Abstract
TIR domain-containing adaptor-inducing interferon-β (TRIF), a cytosolic adaptor protein, plays a key role in the mammalian toll-like receptor-mediated signaling pathway. However, the role of TRIF in large yellow croaker (LcTRIF) remains poorly understood. The main objective of this study was to explore the role of LcTRIF in triggering antiviral immune responses and the potential function of LcTRIF in regulating lipid metabolism. In the present study, the full-length coding sequence of TRIF from large yellow croaker was cloned and characterized. In vivo, upon poly (I:C) stimulation, the transcriptional levels of LcTRIF were rapidly elevated in immune-related tissues at the early stage of injection. In vitro, the MRNA expression of LcTRIF was significantly but not dramatically upregulated in macrophages treated with poly (I:C). Activation of LcTRIF by poly (I:C) significantly increased the transcription of genes involved in inflammatory responses, and this induction was blocked by knockdown of LcTRIF. Moreover, the ability of LcTRIF to induce inflammatory responses may partially be attributed to the promotion of mRNA expression of IFNh and NF-κB pathway genes. In addition, activation of the LcTRIF-mediated pathway inhibited the increase in hepatic stearoyl-coenzyme A (CoA) desaturase 1 induced by palmitic acid and subsequently alleviated lipid accumulation in hepatocytes. These results revealed the crucial role of LcTRIF in triggering antiviral immune responses and the unconventional metabolic function of LcTRIF in regulating hepatic lipogenesis in large yellow croaker.
Keywords: Larimichthys crocea, TRIF, poly (I:C), antiviral immune responses, hepatic lipogenesis
Introduction
Toll-like receptors (TLRs), one of the most extensively studied pattern-recognition receptors (PRRs), play a crucial role in both the innate immune system and the adaptive immune system by recognizing conserved components of pathogens referred to as pathogen associated molecular patterns (PAMPs) (1, 2). Upon stimulation with PAMPs, TLRs selectively recruit distinct TIR domain-containing adaptor molecules and induce specific immune responses to kill invading pathogens (3, 4). To date, six adaptors have been identified in mammals, including myeloid differentiation primary response gene 88 (MyD88), TIR domain-containing adaptor-inducing interferon-β (TRIF), TIR domain-containing adaptor protein (TIRAP), TRIF-related adaptor molecule (TRAM), sterile α and armadillo motif-containing protein 1 (SARM1), and B cell adaptor for phosphoinositide 3-kinase (BCAP).
TRIF, also known as TIR domain-containing adapter molecule 1 (TICAM-1), is unique to TLR3-and TLR4-mediated signaling pathways, which activate interferon (IFN) regulatory factors 3/7 (IRF3/7) and nuclear factor-kappa B (NF-κB) and induce the production of type I IFN and inflammatory cytokines, leading to direct killing of invading pathogens (5–7). An increasing number of studies have recognized that apart from its typical role in triggering antiviral immune responses, the TRIF-dependent TLR pathway also plays a distinct role in lipid metabolism that is associated with various metabolic diseases in metabolic cells (8–10). In mammals, TRIF possesses proline-rich domains on the N- and C-terminal regions and a highly conserved TIR domain. Each domain plays a distinct role in signal transduction. The N-terminal domain contains a binding site for tumor necrosis factor receptor-associated factor (TRAF) proteins, which is crucial for IRF3 activation (6, 11). The C-terminal domain harbors receptor-interacting protein 1 (RIP1) interaction motif (RHIM), which is crucial for NF-κB activation and apoptosis (12–14). The TIR domain of TRIF is responsible for interacting with the TIR domain of TLR3 as well as the TLR4–bridging adaptor TRAM (5, 7, 15). However, in teleosts, TRIF lacks both the N-terminal and the C-terminal proline-rich domains and the proline in box2 of the TIR domain, which suggests that fish TRIF activates type I IFN and NF-κB through different ways with its mammalian TRIF orthologs (16–19). These findings revealed a real difference in TRIF-mediated antiviral immune responses between teleosts and mammals. To date, TRIF homologs have been identified in several fish species, and their roles in antiviral immune responses have been investigated (16, 18–20). However, the unconventional function of the TRIF-mediated pathway in regulating lipid metabolism has not been reported in fish.
Large yellow croaker (Larimichthys crocea) is an economically important mariculture fish species in China. The production of this fish species was 177640 t in China in 2017, which has continued to hold first place in the mariculture output for several years (21). A high-fat diet (HFD) has been commonly used in the culture of large yellow croaker owing to its protein sparing effect. However, long-term intake of HFD often leads to abnormal lipid accumulation accompanied by low-grade, chronic inflammation (22, 23), which is similar to the hepatic steatosis observed in mammals, and results in a high mortality rate and diminishes the health benefits of fish for human consumption. In the present study, our main objective was to explore the role of LcTRIF in triggering antiviral immune responses and the potential function of LcTRIF in regulating lipid metabolism. Understanding the functions of TRIF in large yellow croaker may contribute to the development of management strategies for defense against virus infections, and alleviate the abnormal lipid accumulation and inflammation induced by a HFD.
Materials and Methods
Fish, Challenge, and Sample Collection
Juveniles of large yellow croaker, Larimichthys crocea (50.6 g ± 7.8 g) obtained from Ningbo, China, were reared in a circulating seawater system at 24–28°C. After 1 week of acclimation, the fish were used for the induction experiments. Fish were injected intraperitoneally with 0.25 mL of 1 mg/mL polyinosinic: polycytidylic acid [poly (I:C), Sigma, USA] or 0.25 mL of phosphate buffered saline (PBS, pH 7.4, Gibco). Tissues including the gill, head kidney, liver and spleen were collected from four fish at different time points (0, 6, 12, 24, and 48 h), and frozen immediately in liquid nitrogen, and then stored at −80°C for further analysis. Husbandry and handling of fish in the present study were performed strictly according to the Management Rule of Laboratory Animals (Chinese Order No. 676 of the State Council, revised 1 March 2017) and the Guidelines for the Use of Fishes in Research (24).
Cells Culture and Reagents
Macrophages were isolated from the head kidney of large yellow croaker by using Percoll (Pharmacia, USA) density gradients according to our previous report (25). Macrophages were seeded into 6-well plates at a density of 2 × 106 cells/mL and were cultured in 95% DMEM/F12 medium (BI, Israel) supplemented with 5% FBS (BI, Israel) and antibiotics (Gibco, USA) in a 5% CO2 atmosphere at 26°C. After 24 h of cultivation, macrophages were washed with PBS, transfected with or without small interfering RNAs (siRNAs) for 48 h, and then incubated with 5 μg/mL poly (I:C) for 2 h on the basis of the inflammatory macrophage model previously established by ourselves (26).
Primary hepatocytes of large yellow croaker were isolated and cultivated based on our previous report (27). Hepatocytes were seeded into 6-well plates at a density of 2 × 106 cells/mL and were maintained in DMEM/F12 medium containing 15% FBS and antibiotics in a 5% CO2 atmosphere at 28°C. After 2 d of cultivation, the cells were washed with PBS and incubated with FBS free medium prior to treatment with or without 800 μM palmitic acid (PA) conjugated with fatty acid free BSA (Equitech-Bio, USA) and 5 μg/mL poly (I:C) for 6 h (for gene expression) or 12 h (for lipid droplet visualization and TAG quantification).
HEK293T cells were cultured in 90% DMEM, high glucose medium (BI, Israel) supplemented with 10% FBS and antibiotics in a 5% CO2 atmosphere at 37°C.
Cloning and Sequence Analysis of the LcTRIF Gene
Primers for the amplification of partial cDNA were designed according to the large yellow croaker genomic sequence and the predicted sequence of LcTRIF (Supplementary Table 1). RACE primers were designed based on the partial LcTRIF cDNA sequence to clone the 3' and 5'-end sequences through nest-PCR using the SMARTerTM RACE cDNA Amplification Kit (Clontech, USA) (Supplementary Table 1). The full length of LcTRIF was assembled by overlapping the 5'-end sequence, partial cDNA sequence and 3'-end sequence and then was confirmed by sequencing the PCR product amplified by primers TRIF-FL-F and TRIF-FL-R (Supplementary Table 1). The nucleotide and deduced amino acid sequences of LcTRIF were analyzed by DNAMAN. Sequence similarity analysis was conducted using the BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The prediction of the protein domain was performed by SMART (http://smart.embl-heidelberg.de/). Multiple alignments of amino acid sequences from different species were analyzed using DNAMAN, and the phylogenetic tree was constructed by the neighbor-joining method in MEGA.
Quantification of Gene Expression
Total RNA extraction, cDNA synthesis and quantitative real-time PCR (qRT-PCR) were carried out according to our previous report (28). The primer sequences for TRIF, stearoyl–coenzyme A (CoA) desaturase 1 (SCD1), fatty acid synthase (FAS), sterol-regulatory element binding protein 1 (SREBP1), diacylgycerol acyltransferase 2 (DGAT2), acetyl-CoA carboxylase 1 (ACC1), IFNh, IRF3, interleukin 1β (IL-1β), tumor necrosis factor α (TNFα), and β-actin are listed in Supplementary Table 2. β-actin, GAPDH, 18S rRNA, and ubiquitin were selected to test their suitability for the normalization of gene expression levels in various tissues of large yellow croaker. NormFinder algorithms and geNorm were further used to verify the stability and suitability of these genes (29, 30). No significant differences in β-actin expression were detected among all treatments, suggesting that β-actin could be used as a reference gene in the present study. The mRNA expression of each gene was normalized to that of β-actin using the 2−ΔΔt method (31).
Plasmid Construction
PCR was performed with primers listed in Supplementary Table 3 for various plasmid construction. In detail, for the expression plasmid, the coding sequence (CDS) of LcTRIF was cloned from large yellow croaker cDNA, the 5′ and 3′ ends homologous arms of the pCS2+ vector (5′ ends: 5′-CGATTCGAATTCAAGGCCTCTCGAG- 3′; 3′ ends: 5′-CTCACTATAGTTCTAGAGGCTCGAG- 3′) designed by TSINGKE Biotech Co., Ltd. (Qingdao, China) were added to the LcTRIF CDS by PCR. A linearized pCS2+ vector was obtained by digestion with QuickCut™ XhoI (Takara, Japan). Then, the CDS of LcTRIF was inserted into the XhoI site of the pCS2+ vector by homologous recombination using the ClonExpress® II One Step Cloning Kit (Vazyme Biotech Co., Ltd, Nanjing, China) to construct the pCS2-TRIF expression plasmid. For reporter plasmids, the IFNh promoter (1,324 bp genomic fragment, GenBank Accession No: KU144879.1), NF-κB response promoter (2,208 bp genomic fragment, GenBank Accession No: NM_001303377.1) and SCD1 promoter (2,911 bp genomic fragment, GenBank Accession No: KP202156.1) were cloned, the 5' and 3' ends homologous arms of the pGL3 Basic vector (5′ ends: 5′-TCTTACGCGTGCTAGCCCGGGC- 3′; 3′ ends: 5′-GCCAAGCTTACTTAGATCGCAGATC- 3') were added to the promoter sequences by PCR. Then, the promoters of IFNh, NF-κB and SCD1 were each subcloned into the XhoI site of the linearized pGL3-basic vector to construct the pGL3-IFNh, pGL3-NF-κB, and pGL3-SCD1 plasmids, respectively. For the subcellular localization studies, truncated forms of TRIF including the N terminus (1–350 aa), the TIR domain (351–471 aa), the C terminus (472–601 aa), ΔC (1–471 aa), and ΔN (351–601 aa) were subcloned into pcDNA3.1-EGFP vector. Plasmids used for transfection in the present study were purified using the EasyPure HiPure Plasmid MiniPrep Kit (TransGen Biotech Co., Ltd, Beijing, China) and were tested by sequencing at Sangon Biotech Co., Ltd. (Shanghai, China).
RNA Interference
To knockdown the expression of LcTRIF, an RNA interference assay was conducted by transfecting siRNAs targeting the LcTRIF mRNA. Four siRNA sequences (Supplementary Table 4) were designed and synthesized by GenePharma (Shanghai, China). Macrophages were mock-transfected or transfected with siRNAs using Lipofectamine® RNAiMAX Transfection Reagent (Invitrogen, USA) for 48 h according to the manufacturer's instructions, and the knockdown efficiencies of the siRNAs were examined by qRT-PCR (Supplementary Figure 1).
Dual-Luciferase Reporter Assays
HEK293T cells (5 × 105 cells/mL) were cultured in 24-well plates overnight and transiently cotransfected with the IFNh promoter luciferase reporter plasmid, NF-κB response promoter luciferase reporter plasmid or SCD1 promoter luciferase reporter plasmid, together with the LcTRIF expression plasmid using Lipofectamine 3000 Reagent (Invitrogen, USA). After 24 h of transfection, the cells were stimulated with or without poly (I:C). After 24 h incubation, cells were lysed and analyzed for luciferase activity by the Dual-Luciferase Reporter Assay System (TransGen Biotech Co., Ltd, Beijing, China).
BODIPY 493/503 Staining and Triglyceride (TAG) Content Quantification
Hepatocytes isolated from large yellow croaker were fixed with 4% paraformaldehyde (Solarbo, China) and incubated with BODIPY 493/503 (Invitrogen, USA) in the dark for 15 min. Then, lipid droplets were visualized by fluorescence microscopy (Leica, Germany). TAG content in the hepatocytes were quantified by a TAG Assay Kit according to the manufacturer's instructions (Applygen Technologies Inc., Beijing, China).
Confocal Laser Microscopy Imaging
For the localization of LcTRIF, HEK293T cells (1.0 × 105 cells/well) were seeded into coverslips in 24-well plates. The following day, cells were transiently transfected with pcDNA3.1-EGFP-TRIF vector or various truncation vectors using Lipofectamine 3000 Reagent (Invitrogen, USA). After 24 h, the cells were fixed with 4% paraformaldehyde and stained with DAPI (Solarbio, China) in the dark for 10 min. Thereafter, cells on coverslips were mounted onto a slide glass using PBS containing 2.3% 1,4-diazabicyclo [2.2.2]octane (DABCO) and 50% glycerol. Cells were visualized at × 60 magnification under an NIKON A1+ Confocal Laser Microscope (NIKON, Japan).
Statistical Analysis
SPSS 20.0 was used to perform the statistical analysis. Values are presented as mean ± SD of three independent experiments with 4 technical replicates for each experiment. Data were processed using one-way ANOVA, followed by Tukey's Test. A value of P < 0.05 was considered statistically significant.
Results
Identification of the TRIF Gene in Large Yellow Croaker
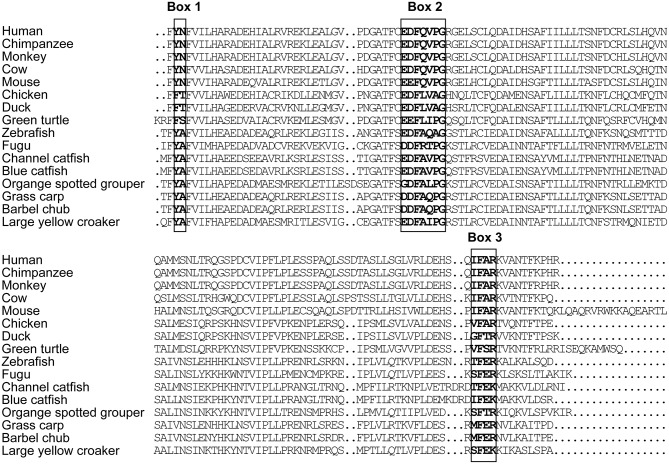
The full-length cDNA of LcTRIF (GenBank Accession No: MH820380.1) was 3,012 bp and contained a 5′ untranslated terminal region (UTR) of 438 bp, a 3′ UTR of 768 bp, and an open reading frame (ORF) of 1,806 bp encoding a polypeptide of 601 amino acids (aa), which exhibited the typical characteristics of a TIR domain (351–471 aa) close to its C terminus. Two polyadenylation signals (AATAAA) were located 16 and 584 bp upstream of the poly(A) tail, respectively, and two instability signals (ATTTA) were found in the 3′ UTR (Figure 1). Multiple sequence alignment of the TIR domain of large yellow croaker TRIF with that of other species found that LcTRIF had three highly conserved regions: box 1 (YN), box 2 (EDFQVPG), and box 3 (IFAR) (Figure 2). The phylogenetic tree constructed based on the TIR domain of TRIF from different species showed that fish TRIF members formed an independent cluster, and LcTRIF was genetically closest to fugu TRIF (Figure 3).
Figure 1.
Nucleotide and deduced amino acid sequences of TRIF in large yellow croaker. The TIR domain is underlined. The two poly(A) signals (AATAAA) are highlighted in bold, and the two instability signals (ATTTA) are marked by boxes.
Figure 2.
Alignment of TRIF TIR domain sequences from human (accession number NP_891549.1), chimpanzee (accession number NP_001123604.1), monkey (accession number AAS20428.1), cow (accession number NP_001025472.1), mouse (accession number AAH33406.1), chicken (accession number NP_001074975.1), duck (accession number NP_001297720.1), green turtle (accession number XP_007072279.1), zebrafish (accession number ABP04048.1), fugu (accession number NP_001106665.1), channel catfish (accession number NP_001187154.1), blue catfish (accession number ABH10662.1), orange spotted grouper (accession number AEX01719.1), grass carp (accession number AGW25589.1), and barbell chub (accession number AMP81962.1).
Figure 3.
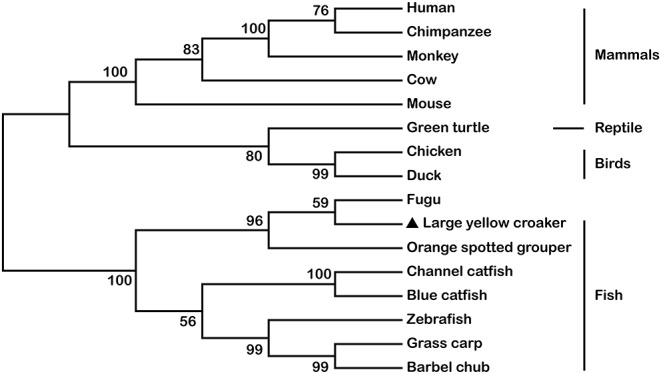
Phylogenetic tree based on the sequences of TRIF TIR domains from mammals, reptiles, birds, and other fish species. The accession numbers for the TIR domains of TRIF are shown in Figure 2.
Modulation of LcTRIF mRNA Expression in Response to Poly (I:C) Stimulation in vivo
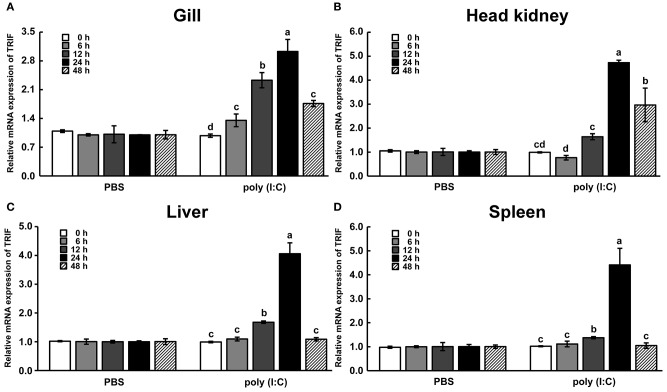
To investigate the temporal expression of LcTRIF in response to poly (I:C) (a dsRNA analog) stimulation, the mRNA levels of LcTRIF in immune-related tissues (gill, head kidney, liver, and spleen) were detected at different time points post injection (0, 6, 12, 24, and 48 h). In the gill, the mRNA levels of LcTRIF were significantly elevated at 6 h and reached maximum levels at 24 h (P < 0.05). In the head kidney, liver, and spleen, LcTRIF mRNA expression was obviously upregulated by poly (I:C) stimulation at 12 h and reached maximum levels at 24 h (P < 0.05) (Figure 4). No significant differences in LcTRIF mRNA expression were found in those tissues at different time points after injection with PBS (P > 0.05).
Figure 4.
LcTRIF mRNA expression in immune-related tissues: gill (A), head kidney (B), liver (C), and spleen (D) at different time points post poly (I:C) stimulation in large yellow croaker. Values (mean ± SD of three independent experiments with 4 technical replicates each) in bars that have the same superscript letter are not significantly different (P > 0.05, Tukey's test).
Activation of LcTRIF by Poly (I:C) Induced Inflammatory Response in vitro
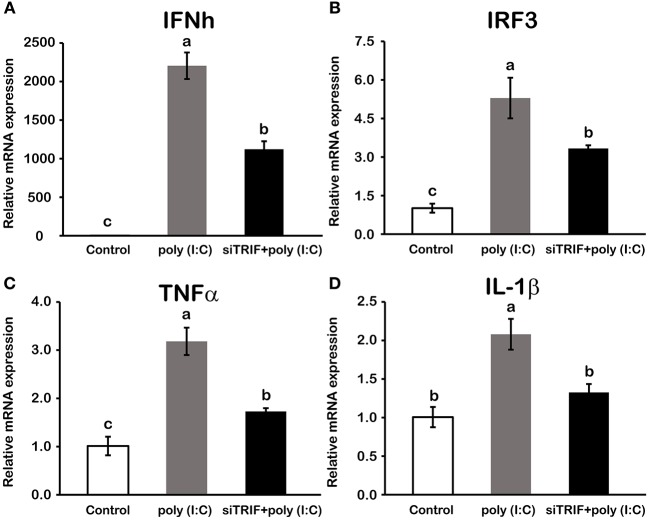
In macrophages, mRNA expression of LcTRIF was rapidly increased in response to poly (I:C) stimulation at 2 h (P < 0.05), then gradually decreased over time, and returned to the control level at 12 h (Figure 5). In addition, activation of LcTRIF by poly (I:C) significantly elevated the transcription of genes involved in inflammatory responses, including IFNh, IRF3, TNFα, and IL-1β (P < 0.05), and the ability of poly (I:C) to induce the expression of these genes in macrophages was blocked by knockdown of LcTRIF with siRNA (P < 0.05) (Figure 6).
Figure 5.
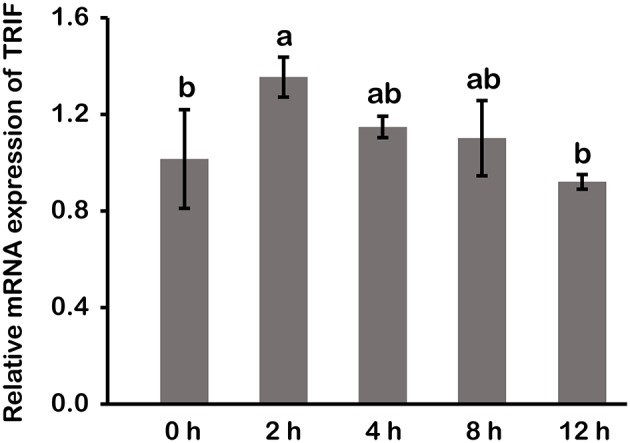
mRNA expression of LcTRIF in macrophages of large yellow croaker at different time points after poly (I:C) stimulation. Values (mean ± SD of three independent experiments with 4 technical replicates each) in bars that have the same superscript letter are not significantly different (P > 0.05, Tukey's test).
Figure 6.
Activation of LcTRIF by poly (I:C) induced the mRNA expression of genes involved in the macrophage inflammatory response of large yellow croaker. Macrophages cultured in 6-well plates overnight were transfected with or without siTRIF and control siRNA. After 48 h, cells were stimulated with 5 μg/mL poly (I:C) for 2 h. The mRNA expression of IFNh (A), IRF3 (B), TNFα (C), and IL-1β (D) were examined by qRT-PCR. Values (mean ± SD of three independent experiments with 4 technical replicates each) in bars that have the same superscript letter are not significantly different (P > 0.05, Tukey's test). IFNh, interferon h; IRF3, interferon regulatory factors 3; TNFα, tumor necrosis factor α; IL-1β, interleukin 1β.
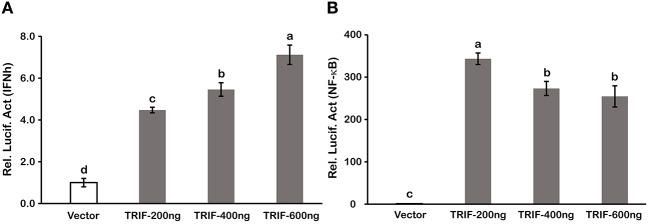
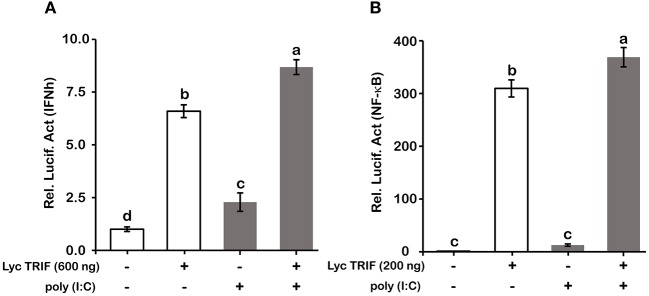
Type I IFN and NF-κB are commonly regarded as important inflammatory factors involved in virus-induced innate immune responses. To further elucidate the mechanisms of LcTRIF in inducing the inflammatory response, a luciferase reporter assay was carried out to test the promoter activities of IFNh and NF-κB in response to LcTRIF overexpression. The results showed that IFNh promoter activity was significantly enhanced in HEK293T cells transfected with 200 ng LcTRIF plasmid, and this enhancement increased with the increasing dose of LcTRIF plasmid, with promoter activity reaching a peak at the 600 ng dose (Figure 7A). However, LcTRIF overexpression induced the activity of the NF-κB response promoter in a different way, whereby the NF-κB promoter activity was inhibited after initial enhancement by 200 ng of LcTRIF plasmid (Figure 7B). Moreover, the enhancement of IFNh and NF-κB promoter activities by LcTRIF overexpression was more pronounced in the case of poly (I:C) stimulation (Figure 8). These results indicated that LcTRIF plays an important role in the immune responses triggered by dsRNA virus infections in macrophages of large yellow croaker.
Figure 7.
LcTRIF overexpression induced promoter activities of IFNh (A) and NF-κB (B). HEK293T cells cultured in 24-well plates overnight were cotransfected with PCS2 empty vector or increasing amounts (200, 400, and 600 ng) of TRIF expression vector together with 200 ng of IFNh or NF-κB promoter luciferase reporter vector and 20 ng of pRL-TK. Twenty-four hours later, the luciferase activities were measured. Values (mean ± SD of three independent experiments with 4 technical replicates each) in bars that have the same superscript letter are not significantly different (P > 0.05, Tukey's test).
Figure 8.
Poly (I:C) stimulation enhanced the promoter activities of IFNh (A) and NF-κB (B) upon LcTRIF overexpression. HEK293T cells cultured in 24-well plates overnight were cotransfected with TRIF expression vector together with the IFNh or NF-κB promoter luciferase reporter vector and 20 ng of pRL-TK. Twenty-four hours later, the cells were stimulated with poly (I:C). The luciferase activities were measured after 24 h post stimulation. Values (mean ± SD of three independent experiments with 4 technical replicates each) in bars that have the same superscript letter are not significantly different (P > 0.05, Tukey's test).
Activation of LcTRIF by Poly (I:C) Inhibited PA-Induced Lipid Accumulation in Hepatocytes
The TRIF-dependent TLR pathway is generally believed to play a crucial role in regulating the inflammatory response in immune cells. Recently, increasing number of studies have suggested that the TRIF-dependent TLR pathway also plays a distinct role in lipid metabolism in metabolic cells (8–10). To investigate the specific role of LcTRIF in lipid metabolism, hepatocytes from large yellow croaker were incubated with or without PA and poly (I:C). BODIPY 493/503 staining showed that more lipid droplets accumulated in the hepatocytes incubated with PA than in the control group, whereas activation of the TRIF-mediated pathway by poly (I:C) inhibited the hepatic lipid accumulation induced by PA (Figure 9A). Quantification of extracted hepatic lipids showed that poly (I:C) stimulation significantly suppressed the PA-induced increase in TAG content (Figure 9B) (P < 0.05).
Figure 9.
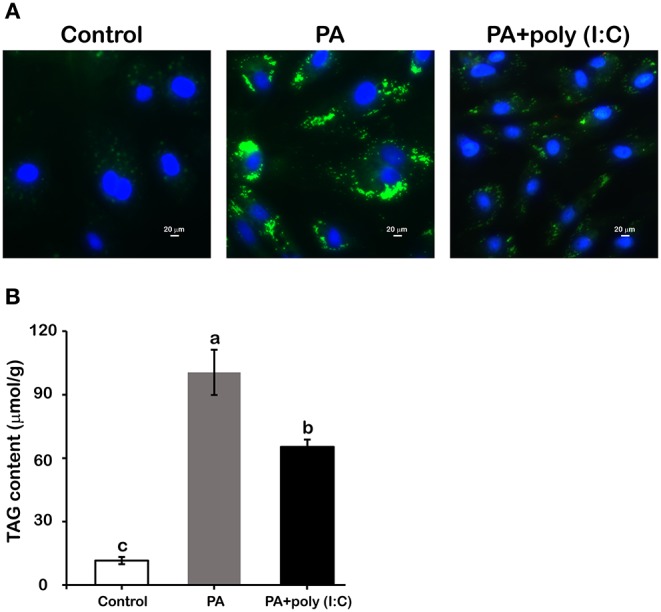
Activation of LcTRIF by poly (I:C) inhibited palmitic acid (PA)-induced hepatic lipid accumulation. Hepatocytes from large yellow croaker were incubated with or without PA (800 μM) and poly (I:C) (5 μg/mL) for 12 h. BODIPY 493/503 staining (A) and hepatic triglyceride content quantification (B) were performed. Values (mean ± SD of three independent experiments with 4 technical replicates each) in bars that have the same superscript letter are not significantly different (P > 0.05, Tukey's test).
LcTRIF Inhibited PA-Induced Hepatic Lipid Accumulation by Suppressing SCD1 Expression
To further investigate the mechanism of how LcTRIF affected hepatic lipid accumulation in large yellow croaker, the expression of genes involved in lipogenesis was detected. The results showed that PA incubation significantly increased SCD1 expression in hepatocytes, and the activation of LcTRIF by poly (I:C) attenuated this induction (P < 0.05). However, PA and/or poly (I:C) treatment had no effect on the expression of other lipogenesis-related genes, such as ACC1, SREBP1, FAS, or DGAT2 (P > 0.05) (Figure 10). Since LcTRIF inhibited SCD1 expression, we next examined whether this effect occurred at the promoter region of SCD1. The results from the luciferase report assay demonstrated that overexpression of LcTRIF in HEK293T cells significantly inhibited the promoter activity of SCD1, and this inhibition was more pronounced in poly (I:C) stimulation condition (Figure 11). These results indicated that the inhibition of hepatic lipid accumulation by LcTRIF might be partially attributed to the suppression of SCD1 expression.
Figure 10.
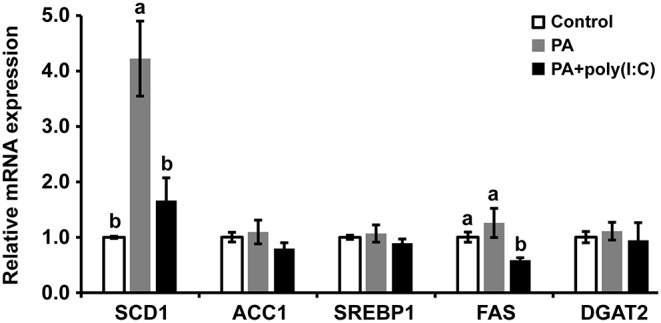
Activation of LcTRIF by poly (I:C) inhibited palmitic acid (PA)-induced hepatic SCD1 mRNA expression. Hepatocytes from large yellow croaker were incubated with or without PA (800 μM) and poly (I:C) (5 μg/mL) for 6 h, and the mRNA expression of hepatic SCD1, ACC1, SREBP-1, FAS, and DGAT2 was examined by qRT-PCR. Values (mean ± SD of three independent experiments with 4 technical replicates each) in bars that have the same superscript letter are not significantly different (P > 0.05, Tukey's test). ACC1, Acetyl-CoA carboxylase 1; DGAT2, Diacylgycerol acyltransferase 2; FAS, Fatty acid synthase; SCD1, Stearoyl–coenzyme A (CoA) desaturase 1, SREBP1, Sterol-regulatory element binding protein 1.
Figure 11.
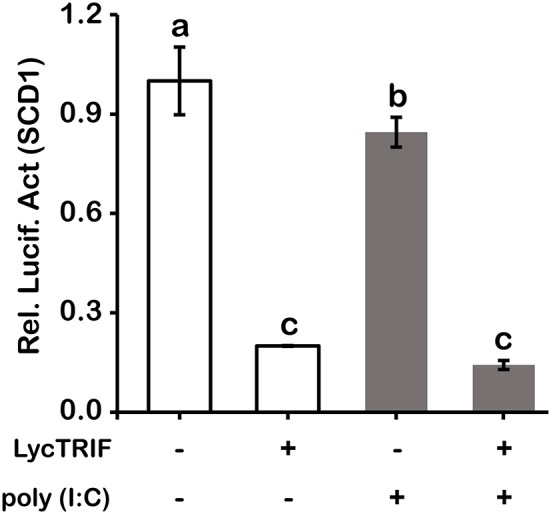
Overexpression of LcTRIF inhibited the promoter activity of SCD1. HEK293T cells cultured in 24-well plates overnight were cotransfected with the TRIF expression vector together with the SCD1 promoter luciferase reporter vector and 20 ng of pRL-TK. Twenty-four hours later, the cells were treated with or without poly (I:C) stimulation. The luciferase activities were measured after 24 h post stimulation. Values (mean ± SD of three independent experiments with 4 technical replicates each) in bars that have the same superscript letter are not significantly different (P > 0.05, Tukey's test).
LcTRIF Was Distributed in the Cytoplasm
The subcellular localization of LcTRIF was predicted by online software (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/), which indicated that LcTRIF was localized in the cytoplasm. To further investigate whether LcTRIF was localized in the cytoplasm, HEK293T cells were transfected with the pcDNA3.1-EGFP-TRIF fusion vector and then visualized by confocal microscopy. The results shown that LcTRIF was diffusely present in the cytoplasm. However, the truncated LcTRIF segments had different localizations. The truncated segments with only the C terminus or the TIR domain was distributed in the entire cell. The segment spanning the C terminus and the TIR domain (ΔN) was localized in the nucleus. The truncated segment with only the N terminus or the segment spanning the N terminus and the TIR domain (ΔC) was exclusively distributed in the cytoplasm, which showed the same subcellular localization as the full length protein (Figure 12). These results revealed that large yellow croaker TRIF was a cytoplasm-localized protein and that the N terminal sequence might contribute to its unique subcellular localization.
Figure 12.
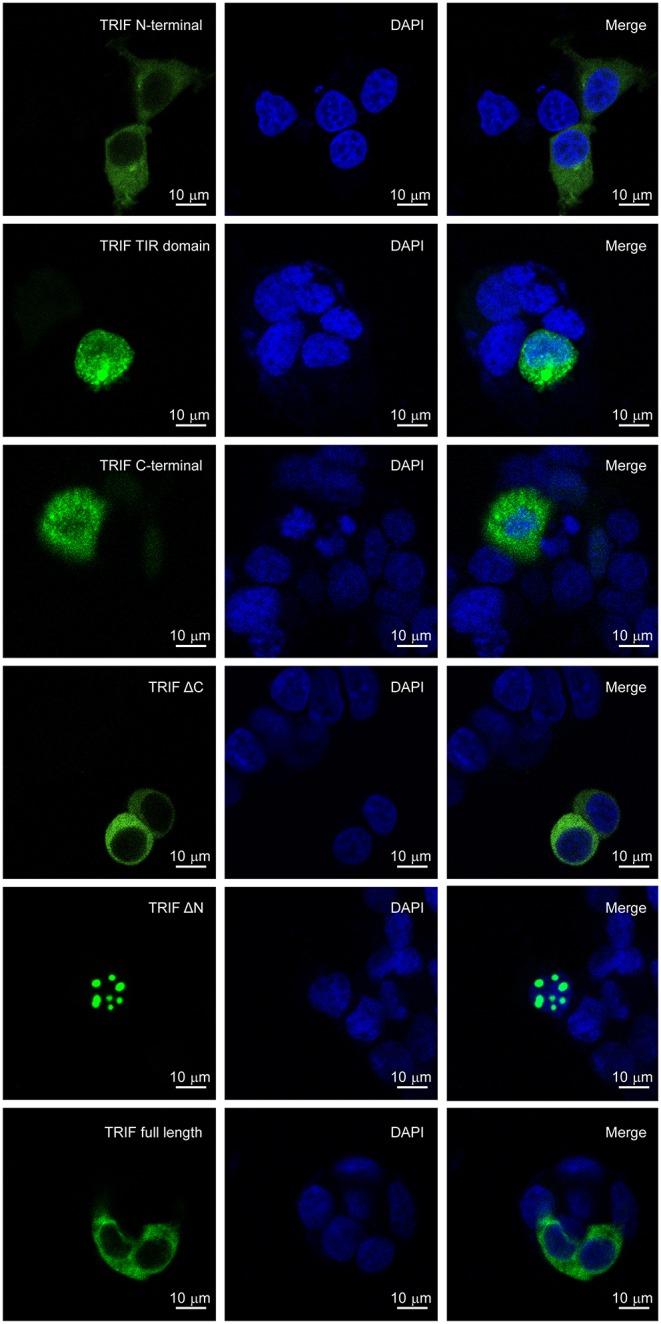
Localization of truncated forms of LcTRIF. Five truncated forms of pcDNA3.1-EGFP-TRIF fusion vectors including the TRIF N terminus (1–350 aa), TIR domain (351–471 aa), C terminus (472–601 aa), ΔC (1–471 aa), ΔN (351–601 aa), or TRIF (1–601 aa) were transfected into the HEK293T cells, respectively. After 24 h, the cells were stained with DAPI and then visualized by confocal laser microscopy.
Discussion
In the current study, the TRIF gene of large yellow croaker was cloned and characterized and was shown to share 35.30% identity with its counterpart in zebrafish. The deduced amino acid sequence of LcTRIF displayed a typical TIR domain that was highly conserved with the TIR domains of other fish species. All the teleost TRIF clustered together and separated from those of birds, reptiles, and mammals in a phylogenetic tree constructed based on the TIR domains. Compared with mammalian homologs, large yellow croaker TRIF contained proline in box 2 of the TIR domain, which is conserved in mammalian TRIF, TIRAP, and MyD88 and is essential for TRIF- and MyD88-mediated signal transduction (5, 32). However, the proline-rich domains close to the N terminus and C terminus in mammalian TRIF were not found in large yellow croaker TRIF. The lack of N-terminal and C-terminal proline-rich domains has also been reported for the zebrafish TRIF and orange spotted grouper TRIF (18, 19). Previous studies have demonstrated that TLRs and its adaptors had specific localization in the cells for their functions performed (18, 33, 34). To determine which sequence motif of LcTRIF is responsible for its localization, TRIF-EGFP fusion vectors and its truncated forms were constructed. The confocal laser microscopy analysis showed that large yellow croaker TRIF was uniformly distributed over the cytoplasm, and the N terminal sequences might contribute to its subcellular localization.
In vivo, upon poly (I:C) stimulation, the mRNA expression of LcTRIF was significantly upregulated in immune-related tissues at the early stage of injection. In vitro, the mRNA expression of LcTRIF was significantly but not dramatically upregulated in macrophages treated with poly (I:C). A similar expression pattern of TRIF in response to poly (I:C) stimulation has also been reported in kidney cells of grass carp (20). The activation of LcTRIF by poly (I:C) significantly elevated the mRNA levels of genes involved in inflammatory responses, and LcTRIF knockdown suppressed the poly (I:C) induced increase in the expression of these genes, which indicated that LcTRIF plays an important role in the immune responses triggered by dsRNA virus infections in macrophages of large yellow croaker. In addition, the results from the luciferase reporter assay showed that the promoter activities of IFNh and NF-κB were significantly increased in LcTRIF overexpression cells, and this increase was more pronounced in the case of poly (I:C) stimulation, which illustrated that LcTRIF triggers both the IFNh and NF-κB pathways in response to virus infections. These results agreed well with previous studies in mammals (5, 15) and fish (33, 34) demonstrating that TRIF-dependent pathways activate both type I IFN and NF-κB responses to protect the host from virus infections. Moreover, LcTRIF induced IFNh and NF-κB promoter activities in two different ways, which suggested that there was a negative feedback effect of the large yellow croaker TRIF-mediated NF-κB pathway.
Of particular interest was the distinct role of LcTRIF in hepatic lipogenesis of large yellow croaker. To date, TRIF homologs have been identified in several fish species, and their functions in antiviral immune responses have been investigated (16, 18–20). However, the potential role of TRIF in the lipid metabolism of fish has not been reported. In the present study, the activation of LcTRIF by poly (I:C) suppressed PA-induced hepatic lipid accumulation, and the inhibitory effects of LcTRIF on lipid accumulation might be due to the suppression of SCD1 expression. SCD1 is a key enzyme in lipogenesis, and catalyzes the formation of mono-unsaturated fatty acids (MUFA) from saturated fatty acids (SFA). MUFA are major substrates for TAG biosynthesis. Studies have showed that SCD1-deficient mice have significant lower hepatic TAG content than wild-type mice (35–38). The expression of SCD1 is generally regulated by several key metabolic transcription factors, including peroxisome proliferator-activated receptor α, sterol regulatory element-binding protein transcription factor 1c and liver X receptor (39, 40). Apart from these metabolic transcription factors, IRF3, a downstream transcription factor of TRIF, also plays a pivotal role in regulating SCD1 expression at the transcriptional level. Activation of TRIF by poly (I:C) induces phosphorylation of IRF3, which directly binds to the SCD1 promoter and blocks its transcription. The attenuation of SCD1 expression ameliorates hepatic lipid accumulation (10, 41). It has been speculated that the inhibitory effects of LcTRIF on SCD1 promoter activity in large yellow croaker is also mediated by IRF3. Further studies will be performed to verify this speculation. Moreover, the suppression of SCD1 expression by the LcTRIF-mediated pathway may also be considered an immune response, which limits viral assembly by counteracting virus-hijacked host lipogenesis (42, 43).
In summary, this study reported the identification and characterization of the TRIF gene from large yellow croaker, which had certain unique characteristics compared with its mammalian homologs. Activation of LcTRIF by poly (I:C) induced inflammatory response by promoting activation of the IFNh and NF-κB pathways in macrophages. In addition, LcTRIF activation inhibited PA-induced hepatic lipid accumulation by suppressing the SCD1 expression, suggesting a potential role of the TRIF-mediated signaling pathway in regulating lipogenesis in non-immune cells of fish. These results may contribute to the development of management strategies for defense against virus infections and alleviate abnormal lipid accumulation and inflammation induced by the use of HFD in teleosts.
Data Availability Statement
All datasets generated for this study are included in the manuscript/Supplementary Files.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Ocean University of China.
Author Contributions
KM, QA, and SZ designed the research. SZ and XXu conducted the research. SZ and XXi analyzed the data. SZ wrote the paper. SG provided language help. All authors reviewed and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Qiangde Liu for technical assistance in confocal laser microscopy imaging; Qingfei Li and Peng Tan for their valuable suggestions and careful reviews of this manuscript.
Footnotes
Funding. This work was supported by the National Science Fund for Distinguished Young Scholars of China (Grant no. 31525024) and the Agriculture Research System of China (Grant no. CARS-47-11).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02506/full#supplementary-material
References
- 1.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. (2001) 2:675–80. 10.1038/90609 [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. (2002) 20:197–216. 10.1146/annurev.immunol.20.083001.084359 [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. (2006) 124:783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- 4.O'Neill LAJ, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. (2007) 7:353–64. 10.1038/nri2079 [DOI] [PubMed] [Google Scholar]
- 5.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3–mediated interferon-β induction. Nat Immunol. (2003) 4:161–7. 10.1038/ni886 [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, et al. LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the toll adapters TRAM and TRIF. J Exp Med. (2003) 198:1043–55. 10.1084/jem.20031023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seya T, Oshiumi H, Sasai M, Akazawa T, Matsumoto M. TICAM-1 and TICAM-2: toll-like receptor adapters that participate in induction of type 1 interferons. Int J Biochem Cell B. (2005) 37:524–9. 10.1016/j.biocel.2004.07.018 [DOI] [PubMed] [Google Scholar]
- 8.Sharifnia T, Antoun J, Verriere TGC, Suarez G, Wattacheril J, Wilson K, et al. Hepatic TLR4 signaling in obese NAFLD. Am J Physiol Gastrointest Liver Physiol. (2015) 309:G270–8. 10.1152/ajpgi.00304.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, Miura K, Zhang B, Matsushita H, Yang YM, Liang S, et al. TRIF differentially regulates hepatic steatosis and inflammation/fibrosis in mice. Cell Mol Gastroenterol Hepatol. (2017) 3:469–83. 10.1016/j.jcmgh.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Li J, Yiu JHC, Lam JKW, Wong CM, Dorweiler B, et al. TRIF-dependent Toll-like receptor signaling suppresses Scd1 transcription in hepatocytes and prevents diet-induced hepatic steatosis. Sci Signal. (2017) 10:eaal3336. 10.1126/scisignal.aal3336 [DOI] [PubMed] [Google Scholar]
- 11.Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, et al. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. (2003) 171:4304–10. 10.4049/jimmunol.171.8.4304 [DOI] [PubMed] [Google Scholar]
- 12.Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, et al. RIP1 is an essential mediator of Toll-like receptor 3–induced NF-κB activation. Nat Immunol. (2004) 5:503–7. 10.1038/ni1061 [DOI] [PubMed] [Google Scholar]
- 13.Han KJ, Su X, Xu LG, Bin LH, Zhang J, Shu HB. Mechanisms of the TRIF-induced interferon-stimulated response element and NF-κB activation and apoptosis pathways. J Biol Chem. (2004) 279:15652–61. 10.1074/jbc.M311629200 [DOI] [PubMed] [Google Scholar]
- 14.Kaiser WJ, Offermann MK. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J Immunol. (2005) 174:4942–52. 10.4049/jimmunol.174.8.4942 [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, et al. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-β promoter in the Toll-like receptor signaling. J Immunol. (2002) 169:6668–72. 10.4049/jimmunol.169.12.6668 [DOI] [PubMed] [Google Scholar]
- 16.Baoprasertkul P, Peatman E, Somridhivej B, Liu Z. Toll-like receptor 3 and TICAM genes in catfish: species-specific expression profiles following infection with Edwardsiella ictaluri. Immunogenetics. (2006) 58:817–30. 10.1007/s00251-006-0144-z [DOI] [PubMed] [Google Scholar]
- 17.Sullivan C, Postlethwait JH, Lage CR, Millard PJ, Kim CH. Evidence for evolving Toll-IL-1 receptor-containing adaptor molecule function in vertebrates. J Immunol. (2007) 178:4517–27. 10.4049/jimmunol.178.7.4517 [DOI] [PubMed] [Google Scholar]
- 18.Fan S, Chen S, Liu Y, Lin Y, Liu H, Guo L, et al. Zebrafish TRIF, a Golgi-localized protein, participates in IFN induction and NF-κB activation. J Immunol. (2008) 180:5373–83. 10.4049/jimmunol.180.8.5373 [DOI] [PubMed] [Google Scholar]
- 19.Wei J, Zhang X, Zang S, Qin Q. Expression and functional characterization of TRIF in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immun. (2017) 71:295–304. 10.1016/j.fsi.2017.09.063 [DOI] [PubMed] [Google Scholar]
- 20.Yang C, Li Q, Su J, Chen X, Wang Y, Peng L. Identification and functional characterizations of a novel TRIF gene from grass carp (Ctenopharyngodon idella). Dev Comp Immunol. (2013) 41:222–9. 10.1016/j.dci.2013.05.018 [DOI] [PubMed] [Google Scholar]
- 21.Guo Y, Zhao W. China Fishery Statistical Yearbook. Beijing: Ministry of Agriculture Press; (2017). p. 22. [Google Scholar]
- 22.Yan J, Liao K, Wang T, Mai K, Xu W, Ai Q. Dietary lipid levels influence lipid deposition in the liver of large yellow croaker (Larimichthys crocea) by regulating lipoprotein receptors, fatty acid uptake and triacylglycerol synthesis and catabolism at the transcriptional level. PloS ONE. (2015) 10:e0129937. 10.1371/journal.pone.0129937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T, Yan J, Xu W, Ai Q, Mai K. Characterization of Cyclooxygenase-2 and its induction pathways in response to high lipid diet-induced inflammation in Larmichthys crocea. Sci. Rep. (2016) 6:19921. 10.1038/srep19921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins JA, Bart HL, Jr, Bowker JD, MacMillan JR, Nickum JG, Rose JD, et al. Guidelines for the Use of Fishes in Research. Bethesda, MD: American Fisheries Society; (2014). p. 17. [Google Scholar]
- 25.Li Q, Ai Q, Mai K, Xu W, Zheng Y. A comparative study: in vitro effects of EPA and DHA on immune functions of head-kidney macrophages isolated from large yellow croaker (Larmichthys crocea). Fish Shellfish Immun. (2013) 35:933–40. 10.1016/j.fsi.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 26.Li Q. Effects of LPS or poly(I:C) on pathogen recognition and inflammation related gene expressions of head kidney macrophage derived from large yellow croaker (Doctoral thesis). Ocean University of China, Qingdao, China (2018). [Google Scholar]
- 27.Li S, Monroig Ó, Wang T, Yuan Y, Navarro JC, Hontoria F, et al. Functional characterization and differential nutritional regulation of putative Elovl5 and Elovl4 elongases in large yellow croaker (Larimichthys crocea). Sci. Rep. (2017) 7:2303. 10.1038/s41598-017-02646-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo R, Ai Q, Mai K, Xu W, Wang J, Xu H, et al. Effects of dietary n-3 highly unsaturated fatty acids on growth, nonspecific immunity, expression of some immune related genes and disease resistance of large yellow croaker (Larmichthys crocea) following natural infestation of parasites (Cryptocaryon irritans). Fish Shellfish Immun. (2012) 32:249–58. 10.1016/j.fsi.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 29.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. (2002) 3:research0034.1. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. (2004) 64:5245–50. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. (2001) 25:402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 32.Xu Y, Tao X, Shen B, Horng T, Medzhitov R, Manley JL, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. (2000) 408:111–15. 10.1038/35040600 [DOI] [PubMed] [Google Scholar]
- 33.Matsuo A, Oshiumi H, Tsujita T, Mitani H, Kasai H, Yoshimizu M, et al. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J Immunol. (2008) 181:3474–85. 10.4049/jimmunol.181.5.3474 [DOI] [PubMed] [Google Scholar]
- 34.Ji J, Rao Y, Wan Q, Liao Z, Su J. Teleost-specific TLR19 localizes to endosome, recognizes dsRNA, recruits TRIF, triggers both IFN and NF-κB pathways, and protects cells from grass carp reovirus infection. J Immunol. (2018) 200:573–85. 10.4049/jimmunol.1701149 [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki M, Kim YC, Gray-Keller MP, Attie AD, Ntambi JM. The biosynthesis of hepatic cholesterol esters and triglycerides is impaired in mice with a disruption of the gene for stearoyl-CoA desaturase 1. J Biol Chem. (2000) 275:30132–8. 10.1074/jbc.M005488200 [DOI] [PubMed] [Google Scholar]
- 36.Miyazaki M, Kim YC, Ntambi JM. A lipogenic diet in mice with a disruption of the stearoyl-CoA desaturase 1 gene reveals a stringent requirement of endogenous monounsaturated fatty acids for triglyceride synthesis. J Lipid Res. (2001) 42:1018–24. [PubMed] [Google Scholar]
- 37.Miyazaki M, Dobrzyn A, Man WC, Chu K, Sampath H, Kim HJ, et al. Stearoyl-CoA desaturase 1 gene expression is necessary for fructose-mediated induction of lipogenic gene expression by sterol regulatory element-binding protein-1c-dependent and-independent mechanisms. J Biol Chem. (2004) 279:25164–71. 10.1074/jbc.M402781200 [DOI] [PubMed] [Google Scholar]
- 38.Ntambi JM, Miyazaki M, Stoehr JP, Lan H, Kendziorski CM, Yandell BS, et al. Loss of stearoyl–CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci USA. (2002) 99:11482–6. 10.1073/pnas.132384699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu K, Miyazaki M, Man WC, Ntambi JM. Stearoyl-coenzyme A desaturase 1 deficiency protects against hypertriglyceridemia and increases plasma high-density lipoprotein cholesterol induced by liver X receptor activation. Mol Cell Biol. (2006) 26:6786–98. 10.1128/MCB.00077-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hebbachi AM, Knight BL, Wiggins D, Patel DD, Gibbons GF. Peroxisome proliferator-activated receptor α deficiency abolishes the response of lipogenic gene expression to re-feeding restoration of the normal response by activation of liver X receptor alpha α. J Biol Chem. (2008) 283:4866–76. 10.1074/jbc.M709471200 [DOI] [PubMed] [Google Scholar]
- 41.Wang XA, Zhang R, She ZG, Zhang XF, Jiang DS, Wang T, et al. Interferon regulatory factor 3 constrains IKKβ/NF-κB signaling to alleviate hepatic steatosis and insulin resistance. Hepatology. (2014) 59:870–85. 10.1002/hep.26751 [DOI] [PubMed] [Google Scholar]
- 42.Li Q, Pène V, Krishnamurthy S, Cha H, Liang TJ. Hepatitis C virus infection activates an innate pathway involving IKK-α in lipogenesis and viral assembly. Nat Med. (2013) 19:722. 10.1038/nm.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen LN, Lim YS, Pham LV, Shin HY, Kim YS, Hwang SB. Stearoyl coenzyme A desaturase 1 is associated with hepatitis C virus replication complex and regulates viral replication. J Virol. (2014) 88:12311–25. 10.1128/JVI.01678-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the manuscript/Supplementary Files.