Abstract
Cardiac Valve Disease is one of the most common heart disorders with an emerging epidemic of cardiac valve degeneration due to aging. Zebrafish can regenerate most of their organs, including their heart. We aimed to explore the regenerative potential of cardiac valves and the underlying molecular mechanisms involved. We used an inducible, tissue-specific system of chemogenetic ablation and showed that zebrafish can also regenerate their cardiac valves. Upon valvular damage at larval stages, the intracardiac flow pattern becomes reminiscent of the early embryonic stages, exhibiting an increase in the retrograde flow fraction through the atrioventricular canal. As a result of the altered hemodynamics, notch1b and klf2a expression are ectopically upregulated, adopting the expression pattern of earlier developmental stages. We find that Notch signaling is re-activated upon valvular damage both at larval and adult stages and that it is required during the initial regeneration phase of cardiac valves. Our results introduce an animal model of cardiac valve specific ablation and regeneration.
Subject terms: Disease model, Zebrafish, Regeneration
Introduction
Zebrafish valve development is a unique system where we can study the interactions between morphogenesis and function of the heart. Cardiac valves develop after the heart starts beating and since zebrafish hearts have a single atrium and ventricle the cells are easy to trace and follow at single-cell resolution1. Zebrafish embryos can survive even in the absence of a fully functional cardiovascular system for several days because they receive enough oxygen by passive diffusion from the water they grow. This feature enables the study of severe heart mutations that would be instantly lethal in mammalian embryos and provide the ability to manipulate cardiac function and intracardiac flow dynamics2. Taking advantage of surgical and pharmacological manipulations as well as of mutants with defective myocardial contractility (silent heart, weak atrium) and altered intracardiac flow dynamic due to changes in heart geometry (southpaw), zebrafish has been pivotal in studying these interactions in vivo3–8. These studies are largely facilitated by the ability to do high-resolution imaging at the cellular level, using high-speed cameras and/or Selective Plane Illumination Microscopy (SPIM) imaging and image reconstruction8,9, allowing a better understanding of the effects of shear-stress on cardiac valve cells.
Notch is one of the most ancient and conserved signaling pathways10 and has been connected to cardiac and, more specifically, valve development in several occasions. Mutations in NOTCH signaling elements cause congenital heart defects and NOTCH1 heterozygous Humans show increased risk of bicuspid aortic valve and aortic valve calcification11. Notch remains active in the valve region throughout the lifetime of an organism (including zebrafish) in the high shear-stress regions and acts to suppress signaling pathways that could lead to valve calcification12,13. Aging is the highest risk factor for valve calcification and in fact mice with shortened telomeres exhibit the age-dependent human phenotypes from neonatal stages14. Zebrafish emerged as a valuable model to study organ regeneration. Several tissues such as the fin15, the myocardium16, the pancreas17,18 and most other organs tested19 showed regenerative capability and the signaling pathways and mechanisms identified are now being tested in mammalian systems.
Tissue-engineered heart valves with regenerative capacities are expected to be a promising alternative to the current surgery treatments, particularly for young patients20. We set out to explore the regenerative capacity of cardiac valves and identify signaling pathways that are necessary for this. Since zebrafish cardiac valve cells even in adult stages are not available to surgery, and there are no available valve specific promoters, we developed an inducible two component system using specific GAL4 driver lines to express the Nitroreductase gene in valve cells and induce their damage by adding Metronidazole in the fish water. We found that reactivation of the Notch signaling pathway, following valvular damage is necessary for the initiation of valve regeneration in both larval and adult stages.
Results
An inducible valve ablation system reveals the regenerative potential of zebrafish cardiac valves
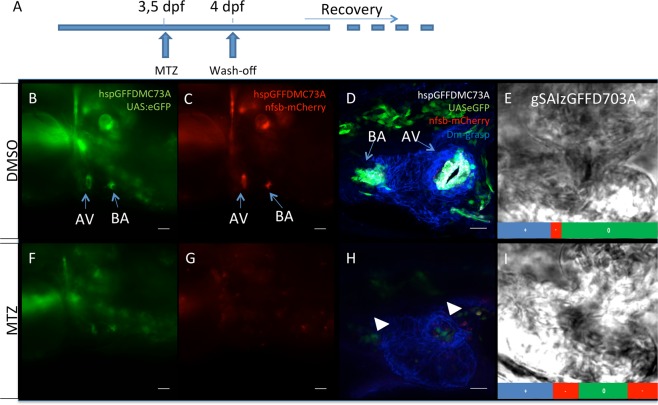
There is currently a lack of well-characterized promoters for valve specific expression pattern. We have conducted a large-scale screen for the GAL4 transgenic fish using the gene trap and enhancer trap methods at the National Institute of Genetics, Mishima, Japan. In the course of the screen, we identified two cardiac valve driver lines with different pattern and intensity of expression within the cardiac valves Tg(hspGFFDMC73A) and Tg(gSAIzGFFD703A). Tg(hspGFFDMC73A) drives, at 72hpf, robust expression in both Valve Endocardial Cells (VECs) (cuboidal cells at the AV boundary, arrowheads in Supplementary Fig. 1B) and future Valve Interstitial Cells (VICs) which at these stages largely overlap with the TCF positive, mesenchymal looking cells (arrows in Supplementary Fig. 1C) and this expression pattern remains on up to adulthood. Tg(gSAIzGFFD703A) shows weaker expression and is restricted to VECs at 72hpf (Supplementary Fig. 1D–F). We crossed both driver lines to the Tg(UAS-E1b:NfsB-mCherry) to test the regenerative potential of cardiac valves initially at larval stages. We optimized a metronidazole-induced ablation of valve cells at 96 hours post fertilization (hpf) (Fig. 1A), which resulted in the reproducible ablation of >80% NTR positive cells (Fig. 1 and Supplementary Fig. 2A). In order to test if cardiac valve cell ablation actually had the predicted effect on cardiac function, we quantified the intracardiac blood flow dynamics and confirmed an increase (4,91 folds) in the retrograde flow fraction of the hemodynamic pattern (Supplementary video 1 untreated, 2 ablated quantified in Fig. 1E,I and Supplementary Fig. 3A). The flow pattern of the embryos with valvular ablation is reminiscent of earlier stages (72 hpf) during valve development when the valve cells are few and they are not capable of fully preventing retrograde blood flow6. Chemogenetic ablation of valve cells in the Tg(hspGFFDMC73A) showed a higher increase of the retrograde flow fraction in accordance with the number of mCherry positive cells that were ablated (Supplementary Fig. 1C, compare with Supplementary Fig. 1F). The ablation was confirmed to be mediated via apoptosis, as detected in the MTZ treated embryos with TUNEL assay (Supplementary Fig. 4).We washed off metronidazole and followed larvae for the following eight days. We identified that GFP and mCherry positive cells started reappearing already at 2 days post ablation (Fig. 2A,B compare with 2D,E) and they were comparable to the untreated larvae by eight days post ablation (Fig. 2C compare with 2F and quantified in Supplementary Fig. 2B). Concerning the flow pattern at this developmental stage, no differences were observed between retrograde blood flows for both untreated (Supplementary video 3) and recovering from treatment embryos (Supplementary video 4 and quantified in Supplementary Fig. 3D).
Figure 1.
Chemically induced genetic cell ablation of zebrafish larval cardiac valves. (A) Outline of chemical treatment of UAS-E1b:NfsB-mCherry transgenic embryos with Metronidazole (MTZ). Metronidazole was applied at 3.5 dpf embryos and washed-off at 4 dpf. Embryos were then left for recovery in order to observe regeneration process. (B,C) Gfp and mCherry Gal4 driven expression in 4 dpf cardiac valves (Arrows). AV: atrioventricular canal, (BA): bulbus arteriosus. Scale bars: 100 μm. (D) Confocal z stack projection of a double transgenic Tg(hspGFFDMC73A/UAS-E1b:NfsB-mCherry) embryonic heart stained with cell-cell adhesion marker Dm:grasp (blue). Scale bar: 20 μm (F,G) Gfp and mCherry Gal4 driven expression in 4 dpf cardiac valves of MTZ treated embryos. Injury is observed at AV canal and BA nfsb-mCherry+ cells that have been ablated. Scale bars: 100 μm. (H) Confocal z-stack projection of a double transgenic Tg(hspGFFDMC73A/UAS-E1b:NfsB-mCherry) heart treated with MTZ and stained with cell-cell adhesion marker Dm-grasp (blue). Arrowheads depict the position where AV canal and BA differentiated cells should be. Scale bar: 20 μm. (E–I) Snapshots of high frame live videos of a DMSO treated and a MTZ-treated Tg(gSAIzGFFD703A/UAS-E1b:NfsB-mCherry) embryo 4 dpf, respectively (S. Movies 3 and 4). Bars show the haemodynamic flow patterns at the atriovantricular canal of uninjured and injured valves per heartbeat. The number of frames per total frames of a heartbeat was measured for forward flow (+) no flow (0) and reverse flow (−). The reverse flow fraction is increased 4,91 times (quantified from n = 8 ctrl embryos and 8 MTZ-treated embryos. p < 0.001 using paired t-test) upon valve ablation. +: forward fraction. 0: null fraction. −: reverse fraction.
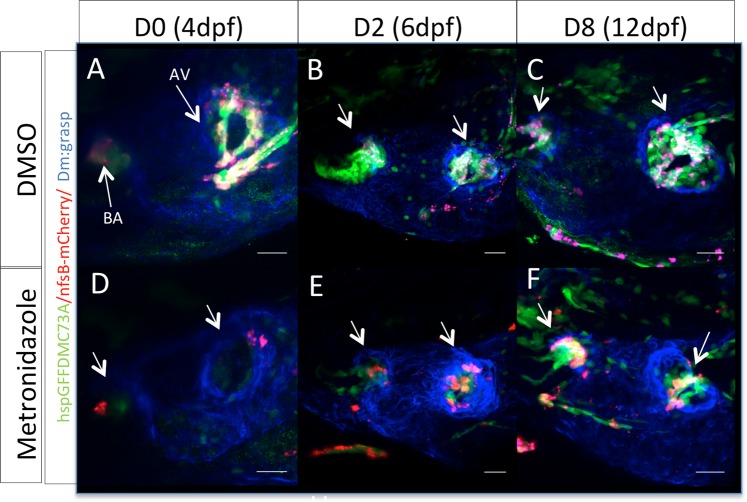
Figure 2.
Zebrafish embryonic valves can regenerate. (A–C) Confocal z-stacks of untreated Tg(hspGFFDMC73A/UAS-E1b:NfsB-mCherry) larvae at 4 dpf, 6 dpf and 12 dpf respectively. (D–F) Confocal z-stacks of MTZ treated Tg(hspGFFDMC73A/UAS-E1b:NfsB-mCherry) and larvae at 4 dpf, 6 dpf and 12 dpf respectively. Arrows show the ablation of mCherry+ cells at the AV canal and OFT at 4 dpf. From 6 dpf more mCherry+ cells could be observed at the AV canal. By 12 dpf the regeneration process could be imaged, as mCherry+, AV differentiated cells were present at both valves. Scale bars: 20 microns. Each experiment was carried out at least 3 independent times with n = 10 embryos per experimental group.
Notch1 is reactivated ectopically upon valvular damage and is necessary for valve regeneration
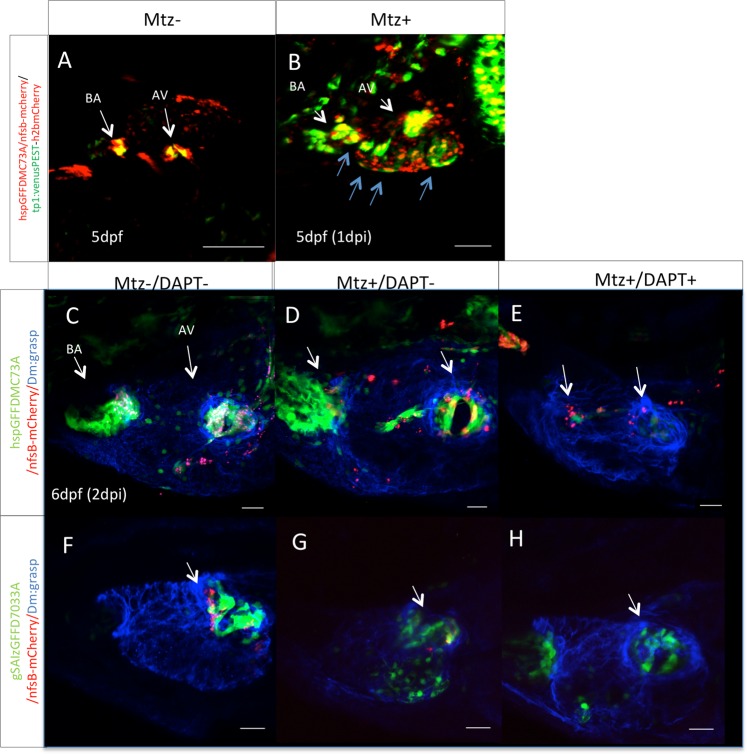
In order to understand the underlying mechanisms of cardiac valve regeneration, we used several transgenic lines and in situ hybridization experiments. We identified that the Notch reporter line showed ectopic upregulation of expression following valvular damage. Both the shear-stress sensitive kruppel-like factor 2a (klf2a) and notch1b are ectopically upregulated 24 hours following valvular ablation (Supplementary Fig. 5,B, D compare with 5,A,C). In the destabilized version of the notch reporter line Tg(TP1:VenusPEST), Notch signaling was activated in endocardial cells adjacent to the damaged area (Fig. 3B), while in the Tg(TP1:h2bmCherry) the expression domain of mCherry positive cells is expanded throughout the endocardium (Fig. 3B compare with 3A), since it represents the accumulating activation of ectopic Notch signaling over time. When DAPT was added in the water during the regeneration phase, no ectopic Notch activation was observed as expected. In addition, the regeneration process also halted, as monitored by the lack of reappearing UAS-E1b:NfsB-mCherry positive cells at the valve region (Fig. 3E compare with 3D, 3C and 3H, compare with 3G, 3F and quantified in Supplementary Fig. 6).
Figure 3.
Notch signaling is activated during valve regeneration. (A) Confocal z-stack of a Tg(TP1:VenusPEST)/hspGFFDMC73A: UAS-E1b:NfsB-mCherry double transgenic embryo at 5 dpf. Tg(TP1:VenusPEST) signal of activated Notch signaling is restricted to the valves. Scale bar: 100 microns. (B) 1 day post MTZ treatments. Expansion of activation of Notch signaling throughout the ventricle is observed (blue arrows) Scale bar: 50microns. Confocal z-stacks of untreated Tg(hspGFFDMC73A: UAS-E1b:NfsB-mCherry) (C) and Tg(GSAIzGFFD703A: UAS-E1b:NfsB-mCherry) (F) double transgenic embryos at 6 dpf. Confocal z-stack of MTZ treated Tg(hspGFFDMC73A: UAS-E1b:NfsB-mCherry) (D), and Tg(GSAIzGFFD703A: UAS-E1b:NfsB-mCherry) (G), at 6 dpf. Confocal z-stack of MTZ treated and then during regeneration 48 hours DAPT treated Tg(hspGFFDMC73A: UAS-E1b:NfsB-mCherry) (E), and Tg(GSAIzGFFD703A: UAS-E1b:NfsB-mCherry) (H), at 6 dpf. (I) Each experiment was carried out at least 3 independent times with n = 10 embryos per experimental group. Scale bars: 20 micron.
Notch signaling is upregulated following adult cardiac valve ablation and is necessary for their regeneration
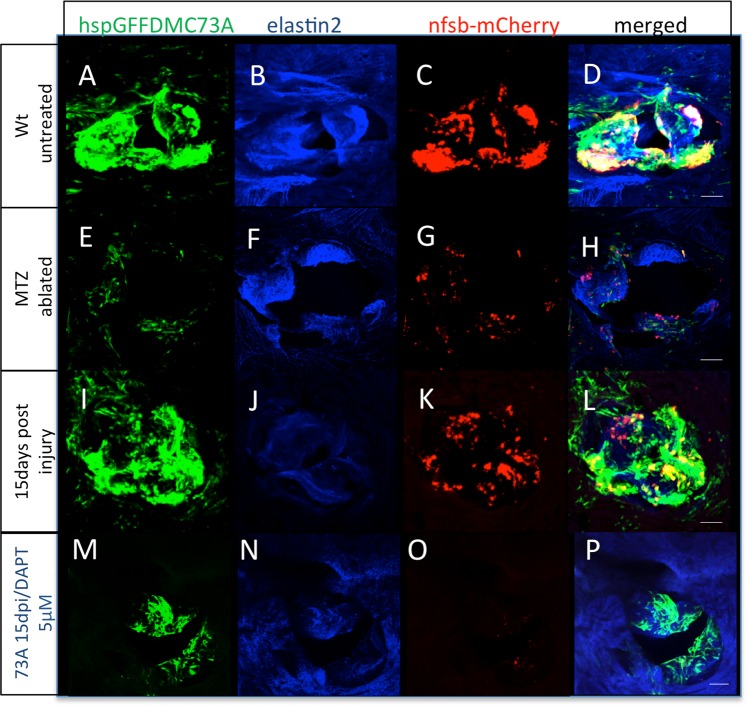
The Tg(hspGFFDMC73A) driver line remains active also at adult stages in both VECs and VICs (Fig. 4A–D, Supplementary Movie 5). We added metronidazole for 12 hours and dissected hearts following the treatment to show that most of the UAS-E1b:NfsB-mCherry positive cells were ablated (Figure E-H, Supplementary Movie 6). Again, at this developmental stage, apoptosis was detected in MTZ treated adult valves with the TUNEL assay (Supplementary Fig. 7). We also dissected Tg(hspGFFDMC73A/UAS-E1b:NfsB-mCherry) hearts that carry the Tg(TP1:VenusPEST) transgene and showed that Notch is upregulated at the valve region (Supplementary Movies 7,8 and Supplementary Fig. 8,C,D compare with S8A,B, quantified in S8E). We allowed the fish to recover in system water or system water with 5 μM DAPT. Adult animals that were allowed to recover showed reappearance of UAS-E1b:NfsB-mCherry positive cells within 15 days following injury (Fig. 4I–L, Supplementary Movie 9) while we observed that Notch signaling inhibition significantly hampered the regenerative potential of adult cardiac valves (Fig. 4M–P, Supplementary Movie 10).
Figure 4.
Zebrafish adult valves retain the ability to regenerate through Notch signaling reactivation. (A–D) Z-stack projection of a wt untreated adult valve of an hspGFFDMC73A/UAS-E1b:NfsB-mCherry transgenic fish stained with elastin2. (E–H) Valvular damage of an adult heart of a hspGFFDMC73A/UAS-E1b:NfsB-mCherry transgenic immediately after completion of the MTZ treatment (n = 10). (I–L) Regenerating valve 15 days post chemogenetic valve cell ablation (n = 13). (M–P) Inhibition of regeneration process in the heart of an adult heart of a hspGFFDMC73A/UAS-E1b:NfsB-mCherry transgenic after MTZ treatment and 15 day treatment with DAPT (n = 12). Scale bars: 50 micron.
Discussion
In this study, we describe the ability of cardiac valves to regenerate after chemogenetic ablation in larval and adult zebrafish. Moreover, we introduce 2 new transgenic zebrafish lines with valve expression promoters. These lines could further contribute to valve specific expression or silencing of genes of interest. Recent advances on imaging and the identification of novel cardiac valve mutants and gene networks, have helped deciphering the effect of intracardiac blood flow dynamics and shear stress on the endothelial cells that are destined to become valvular. Klf2a is the best-characterized flow sensitive transcription factor to date6,21,22. Some of its key downstream targets include the Cerebral Cavernous Malformations proteins (CCM)23,24 and Notch that are very important for proper valve morphogenesis1,11,12,25–27. Endocardial notch1b activation requires functional primary cilia28 and klf2a6,21,22. Klf2a has been recently shown to be also required for myocardial trabeculation integrity during development via the modulation of Fgf signaling29 as well as for myocardial reprogramming via its well-established endocardial hemodynamic response and Notch mediated function30. The CCM proteins appear to function antagonistically to the activation of β1 integrin by shear stress31. Knockdown of β1 integrin suppresses the cardiovascular defects of ccm mutant embryos23. In addition to the intracardiac flow dynamics and endocardial/myocardial interactions at the valve-forming region, it is worth noticing that there is extensive extracellular matrix components (ECM) also known as cardiac jelly. One of its major components is hyaluronic acid, produced by the Has2 enzyme that is tightly regulated during AV development to restrict the AV region via the BMP signaling pathway32 as well as via mir2333. Wnt signaling is also required for valve development34,35. Fibronectin synthesis has been shown to be flow dependent and downstream of Klf2a, coupling the mechanosensory system to ECM composition36.
It is becoming clear that valve regeneration would require several steps, including the proliferation of endocardial cells, their transformation to interstitial cells and the tightly regulated production of several ECM components. Therefore, cardiac valve tissue engineering (CVTE) would require the combination of optimized biomaterials with different cell types and conditioning protocols making it a very challenging process. Despite the challenges, CVTE is expected to be a promising therapeutic alternative to mechanical and bioprosthetic valves. Both of these types require life-long anticoagulative therapies and accumulate damages due to the stressful hemodynamic microenvironment of cardiac valves, since they do not contain any living cells with adaptive or regenerative potential. Such potential is particularly vital in pediatric patients whose cardiac valves need to adapt to the growing size of their hearts. The optimization of recellularization protocols to accommodate the need for replacement valves that could grow/adapt with somatic outgrowth requires the best understanding of valve development and regeneration mechanisms20,37. Here we addressed the initial step of valve regeneration that results from the functional consequence of a dysfunctional valve: the increase of retrograde blood flow.
We propose a system where the immature flow patterns are sensed by the endocardial cells, and the increase of the retrograde intracardiac flow pattern, due to a dysfunctional valve, could be the stimulus for its regeneration. Following injury, the mechanosensitive transcription factor klf2a is upregulated, and a Notch dependent developmental program is activated for valve regeneration. This is reminiscent of the embryonic expression pattern of these signaling pathways (Fig. 5). Recapitulating development is a recurrent scenario during the regeneration process of different organs. Studies of in vitro cell systems that incorporate biomechanical stress and conditioning as well as in silico computational modeling of hemodynamics for tissue-engineered heart valve shape optimization37 would greatly benefit from in vivo experimental systems, such as the zebrafish, that show endogenous regenerative potential of their cardiac valves.
Figure 5.
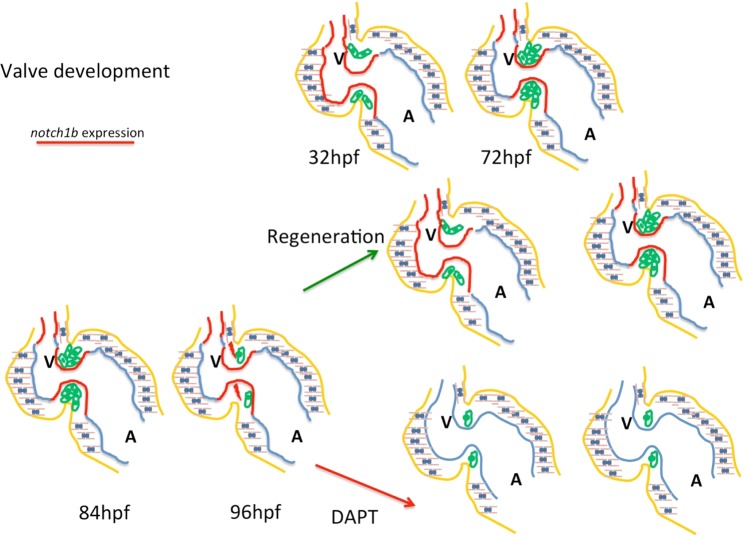
Sensing of immature intracardiac flow patterns activates a Notch mediated regenerative program, recapitulating cardiac valve development. At embryonic developmental stages a feedback loop where intracardiac flow dynamics regulate klf2a and notch1b results in the restricted expression of notch1b at the high sheer-stress areas of the heart: the atrioventricular valve and the outflow tract. The intracardiac flow pattern following valvular damage, is disturbed causing an increase in the retrograde blood flow fraction, reminiscent of the flow pattern of earlier stage of embryogenesis. As a result, notch1b is re-activated ectopically the ventricular endocardium. When cardiac valves regenerate and flow dynamics are restored, notch1b expression gets restricted at the atrioventricular canal and the outflow tract.
Methods
Zebrafish transgenic lines
gSAIzGFFD703A and hspGFFDMC73A zebrafish lines were generated during a large-scale screen for zebrafish transgenic Gal4 lines by the gene trap and enhancer trap methods at the Kawakami lab38,39. More information on the insertion site and expression pattern can be found at http://kawakami.lab.nig.ac.jp/ztrap/. The transgenic lines Tg(UAS:eGFP1A)38 and Tg(UAS-E1b:NfsB-mCherry)c26418, which we will refer to as NfsB-mCherry for short in figures, were crossed with the above lines to create double or triple transgenic lines. hspGFFDMC73A/Tg(UAS-E1b:NfsB-mCherry)c264 double transgenics were crossed with Tg(Tp1:venus-PEST) and/or Tg(Tp1:h2B:mCherry)40 in order to create appropriate Green/Red combinations of the transgenics to distinguish Notch signaling reporter expression following valve cell ablation. Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 line35 was also used to image expression pattern and the overlap of mesenchymal-like TCF positive cells with gSAIzGFFD703A line.
Metronidazole (Mtz) treatment of zebrafish embryos and adults
Mtz (Sigma) was diluted in embryo water/0,2%DMSO in a final concentration of 4 mM for hspGFFDMC73A/UAS-E1b:NfsB-mCherry 3 dpf embryos and at 10 mM for gSAIzGFFD703A/UAS-E1b:NfsB-mCherry 3 dpf embryos. 0,2% DMSO in embryo water was used as a control. Mtz treatment (concentration and exposure time) was optimized for the two lines and the different developmental stages to ensure reproducibility between larvae and adults as well as >80% of successful elimination of NTR+ cells. Embryos were treated for 12 and 20 hours for the 73 A and 703 A lines, respectively. Embryos were then washed-off Mtz and left for recovery for 8days. For adult transgenic fish hspGFFDMC73A/UAS-E1b:NfsB-mCherry Mtz treatments, fish were treated with 5 mM in system water for 12 hours and left for the mentioned interval to recover before proceeding to heart extractions. Euthanasia was carried out by prolonged immersion in water with an overdose of tricaine methane sulfonate (MS222, 300 mg/l).
DAPT treatments
4 dpf fish were treated in 70 μΜ DAPT diluted in EW/1%DMSO until 6 dpf, where they were fixed and immunostained. Adult DAPT treatments took place as previously described with 5 μΜ DAPT final concentration in system water.
Immunofluorescence
Zebrafish embryos were fixed in 4% PFA and stained using the Zn5 (Dm:grasp, Alcama) Ab with Alexa anti-mouse 633 as a secondary antibody. Embryos were then embedded in 4% agarose and sectioned in a vibratome at 180 μΜ sections. Adult hearts were extracted, fixed in 4%PFA, embedded, sectioned and then stained on 180 μΜ vibratome sections. Tropoelastin Ab was previously described41 with Alexa anti-rabbit 633 used as the secondary Ab.
Zebrafish embryos’ live imaging and retrograde flow fraction quantification
5 dpf embryos were anesthetized in tricaine, embedded in 1% low melting agarose and imaged under a Leica confocal microscope. For brightfield videos of 4 dpf embryos ORCA Hamamatsu camera was used and quantification of flow fractions during independent heart beats was measured as previously described7. Bitplane Imaris was used to get 3D videos of stacks of adult zebrafish cardiac valve confocal images.
In situ hybridization
Notch1b and klf2a probes were used to check RNA expression, as previously described7, following MTZ treated embryos.
TUNEL assay
For the combined immunocytochemistry-TUNEL assay, 96hpf embryos and adult fish hearts were fixed overnight in 4%PFA-PBS, embedded in 4% agarose and vibrotome sectioned (130 μm thick sections). Sections were fixed again for 10 min in 4%PFA-PBS, washed with PBS and permeabilized with proteinase K (10 μg/ml) for 10 min. After washing (3 × 5 min) with PBST (1× PBS/0.1% Triton) and blocking with 4%BSA in PBST for 2 hours, sections were incubated with primary antibodies overnight at 4 °C (zn5: Zebrafish International Stock Center, 1:10 for embryos or Eln-2, 1:500 for adult fish). The next day, tissue was washed with PBST (3 × 10 min), rinsed with PBS and stained with TUNEL (In Situ Cell Death Detection Kit, Fluorescein, Roche) for 1 h at 37 °C according to the manufacturer’s instructions. After staining, sections were washed with PBST (3 × 5 min) and incubated with secondary antibodies (Alexa anti-mouse 633 for ZN5 or Alexa anti-rabbit 633 for Eln-2). Before imaging, cells were counterstained with 40, 6-diamidino-2-phenylindole (DAPI).
Quantification and statistical analyses
Statistical details of experiments and exact values of the number of animals used for the analyses can be found at the respective figure legends. Initiation of regeneration was defined as the reoccurrence of UAS-E1b:NfsB-mCherry signal in regenerating valves after MTZ treatments.
Ethical statement
The adult zebrafish regeneration protocol was approved by the BRFAA ethics review board and the Attica Veterinary Department according to the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes (EL25BIO003/5520).
Supplementary information
Acknowledgements
We would like to thank Prof. Fred Keely (SickKids hospital and University of Toronto, Canada) for sharing the elastin2 antibody and Dr. Stamatis Pagakis from the BioImaging facility of BRFAA. This work was supported by the Greek General Secreteriat for Research and Development European Social Fund, the European Union and National Funds (Aristia I, zfValves -270 grant to DB); the Fondation Sante (Research Grant to DB); NBRP; NBRP/Genome Information upgrading program from Amed (KK), and the NIG-Joint 2013-A80 (to DB, PK and KK).
Author contributions
D.B. and K.K. conceived this study. P.K., A.A., D.B., were involved in the experimental set-up and analyzed the data. D.B. wrote the manuscript, P.K., A.A., and K.K. commented and edited the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-52558-y.
References
- 1.Beis D, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132(18):4193–204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 2.Bournele D, Beis D. Zebrafish models of cardiovascular disease. Heart Fail Rev. 2016;21(6):803–813. doi: 10.1007/s10741-016-9579-y. [DOI] [PubMed] [Google Scholar]
- 3.Hove JR, et al. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421(6919):172–177. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- 4.Bartman T, et al. Early myocardial function affects endocardial cushion development in zebrafish. PLoS Biol. 2004;2(5):e0020129. doi: 10.1371/journal.pbio.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DYR. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135(6):1179–1187. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- 6.Vermot J, et al. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 2009;7(11):e1000246. doi: 10.1371/journal.pbio.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalogirou S, et al. Intracardiac flow dynamics regulate atrioventricular valve morphogenesis. Cardiovasc Res. 2014;104(1):49–60. doi: 10.1093/cvr/cvu186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pestel J, et al. Real-time 3D visualization of cellular rearrangements during cardiac valve formation. Development. 2016;143(12):2217–2227. doi: 10.1242/dev.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mickoleit M, et al. High-resolution reconstruction of the beating zebrafish heart. Nat Methods. 2014;11(9):919–922. doi: 10.1038/nmeth.3037. [DOI] [PubMed] [Google Scholar]
- 10.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 11.Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437(7056):270–4. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 12.Theodoris CV, et al. Human disease modeling reveals integrated transcriptional and epigenetic mechanisms of NOTCH1 haploinsufficiency. Cell. 2015;160(6):1072–86. doi: 10.1016/j.cell.2015.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White MP, et al. NOTCH1 regulates matrix gla protein and calcification gene networks in human valve endothelium. J Mol Cell Cardiol. 2015;84:13–23. doi: 10.1016/j.yjmcc.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theodoris CV, et al. Long telomeres protect against age-dependent cardiac disease caused by NOTCH1 haploinsufficiency. J Clin Invest. 2017;127(5):1683–1688. doi: 10.1172/JCI90338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfefferli C, Jaźwińska A. The art of fin regeneration in zebrafish. Regeneration. 2015;2015(2(2)):72–83. doi: 10.1002/reg2.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González-Rosa JM, Burns CE, Burns CG. Zebrafish heart regeneration: 15 years of discoveries. Regeneration (Oxf). 2017;4(3):105–123. doi: 10.1002/reg2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curado S, et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236(4):1025–35. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 18.Davison JM, et al. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013;29(11):611–20. doi: 10.1016/j.tig.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Usprech J, Chen WL, Simmons CA. Heart valve regeneration: the need for systems approaches. Wiley Interdiscip Rev Syst Biol Med. 2016;8(2):169–82. doi: 10.1002/wsbm.1329. [DOI] [PubMed] [Google Scholar]
- 21.Heckel E, et al. Oscillatory Flow Modulates Mechanosensitive klf2a Expression through trpv4 and trpp2 during Heart Valve Development. Curr Biol. 2015;25(10):1354–61. doi: 10.1016/j.cub.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 22.Duchemin AL, Vignes H, Vermot J. Mechanically activated piezo channels modulate outflow tract valve development through the Yap1 and Klf2-Notch signaling axis. eLife. 2019;8:e44706. doi: 10.7554/eLife.44706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renz M, et al. Regulation of β1 Integrin-Klf2-Mediated Angiogenesis by CCM Proteins. Dev Cell. 2015;32(2):181–190. doi: 10.1016/j.devcel.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 24.Donat S, Lourenço M, Paolini A, Otten C, Renz M. Abdelilah-Seyfried S. Heg1 and Ccm1/2 proteins control endocardial mechanosensitivity during zebrafish valvulogenesis. eLife. 2018;7:e28939. doi: 10.7554/eLife.28939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Timmerman LA, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torregrosa-Carrión R, et al. NOTCH Activation Promotes Valve Formation by Regulating the Endocardial Secretome. Mol Cell Proteomics. 2019;18(9):1782–1795. doi: 10.1074/mcp.RA119.001492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu JJ, et al. Contractile and hemodynamic forces coordinate Notch1b-mediated outflow tract valve formation. JCI Insight. 2019;5:124460. doi: 10.1172/jci.insight.124460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samsa LA, et al. Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish. Development. 2015;142(23):4080–4091. doi: 10.1242/dev.125724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasouli SJ, et al. The flow responsive transcription factor Klf2 is required for myocardial wall integrity by modulating Fgf signaling. eLife. 2018;7:e38889. doi: 10.7554/eLife.38889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gálvez-Santisteban M, et al. Hemodynamic-mediated endocardial signaling controls in vivo myocardial reprogramming. eLife. 2019;8:e44816. doi: 10.7554/eLife.44816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macek Jilkova Z, et al. CCM proteins control endothelial 1 integrin dependent response to shear stress. Biol Open. 2014;3(12):1228–35. doi: 10.1242/bio.201410132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith KA, et al. Transmembrane protein 2 (Tmem2) is required to regionally restrict atrioventricular canal boundary and endocardial cushion development. Development. 2011;138(19):4193–4198. doi: 10.1242/dev.065375. [DOI] [PubMed] [Google Scholar]
- 33.Lagendijk AK, Goumans MJ, Burkhard SB, Bakkers J. MicroRNA-23 restricts cardiac valve formation by inhibiting has2 and extracellular hyaluronic acid production. Circ Res. 2011;109(6):649–657. doi: 10.1161/CIRCRESAHA.111.247635. [DOI] [PubMed] [Google Scholar]
- 34.Hurlstone AF, et al. The Wnt/β-catenin pathway regulates cardiac valve formation. Nature. 2003;425(6958):633–637. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- 35.Moro E, et al. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev Biol. 2012;366(2):327–340. doi: 10.1016/j.ydbio.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Steed E, et al. Klf2a couples mechanotransduction and zebrafish valve morphogenesis through fibronectin synthesis. Nat Commun. 2016;7:11646. doi: 10.1038/ncomms11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emmert MY, et al. Computational modeling guides tissue-engineered heart valve design for long-term in vivo performance in a translational sheep model. Sci Transl Med. 2018;10(440):eaan4587. doi: 10.1126/scitranslmed.aan4587. [DOI] [PubMed] [Google Scholar]
- 38.Asakawa K, et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA. 2008;105:1255–1260. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawakami K, et al. zTrap: zebrafish gene trap and enhancer trap database. BMC Developmental Biology. 2010;10(1):105. doi: 10.1186/1471-213X-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ninov N, Borius M, Stainier DYR. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139(9):1557–1567. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miao M, Bruce AEE, Bhanji T, Davis EC, Keeley FW. Differential expression of two tropoelastin genes in zebrafish. Matrix Biol. 2007;26(2):115–124. doi: 10.1016/j.matbio.2006.09.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.