHS is important. False positives have the immediate effect of not only unnecessarily worsening parental anxiety21 and increasing health-care costs but also may lead to a decrease in follow up (discussed in a separate section below). Increasing the PPV should be a goal to improve the current physiologic NBHS but is not an inherent benefit of adding genetic screening.
Types of deafness that are not identified by the current NBHS
The effect of mild-to-moderate hearing loss on language development has been heavily debated, but new data and review of previous studies indicate that it is in fact a significant risk factor for communication difficulties.22 Data from the CDC show that the majority (56.1%) of bilateral deafness in newborns is mild to moderate,8 however an even larger percentage of newborns may be affected, as very mild deafness can go undetected by NBHS (depending on the thresholds and testing methods that are used). Although implementing physiologic screening at lower hearing levels would detect more mild-to-moderate deafness, it would also increase false positive rates and burden confirmatory diagnostic audiology evaluation. The precise sound intensity used in the NBHS is difficult to define accurately because screening equipment manufacturers choose stimulus levels and characteristics for their own equipment that cannot be adjusted by the operator. The screening level of AABR is typically programmed into the screening equipment; this level is based on research indicating the optimal screening level for AABR. There are no calibration standards for transient stimuli or for determining the actual stimulus levels in ear canals of newborns, which may be greater than specified by the manufacturer because of the small size of a newborn ear canal, possibly leading to false negative results. Identification of infants with permanent and/or progressive mild deafness is integral to improving the current physiologic NBHS.
Children who develop deafness after the newborn period will not be detected by the current NBHS. This limitation is important as the number of children with significant hearing loss increases throughout childhood from a congenital prevalence of 1.7 per 1000 births8 to a best-estimated prevalence during childhood of 31 per 1000 (ref. 23). Studies of the prevalence of childhood deafness are primarily based on surveys and although further research in this area is needed, particularly in regard to prevalence in preschool-aged children; it is clear that the prevalence of deafness increases with age during childhood.23 Importantly, a significant number of children who develop prelingual deafness following the newborn period are not detected by the currently implemented NBHS. Dedhia and coauthors estimated that nearly 25% of all children with SNHL are not identified by the NBHS and that nearly two-thirds of these had severe-to-profound deafness.24 Young et al. found that 30% of all cochlear implant candidates passed their NBHS and these children had significantly delayed treatment.25 It is unclear based on current studies whether these children had congenital deafness or deafness that began soon after the newborn period and further study is needed to clarify this point. However, because these children are not identified by the current universal NBHS, the detection of their deafness currently occurs through a patchwork of school-age hearing screening programs that vary significantly by state. Establishing a universal NBHS that is able to identify children with deafness that occurs outside the newborn period would prevent a delay in diagnosis and treatment.
Auditory neuropathy spectrum disorder (ANSD) is characterized by absent or severely abnormal inner hair cell (IHC), synaptic, and/or spiral ganglion function as measured by ABR testing, with preservation of outer hair cell (OHC) function as measured by OAE testing.26 Therefore, a newborn with ANSD manifesting as severe-to-profound deafness may go undetected by OAE screening when OAE screening is performed alone. The presentation of ANSD is also highly variable, with some infants and children with ANSD having asymmetric or unilateral deafness. At older ages, ANSD is accompanied by poor speech discrimination and poor word understanding, especially in the presence of noise. The prevalence of ANSD is reported to be 2.7% of DHH newborns identified by NBHS programs based on data from the CDC.8 Other reported data indicate the prevalence of ANSD to be 1.2%, 5.1%, or 8.4% depending on the population.27–29 Forty percent (40%) of ANSD is estimated to have a genetic basis, with the remainder due to acquired causes like hypoxia, prematurity, and jaundice, underscoring the increased rate of ANSD in neonatal intensive care units. The list of causative genes for ANSD includes DIAPH3, OTOF, PJVK, and mitochondrial DNA (mtDNA) variants (m.1095T>C) for nonsyndromic ANSD, and AIFM1, DDDP, MPZ, OPA1, PMP22, and TMEM126A for syndromic ANSD, although based on prevalence data there are likely other genes involved. The gene most frequently implicated in nonsyndromic ANSD is OTOF, which is estimated to be responsible for 0.5–3.5% of prelingual deafness across multiethnic cohorts.30–32 NBHS that relies only on OAE will not identify these babies, but fortunately the majority of neonatal intensive care units (where the rate of ANSD is highest) perform screening with AABR, as recommended by JCIH, and half of all NBHS is now performed with AABR. The actual number of newborns missed by the current NBHS due to ANSD is not known but ensuring these babies are screened is key to improving screening outcomes.
The current NBHS does not screen for a relatively common risk factor that can cause deafness in the newborn period. Several mitochondrial DNA variants lead to exquisite sensitivity to aminoglycoside-induced deafness. Aminoglycosides, in particular gentamicin, are commonly used in the neonatal period due to low cost and effectiveness against Gram-negative bacteria.33 Newborns who carry certain genetic variants in the mitochondrial gene MT-RNR1 can experience significant deafness with a single dose of these commonly used antibiotics. In addition, animal studies show that there is a synergistic effect of these genetic variants, aminoglycosides, and noise, which further predisposes these newborns to deafness. Prevalence of these mitochondrial variants was found to be 0.2% in one study of 703 children from a neonatal intensive care unit in the United States and 0.19% in a study of 58,397 Chinese children.34,35 Identifying these newborns with screening could have immediate treatment implications. In addition, diagnosis could prevent aminoglycoside-induced later-onset deafness in vulnerable individuals and lead to targeted cost-effective evaluation in maternal relatives after positive screening results from one individual.
Limited etiological information is included in the current NBHS
The current NBHS provides a simple result: “pass” or “refer.” Subsequent audiologic and diagnostic evaluation provides information on degree of deafness and etiology. An extremely important goal of diagnostic testing for deafness in children is to identify etiologies that have further diagnostic and treatment implications. Among these are syndromic forms of deafness, the most common of which are described in Table S2 and include Usher syndrome (deafness–blindness), Pendred syndrome (deafness including inner ear malformations and thyroid goiter), branchiootorenal syndrome (BOR, branchial cleft anomalies, deafness, and renal abnormalities), Jervell and Lange-Nielsen syndrome (deafness and cardiac arrhythmias), and Alport syndrome (deafness, renal disease, and eye abnormalities). These syndromic forms of deafness are referred to as nonsyndromic hearing loss (NSHL) mimics because they present at birth as nonsyndromic deafness (no other associated abnormalities). Recent data show that these syndromes are more common than previously reported. In a cohort of 2460 individuals of all ages with deafness, the diagnoses of NSHL mimics totaled 25% of all diagnoses and most commonly included Usher syndrome (10%), followed by Pendred syndrome (5%), deafness–infertility syndrome (4%), and branchiootorenal syndrome (2%) (R.J.S. unpublished data). Further data are required on the clinical and ethical repercussions of early diagnosis of syndromic forms of hearing loss. Early genetic diagnosis of syndromic forms of deafness would significantly reduce other testing and provide opportunities for early intervention.
Identification of a genetic etiology has not only the potential to inform prognostic monitoring of deafness and/or associated syndromic features, but may also refine estimates of recurrence risk for family members. The information may influence family reproductive and financial planning.
There are a large number of newborns lost to follow up in the current NBHS
A significant number of newborns are lost to diagnostic follow up in the current NBHS protocol. The most recent data from 2015 showed that 27.9% of newborns who referred on NBHS were lost to follow up prior to diagnostic audiology.8 Of these newborns, 54.7% were due to unknown reasons; 14.7% were cases in which there was an inability to contact the newborn’s guardian; and in the remaining 30.6% of cases, the parents or family were contacted but were not responsive to follow-up requests.8 There are several reasons that children are lost to follow up including social, economic, and geographic factors.36 Data show that altering testing methodologies, including a screen–rescreen policy as well as increasing time from birth can improve positive predictive value, but further research is needed in this area37–39 It is not unreasonable to assume that by providing more etiologic information to parents during the screening process and improving the positive predictive value of screening, fewer children would be lost to follow up. Providing an etiologic diagnosis sooner may motivate parents to seek early intervention services but further study in this area is needed.
Incorporating genetic screening into the NBHS
Our understanding of the molecular physiology of hearing and deafness has improved dramatically in the past two decades, in part through the study of genetic causes of deafness. The first gene to be implicated in nonsyndromic human deafness, GJB2, was discovered in 1997.40 Since that time, over 130 genes and nearly 8000 genetic variations have been identified that are associated with nonsyndromic deafness (Table S1). These variations differ significantly based on ethnic population examined. In addition, there are more than 600 clinical syndromes that include deafness; in several, deafness is the first presenting clinical feature (NSHL mimics) (Table S2).
The extreme genetic heterogeneity of deafness has made single-gene targeted genetic testing inefficient compared with its use in disorders where a single primary underlying genetic cause predominates (e.g., testing the CFTR gene to diagnose cystic fibrosis). In addition, with respect to deafness, sequential single-gene sequencing tests are costly, time-consuming, and relatively low in their diagnostic yield.41 The first clinical diagnostic genetic test for deafness, evaluation for variants in GJB2, began in 1999. Initial tests were based on variant detection assays, but these assays quickly gave way to single-gene Sanger sequencing, which has now been replaced by comprehensive genetic testing using massively parallel sequencing techniques. Multiple genes are sequenced in parallel to provide a complete analysis of the genetic landscape, which is essential for a heterogeneous condition like deafness. Massively parallel sequencing was first demonstrated for genetic testing for deafness in 2010 and was shown to have an analytical sensitivity and specificity of >99% for single-nucleotide variant detection making it suitable for clinical genetic testing.42 Massively parallel sequencing is now the standard of care for the genetic evaluation of children with deafness as recommended by the American College of Medical Genetics and Genomics and the International Pediatric Otolaryngology Working Group.11,12
Recent data show that when a comprehensive genetic testing platform is used to evaluate persons with deafness, including all ages and types of deafness with no exclusionary criteria, a diagnosis is provided in ~40% of cases.10,30 Data from 2460 individuals with deafness run on the OtoSCOPE platform from the University of Iowa Molecular Otolaryngology and Renal Research Labs (MORL) show a diagnostic rate of 39.9% (R.J.S., unpublished data), which is consistent with a review of other published studies.10 This diagnostic rate varies based on gene content, variant interpretation standards, and patient population studied. In comparison, data from the Laboratory of Molecular Medicine (LMM) at Partners HealthCare show a diagnostic rate of 25.2%, an inconclusive rate of 57.8%, and a negative rate of 17.0% in a cohort of 1156 patients presenting for multigene panel testing (S.A. and J.S., unpublished data).
The number of individuals who are DHH that are provided a diagnosis from genetic testing varies based on clinical history such as the type of deafness, age of patient, family history, and ethnicity.30 For the large cohort of clinical samples from the University of Iowa, the diagnostic rate for those with congenital deafness was 53.6% (R.J.S., unpublished data). Published data show that in those affected by congenital deafness, if there is also a family history consistent with dominant or recessive inheritance, the diagnostic rates are 67% and 55%, respectively. It is likely that these percentages will increase as additional genetic causes of deafness are identified. Data from the University of Iowa show that diagnostic rates decrease with age of onset of deafness, and are 39.6% at 3–6 years and 27.3% at 12–18 years, paralleling the increase in nongenetic causes of deafness with increasing age (rates based on age at testing as precise data on age of onset are not available).
In comparison, other commonly used diagnostic tests for evaluation of children with deafness have lower diagnostic rates: computed tomography imaging, 30% (ref. 43); magnetic resonance imaging, 26% (ref. 44); ophthalmologic evaluation, 8%; renal ultrasound, 4%; and electrocardiogram, 1% (ref. 45). Genetic testing therefore has the highest diagnostic rate of any test used to identify the etiological cause of childhood deafness and is now recommended as the first-line test in evaluation of children with documented bilateral SNHL.10
There are several commercially available comprehensive genetic testing platforms (Genetic Testing Registry). The number of genes included, cost, and turnaround time vary, but in general most platforms rely on targeted genomic enrichment and massively parallel sequencing technologies, which allow sequencing of many genes simultaneously.46 Average turnaround time per test is 2–3 months and listed prices range from ~$1500 to $5000 per test. More importantly, however, is the downstream analysis, which can result in differences at the level of the identification and/or interpretation of genetic variants.
The inclusion of copy-number variant (CNV) analysis in comprehensive genetic testing is crucial, as has been illustrated in a study of 686 patients in which at least one CNV was identified in 104 persons (15.2%) and implicated in the genetic diagnosis 18.7% of the time.47 CNVs were most commonly identified in STRC (73% of CNVs identified) followed by OTOA (13% of CNVs identified). More recent, unpublished data confirm that CNVs in STRC are one of the most commonly detected pathogenic variants causing deafness and the second most common gene implicated in deafness behind GJB2 (Table 2, 3). If only mild-to-moderate deafness is considered, STRC deletion is the most common cause of deafness.
Table 2.
Ten most commonly identified genes causing deafness
| Gene | Count | % of Diagnoses |
|---|---|---|
| MORL | ||
| GJB2 | 210 | 21.4 |
| STRC | 140 | 14.3 |
| SLC26A4 | 58 | 5.9 |
| MYO7A | 49 | 5.0 |
| TECTA | 41 | 4.2 |
| MYO15A | 42 | 4.3 |
| CDH23 | 38 | 3.9 |
| USH2A | 40 | 4.1 |
| ADGRV1 | 18 | 1.8 |
| WFS1 | 18 | 1.8 |
| MORL total | 654 | 66.6 |
| LMM | ||
| GJB2 | 270 | 36.6 |
| STRC | 66 | 9.0 |
| USH2A | 66 | 9.0 |
| MYO7A | 58 | 7.9 |
| SLC26A4 | 45 | 6.1 |
| CDH23 | 19 | 2.6 |
| WFS1 | 19 | 2.6 |
| TMPRSS3 | 17 | 2.3 |
| OTOF | 15 | 2.0 |
| TMC1 | 12 | 1.6 |
| LMM total | 587 | 79.6 |
Ten genes most commonly identified as causes of deafness in two clinical testing laboratories: Molecular Otolaryngology & Renal Research Labs (MORL, Iowa City, IA) and the Laboratory for Molecular Medicine (LMM, Cambridge, MA). MORL data are from 2460 individuals from the US population with deafness who presented for diagnostic comprehensive genetic testing with the OtoSCOPE platform. The total number of diagnoses provided is 982, or 39.9% of 2460. All ethnicities are included and no exclusions were made based on type of deafness (R.J.S., unpublished data). LMM data are from 737 positively diagnosed cases mainly from the US population with deafness who presented for diagnostic genetic testing at the LMM. GJB2-related deafness was overrepresented because 104 cases were only tested for GJB2 including deletions of 5’ upstream regulatory regions involving GJB6. Data from MORL are adapted from Sloan-Heggen et al. (2016);13 data from LMM are unpublished (contributed by S.A. and J.S.).
Table 3.
Ten most commonly identified genetic variants causing deafness
| Gene | Variant | Diagnostic frequency (n alleles) | % of diagnosed variants |
|---|---|---|---|
| MORL | |||
| STRC | CNV—partial/whole-gene deletion | 84 | 11.8% |
| GJB2 | c.35delG, p.Gly12fs | 51 | 7.1% |
| GJB2 | c.109G>A, p.Val37Ile | 27 | 3.8% |
| GJB2 | c.101T>C, p.Met34Thr | 20 | 2.8% |
| OTOA | CNV—partial/whole-gene deletion | 7 | 1.0% |
| USH2A | c.4714C>T, p.Leu1572Phe | 7 | 1.0% |
| USH2A | c.2299delG, p.Glu767fs | 7 | 1.0% |
| SLC26A4 | c.1001+1G>A | 6 | 0.8% |
| GJB2 | c.167delT, p.Leu56fs | 5 | 0.7% |
| MYO7A | c.3719G>A, p.Arg1240Gln | 4 | 0.6% |
| MORL total | 218 | 30.5% | |
| LMM | |||
| GJB2 | c.35delG, p.Gly12fs | 196 | 14.3% |
| GJB2 | c.109G>A, p.Val37Ile | 131 | 9.6% |
| STRC | CNV—partial/whole-gene deletion | 110 | 8.0% |
| GJB2 | c.101T>C, p.Met34Thr | 49 | 3.6% |
| USH2A | c.2299delG, p.Glu767fs | 38 | 2.8% |
| GJB2 | c.167delT, p.Leu56fs | 17 | 1.2% |
| GJB2 | c.−23+1G>A | 13 | 1.0% |
| GJB2 | c.269T>C, p.Leu90Pro | 13 | 1.0% |
| GJB2 | c.313_326del, p.Lys105fs | 11 | 0.8% |
| GJB2 | GJB6-deletion upstream | 10 | 0.7% |
| LMM total | 588 | 43.0% |
Ten variants most commonly identified as causes of deafness in two clinical testing laboratories: Molecular Otolaryngology & Renal Research Labs (MORL, Iowa City, IA) and the Laboratory for Molecular Medicine (LMM, Cambridge, MA). MORL data are from 2460 individuals from the US population with deafness who presented for diagnostic comprehensive genetic testing with the OtoSCOPE platform. The total number of diagnoses provided is 982, or 39.9% of 2460. All ethnicities are included and no exclusions were made based on type of deafness (R.J.S., unpublished data). LMM data are from 737 diagnosed cases mainly from the US population with deafness who presented for diagnostic genetic testing at the LMM. GJB2-related deafness was overrepresented because 104 cases were only tested for GJB2 including deletions of 5’ upstream regulatory regions involving GJB6. Data from MORL are adapted from Sloan-Heggen et al. (2016);13 data from LMM are unpublished (contributed by S.A. and J.S.).
CNV copy-number variant.
Importantly, comprehensive genetic testing for deafness also includes testing for NSHL mimics. These diagnoses comprise up to 25% of all genetic diagnoses in children undergoing genetic testing for deafness (R.J.S., unpublished data). Identification of these syndromic forms of deafness in newborns directly affects follow-up testing and care and reduces time to diagnosis. In addition, comprehensive genetic testing evaluates for mitochondrial variants that predispose to aminoglycoside-induced deafness and therefore can aid in prevention of this medication-related side effect.
Comprehensive genetic testing has several strengths that make it particularly valuable for the evaluation of newborns with deafness: (1) it provides an etiological diagnosis, (2) it tests for all severities of deafness (mild–profound), (3) it tests for syndromic forms of deafness, and (4) it can assess for risk of aminoglycoside-induced deafness. Therefore, incorporating some form of genetic testing as an additional screening component in the NBHS would (1) identify additional newborns and young children with deafness and at risk for deafness, (2) improve time to diagnosis and intervention, (3) identify comorbidities of congenital deafness that have important implications for medical management of children who are DHH, and (4) possibly reduce those lost to follow up.
Currently available comprehensive genetic tests used in the diagnostic evaluation of deafness are not suitable for the NBHS because of (1) high cost, (2) long turnaround time (weeks to months), (3) complexity of bioinformatics (including the identification of CNVs), and (4) challenges in variant interpretation (Table 4). Notwithstanding these challenges, genetic screening would add opportunities to improve detection of deafness thus justifying strategies to integrate its use into the NBHS. Three strategies—limited genetic evaluation for screening, comprehensive genetic sequencing, and genome evaluation—are discussed below.
Table 4.
Challenges to the integration of a genetic screening into the universal newborn hearing screen
| Barrier | Impact | Proposed immediate step | Proposed long-term strategy |
|---|---|---|---|
| Expense | Prohibits incorporation into population-based screening | Implement a small screening panel capturing a limited number of targeted variants | More cost-effective sequencing technology; validation of targeted genetic panels in the US population |
| Variant interpretation | Requirement for expert variant interpretation prohibits population-based screening | Include only known pathogenic variants with automated variant interpretation | Improved understanding of variant effect on hearing and deafness through algorithmic approaches |
| Expressivity/penetrance | Limited understanding of permanent impact of some genetic variants can preclude confident interpretation of results | Include only variants with well-understood expressivity and penetrance | Continued evaluation of long-term consequence of genetic variants on hearing |
| Secondary and incidental findings | Identified genetic variants may cause other diseases and raise ethical dilemmas and uncertainty in interpretation | Limit to pathogenic variants known to cause deafness | Establish guidelines for handling incidental and secondary genetic variants in universal screening tests for deafness |
Limited genetic screening for NBHS
Implementation of limited genetic screening requires the selection of specific genes and/or variants, a challenging task given the extreme genetic heterogeneity of deafness. For example, in a cohort of 1119 patients with deafness, the gene most frequently implicated in deafness was GJB2.23 It accounted for 21.6% of diagnoses. In aggregate, however, ten genes accounted for 72.3% of all diagnoses (Table 2). At the variant level, in comparison, the ten most frequently encountered causative variants accounted for only 30.4% of diagnoses. The most commonly identified single causative variant in this cohort was a deletion involving STRC, which accounted for 11.8% of causative variants in individuals who were identified to have genetic hearing loss and can only be detected by CNV analysis. This type of genetic change cannot be identified using a nucleotide variant detection assay and would require a secondary screening assay.48
Data from 2460 individuals with deafness from a clinical genetic testing lab show the diminishing returns associated with sequencing an ever-higher number of genes (R.J.S., unpublished data, Figure S1). In this group of 2460 individuals, 79 different genes were identified as causative, with 34 genes accounting for only one or two diagnoses each. Therefore, there appear to be several genes that could be selected for targeted genetic screening, but there are diminishing returns as more genes are added (Figure S1). A limited panel of genes may provide an avenue for developing genetic NBHS. However, as described below, the logistics of this type of screening would prove challenging.
Several groups have studied the value of limited panels of genes or variants to augment NBHS in large cohorts. In a recent study of 5173 Chinese newborns, screening of four genetic variants resulted in identification of 46 newborns who passed the physiologic NBHS but had a genetic cause of deafness identified and therefore were not detected by physiologic NBHS.49 This study is limited in that long-term follow up to evaluate the degree of deafness in these children was not provided and so the exact type of deafness caused by these genetic variants is not known. In addition, one of the variants evaluated, GJB2 p.V37I, while very common particularly in Asian populations, has variable penetrance and causes mild-to-moderate hearing loss, making definitive interpretation of screening results difficult. A larger study of 58,397 Chinese newborns who were screened for 20 common genetic variants identified a genetic carrier result in 5.28% and the identification of five newborns with a positive genetic diagnosis of deafness who passed their NBHS in both ears.35 Another recent study that included concurrent genetic and physiologic screening in 1716 Chinese newborns found that 47% of those with a definitive GJB2 genetic diagnosis passed their physiologic screening.50 In contrast, three similar studies of 14,913, 10,043, and 2500 newborns that used a combination of sequencing approaches to detect common variants found no newborns who passed their physiologic NBHS but had a positive genetic diagnosis of deafness.51–53 The largest study to date of 142,417 Chinese newborns identified a carrier rate of 3.01% for the four most common genetic causes of deafness but did not include audiologic data to determine how genetic screening could augment physiologic screening.54 This variability in detection rates might reflect differences in sample sizes, variants screened, and screening methodologies; however, the data do indicate that even limited genetic screening can identify newborns who otherwise would have been missed by physiologic screening alone.
Limited genetic screens have inherent ethnic bias, which is an important drawback in a racially and ethnically diverse population like that of the United States. Table S3 shows the top 20 most commonly encountered pathogenic variants in GJB2, illustrating the significant differences among populations with respect to the most commonly encountered pathogenic variants. Note that in these six populations, there are 15 different variants that are rarely shared between populations. Of the top five variants, only one is shared among the East Asian, Latino, European, and African populations. These data underscore the challenge in implementing limited genetic variant screening in an ethnically diverse population. Screening for deafness genes and not variants could circumvent some of this ethnic bias because many genetic variants are ethnically specific. However, the candidate genes or variants for screening would need to be thoughtfully selected to provide the best screening for a given population.
A limited genetic variant screening tool would ideally use only a single methodology. Polymerase chain reaction followed by restriction fragment length polymorphism (PCR-RFLP) analysis, single-nucleotide polymorphism (SNP) microarray, and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) have all been used for genetic screening, each with its own advantages and disadvantages. New methods of screening for deafness variants continue to be developed. A recent study using a custom PCR-based, multiplex ligation assay to screen for 115 variants cost $25 per sample.55 Another recently developed method using real-time PCR and melting curve analysis to detect 12 variants costs less than $10 (ref. 56). A third method that relies on multiplex PCR followed by a “reverse dot blot assay” can screen for 20 variants at a reported cost of $3 per sample.57 Finally, a fluidic microarray-based screening for nine common variants for deafness in patients of European ancestry has been validated and costs $30 per sample.58 All methods would need to be compared with gold standard genetic diagnostics prior to implementation as part of a genetic NBHS. In addition, the variants used for screening would need to be carefully selected based on goals of the screening and the population to be screened.
These data can be summarized as follows: screening for a handful of deafness-causing genetic variants would be cost effective (theoretically ~$10–30 per sample) but is hampered by inherent ethnic bias, which is critical to consider when screening an ethnically diverse population. A genetic screen limited to the ten most common variants causing deafness in the US population would identify less than one-third of those with genetic hearing loss (30.5–40%, Table 3). There are several methods by which this screening rate could be improved: (1) variant screening panels based on ethnicity, (2) variant screening panels based on phenotype (i.e., severe-to-profound versus mild-to-moderate hearing loss), or (3) variant screening at the gene level (i.e., sequencing of a select number of genes). Research is required to determine which of these methods would be most successful, but gene-based genetic screening is discussed in the next section.
Comprehensive genetic screening for NBHS
Targeted genomic enrichment followed by massively parallel sequencing has formed the cornerstone of diagnostic comprehensive genetic testing in the clinical evaluation of deafness since its development in 2010.42 This method allows for isolation of targeted genomic regions, typically coding and splicing regions, which are subjected to sequencing followed by downstream bioinformatics analysis. The method can be scaled from a handful of genes to include every gene in the human genome (exome sequencing). On a clinical basis, these tests typical cost $1500–5000 per test (current pricing available at the Genetic Testing Registry, https://www.ncbi.nlm.nih.gov/gtr/). The primary costs are the initial sample processing, which includes DNA extraction, targeted enrichment, sequencing, bioinformatics, and interpretation of results. The greatest costs, particularly when scaled to the population level, are clinical interpretation of results. As the number of genes increases, the cost for targeted genomic enrichment essentially remains fixed. Therefore, the cost difference between screening 10 genes and 100 genes (the approximate number of genes implicated in nonsyndromic deafness), particularly on a large number of individuals, becomes negligible. As a limited screening, it is tempting to design a panel to sequence the ten genes most frequently involved in deafness (Figure S1, Table 2), however costs are currently prohibitive with today’s technology to incorporate this option into a universal screen. Even when the technological costs are reduced, interpretation of results would be burdensome because this step currently requires expert analysis. An automated pipeline would be needed to scale genetic testing to universal screening. In addition, as the number of genes increases so too does the number of incidental findings and the number of variants of uncertain significance.
Genome sequencing for NBHS
An attractive approach, which is currently an active area of research, is genome sequencing (GS) with automated variant analysis for genetic NBHS.59 The Human Genome Project was completed in 2003 and since that milestone, with the rapid advances in sequencing technology, a genome can be sequenced in <48 hours at a cost of less than $1000. GS requires fewer steps for sample processing (targeted genomic enrichment is not required) and variant analysis, including CNV and structural variant analysis, can be included. The diagnosis of cCMV would also be feasible as the viral DNA of CMV is coisolated with human genomic DNA from peripheral blood and its sequencing would allow for diagnosis. Because GS is essential to the diagnosis of some disorders, it is currently used for diagnosis of acutely ill children in the neonatal intensive care unit (preprint available at https://www.biorxiv.org/content/10.1101/253534v1.abstract). GS is equally useful in screening for metabolic disorders and other severe genetic diseases, and as the cost of sequencing is further decreasing more widespread GS is likely to become a reality. Performing GS on newborns would have broad implication for their medical care, as specific genes associated with diseases of interest could be examined as the need arose without the requirement for repeated sequencing. When implemented, focused analysis of known deafness genes could become routine and would be integrated into the interpretation of results generated by the physiologic NBHS.
Technical barriers to implementation of any genetic screening method
Regardless of the screening method, there are important features of genetic NBHS that must be addressed prior to its integration into NBHS. Challenges include (1) variant interpretation, (2) incidental findings, and (3) integration of the physiologic NBHS with the genotypic data. Results from two different simultaneous screening tests—one physiologic and one genetic—would be available concurrently under ideal conditions.
Variant interpretation is essential to genetic testing. There are 876,135 genetic variants found within the 152 known nonsyndromic and syndromic deafness genes, of which 7474 have been classified as pathogenic, implying a causal association with deafness (http://deafnessvariationdatabase.org). Labeling a variant as pathogenic is complex. Interpretation of variant effect relies on data from functional experiments, familial information including segregation of the variant with deafness, computer algorithms and variant frequency data from large population databases, as well as an understanding of the molecular physiology of the gene and protein of interest. Genetic variants are classified as pathogenic (P), likely pathogenic (LP), benign (B), likely benign (LB), or a variant of uncertain significance (VUS). Typically, when comprehensive genetic testing is ordered as a diagnostic test, variant interpretation is considered in the context of the clinical data, including the audiogram and family history, and ideally interpreted by a multidisciplinary panel of experts. This degree of clinical data is typically not available as part of a screening test, although the value of both the physiologic NBHS and the genetic NBHS would be enhanced if the interpretation of these two data sets were integrated to generate a consensus combined report.
An example of integration of the genetic and physiologic NBHSs is shown in Table 5. The genetic NBHS includes three possible results: “negative” (no VUS, LP, or P variants identified), “refer” (or positive, implying the identification of known LP/P variants that predict a deafness phenotype), or “uncertain” (the identification of VUS or a known LP/P variant not predicted to be causative of deafness, i.e., heterozygous for a single P/LP variant implicated in autosomal recessive deafness) (Table 5). Newborns who are referred or have an uncertain result on their genetic screen may require diagnostic genetic testing and genetic counseling of family members. This burden on genetic counselors and geneticists needs to be considered prior to integration of any genetic screen, particularly if a large number of genes are screened, as the number of VUS is directly proportional to the number of genes included in the screen. Newborns who pass their physiologic NBHS and refer on genetic NBHS (conflicting screening results) would undergo diagnostic audiology and diagnostic genetic testing. The goal would be to use genetic screening programs to complement physiologic screening, thereby improving early diagnostic precision and early intervention. This extra screening and diagnostic testing could be incorporated into the current 1-3-6 recommended screening timeline (Table 6).
Table 5.
Coordination and recommended evaluation based on a combined physiologic and genetic NBHS with a limited genetic screening panel
| Genetic newborn hearing screening result | ||||
|---|---|---|---|---|
| Negative (B/LB variants) | Uncertain (VUS; single P/LP variant not predicting a deafness phenotype, i.e., associated with AR deafness) | Positive (P/LP variants predicting deafness phenotype) | ||
| Physiologic newborn hearing screening result | Negative (Pass) | - Routine hearing surveillanceb |
- Routine hearing surveillance - Genetic counseling if indicated by other symptoms or concerns |
- Diagnostic audiometry - Long-term follow up - Genetic counseling |
| Positivea (Fail/did not pass) |
- Diagnostic audiometry - Further diagnostic deafness evaluation |
- Diagnostic audiometry - Further diagnostic deafness evaluation - Genetic counseling |
- Diagnostic audiometry - Long-term follow up - Genetic counseling |
|
Screening tests are administered concurrently.
AR autosomal recessive, B/LB benign/likely benign, NBHS newborn hearing screening, P/LP pathogenic/likely pathogenic, VUS variant of uncertain significance.
aBased on available data, we recommend congenital cytomegalovirus (cCMV) testing for all newborns who refer on physiologic NBHS.
bRoutine hearing surveillance based on Joint Committee on Infant Hearing (JCIH) guidelines.
Table 6.
Proposed timelines for NBHS that integrates physiologic, genetic and CMV screening, confirmatory testing, and habilitation/intervention
| Stage | Time frame | Audiologic evaluation | Genetic evaluation | cCMV evaluation | Clinical evaluation |
|---|---|---|---|---|---|
| Screening | Prior to discharge from hospital | Physiologic hearing screening | Heel-stick blood sample collected, processed, screening begun | Saliva or urine sample collected for CMV testing | Clinical exam by newborn provider |
| Confirmation | <1 month of age | Refer with positive physiologic screen results (did not pass) | Genetic results returned (at 1–2 months) | CMV confirmatory testing | Routine examination by primary care provider |
| Diagnosis | <3 months of age | Diagnostic audiometric testing and evaluation results | Genetic counseling with further genetic testing if indicated | Consider treatment with antivirals if indicated | Examination by otolaryngologist and evaluation/fitting for assistive devices; referral to clinical geneticists and specialists as indicated by genetic testing |
| Habilitation/intervention | <6 months of age | Repeat audiometry as indicated | Evaluate outcomes of antiviral treatment |
Confirm placement of assistive devices Enroll in early intervention programs |
CMV cytomegalovirus, NBHS newborn hearing screening.
Incidental findings are routinely encountered during any genetic test and represent findings outside of the intended diagnostic scope.60 In contrast, secondary findings are results that are intentionally sought outside the purpose of the test. Examples of incidental findings include genetic risk factors for cardiac disease and carrier status for diseases not related to deafness. Secondary findings are typically sought based on recommendation of an expert panel. The wider the scope of the genetic test, the greater is the risk for incidental findings. For example, a genetic screen of 20 hand-selected genetic variants will have few, if any, incidental findings. The number of incidental findings increases if a whole gene or several genes are sequenced, as genetic disorders other than deafness may be associated with the same genes. Exome and genome sequencing will inherently uncover incidental findings. The risk of incidental findings should be offered to be disclosed to any individual undergoing genetic testing or screening. The American College of Medical Genetics and Genomics (ACMG) has released guidelines on reporting of incidental and secondary findings to guide clinicians.61 Any genetic screening method should therefore include planning for offering the reporting of incidental and secondary findings and disclosure of these findings to those being screened prior to testing.
Genetic screening conclusions: an interim step
If comprehensive NBHS with integrated genetic screening is to be implemented in the near future, then compromises must be made regarding the variants or genes to be screened while taking into context the population to be screened, costs of screening, and difficulty with interpretation of results. While it is likely that in the next several years, with decreasing costs and improved automatic analysis methods, genome sequencing will become more routine, at this point in time variant-based screening panels provide the best interim step toward comprehensive genetic NBHS. Regardless of the genetic screening method chosen, an investment in training laboratory personnel will be required to allow accurate interpretation of identified genetic variants.
Multiple variant-based screening panels would provide a method of implementing limited genetic screening for deafness that is inexpensive and easily interpretable. However, platforms must be thoughtfully designed based on specific goals and the target population. For example, to augment the established physiologic NBHS, a platform could target the most commonly implicated pathogenic variants causing deafness in a given population, or alternatively be tailored to specific degrees of deafness or genes/variants (i.e., a platform detecting pathogenic variants that cause mild deafness versus a platform detecting pathogenic variants that cause Usher syndrome).62 Based on technology available, associated costs, and goals of NBHS, genetic screening of a handful of deafness genes could also be performed.63 Designers of these screens should be cognizant of the increased analysis and interpretation burden of incidental and secondary findings, as well as variants of uncertain significance identified as whole-gene screening is added.
In an effort to centralize the knowledge base of genes and variants relevant to syndromic and nonsyndromic hearing loss and standardize their interpretations in the clinical genetic testing community, the ClinGen Hearing Loss Clinical Domain Working Group (HLWG) was launched in 2016 as part of the Clinical Genome Resource (ClinGen, http://www.clinicalgenome.org). The HLWG represents an international consortium of hearing loss experts with a diverse array of backgrounds, including physicians (clinical geneticists, otolaryngologists, neuro-otologists), clinical laboratory diagnosticians, clinical and basic research scientists, and genetic counselors. The aims of the HLWG are (1) to curate genes associated with nonsyndromic and syndromic deafness and determine their clinical validity by evaluating the strength of evidence supporting or refuting causality for each gene, and (2) to standardize the interpretation and classification of variants in hearing loss genes by adapting existing ACMG/Association for Molecular Pathology (AMP) guidelines to account for current knowledge of the genotypic and phenotypic heterogeneity unique to hearing loss. An early release of the hearing loss–specific modification of the ACMG/AMP guidelines developed by the HLWG has been made publicly available (see https://www.biorxiv.org/content/early/2018/05/08/313734).
These efforts can inform which genes and/or variants are most clinically relevant for inclusion on an NBHS screening panel. For example, evidence-based gene curations performed by the HLWG revealed that many genes with a published hearing loss association have “limited” clinical validity. Detection of novel variants in genes with “limited” evidence supporting an association to hearing loss would likely yield an uncertain significance classification, adding unnecessary ambiguity to a NBHS test result. Supplemental Table 4 lists the clinical validity classifications for 91 genes commonly found in diagnostic hearing loss panels, 10 of which have a “limited” classification and 1 of which has conflicting evidence disputing its association with hearing loss. Further curation of causative deafness variants by expert panels, including ClinGen as well as the Deafness Variation Database (DVD, http://deafnesssvariationdatabase.org) will assist with reduction in variant interpretation burden for any genetic NBHS. Importantly, these resources are free and unrestricted to medical and lab professionals, as well as to deaf individuals, their families, or expecting parents, providing an educational tool to clarify hearing loss genetic etiologies and promote the utility of genetic testing and its incorporation into current NBHS strategies.
Any genetic screen would require clinical validation prior to implementation, and would have limitations as well as specific strengths and weaknesses that must be clearly understood by the medical and lay communities. Importantly, a primary weakness of these limited genetic platforms is that they cannot reliably detect CNVs, and as mentioned a CNV in STRC is the first or second most commonly diagnosed single causative variant in the United States (Table 3). While individual assays could be developed and performed in parallel to address this problem, that requires adding an additional layer of complexity to the screen.64
Addition of a genetic screen to the NBHS would provide valuable etiologic information to parents and providers, and would likely reduce time to diagnosis, intervention, and habilitation, although pilot studies are needed to address these challenges.
Screening for cCMV-related deafness
The leading nongenetic cause of deafness at birth is congenital cytomegalovirus infection (cCMV), which is estimated to underlie up to 10% of all congenital deafness and 15–20% of all childhood deafness.14,15 Of newborns who fail physiologic NBHS, ~6.0% will test positive for cCMV.65,66
cCMV infection occurs in 0.64% (~1 in 200) of all live births in the United States and can lead to permanent disability, including cognitive impairment, cerebral palsy, developmental delay, and hearing and vision loss.67 Risk for cCMV infection is 32% following a primary maternal infection during pregnancy, however in CMV-positive mothers, the risk of maternal-to-fetal transmission is much lower and is estimated at only ~1.4 % (ref. 67). About 10% of cCMV-infected babies are obviously symptomatic at birth with signs and symptoms of infection that include intrauterine growth restriction, microcephaly, and jaundice (symptomatic infection); SNHL is present in approximately 30% of this cohort. The remainder of babies born with cCMV display no obvious outward signs of infection and are classified as having asymptomatic CMV. Approximately 14% of these babies develop SNHL by 5 years old, with 25% developing SNHL by age 18; however, the risk of developing SNHL beyond 5 years old is not statistically different between cCMV-positive and cCMV-negative groups.16
The defining characteristic of deafness due to cCMV is its variability. In children born with asymptomatic cCMV, it is most frequently mild, unilateral, fluctuating, and progressive. In children born with symptomatic cCMV, the deafness is more likely to be bilateral and moderate to severe/profound in degree but is still frequently progressive and fluctuating. Presumably, it is for these reasons that in a recent study of 99,945 infants screened for cCMV, 43% of infants with cCMV and deafness at birth were not identified by NBHS.68
As a step toward better cCMV detection, selected hospitals with birthing centers, as well as several states, now implement targeted cCMV screening programs for infants who refer on NBHS.65 Although these programs have been successful, they are not universal and only identify newborns who refer on NBHS and are then tested for cCMV. One recent study of 10,964 newborns found that in a targeted saliva-based cCMV screen of the 171 newborns who failed physiologic screening, only 3 screened positive for cCMV.68 Another recent study performed universal cCMV screening in 1716 newborns using quantitative real-time PCR on dried blood spots, and only detected 3 positive cases who all passed physiologic NBHS. This result confirms the low sensitivity of the screening method using dried blood spots. Saliva-based tests are more sensitive but require additional sample collection and processing, thus increasing the screening cost. Further research is required to evaluate the relative merits of universal versus targeted testing for cCMV.
Antiviral therapy has been successful in improving or halting SNHL in symptomatic children with cCMV.69,70 A recent retrospective review also has shown that antiviral treatment in children with asymptomatic cCMV and hearing loss can prevent progression of deafness; however, the study is limited by its retrospective nature and lack of long-term follow up.71 Prospective clinical trials to determine whether antiviral therapy benefits children with asymptomatic cCMV are ongoing.67,72
cCMV detection can be incorporated into the NBHS by providing either universal screening or targeted screening after a failed NBHS. While targeted screening for cCMV is less costly, as noted earlier a significant number of cCMV-positive children are not identified. Universal screening for cCMV would identify these newborns and as such would be a valuable addition to a comprehensive NBHS. In particular, the prospect of identifying and treating hearing loss in children with asymptomatic cCMV makes screening for cCMV of added importance.
PATIENT AND FAMILY PERSPECTIVES
Implementation of genetic NBHS as a component of physiologic NBHS must be culturally sensitive. Parents of deaf children and members of the Deaf community have expressed a desire to have their opinions represented in the development of public policies governing genetic NBHS; given their unique perspective, they may offer valuable insight on strategies to enhance mainstream acceptance of these tests. Health-care providers interacting with individuals with deafness should remain cognizant and sensitive to the importance of Deaf culture. This culture is comprised of unique social and societal attributes. Members of the Deaf community (i.e., the Deaf) do not consider themselves to be hearing “impaired” nor do they feel that they have a hearing “loss.” Their deafness is not considered “pathology” or a “disease” to be treated or cured. For a Deaf person, Deaf is a state of being that reflects completeness, linguistic differences (the use of signed languages), and the living of full rewarding lives with no need to compensate for being “incomplete.” The phenomenon of the Deaf community, more recently termed Deaf World, has existed for over two centuries.73,74 Initially, the shared sense of bonding of the Deaf community focused on whether people were deaf or could hear. Over time, the broadening of the concept of Deaf community incorporated users of signed languages, hard-of-hearing and oral deaf people, and hearing people who shared the common goals of communication access, linguistic differences, and respect for individual needs.75
Extending a family-centered approach to genetic NBHS will require access to genetic professionals and resources. Pretesting and post-testing counseling must convey the utility and benefit of genetic testing for deafness as a part of any NBHS strategy, addressing potential outcomes and implications of such testing, as well as its limitations.12 To provide meaningful counseling and support, an understanding of the cultural, linguistic, and psychosocial needs of parents and families is essential, particularly for families who identify with the Deaf community. Information on resources and support needs to be readily available when meeting with families.
Several studies have assessed attitudes toward genetic testing by parents of children with deafness and by adults who are deaf or hard-of-hearing. One study reported that roughly three-fourths of participants show interest in genetic testing, for both parents identifying with hearing communities as well as parents and/or adults identifying with the Deaf community.75 However, persons identifying with the Deaf community tend to have less positive attitudes toward genetic testing, which may be mitigated by providing culturally sensitive counseling.76,77
In a small survey of 30 parents of deaf children regarding the decision to pursue genetic testing, those electing to do so indicated that the knowledge and support of the pediatrician was a major factor.78 Another survey of parental-perceived benefits of genetic testing following pre- and post-test counseling reported that following pretest counseling, all parents perceived benefits; however, this perception continued for parents receiving positive genetic results and declined for parents receiving inconclusive or negative results.79 These studies highlight the role of the clinical team in influencing the decision to pursue genetic testing and in setting expectations on potential outcomes. Inconclusive and negative results add a level of uncertainty and confusion to the diagnostic process that can exacerbate anxiety and frustration for families.
Genetic testing can have social and ethical implications that families must recognize.77 A major concern is the privacy and confidentiality of genetic testing results and the social and medical implications of any positive findings. Pretest counseling should include information on legislation and policies to protect individuals from discrimination by employers or health insurers based on genetic results. By using a family-centered and culturally sensitive approach, clinicians can provide excellent care of those with deafness and children who are deaf or hard-of-hearing as they navigate screening and testing.
CONCLUSIONS AND FUTURE DIRECTIONS
NBHS has had a dramatic impact on children with deafness. Data from the past 20 years show that early identification and intervention reduce differences in development and academic achievement between children with deafness and their hearing peers. In support of a 1-3-6 screening/diagnosis/intervention model, it has been shown that children identified by 6 months demonstrate significantly better language scores than those identified later.1,2 When universal NBHS is compared with a risk factor–based screening program, three quasi-randomized controlled trials have shown that outcomes are better for children who undergo universal NBHS because this cohort is identified and treated sooner than are children with deafness identified by risk factor criteria.80,81
Given advances in our understanding of the genetics of deafness and the technology to integrate genomic medicine into health care, it is time to assess the existing NBHS model to determine the role of genetic screening. Genetic screening can improve the physiologic NBHS by increasing the number of children with deafness who are identified, reducing the time to intervention, and providing etiologic information including risk for syndromic and aminoglycoside-sensitive deafness. We also recommend incorporating cCMV screening due to convincing data showing feasibility as well as clear benefits to newborns with deafness and their families. A clearer view then emerges of a comprehensive NBHS that incorporates physiologic, genetic, and cCMV screening. At least one study has demonstrated the feasibility of this method with improved detection of newborns at risk for deafness.50
A comprehensive NBHS that incorporates a carefully designed multiple-variant genetic screening panel coupled with targeted cCMV screening would be easiest and most cost effective to implement today (Box 2 and 3). With further advances in more broad-based genetic medicine and GS as a cornerstone of individualized precision medicine, comprehensive genetic NBHS and universal cCMV screening could complement physiologic NBHS. We identified several important research questions (Box 3) and resources (Box 4) that should guide further inquiry in to this important topic. A key step in the implementation of molecular testing in the NBHS will be input from stakeholders including parents and DHH individuals themselves. In our opinion, comprehensive NBHS would positively impact the diagnosis and early intervention for deafness in all children in the United States, improve our understanding of childhood deafness, and lay an important and critical foundation for future molecular therapies for deafness.
Box 1 Definitions.
Auditory brainstem response (ABR): an electrical potential measured in response to auditory stimulus using electrodes placed on the scalp
Conductive hearing loss (CHL): hearing loss due to inability or inefficiency of sound transmission through the external ear canal, across the tympanic membrane and through the ossicles to the inner ear
Massively parallel sequencing: sequencing of DNA using techniques that allow simultaneous (parallel) analysis of millions of fragments of DNA and allow for rapid sequencing of large portions of the human genome (also known as next-generation sequencing)
Nonsyndromic deafness mimics: syndromic forms of deafness that present at birth as nonsyndromic deafness (no other associated abnormalities) and therefore may initially clinically appear to be isolated hearing loss; examples include Usher syndrome and Jervell and Lange-Nielsen syndrome
Nonsyndromic hearing loss (NSHL): hearing loss that is isolated an not associated with other clinical features
Otoacoustic emission (OAE): a measurement of the response of outer hair cells to sound
Pathogenic variant: a genetic variant with confirmed evidence relating it to a human genetic disease or disorder
Sensorineural hearing loss (SNHL): hearing loss due to damage or malfunctioning of the sensory portion of the inner ear, organ of Corti, or auditory nerve
Variant: a difference in the human genetic code from the reference sequence that may be inherited and may cause disease, put an individual at risk for disease, or may have no effect on health
Variant of uncertain significance (VUS): a genetic variant with an unclear relation to a human genetic disease or disorder
Box 2 Key points.
Universal newborn hearing screening using physiologic testing of auditory function has dramatically improved early identification and intervention for infants with permanent deafness.
Nevertheless, physiologic newborn hearing screening as currently implemented has limitations that include (1) a low positive predictive value; (2) a high number of children lost to follow up; (3) inability to detect those types of deafness that are not screened for including many cases of mild deafness, auditory neuropathy spectrum disorder, and children with onset of deafness outside the immediate newborn period; and (4) limited etiologic information.
Comprehensive genetic testing for deafness using massively parallel sequencing has become the most valuable etiological diagnostic test in the evaluation of children with deafness but is not currently suitable for universal screening due to cost and complexity.
Incorporating some form of genetic testing into universal newborn hearing screen would (1) identify additional newborns with deafness, (2) improve time to diagnosis and intervention, (3) identify comorbidities of congenital deafness that have important implications for medical management, and (4) possibly reduce newborns lost to follow up.
Genetic screening could be incorporated into the current newborn hearing screen by using a limited genetic panel and could be further integrated with testing for cCMV to improve the current NBHS.
Familial experiences and expectations with how and when newborn hearing screening and testing are offered are variable.
In the future genome sequencing will be routine and targeted analysis of all genes implicated in deafness will be available as the basis for molecular newborn hearing screening.
Box 3 Identified research questions.
What is the true false negative rate for the physiologic NBHS (i.e., which children with deafness are missed by physiologic NBHS)?
What method would best improve the positive predictive value of the physiologic NBHS?
What is the optimal method of population-wide genetic screening for deafness?
What are the relative merits of universal versus targeted cCMV screening in the newborn population?
What is the best method for screening for cCMV—urine, saliva, cord blood PCR versus culture?
What is the prevalence of hearing loss in preschool-age children?
What is the prevalence of mild (slight) hearing loss in newborns?
What are the clinical and ethical repercussions of early diagnosis of syndromic hearing loss (nonsyndromic hearing loss mimics)?
Box 4 Identified resources.
Web resources
ClinGen: http://www.clinicalgenome.org
Deafness Variation Database: http://deafnessvariationdatabase.org
Genetic Testing Registry: https://www.ncbi.nlm.nih.gov/gtr/
Hereditary Hearing Loss Homepage: http://hereditaryhearingloss.org
OMIM: https://omim.org/
Supplementary information
Acknowledgements
This study was coordinated by the Newborn Hearing Screening Working Group of the National Coordinating Center for the Regional Genetics Networks. In addition to the authors of the manuscript, the working group included the following members who all contributed to conceptualization, critical commentary, and review of the final manuscript: Ahmad Abou Tayoun, PhD, FACMG, Division of Genomic Diagnostics, Children’s Hospital of Philadelphia, Philadelphia, PA; Kathleen Arnos, PhD, Department of Science, Technology and Math, Gallaudet University, Washington, DC; Gail Demmler-Harrison, MD, Department of Pediatrics and Pathology & Immunology, Baylor College of Medicine, Houston, TX; Terese Finitzo, PhD, F-ASHA, FAAA, Oz Systems, Arlington, TX; David Flannery, MD, American College of Medical Genetics and Genomics, Bethesda, MD; Aaron Goldenberg, PhD, MPH, Department of Bioethics, Case Western Reserve University, Cleveland, OH; Cathy Harbison, RN, Missouri Newborn Hearing Screening Program, Missouri Department of Health and Senior Services, Jefferson City, MO, Nannette Nicholson, PhD, CCC-A, Department of Audiology and Speech Pathology, University of Arkansas for Medical Sciences, Little Rock, AR; Teresa Nold, South Dakota Parent Connection, Sioux Falls, SD; Arti Pandya, MD, MBA, Division of Genetics and Metabolism, Department of Pediatrics, University of North Carolina Chapel Hill, Chapel Hill, NC; Nathaniel Robin, MD, Department of Genetics, University of Alabama at Birmingham School of Health Professionals, Birmingham, AL; Michael Watson, PhD, MS, FACMG, American College of Medical Genetics and Genomics, Bethesda, MD; Karl White, PhD, National Center for Hearing Assessment and Management, Utah State University, Logan, UT; Lindsey Woodard, Arkansas Hands and Voices, Conway, AR. This project was supported by the Health Resources and Services Administration (HRSA) of the US Department of Health and Human Services (HHS) under #U22MC24100 (National Coordinating Center for the Regional Genetic Services Collaboratives) for $100,000. This information or content and conclusions are those of the author and should not be construed as the official position or policy of, nor should any endorsements be inferred by HRSA, HHS, or the US Government. Other funding received: NIDCD R01s DC003544, DC002842, and DC012049 to R.J.S.; NIDCD R01DC015052 to C.C.M.
Disclosure
The authors declare no conflicts of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/18/2019
In the original version of this Article, several individuals were erroneously acknowledged in the acknowledgements. The Acknowledgement section in the PDF and HTML versions of the Article has have now been corrected.
Change history
6/18/2019
In the original version of this Article, several individuals were erroneously acknowledged in the acknowledgements, they have been removed. The Acknowledgement section in the PDF and HTML versions of the Article has now been corrected to the following:
Contributor Information
Cynthia C. Morton, Email: cmorton@bwh.harvard.edu.
Richard J. Smith, Email: richard-smith@uiowa.edu.
Supplementary information
The online version of this article (10.1038/s41436-019-0563-5) contains supplementary material, which is available to authorized users.
References
- 1.Yoshinaga-Itano C, Sedey AL, Coulter DK, Mehl AL. Language of early- and later-identified children with hearing loss. Pediatrics. 1998;102:1161–1171. doi: 10.1542/peds.102.5.1161. [DOI] [PubMed] [Google Scholar]
- 2.Yoshinaga-Itano C, Sedey AL, Wiggin M, Chung W. Early hearing detection and vocabulary of children with hearing loss. Pediatrics. 2017;140:e20162964. doi: 10.1542/peds.2016-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korver AMH, Konings S, Dekker FW, et al. Newborn hearing screening vs later hearing screening and developmental outcomes in children with permanent childhood hearing impairment. JAMA. 2010;304:1701–1708. doi: 10.1001/jama.2010.1501. [DOI] [PubMed] [Google Scholar]
- 4.Ching TYC, Dillon H, Button L, et al. Age at intervention for permanent hearing loss and 5-Year language outcomes. Pediatrics. 2017;140:e20164274. [DOI] [PMC free article] [PubMed]
- 5.Joint Committee on Infant Hearing. 1994 Position statement. http://www.jcih.org/JCIH1994.pdf. [PubMed]
- 6.White KR. The current status of EHDI programs in the United States. Ment Retard Dev Disabil Res Rev. 2003;9:79–88. doi: 10.1002/mrdd.10063. [DOI] [PubMed] [Google Scholar]
- 7.Joint Committee on Infant Hearing. Year 2007 position statement: principles and guidelines for early hearing detection and intervention programs. Pediatrics. 2007;120:898–921. doi: 10.1542/peds.2007-2333. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. Hearing loss in children. 2 March 2018. https://www.cdc.gov/ncbddd/hearingloss/ehdi-data2015.html. Accessed 18 May 2018.
- 9.White KR, Forsman I, Eichwald J, Munoz K. The evolution of early hearing detection and intervention programs in the United States. Semin Perinatol. 2010;34:170–179. doi: 10.1053/j.semperi.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Shearer AE, Smith RJH. Massively parallel sequencing for genetic diagnosis of hearing loss: the new standard of care. Otolaryngol Head Neck Surg. 2015;153:175–182. doi: 10.1177/0194599815591156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liming BJ, Carter J, Cheng A, et al. International Pediatric Otolaryngology Group (IPOG) consensus recommendations: hearing loss in the pediatric patient. Int J Pediatr Otorhinolaryngol. 2016;90(C):251–258. doi: 10.1016/j.ijporl.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 12.Alford RL, Arnos KS, Fox M, et al. American College of Medical Genetics and Genomics guideline for the clinical evaluation and etiologic diagnosis of hearing loss. Genet Med. 2014;16:347–355. doi: 10.1038/gim.2014.2. [DOI] [PubMed] [Google Scholar]
- 13.Sloan-Heggen CM, Bierer AO, Shearer AE, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135:441–450. [DOI] [PMC free article] [PubMed]
- 14.Morton CC, Nance WE. Newborn hearing screening—a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 15.Grosse SD, Ross DS, Dollard SC. Congenital cytomegalovirus (CMV) infection as a cause of permanent bilateral hearing loss: a quantitative assessment. J Clin Virol. 2008;41:57–62. doi: 10.1016/j.jcv.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Lanzieri TM, Chung W, Flores M, et al. Hearing loss in children with asymptomatic congenital cytomegalovirus infection. Pediatrics. 2017;139:e20162610. doi: 10.1542/peds.2016-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marazita ML, Ploughman LM, Rawlings B, Remington E, Arnos KS, Nance WE. Genetic epidemiological studies of early‐onset deafness in the US school‐age population. Am J Med Genet. 1993;46:486–491. doi: 10.1002/ajmg.1320460504. [DOI] [PubMed] [Google Scholar]
- 18.Feuchtbaum L, Lorey F, Faulkner L, et al. California’s experience implementing a pilot newborn supplemental screening program using tandem mass spectrometry. Pediatrics. 2006;117:S261–S269. doi: 10.1542/peds.2005-2633E. [DOI] [PubMed] [Google Scholar]
- 19.Clemens CJ, Davis SA, Bailey AR. The false-positive in universal newborn hearing screening. Pediatrics. 2000;106:e7. doi: 10.1542/peds.106.1.e7. [DOI] [PubMed] [Google Scholar]
- 20.Matern D, Tortorelli S, Oglesbee D, Gavrilov D, Rinaldo P. Reduction of the false-positive rate in newborn screening by implementation of MS/MS-based second-tier tests: the Mayo Clinic experience (2004–2007) J Inherit Metab Dis. 2007;30:585–592. doi: 10.1007/s10545-007-0691-y. [DOI] [PubMed] [Google Scholar]
- 21.Khairi MDM, Rafidah KN, Affizal A, Normastura AR, Suzana M, Normani ZM. Anxiety of the mothers with referred baby during universal newborn hearing screening. Int J Pediatr Otorhinolaryngol. 2011;75:513–517. doi: 10.1016/j.ijporl.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Tomblin JB, Harrison M, Ambrose SE, Walker EA, Oleson JJ, Moeller MP. Language outcomes in young children with mild to severe hearing loss. Ear Hear. 2015;36:76S–91S. doi: 10.1097/AUD.0000000000000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehra S, Eavey RD, Neck DKJO-HA. The epidemiology of hearing impairment in the United States: newborns, children, and adolescents. Otolaryngol Head Neck Surg. 2009;140:461–472. doi: 10.1016/j.otohns.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Dedhia K, Kitsko D, Sabo D, Chi DH. Children with sensorineural hearing loss after passing the newborn hearing screen. JAMA Otolaryngol Head Neck Surg. 2013;139:119–123. doi: 10.1001/jamaoto.2013.1229. [DOI] [PubMed] [Google Scholar]
- 25.Young NM, Reilly BK, Burke L. Limitations of universal newborn hearing screening in early identification of pediatric cochlear implant candidates. Arch Otolaryngol Head Neck Surg. 2011;137:230–234. doi: 10.1001/archoto.2011.4. [DOI] [PubMed] [Google Scholar]
- 26.Moser T, Starr A. Auditory neuropathy—neural and synaptic mechanisms. Nat Rev Neurol. 2016;12:135–149. doi: 10.1038/nrneurol.2016.10. [DOI] [PubMed] [Google Scholar]
- 27.Foerst A, Beutner D, Lang-Roth R, Huttenbrink K-B, Wedel von H, Walger M. Prevalence of auditory neuropathy/synaptopathy in a population of children with profound hearing loss. Int J Pediatr Otorhinolaryngol. 2006;70:1415–1422. doi: 10.1016/j.ijporl.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Penido RC, Isaac ML. Prevalence of auditory neuropathy spectrum disorder in an auditory health care service. Braz J Otorhinolaryngol. 2013;79:429–433. doi: 10.5935/1808-8694.20130077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vignesh SS, Jaya V, Muraleedharan A. Prevalence and audiological characteristics of auditory neuropathy spectrum disorder in pediatric population: a retrospective study. Indian J Otolaryngol Head Neck Surg. 2014;68:196–201. doi: 10.1007/s12070-014-0759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sloan-Heggen CM, Bierer AO, Shearer AE, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135:441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan D, Tekin D, Bademci G, et al. Spectrum of DNA variants for nonsyndromic deafness in a large cohort from multiple continents. Hum Genet. 2016;135:953–961. doi: 10.1007/s00439-016-1697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Ballesteros M, Reynoso R, Olarte M, et al. A multicenter study on the prevalence and spectrum of mutations in the otoferlin gene (OTOF) in subjects with nonsyndromic hearing impairment and auditory neuropathy. Hum Mutat. 2008;29:823–831. doi: 10.1002/humu.20708. [DOI] [PubMed] [Google Scholar]
- 33.Zimmerman E, Lahav A. Ototoxicity in preterm infants: effects of genetics, aminoglycosides, and loud environmental noise. J Perinatol. 2013;33:3–8. doi: 10.1038/jp.2012.105. [DOI] [PubMed] [Google Scholar]
- 34.Ealy M, Lynch KA, Meyer NC, Smith RJH. The prevalence of mitochondrial mutations associated with aminoglycoside-induced sensorineural hearing loss in an NICU population. Laryngoscope. 2011;121:1184–1186. doi: 10.1002/lary.21778. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Wang P, Han B, et al. Newborn hearing concurrent genetic screening for hearing impairment—a clinical practice in 58,397 neonates in Tianjin, China. Int J Pediatr Otorhinolaryngol. 2013;77:1929–1935. doi: 10.1016/j.ijporl.2013.08.038. [DOI] [PubMed] [Google Scholar]
- 36.Liu C-L, Farrell J, MacNeil JR, Stone S, Barfield W. Evaluating loss to follow-up in newborn hearing screening in Massachusetts. Pediatrics. 2008;121:e335–e343. doi: 10.1542/peds.2006-3540. [DOI] [PubMed] [Google Scholar]
- 37.Clemens CJ, Davis SA. Minimizing false-positives in universal newborn hearing screening: a simple solution. Pediatrics. 2001;107:E29. doi: 10.1542/peds.107.3.e29. [DOI] [PubMed] [Google Scholar]
- 38.da Mata Lupoli L, Garcia L, Anastasio ART, Fontana AC. Time after birth in relation to failure rate in newborn hearing screening. Int J Pediatr Otorhinolaryngol. 2013;77:932–935. doi: 10.1016/j.ijporl.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Caluraud S, Marcolla-Bouchetemblé A, de Barros A, et al. Newborn hearing screening: analysis and outcomes after 100,000 births in Upper-Normandy French region. Int J Pediatr Otorhinolaryngol. 2015;79:829–833. doi: 10.1016/j.ijporl.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 40.Kelsell DP, Dunlop J, Stevens HP, et al. Connexin 26 mutations in hereditary nonsyndromic sensorineural deafness. Nature. 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- 41.Hilgert N, Smith RJH, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681:189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shearer AE, DeLuca AP, Hildebrand MS, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen JX, Kachniarz B, Shin JJ. Diagnostic yield of computed tomography scan for pediatric hearing loss: a systematic review. Otolaryngol Head Neck Surg. 2014;151:718–739. doi: 10.1177/0194599814545727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kachniarz B, Chen JX, Gilani S, Shin JJ. Diagnostic yield of MRI for pediatric hearing loss: a systematic review. Otolaryngol Head Neck Surg. 2014;152:5–22. doi: 10.1177/0194599814555837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin JW, Chowdhury N, Mody A, et al. Comprehensive diagnostic battery for evaluating sensorineural hearing loss in children. Otol Neurotol. 2011;32:259–264. doi: 10.1097/MAO.0b013e31820160fa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shearer AE, Hildebrand MS, Sloan CM, Smith RJH. Deafness in the genomics era. Hear Res. 2011;282:1–9. doi: 10.1016/j.heares.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shearer AE, Kolbe DL, Azaiez H, et al. Copy number variants are a common cause of nonsyndromic hearing loss. Genome Med. 2014;6:37. doi: 10.1186/gm554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amr SS, Murphy E, Duffy E, et al. Allele-specific droplet digital PCR combined with a next-generation sequencing-based algorithm for diagnostic copy number analysis in genes with high homology: proof of concept using stereocilin. Clin Chem. 2018;64:705–714. doi: 10.1373/clinchem.2017.280685. [DOI] [PubMed] [Google Scholar]
- 49.Wu C-C, Tsai C-H, Hung C-C, et al. Newborn genetic screening for hearing impairment: a population-based longitudinal study. Genet Med. 2017;19:6–12. doi: 10.1038/gim.2016.66. [DOI] [PubMed] [Google Scholar]
- 50.Lu CY, Tsao PN, Ke YY, et al. Concurrent hearing, genetic, and cytomegalovirus screening in newborns. J Pediatr. 2018;199:144–150. [DOI] [PubMed]
- 51.Wang Q-J, Zhao Y-L, Rao S-Q, et al. Newborn hearing concurrent gene screening can improve care for hearing loss: a study on 14,913 Chinese newborns. Int J Pediatr Otorhinolaryngol. 2011;75:535–542. doi: 10.1016/j.ijporl.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z, Ding W, Liu X, et al. Auditory screening concurrent deafness predisposing genes screening in 10,043 neonates in Gansu province, China. Int J Pediatr Otorhinolaryngol. 2012;76:984–988. doi: 10.1016/j.ijporl.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 53.He X, Li X, Guo Y, et al. Newborn screening of genetic mutations in common deafness genes with bloodspot-based gene chip array. Am J Audiol. 2018;27:57–66. doi: 10.1044/2017_AJA-17-0042. [DOI] [PubMed] [Google Scholar]
- 54.Hao Z, Fu D, Ming Y, et al. Large scale newborn deafness genetic screening of 142,417 neonates in Wuhan, China. PLoS One. 2018;13:e0195740–15. doi: 10.1371/journal.pone.0195740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du W, Cheng J, Ding H, Jiang Z, Guo Y, Yuan H. A rapid method for simultaneous multigene mutation screening in children with nonsyndromic hearing loss. Genomics. 2014;104:264–270. doi: 10.1016/j.ygeno.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 56.Wang X, Hong Y, Cai P, et al. Rapid and reliable detection of nonsyndromic hearing loss mutations by multicolor melting curve analysis. Sci Rep. 2017;7:42894. doi: 10.1038/srep42894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S, Peng Q, Li W, Ma Q, Lu X. A reverse dot blot assay for the screening of twenty mutations in four genes associated with NSHL in a Chinese population. PLoS One. 2017;12:e0177196.. doi: 10.1371/journal.pone.0177196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan D, Xiang G, Chai X, et al. Screening of deafness-causing DNA variants that are common in patients of European ancestry using a microarray-based approach. PLoS One. 2017;12:e0169219.. doi: 10.1371/journal.pone.0169219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen J, Morton CC. Next-generation newborn hearing screening. In: Vona B, Haaf T, editors. Genetics of Deafness. Basel: Karger Publishers; 2016. p. 30–39.
- 60.Solomon BD, Hadley DW, Pineda-Alvarez DE, et al. Incidental medical information in whole-exome sequencing. Pediatrics. 2012;129:e1605–e1611. doi: 10.1542/peds.2011-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baux D, Vaché C, Blanchet C, et al. Combined genetic approaches yield a 48% diagnostic rate in a large cohort of French hearing-impaired patients. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-16846-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abou Tayoun AN, Turki AlSH, Oza AM, et al. Improving hearing loss gene testing: a systematic review of gene evidence toward more efficient next-generation sequencing-based diagnostic testing and interpretation. Genet Med. 2016;18:545–553. doi: 10.1038/gim.2015.141. [DOI] [PubMed] [Google Scholar]
- 64.Diener ML, Zick CD, McVicar SB, Boettger J, Park AH. Outcomes from a hearing-targeted cytomegalovirus screening program. Pediatrics. 2017;139:e20160789–12. doi: 10.1542/peds.2016-0789. [DOI] [PubMed] [Google Scholar]
- 65.Rawlinson WD, Palasanthiran P, Hall B, et al. Neonates with congenital cytomegalovirus and hearing loss identified via the universal newborn hearing screening program. J Clin Virol. 2018;102:110–115. doi: 10.1016/j.jcv.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 66.Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol. 2007;17:253–276. doi: 10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- 67.Fowler KB, McCollister FP, Sabo DL, et al. A targeted approach for congenital cytomegalovirus screening within newborn hearing screening. Pediatrics. 2017;139:e20162128. doi: 10.1542/peds.2016-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vancor E, Shapiro ED, Loyal J. Results of a targeted screening program for congenital cytomegalovirus infection in infants who fail newborn hearing screening. J Pediatr Infect Dis Soc. 2018;354:2151–2155. doi: 10.1093/jpids/pix105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.James SH, Kimberlin DW. Advances in the prevention and treatment of congenital cytomegalovirus infection. Curr Opin Pediatr. 2016;28:81–85. doi: 10.1097/MOP.0000000000000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pasternak Y, Ziv L, Attias J, Amir J, Bilavsky E. Valganciclovir is beneficial in children with congenital cytomegalovirus and isolated hearing loss. J Pediatr. 2018;199:166–170. [DOI] [PubMed]
- 71.Grosse SD, Dollard SC, Kimberlin DW. Screening for congenital cytomegalovirus after newborn hearing screening: what comes next? Pediatrics. 2017;139:e20163837–5. doi: 10.1542/peds.2016-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kimberlin DW, Jester PM, Sánchez PJ, et al. Valganciclovir for symptomatic congenital cytomegalovirus disease. N Engl J Med. 2015;372:933–943. doi: 10.1056/NEJMoa1404599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Woll B, Ladd P. Deaf Communities. In: Marschark M, Spencer PE, editors. The Oxford Handbook of Deaf Studies, Language, and Education, Second Edition. Oxford: Oxford University Press; 2011. p. 151–163.
- 74.Parasnis I. On Interpreting the Deaf Experience within the Context of Cultural and Language Diversity. In: Parasnis I, editor. Cultural and Language Diversity and the Deaf Experience. Cambridge: Cambridge University Press; 1996. p. 3–19.
- 75.Withrow KA, Tracy KA, Burton SK, et al. Impact of genetic advances and testing for hearing loss: results from a national consumer survey. Am J Med Genet A. 2009;149A:1159–1168. doi: 10.1002/ajmg.a.32800. [DOI] [PubMed] [Google Scholar]
- 76.Arnos KS. Ethical and social implications of genetic testing for communication disorders. J Commun Disord. 2008;41:444–457. doi: 10.1016/j.jcomdis.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Palmer CGS, Lueddeke JT, Zhou J. Factors influencing parental decision about genetics evaluation for their deaf or hard-of-hearing child. Genet Med. 2009;11:248–255. doi: 10.1097/GIM.0b013e318195aad9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palmer CGS, Martinez A, Fox M, et al. A prospective, longitudinal study of the impact of GJB2/GJB6 genetic testing on the beliefs and attitudes of parents of deaf and hard‐of‐hearing infants. Am J Med Genet A. 2009;149A:1169–1182. doi: 10.1002/ajmg.a.32853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kennedy CR, McCann DC, Campbell MJ, et al. Language ability after early detection of permanent childhood hearing impairment. N Engl J Med. 2006;354:2131–2141. doi: 10.1056/NEJMoa054915. [DOI] [PubMed] [Google Scholar]
- 80.Korver AMH, Smith RJH, Van Camp G, et al. Congenital hearing loss. Nat Rev Dis Primers. 2017;3:16094. doi: 10.1038/nrdp.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wake M, Ching TYC, Wirth K, et al. Population outcomes of three approaches to detection of congenital hearing loss. Pediatrics. 2016;137:e20151722. doi: 10.1542/peds.2015-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


