Abstract
The anadromous Hilsa shad (Tenualosa ilisha) live in the Bay of Bengal and migrate to the estuaries and freshwater rivers for spawning and nursing of the juveniles. This has led to two pertinent questions: (i) do all Hilsa shad that migrate from marine to freshwater rivers come from the same population? and (ii) is there any relationship between adults and juveniles of a particular habitat? To address these questions, NextRAD sequencing was applied to genotype 31,276 single nucleotide polymorphism (SNP) loci for 180 individuals collected from six strategic locations of riverine, estuarine and marine habitats. FST OutFLANK approach identified 14,815 SNP loci as putatively neutral and 79 SNP loci as putatively adaptive. We observed that divergent local adaptations in differing environmental habitats have divided Hilsa shad into three genetically structured ecotypes: turbid freshwater (Western Riverine), clear freshwater (Eastern Riverine) and brackish-saline (Southern Estuarine-Marine). Our results also revealed that genes involved in neuronal activity may have facilitated the juveniles’ Hilsa shad in returning to their respective natal rivers for spawning. This study emphasized the application of fundamental population genomics information in strategizing conservation and management of anadromous fish such as Hilsa shad that intersect diverse ecotypes during their life-history stages.
Subject terms: Molecular ecology, Population genetics
Introduction
Anadromous fish migrate from a freshwater breeding habitat to a marine feeding habitat and back to freshwater for spawning and their level of genetic diversity and population divergence is intermediate to that of marine and freshwater species1–3. The Hilsa shad (Tenualosa ilisha, Hamilton, 1822) is an anadromous clupeid species with a diverse range of distribution extending from freshwater rivers, estuaries, foreshore areas and seas of the Indo-Pacific region. It is the most important commercial trans-boundary species of which Bangladesh has the major share (86%), followed by India (8%), Myanmar (4%) and other countries within the Bay of Bengal region4. Hilsa provides the nutrition of 260 million Bengali people living in Bangladesh, some states of India and others living around the world, and it also support the livelihood of 2.5 million fishers and related people in the value chain alone in Bangladesh5. It is the single-most dominating food fish in the Bay of Bengal region with an average of 44% contribution to capture fisheries in Bangladesh6.
The Hilsa shad mainly inhabits in the Bay of Bengal including lower regions of the estuaries and foreshore areas, but migrates to the upstream rivers, mainly the Ganges-Brahmaputra-Meghna river systems during the spawning season and returns to the original habitat after spawning7. There are some exceptions to this behavior as two other subtypes of the species -a marine type and a fluvial potamodromous type have also been reported8–10. The marine subtype inhabited nearshore coastal and/or sea habitats and relies on downstream estuarine waters for spawning without migrating to freshwater9–11. The potamodromous types appear to stay in the middle reaches of the major river system and complete its life cycle within freshwater without migrating to the sea8,10. However, it is not truly known whether the anadromous T. ilisha mix and breed with two others minor subtypes during migration or whether they pass each other spatially and temporally. Therefore, understanding of stock structure and divergence in Hilsa shad population across its range of different habitats (sea, estuary and rivers) is still in dispute.
Understanding the genetic background and population structure of a species is crucial for planning and implementing conservation and management schemes, and needs to be explicitly considered for long-term sustainability. It is highly essential for fishery managers to know whether they are dealing with single or multiple spawning populations to design a sustainable management plan. However, revealing the genetic structure of an anadromous fish is challenging for conservation and management purposes12. This is mainly because of the highly connected diverged population with large effective population size of anadromous fish often show very weak genetic differentiation, thus decreasing the power of genetic tools in assigning individuals to their origins and defining the management units. However, the recent advances in next-generation sequencing (NGS) genotyping methods have expanded the prospects for exploring adaptive genetic markers to finely define weakly structured population, which ultimately improve our understanding of the genetic basis of fitness traits13.
Past research on the population genetic structure of this Hilsa shad mostly focused to investigate whether or not the species belongs to a single stock that uses rivers, brackish water estuaries and marine waters. The previous studies were mostly conducted through the use of allozyme markers14–16, Restriction Fragment Length Polymorphism (RFLP)17,18, Random Amplification of Polymorphic DNA (RAPD) markers19–22, and mitochondrial DNA cytochrome b gene nucleotide sequencing23,24. However, results from all of these studies are contradictory and inconclusive. The allozyme marker based studies reported that there is only one overall panmictic population of Hilsa shad in the Gangetic river systems15 and the Bay of Bengal region including Bangladesh, India and Myanmar16, but the Hilsa shad of the Bay of Bengal was genetically distinct from Kuwait and Indonesian Hilsa shad population16. Similarly, study-using mitochondrial DNA cytochrome b region also reported that the Hilsa shad population of the Ganga and the Hoogly rivers are the same population23. In contrast, studies using RAPD markers indicated that there is more than one gene pool of Hilsa shad in Bangladesh waters19,20. Similarly, it was reported that Hilsa shad has genetically two different populations from two major inland rivers of Bangladesh, the Padma and the Meghna20. PCR-RFLP analysis of the mitochondrial DNA D-loop region provided the same conclusion, suggesting that the Hilsa shad population has at least two differentiated populations in Bangladesh waters, which were subsequently divided into three, corresponding to the riverine, estuarine and marine populations18.
The inconclusive results of the past research about the population structure of the Hilsa shad can be addressed by using the genome-wide approach, which allows the genotyping of thousands of markers to detect genetic structure of this species at a finer spatial scale25. NGS-based restriction-site-associated DNA (NextRAD) techniques is a modern approach of high-throughput sequencing techniques to study population genetic structures that simultaneously facilitate both genotyping-by sequencing and discovering a large number of single-nucleotide polymorphisms (SNPs) across the genome26. The NextRAD approach includes surveys of both neutral and adaptive panels of SNP loci that can be used to determine the levels of genetic differentiation and genetic diversity. This approach has already been successfully applied in several fish species and has proven to be effective at identifying the genomic basis of fitness traits27–29. Our application of nextRAD sequencing alleviates the problem of absent reference genome sequence of this non-model species and facilitates the development of SNP markers, with the aim of identification of candidate adaptive loci. The FST OutFLANK approach has been proven as a popular means for identifying the adaptive markers of a fish species in which annotated reference genome is not readily available30,31. As Hilsa shad is a highly dispersive, anadromous fish, represents an ideal candidate for identifying putatively adaptive panel of SNP loci with genome scan and outlier test.
Until today, two important questions about the population genetic structure and parental assignment remained unclear: (1) What is the degree of genetic differences among the Hilsa shad populations available in different migratory routes of the species? (2) Are the juveniles of Hilsa shad (locally known as Jatka) of a particular habitat return to their respective natal rivers (rivers where they were born/nursed) for spawning as adults? To unravel the answers of the above two questions, the present study was conducted by identifying a set of neutral and adaptive genetic markers. The NextRAD sequencing technique was employed to genotype 31,276 SNP loci for 180 individuals of the species collected from six strategic aquatic habitats for revealing parental assignment and stock structure of the Hilsa shad population in Bangladesh waters. We applied the FST OutFLANK approach to determine adaptive vs. neutral SNP loci, and additional analysis were employed to verify the significance of these putatively adaptive loci under selection in the context with different migratory routes and habitats.
Material and Methodology
Sample collection
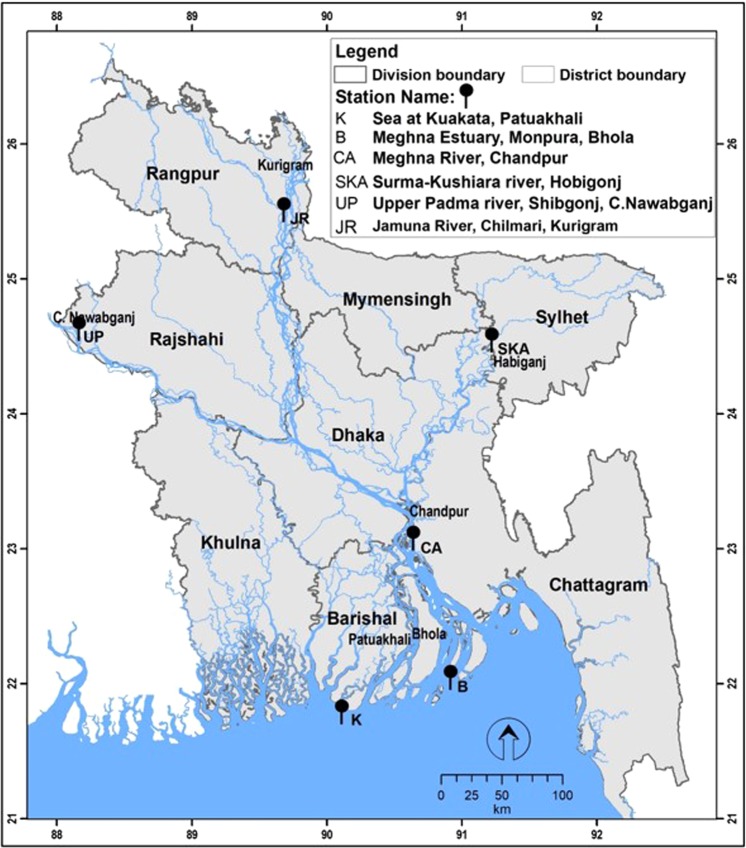
Juveniles and adults of T. ilisha were collected from the riverine, estuarine and coastal systems in Bangladesh, comprising six sampling sites (Fig. 1; Table 1). Two life stages were represented to address our research questions related to migration route and origins of these individuals with their respective populations. A total of 30 individuals were sampled from each population, with the life stages ratio dependent on their availability. Fin clips were preserved in absolute ethanol prior to DNA extraction. DNA isolations were performed using Promega DNA purification system (Promega, Madison, WI, USA) according to the manufacturer’s protocol. DNA quantifications were conducted using real-time PCR fluorescence measurements of double stranded DNA32 and the Quant-it kit (Life Technologies, Foster City, CA). All samples were collected under Bangladesh’s government permit and in accordance with animal care protocol (CVASU20160422) as approved by the Chattogram Veterinary and Animal Sciences University’s Animal Care and Biosafety Committee.
Figure 1.
Info-map showing migration of three stocks of Tenualosa ilisha with their labels as in Table 1.
Table 1.
Summary of sampling information for Tenualosa ilisha for nextRAD sequencing.
| Site | Area | Year of collection | Latitude | Longitude | Sample sizes (N) | |
|---|---|---|---|---|---|---|
| Juvenile | Adult | |||||
| K | Sea at Kuakata Patuakhali | 2017 | 21° 47′33.77″ | 90° 06′ 43.32″ | 15 | 15 |
| B | Meghna Estuary Monpura, Bhola | 2017 | 22° 02′ 53.41″ | 90° 55′ 07.65″ | 30 | 0 |
| CA | Meghna River, Chandpur Sadar | 2017 | 23° 04′ 49.91″ | 90° 38′ 28.53″ | 15 | 15 |
| SKA | Surma-Kushiara River, Hobigonj | 2016 | 24° 32′ 57.75″ | 91° 13′ 32.01″ | 26 | 4 |
| UP | Upper Padma River, Shibgonj, Chapai Nawabganj | 2016 | 24° 38′ 01.62″ | 88° 03′ 29.51″ | 21 | 9 |
| JR | Jamuna River Chilmari, Kurigram | 2017 | 25° 30′ 49.61″ | 89° 40′ 57.77″ | 18 | 12 |
nextRAD genotyping
Genomic DNA was converted into nextRAD genotyping-by-sequencing libraries (SNPsaurus, LLC, USA). Genomic DNA was first fragmented with Nextera DNA Flex Library Prep Kit (Illumina, Inc, USA), which also ligates short adapter sequences to the ends of the fragments. The Nextera reaction was scaled for fragmenting 10 ng of genomic DNA, although 20 ng of genomic DNA was used for input to compensate for the amount of degraded DNA in the samples and to increase fragment sizes. Fragmented DNA was then amplified for 25 cycles at 72 °C, with one of the primers matching the adapter and extending 8 nucleotides into the genomic DNA with the selective sequence GTGTAGAG. Thus, only fragments starting with a sequence that can be hybridized by the selective sequence of the primer will be efficiently amplified. The nextRAD libraries were sequenced on a HiSeq 4000 with eight lanes of 150 bp reads (University of Oregon, USA). The resulting fragments are fixed at the selective end, and have random lengths depending on the initial Nextera fragmentation.
Data quality control and filtering
Reads were trimmed using bbduk in custom scripts of SNPsaurus, LLC (BBMap tools, http://sourceforge.net/projects/bbmap/):bbmap/bbduk.shin = reads/run_1612/1612_GTAGAGGACTAAGCCT_S483_L006_R1_001_subset.fastq.gzout = reads/run_1612/1612_GTAGAGGACTAAGCCT_S483_L006_R1_001_t.fastq.gz ktrim = r k = 17 hdist = 1 mink = 8 ref = bbmap/resources/nextera.fa.gz minlen = 100 ow = t qtrim = r trimq = 10. The original shadstringent.vcf file contained data for 31, 276 SNP loci existing within a catalog of 92, 721 consensus NextRAD tagged sequences of 150 bases each. A column containing a unique ID for each SNP locus was added to the unfiltered vcf file using a custom perl script to remove redundancy among loci. Further filtering steps included the removal of complex SNPs with more than two alleles, less than 5% overall minor allele frequency, and less than 80% completeness of data among samples. Likewise, samples containing less than 80% completeness of data among the remaining loci were removed from the dataset. Additionally, where there were multiple SNPs located in a single NextRAD sequence tag only the first cataloged SNP was kept in the dataset so as to avoid the possibility of a single NextRAD locus having a disproportionate effect on the analyses. The unique IDs for SNP loci passing quality control standards were passed to a whitelist, which was used to filter the original vcf file using a custom perl script. After all filtering steps a total of 14, 894 individual SNP loci remained in the dataset. An additional custom perl script was used to convert the filtered vcf file to genepop format.
SNP discovery and gene annotation
A de novo reference was created by collecting 10 million reads in total, evenly from the samples, and excluding reads that had counts of <7 or >1000. The remaining loci were then aligned to each other to identify allelic loci and collapse allelic haplotypes to a single representative. All reads were mapped to the reference with an alignment identity threshold of 0.95 using bbmap (BBMap tools). Genotype calling was done using Samtools and bcftools (samtools mpileup -gu -Q 15 -t DP, DPR -f ref.fasta -b samples.txt | bcftools call -cv - >genotypes.vcf). The vcf was filtered to remove alleles with a population frequency of <3%. Loci that were heterozygous in all samples or had more than 2 alleles in a sample (suggesting collapsed paralogs) were removed. The absence of artifacts was checked by counting SNPs at each read nucleotide position and determining that SNP number did not increase with reduced base quality at the end of the read. In addition, each haplotype from all nextRAD-tags that contained putatively adaptive loci were subject to a BLASTn33 search of all sequences in the NCBI non-redundant database (word size = 11; mismatch scores = 2, −3; maximum e-value = 15). To reduce annotations to repetitive sequences in the database, each locus in the vcf file had the associated sequence annotated by blast, using: blastn -db ncbi-blast-2.4.0+/db/nt and filtering for those blast hits with a significance of e-15 or better.
Population genetic analysis
The filtered data was imported as a genind object into R and analyzed largely using the adegenet package34. After a first look revealed little variation among collections, an outlier locus approach was taken using the R package OUTFLANK35 which calculates a neutral distribution of Fst values and then uses this distribution to assign q-values to each locus to detect adaptive loci which are putatively influenced by selection. Default parameters were used for OUTFLANK analysis and the “number_of_samples” parameter was set to 6 (a number equal to the collections sampled). Output from this analysis was used to create a neutral loci dataset and an adaptive loci dataset for further analysis. The complete, outlier, and neutral datasets were further analyzed using the GenePop R package36. Significance testing for deviation from Hardy-Weinberg Equilibrium (HWE) test was conducted on the complete dataset using default settings. This data was not used for filtering of loci from either dataset since the collections don’t represent single populations and loci under divergent selection are expected to violate HWE. Neighbor-joining trees were generated using both the neutral and outlier datasets using Nei’s genetic distance method.
The R package ‘poppr’ was used to conduct Analysis of Molecular Variance (AMOVA) on both datasets37. The GenePop R package was also used for significance testing of pairwise FST to determine the genetic differences between collection sites using the neutral and outlier datasets using default settings with the samples grouped by collection site. Isolation by Distance analysis using the outlier dataset was done using the ‘adegenet’ R package using a pairwise distance matrix for all collection sites. Significance testing for isolation by distance was done by Mantel test using an Edward’s genetic distance matrix and a physical distance matrix between the collection sites with 9,999 iterations (also using the ‘adegenet’ package).
Principal Component Analysis (PCA) was done using the adegenet R package on both neutral dataset (14,815 SNPs) and outlier dataset (79 SNPs). The Bayesian clustering method implemented in the STRUCTURE software v. 2.3.438 was used to genetically assign individuals to clusters. Simulations were run for 100,000 steps following a burn-in period of 100,000 steps, considering values of K (number of clusters) from one to 15, with 10 replications for each value of K. The analysis was performed using admixture, correlated allele frequencies and no prior information regarding sampling location or morphological species. For each individual the program identifies the fraction of the genome that belongs to each one of the clusters. The rate of change in the log likelihood between successive K values39 was also estimated. The calculations were performed with STRUCTURE HARVESTER40. The clusters of the estimated population structure were visualized by using CLUMPAK41. Clustering analysis using Discriminate Analysis of Principal Components (DAPC) using the outlier dataset was also done using ‘adegenet’ in order to reveal possible genetic clustering among samples without grouping by collection sites.
Data accessibility
Our raw data with SRA accession number PRJNA503852 are available online in the NCBI sequence read archive (https://www.ncbi.nlm.nih.gov/sra/PRJNA503852, data publicly released on 31st December 2018).
Result
NextRAD Sequencing, Annotation and GO Categorization of Neutral and Adaptive Loci
On average, 2 million reads of 150 bp per individual were generated from the 180 nextRAD-genotyped T. ilisha. Out of the 180 individual fish, 13 individual sequences with greater than 20% missing genotypes were removed from the dataset, while the remaining 167 samples were used for all downstream analyses. Out of a total of 46,307 loci within a catalog of 92,971 consensus sequences, only 31,276 loci were remained when indels and SNP sites with less than 5% minor allele frequency have been removed from the original dataset. Further filtering for completeness of data at or above 80% for both samples and loci left a final set of 14,894 loci for analysis. As no reference genome was available for this species during the analyses; a de novo assembly was constructed using a custom script generated by SNPsaurus. We conducted homolog search of each contig in the assembly in the RefSeq database, and discovered that only 1.95% (1,814 of 92,971) of the contigs returned matching coding regions.
Hardy-Weinberg (HWE) tests of the 14,894 loci revealed that an approximately 15% of these loci were significantly deviated from the HWE expectations. Some of the loci with a significant heterozygote deficit may be linked to adaptive variations and population structuring between sampling sites. Outlier analysis run in R Package OUTFLANK identified 79 putatively adaptive SNPs as being FST outliers and candidates for positive selection loci (P > 0.995) (Fig. 2). Neutral loci were determined using a conservative threshold range of probabilities between 0.10 and 0.90. There were 14,815 putatively neutral SNPs that fell within these probability levels.
Figure 2.

Result from the R Package OUTFLANK FST outlier analysis for 14,815 SNPs. Among these, 79 SNPs (blue open circles) are above a 0.995 probability and were considered as putatively adaptive loci under selection with highest FST values. The remaining SNPs (black open circles) were considered neutral.
From the 14,894 loci being analyzed, only 1,814 loci (0.06%) were significantly matched to known genes in the public Nr databases and has been categorized into 51 groups with general functions of biological processes, molecular functions, cellular components and systemic functions (Fig. 3). 27 groups have shown a significant percentage from 1% up to 24%, while the remaining 24 groups remain less than 1%. Out of 79 identified putatively adaptive loci, only 14 loci were observed to be in the coding region and their gene functions are depicted in Table 2. Among the 14 putatively adaptive SNP loci, three loci (19201_25, 12546_217 and 74884_159) encodes the genes mostly involved in mitochondrial function and are mainly responsible for energy metabolism (Table 2). Other three loci (82109_13, 52941_15 and 44093_43) were found to be involved in neuronal activity important for neural communication and responsible to control a range of behavioral phenotypes. Other loci encode the gene mainly involved in transcription (9754_9 and 51360_15), reproduction (8853_12), cell growth (74886_11) and different signaling pathways (5615_159, 35109_9 and 44450_13).
Figure 3.
Gene ontology classification of 1,814 assembled contigs of Tenualosa ilisha
Table 2.
Summary of gene functions and annotation based on reference genome for 14 putatively adaptive loci that differentiates the Tenualosa ilisha populations in Bangladesh.
| Locus | GenBank Accession Number | Gene | Species | Gene Function |
|---|---|---|---|---|
| 9754_9 | XM_012816864.1 | KAT8 regulatory NSL complex subunit | Clupea harengus | Transcription: histone acetylation |
| 8853_12 | XM_012824429.1 | leucine-rich repeats and guanylate | Clupea harengus | Reproduction: spermatogenesis and cell differentiation |
| 82109_13 | XM_012814935.1 | Dmx-like 2 (dmxl2), mRNA | Clupea harengus | Neuronal activity: a key controller of neuronal and endocrine homeostatic processes |
| 74886_11 | XM_012821414.1 | FYN proto-oncogene, Src family tyrosine | Clupea harengus | Cell growth |
| 74884_159 | AP011610.1 | mitochondrial DNA, complete genome | Tenualosa ilisha | Mitochondrial function |
| 5615_159 | XM_010865640.1 | MAP/microtubule affinity-regulating kinase | Esox lucius | Signalling: cytoskeletal signalling, glucose or energy metabolism |
| 52941_15 | XM_006792985.1 | neurexin-3b-like | Neolamprologus brichardi | Neuronal cell surface protein associated with a range of behavioural phenotypes |
| 51360_15 | XM_012823915.1 | cleavage and polyadenylation specific | Clupea harengus | Transcription: plays a central role in 3-prime processing of pre-mRNAs |
| 44450_13 | XM_017701045.1 | inositol-trisphosphate 3-kinase | Pygocentrus nattereri | Protein-protein interactions: involved in inositol metabolic process, signal transduction and cytoskeleton organization |
| 44093_43 | XM_016260991.1 | glutamate receptor ionotropic | Sinocyclocheilus grahami | Neuronal activity: important for neural communication, memory formation, learning, and regulation |
| 35109_9 | XM_013137472.1 | ryanodine receptor 3-like | Esox lucius | Signalling: cellular mediator of Ca2+ release channels and thereby plays a role in triggering muscle contraction |
| 19201_25 | AP011611.1 | mitochondrial DNA, complete genome | Tenualosa ilisha | Mitochondrial function |
| 16116_52 | XM_012835890.1 | C-type lection lectoxin-Enh3-like | Clupea harengus | Mannose-binding lectin and agglutinates a variety of animal cells |
| 12546_217 | FJ582830.1 | cytochrome oxidase subunit 1 | Amphiprion sebae | Mitochondrial function: involved in energy metabolism and aerobic respiration |
Population structure analysis based on phylogenetic analysis
Neighbor-joining (NJ) analysis of 14,815 putatively neutral loci (Fig. 4a) revealed weak structure, and showed that the populations were not delineated strictly into their respective units as observed in the outlier loci dataset. Two main groupings were formed: (i) (Surma-Kushiara and Kuakata = 88.2% bootstrap support), (ii) (Upper Padma and Jamuna = 86.1% bootstrap support), and the remainder of the collections branched individually (Chandpur and Bhola). The phylogenetic analyses of the 79 putatively adaptive loci showed the following three main groupings of collections: Marine and Estuary (Kuakata and Bhola = 99.9% bootstrap support), North-Western Riverine (Upper Padma and Jamuna = 100% bootstrap support), and North-Eastern Riverine (Surma-Kushiara and Chandpur = 99.9% bootstrap support) (Fig. 4b). Overall, the terminal branch lengths were longer in the NJ tree based on adaptive loci when compared to the NJ tree generated from neutral loci dataset, suggesting substantial genetic variations among populations in their respective clusters (Fig. 4).
Figure 4.
Neighbor-joining trees based on Nei’s genetic distances using the following sets of loci: (A) putatively neutral panel of SNPs, and (B) the putatively adaptive panel of SNPs. Branch nodes are denoted as the percentage of bootstrap support that was generated with 1,000 replicates. Collection codes correspond to those listed in Table 1.
Demographic inferences from AMOVA, FST statistics, PCA and DAPC analysis
For the AMOVA analysis, 27 loci were dropped from the outlier dataset and 5,593 loci were dropped from the neutral dataset (approximately 1/3 of the loci in each set) (Table 3, Fig. S1). In both cases, variation between populations was less than observed variation among samples within collection sites. Consistent with the NJ clustering analysis, AMOVA analysis of the outlier dataset also showed best support for the population differentiation, yielded an overall FST of 0.1826 (P = 0.001). The largest component of genetic variability was explained at the individual level (63.8%). The portion of variation captured among the six populations was 18.6% (P = 0.0001), while the remaining variation among individuals within population was 17.6% (P = 0.0001). An AMOVA analysis based on these neutral loci dataset yielded a mean FST of 0.0029 (P = 0.001). The highest genetic variation was observed among individuals (86.8%), with little genetic variation among individuals within population (12.9%, P = 0.0001) and even significantly smaller variation between populations (0.3%, P = 0.0001).
Table 3.
Analysis of Molecular Variance (AMOVA) among 6 sampling locations of Tenualosa ilisha distributed in aquatic ecosystem of Bangladesh.
| Source of variation | Sigma | % of variation | P-value |
|---|---|---|---|
| For neutral SNPs loci | |||
| Between population | 7.523 | 0.2901 | 0.0001 |
| Among samples within population | 334.749 | 12.911 | 0.0001 |
| Among samples | 2250.472 | 86.798 | — |
| For adaptive SNPs loci | |||
| Between population | 3.474 | 18.607 | 0.0001 |
| Among samples within population | 3.294 | 17.642 | 0.0001 |
| Among samples | 11.90413 | 63.751 | — |
The pairwise FST values for the adaptive loci dataset were markedly higher than the FST values for the neutral loci dataset, supporting genetic distinctiveness between the Hilsa shad populations (Table 4). For the 14,815 putatively neutral loci, pairwise FST values ranged from 0.0109 to 0.0156 and averaged 0.0132 (Table 1). The pairwise FST analysis for 79 putatively adaptive loci showed an above average range across all comparisons ranging from 0.0289 to 0.1919 and averaged at 0.1100. Individuals from Estuary (B) and Jamuna River (JR) were observed to have the highest pairwise FST values (FST = 0.19192, P = 0.001; Table 2). However, all of the pairwise FST values for both neutral and adaptive panels of SNP loci showed significant difference between them (Table 2). We also investigated the correlation of population differentiation (FST) and geographic distance. Isolation-by-Distance (IBD) analysis of neutral loci generated a regression slope of 0.0565, and the Mantel test was insignificant (P > 0.05). We found a statistically significant relationship between increasing degree of geographic separation and increasingly large FST for the putatively adaptive loci, in which the IBD slope was significantly higher at R2 = 0.1722 (P = 0.0134) (Fig. 5).
Table 4.
Pairwise FST values for the putatively neutral SNPs (above diagonal) and putatively adaptive SNPs (below diagonal) in Hilsa shad.
| K (SS) | B (ME) | CA (MR) | SKA | UP | JR | |
|---|---|---|---|---|---|---|
| K (SS) | — |
0.01422 (0.001) |
0.01309 (0.001) |
0.01094 (0.003) |
0.01106 (0.002) |
0.01194 (0.001) |
| B1 (ME) |
0.02888 (0.019) |
— |
0.01561 (0.001) |
0.01437 (0.001) |
0.01432 (0.001) |
0.01421 (0.001) |
| CA (MR) |
0.06975 (0.002) |
0.12462 (0.001) |
— |
0.01296 (0.001) |
0.01312 (0.001) |
0.01309 (0.001) |
| SKA |
0.08507 (0.001) |
0.15021 (0.001) |
0.04765 (0.016) |
— |
0.01113 (0.002) |
0.01173 (0.001) |
| UP |
0.06401 (0.001) |
0.10978 (0.001) |
0.16010 (0.001) |
0.15341 (0.001) |
— |
0.01100 (0.002) |
| JR |
0.11544 (0.001) |
0.19192 (0.001) |
0.18607 (0.001) |
0.17230 (0.001) |
0.03931 (0.006) |
— |
Collection codes correspond to those listed in Table 1. P value are shown within the parenthesis for each pairwise FST value.
Figure 5.
Correlation of pairwise FST value with pairwise geographical distances among Tenualosa ilisha collection. The red triangles represent the putatively adaptive loci FST and the blue circles are the neutral loci FST. The Mantel test showed simulated p-value of 0.0134 based on 9,999 replicates indicating significance effect.
We have also conducted a pairwise comparison of FST values based on 79 putatively adaptive loci between adult and juveniles for five sampling sites except individuals from Meghna estuary in Bhola (Table 5). Notably, juveniles from three sampling sites (Surma-Kushiara, Upper Padma and Jamuna) were observed to show extremely weak genetic differentiation with their respective adults with non-significant zero FST values. Interestingly, the genetic variation between adults from Meghna River in Chandpur (CA-A) and juveniles from Surma-Kushiara (SKA-J) was also found to be zero (FST values = 0.0000). However, juveniles from these three sites showed a significant higher genetic differentiation with the adults of the other collection sites. In contrast, small, but significant distinction was seen between adults and juveniles in Meghna River at Chandpur (FST values = 0.06296, P = 0.003) and sea at Kuakata (FST values = 0.07787, P = 0.002).
Table 5.
Pairwise FST values based on 79 putatively adaptive loci between adult and juveniles for five sampling sites in Bangladesh.
| K-J | CA-J | SKA-J | UP-J | JR-J | |
|---|---|---|---|---|---|
| K-A | 0.07787 (0.002) | 0.21494 (0.001) | 0.11007 (0.001) | 0.14356 (0.001) | 0.19765 (0.001) |
| CA-A | 0.05743 (0.004) | 0.06296 (0.003) | 0.00000 (0.594) | 0.22993 (0.001) | 0.26771 (0.001) |
| SKA-A | 0.02957 (0.028) | 0.15486 (0.001) | 0.00000 (0.582) | 0.07906 (0.002) | 0.23532 (0.001) |
| UP-A | 0.03346 (0.016) | 0.13463 (0.001) | 0.12698 (0.001) | 0.00000 (0.592) | 0.09103 (0.001) |
| JR-A | 0.11087 (0.001) | 0.13409 (0.001) | 0.32294 (0.001) | 0.02339 (0.032) | 0.00000 (0.564) |
Collection codes correspond to those listed in Table 1. “J” indicates juvenile Hilsa shad and “A” indicates adult Hilsa shad. P value are shown within the parenthesis for each pairwise FST value.
Principal component analyses on both outlier and neutral SNPs revealed an overall different population structure (Fig. 6). PCA of the neutral SNPs dataset supported a single homogenous population, consistent with the low genetic differentiation as indicated by pairwise FST values. On the other hand, PCA based on the adaptive SNPs showed three major clusters (Marine and Estuary, North-Western Riverine and North-Eastern Riverine) without clear separation of the populations according to the collection sites, which is found to be consistent with the NJ clustering, AMOVA analysis and FST values. Similarly, STRUCTURE analysis (Fig. 7) have also shown similar clustering pattern of the six Hilsa shad populations into three main genetic clusters based on the delta K statistic (K = 3). In contrast with the PCA, NJ clustering and STRUCTURE analyses, our DAPC analysis based on the adaptive SNPs dataset identified four distinct partitioning of populations, although the population structure was not explainable by the collection groups (Fig. 8). However, based on the DAPC output, we further unravel the assignment of individuals to their respective clusters (Fig. 9). The distinctive fourth clusters observed in non-model-based spatial analysis (DAPC) were embedded in a major cluster of the model-based analysis (NJ tree), as the North-Eastern Riverine (Surma-Kushiara and Chandpur). The DAPC assignment analysis showed that Surma-Kushiara and Chandpur was further divided into two separate clusters.
Figure 6.
Scatterplots showing the Principal Components Analysis (PCA) for: (A) the putatively neutral panel of SNP loci and (B) the putatively adaptive panels of SNP loci. Collection codes correspond to those listed in Table 1. PCAs determined optimal clustering of three groupings (1) marine/brackish represented by two closest relatives at Kuakata sea (K) and the Meghna Estuary, Bhola (B), (2) muddy freshwater represented by two closest relatives at the Upper Padma (UP) and the Jamuna (JR), and (3) clear freshwater types represented by the Meghna River (CA) and its upstream tributaries, Surma-Kushiara (SKA). Both the neutral and outlier datasets were highly significant for population differentiation via GenePop test.
Figure 7.
Bayesian STRUCTURE bar plot based on the (A) the putatively 14,815 neutral SNP loci and (B) the putatively 79 adaptive SNP loci of 180 Tenualosa ilisha individuals. Black lines separate individuals of different populations. Each vertical line represents an individual. The colors represent the proportion of inferred ancestry from K ancestral populations. Based on the delta K statistic, the best supported number of a posteriori genetic clusters was K = 3 for the standard admixture model (Table S1).
Figure 8.

Scatterplots showing Discriminant Analysis of Principal Components (DAPC) of genetic differentiation among the 180 Tenualosa ilisha for the 79 putatively adaptive panels of SNPs loci. Ovals are the inertial ellipse, dot represent individual genotypes and the line extends to centroids of each population. DAPC grouped the collection sites of T. ilisha into four major clusters.
Figure 9.

Bar graph showing Tenualosa ilisha collection sites-wise DAPC grouping. Collection codes in X-axis correspond to those listed in Table 1.
Discussion
The use of genomic approaches to study population structuring and species demographics has become increasingly common with high throughput sequencing platform, supplementing findings from previous studies that have used traditional genetic markers such as allozyme, mitochondrial DNA and microsatellites42. We used nextRAD sequencing to generate a panel of genome-wide SNPs to examine fine-scale population structuring in T. ilisha in their native distribution regions throughout Bangladesh.
Our AMOVA and FST analyses of 79 putatively adaptive loci indicate substantial levels of genome-wide differentiation among the six populations. The neighbor-joining tree (NJ) and spatial analyses (PCA, STRUCTURE and DAPC) generated by small panel of highly differentiated loci (79 putatively adaptive loci) provided strong evidence for three major genetically separated groups of Hilsa shad in Bangladesh: Marine and Estuary (Kuakata and Bhola), Western Riverine (Upper Padma and Jamuna), and Eastern Riverine (Surma-Kushiara and Meghna). This is consistent with the findings of19 based on RAPD markers, of17 based on RFLP of mitochondrial DNA, and of18 based on PCR-RFLP of mitochondrial D-loop region. These studies came to a consensus that Hilsa shad populations were genetically diverged by three different environments. This broad structuring in three independent groups in turn mirrors the spatial structuring which is likely maintained by environmental differences such as turbidity and salinity levels in the marine or brackish, muddy/turbid freshwater (Western Riverine) and clear freshwater (Eastern Riverine) habitat43. Fishes of Western Riverine habitats are of bright silvery in color with thicker structure that could be the influence of the whitish silty turbid water, while fishes of Eastern Riverine are thinner, a bit darker and elongated that could be the influence of dark colored water43. Likewise, biometrical comparisons have also delineated Hilsa shad into three stocks according to different ecotypes44–46. Our spatial analysis and low FST value (0.02888) did reveal that population of Kuakata and Bhola appear to be intermediate which may be indicative of gene flow and admixture events, as both locations seems to be adjacently located to each other in the further southern coastal regions (Fig. 1). In fact, there was evidence stating the ability of T. ilisha, to spawn in the estuary, although no report has indicated any marine spawning in Bangladesh47. Therefore, fishes from Kuakata may have migrated to Bhola to spawn, but not to any other rivers, as significant differentiations between Kuakata and other river populations have been observed. Two clusters which were found within the North-Eastern and Eastern Riverine group (Surma-Kushiara and Chandpur) may likely represent two separate breeding grounds within the same riverine system, which is also congruent with previous studies48. Besides, the presence of population structuring in Hilsa shad was also reported by studies conducted using allozymes markers14,49,50 and RAPD analysis20,21.
On the other hand, there was no evidence of genetic differentiation between all studied populations based on multiple analyses of 14,815 neutral loci dataset in this study. Our results were consistent with findings from other studies using various markers and techniques such as allozymes15,16,51, otolith microchemistry and morphometrics8, mitochondrial cytochrome b21,23 and mitochondrial control region52. The lack of detectable population structural differences among the Hilsa shad in their distributed regions in Bangladesh suggests that there was extensive movement and mixing of Hilsa shad throughout Bangladesh for breeding purpose, given its anadromous nature53. Indeed, reproductive isolation may be likely impeded by the influx of large quantities of freshwater during monsoon, seasonal water circulation and continuous migration of Hilsa shad54. Therefore, these populations should be considered as belonging to a single gene pool and managed as a panmictic population8.
However, the lack of population genetic structure does not preclude a genetic basis for the presence of genetic differentiation as postulated by the presence of putatively adaptive loci, which have clearly separated the populations. It is highly possible that the neutral loci and previously used methods were unable to detect the smaller-scale genetic difference between the three ecotypes identified by putatively adaptive loci. Moreover, we also hypothesized that the lower water discharge from the upstream river flow from Farakka barrage (in the West Bengal, India) with constant heavy siltation may have disrupted the migration routes of the anadromous Hilsa shad ascending tributaries in the Western Riverine habitats11. Previous meristic and morphometric studies have also revealed some morphological differences among the Hilsa stocks from different environments55. Owing to the distinctive differences observed in the body height, the Hilsa stocks were divided into Western Riverine’s ‘Broad type’ and Eastern Riverine’s ‘Slender type’56–58. The different seasonal spawning between these two morphotypes: the “broad type” during monsoon and “slender type” during winter, and each with different fecundity level, may have also contributed to the limited natal and breeding dispersal, and thus population divergence of Hilsa shad59.
We identified that some genes of the 14 putatively adaptive loci codes for multiple transcript variants encoding different isoforms, where genetic variation at this locus has been associated and may result with a range of behavioral phenotypes60. Isoforms are messenger RNAs resulted from transcription of the same gene locus but at a different transcription start sites (TSSs) or untranslated regions (UTRs), which results in alteration of gene functions61. This molecular complexity may lead to phenotypic differences and genetic variation between individuals at cellular level and thus deliberating the potential of population divergence in Hilsa shad62,63. From the match analysis of putatively adaptive loci, gene from the 51,360_15 locus encodes cleavage and polyadenylation specificity factor (CPSF), which is involved in the cleavage of the 3’ signaling region from a newly synthesized pre-messenger RNA (pre-mRNA) molecule in the process of gene transcription (Table 2). On a similar study of population genomics, through transcriptome analysis it was found that intron retention and alternative polyadenylation is largely independent along the transcript, leading to thousands of novel isoforms. With that, the study was concluded by proposing that the independent combination of alternative RNA processing events has contributed to complex isoform evolution in the organism, which provides a new foundation for the study of phenotypic difference among the population64, where similar mechanism can be expected in the phenotypic variations in the Hilsa shad population species in Bangladesh.
Three genes from the match value (19201_25, 12546_217 and 74884_159) associates with the mitochondrial DNA, function and mitochondrial cytochrome oxidase subunit 1 (Table 2). Various studies have shown the relation of mitochondrial DNA and diversity in a population and the study of this analysis has demonstrated large values of nucleotide and a set of genetic determinants located on a single chromosome called haplotype. The presence of shared haplotypes in most populations is either the restricted gene flow among local populations or the predominance of ancestral haplotypes65. Although mitochondrial COI gene have been widely used for species identification studies, it is also used for estimation of inter- and intra-specific diversity in many organisms65,66. A significant weightage of greater than 5% were morphology (skin and scale pattern), enzymatic activity, immunity, signaling, nervous system and neuronal activity (Fig. 3) may explain the variations between the Hilsa shad individuals of three different environments with a vast diverse of water parameters such as salinity and turbidity. Based on the functional annotations, the two main categories, the morphology (skin and scale pattern) and immunity may also play an important role in the distribution of Hilsa shad individuals based on their adaptability to external environment, prey and predation, immunity and disease resistance over a certain period.
While Hilsa shad populations were found to be genetically differentiated by three major groups due to geographical separation, juveniles and adults from each populations seems to be sharing the same genetic structure, except for individuals from Kuakata and Chandpur. The significant weak genetic structuring in three sampling locations (Surma-Kushiara, Upper Padma, and Jamuna) as revealed by zero FST values indicates that the juveniles are offspring of the existing adults and the adults may potentially return to their natal ground for spawning purposes. Given that the three rivers are geographically isolated, admixture events are not likely to happen. Contradicting genetic variation patterns were observed in the Surma-Kushiara River (SKA), in which adults from Meghna River in Chandpur (CA) were genetically similar to juveniles from Surma-Kushiara, which likely reflect the migratory behavior of some of the Chandpur River’s adults to the spawning ground in the upper stream of Surma-Kushiara River to breed. This finding was consistent with the major groupings in the above-mentioned analyses, in which individuals of Chandpur and Surma-Kushiara River were clustered as Eastern Riverine group. Unexpectedly, similar trend was not clearly depicted in both Chandpur (CA) and Kuakata (K), in which the adults and juveniles showed low genetic differentiation, but not sharing the same genetic structure as Surma-Kushiara, Upper Padma and Jamuna. This might be because that these two sites are the common routes of Hilsa shad to migrate from downstream to the geographically separated upstream rivers, thereby frequent admixture events may likely happen, and some adults may have been mixed during sampling activities. However, these observations may still allow us to conclude that Hilsa shad do return to their respective natal grounds for spawning, in which they are geographically separated. Fascinatingly, some putatively adaptive SNP loci (82109_13, 52941_15 and 44093_43) encode the genes, which are involved in neuronal function such as neural communication, memory formation and learning, in which lead to us to suggest that these genes might have played an important role in facilitating Hilsa shad to reach their natal ground in subsequent years for spawning.
Conclusion and management implications
Although neutral loci did not reveal much genetic structural differences among the Hilsa shad populations, by using 79 adaptive loci, we were still able to observe genetic differences which are so distinctive that they warrant the division of Hilsa shad into three separate ecotypes in Bangladesh waters: (1) marine/brackish represented by two closest relatives at Kuakata and the Meghna Estuary, Bhola, (2) muddy freshwater represented by two closest relatives in the Upper Padma and the Jamuna, and (3) clear freshwater types represented by the Meghna River and its upstream tributaries, Surma-Kushiara. In a conservation management context, it is important to maintain the genetic diversity of Hilsa shad in each location, given that our results have suggested high levels of gene flow within populations but lower variation across most of their range. The most important phenomenon revealed from this study that has great management implications is the fact that Hilsa shad returns to their natal rivers for spawning. Hilsa fishers try to catch juveniles before they move to the sea for maturation, because of the fear that they might not be able to uphold their catch yield. This finding will help in motivating fishers not to catch the juveniles, as they will get the assurance of getting back the juvenile as adult in the respective rivers. Our findings also indicated the importance of incentive-based juvenile protection activities involving all stakeholders, including the fishers for each of the rivers where Hilsa shad spawn and juveniles spend their early life. We note that empirically testing these hypotheses will require further characterization of the remaining putatively adaptive loci based on a more complete whole-genome sequence and at a larger regional level constituting the native distribution of Hilsa shad. Ultimately, isolating mechanisms, environmental factors, and the temporal stability of these patterns in structuring these groups in Bangladesh will require further study to help fishery managers in formulating ecotypes oriented management strategies for sustainable fisheries of two million fishers who participate in the Hilsa shad fishery. We suggest that this innovative work provides a model that may be of value in other aquatic species, especially in anadromous fish species that intersect diverse ecotypes during their life-history stages.
Supplementary information
Acknowledgements
This work was undertaken as a part of the CGIAR Research Program on Fish Agri-Food Systems (FISH). The authors acknowledge the financial support provided by USAID through the Enhanced Coastal Fisheries in Bangladesh (ECOFISH-BD) Activity, jointly implemented by WorldFish, Bangladesh and South Asia Office and the Department of Fisheries (DOF), Bangladesh. Thanks are extended to the Chattogram Veterinary and Animal Sciences University (CVASU) and University Malaysia Terengganu (UMT) for their participation in this collaborative research. Special thanks are due to Prof. Dr. John Benzie, WorldFish HQ, Penang, Malaysia for his interest in this research and valuable suggestion. Thanks are extended to the research associates and research assistants of the ECOFISH-BD Project for their efforts in collection of Hilsa samples. The authors alone are responsible for the opinions expressed in this article.
Author contributions
Conceived and designed the research: L.L.W. and M.A. Performed field work and collected data: M.J.R and M.N. Performed spatial analysis and interpreted the analysis: Y.I and S.A. Analyzed the sequenced data, performed the annotation analysis, interpreted the data and wrote the paper: L.L.W. and M.A. Critically reviewed and edited the manuscript: M.A.W. and M.W.D. All authors approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-52465-2.
References
- 1.McDowall RM. Anadromy and homing: two life-history traits with adaptive synergies in salmonid fishes? Fish and Fisheries. 2001;2:78–85. doi: 10.1046/j.1467-2979.2001.00036.x. [DOI] [Google Scholar]
- 2.Skovrind M, et al. Genomic population structure of freshwater‐resident and anadromous ide (Leuciscus idus) in north‐western. Europe. Ecology and evolution. 2016;6(4):1064–1074. doi: 10.1002/ece3.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McBride MC, Hasselman DJ, Willis TV, Palkovacs EP, Bentzen P. Influence of stocking history on the population genetic structure of anadromous alewife (Alosa pseudoharengus) in Maine rivers. Conservation genetics. 2015;16(5):1209–1223. doi: 10.1007/s10592-015-0733-1. [DOI] [Google Scholar]
- 4.Rahman MJ, Wahab MA, Amin SN, Nahiduzzaman M, Romano N. Catch Trend and Stock Assessment of Hilsa Tenualosa ilisha Using Digital Image Measured Length-Frequency Data. Marine and Coastal Fisheries. 2018;10(4):386–401. doi: 10.1002/mcf2.10034. [DOI] [Google Scholar]
- 5.Department of Fisheries Fish week compendium 2016. Ministry of Fisheries and Livestock and Department of Fisheries, Dhaka, Bangladesh (2016).
- 6.Department of Fisheries Yearbook of fisheries statistics of Bangladesh 2016–17. Ministry of Fisheries and Livestock and Department of Fisheries, Dhaka, Bangladesh (2017).
- 7.Hossain MS, Sarker S, Sharifuzzaman SM, Chowdhury SR. Discovering spawning ground of Hilsa Shad (Tenualosa ilisha) in the coastal waters of Bangladesh. Ecological Modelling. 2014;282:59–68. doi: 10.1016/j.ecolmodel.2014.03.001. [DOI] [Google Scholar]
- 8.Milton DA, Chenery SR. Can Otolith chemistry detect the population structure of the shad Hilsa Tenualosa ilisha? Comparison with the results of genetic and morphological studies. Marine Ecology Progress Series, 2001;222:239–251. doi: 10.3354/meps222239. [DOI] [Google Scholar]
- 9.Milton DA, Chenery SR. Movement patterns of the tropical shad (Tenualosa ilisha) inferred from transects of 87Sr/86Sr isotope ratios in their otoliths. Canadian. Journal of Fisheries and Aquatic Science. 2003;60:1376–1385. doi: 10.1139/f03-133. [DOI] [Google Scholar]
- 10.Hossain MS, Sharifuzzaman SM, Chowdhury SR. Habitats across the life cycle of Hilsa shad (Tenualosa ilisha) in aquatic ecosystem of Bangladesh. Fisheries Management and Ecology. 2016;23:450–462. doi: 10.1111/fme.12185. [DOI] [Google Scholar]
- 11.Blaber SJM, Milton DA, Chenery SR, Fry GC. New insights into the life history of Tenualosa ilisha and fishery implications. American Fisheries Society Symposium. 2003;35:223–240. [Google Scholar]
- 12.FW A, Hohenlohe PA, Luikart G. Genomics and the future of conservation genetics. Nature Reviews Genetics. 2010;11(10):697–709. doi: 10.1038/nrg2844. [DOI] [PubMed] [Google Scholar]
- 13.Davey JW, et al. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nature Reviews Genetics. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- 14.Rahman M, Naevdal G. Population genetics studies of Hilsa shad, Tenualosa ilisha (Hamilton), in Bangladesh waters: evidence for the existence of separate gene pools. Fisheries Management and Ecology. 2000;7:401–412.. doi: 10.1046/j.1365-2400.2000.00211.x. [DOI] [Google Scholar]
- 15.Lal KK, Kumar D, Srivastava SK, Mukherjee A. Generic variation in Hilsa shad (Tenualosa ilisha) population in river Ganges. Indian Journal of Fisheries. 2004;51:33–42. [Google Scholar]
- 16.Salini JP, Milton DA, Rahman MJ, Hussain MG. Allozyme and morphological variation throughout the geographic range of the tropical shad, Hilsa Tenualosa ilisha. Fisheries Research. 2004;66:53–69. doi: 10.1016/S0165-7836(03)00124-3. [DOI] [Google Scholar]
- 17.Ahmed ASI, Islam MS, Azam MS, Khan MMR, Alam MS. RFLP analysis of the mtDNA D-loop region in Hilsa shad (Tenualosa ilisha) population from Bangladesh. Indian Journal of Fisheries. 2004;51(1):25–31. [Google Scholar]
- 18.Mazumder SK, Alam MS. High levels of genetic variability and differentiation in Hilsa shad, Tenualosa ilisha (Clupeidae, Clupeiformes) populations revealed by PCR-RFLP analysis of the mitochondrial DNA D-loop region. Genetics and Molecular Biology. 2009;32:190–196. doi: 10.1590/S1415-47572009005000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahle G, Rahman M, Eriksen AG. RAPD finger printing used for discriminating among three populations of Hilsa shad (Tenualosa ilisha) Fisheries Research. 1997;32:263–269. doi: 10.1016/S0165-7836(97)00058-1. [DOI] [Google Scholar]
- 20.Shifat R, Begum A, Khan H. Use of RAPD fingerprinting for discriminating two populations of Hilsa shad (Tenualosa ilisha Ham.) from inland rivers of Bangladesh. Journal of Biochemistry and Molecular Biology. 2003;36:643–652. doi: 10.5483/bmbrep.2003.36.5.462. [DOI] [PubMed] [Google Scholar]
- 21.Brahmane MP, et al. Use of RAPD fingerprinting for delineating populations of Hilsa shad Tenualosa ilisha (Hamilton, 1822) Genetics and Molecular Research. 2006;5:643–652. [PubMed] [Google Scholar]
- 22.Jorfi E, Farhad A, Ghorezshi SA, Mortezaei SRS. A study on population genetic of Hilsa shad, Tenualosa iliisha in Khoozestan, Iran using molecular method (RAPD) Iranian Scientific Fisheries Journal. 2008;17(1):31–44. [Google Scholar]
- 23.Brahmane MP, Kundu SN, Das MK, Sharma AP. Low genetic diversity and absence of population differentiation of Hilsa (Tenualosa ilisha) revealed by mitochondrial DNA cytochrome b region in Ganga and Hooghly rivers. African Journal of Biotechnology. 2013;12:3383–3389. [Google Scholar]
- 24.Behera BK, et al. Population genetic structure of Indian shad, Tenualosa ilisha Inferred from variation in mitochondrial DNA sequences. Journal of Environmental Biology. 2015;36:1193–1197. [PubMed] [Google Scholar]
- 25.Carreras C, et al. Population genomics of an endemic Mediterranean fish: differentiation by fine scale dispersal and adaptation. Scientific Reports. 2017;7:43417. doi: 10.1038/srep43417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA. Genotyping-by-sequencing in ecological and conservation genomics. Molecular Ecology. 2013;22:2841–2847. doi: 10.1111/mec.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hohenlohe PA, et al. Population genomics of parallel adaptation in three spine stickleback using sequenced RAD tags. PLoS Genetics. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goetz F, et al. A genetic basis for the phenotypic differentiation between siscowet and lean lake trout (Salvelinus namaycush) Molecular Ecology. 2010;19:176–196. doi: 10.1111/j.1365-294X.2009.04481.x. [DOI] [PubMed] [Google Scholar]
- 29.Hess JE, Campbell NR, Close DA, Docker MF, Narum SR. Population genomics of Pacific lamprey: adaptive variation in a highly dispersive species. Molecular Ecology. 2012;22:2898–2916. doi: 10.1111/mec.12150. [DOI] [PubMed] [Google Scholar]
- 30.Bierne N, Welch J, Loire E, Bonhomme F, David P. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Molecular Ecology. 2011;20:2044–2072. doi: 10.1111/j.1365-294X.2011.05080.x. [DOI] [PubMed] [Google Scholar]
- 31.Le Corre V, Kremer A. The genetic differentiation at quantitative trait loci under local adaptation. Molecular Ecology. 2012;21:1548–1566. doi: 10.1111/j.1365-294X.2012.05479.x. [DOI] [PubMed] [Google Scholar]
- 32.Blotta I, Prestinaci F, Mirante S, Cantafora A. Quantitative assay of total dsDNA with PicoGreen reagent and real-time fluorescent detection. Annali dell’Istituto superiore di sanita. 2005;41(1):119–23. [PubMed] [Google Scholar]
- 33.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jombart T. adegenet: An R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129>. [DOI] [PubMed] [Google Scholar]
- 35.Whitlock MC, Lotterhos KE. Reliable Detection of Loci Responsible for Local Adaptation: Inference of a Null Model through Trimming the Distribution of FST. The American Naturalist. 2015;186:S24–S36. doi: 10.1086/682949. [DOI] [PubMed] [Google Scholar]
- 36.Rousset F. Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 37.Kamvar ZN, Tabima JF, Grünwald NJ. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2014;2:e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 40.Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 41.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cristescu ME. Genetic reconstructions of invasion history. Molecular Ecology. 2015;24:2212–2225. doi: 10.1111/mec.13117. [DOI] [PubMed] [Google Scholar]
- 43.Mojumdar CH. Culture of Hilsa. Mod. Rev. 1939;66:293–295. [Google Scholar]
- 44.Pillay SR, Rosa H. Synopsis of biological data on Hilsa, Hilsa ilishia (Hamilton 1882) FAO Fisheries Biology Synopsis, 1963;25(1):l–6. [Google Scholar]
- 45.Dutt S. The Indian shad, Hilsa shad (Hamilton) in the sea. Current Science. 1966;35(13):329–330. [Google Scholar]
- 46.Raja, B. T. A. A review of the biology and fisheries of Hilsa ilisha in the upper Bay of Bengal. Bay of Bengal Programme, BOBP/WP/37. Marine Fishery Resource Management in the Bay of Bengal, 66 pp. (1985)
- 47.BOBPHilsa investigation in Bangladesh. Bay of Bengal Programme (BOBP). UNDP, Colombo, 113 pp. (1987).
- 48.Mazid, M. A. & Islam, M. S. Hilsa Fishery Development and Management (in Bengali). A report published by Fisheries Research Institute, Mymenshing, Bangladesh, June 1991 (1991).
- 49.Rahman M, Nævdal G. Studies on hemoglobins of Hilsa shad by isoelectric focusing. Report series, Department of Fisheries and Marine Biology, University of Bergen (IFM Rapport) 1997;16:6. [Google Scholar]
- 50.Rahman M, Nævdal G. Identification of juvenile Hilsa shad in Bangladesh by genetic methods. Fisheries Management and Ecology. 1998;5:255–260. doi: 10.1046/j.1365-2400.1998.00103.x. [DOI] [Google Scholar]
- 51.Hussain, M. G., Salini, J. P., Islam, M. S. & Mazid, M. A. Genetic structure of Hilsa shad populations: preliminary results using starch gel allozymes. In: Hilsa Fisheries Research in Bangladesh. (eds Mazid, M. A., Blaber), pp 37–42. Bangladesh Fisheries Research Institute, Mymensingh, Bangladesh (1998).
- 52.Verma, R, Singh, M & Kumar, S. Unraveling the Limits of Mitochondrial Control Region to Estimate the Fine Scale Population Genetic Differentiation in Anadromous Fish Tenualosa ilisha. Scientifica, Article ID 2035240, 1–9, 10.1155/2016/2035240 (2016). [DOI] [PMC free article] [PubMed]
- 53.Santos MCF, Ruffino ML, Farias JP. High levels of genetic variability and panmixia of the tambaqui Colossoma macropomum (Cuvier, 1816) in the main channel of the Amazon River. Journal of Fish Biology. 2007;71(Suppl):33–44. doi: 10.1111/j.1095-8649.2007.01514.x. [DOI] [Google Scholar]
- 54.De DK, Saigal BN. Spawning of Hilsa, Tenualosa ilisha (Ham) in the Hooghly estuary. Journal of the Inland Fisheries Society of India. 1989;21:46–48. [Google Scholar]
- 55.Quddus MMA, Shimizu M, Nose Y. Meristic and morphometric differences in two types of Hilsa ilisha in Bangladesh waters. Bulletin of the Japanese Society of Scientific Fisheries. 1984;50(1):43–49. doi: 10.2331/suisan.50.43. [DOI] [Google Scholar]
- 56.Ghosh AN, Bhattacharya RK, Rao KV. On the identification of the sub-populations of Hilsa ilisha (Ham.) in the Gangetic system with a note on their distribution. Proceedings of the National Academy of Sciences, India, Section B: Biological Sciences. 1968;34:44–57. [Google Scholar]
- 57.Shafi M, Quddus MMA, Hossain H. A morphometric study of the population of Hilsa ilisha (Hamilton-Buchanan) from the river Meghna. Proceeding Second Bangladesh Science Conferencing. 1977;2:22–24. [Google Scholar]
- 58.Quddus, M. M. A. Two types of Hilsa ilisha and their population biology from Bangladesh waters. Thesis submitted for Doctor of Agricultural Science at the University of Tokyo. Japan, 180 pp. (1982).
- 59.Pillay SR, Rao KV, Mathur PK. Preliminary report on the tagging of the Hilsa, Hilsa ilisha (Hamilton) Proceedings of Indo-Pacific Fisheries Council. 1962;10:28–36. [Google Scholar]
- 60.Liu T, Lin K. The distribution pattern of genetic variation in the transcript isoforms of the alternatively spliced protein-coding genes in the human genome. Molecular Biosystem. 2015;11:1378–1388. doi: 10.1039/C5MB00132C. [DOI] [PubMed] [Google Scholar]
- 61.Mitchell P, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 62.Kwan T, et al. Tissue Effect on Genetic Control of Transcript Isoform Variation. Plos Genetics. 2009;5(8):e1000608. doi: 10.1371/journal.pgen.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braunschweig U, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Research. 2014;24(11):1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang S, et al. Isoform Evolution in Primates through Independent Combination of Alternative RNA Processing Events. Molecular Biology and Evolution. 2017;34(10):2453–2468. doi: 10.1093/molbev/msx212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosetti N, Remis M. Spatial Genetic Structure and Mitochondrial DNA Phylogeography of Argentinean Populations of the Grasshopper Dichroplus elongatus. Plos ONE. 2012;7(7):e40807. doi: 10.1371/journal.pone.0040807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Y, Gentekaki E, Yi Z, Lin X. Genetic Differentiation of the Mitochondrial Cytochrome Oxidase c Subunit I Gene in Genus Paramecium (Protista, Ciliophora) Plos ONE. 2013;8(10):e77044. doi: 10.1371/journal.pone.0077044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our raw data with SRA accession number PRJNA503852 are available online in the NCBI sequence read archive (https://www.ncbi.nlm.nih.gov/sra/PRJNA503852, data publicly released on 31st December 2018).