Abstract
Metformin is one of the treatments used for PCOS pathology decreasing body weight, plasma androgen, FSH and glucose levels. Unfortunately, there is little known about metformin’s effects on lipid metabolism, a crucial process in PCOS pathology. We have employed a lipidomic approach to explore alterations in the plasma lipid profile of patients with PCOS following metformin treatment. The aim is to offer new insights about the effect of metformin in PCOS patients. Plasma samples were obtained from 27 subjects prior to and following 12 weeks of metformin treatment. A detailed biochemical characterization and lipidomic profile was performed. Metformin reduces BMI, HOMA-IR, FSH and androstenedione and increases DHEA-S but no changes were found in glucose levels after treatment. Multivariate statistics revealed a specific lipidomic signature due to the effect of 12 weeks of metformin treatment in PCOS patients. This signature includes changes in sphingolipid metabolism suggesting a crosstalk between these lipid species and the androgenic metabolism and a decrease in oxidized lipids reinforcing that metformin treatment improves oxidative stress status. Our study confirms the specific effect of metformin in lipid metabolism on women with PCOS after 12 weeks of treatment.
Subject terms: Lipidomics, Endocrine reproductive disorders
Introduction
Polycystic ovary syndrome (PCOS) is a multifactorial disorder that affects 7–9% of women of a reproductive age, and is characterized by clinical/biochemical hyperandrogenism, polycystic ovarian morphology (during ultrasound) and chronic oligo-anovulation. It is a multifaceted disease in which uncontrolled ovarian steroidogenesis, excessive oxidative stress, aberrant insulin signaling, and genetic/environmental factors play a role1. In addition, androgen levels are an intrinsic factor2.
PCOS is a pathology specifically associated with insulin resistance (IR) in which both the receptor and the mechanism of action of insulin are affected in different target tissues3. Although obesity seems to be heavily implicated in the pathogenesis of PCOS, affecting 40–70% of patients4, a considerable proportion of non-obese PCOS patients (30%) have IR5. Although the causal factor that leads to IR in PCOS has not been fully deciphered, a marked correlation between IR and an inappropriate accumulation of lipid species in insulin target tissues have been described. In line with this, there is evidence indicating that lipids act as signalling molecules and mediates IR6,7. IR is considered to underlie other aberrations that affect the health of PCOS patients8. For example, it influences lipid profile, with 70% of women with PCOS exhibiting at least one abnormal lipid constituent9. Furthermore, female PCOS patients with obesity often have dyslipidaemia, with higher triglyceride (TG) levels and lower high-density lipoprotein cholesterol (HDL-C)10 with respect to the healthy population. Studies have reported significantly lower levels of HDL-C in PCOS women versus weight-matched controls11. Approximately one-third of women with PCOS have been shown to also exhibit metabolic syndrome12. Indeed, PCOS is considered one of the ovarian manifestations of metabolic syndrome13. Alternatively, inflammation may also play a crucial role in the development of IR in women with PCOS14,15 and has previously been associated with other PCOS associated pathologies, including IR16. In relation to this, previous studies with PCOS patients have shown that acute hyperglycemia and IR enhance levels of proinflammatory transcription factor nuclear kappa B (NF-kB)17 and those of proinflammatory cytokines17–19.
In addition to hormonal derangements and altered glucose metabolism, PCOS has traditionally been linked with atherogenic dyslipidemia, a major determinant of cardiovascular diseases20. However, a recent study performed with PCOS patients without associated pathologies and presenting a non-pathogenic lipid profile (Total cholesterol, LDL-C, HDL-C and TG) describes the presence of a PCOS lipidomic fingerprint, in which glycerolipid, glycerophospholipid and sphingolipid metabolism is affected21, suggesting that these molecules are related to the physiopathology of PCOS and opening up new scenarios in the search for new drugs. Therefore, the lipid profile characterized by high triglycerides22, low HDL cholesterol and increased LDL cholesterol, which has been related to androgen excess22,23, is a classic profile linked to IR rather than PCOS. Further lipidomic studies that analyse 325 lipid species in women with or without PCOS show an association of BMI with 12 classes of lipid species including phospholipids, ceramides, gangliosides and acylglycerols, and a free androgen index with 8 classes of lipids, namely ceramides, phospholipids and acylglycerols, thus supporting prior findings that adiposity is a key driver of dyslipidaemia in PCOS24.
The underlying pathophysiological mechanisms of PCOS are not fully elucidated; consequently current therapeutic options for PCOS women mostly treat symptoms. Metformin is a biguanide used as the first choice oral anti-hyperglycemic drug to treat type 2 diabetes. Its mechanism of action is twofold: it promotes a reduction in glucose production and it improves insulin sensitivity, the latter being a consequence of changes in lipid metabolism25. The changes arise through the mitochondrial electron transport chain becoming inhibited by metformin and the consequent activation of 5′-AMP-activated protein kinase (AMPK), which inhibits fatty acid (FA) synthesis. In this way, metformin prevents lipid storage and the impairment of FA oxidation in insulin-sensitive tissues, both of which processes are described in IR26. Recently, metformin has also been used to management of hirsutism, acne and IR during PCOS27,28. Concerning IR, metformin exerts multiple actions on different insulin-sensitive tissues, including skeletal muscle, adipose tissue, liver, the endothelium and the ovaries29, and seems to improve the long-term health of PCOS women by prevent endometrial cancer, diabetes and cardiovascular disease30.
The expanding development of omics approaches has allowed a wider view of the molecular mechanisms underlying human pathology. Indeed, we have recently confirmed a specific plasma lipidomic signature in women with PCOS with respect to healthy individuals21. This signature implies changes in the metabolism of glycerolipids, glycerophospholipids and sphingolipids, pointing to changes in the glycerophospholipid biosynthetic pathway and cell signalling. Besides the documented role of metformin in lipid metabolism, the potential lipidomic changes in response to metformin treatment in PCOS patients has never been addressed and it would be interesting to determine if metformin can restore the previously reported lipidomic profile that is altered during PCOS21.
The present study aims to clarify whether metformin treatment in PCOS patients induces a specific lipidome profile in plasma, thus providing new insight into the management and treatment of this metabolic and endocrine disease.
Results
Anthropometric and metabolic parameters
Table 1 shows the anthropometric and metabolic parameters of our PCOS patients before treatment (pre-treatment, PRE) and after 12 weeks of metformin (post-treatment, POST). No statistical significance was found when comparing anthropometric parameters between groups for all the parameters except weight and BMI, significantly lower after 12-weeks of metformin treatment. Among subjects, 11 presented IR (HOMA ≥ 2.6, 2 overweight (BMI ≥ 25) and 12 obesity (BMI ≥ 30) before treatment. Parameters related to lipid metabolism (total cholesterol, LDL-c, HDL-c and triglycerides) were also similar in PCOS patients after metformin treatment. Glucose and insulin levels showed a trend to decrease after 12-weeks of metformin treatment. Although differences in glucose and insulin are not statistically significant, the HOMA-IR value is significantly lower after treatment with a reduction of 0.5 points (p < 0.05). We observed that the endocrine profile of PCOS subjects changed after 12 weeks of metformin treatment, as represented by decreased FSH and androstenedione levels and increased DHEA-S levels (p < 0.05 in all cases).
Table 1.
Anthropometric, biochemical and endocrinological data of PCOS patients before treatment with metformin (pre-treatment, PRE) and at the end of 12-weeks follow-up (post-treatment, POST).
| PRE | POST | p-value B.H. | |
|---|---|---|---|
| n | 27 | 27 | |
| Age (years) | 24.7 ± 6.4 | — | |
| Weight (kg) | 78.9 ± 4.7 | 76.3 ± 4.4 | 0.011 |
| BMI (kg/m2) | 29.2 ± 1.8 | 28.2 ± 1.6 | 0.010 |
| Waist circumference (cm) | 94.9 ± 3.8 | 94.7 ± 3.4 | 0.882 |
| SBP (mmHg) | 119.5 ± 3.3 | 114.0 ± 3.2 | 0.100 |
| DBP (mmHg) | 73.4 ± 2.8 | 70.7 ± 2.1 | 0.220 |
| Total cholesterol (mg/dl) | 176.0 ± 5.8 | 172.1 ± 6.0 | 0.308 |
| LDL-c (mg/dl) | 112.5 ± 4.9 | 107.6 ± 5.3 | 0.106 |
| HDL-c (mg/dl) | 44.6 ± 2.1 | 44.7 ± 1.7 | 0.931 |
| Triglycerides (mg/dl) | 94.6 ± 11.6 | 101.1 ± 12.0 | 0.242 |
| Glucose (mg/dl) | 83.4 ± 1.9 | 79.5 ± 1.7 | 0.078 |
| Insulin (μUI/ml) | 14.05 ± 1.8 | 12.42 ± 1.4 | 0.071 |
| HOMA-IR | 2.97 ± 0.4 | 2.47 ± 0.3 | 0.034 |
| FSH (mIU/ml) | 4.89 ± 0.3 | 4.06 ± 0.3 | 0.014 |
| LH (mIU/ml) | 7.95 ± 2.3 | 5.63 ± 0.8 | 0.302 |
| Free Androgen Index | 0.79 ± 0.1 | 0.72 ± 0.1 | 0.814 |
| Testosterone (ng/ml) | 0.80 ± 0.1 | 0.73 ± 0.1 | 0.228 |
| Androstenedione (ng/ml) | 4.52 ± 0.4 | 3.70 ± 0.4 | 0.013 |
| SHBG (nmol/l) | 57.20 ± 8.5 | 45.25 ± 4.6 | 0.120 |
| DHEA-S (µg/dl) | 293.8 ± 49.8 | 335.69 ± 57.4 | 0.027 |
Data are expressed as mean ± SEM. Statistical significance (p < 0.05) was considered when compared by a paired t-test after Benjamini-Hochberg correction.
Plasma lipidome of PCOS patients under metformin treatment
In order to assess how 12 weeks of metformin treatment effected the global plasma lipidomic profile of PCOS patients, we carried out a non-targeted lipidomic approach, focusing on low molecular-weight ionizable lipid molecules (m/z of 300–3000). In the case of both polarities (negative and positive), 1950 features were aligned after applying the MFE algorithm31. Then, we selected only those features present in at least 50% of the samples of the same group (312 lipid species ionized with positive polarity and 64 lipid species in negative polarity).
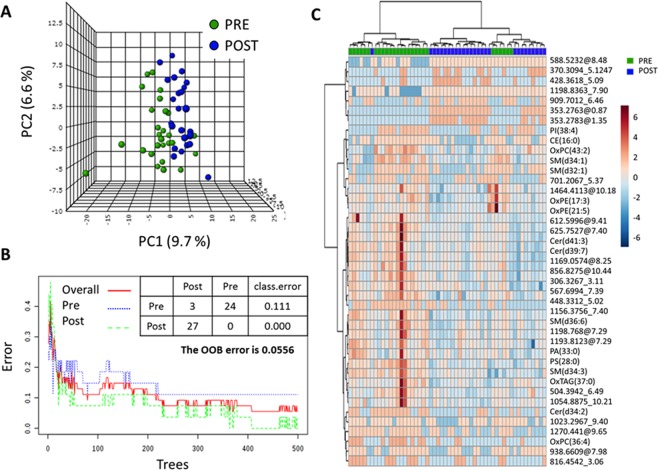
To determine whether the detected lipidome was altered in the PCOS patients after the 12-week of treatment multivariate statistics were applied (Fig. 1 and Fig. S1). All the analyses reflected in Fig. 1 were carried out with lipid species ionized with a positive ESI polarity, while the results of the multivariate analyses of lipidome with negative ESI are represented in Fig. S1. Non-supervised PCA (Fig. 1A) revealed clustering of both groups, thus suggesting that the effect of metformin in PCOS patients has a specific plasma lipidomic signature. In line with this, the machine learning algorithm Random Forest correctly classified all of the patients after the treatment (classification error PRE: 0.111, classification error POST: 0.000, out of bag error: 0.0556) based on the lipidome detected using the positive polarity mode (Fig. 1B). All of the multivariate analyses performed with the negatively charged lipid species showed no differences attributable to the metformin treatment in PCOS patients (Fig. S1).
Figure 1.
Multivariate statistics and machine learning show a specific lipidomic signature after 12-weeks metformin treatment in PCOS patients. (A) Unsupervised Principal Component Analyses (PCA) and (B) Random Forest (RF) classification indicate that it is possible to discriminate PCOS patients before (PRE) and after (POST) 12-weeks of metformin basing on their plasma lipidome. (C) Heat map representations of hierarchical clustering analyses using 42 most statistical significant lipids species (T-test Paired with Benjamini Hochberg Correction) between PCOS patients before and after 12-weeks metformin treatment. Unknown identities are represented as exact mass_ retention time. Metabolite identification is based on exact mass, retention time, isotopic distribution and/or MSMS spectrum. Each line of this graphic represents an accurate mass ordered by retention time, coloured by its abundance intensity normalized to internal standard and baselining to median/mean across the samples. The scale from −6 (blue) to 6 (red) represents this normalized abundance in arbitrary units. All the analyses have been performed with the 312 features detected and filtered in ESI positive polarity.
To better characterize the effect of metformin on the plasma lipidome we performed a parametric paired t-test for equal variances. Of the 376 lipid species detected, 45 were found to differ in a statistically significant manner between groups. The hierarchical analysis of these 45 lipid species clearly showed a differential regulation after and before metformin treatment (Fig. 1C). Interestingly, all of the 18 identified lipid species (based on exact mass, retention time, isotopic distribution and/or MS/MS spectrum) were decreased after metformin treatment (Table 2), while the remaining 27 were unidentified (Table 3).
Table 2.
Identified compounds statistically different between groups (paired t-test with Benjamini Hochberg correction).
| Lipid Category | Compound | FC | p (BH Corr) | Change post treatment | AUC ROC | Specificity | Sensitivity | m/z | RT | Adduct |
|---|---|---|---|---|---|---|---|---|---|---|
| GL | OxTAG(37:0)b | −1.06 | 3.05E-02 | down | 0.68724 | 0.7 | 0.7 | 700.6212 | 9.89 | M + NH4+ |
| OxPE(17:3)b | −1.40 | 5.85E-05 | down | 0.90123 | 0.9 | 0.9 | 536.1593 | 7.44 | M + H+ | |
| PA(33:0)a | −1.08 | 1.70E-03 | down | 0.79012 | 0.9 | 0.6 | 663.4474 | 7.97 | M + H+ | |
| PS(28:0)a | −1.04 | 4.74E-02 | down | 0.72702 | 0.7 | 0.7 | 680.4734 | 7.96 | M + H+ | |
| GP | OxPC(43:2)b | −1.19 | 2.01E-03 | down | 0.71605 | 0.9 | 0.6 | 758.5637 | 7.63 | M + H+ |
| OxPC(36:4)b | −41.39 | 1.99E-02 | down | 0.71811 | 0.9 | 0.5 | 812.5046 | 3.06 | M + H+ | |
| PI(38:4)c | −43.45 | 3.09E-02 | down | 0.71331 | 0.8 | 0.6 | 887.5621 | 6.35 | M + H+ | |
| PE(P-38:6)c | −54.66 | 4.47E-02 | down | 0.70096 | 0.9 | 0.6 | 746.5292 | 8.23 | M + H+ | |
| OxPE(21:5)b | −1.17 | 4.70E-02 | down | 0.83676 | 0.7 | 0.9 | 610.1774 | 7.93 | M + Na+ | |
| Cer(d34:2)a | −99.43 | 4.48E-03 | down | 0.74623 | 0.8 | 0.7 | 536.4794 | 6.05 | M + H+ | |
| Cer(d39:7)a | −1.12 | 3.48E-02 | down | 0.72291 | 0.9 | 0.6 | 596.5207 | 8.28 | M + H+ | |
| Cer(d41:3)a | −1.19 | 1.23E-02 | down | 0.8107 | 0.7 | 0.8 | 632.591 | 8.27 | M + H+ | |
| SP | SM(d32:1)c | −65.34 | 2.32E-02 | down | 0.76818 | 0.7 | 0.7 | 675.5355 | 6.41 | M + H+ |
| SM(d34:3)a | −1.12 | 1.65E-04 | down | 0.84088 | 0.9 | 0.7 | 699.5899 | 9.16 | M + H+ | |
| SM(d34:1)a | −1.54 | 8.25E-04 | down | 0.83813 | 0.9 | 0.7 | 703.5677 | 7.24 | M + H+ | |
| SM(d36:6)a | −1.08 | 8.69E-05 | down | 0.90672 | 0.9 | 0.8 | 721.5014 | 7.97 | M + H+ | |
| SM(d38:2)c | −1.06 | 1.16E-02 | down | 0.77778 | 0.9 | 0.7 | 757.6426 | 9.17 | M + H+ | |
| ST | CE(16:0)a | −35.61 | 4.98E-02 | down | 0.738 | 0.8 | 0.7 | 642.6079 | 10.77 | M + NH4+ |
aLipid species identified by exact mass and retention time and MS/MS spectrum.
bLipid species identified by exact mass and retention time and confirmed with LipidMatch libraries.
cLipid species with a possible identity based on exact mass and retention time.
Table 3.
Unidentified compounds statistically different between groups (Paired t-test with Benjamini Hochberg correction).
| Compound | FC | p (BH Corr) | Change post treatment | mass | RT | ESI |
|---|---|---|---|---|---|---|
| 307.3267_3.11 | −1.22 | 1.70E-03 | down | 307.3267 | 3.11 | POS |
| 329.1986_2.74 | −83.21 | 4.47E-02 | down | 329.1986 | 2.74 | NEG |
| 353.2763_0.87 | 33,497.97 | 2.03E-11 | up | 353.2763 | 0.87 | POS |
| 353.2783_1.35 | 4,901.52 | 6.69E-07 | up | 353.2783 | 1.35 | POS |
| 371.3094_5.12 | 201.58 | 9.70E-03 | up | 371.3094 | 5.12 | POS |
| 428.3618_5.09 | 258.04 | 2.93E-02 | up | 428.3618 | 5.09 | POS |
| 448.3312_5.02 | −1.27 | 3.69E-04 | down | 448.3312 | 5.02 | POS |
| 504.3942_6.49 | −1.09 | 9.70E-03 | down | 504.3942 | 6.49 | POS |
| 555.353_2.75 | −210.03 | 1.01E-02 | down | 555.353 | 2.75 | NEG |
| 567.6994_7.39 | −1.18 | 1.51E-02 | down | 567.6994 | 7.39 | POS |
| 588.5232_8.48 | 506.49 | 2.13E-03 | up | 588.5232 | 8.48 | POS |
| 612.5996_9.41 | −1.15 | 9.70E-03 | down | 612.5996 | 9.41 | POS |
| 625.7527_7.39 | −1.24 | 2.41E-03 | down | 625.7527 | 7.4 | POS |
| 701.2067_5.37 | −32.21 | 4.45E-02 | down | 701.2067 | 5.37 | POS |
| 816.4542_3.06 | −27.2 | 3.32E-02 | down | 816.4542 | 3.06 | POS |
| 856.8275_10.44 | −1.16 | 8.16E-03 | down | 856.8275 | 10.44 | POS |
| 909.7012_6.46 | 62.93 | 1.96E-02 | up | 909.7012 | 6.46 | POS |
| 938.6609_7.98 | −48.1 | 1.23E-02 | down | 938.6609 | 7.98 | POS |
| 1023.2967_9.4 | −36.31 | 3.89E-02 | down | 1023.2967 | 9.4 | POS |
| 1054.8875_10.21 | −1.09 | 3.32E-02 | down | 1054.8875 | 10.21 | POS |
| 1156.3756_7.4 | −98.37 | 2.04E-02 | down | 1156.3756 | 7.4 | POS |
| 1169.0574_8.25 | −1.31 | 2.13E-03 | down | 1169.0574 | 8.25 | POS |
| 1193.8123_7.29 | −1.14 | 1.44E-02 | down | 1193.8123 | 7.29 | POS |
| 1198.768_7.29 | −1.16 | 3.16E-03 | down | 1198.768 | 7.29 | POS |
| 1198.8363_7.9 | 32.52 | 1.16E-02 | up | 1198.8363 | 7.9 | POS |
| 1270.441_9.65 | −234.77 | 3.97E-03 | down | 1270.441 | 9.65 | POS |
| 1464.4113_10.18 | −1.41 | 5.74E-03 | down | 1464.4113 | 10.18 | POS |
NEG: negative ESI mode; POS: positive ESI mode.
Compounds are represented as exactmass_retentiontime.
Among the lipid species that could be identified, 8 belonged to the glycerophospholipid family, four of which (ethanolamine and choline GP-based on) were oxidized and one was a plasmalogen polyunsaturated species. We also identified 8 sphingolipids with a differential regulation due to the metformin treatment. Specifically, three polyunsaturated ceramides and five sphingomyelins, two of which were monounsaturated and three polyunsaturated. Finally, levels of one oxidized triacylglycerol and one cholesterol ester were also found to be lower after the treatment.
In order to further determine to what extent these metabolites can predict the effect of treatment of PCOS with metformin, ROC analyses were performed using MS peak areas. The details of the area under the curve, as well as the specificity and sensitivity of each lipid species, are presented in Table 2. The lipid species with the highest AUC were oxPE(17:3), with a value of 0.90123 and 90% specificity and sensitivity, and SM(d36:6), with 0.90672 AUC, 90% specificity and 80% sensitivity.
When plasma fatty acid composition was analysed, differences were found due to the effect of the 12-week metformin treatment (Table 4). There was a significant increase in the content of 20:2n-6, whereas 20:3n-6, 20:4n-6, 22:1n-9 and 22:5n-3 were found to have decreased. This led to a slight but significant alteration of the average chain length (ACL). When we evaluated other parameters based on fatty acid composition, such as SFA, UFA, MUFA, PUFA, PUFAn-3, PUFAn-6, DBI, PI, and AI, no differences were found in PCOS patients after the metformin treatment.
Table 4.
Total fatty acid composition of plasma from subjects at the baseline and at the end of 12-weeks follow-up.
| PRE-TREATMENT | POST-TREATMENT | p-value | |
|---|---|---|---|
| C14:0 | 0.668 ± 0.007 | 0.725 ± 0.009 | 0.23 |
| C16:0 | 22.475 ± 0.06 | 22.614 ± 0.054 | 0.701 |
| C16:1(n-7) | 1.505 ± 0.019 | 1.456 ± 0.017 | 0.533 |
| C18:0 | 8.103 ± 0.032 | 8.119 ± 0.044 | 0.927 |
| C18:1(n-9) | 20.948 ± 0.125 | 21.203 ± 0.137 | 0.641 |
| C18:1(n-7) | 1.647 ± 0.007 | 1.597 ± 0.006 | 0.257 |
| C18:2(n-6) | 31.644 ± 0.148 | 32.242 ± 0.177 | 0.497 |
| C18:3(n-3) | 0.285 ± 0.003 | 0.324 ± 0.005 | 0.169 |
| C18:4(n-3) | 0.144 ± 0.001 | 0.152 ± 0.001 | 0.11 |
| C20:0 | 0.149 ± 0.001 | 0.182 ± 0.004 | 0.127 |
| C20:1(n-9) | 0.222 ± 0.002 | 0.237 ± 0.004 | 0.56 |
| C20:2(n-6) | 0.034 ± 0 | 0.054 ± 0.002 | 0.042 |
| C20:3(n-6) | 1.615 ± 0.015 | 1.472 ± 0.013 | 0.015 |
| C20:4(n-6) | 5.976 ± 0.046 | 5.597 ± 0.044 | 0.006 |
| C20:5(n-3) | 0.04 ± 0.001 | 0.048 ± 0.001 | 0.067 |
| C22:0 | 0.253 ± 0.002 | 0.26 ± 0.002 | 0.505 |
| C22:1(n-9) | 2.074 ± 0.035 | 1.605 ± 0.019 | 0.003 |
| C22:4(n-6) | 0.177 ± 0.001 | 0.171 ± 0.001 | 0.077 |
| C22:5(n-6) | 0.143 ± 0.003 | 0.116 ± 0.001 | 0.114 |
| C22:5(n-3) | 0.251 ± 0.002 | 0.212 ± 0.002 | 0.002 |
| C24:0 | 0.111 ± 0.002 | 0.099 ± 0.001 | 0.384 |
| C22:6(n-3) | 1.226 ± 0.016 | 1.176 ± 0.013 | 0.427 |
| C24:1(n-9) | 0.268 ± 0.002 | 0.262 ± 0.002 | 0.514 |
| C26:0 | 0.039 ± 0.001 | 0.076 ± 0.006 | 0.286 |
| ACL | 17.845 ± 0.004 | 17.811 ± 0.003 | 0.012 |
| SFA | 31.798 ± 0.067 | 32.075 ± 0.071 | 0.527 |
| UFA | 68.202 ± 0.067 | 67.925 ± 0.071 | 0.527 |
| MUFA | 26.665 ± 0.133 | 26.36 ± 0.141 | 0.639 |
| PUFA | 41.537 ± 0.149 | 41.565 ± 0.176 | 0.974 |
| PUFA(n-3) | 1.947 ± 0.015 | 1.912 ± 0.015 | 0.586 |
| PUFA(n-6) | 39.59 ± 0.141 | 39.653 ± 0.175 | 0.941 |
| DBI | 130.443 ± 0.263 | 128.959 ± 0.278 | 0.282 |
| PI | 73.754 ± 0.31 | 71.893 ± 0.287 | 0.138 |
| AI | 48.762 ± 0.304 | 49.009 ± 0.31 | 0.862 |
| Ratio MUFA/SFA | 0.841 ± 0.005 | 0.822 ± 0.004 | 0.394 |
Abbreviations: ACL, average chain length; SFA, saturated fatty acids; UFA, unsaturated fatty acids; MUFA, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; PUFAn-3, polyunsaturated fatty acids series n-3; PUFAn-6, polyunsaturated fatty acids series n-6; DBI, double bond index; PI, peroxidizability index; AI, anti-inflammatory index.
Discussion
PCOS is a heterogenic and multigenic metabolic disorder which affects women of a reproductive age and is characterized by hyperandrogenism and is commonly associated with other pathologies such as IR and obesity32. Currently, there is no universal treatment for PCOS patients, and most available drugs are directed at treating symptoms such as androgen excess, IR or oligo-ovulation32,33. Among available insulin-sensitizer drugs, metformin is one of the most used, having similar effects as lifestyle changes in terms of decreasing body weight/BMI and more marked effects in terms of decreasing androgen concentrations34. Furthermore, previous studies have demonstrated that metformin reduces plasma glucose levels and FSH in PCOS subjects, and also improves the oxidative stress status of patients, among other benefits18. By decreasing gluconeogenesis and lipogenesis and enhancing glucose uptake in the liver, skeletal muscle, adipose tissue and ovaries metformin increases insulin sensitivity. One potential mechanism proposed for the glucose-lowering effect of metformin is the inhibition of mitochondrial complex I and the consequent stimulation of AMP kinase18,25,34. On the other hand, the intracellular lipid-lowering effects of metformin have been associated with an increase in the mitochondrial channelling of fatty acids. Treatment with metformin enhances the mitochondrial β-oxidation process, directing excess intracellular FA towards β-oxidation, which reduces the supply of substrates for the synthesis of bioactive lipids, which in turn affects the insulin signalling pathway35. Furthermore, metformin blocks increments in the fatty acid transport protein CD36 and in aberrant ceramide and diacylglycerol content in the skeletal muscle of diabetic rats36. However, the molecular mechanisms through which metformin induces these changes are not fully understood. As far as we know, the effect of metformin on plasma lipid profile in PCOS has been basically focused on the study of lipoprotein profile. Therefore, the objective of this study was to characterize how a 12-week metformin treatment affects the plasma lipidomic profile of women with PCOS.
Anthropometric and metabolic parameters in PCOS patients prior to and following 12 weeks of metformin treatment showed a decrease in BMI values, whereas no changes in total triglycerides, total cholesterol, LDLc and HDLc levels were detected. This reduction in body weight could be partially responsible for the lipidomic changes in plasma described after metformin treatment. In terms of insulin resistance parameters, we found a slight decrease in plasma glucose and insulin levels after treatment, though these changes were not statistically significant. However, the HOMA-IR index decreased after the 12-week treatment, thus indicating an improvement in insulin resistance. These results are in line with most previously published studies on the subject and confirm the beneficial effect of metformin on body weight and insulin sensitivity25.
Endocrine measurements revealed several rearrangements after metformin treatment; namely, decreased levels of FSH and androstenedione and increased levels of DHEA-S. The FSH levels found in plasma patients after treatment were very similar to those detected previously in non-PCOS women, suggesting a reversion of PCOS pathology after metformin treatment21. With respect to androstenedione levels, after treatment we found very similar levels to those detected in “pure” PCOS women, suggesting that this parameter is affected by PCOS-associated pathologies21. Total testosterone concentrations are very much influenced by SHBG concentration, as 65% of plasma testosterone is bound to SHBG37. Previous studies in PCOS patients without associated pathologies showed no changes in SHBG and testosterone levels compared with control subjects, thus reinforcing the idea that PCOS-associated pathologies could be partially responsible for the changes observed in androgenic metabolism21. In the present study, we did not observe differences in testosterone and SHBG levels after metformin treatment.
Once we characterized the classical parameters of PCOS patients, we analyzed the whole plasma lipidomic profile using an LC-MS-based technique. Furthermore, fatty acid composition was also measured in order to better characterize the composition of the plasma lipidome. The results revealed the existence of a plasma lipidomic profile specifically associated with a 12-week metformin treatment in PCOS patients, which is the first time this has been reported. This signature is defined mostly by glycerophospholipids and sphingolipids; we detected 45 lipid molecular species whose concentration was altered by the effect of metformin treatment (7 lipid species were up-regulated, while 38 were down-regulated).
The interaction between sphingolipids and steroid hormones has previously been described, and this interaction modulates the steroidogenic signaling pathway. Specifically, these lipids can modulate steroidogenesis, acting at different levels as a second messengers, paracrine/autocrine regulators and/or ligands for nuclear receptors38. For example, ceramides have been shown to regulate progesterone and testosterone production, although the precise molecular mechanisms underlying this process are unclear and require further study39.
In the present work, we described lower levels of 3 ceramides and 4 sphingomyelins after metformin treatment, suggesting that the effect of metformin on androgenic metabolism is partially mediated by sphingolipid metabolism regulation. Moreover, the role of sphingolipids, and specially ceramides, in the mediation of insulin resistance40–42 has been reported previously, so the effect of metformin on parameters of insulin resistance in women with PCOS could also be a result of the interaction of these species with the insulin signaling pathway.
Concerning glycerophospholipids, we identified 8 species that were down-regulated after metformin treatment, 4 of which were oxidized. Surprisingly, we also identified an oxidized TAG. The effect of metformin on cell redox status has been described previously18. The present results reaffirm the improvement of oxidative stress in PCOS patients after metformin treatment and reveal that an improvement also takes place in the lipid metabolism, thus decreasing lipoxidative damage.
Globally, fatty acid analyses indicated minor changes in fatty acid composition after metformin treatment. Although we observed changes in C20-based polyunsaturated fatty acids (C20:2 (n-6), C20:2 (n-6), C20:6 (n-6)), C22:1 (n-9) and C22:5(n-3), the variations only represented about 0.2% of ACL reduction, while no changes were detected in the other parameters calculated. The present results may indicate a modulation of elongase and desaturase activity, especially in the elongases ELOVL3, 5 and 2 and desaturases Δ8 and 543. Fatty acid elongation and desaturation are crucial processes in the biosynthesis of saturated, monounsaturated and polyunsaturated fatty acids. Furthermore, the deregulation of these enzymes’ activity has been previously related to IR and diabetes44,45. Specifically, previous studies in mice have reported a relationship between low ELOVL5 activity in the liver and glucose intolerance and insulin resistance46,47. In line with this, our results suggest that the minor alterations found in some PUFA could have been a response to slight changes in insulin levels and insulin sensitivity induced by metformin.
The principle limitation of the present study is the number of patients (n = 27) and the fact that they came from a reduced geographical area. Furthermore, we have used the Rotterdam criteria for this study, and not the last international guideline, due to the fact that our patients were part of a previously recruited cohort. Moreover, as we have stated before, the PCOS condition is usually associated with other pathologies, such as IR or obesity. In the present study we included 11 IR patients and 6 non-IR patients, and so we cannot be sure that IR was not a cofounding factor. Further studies with a larger cohort would guarantee a “purer” PCOS population. In addition to these population and clinical factors, our study has not enabled us to unravel the biological significance of this compositional complexity. Moreover, although sampling was scheduled to minimize the potential influence of diet on the lipidome, we cannot rule out the possibility that some compounds appearing as a result of the metabolism of nutrients affected the plasma lipidome in general, although the fact that our cohort was from a small geographic area would suggest that they were relatively homogeneous in terms of lifestyle and dietary habits.
All in all, we can conclude that metformin treatment induces a specific plasma lipidomic profile in PCOS women that is characterized mainly by a decrease in sphingolipids and glycerophospholipids and a reduction of lipoxidative species. We believe that this work could aid future research in the exploration of the molecular mechanisms involved in sphingolipid – steroid interaction. Our study confirms that lipid profile highlights a specific effect of metformin on PCOS women after 12 weeks of treatment.
Methods
Subjects
This study was carried out in the Service of Endocrinology at the University Hospital Dr. Peset (Valencia, Spain). Plasma samples of twenty-seven women with PCOS before and after treatment for 12 weeks with metformin were analysed. Following the criterion of our Hospital and according to a previous study published18 treatment with metformin was initiated at 500 mg per day (during the first 2 weeks). After 2 weeks it was increased to 1000 mg/day (weeks 3 and 4) and then to 1500 mg/d during weeks 5–12. Patients did not take any other medication.
Diagnosis of PCOS was confirmed using the Rotterdam criteria48. In brief, presence of oligoovulation (cycles longer than 35 days or less than 26 days)49; free testosterone levels higher than 0.5 ng/dl (this cut-off level was estimated as the mean ± 2 SD according to the levels in healthy women); hirsutism (Ferriman-Gallwey score > 7) and polycystic ovaries (presence of at least 12 small −2 to 9 mm- follicles in each ovary), assessed by trans-vaginal ultrasonography. Ultrasound scans were performed and scored independently by one of two experienced and blinded reviewers.
None of the subjects had any condition affecting her reproductive physiology or any systemic or endocrine disease or galactorrhea. Exclusion criteria were malignant neoplasia, active infectious diseases, anemia, diabetes mellitus, history of ischaemic heart disease, thromboembolism, stroke and the taking of antihypertensive or lipid-lowering drugs. It was confirmed for all participants the absence of any medication that might have affected the hypothalamic-pituitary-gonadal axis during the previous semester. Approval by the ethics committee of the University Hospital Dr. Peset was obtained and the study was performed in accordance with the declaration of Helsinki. All participants provided their informed consent as required by these institutions.
Biochemical determinations
All participants were subject to an anthropometric evaluation to measure weight (kg), height (m) and waist circumference (cm) and to calculate the body mass index (BMI = weight (kg)/height (m)2). The weight was determined without footwear and with light clothing using electronic scales with an approximation of 0.1 kg and a capacity of up to 200 kg. The height was measured with a stadiometer with an approximation of 0.5 cm. The BMI was calculated by dividing the weight in kilograms by the square of the height in metres. The circumference of the waist was measured at the natural indentation between the 10th rib and the iliac crest and the circumference of the hips at the height of the major trochanter, using a metric tape with approximations of 0.5 cm. All measurements were made by the same nurse-study. Blood was collected from the antecubital vein on the follicular phase of the menstrual cycle or after 3 months of amenorrhea; after 12 hours of fasting. To separate serum and plasma from blood cells, samples were immediately centrifuged at 1500 g for 10 min at 4 _C. Fresh samples were used to measure biochemical parameters and the remaining aliquots were stored at −80 °C for subsequent measurement of lipidomic parameters.
Total cholesterol and triglycerides were measured using enzymatic assays (Beckman Coulter, La Brea, CA, USA). High density lipoprotein cholesterol (HDLc) levels were obtained by a direct method using a Beckman LX-20 autoanalyser (Beckman Coulter, La Brea, CA, USA). The intraserial variation coefficient was < 3.5% for all determinations. Levels of low density lipoprotein cholesterol (LDLc) were calculated using the Friedewald formula50. Insulin concentration was determined by means of an enzymatic luminescence technique. Glucose levels were obtained with a Dax-72 autoanalyzer using enzymatic techniques (Bayer Diagnostic, Tarrytown, NY, USA). The homeostasis model assessment of insulin resistance (HOMA-IR) was calculated using baseline glucose and insulin levels:51 HOMA = (fasting insulin (μU/ml) × fasting glucose (mmol/L)/22.5. Concentration of high sensitivity C-reactive protein (hsCRP) was assessed by an immunonephelometric assay (Behring Nephelometer II, Dade Behring, Inc., Newark, DE, USA) which had an intra-assay coefficient of variation of 8.7% and a sensitivity of 0.01 mg/L. Follicle-stimulating hormone (FSH) and luteinizing hormone (LH) were measured using a 2-site monoclonal non-isotopic system (Architect, Abbott Laboratories, Abbott Park, IL). Androstenedione, testosterone and sex hormone binding globulin (SHBG) were measured in our hospital’s Clinical Analysis Service using specialized chemiluminiscence techniques.
Differences among anthropometric and metabolic parameters were analyzed by paired paired t-test for equal variances (p < 0.05 with Benjamini- Hochberg Multiple Testing Correction) and differences were considered significant when p < 0.05).
Lipidomic analysis
Chemicals
Synthetic lipids were from Avanti Polar Lipids Inc. (Alabaster, AL, USA) and Sigma-Aldrich (Madrid, Spain). Fatty acid methyl ester standards were from Larodan Fine Chemicals (Mälmo, Sweden) and from Sigma-Aldrich (Madrid, Spain). Methyl tert-butyl ether (MTBE) LC-MS, acetonitrile LC-MS, isopropanol LC-MS, potassium chloride, chloroform, ammonium formate and ammonium hydroxide were purchased from Sigma-Aldrich (Madrid, Spain); methanol was from Carlo Erba (Milano, Italy); acetone was from Riedel-de-Häen (Seelze, Germany); and LC/MS-grade isopropanol and formic acid were from Baker (Phillipsburg, NJ, USA).
Untargeted lipidomic analysis: Global lipidomic profile
Preparation of lipid standards
For external and internal standardization lipid standards consisting of labeled lipids (see Supplementary Table 1) were used. Stock solutions were prepared by dissolving lipid standards in MTBE at a concentration of 1 mg/mL and working solutions were diluted to 2.5 μg/ mL in MTBE.
Lipid extraction
A lipidomic analysis was performed to the plasma sample based on a previously validated method52. Briefly, to precipitate the protein fraction, 5 μl of miliQ water and 20 μl of methanol were added to 10 μl of plasma sample. After the addition, samples were shaken for 2 min. Then, 250 μl of MTBE plus internal standards were added and samples were ultra-sounded in a water bath (ATU Ultrasonidos, Valencia, Spain) with a frequency and power of 40 kHz and 100 W, respectively, at 10 °C for 30 min. Then, 75 μL of miliQ water were added to the mixture and organic phase was separated by centrifugation (1,400 g) at 10 °C for 10 min. The upper phase, containing all the extracted lipid species, was collected and subjected to mass-spectrometry. A pool of all lipid extracts was prepared and used as quality controls (QC) as previously described53.
LC-MS/MS method
Lipid extracts were subjected to liquid chromatography coupled to mass-spectrometry (LC-MS) using an Agilent UPLC 1290 coupled to the Q-TOF MS/MS 6520 (Agilent Technologies, Barcelona, Spain) basing on previously published method54. Sample compartment was refrigerated at 4 °C and, for each sample, 10 μl of lipid extract was applied onto 1.8 μm particle 100 × 2.1 mm id Waters Acquity HSS T3 column (Waters, Mildord, MA, USA) heated at 55 °C. The flow rate was 400 μl/min with solvent A composed of 10 mM ammonium acetate in acetonitrile-water (40:60, v/v) and solvent B composed of 10 mM ammonium acetate in acetonitrile-isopropanol (10:90, v/v). The gradient started at 40% B and reached 100% B in 10 min and held for 2 min. Finally, the system was switched back to 60% B and equilibrated for 3 min. Duplicate runs of the samples were performed to collect positive and negative electrospray ionized lipid species in a TOF mode, operated in full-scan mode at 100 to 3000 m/z in an extended dynamic range (2 GHz), using N2 as nebulizer gas (5 L/min, 350 °C). The capillary voltage was set 3500 V with a scan rate of 1 scan/s. Continuous infusion using a double spray with masses 121.050873, 922.009798 (positive ion mode) and 119.036320, 966.000725 (negative ion mode) was used for in-run calibration of the mass spectrometer. For MS/MS analyses, we applied a previously described method55.
Data analyses
The MassHunter Data Analysis Software (Agilent Technologies, Barcelona, Spain) was used to collect the results and the MassHunter Qualitative Analysis Software (Agilent Technologies, Barcelona, Spain) to obtain the molecular features of the samples, representing different, co-migrating ionic species of a given molecular entity (i.e. ion adducts) using the Molecular Feature Extractor algorithm (Agilent Technologies, Barcelona, Spain)31. We selected samples with a minimum absolute abundance of 5000 counts and with a minimum of 2 ions. Compounds from different samples were aligned using a RT window of 0.1% ± 0.15 min and a mass window of 10.0 ppm ± 2.0 mDa. Only common features (found in at least 50% of the samples of the same condition) were analyzed, correcting for individual bias and excluding possible contaminants and artefacts. Finally, MassHunter Mass Profiler Professional Software (Agilent Technologies, Barcelona, Spain) was used to perform a non-targeted lipidomic analysis over the extracted features. Only common features (found in at least 50% of the samples of the same condition) were taken into account to correct for individual bias. Multivariate statistics (Hierarchical Clustering, PCA and Random Forest analyses) and biomarker analysis were done using both MassHunter Mass Profiler Professional and Metaboanalyst softwares56,57. The masses representing significant differences by the paired t-test for equal variances (p < 0.05 with Benjamini- Hochberg Multiple Testing Correction) were searched against the LIPID MAPS database (exact mass ppm < 20) and identified with the R-based tool LipidMatch58. Finally, the MS/MS spectra were checked using the LipidBlast software59.
Targeted Lipidomic analysis: plasma fatty acids composition
Fatty acid preparation
After lipid extraction, fatty acyl groups were analyzed as methyl esters derivatives by gas chromatography (GC)55. Briefly, fatty acids were transesterified by incubation in 2 ml of 5% methanolic HCl at 75 °C for 90 min. The resulting fatty acid methyl esters (FAMEs) were extracted by adding 2 ml of n-pentane and 1 ml of saturated NaCl solution. The n-pentane phase was separated, evaporated under N2 gas, re-dissolved in 80 μl of carbon disulfide and 2 μl were used for GC analysis.
GC method
The analysis was performed on a GC System 7890A with a Series Injector 7683B and a flame ionization detector (FID) (Agilent Technologies Inc., Barcelona, Spain) equipped with a DBWAX capillary column (length 30 m × inner diameter 0.25 mm × film thickness 0.20 μm; Agilent Technologies Inc., Barcelona, Spain). The injections were performed in the splitless mode. The temperature of the injector was 220 °C. The flow rate of helium (99.99%) carrier gas was maintained at a constant rate of 1.8 ml/min. The column temperature was held at 145 °C for 5 min; subsequently, the column temperature was increased by 2 °C/min to 245 °C for 50 min, and held at 245 °C for 10 min, and with a post-run of 250 °C for 10 min.
Data analysis
Identification of the twenty-four FAMEs was made by comparison with authentic standards. Results were expressed as mol%. The fatty acid profile detected, identified and quantified represents more than 95% of the total chromatogram. The following fatty acid indexes were calculated: saturated fatty acids (SFA); unsaturated fatty acids (UFA); monounsaturated fatty acids (MUFA); polyunsaturated fatty acids (PUFA) from n-3 and n-6 series (PUFAn-3 and PUFAn-6); average chain length (ACL) = [(Σ%Total14 × 14) + (Σ%Total16 × 16) + (Σ%Total18 × 18) + (Σ%Total20 × 20) + (Σ%Total22 × 22) + (Σ%Total24 × 24)]/100]; double bond index (DBI) = [(1 × Σmol% monoenoic) + (2 × Σmol% dienoic) + (3 × Σmol% trienoic) + (4 × Σmol% tetraenoic) + (5 × Σmol% pentaenoic) + (6 × Σmol% hexaenoic)]; peroxidizability index (PI) = [(0.025 × Σmol% monoenoic) + (1 × Σmol% dienoic) + (2 × Σmol% trienoic) + (4 × Σmol% tetraenoic) + (6 × Σmol% pentaenoic) + (8 × Σmol% hexaenoic)]; and anti-inflammatory index (AI): [[(20:3n-6) + (20:5n-3) + (22:6n-3)]/ (20:4n-6)] * 100. Comparisons between groups were analyzed with a paired t-test for equal variances. The level of statistical significance was set at p < 0.05 in all the analyses.
Supplementary information
Acknowledgements
We thank Maria Rosa Gomez and David Argiles for technical support. This research was funded by grants from Carlos III Health Institute (ISCIII) (PI16/1083, PI16/0301) to V.M.V and M.R, respectively and CIBERehd (CB06/04/0071)−initiatives of the ISCIII to M.R and V.M.V, the European Regional Development Fund (ERDF “A way to build Europe”). Unrestricted grant from Menarini S.A to M.R. V.M.V. and M.R. are recipients of contracts from the Ministry of Health of the Valencian Regional Government and Carlos III Health Institute (CES10/030 and CP10/0360, respectively). R. P. is recipient of contracts from the Spanish Ministry of Economy and Competitiveness, Institute of Health Carlos III (grant number PI14/00328); Spanish Ministry of Science, Innovation and Universities (grant number RTI2018-099200-B-I00), and the Generalitat of Catalonia, Department of Health (grant number SLT002/16/00250) and Department of Business and Knowledge (grant number 2017SGR696). This study has been co-financed by FEDER funds from the European Union (‘A way to build Europe’). I.P. received a predoctoral fellowship from the Lleida University. S.R-LL. is recipient of a Juan de la Cierva contract from Spanish Ministry of Economy and Competitiveness (FJCI-2015-25040). C.B. is recipient of a Sara Borrell contract from Institute of Health Carlos III (CD14/00043). M.J is a Serra Húnter Fellow.
Author contributions
M.J., V.M.V. and R.P. designed the experiments. I.P., S.R.-L.L., A.N., C.B., M.R and M.J. performed the experiments and data analysis. M.J., V.M.V. and R.P. supervised the design and data interpretation. The manuscript was written by I.P., A.H.-M., M.J., V.M.V. and R.P. and edited by M.J. and V.M.V. All authors discussed the results and commented on the manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Antonio Hernandez-Mijares, Reinald Pamplona, Victor M. Victor and Mariona Jové.
Contributor Information
Victor M. Victor, Email: victor.victor@uv.es
Mariona Jové, Email: mariona.jove@udl.cat.
Supplementary information
is available for this paper at 10.1038/s41598-019-52263-w.
References
- 1.El Hayek S, Bitar L, Hamdar LH, Mirza FG, Daoud G. Poly Cystic Ovarian Syndrome: An Updated Overview. Front. Physiol. 2016;7:124. doi: 10.3389/fphys.2016.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diamanti-Kandarakis E, Dunaif A. Insulin Resistance and the Polycystic Ovary Syndrome Revisited: An Update on Mechanisms and Implications. Endocr. Rev. 2012;33:981–1030. doi: 10.1210/er.2011-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diamanti-Kandarakis E, Papavassiliou AG. Molecular mechanisms of insulin resistance in polycystic ovary syndrome. Trends Mol. Med. 2006;12:324–332. doi: 10.1016/j.molmed.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Yildiz BO, Knochenhauer ES, Azziz R. Impact of Obesity on the Risk for Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2008;93:162–168. doi: 10.1210/jc.2007-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Randeva HS, et al. Cardiometabolic Aspects of the Polycystic Ovary Syndrome. Endocr. Rev. 2012;33:812–841. doi: 10.1210/er.2012-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGarry JD. What if Minkowski had been ageusic? An alternative angle on diabetes. Science. 1992;258:766–70. doi: 10.1126/science.1439783. [DOI] [PubMed] [Google Scholar]
- 7.Petersen KF, Shulman GI. Etiology of insulin resistance. Am. J. Med. 2006;119:S10–6. doi: 10.1016/j.amjmed.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apridonidze T, Essah PA, Iuorno MJ, Nestler JE. Prevalence and Characteristics of the Metabolic Syndrome in Women with Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2005;90:1929–1935. doi: 10.1210/jc.2004-1045. [DOI] [PubMed] [Google Scholar]
- 9.Legro RS, Kunselman AR, Dodson WC, Dunaif A. Prevalence and predictors of risk for type 2 diabetes mellitus and impaired glucose tolerance in polycystic ovary syndrome: a prospective, controlled study in 254 affected women. J Clin Endocrinol Metab. 1999;84:165–9. doi: 10.1210/jcem.84.1.5393. [DOI] [PubMed] [Google Scholar]
- 10.Bickerton AST, et al. Cardiovascular risk in women with polycystic ovarian syndrome (PCOS) J Clin Pathol. 2005;58:151–154. doi: 10.1136/jcp.2003.015271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway GS, Agrawal R, Betteridge DJ, Jacobs HS. Risk factors for coronary artery disease in lean and obese women with the polycystic ovary syndrome. Clin. Endocrinol. (Oxf). 1992;37:119–25. doi: 10.1111/j.1365-2265.1992.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 12.Ehrmann DA, et al. Prevalence and Predictors of the Metabolic Syndrome in Women with Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2006;91:48–53. doi: 10.1210/jc.2005-1329. [DOI] [PubMed] [Google Scholar]
- 13.Yang C, Geng Y, Li Y, Chen C, Gao Y. Impact of ovarian endometrioma on ovarian responsiveness and IVF: a systematic review and meta-analysis. Reprod. Biomed. Online. 2015;31:9–19. doi: 10.1016/j.rbmo.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Gregor MF, Hotamisligil GS. Thematic review series: Adipocyte Biology. Adipocyte stress: the endoplasmic reticulum and metabolic disease. J. Lipid Res. 2007;48:1905–1914. doi: 10.1194/jlr.R700007-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Hotamisligil GS. Endoplasmic Reticulum Stress and the Inflammatory Basis of Metabolic Disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duleba AJ, Dokras A. Is PCOS an inflammatory process? Fertil. Steril. 2012;97:7–12. doi: 10.1016/j.fertnstert.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González F, Rote NS, Minium J, Kirwan JP. Increased Activation of Nuclear Factor κB Triggers Inflammation and Insulin Resistance in Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2006;91:1508–1512. doi: 10.1210/jc.2005-2327. [DOI] [PubMed] [Google Scholar]
- 18.Victor VM, et al. Effects of metformin on mitochondrial function of leukocytes from polycystic ovary syndrome patients with insulin resistance. Eur. J. Endocrinol. 2015;173:683–691. doi: 10.1530/EJE-15-0572. [DOI] [PubMed] [Google Scholar]
- 19.Victor VM, et al. Mitochondrial Complex I Impairment in Leukocytes from Polycystic Ovary Syndrome Patients with Insulin Resistance. J. Clin. Endocrinol. Metab. 2009;94:3505–3512. doi: 10.1210/jc.2009-0466. [DOI] [PubMed] [Google Scholar]
- 20.Lizneva D, et al. Criteria, prevalence, and phenotypes of polycystic ovary syndrome. Fertil. Steril. 2016;106:6–15. doi: 10.1016/j.fertnstert.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Jové M, et al. Lipidomics reveals altered biosynthetic pathways of glycerophospholipids and cell signaling as biomarkers of the polycystic ovary syndrome. Oncotarget. 2018;9:4522–4536. doi: 10.18632/oncotarget.23393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wild RA, Rizzo M, Clifton S, Carmina E. Lipid levels in polycystic ovary syndrome: systematic review and meta-analysis. Fertil. Steril. 2011;95:1073–1079.e11. doi: 10.1016/j.fertnstert.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 23.Mudali S, et al. Endogenous Postmenopausal Hormones and Serum Lipids: The Atherosclerosis Risk in Communities Study. J. Clin. Endocrinol. Metab. 2005;90:1202–1209. doi: 10.1210/jc.2004-0744. [DOI] [PubMed] [Google Scholar]
- 24.Moran LJ, Mundra PA, Teede HJ, Meikle PJ. The association of the lipidomic profile with features of polycystic ovary syndrome. J. Mol. Endocrinol. 2017;59:93–104. doi: 10.1530/JME-17-0023. [DOI] [PubMed] [Google Scholar]
- 25.Pernicova I, Korbonits M. Metformin—mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- 26.Collier CA, Bruce CR, Smith AC, Lopaschuk G, Dyck DJ. Metformin counters the insulin-induced suppression of fatty acid oxidation and stimulation of triacylglycerol storage in rodent skeletal muscle. Am. J. Physiol. Metab. 2006;291:E182–E189. doi: 10.1152/ajpendo.00272.2005. [DOI] [PubMed] [Google Scholar]
- 27.Sivalingam VN, Myers J, Nicholas S, Balen AH, Crosbie EJ. Metformin in reproductive health, pregnancy and gynaecological cancer: established and emerging indications. Hum. Reprod. Update. 2014;20:853–868. doi: 10.1093/humupd/dmu037. [DOI] [PubMed] [Google Scholar]
- 28.Diamanti-Kandarakis E, Christakou CD, Kandaraki E, Economou FN. Metformin: an old medication of new fashion: evolving new molecular mechanisms and clinical implications in polycystic ovary syndrome. Eur. J. Endocrinol. 2010;162:193–212. doi: 10.1530/EJE-09-0733. [DOI] [PubMed] [Google Scholar]
- 29.Palomba S, Falbo A, Zullo F, Orio F. Evidence-Based and Potential Benefits of Metformin in the Polycystic Ovary Syndrome: A Comprehensive Review. Endocr. Rev. 2009;30:1–50. doi: 10.1210/er.2008-0030. [DOI] [PubMed] [Google Scholar]
- 30.Wetmore JB, et al. Polycystic Kidney Disease and Cancer after Renal Transplantation. J. Am. Soc. Nephrol. 2014;25:2335–2341. doi: 10.1681/ASN.2013101122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sana TR, Roark JC, Li X, Waddell K, Fischer SM. Molecular formula and METLIN Personal Metabolite Database matching applied to the identification of compounds generated by LC/TOF-MS. J. Biomol. Tech. 2008;19:258–66. [PMC free article] [PubMed] [Google Scholar]
- 32.Escobar-Morreale HF. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat. Rev. Endocrinol. 2018;14:270–284. doi: 10.1038/nrendo.2018.24. [DOI] [PubMed] [Google Scholar]
- 33.Radosh L. Drug treatments for polycystic ovary syndrome. Am. Fam. Physician. 2009;79:671–6. [PubMed] [Google Scholar]
- 34.Naderpoor N, et al. Metformin and lifestyle modification in polycystic ovary syndrome: systematic review and meta-analysis. Hum. Reprod. Update. 2015;21:560–574. doi: 10.1093/humupd/dmv025. [DOI] [PubMed] [Google Scholar]
- 35.Zabielski P, et al. The effect of high fat diet and metformin treatment on liver lipids accumulation and their impact on insulin action. Sci. Rep. 2018;8:7249. doi: 10.1038/s41598-018-25397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holland WL, Summers SA. Sphingolipids, Insulin Resistance, and Metabolic Disease: New Insights from in Vivo Manipulation of Sphingolipid Metabolism. Endocr. Rev. 2008;29:381–402. doi: 10.1210/er.2007-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karakas SE. New biomarkers for diagnosis and management of polycystic ovary syndrome. Clin. Chim. Acta. 2017;471:248–253. doi: 10.1016/j.cca.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Lucki NC, Sewer MB. The interplay between bioactive sphingolipids and steroid hormones. Steroids. 2010;75:390–9. doi: 10.1016/j.steroids.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santana, P. et al. Interleukin-IP Stimulates Sphingomyelin Hydrolysis in Cultured Granulosa Cells: Evidence for a Regulatory Role of Ceramide on Progesterone and Prostaglandin Biosynthesis*. Endocrinology137 (1996). [DOI] [PubMed]
- 40.Chaurasia B, Summers SA. Ceramides – Lipotoxic Inducers of Metabolic Disorders. Trends Endocrinol. Metab. 2015;26:538–550. doi: 10.1016/j.tem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Gancheva S, Jelenik T, Álvarez-Hernández E, Roden M. Interorgan Metabolic Crosstalk in Human Insulin Resistance. Physiol. Rev. 2018;98:1371–1415. doi: 10.1152/physrev.00015.2017. [DOI] [PubMed] [Google Scholar]
- 42.Aburasayn H, Al Batran R, Ussher JR, Batran AR. Targeting ceramide metabolism in obesity. Am J Physiol Endocrinol Metab. 2016;311:423–435. doi: 10.1152/ajpendo.00133.2016. [DOI] [PubMed] [Google Scholar]
- 43.Naudí A, et al. Membrane lipid unsaturation as physiological adaptation to animal longevity. Front. Physiol. 2013;4:372. doi: 10.3389/fphys.2013.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poisson J-PG, Cunnane SC. Long-chain fatty acid metabolism in fasting and diabetes: relation between altered desaturase activity and fatty acid composition. J. Nutr. Biochem. 1991;2:60–70. doi: 10.1016/0955-2863(91)90030-9. [DOI] [Google Scholar]
- 45.Wang Y, et al. Regulation of hepatic fatty acid elongase and desaturase expression in diabetes and obesity. J. Lipid Res. 2006;47:2028–2041. doi: 10.1194/jlr.M600177-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tripathy S, Torres-Gonzalez M, Jump DB. Elevated hepatic fatty acid elongase-5 activity corrects dietary fat-induced hyperglycemia in obese BL/6J mice. J. Lipid Res. 2010;51:2642–2654. doi: 10.1194/jlr.M006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yashiro H, et al. A Novel Selective Inhibitor of Delta-5 Desaturase Lowers Insulin Resistance and Reduces Body Weight in Diet-Induced Obese C57BL/6J Mice. PLoS One. 2016;11:e0166198. doi: 10.1371/journal.pone.0166198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum. Reprod. 19, 41–7 (2004). [DOI] [PubMed]
- 49.Azziz R. Diagnostic criteria for polycystic ovary syndrome: a reappraisal. Fertil. Steril. 2005;83:1343–6. doi: 10.1016/j.fertnstert.2005.01.085. [DOI] [PubMed] [Google Scholar]
- 50.Friedewald, W. T., Levy, R. I. & Fredrickson, D. S. Estimation of the Concentration of Low-Density Lipoprotein Cholesterol in Plasma, Without Use of the Preparative Ultracentrifuge. Clin. Chem. 18 (1972). [PubMed]
- 51.Matthews DR, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–9. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 52.Pizarro C, Arezana-Rámila I, Pérez-del-Notario N, Pérez-Matute P, González-Sáiz JM. Plasma Lipidomic Pro fi ling Method Based on Ultrasound Extraction and Liquid Chromatography Mass Spectrometry. Anal. Chem. 2013;85:12085–12092. doi: 10.1021/ac403181c. [DOI] [PubMed] [Google Scholar]
- 53.Want EJ, et al. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2012;8:17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
- 54.Castro-Perez JM, et al. Comprehensive LC−MS E Lipidomic Analysis using a Shotgun Approach and Its Application to Biomarker Detection and Identification in Osteoarthritis Patients. J. Proteome Res. 2010;9:2377–2389. doi: 10.1021/pr901094j. [DOI] [PubMed] [Google Scholar]
- 55.Jové M, et al. Plasma lipidomics discloses metabolic syndrome with a specific HDL phenotype. FASEB J. 2014;28:5163–71. doi: 10.1096/fj.14-253187. [DOI] [PubMed] [Google Scholar]
- 56.Xia, J. & Wishart, D. S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. In Current Protocols in Bioinformatics 14.10.1–14.10.91 (John Wiley & Sons, Inc., 10.1002/cpbi.11 2016). [DOI] [PubMed]
- 57.Chong J, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koelmel JP, et al. LipidMatch: an automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinformatics. 2017;18:331. doi: 10.1186/s12859-017-1744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kind T, et al. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.