Summary
Background/Objectives:
Low-grade inflammation is an underlying feature of obesity and identifying inflammatory markers is crucial to understanding this disease. Therefore, the purpose of this study was twofold: (i) to perform a global microarray analysis and (ii) to investigate the role of lactoferrin (LTF), one of the most altered genes, in relation to obesity in Latino youth.
Methods:
Non-diabetic Latino youth (71 males/92 females; 15.6 ± 3.2 years) were studied. A subset of 39 participants was randomly selected for global microarray analysis profiling from the whole blood sample. Serum LTF was compared between lean (n = 78) and overweight/obese (n = 85) participants.
Results:
Microarray analysis revealed that a total of 1870 probes were altered in expression ≥1.2-fold and P < 0.05 in overweight/obese participants compared with lean. KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis revealed significant enrichment for pathways including toll-like receptor (TLR) and B cell receptor signalling pathways. LTF and TLR5 were increased in expression by 2.2 and 1.5 fold, respectively, in the overweight/obese participants. Increased LTF concentrations were significantly associated with high risk of obesity-related phenotypes (all P < 0.05).
Conclusions:
Our data suggest that increased LTF is associated with obesity risk among Latino youth. This finding is discordant to what has been shown in adults and suggests that age may modulate the association between LTF and obesity-related health.
Keywords: Adiposity, gene expression, inflammation, paediatric
Introduction
Childhood obesity is a global epidemic that, along with type 2 diabetes and cardiovascular disease, has emerged as a critical health concern among youth (1). One of the hypothesized links between obesity and cardiometabolic disease is the presence of a chronic low-grade inflammation (2). In response to increased adiposity, the body produces higher circulating levels of inflammatory cytokines, such as tumour necrosis factor-α (TNF-α), interleukin (IL)-6 and C-reactive protein (CRP), and a reduction in the anti-inflammatory cytokine adiponectin (3). The production of inflammatory cytokines from adipose tissue and circulating blood cells is considered an important process that contributes to metabolic disturbances associated with increased body weight (2). Efforts to identify which inflammatory markers are most important and how they may be associated with obesity-related disorders are necessary.
Recent studies suggest that lactoferrin (LTF) may be an important modulator of the inflammatory response through regulation of proinflammatory cytokines (4). LTF is present in epithelial secretions and in the secondary granules of neutrophils, which are considered as the first line of host-defence against infections (5). In addition, LTF has an important role in regulating the inflammatory process in both animals (6) and humans (7). Furthermore, several studies in adults have shown that circulating LTF concentration is associated with dyslipidaemia (8) and chronic low-grade inflammation coinciding with insulin resistance (9). Moreover, the authors found that LTF gene expression was negatively associated with body mass index (BMI), fat mass, fasting plasma glucose and triglycerides concentrations (10).
To our knowledge, it is not known whether circulating LTF levels are associated with obesity-related risk in youth. Given that LTF levels may vary across gender and/or age (11), it is necessary to confirm previous findings in adults in younger populations. Therefore, the purpose of our study was (i) to perform a global microarray analysis to identify novel targets of obesity and (ii) to investigate the association between circulating LTF levels with obesity-related phenotypes and glycaemic markers in Latino youth.
Methods
Participants
Data from 163 non-diabetic Latino youth (8–21 years) who participated in the Arizona Insulin Resistance Registry were used in the present analysis. Details of the study are described elsewhere (12). The study protocol was approved by the Institutional Review Board of the Arizona State University.
Phenotypic characterization
Measurements of phenotypes are described in the Supporting Information. For comparison purposes, participants were divided into two groups based on BMI categories (normal weight vs. overweight/obese). BMI status was classified using both absolute BMI value (< or ≥25 kg m−2) and BMI percentile for age and sex (< or ≥85th percentile), which were taken from the Centers for Disease Control and Prevention. In addition, the lean (normal weight) group was used as a reference in the global gene expression profiling to identify the genes that were up- or down-regulated in the overweight/obese group.
Global gene expression profiling
We performed global gene expression profiling on a subset of 39 randomly selected Latino youth from the registry. The Supporting Information provides additional information of the overall process of RNA isolation and description of microarray processing/analysis.
Quantitative real-time polymerase chain reaction
Blood expression of various genes was determined using quantitative real-time polymerase chain reaction (Q-RT-PCR) on the 7900HT sequence detection system as described in the Supporting Information.
Statistical analysis
Independent sample t-tests and χ2 analyses were used to compare descriptive characteristics by sex and BMI status. Analysis of covariance was used to compare phenotypes after adjusting for age, sex or BMI. Data that did not meet the assumptions for normality were log10 transformed. Pearson’s correlation analyses were used to estimate correlation between LTF levels and phenotypes. Additional sex-specific associations were analysed using Pearson’s correlation tests. All data were analysed using SPSS (IBM, SPSS, Version 20.0, Chicago, IL, USA) with significance level set at P ≤ 0.05.
Results
Microarray analysis
Figure S1 provides an overview of the steps used in the microarray analysis to identify genes that differed most robustly by weight status. Phenotypic characteristics of those participants are presented in Table S1. Results of comparison analyses among the 39 participants are concordant with the descriptive analyses of the total study population with the exception of the ratios of sex and BMI status (n = 163, Table 1). A total of 1870 probes were altered in expression ≥1.2 fold and P < 0.05 (uncorrected). The 1870 probes were submitted to the database for annotation, visualization and integrated discovery for exploratory pathway analysis, and significant KEGG pathways are shown in Table S2.
Table 1.
Characteristics of participants by sex or BMI status
| Variables | Total (n = 163) | By obesity status | By sex | ||
|---|---|---|---|---|---|
| Normal weight (n = 78) | Overweight/obese (n = 85) | Male (n = 71) | Female (n = 92) | ||
| Age (years) | 15.6 ± 3.2 | 15.5 ± 3.3 | 15.7 ± 3.2 | 15.7 ± 3.5 | 15.4 ± 2.9 |
| Sex (male/female), n (%) | 71 (44)/92 (56) | 28 (36)/50 (64) | 43 (51)/42 (49) | – | – |
| Lean/overweight and obese, n (%) | 79 (48)/85 (52) | – | – | 28 (39)/43(61) | 50 (54)/42 (46) |
| NGT/prediabetes, n (%) | 110 (79)/29 (21) | 57 (88)/8 (12) | 53 (72)/21 (28)* | 48 (80)/12 (20) | 62 (78)/17 (22) |
| BMI (kg m−2) | 25.0 ± 6.7 | 20.0 ± 2.6 | 29.5 ± 5.9** | 24.4 ± 6.9 | 25.6 ± 6.3 |
| Fat mass (kg) | 16.9 ± 11.5 | 9.0 ± 5.0 | 24.2 ± 11.0** | 17.9 ± 11.8 | 15.7 ± 11.1† |
| Waist circumference (cm) | 85.3 ± 17.4 | 72.29 ± 8.4 | 97.4 ± 14.6** | 82.9 ± 17.0 | 88.3 ± 17.5† |
| Hip circumference (cm) | 99.3 ± 15.0 | 89.3 ± 9.2 | 108.7 ± 13.3** | 98.1 ± 15.3 | 100.9 ± 14.6 |
| Systolic blood pressure (mmHg) | 112.5 ± 12.5 | 108.7 ± 10.4 | 116.0 ± 13.3** | 109.3 ± 9.9 | 116.5 ± 14.4†† |
| Diastolic blood pressure (mmHg) | 69.2 ± 8.9 | 67.2 ± 8.9 | 70.9 ± 8.7* | 69.4 ± 8.9 | 68.9 ± 9.1 |
| Triglyceride (mg dL−1) | 97.7 ± 50.9 | 83.7 ± 38.9 | 110.6 ± 57.0** | 97.7 ± 53.6 | 97.8 ± 47.5 |
| HDL (mg dL−1) | 45.1 ± 11.1 | 47.6 ± 11.7 | 42.7 ± 9.9* | 46.1 ± 11.1 | 43.7 ± 10.9 |
| LDL (mg dL−1) | 85.0 ± 23.8 | 80.3 ± 20.1 | 89.3 ± 26.0* | 85.9 ± 24.9 | 83.7 ± 22.4 |
| VLDL (mg dL−1) | 16.8 ± 9.7 | 14.9 ± 9.6 | 18.5 ± 9.5* | 16.4 ± 8.9 | 17.3 ± 10.6 |
| Total cholesterol (mg dL−1) | 150.2 ± 55.4 | 149.5 ± 73.5 | 150.7 ± 31.0 | 155.0 ± 69.7 | 143.9 ± 26.4 |
| ALT (U L−1) | 19.3 ± 13.6 | 16.2 ± 10.7 | 22.3 ± 15.3** | 17.2 ± 14.1 | 22.1 ± 12.6† |
| AST (U L−1) | 22.8 ± 9.3 | 22.8 ± 9.8 | 22.7 ± 8.9 | 21.9 ± 9.7 | 23.9 ± 8.7 |
| HbA1c (%) | 5.5 ± 0.3 | 5.5 ± 0.3 | 5.5 ± 0.3 | 5.5 ± 0.3 | 5.5 ± 0.3 |
| White blood cell (k mm−3) | 6.5 ± 1.5 | 6.1 ± 1.5 | 6.9 ± 1.4** | 6.5 ± 1.4 | 6.5 ± 1.6 |
| Adiponectin (μg mL−1) | 8.1 ± 3.6 | 9.3 ± 3.8 | 7.0 ± 2.9** | 8.7 ± 3.7 | 7.3 ± 3.3† |
| Fasting glucose (mg dL−1) | 90.6 ± 6.3 | 90.6 ± 5.9 | 90.7 ± 6.7 | 89.4 ± 6.3 | 92.4 ± 5.8† |
| Fasting insulin (μU mL−1) | 9.6 ± 7.5 | 7.3 ± 4.3 | 11.9 ± 9.2** | 8.9 ± 5.8 | 10.5 ± 9.3 |
| LTF (ng mL−1) | 1458.3 ± 834.5 | 1292.5 ± 647.8 | 1610.5 ± 953.5* | 1479.8 ± 780.1 | 1430.49 ± 905.0 |
Data are means ± standard deviation, n (%). Glycaemic status data were not available on 24 of the 163 participants.
P < 0.05 after adjusting age and sex.
P < 0.001 after adjusting age and sex.
P < 0.05 after adjusting age and BMI.
P < 0.001 after adjusting age and BMI.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; HDL, high-density lipoprotein; LDL, low-density lipoprotein; LTF, lactoferrin; NGT, normal glucose tolerance; prediabetes, impaired fasting glucose, impaired glucose tolerance; VLDL, very low-density lipoprotein.
In addition to pathway analysis, GeneSpring analysis was performed to identify the genes that differed most robustly and our data revealed 501 probes that were altered in expression ≥1.5 fold and P < 0.05. Of the 501 probes, 144 had unknown gene annotations and the remaining 357 probes represented 261 unique genes. Of the 261 unique genes, 163 were lower in expression while the remaining 98 were higher in expression in the overweight/obese group as shown in Tables S3 and S4, respectively. There were a number of individual genes involved in inflammatory pathways including toll-like receptor (TLR) 5 and lymphocyte antigen 96 (LY96), which were both significantly increased in expression by 1.5-fold compared with lean youth. Moreover, LTF, one of the most robustly altered genes in our dataset, was significantly higher in expression in the overweight/obese participants by 2.2-fold. The LTF microarray data were validated by comparing the changes in mRNA levels of LTF using Q-RT-PCR as shown in Fig. S2. Normalized (log2 transformed signal) microarray results were strongly and inversely correlated with ΔCt values from Q-RT-PCR (r = −0.79, P < 0.001). Moreover, the changes in LTF gene expression determined by Q-RT-PCR were concordant with the changes in gene expression determined by microarray analysis. To further validate our microarray data, an additional two genes (TLR5 and histidine decarboxylase) were quantified using Q-RT-PCR and our results confirmed that gene expression changes captured in the microarray were concordant with the changes in quantified mRNA levels of these genes measured by Q-RT-PCR as shown in Table S5.
Association between lactoferrin and obesity-related phenotypes
The descriptive characteristics of participants are presented in Table 1. Among the 163 Latino youth, 78 participants were of normal weight while 85 were classified as overweight or obese. No differences in age or glycaemic status (normal glucose tolerance [NGT] vs. prediabetes) were noted between the two BMI categories. Lean participants exhibited a healthier profile than the overweight/obese participants in terms of BMI, fat mass, waist and hip circumferences, systolic/diastolic blood pressures and all lipid profiles except very low-density lipoprotein. In the metabolic panels, the overweight/obese participants showed significantly lower adiponectin levels (7.0 ± 2.9 vs. 9.3 ± 3.8 μg mL−1) and higher alanine aminotransferase(22.3 ± 15.3 vs. 16.2 ± 10.7 U L−1), white blood cell counts (6.9 ± 1.4 vs. 6.1 ± 1.5 k mm−3), fasting insulin(11.9 ± 9.2 vs. 7.3 ± 4.3 μU mL−1) and LTF concentrations (1610.5 ± 953.5 vs. 1292.5 ± 647.8 ng mL−1, all P < 0.05) when compared with lean participants. Differences persisted after controlling for age and sex.
In addition, when the data were analysed by sex, there were no differences in age or glycaemic status. However, a trend towards significance in the frequency of obesity was observed between men and women, indicating that the prevalence of overweight and obesity is higher in men than women (61% vs. 46%, P = 0.059). Women exhibited significantly higher waist circumferences (88.3 ± 17.5 vs.83.0 ± 17.0 cm), systolic blood pressure (116.5 ± 14.4 vs. 109.3 ± 9.9 mmHg) and lower fat mass (15.7 ± 11.1 vs. 17.9 ± 11.8 kg, all P < 0.05) as compared with men. LTF levels were not statistically different by sex. All significant differences between men and women remained after controlling for age and BMI.
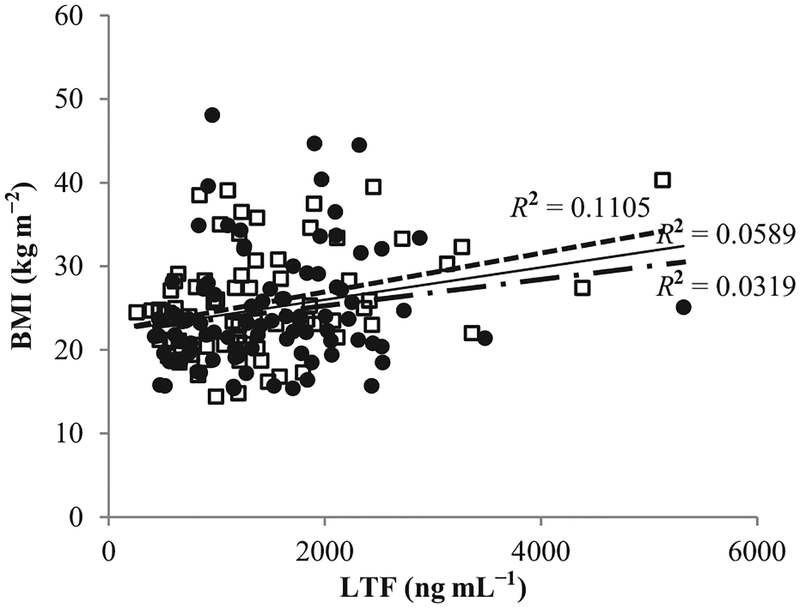
The association between circulating LTF levels and obesity-related phenotypes, including anthropometrics and metabolic characteristics, are presented in Table 2 and Fig. 1. The correlation between LTF concentrations and age was significantly positive (r = 0.23, P < 0.001). This correlation was stronger in men (r = 0.32, P < 0.001) although it shifted to non-significance in women (r = 0.16, P = 0.13). In men, LTF concentrations were found to increase alongside BMI (r = 0.33), fat mass (r = 0.32), waist (r = 0.31) and hip circumferences (r = 0.30, all P < 0.05). These results indicate that a higher obesity risk accompanies higher LTF concentrations for men. However, there were no significant associations between LTF concentrations and obesity-related phenotypes in women. Additionally, white blood cell counts were positively and significantly correlated with LTF concentrations in both men and women (r = 0.46 and r = 0.38, respectively, P < 0.001). When the correlation between LTF levels and obesity-related phenotypes was examined after controlling for white blood cell counts, all significant correlations no longer remained. Fasting markers of glycaemic and insulin dynamics, including HbA1c, fasting plasma glucose and insulin, were not significantly associated with LTF levels.
Table 2.
Correlation between phenotypic variables and circulated LTF concentrations
| Correlation with LTF | |||
|---|---|---|---|
| Total | Male | Female | |
| Age (years) | 0.23** | 0.32** | 0.16 |
| BMI (kg m−2) | 0.24** | 0.33** | 0.18 |
| Fat mass (kg) | 0.22** | 0.32** | 0.13 |
| Waist circumference (cm) | 0.24** | 0.31** | 0.20 |
| Hip circumference (cm) | 0.22** | 0.30* | 0.16 |
| Systolic blood pressure (mmHg) | 0.14 | 0.26* | 0.03 |
| Diastolic blood pressure (mmHg) | 0.21** | 0.22 | 0.20 |
| Triglyceride (mg dL−1) | 0.11 | 0.10 | 0.12 |
| HDL (mg dL−1) | −0.07 | −0.09 | −0.06 |
| LDL (mg dL−1) | 0.16* | 0.07 | 0.24* |
| VLDL (mg dL−1) | 0.07 | 0.03 | 0.12 |
| Total cholesterol (mg dL−1) | 0.04 | 0.05 | 0.04 |
| ALT (U L−1) | 0.14 | 0.18 | 0.12 |
| AST (U L−1) | 0.03 | 0.07 | 0.01 |
| HbA1c (%) | −0.14 | −0.07 | −0.20 |
| White blood cell (k mm−3) | 0.42** | 0.46** | 0.38** |
| Adiponectin (μg mL−1) | −0.07 | −0.12 | −0.04 |
| Fasting glucose (mg dL−1) | 0.07 | 0.04 | 0.11 |
| Fasting insulin (μU mL−1) | −0.00 | 0.08 | −0.11 |
P < 0.05.
P < 0.01.
ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; HDL, high-density lipoprotein; LDL, low-density lipoprotein; LTF, lactoferrin; VLDL, very low-density lipoprotein.
Figure 1.
Correlation between body mass index (BMI) and circulated lactoferrin (LTF) concentrations between men (white square and dashed line) and women (black circle and dash-dotted line). Solid line represents total group.
Discussion
In the present study, we attempted to identify novel genes involved in inflammation that were altered in expression in the whole blood of overweight/obese youth compared with lean counterparts using global gene expression profiling. We identified a number of genes that were differentially expressed in the overweight/obese youth. LTF, which has been shown to play a role in the inflammatory process, was one of the most robustly up-regulated genes. We demonstrated the associations between circulating LTF levels and cardiometabolic risk factors among Latino youth. Specifically, LTF concentrations were higher in overweight/obese youth compared with lean youth and were significantly associated with BMI, fat mass, hip and waist circumferences.
Our global microarray analysis from the 39 randomly selected Latino youth revealed alterations in multiple genes and pathways related to inflammation. TLR5 and LY96 were up-regulated in overweight/obese participants and were of interest because these two genes were simultaneously detected in the TLR signalling pathway. This pathway has been shown to play an important role in inflammatory responses as well as cell proliferation and survival (13). Although the function of TLR5 is less known, other members of the TLR family are well-characterized including TLR2 (14) and TLR4 (15). Vijay-Kumar et al. (16) demonstrated that mice deficient of TLR5 exhibited higher risk for the metabolic syndrome including hyperlipidaemia, hypertension, insulin resistance and increased adiposity suggesting that TLR5 malfunction may lead to defects in insulin receptor signalling. Moreover, LY96 (also known as myeloid differentiation protein-2) has been shown to be associated with lipopolysaccharide (LPS)-mediated activation of innate immune system by cooperating with TLR4 on the cell surface (17). Nagai et al. (18) showed that mice deficient of LY96 displayed hyporesponsiveness to LPS challenge (which is a stimulator of inflammation), indicating that it is essential for LPS recognition of TLR4.
LTF, one of the most robustly up-regulated genes in overweight/obese youth, has also been shown to play a role in the TLR signalling pathway (19) and has recently been studied for its potential therapeutic applications to obesity-associated metabolic disturbances (7,20,21). Growing evidence suggests that LTF is thought to be a modulator of immune system and inflammatory responses (4). A study by Tamano et al. (6) demonstrated that LTF infusion for 40 weeks in male rats significantly reduced serum triglyceride level by 72% compared with control rats, suggesting that LTF may have protective effects against metabolic disease. Similarly, a study in Japanese adults with abdominal obesity investigated the effects of enteric-coated LTF on lipid metabolism and body composition (7). The results of that study demonstrated that individuals given LTF had significant decreases in body weight, BMI and hip circumference, suggesting that LTF is important for reducing visceral fat. Furthermore, Moreno-Navarrete et al. examined the association of circulating LTF concentration with lipids and measures of obesity (8–10,21,22). Among male adults with altered glucose tolerance, it was found that LTF concentration was inversely proportional with fasting triglyceride, BMI and waist-to-hip ratio, and positively correlated with high-density lipoprotein (8,9). They also found that patients with altered glucose tolerance exhibited significantly lower LTF levels compared with patients with NGT (9). Consistent with this, a previous study measured LTF gene expression in adipose tissue and found that LTF expression was negatively associated with BMI, body fat percentage, fasting glucose and triglycerides (10).
However, little is known regarding LTF levels and adiposity or metabolic state in the younger populations. To our knowledge, there is no study that has examined the association between circulating LTF levels (or LTF gene expression) and phenotypes that are related to obesity or cardiometabolic risk in youth. Our data in Latino youth revealed significant positive correlations between circulating LTF concentrations and adiposity. These results are incongruent to what has been described in adult populations and emphasize the need to examine the effects of LTF in younger populations. Because LTF is thought to be actively secreted by increased secondary degranulation of neutrophils into blood during inflammatory processes (23), maintaining a low level of LTF under normal conditions is advantageous. However, it may be physiologically appropriate that increased adiposity leads to elevated LTF levels in youth, as inflammation-related metabolic status may be less robustly impacted (24). In contrast, low LTF levels in obese adults may be due to chronic hyperglycaemia or inflammatory responses that interrupt degranulation of neutrophils, leading to reduced efficiency of LTF secretion (25). Taken together, our data provide a novel description of circulating LTF states in relation to adiposity in youth, which suggests that participants with higher BMI exhibit higher secretion of LTF into the blood stream. To further substantiate this claim, a prospective study that can demonstrate whether the obesity-associated patterns of LTF secretion is changed in relation to participants’ growth patterns and their inflammatory status over time is warranted.
To our knowledge, this is the first study to examine the association between circulating LTF concentrations and obesity-related phenotypes among the paediatric population. We focused on Latino youth who represent a vulnerable population at an increased risk for type 2 diabetes. Moreover, we performed a global microarray study to find up- and down-regulated genes related to inflammation by BMI categories. There are limited data regarding the gene expression profiling in obesity or type 2 diabetes in youth. Hardy et al. (26) performed mRNA quantifications of TLRs and proinflammatory genes (IL-6 and TNF-α) to examine associations between the activation of TLR signalling and metabolic disturbance from overweight adolescents with metabolic syndrome. In addition, Kaizer et al. (27) performed a global microarray analysis on peripheral blood mononuclear cells (PBMCs) among children with type 1 and type 2 diabetes compared with healthy controls. The authors demonstrated that IL-1β was highly expressed in both type 1 and type 2 diabetics, and MYC (V-Myc Avian Myelocytomatosis Viral Oncogene Homolog) was altered only in patients with type 1 diabetes. In the present study, we used PAXgene RNA (BD Diagnostics, Franklin Lakes, NJ) to measure global gene expression levels in our subjects. Although it has been reported that the gene expression levels from PAXgene RNA are relatively lower than fold changes from studies using PBMCs (28), our evidence provides insight into novel targets for obesity in youth, which were validated independently with Q-RT-PCR.
Despite the strengths of our study, we acknowledge potential limitations that should be considered. First, well-established inflammatory makers such as TNF-α, IL-6 and CRP were not available for our analysis. Because LTF is deemed as a modulator of inflammation process by regulating proinflammatory cytokines (4), any association between LTF levels and other inflammatory makers should be addressed. In the present study, we measured the levels of adiponectin, an adipocyte-driven hormone that is considered an anti-inflammatory marker. We observed that overweight/obese youth exhibited significantly lower adiponectin levels than lean youth. However, interestingly, when the correlation between adiponectin and LTF levels is examined, no relationship was observed. Second, a relatively wide spectrum of age is used without measurement of pubertal stage. Previous studies show that puberty is associated with metabolic disorders that may vary by sex (29). Although age is not an optimal surrogate measure of pubertal stage, we did attempt to create a linear regression model that includes age and LTF levels for predicting adiposity in order to minimize the confounding effects of pubertal development. With the exception of hip circumference, LTF concentrations were significantly associated with BMI, fat mass and waist circumference after controlling for age (data not shown). Third, we did not assess neutrophil counts which are responsible for LTF secretion. However, neutrophils are the most abundant type of white blood cell and we were able to confirm a significant correlation between LTF levels and total white blood cell counts. Fourth, serum LTF levels may be affected by neutrophil degranulation which may occur while serum samples were prepared (30). Although the difference in LTF levels between plasma and serum was not tested in this study, we confirmed that significant correlations between LTF levels and obesity-related phenotypes were maintained even after controlling for the levels of myeloperoxidase (a marker of neutrophil degranulation) from a subset of 39 participants (data not shown). Lastly, because of the low prevalence of prediabetes and type 2 diabetes (21% and 0% of our study population, respectively), we opted to focus on obesity-related phenotypes. Future studies will need to recruit much larger cohorts followed over longer periods to definitively test the contribution of LTF to cardiometabolic disease risk.
In summary, we have performed global transcriptomic analyses on Latino youth yielding lists of pathways and genes that are altered by weight status. We highlighted a number of inflammatory genes including LTF, which was one of the most robustly up-regulated genes in the overweight/obese youth compared with lean counterparts. We have demonstrated that high circulating LTF levels were consistently associated with obesity-related characteristics in Latino youth. Further studies to investigate whether the LTF level in the blood is changed in response to growth patterns with obesity in younger populations are warranted.
Supplementary Material
Figure S1. Flow diagram of the steps used in the analysis of the microarray data.
Figure S2. Correlation between circulated lactoferrin concentrations and RT-PCR (Ct value).
Table S1. Characteristics of the 39 participants that were used for microarray analysis and Q-RT-PCR by sex or BMI status.
Table S2. Results of the significant KEGG pathway analysis performed in DAVID on the 1870 probes that were altered in expression ≥1.2 fold and P < 0.05.
Table S3. Results of GeneSpring analysis revealed 501 probes that were altered in expression ≥1.5 fold and P < 0.05. Of the 501 probes, 144 had unknown gene annotations and the remaining 357 probes represented 261 unique genes (multiple probes associated with the same gene). Of the 261 unique genes, 163 were decreased in expression.
Table S4. Results of GeneSpring analysis revealed 501 probes that were altered in expression ≥1.5 fold and P < 0.05. Of the 501 probes, 144 had unknown gene annotations and the remaining 357 probes represented 261 unique genes (multiple probes associated with the same gene). Of the 261 unique genes, 98 were increased in expression.
Table S5. Results of the mRNA expression determined by Q-RT-PCR (with corresponding microarray results shown).
Acknowledgements
DKC conceived the experiments. JYK, LEC, GQS and DKC carried out the experiments and analysed the data. JYK and DKC were involved in writing the article and all authors had final approval of the submitted and published versions. We thank the children and adolescents who participated in this study. We thank Veronica Vital, RN and the rest of the ASU clinical research staff for their excellent care of the participants. This work was supported by Health Research Alliance Arizona and the Center for Metabolic Biology at Arizona State University.
Footnotes
Conflicts of Interest Statement
No potential conflicts of interest relevant to this article were reported.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
References
- 1.Franks PW, Hanson RL, Knowler WC, Sievers ML, Bennett PH, Looker HC. Childhood obesity, other cardiovascular risk factors, and premature death. N Engl J Med 2010; 362: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dandona P, Aljada A, Bandyopadhyay A. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol 2004; 25: 4–7. [DOI] [PubMed] [Google Scholar]
- 3.DeBoer MD. Obesity, systemic inflammation, and increased risk for cardiovascular disease and diabetes among adolescents: a need for screening tools to target interventions. Nutrition 2013; 29: 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legrand D, Elass E, Carpentier M, Mazurier J. Lactoferrin: a modulator of immune and inflammatory responses. Cell Mol Life Sci 2005; 62: 2549–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer S, Lonnerdal B. Lactoferrin, lactoferrin receptors and iron metabolism. Eur J Clin Nutr 1993; 47: 232–241. [PubMed] [Google Scholar]
- 6.Tamano S, Sekine K, Takase M, Yamauchi K, Iigo M, Tsuda H. Lack of chronic oral toxicity of chemopreventive bovine lactoferrin in F344/DuCrj rats. Asian Pac J Cancer Prev 2008; 9: 313–316. [PubMed] [Google Scholar]
- 7.Ono T, Murakoshi M, Suzuki N, et al. Potent anti-obesity effect of enteric-coated lactoferrin: decrease in visceral fat accumulation in Japanese men and women with abdominal obesity after 8-week administration of enteric-coated lactoferrin tablets. Br J Nutr 2010; 104: 1688–1695. [DOI] [PubMed] [Google Scholar]
- 8.Moreno-Navarrete JM, Ortega FJ, Bassols J, Castro A, Ricart W, Fernandez-Real JM. Association of circulating lactoferrin concentration and 2 nonsynonymous LTF gene polymorphisms with dyslipidemia in men depends on glucose-tolerance status. Clin Chem 2008; 54: 301–309. [DOI] [PubMed] [Google Scholar]
- 9.Moreno-Navarrete JM, Ortega FJ, Bassols J, Ricart W, Fernandez-Real JM. Decreased circulating lactoferrin in insulin resistance and altered glucose tolerance as a possible marker of neutrophil dysfunction in type 2 diabetes. J Clin Endocrinol Metab 2009; 94: 4036–4044. [DOI] [PubMed] [Google Scholar]
- 10.Moreno-Navarrete JM, Serrano M, Sabater M, et al. Study of lactoferrin gene expression in human and mouse adipose tissue, human preadipocytes and mouse 3T3-L1 fibroblasts. Association with adipogenic and inflammatory markers. J Nutr Biochem 2013; 24: 1266–1275. [DOI] [PubMed] [Google Scholar]
- 11.Bennett RM, Mohla C. A solid-phase radioimmunoassay for the measurement of lactoferrin in human plasma: variations with age, sex, and disease. J Lab Clin Med 1976; 88: 156–166. [PubMed] [Google Scholar]
- 12.Shaibi GQ, Coletta DK, Vital V, Mandarino LJ. The design and conduct of a community-based registry and biorepository: a focus on cardiometabolic health in Latinos. Clin Transl Sci 2013; 6: 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Jiang S, Tapping RI. Toll-like receptor signaling in cell proliferation and survival. Cytokine 2010; 49: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang HJ, Kim HS, Hwang DH, Quon MJ, Kim JA. Toll-like receptor 2 mediates high-fat diet-induced impairment of vasodilator actions of insulin. Am J Physiol Endocrinol Metab 2013; 304: E1077–E1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyna SM, Ghosh S, Tantiwong P, et al. Elevated toll-like receptor 4 expression and signaling in muscle from insulin-resistant subjects. Diabetes 2008; 57: 2595–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010; 328: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauer S, Kunde YA, Apodaca TA, Goldstein B, Hong-Geller E. Soluble MD2 increases TLR4 levels on the epithelial cell surface. Cell Immunol 2009; 255: 8–16. [DOI] [PubMed] [Google Scholar]
- 18.Nagai Y, Akashi S, Nagafuku M, et al. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat Immunol 2002; 3: 667–672. [DOI] [PubMed] [Google Scholar]
- 19.Puddu P, Latorre D, Carollo M, et al. Bovine lactoferrin counteracts Toll-like receptor mediated activation signals in antigen presenting cells. PLoS ONE 2011; 6: e22504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yagi M, Suzuki N, Takayama T, et al. Lactoferrin suppress the adipogenic differentiation of MC3T3-G2/PA6 cells. J Oral Sci 2008; 50: 419–425. [DOI] [PubMed] [Google Scholar]
- 21.Moreno-Navarrete JM, Ortega F, Sabater M, Ricart W, Fernandez-Real JM. Proadipogenic effects of lactoferrin in human subcutaneous and visceral preadipocytes. J Nutr Biochem 2011; 22: 1143–1149. [DOI] [PubMed] [Google Scholar]
- 22.Moreno-Navarrete JM, Ortega FJ, Ricart W, Fernandez-Real JM. Lactoferrin increases (172Thr)AMPK phosphorylation and insulin-induced (p473Ser)AKT while impairing adipocyte differentiation. Int J Obes (Lond) 2009; 33: 991–1000. [DOI] [PubMed] [Google Scholar]
- 23.Ward PP, Paz E, Conneely OM. Multifunctional roles of lactoferrin: a critical overview. Cell Mol Life Sci 2005; 62: 2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Heredia FP, Gomez-Martinez S, Marcos A. Obesity, inflammation and the immune system. Proc Nutr Soc 2012; 71: 332–338. [DOI] [PubMed] [Google Scholar]
- 25.Stegenga ME, van der Crabben SN, Blumer RM, et al. Hyperglycemia enhances coagulation and reduces neutrophil degranulation, whereas hyperinsulinemia inhibits fibrinolysis during human endotoxemia. Blood 2008; 112: 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardy OT, Kim A, Ciccarelli C, Hayman LL, Wiecha J. Increased Toll-like receptor (TLR) mRNA expression in monocytes is a feature of metabolic syndrome in adolescents. Pediatr Obes 2013; 8: e19–e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaizer EC, Glaser CL, Chaussabel D, Banchereau J, Pascual V, White PC. Gene expression in peripheral blood mononuclear cells from children with diabetes. J Clin Endocrinol Metab 2007; 92: 3705–3711. [DOI] [PubMed] [Google Scholar]
- 28.Min JL, Barrett A, Watts T, et al. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics 2010; 11: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman RP, Vicini P, Sivitz WI, Cobelli C. Pubertal adolescent male-female differences in insulin sensitivity and glucose effectiveness determined by the one compartment minimal model. Pediatr Res 2000; 48: 384–388. [DOI] [PubMed] [Google Scholar]
- 30.Plow EF. Leukocyte elastase release during blood coagulation. A potential mechanism for activation of the alternative fibrinolytic pathway. J Clin Invest 1982; 69: 564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Flow diagram of the steps used in the analysis of the microarray data.
Figure S2. Correlation between circulated lactoferrin concentrations and RT-PCR (Ct value).
Table S1. Characteristics of the 39 participants that were used for microarray analysis and Q-RT-PCR by sex or BMI status.
Table S2. Results of the significant KEGG pathway analysis performed in DAVID on the 1870 probes that were altered in expression ≥1.2 fold and P < 0.05.
Table S3. Results of GeneSpring analysis revealed 501 probes that were altered in expression ≥1.5 fold and P < 0.05. Of the 501 probes, 144 had unknown gene annotations and the remaining 357 probes represented 261 unique genes (multiple probes associated with the same gene). Of the 261 unique genes, 163 were decreased in expression.
Table S4. Results of GeneSpring analysis revealed 501 probes that were altered in expression ≥1.5 fold and P < 0.05. Of the 501 probes, 144 had unknown gene annotations and the remaining 357 probes represented 261 unique genes (multiple probes associated with the same gene). Of the 261 unique genes, 98 were increased in expression.
Table S5. Results of the mRNA expression determined by Q-RT-PCR (with corresponding microarray results shown).