Abstract
Purpose
For neurodevelopmental disorders (NDDs), etiological evaluation can be a diagnostic odyssey involving numerous genetic tests, underscoring the need to develop a streamlined algorithm maximizing molecular diagnostic yield for this clinical indication. Our objective was to compare the yield of exome sequencing (ES) with that of chromosomal microarray (CMA), the current first-tier test for NDDs.
Methods
We performed a PubMed scoping review and meta-analysis investigating the diagnostic yield of ES for NDDs as the basis of a consensus development conference. We defined NDD as global developmental delay, intellectual disability, and/or autism spectrum disorder. The consensus development conference included input from genetics professionals, pediatric neurologists, and developmental behavioral pediatricians.
Results
After applying strict inclusion/exclusion criteria, we identified 30 articles with data on molecular diagnostic yield in individuals with isolated NDD, or NDD plus associated conditions (such as Rett-like features). Yield of ES was 36% overall, 31% for isolated NDD, and 53% for the NDD plus associated conditions. ES yield for NDDs is markedly greater than previous studies of CMA (15–20%).
Conclusion
Our review demonstrates that ES consistently outperforms CMA for evaluation of unexplained NDDs. We propose a diagnostic algorithm placing ES at the beginning of the evaluation of unexplained NDDs.
Keywords: autism, consensus development conference, diagnostic yield, genetic testing, intellectual disability
INTRODUCTION
Neurodevelopmental disorders (NDDs) are a heterogeneous group of conditions that impact brain development and affect various aspects of daily functioning. At least 30% of NDDs are thought to have a genetic basis.1 Among these disorders, global developmental delay (GDD)—a precursor diagnosis to intellectual disability (ID)—and autism spectrum disorder (ASD) are two entities for which there are guidelines for genetic testing.2,3 The 2010 guideline from the American College of Medical Genetics and Genomics (ACMG) suggests that chromosomal microarray (CMA) and fragile X (FXS) testing should be first-tier tests for individuals with unexplained GDD/ID and/or ASD (except for females with ASD and normal cognition, for which FXS testing is not recommended).3 There are also considerations for single-gene testing of MECP2 and PTEN under certain circumstances,2 although gene panels are now typically preferred over single-gene testing. These recommendations and practices exist because of the complex genetic heterogeneity underlying NDDs that sometimes makes diagnostic determinations difficult based on history/examination alone.4 Even when following these guidelines, establishing a diagnosis can require iterative genetic testing. Previous expert guidance on genetic testing for NDDs has advanced testing as standard of care for individuals with NDDs.2,5 Such guidelines also predate the rapid uptake of clinical exome sequencing (ES) and therefore ES is not addressed.
Due to the genetic heterogeneity of NDDs, and the current standard of genome-wide CMA as the first-tier test, we focused on ES in comparison with CMA because it also provides a genome-wide assessment. The advent of ES has elucidated monogenic forms of GDD/ID and/or ASD not detectable by CMA, FXS testing, or single-gene sequencing of MECP2 or PTEN.4,6 For individuals with GDD/ID, ASD, and/or multiple congenital anomalies (MCAs), the molecular diagnostic yield of CMA is up to 15–20% (ref. 5), including more recent cohorts.7–9 CMA typically detects only chromosomal copy-number variants (CNVs) and regions of homozygosity, not single-gene disorders10 (Table 1). Several ES studies have focused on individuals with varying presentations of NDDs and have identified genetic causes in as many as 61% of cases.1,11,12
Table 1.
Types of genetic variants detected and missed by CMA and ES
| Test | Types of variants detected | Types of variants missed |
|---|---|---|
| CMA | Chromosomal copy-number variants (deletions and duplications) at the level of intragenic exons or larger |
Balanced chromosomal rearrangements Low-level mosaicism Trinucleotide repeat expansion disorders (e.g., fragile X syndrome) Single-gene sequence-level variants found by ES |
| ES | Single-gene variants |
Chromosomal copy-number variants (except when specifically included in analysis pipeline) Balanced chromosomal rearrangements Trinucleotide repeat expansion disorders Intronic/noncoding variants Exon-level deletions/duplications Exonic variants not captured or covered well by sequencing platform |
CMA chromosomal microarray,ES exome sequencing.
Much like CMA, ES is useful for patients with atypical presentations of a genetic disorder, early presentations of a disease for which the classic findings have not appeared, genetic disorders with relatively nonspecific presentations, and multiple genetic conditions. One of the main historical limitations of ES was lack of detection of CNVs (Table 1), though advances in depth-of-sequence coverage analysis and of apparent Mendelian segregation errors can afford detection of CNVs from ES.13 With these advances, some clinical laboratories have started to incorporate CNV analysis into their ES platforms.
The value of elucidating a molecular diagnosis is that it can change clinical management in a number of important ways for individuals and families affected by a NDD. Several studies have examined the impact of a diagnostic CMA finding, showing that it leads to direct changes in patient care for up to 55% of individuals with a positive result.14–17 Similarly, a diagnosis via ES can impact patient care by initiation of surveillance for disease-related conditions, discontinuation of repeated rounds of investigation and irrelevant surveillance, and referrals for further evaluation of associated medical conditions.18 Despite the clear benefit of ES from a diagnostic and management standpoint, there are no accepted guidelines for the use of ES in the evaluation of NDDs.
Therefore, our primary objective was to conduct an evidence-based consensus conference to provide recommendations for the use of ES in the diagnostic evaluation of individuals with NDDs.
MATERIALS AND METHODS
We conducted a scoping review (no registered review protocol) as the basis for a consensus development conference to summarize the molecular diagnostic yield of ES for NDDs, specifically compared with CMA. For the conference, in addition to the core group of experts who authored this paper, we also invited outside experts on CNV analysis from ES data.
Scoping review
For this scoping review, we addressed the following question: Among individuals with NDDs tested by ES, what is the molecular diagnostic yield compared with CMA? We selected articles from PubMed focusing on ES and NDDs. We included studies that involved sequencing of protein-coding regions of Mendelian genes, and excluded studies that were gene panels subsampled from exome data. We defined NDD as GDD, ID, and/or ASD. We searched PubMed with a combination of Medical Subject Headings (MeSH) terms and keywords pertaining to NDDs (i.e., GDD, ID, and/or ASD) and ES, with dates from 1 January 2014 to 29 June 2018 (Table S1). Scoping reviews differ from systematic reviews in that their focus is on more broadly defined research questions, charting of themes, and development of inclusion/exclusion criteria at the study selection stage.19
Article selection occurred in two stages. During the first stage, 12 expert reviewers (representing a diverse range of fields including laboratory genetics, clinical genetics, genetic counseling, developmental behavioral pediatrics, and medical students) were divided into dyads for determination of possible inclusion. Articles included at this stage were primary research studies (e.g., cohort studies, case series with ≥10 participants, review articles, or other articles of interest such as expert opinion). Exclusion criteria were as follows: the article did not (1) include the clinical population of interest, (2) discuss ES, (3) include human data (e.g., an animal or in vitro study only), (4) discuss molecular diagnostic yield, or (5) include ≥10 participants. Each dyad of reviewers was blinded to the other’s choices and independently assigned inclusion/exclusion criteria for each article. When a discrepancy occurred, an independent reviewer assessed whether the article should advance to full article review.
During the second stage, a dyad read the full text of each remaining article for possible inclusion to clarify inclusion eligibility. Reasons for exclusion at this stage were tiered by the following criteria: (1) basic science study, (2) study population not exclusively NDD, (3) <10 participants, (4) did not discuss ES, (5) no mention of molecular diagnostic yield, (6) methods or review paper, or (7) not otherwise defined (book chapter or not in English). We established strict phenotypic NDD guidelines to limit the heterogeneity of included articles. Specifically, when describing the population of interest, the article must have used the phrases “global developmental delay,” “intellectual disability,” and/or “autism spectrum disorder” (or its equivalent). The term “developmental delay” lacked enough specificity unless there were additional context clues to suggest it was referring to GDD/ID. We classified included articles as belonging to one of two categories: (1) isolated NDD: the study population pertained to NDD; or (2) NDD plus associated conditions: the study population pertained to NDD plus a specific clinical finding (defined as any additional neurological, systemic, syndromic, or other clinical characteristic, e.g., microcephaly, neutropenia, or Coffin–Siris syndrome).
We conducted two iterative changes outside of the original scoping review. First, during the conference, members nominated nine additional articles for consideration. Select consensus conference members reviewed the articles’ full text and identified one additional article for inclusion that was not ascertained through the MeSH and keyword search terms. Second, although our manual review included “global developmental delay,” we modified our MeSH and keyword search terms to include “global developmental delay” and to use “exome sequencing” or “whole exome sequencing” (instead of just “whole exome sequencing”). The same select consensus conference members reviewed all the abstracts of the articles not previously captured by the initial MeSH and keyword search terms, and read a subset of the full-text articles, ultimately identifying one more article for inclusion.
Consensus development conference
This effort began as an initiative of the Translational Neuroscience Center (TNC) at Boston Children’s Hospital (BCH), following multiple discussions with experts in the field. The idea of investigating whether there was evidence to support use of clinical ES for patients with NDDs was presented to a TNC donor who provided a modest noncommercial unrestricted donation to support travel expenses of working group members to hold an in-person meeting. The core author group from BCH then convened an interdisciplinary group of experts in clinical genetics, laboratory genetics, genetic counseling, child neurology, neurodevelopmental disabilities, and developmental behavioral pediatrics to determine the need for a scoping review on ES in NDDs. We conducted this project outside the scope of professional organizations for two specific reasons: (1) we wanted the group of experts to represent different countries and disciplines, to create diverse perspectives that would minimize bias; and (2) we considered the question of selecting the genetic test with highest diagnostic yield for NDDs to be an urgent issue, and that our approach would be expedient without compromising the validity of conclusions.
This consensus group developed the framework and selected separate experts to conduct the review. Members of the consensus group provided conflicts of interest disclosure in a standard format. Following the scoping review, the consensus group reconvened to review the evidence curated by the expert review group. We assembled this interdisciplinary team based on practice/geographic diversity and assessments of conflicts of interest, and we used a consensus development conference approach.20 This approach is one of the three main approaches to develop consensus and was chosen for its unique strength of allowing committee members a forum to discuss issues.21 The committee reviewed the evidence table, discussed talks on relevant topics presented by a variety of members and two external experts from clinical laboratories performing ES, voted on the use of ES as a first-tier test for NDDs, and developed a consensus algorithm for genetic testing, including ES, in NDDs.
Statistical analysis
We assessed fixed-effects and random-effects models for model fit. We performed a random-effects meta-regression to determine molecular diagnostic yield of full sample, and stratified by category (isolated NDD, NDD plus associated conditions). Category, year of study, number of participants with a NDD, and number of family members included per proband were evaluated as covariates. For number of family members included, we created three categories: (1) proband-only (all cases proband-only), (2) combination (all cases at least proband), and (3) trio (all casesat least trios). Of note, we included two studies in the combination category in which parental samples were obtained post hoc for variant classification.
Meta-regression was performed using a logistic regression model. Between-study heterogeneity was assessed using between-study variance (τ2), I-2 statistic, and Cochran’s Q-test. Akaike information criterion and Bayesian information criterion were assessed to evaluate model fit. A forest plot was used to summarize our findings.
RESULTS
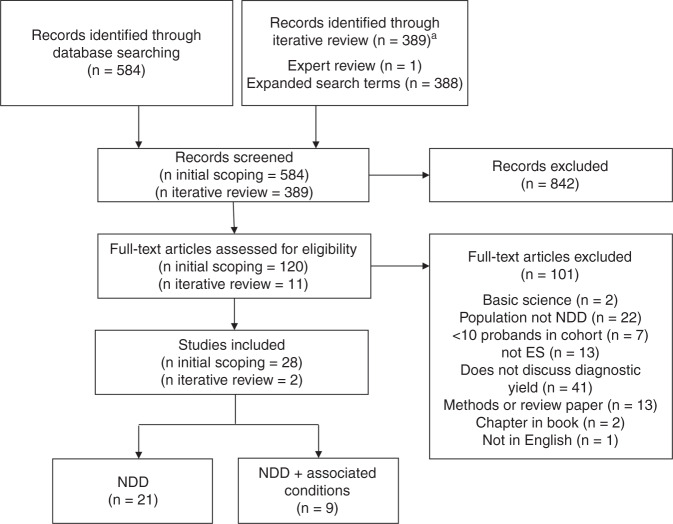
For the scoping review staged selection process, we initially identified 584 unique articles. We eliminated 464 articles after abstract review, and another 92 after full-text review, leaving 28 articles for inclusion. During the iterative review, we identified 389 unique articles, eliminated 378 after abstract review, and another 9 after full-text review, leaving 2 for inclusion. In total, 30 articles were included, which we divided into two categories: those focused on just NDDs or those focused on NDDs plus associated conditions. Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flowchart is shown in Fig. 1.
Fig. 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flowchart of scoping review article inclusion. aRecords identified through iterative review were reviewed by one reviewer at the screening stage.ES exome sequencing,NDD neurodevelopmental disorder.
The 30 articles (Table S2) were diverse in their patient representations. These articles originated from centers around the world, including the United States, Europe, Middle East, and Asia. Among the articles focused on NDD plus associated conditions, the features included features of a clinically defined syndrome (e.g., Coffin–Siris syndrome, DOOR syndrome, Nicolaides–Baraitser syndromes, Rett syndrome, Smith–Magenis syndrome); systemic findings (unexplained metabolic phenotype); associated medical problems (neutropenia); and neurological features (microcephaly, macrocephaly). The types of specialists evaluating the phenotypes of participants represented a diverse variety including clinical geneticists, pediatric neurologists, and developmental pediatricians.
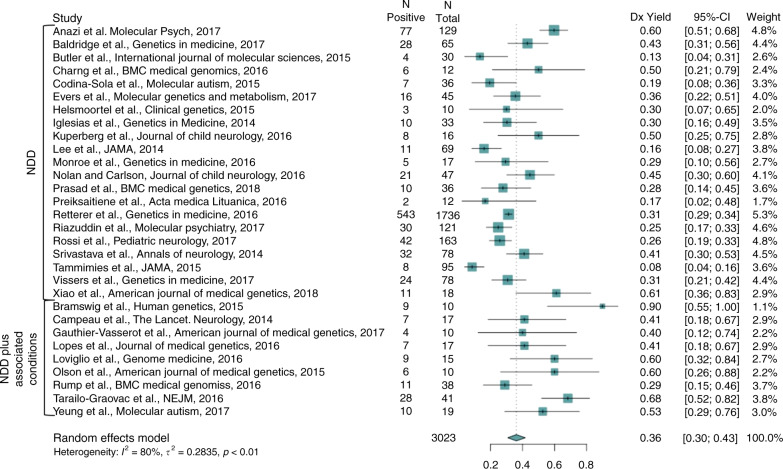
A random-effects meta-analysis of the 30 articles revealed a molecular diagnostic yield of ES of 36% (confidence interval [CI]: 30–43%) weighted by the number of cases in the study (Fig. 2). In the isolated NDD category (n = 21 articles), the yield was 31% (CI: 25–38%), and in the NDD plus associated conditions category (n = 9 articles), the yield was 53% (CI: 41–64%).
Fig. 2.
Forest plot of meta-analysis subcategorized as neurodevelopmental disorder (NDD) and NDD plus associated conditions.CI confidence interval.
Within both the isolated NDD category and the NDD plus associated conditions category, patterns emerged pertaining to molecular diagnostic yield. Among the articles pertaining to the isolated NDD category, there were n = 5 articles including individuals with primarily ASD (molecular diagnostic yield 16%, CI: 11–24%), n = 10 articles that included individuals with primarily ID (yield 39%, CI: 29–50%), n = 6 articles that included individuals with a more heterogeneous mix of ID and/or ASD (yield: 39% (CI: 29%–50%)), and n = 4 articles focused on mostly consanguineous populations (yield: 29%, CI: 23–35%). Meta-regression revealed that year of study, number of family members sequenced, and isolated NDD versus NDD plus associated conditions category did not significantly predict molecular diagnostic yield.
Several studies (n = 6) showed that a presumptive diagnosis by ES could change clinical management (Table S2). Among those with a diagnosis by ES, changes in medical/medication management occurred in 30% (range: 2–46%; n = 6 studies). Four studies discussed impact on reproductive planning and found that 80% (range: 42–100%) of diagnoses were informative for reproductive planning. For those for whom the diagnosis was not informative for reproductive planning, the reasons were not entirely clear. Of note, these numbers on clinical management impact are not necessarily specific to individuals with NDD, since for some studies, we used a subset of the study population to calculate molecular diagnostic yield for NDDs.
DISCUSSION
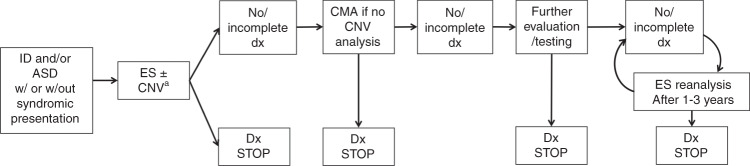
Our scoping review revealed that, among patients with NDDs, ES outperforms the currently accepted first-tier test for NDDs by 10–28%, assuming a range of 30–43% for ES and 15–20% for CMA. Both of these tests are advantageous as unbiased approaches with genetically heterogeneous conditions like NDDs. However, based on this significantly higher molecular diagnostic yield combined with the fact that a diagnosis often impacts clinical management, we recommend ES (including both parents, when feasible and with multiple affected family member analysis, when indicated) as a first-tier test for individuals with unexplained NDDs (Fig. 3). In fact, ES has advantages, such as detection of individuals with multiple monogenic disorders, facilitating characterization of blended phenotypes.22 Multiple prior studies have shown that results from ES impact clinical management for NDDs and other conditions,18,23 which our results support.
Fig. 3.
Diagnostic algorithm incorporating exome sequencing (ES) in the clinical evaluation of individuals with unexplained neurodevelopmental disorders (NDDs) (global developmental delay/intellectual disability [GDD/ID] and/or autism spectrum disorder [ASD]). An incomplete diagnosis represents a diagnosis that explains only part of an individual’s phenotype. Factors such as the turnaround time of test, availability of tests, and availability of genetic counseling may be considerations in application of this algorithm in clinical use. aOr technology that supersedes ES such as genome sequencing. CMA chromosomal microarray, CNV copy-number variant.
If ES is nondiagnostic, we suggest that the next step in the evaluation is CMA (if not already performed, either standalone or as part of ES analysis). If CMA is nondiagnostic, then the clinician can consider further evaluations/tests, including, but not limited to, referral to a clinical geneticist for expert evaluation; periodic (every 1–3 years) reanalysis of ES data, given evidence that doing so may enhance molecular diagnostic yield by 10–16% (refs. 24–26); FXS testing; metabolic testing and/or mitochondrial DNA (mtDNA) sequencing based on clinical presentation (though some laboratories perform mtDNA analysis with ES); and karyotyping to assess for balanced chromosomal rearrangements.
Our algorithm places ES before, or concurrent with, CMA, as the molecular diagnostic yield of ES for NDDs exceeds that of CMA. Estimates for the molecular diagnostic yield of CMA are 15–20%, but this number includes individuals with MCAs, which we did not include in our inclusion criteria unless there was also the presence of GDD/ID and/or ASD. There are certain additional points that will allow this algorithm to remain meaningful as sequencing technology and bioinformatic analyses continue to improve. First, CNV analysis is typically not a standard application of ES across all labs. As a result, the diagnostic yield of CMA and ES for NDDs is approximately the sum of each respective diagnostic yield (i.e., 15–20% for CMA based on prior literature plus 30–43% for ES in our study). Nonetheless, there is increasing movement toward incorporating CNV calling algorithms into ES analysis pipelines.27 For example, within the articles included in our scoping review, three studies called CNVs from exome:1,28,29 two reported CNV yields for NDDs of 16.7% (ref. 28) and 6.4% (ref. 29), and the latter identified two pathogenic CNVs that were not detected on CMA due to poor coverage.29 Second, genome sequencing (GS), by definition, captures more comprehensively all classes of genetic variants in testing (Table 1). We anticipate that GS will eventually supersede ES and CMA in clinical testing algorithms over time, as cost decreases.
It is worthwhile to note comparisons between ES and panel testing. Sensitivity for detection of mosaicism can be lower on ES compared with panel testing, for which read depth and sequence coverage is often greater.30 ES has lower sensitivity to detect single exon–level deletions/duplications, which may be better detected by panels when targeted deletion/duplication analysis is included. However, gene panels require continual curation of a well-defined/comprehensive gene set for phenotypes for which many genes are yet to be discovered.31 Moreover, panels are fixed in the number of genes originally assessed, while ES allows for reanalysis of the data set.
It is also important to note that a diagnosis from ES should not end with laboratory classification of a variant. For instance, the classification of a variant of unknown significance (VUS) from a laboratory should be further evaluated in the clinical setting. With increased knowledge over time of phenotypes related to different genetic disorders, exome reanalysis may change the clinical interpretation of a VUS. Furthermore, a likely pathogenic variant has approximately a 90% true positive rate, so there is approximately a 10% false positive rate as the cause of an individual’s presentation; therefore, clinical judgment is important in establishing a diagnosis.32 Even pathogenic variants may be incompletely penetrant or only partly explain presenting features. Ultimately, final variant interpretation of the ES test result requires review by expert clinicians who can reassess the patient’s phenotype in light of the suggested molecular diagnosis.
Our results demonstrate that the presence of additional clinical features, year of study, and number of family members included in exome analysis did not correlate with molecular diagnostic yield in our meta-regression. Factors such as number of studies pertaining to each of the variables may have contributed to this result, and therefore it still may be the case that the presence of multiple additional phenotypic features may enrich molecular diagnostic yield (as evidenced by the fact there was a higher diagnostic yield in the NDD plus associated conditions category versus the isolated NDD category, though not statistically significant). Additionally, while our findings did not demonstrate higher yield when including more family members, many of the studies were a mixture of proband-only and trio ES, limiting our ability to detect differences in molecular diagnostic yield. Because of this, we expect that the additional yield of trio ES seen in other studies remains true.1,33 Our analysis did not account for history of consanguinity in the meta-regression, though this additional factor may enrich molecular diagnostic yield, based on increased probability of autosomal recessive conditions. Additionally, we were not able to determine whether increased ID severity increased molecular diagnostic yield in our regression, but this factor may be another consideration. In sum, ES demonstrates a higher diagnostic yield, relative to CMA, for individuals with NDDs, with/without additional features. This is attributable to the ability of ES to detect the most prevalent types of pathogenic variants affecting this population (single-nucleotide variants [SNVs]), and that those SNVs represent many loci. This is not surprising because the phenotypic category of NDDs features a high degree of locus heterogeneity and phenotypic variability.
It is worthwhile to consider who would order ES as a first-line test for NDDs. We suggest that test ordering should not be limited to a particular specialty; rather, it is contingent on the ordering provider’s ability to provide informed consent about variant interpretation and secondary findings, and the ability to interpret/report results. Providers should inform patients on possible results, both primary and secondary.34 We have not included assessments of cost-effectiveness and insurance approval for ES in our evaluation.
Laboratory interpretation standards, case review boards, and reanalysis protocols might also affect yield from exome sequencing.24,26,35 We compared diagnostic yield of ES to CMA, and note that CMA results can also be affected in the same ways.36 A recent prospective study examined the use of ES for establishing molecular diagnoses in fetal structural anomalies, involving multidisciplinary input that is important when the phenotype is complex.37
Our analysis had limitations. The scoping review focused on ID and/or ASD, since these are the two NDDs for which there are recommendations already in place for genetic testing. We recognize that our search strategy may have led to exclusion of some articles where phenotype was less specific, such as use of “developmental delay” instead of GDD. For reasons of consistency, we were stricter in our search strategy. Second, we made inferences about the study population inn = 9 studies (e.g., presuming “ID/DD” is ID/GDD). Several studies did not formally define the basis for ASD or ID/GDD. Even in studies belonging to the isolated NDD group, the study population was heterogeneous (e.g., ID plus variable other features though not necessarily plus another specific phenotype). Moreover, although our scoping review had selected articles focusing on GDD, it had not been included in our initial MeSH terms, so we repeated our PubMed search with this change. Further, we excluded certain studies with heterogeneous cohorts for which we could not ascertain the number of individuals with NDD. One such study focused on ID and/or epileptic encephalopathies,38 while another such study focused on NDDs (but broadly defined NDD to include a heterogeneous array of neurological symptoms without clear delineation of subset with GDD/ID and/or ASD).39 Though it may seem surprising that there were more papers on isolated NDD than NDD plus associated conditions, the reason for this is that we only included an article in the NDD plus associated conditions category if all participants in that article had NDDs on top of the additional feature(s). In addition, not all studies used the ACMG variant classification guidelines. In part, this was due to the time period of study inclusion, which began in 2014; the ACMG did not release variant classification guidelines until 2015.40 Another limitation is that we did not include mtDNA sequencing in our analysis. We also recognize that the data were drawn from a variety of both clinical and research settings, and that specific analysis approaches may differ in terms of variant filtering and technical platform (e.g, trio-based ES versus proband-only ES). It is expected that by looking at meta-analysis we account for some of this variability. The second to last limitation is that we acknowledge additional cost analyses associated with the genetic testing technologies will also be important, though these were not part of the original purpose of our study, which focused on molecular diagnostic yield. Finally, while there may be publication bias favoring positive results, this effect is diminished by the large cohorts in our analysis and multiple studies involving consecutively ascertained cases. Publication bias should apply equally to studies on yield of CMA and ES.
Our work establishes that, among patients with NDD, ES produces a higher molecular diagnostic yield than CMA, so by this criterion and as outlined in our algorithm (Fig. 3), it should become a first-tier test. As our recommended algorithm is adopted, we anticipate numerous additional health and economic impact studies about changes in patient-related outcomes enabled by genome sequence–based diagnosis. While step-wise testing has historically had a role in identifying a genetic diagnosis, simplicity is ideal, and a diagnosis can now be accomplished for many patients in a single test with an already impressively high molecular diagnostic yield. Such clinical diagnostic yields will increase with reannotation of the existing data compared with new data sets and with new computational tools.32 This report will guide geneticists, neurologists, developmental pediatricians, child psychiatrists, and other clinicians in incorporating clinical exome or genome sequencing into the management of individuals with NDDs.
Supplementary information
Acknowledgements
The authors acknowledge members of the Exome Scoping Review Work Group participating in the literature scoping review: Bibiana Restrepo and Suma Shankar (MIND Institute, Department of Pediatrics, University of California Davis), Erin Rooney Riggs (Autism & Developmental Medicine Institute, Geisinger), Pete Constantinou (West of Scotland Regional Genetics Service, Glasgow, UK), Anne Reed-Weston (Columbia University College of Physicians & Surgeons), R. Spencer Tong (The Hospital for Sick Children), Jennifer Howe and Janet Buchanan (The Hospital for Sick Children), Rachel Fisher (University of Michigan), and Sonal Mahida (Epilepsy Genetics, Boston Children’s Hospital [BCH]). We also acknowledge the Wade Family Foundation for their financial support of the consensus conference, Caterina Stamoulis (Departments of Medicine and Neurology, BCH) for review of statistical methodology, and librarian Meaghan Muir (BCH) for PubMed and scoping review methodology guidance.
Disclosure
All authors completed the International Committee of Medical Journal Editors conflict of interest disclosure forms, which were reviewed by the senior authors. H.V.F. reports personal fees from UpToDate, outside the submitted work. T.F. has received federal funding or research support from, acted as a consultant to, received travel support from, and/or received a speaker's honorarium from the Cole Family Research Fund, Simons Foundation, Ingalls Foundation, Forest Laboratories, Ecoeos, IntegraGen, Kugona LLC, Shire Development, Bristol-Myers Squibb, Roche Pharma, National Institutes of Health, and the Brain and Behavior Research Foundation, all unrelated to this project. S.W.S. is on the Scientific Advisory Committees of Population Bio and Deep Genomics, and intellectual property from his research held at the Hospital for Sick Children is licensed to Athena Diagnostics, and separately Lineagen. These relationships did not influence data interpretation or presentation during this study, but are being disclosed for potential future considerations. M.S. reports grant support from Novartis, Roche, Pfizer, Ipsen, LAM Therapeutics and Quadrant Biosciences, and he has served on Scientific Advisory Boards for Sage, Roche, Celgene, and Takeda, all unrelated to this project. The other authors declare no conflicts of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Siddharth Srivastava, Jamie A. Love-Nichols.
Change history
7/29/2020
In our meta-analysis, we utilized incorrect numbers of individuals for one publication (Retterer et al., 2016) due to the fact the numbers for ASD and ID groups were not independent representations. We have updated our analysis using corrected numbers based on correspondence with the first author of this paper (diagnostic yield for NDD = 543/1736 as opposed to 570/2063). The updated analysis leads to the same (rounded) weighted diagnostic yield and confidence intervals (CI) as the initial publication (36% [30%–43%]). The updated analysis results in the following updated values in Figure 2 -- Retterer study values: N positive = 543, N total = 1736, study weight = 5.3% and meta-analysis statistics: I2 = 80%, t2 = 0.2835, p < 0.01. The study is also included in two subanalyses reported in the Results. The isolated NDD subcategory (n = 21 articles), updated analysis leads to same (rounded) weighted diagnostic yield and confidence intervals as published (31% [25%–38%]). For the sub-analysis of mix of ID and/or ASD, the initial yield was 37% (CI: 29%–46%). Following updated analysis, the yield of this subset is 39% (CI: 29%–50%).
Change history
7/29/2020
A Correction to this paper has been published: 10.1038/s41436-020-0913-3
Supplementary information
The online version of this article (10.1038/s41436-019-0554-6) contains supplementary material, which is available to authorized users.
References
- 1.Retterer K, Juusola J, Cho MT, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 2.Schaefer GB, Mendelsohn NJ, Professional Practice and Guidelines Committee. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med. 2013;15:399–407. doi: 10.1038/gim.2013.32. [DOI] [PubMed] [Google Scholar]
- 3.Manning M, Hudgins L, Professional Practice and Guidelines Committee. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12:742–745. doi: 10.1097/GIM.0b013e3181f8baad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vissers LELM, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 5.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vorstman JAS, Parr JR, Moreno-De-Luca D, Anney RJL, Nurnberger JI, Hallmayer JF. Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet. 2017;18:362–376. doi: 10.1038/nrg.2017.4. [DOI] [PubMed] [Google Scholar]
- 7.Jang W, Kim Y, Han E, et al. Chromosomal microarray analysis as a first-tier clinical diagnostic test in patients with developmental delay/intellectual disability, autism spectrum disorders, and multiple congenital anomalies: a prospective multicenter study in Korea. Ann Lab Med. 2019;39:299–310. doi: 10.3343/alm.2019.39.3.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng SSW, Chan KYK, Leung KKP, et al. Experience of chromosomal microarray applied in prenatal and postnatal settings in Hong Kong. Am J Med Genet C Semin Med Genet. 2019 Mar 23; doi:10.1002/ajmg.c.31697 [Epub ahead of print]. [DOI] [PubMed]
- 9.Battaglia A, Doccini V, Bernardini L, et al. Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features. Eur J Paediatr Neurol. 2013;17:589–599. doi: 10.1016/j.ejpn.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Waggoner D, Wain KE, Dubuc AM, et al. Yield of additional genetic testing after chromosomal microarray for diagnosis of neurodevelopmental disability and congenital anomalies: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2018;20:1105–1113. doi: 10.1038/s41436-018-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao B, Qiu W, Ji X, et al. Marked yield of re-evaluating phenotype and exome/target sequencing data in 33 individuals with intellectual disabilities. Am J Med Genet A. 2018;176:107–115. doi: 10.1002/ajmg.a.38542. [DOI] [PubMed] [Google Scholar]
- 12.Loviglio MN, Beck CR, White JJ, et al. Identification of a RAI1-associated disease network through integration of exome sequencing, transcriptomics, and 3D genomics. Genome Med. 2016;8:105. doi: 10.1186/s13073-016-0359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfundt R, Del Rosario M, Vissers LELM, et al. Detection of clinically relevant copy-number variants by exome sequencing in a large cohort of genetic disorders. Genet Med. 2017;19:667–675. doi: 10.1038/gim.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulter ME, Miller DT, Harris DJ, et al. Chromosomal microarray testing influences medical management. Genet Med. 2011;13:770–776. doi: 10.1097/GIM.0b013e31821dd54a. [DOI] [PubMed] [Google Scholar]
- 15.Ellison JW, Ravnan JB, Rosenfeld JA, et al. Clinical utility of chromosomal microarray analysis. Pediatrics. 2012;130:e1085–1095. doi: 10.1542/peds.2012-0568. [DOI] [PubMed] [Google Scholar]
- 16.Riggs ER, Wain KE, Riethmaier D, et al. Chromosomal microarray impacts clinical management. Clin Genet. 2014;85:147–153. doi: 10.1111/cge.12107. [DOI] [PubMed] [Google Scholar]
- 17.Henderson LB, Applegate CD, Wohler E, Sheridan MB, Hoover-Fong J, Batista DAS. The impact of chromosomal microarray on clinical management: a retrospective analysis. Genet Med. 2014;16:657–664. doi: 10.1038/gim.2014.18. [DOI] [PubMed] [Google Scholar]
- 18.Tan TY, Dillon OJ, Stark Z, et al. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 2017;171:855–862. doi: 10.1001/jamapediatrics.2017.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colquhoun HL, Levac D, O’Brien KK, et al. Scoping reviews: time for clarity in definition, methods, and reporting. J Clin Epidemiol. 2014;67:1291–1294. doi: 10.1016/j.jclinepi.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Fink A, Kosecoff J, Chassin M, Brook RH. Consensus methods: characteristics and guidelines for use. Am J Public Health. 1984;74:979–983. doi: 10.2105/ajph.74.9.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy MK, Black NA, Lamping DL, et al. Consensus development methods, and their use in clinical guideline development. Health Technol Assess. 1998;2:i-iv–1-88. [PubMed] [Google Scholar]
- 22.O’Donnell-Luria AH, Miller DT. A clinician’s perspective on clinical exome sequencing. Hum Genet. 2016;135:643–654. doi: 10.1007/s00439-016-1662-x. [DOI] [PubMed] [Google Scholar]
- 23.Dixon-Salazar TJ, Silhavy JL, Udpa N, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med. 2017;19:209–214. doi: 10.1038/gim.2016.88. [DOI] [PubMed] [Google Scholar]
- 25.Al-Nabhani M, Al-Rashdi S, Al-Murshedi F, et al. Reanalysis of exome sequencing data of intellectual disability samples: yields and benefits. Clin Genet. 2018;94:495–501. doi: 10.1111/cge.13438. [DOI] [PubMed] [Google Scholar]
- 26.Salmon LB, Orenstein N, Markus-Bustani K, et al. Improved diagnostics by exome sequencing following raw data reevaluation by clinical geneticists involved in the medical care of the individuals tested. Genet Med. 2018 Oct 31; doi:10.1038/s41436-018-0343-7 [Epub ahead of print]. [DOI] [PubMed]
- 27.Retterer K, Scuffins J, Schmidt D, et al. Assessing copy number from exome sequencing and exome array CGH based on CNV spectrum in a large clinical cohort. Genet Med. 2015;17:623–629. doi: 10.1038/gim.2014.160. [DOI] [PubMed] [Google Scholar]
- 28.Charng W-L, Karaca E, Coban Akdemir Z, et al. Exome sequencing in mostly consanguineous Arab families with neurologic disease provides a high potential molecular diagnosis rate. BMC Med Genomics. 2016;9:42. doi: 10.1186/s12920-016-0208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LELM Vissers, van Nimwegen KJM, Schieving JH, et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet Med. 2017;19:1055–1063. doi: 10.1038/gim.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y, Ruivenkamp CAL, Hoffer MJV, et al. Next-generation diagnostics: gene panel, exome, or whole genome? Hum Mutat. 2015;36:648–655. doi: 10.1002/humu.22783. [DOI] [PubMed] [Google Scholar]
- 31.Xue Y, Ankala A, Wilcox WR, Hegde MR. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med. 2015;17:444–451. doi: 10.1038/gim.2014.122. [DOI] [PubMed] [Google Scholar]
- 32.Wright CF, FitzPatrick DR, Firth HV. Paediatric genomics: diagnosing rare disease in children. Nat Rev Genet. 2018;19:253–268. doi: 10.1038/nrg.2017.116. [DOI] [PubMed] [Google Scholar]
- 33.Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SFv2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 35.Harrison SM, Dolinsky JS, Knight Johnson AE, et al. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet Med. 2017;19:1096–1104. doi: 10.1038/gim.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J-C, Radcliff J, Coe SJ, Mahon LW. Effects of platforms, size filter cutoffs, and targeted regions of cytogenomic microarray on detection of copy number variants and uniparental disomy in prenatal diagnosis: results from 5026 pregnancies. Prenat Diagn. 2019;39:137–156. doi: 10.1002/pd.5375. [DOI] [PubMed] [Google Scholar]
- 37.Petrovski S, Aggarwal V, Giordano JL, et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet. 2019;393:758–767. doi: 10.1016/S0140-6736(18)32042-7. [DOI] [PubMed] [Google Scholar]
- 38.Thevenon J, Duffourd Y, Masurel-Paulet A, et al. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet. 2016;89:700–707. doi: 10.1111/cge.12732. [DOI] [PubMed] [Google Scholar]
- 39.Soden SE, Saunders CJ, Willig LK, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anazi S, Maddirevula S, Faqeih E, et al. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol Psychiatry. 2017;22:615–624. doi: 10.1038/mp.2016.113. [DOI] [PubMed] [Google Scholar]
- 42.Baldridge D, Heeley J, Vineyard M, et al. The Exome clinic and the role of medical genetics expertise in the interpretation of exome sequencing results. Genet Med. 2017;191040–1048 [DOI] [PMC free article] [PubMed]
- 43.Butler MG, Rafi SK, Hossain W, Stephan DA, Manzardo AM. Whole exome sequencing in females with autism implicates novel and candidate genes. Int J Mol Sci. 2015;16:1312–1335. doi: 10.3390/ijms16011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Codina-Solà M, Rodríguez-Santiago B, Homs A, et al. Integrated analysis of whole-exome sequencing and transcriptome profiling in males with autism spectrum disorders. Mol Autism. 2015;6:21. doi: 10.1186/s13229-015-0017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evers C, Staufner C, Granzow M, et al. Impact of clinical exomes in neurodevelopmental and neurometabolic disorders. Mol Genet Metab. 2017;121:297307. doi: 10.1016/j.ymgme.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Helsmoortel C, Vandeweyer G, Ordoukhanian P, Van Nieuwerburgh F, Van der Aa N, Kooy RF. Challenges and opportunities in the investigation of unexplained intellectual disability using family-based whole-exome sequencing. Clin Genet. 2015;88:140–148. doi: 10.1111/cge.12470. [DOI] [PubMed] [Google Scholar]
- 47.Iglesias A, Anyane-Yeboa K, Wynn J, et al. The usefulness of whole-exome sequencing in routine clinical practice. Genet Med. 2014;16:922–931. doi: 10.1038/gim.2014.58. [DOI] [PubMed] [Google Scholar]
- 48.Kuperberg M, Lev D, Blumkin L, et al. Utility of whole exome sequencing for genetic diagnosis of previously undiagnosed pediatric neurology patients. J Child Neurol. 2016;31:1534–1539. doi: 10.1177/0883073816664836. [DOI] [PubMed] [Google Scholar]
- 49.Monroe GR, Frederix GW, Savelberg SMC, et al. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet Med. 2016;18:949–956. doi: 10.1038/gim.2015.200. [DOI] [PubMed] [Google Scholar]
- 50.Nolan D, Carlson M. Whole exome sequencing in pediatric neurology patients: clinical implications and estimated cost analysis. J Child Neurol. 2016;31:887–894. doi: 10.1177/0883073815627880. [DOI] [PubMed] [Google Scholar]
- 51.Prasad A, Sdano MA, Vanzo RJ, et al. Clinical utility of exome sequencing in individuals with large homozygous regions detected by chromosomal microarray analysis. BMC Med Genet. 2018;19:46. doi: 10.1186/s12881-018-0555-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Preikšaitienė E, Ambrozaitytė L, Maldžienė Ž, et al. Identification of genetic causes of congenital neurodevelopmental disorders using genome wide molecular technologies. Acta Med Litu. 2016;23:73–85. doi: 10.6001/actamedica.v23i2.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riazuddin S, Hussain M, Razzaq A, et al. Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol Psychiatry. 2017;22:1604–1614. doi: 10.1038/mp.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossi M, El-Khechen D, Black MH, Farwell Hagman KD, Tang S, Powis Z. Outcomes of diagnostic exome sequencing in patients with diagnosed or suspected autism spectrum disorders. Pediatr Neurol. 2017;70:34–43.e2. doi: 10.1016/j.pediatrneurol.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 55.Srivastava S, Cohen JS, Vernon H, et al. Clinical whole exome sequencing in child neurology practice. Ann Neurol. 2014;76:473–483. doi: 10.1002/ana.24251. [DOI] [PubMed] [Google Scholar]
- 56.Tammimies K, Marshall CR, Walker S, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 57.Bramswig NC, Lüdecke H-J, Alanay Y, et al. Exome sequencing unravels unexpected differential diagnoses in individuals with the tentative diagnosis of Coffin-Siris and Nicolaides-Baraitser syndromes. Hum Genet. 2015;134:553–568. doi: 10.1007/s00439-015-1535-8. [DOI] [PubMed] [Google Scholar]
- 58.Campeau PM, Kasperaviciute D, Lu JT, et al. The genetic basis of DOORS syndrome: an exome-sequencing study. Lancet Neurol. 2014;13:44–58. doi: 10.1016/S1474-4422(13)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gauthier-Vasserot A, Thauvin-Robinet C, Bruel A-L, et al. Application of whole-exome sequencing to unravel the molecular basis of undiagnosed syndromic congenital neutropenia with intellectual disability. Am J Med Genet A. 2017;173:62–71. doi: 10.1002/ajmg.a.37969. [DOI] [PubMed] [Google Scholar]
- 60.Lopes F, Barbosa M, Ameur A, et al. Identification of novel genetic causes of Rett syndrome-like phenotypes. J Med Genet. 2016;53:190–199. doi: 10.1136/jmedgenet-2015-103568. [DOI] [PubMed] [Google Scholar]
- 61.Olson HE, Tambunan D, LaCoursiere C, et al. Mutations in epilepsy and intellectual disability genes in patients with features of Rett syndrome. Am J Med Genet A. 2015;167A:2017–2025. doi: 10.1002/ajmg.a.37132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rump P, Jazayeri O, van Dijk-Bos KK, et al. Whole-exome sequencing is a powerful approach for establishing the etiological diagnosis in patients with intellectual disability and microcephaly. BMC Med Genomics. 2016;9:7. doi: 10.1186/s12920-016-0167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarailo-Graovac M, Shyr C, Ross CJ, et al. Exome sequencing and the management of neurometabolic disorders. N Engl J Med. 2016;374:2246–2255. doi: 10.1056/NEJMoa1515792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeung KS, Tso WWY, Ip JJK, et al. Identification of mutations in the PI3K-AKT-mTOR signalling pathway in patients with macrocephaly and developmental delay and/or autism. Mol Autism. 2017;8:66. doi: 10.1186/s13229-017-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.