Abstract
Over the last few decades, evolutionarily conserved molecular networks have emerged as important regulators in the expression and function of eukaryotic genomes. Recently, miRNAs (miRNAs), a large family of small, non-coding regulatory RNAs were identified in these networks as regulators of endogenous genes by exerting post-transcriptional gene regulation activity in a broad range of eukaryotic species. Dysregulation of miRNA expression correlates with aberrant gene expression and can play an essential role in human health and disease. In the context of the lung, miRNAs have been implicated in organogenesis programming, such as proliferation, differentiation, and morphogenesis. Gain- or loss-of-function studies revealed their pivotal roles as regulators of disease development, potential therapeutic candidates/targets, and clinical biomarkers. An altered microRNAome has been attributed to several pulmonary diseases, such as asthma, chronic pulmonary obstructive disease, cystic fibrosis, lung cancer, and idiopathic pulmonary fibrosis. Considering the relevant roles and functions of miRNAs under physiological and pathological conditions, they may lead to the invention of new diagnostic and therapeutic tools. This review will focus on recent advances in understanding the role of miRNAs in lung development, lung health, and diseases, while also exploring the progress and prospects of their application as therapeutic leads or as biomarkers.
Keywords: microRNA dysregulation, COPD, cystic fibrosis, lung cancer, asthma, idiopathic pulmonary fibrosis, antagomiR, aptamer
Main Text
MicroRNAs (miRNAs) have provided a new and outstanding molecular biology paradigm to understand the molecular mechanisms underlying pulmonary diseases. In the last few decades, non-coding RNAs (ncRNAs) such as miRNA have been identified as emerging mediators in human lung disease. miRNA have been shown to play crucial roles in fundamental biological mechanisms by post-transcriptional regulation of their cognate mRNA, impacting cellular events such as metabolism, growth, cell differentiation, and development (thereby regulating organogenesis), apoptosis, inflammation, and cell signaling.1 Dysregulation of miRNA in diseased states often produces signature microRNA profiles that can be used as biomarkers for specific diseases.2 The role of an aberrant microRNAome in the pathophysiology of diseases has identified miRNA as biomarkers, therapeutic targets, or indicators of prognosis. The first miRNA identified was Lin-4 in Caenorhabditis elegans, followed by the discovery of the let-7 family of miRNA, which are crucial regulators in the development of C. elegans.3, 4 Since then, around 2,000 validated miRNA have been identified in the human genome.5, 6 About 60% of human protein-coding genes are now known to be subject to miRNA-mediated post-transcriptional regulation.7 The relation between an aberrant lung microRNAome and lung diseases is becoming increasingly evident, introducing novel paradigms in the molecular mechanisms underlying lung diseases, such as lung cancer, pulmonary fibrosis, chronic obstructive pulmonary disease (COPD), asthma, and pulmonary hypertension.8, 9, 10, 11, 12 As potential regulators of an immune response, miRNA play a vital role in lung immunity. Hence they can also determine the magnitude of inflammation and tissue damage in lung diseases.13, 14 This review focuses on the recent discoveries regarding the influence and contribution of miRNA in inflammatory airway diseases and lung cancer with specific emphasis on therapeutic development.
Biogenesis of MicroRNA
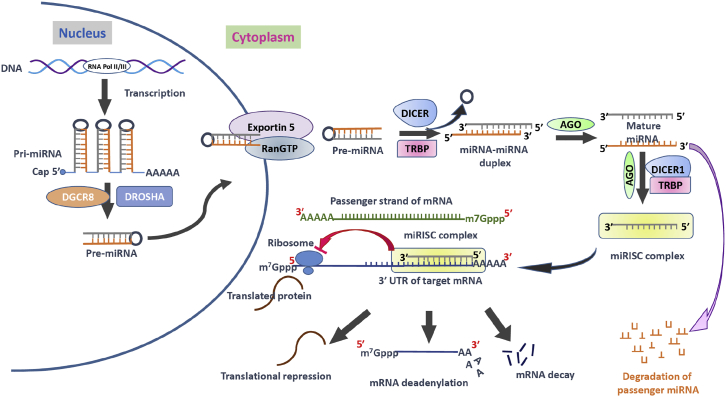
The discovery of miRNA-mediated post-transcriptional gene regulation was one of the watershed events in the field of gene regulation. miRNA are short, single-stranded, non-coding RNA molecules about 22 nt in length. In early 1960, Britten and Davidson15 first proposed that the regulatory mechanisms in higher organisms are controlled by activator RNAs. These activator RNAs are found in the nucleus and transcription product of the redundant genome. They also suggested that redundant genome sequences were found among the integrator genes and that the activator RNAs move from their synthesis site to active transcription of the producer genes (presently called exon). Since the first report of the discovery of lin-4 miRNA by Lee et al., 2,000 different miRNA have been identified and validated to play essential roles in different developmental stages and pathophysiological processes.4 miRNA are commonly encoded either within protein-coding genes called introns or as independent genes in intronic regions.16 Some other studies demonstrated that they are also derived from transposon elements or across two tandem, inverted repetitive elements.17 miRNA are first transcribed as much longer RNAs called primary miRNA, or pri-miRNA.18 Biogenesis of miRNA involves two distinct steps involving both nuclear and cytoplasmic events (Figure 1). In the nucleus, the long pri-miRNA transcript is initially altered with a 5′ 7-methylguanosine cap and a 3′ poly(A) tail and transcribed by RNA polymerase II (Pol II) or III.19, 20, 21 This is subsequently processed by DGCR8 and Drosha to form short hairpin structures of 70–90 nt called a precursor of mature microRNA (pre-miRNA). Exportin-5 exports pre-miRNA to the cytoplasm, where it is further processed by RNase III enzyme Dicer and trans-activating response RNA binding protein (TRBP) to form mature miRNA.22, 23 Subsequently, Argonaute proteins (Ago1 and Ago2) recruit miRNA in the miRNA-induced silencing complex (miRISC) (Figure 1). The loaded RISC complex binds selectively to the miRNA recognition elements (MREs) within the 3′ UTR of target mRNA transcripts.24, 25 To bind selectively to the target sequence within mRNA, the miRNA uses a 2- to 8-nt long “seed sequence,” which is present at the 5′ region of the miRNA.26 Target genes with longer 3′ UTR generally have the maximum density for miRNA binding sites rather than the shorter 3′ UTR. Most of the miRNA-mRNA interaction occurs at the 3′ UTR region, although targeting and silencing of mRNA have also been reported to occur at the 5′ UTR with similar efficiency.14, 27 miRNA:mRNA interaction inhibits protein translation by suppressing translation or degrading the target mRNA, and subsequently plays a critical role in cellular growth and differentiation.
Figure 1.
Mechanism of MicroRNA Processing and Their Inhibitory Mechanism
The microRNA (miRNA) processing pathway begins with transcription of their genes with the help of RNA polymerase II (Pol II) or polymerase III (Pol III) to produce pri-miRNAs in the nucleus. Then a microprocessor complex, composed of RNA-binding protein DGCR8 and type III RNase Drosha, cleaves pri-miRNA into a ∼85-nt stem-loop structure called pre-miRNA. The exportin 5-RAN/GTP complex mediates the transport of pre-miRNA from the nucleus into the cytoplasm. The RNase DICER in complex with double-stranded RNA-binding protein TRBP cleaves the pre-miRNA hairpin to a ∼20- to 22-nt miRNA/miRNA duplex. After the duplex is unwound, the functional strand of the mature miRNA (the guide strand) is loaded into the miRISC-containing DICER1, TRBP, and Argonaute (AGO) proteins. This miRISC silences/inhibits the target mRNAs expression/function through mRNA cleavage, translational repression, or deadenylation. The passenger strand of the miRNA is degraded. AGO, Argonaute proteins; DGCR8, DiGeorge syndrome critical region gene 8; m7G cap, 7-methylguanosine; miRISC, miRNA-induced silencing complex; miRNA, microRNA; pre-miRNA, miRNA precursor; pri-miRNA, primary miRNA; RAN-GTP, Ras-related nuclear protein coupled with guanosine-5′-triphosphate; TRBP, transactivating response RNA-binding protein.
Function and Mechanism of miRNA
miRNA can regulate multiple genes and vice versa, i.e., each mRNA may be targeted by multiple miRNA. miRNA mediate their function as part of an effector unit containing an Argonaute protein and is known as miRNP, miRgonaute or miRISC.28 Several factors affect the biological outcome of a miRNA:mRNA interaction, such as the binding of the miRNA seed region with the target site via base pairing, the number and relative positions of multiple target sites on the same miRNA, target site accessibility as a function of the secondary structure, and flanking target sequences of other miRNA.29, 30, 31 miRNA can mediate transcriptional or post-transcriptional gene silencing.7 Transcriptional gene silencing involves a unique cellular complex called RNA-induced transcriptional silencing (RITS) that contains argonaute molecules responsible for chromatin remodeling.32 Post-transcriptional gene silencing involves suppression of translation and degradation of target mRNA transcripts. Target mRNA degradation by miRNA involves 5′ end decapping and 3′ deadenylation followed by degradation by several endo- and exo-nucleolytic nucleases such as polysomal ribonuclease 1 (PMR1) and Xrn1p, respectively.32, 33, 34 Wakiyama et al.35 have shown that the closed-loop structure of mRNA enhances translation, but by deadenylating the poly(A) tail, miRNA prevent binding of cytoplasmic poly(A)-binding protein (PABPC1) and consequently repress translation. Argonaute protein can also directly repress translation of the target mRNAs by competing with the eukaryotic transcription factor, elF4E, that directs the ribosomes to the target mRNAs.36 Chendrimada et al.37 demonstrated that Ago2 binds to eIF6, preventing the binding of the large ribosomal subunit to the small ribosomal subunit and inhibiting translation. On the other hand, the RNA-binding protein HuR can relieve miRNA-mediated repression. Under normal conditions, HuR protein is primarily localized in the nucleus, but upon several stress conditions, it relocates to the cytoplasm and plays a potential regulatory role by relieving the miRNA-mediated repression.28 Bhattacharyya et al.38 have reported that the miR-122-mediated translational suppression of cationic amino acid transporter 1 (CAT-1) is relieved by binding of HuR protein to the 3′ UTR of CAT-1 mRNA.
Regulation of MicroRNA Expression
Most of the miRNA genes are found in intergenic locations or antisense orientation to the annotated gene, implying that they have their transcription machinery.39, 40 Lee et al.20 first demonstrated that miRNA are transcribed by Pol II, although recently other studies have found that miRNA transcription is also mediated by Pol III.21 Saito et al.41 first showed that chromatin remodeling and epigenetic alterations by DNA methylation and histone tail modifications could regulate the expression of several miRNA with consequent effects on cellular functions. To examine miRNA expression in human cancer cells, they treated the cells with a DNA-demethylating drug named 5-Aza-CdR and histone deacetylase inhibitor named 4-phenyl butyric acid, and found that miR-127 was upregulated significantly among other miRNA targeting the proto-oncogene BCL-6 that is upregulated in cancer cells.42, 43 This study was supported by another study that showed that using the histone deacetylase (HDAC) inhibitor named LAQ824 in breast cancer cell line SKBr3 caused a significant change in 40% of the different miRNA species.44 Most of the miRNA are intragenic or found in the introns of the protein-coding genes and are reasonable to demonstrate that transcription of miRNA is cooperatively regulated with the host genome. Because miRNA have their promoter, it is believed that CpG islands of host promoter also found within the same intron and transcription of both protein-coding gene and miRNA are regulated by DNA methylation.45, 46
Apart from the epigenetic modulation, several other nuclear proteins or factors are responsible for regulating the miRNA expression. Fukuda et al.47 demonstrated that DEAD-box RNA helicases p68 and p72, components of a large Drosha-mediated processing complex, could interact with several transcription factors such as Smads, p53, and estrogen receptor to correctly recognize and bind to a subset of pri-mRNAs, and initiate the cleavage to form pre-miRNA. Guil and Cáceres48 reported that heterogeneous nuclear ribonucleoproteins (hnRNP proteins), RNA-binding proteins, play a role in the processing of endogenous pri-miR-18a, which is context dependent and regulating the activity of miR-18a. Importantly, they found that depletion of hnRNP A1 affects able processing of miR-18a, which increases cell proliferation and promotes the anchorage-independent growth of cancer cells.49 KH-type splicing regulatory protein (KSRP), a multifunctional single-strand RNA-binding protein, binds to the conserved G-rich elements in the terminal loop (TL) of a cohort of miRNA precursors and interacts with both Drosha and Dicer to promote miRNA maturation.50 KSRP interacts with heterogeneous nuclear RNA-binding proteins (hnRNPs), which are involved in mRNA maturation, as well as acts as an auxiliary factor for the Drosha-mediated processing of a microRNA precursor by binding to the TL of a group of pri-miRNA.48, 51 High mobility group A (HMGA) proteins are extensively synthesized during the early embryonic stage, as well as growth and development, and also are involved in regulating miRNA expression by regulating chromatin structure and gene expression.52, 53
Role of miRNA in Lung Development
Lung development and maturation is a complex and vital morphogenetic process that is temporally and spatially regulated by a defined set of genes.54 In the fetus, lung development goes through six defined stages: embryonic, glandular, canalicular, saccular, alveolar, and vascular expansion.55 Several cytokines and their signaling pathways such as transforming growth factor-beta (TGF-β), fibroblast growth factors (FGFs), sonic hedgehog (Shh), and wingless-type MMTV integration site family (WNT)/β-CATENIN are involved in lung development.56, 57, 58 Stage-specific and tissue-specific miRNA expression are crucial for lung development and in maintaining lung homeostasis.59
For instance, members of the miR-17-92 cluster (miR-17, -18a, -19a, -20a, -19b-1, and -92-1) are highly expressed in embryonic lungs.60 Expression of these same miRNA decreases during lung maturation. Conversely, the let-7 family miRNA are elevated in the adult lung compared with early embryonic stages.61 Hayashi et al.62 reported that the expression of miR-21 is required and has a crucial role in branching morphogenesis, a primary developmental process in the lung. Furthermore, expression of miR-142-3p and miR-326 regulates the proper differentiation and proliferation of mesenchymal cells by WNT signaling and Shh signaling pathways, respectively.63, 64 In the development of vascular smooth muscle cells (vSMCs), the role of miRNA and proteins involved in the miRNA pathway was studied extensively. The cooperative role of both miRNA (miR-145 and miR-143) was reported in maintaining proper SMC phenotype, whereas miR-133 and miR-206 play crucial roles in the proliferation, migration, and development of vSMCs by targeting transacting transcription factor-1 and Notch3, respectively.65 Inhibition of miRNA processing by conditional inactivation of DICER, during the embryonic stage, resulted in deformed lung development, and excessive epithelial cell death was reported.66 At the embryonic stage, reduced expression of Ago1 and Ago2 in the distal epithelium and mesenchyme, respectively, suggested that miRNA-regulated gene expression is involved in the lung developmental processes.67, 68
Role of miRNA in Lung Health and Disease
miRNA play an important role in lung health and diseases. Dysregulation of miRNA plays an important role in pathological hallmarks of several lung diseases (Figure 2). Several studies identified altered miRNA expression profiles, which may be associated with pathological processes within the lung and lead to the development of several respiratory diseases, ranging from inflammatory diseases (chronic airway diseases such as COPD, asthma, and cystic fibrosis [CF]) to lung cancers.69, 70 A group of miRNA has been identified to play a role in inflammatory responses in chronic airway diseases, such as COPD, asthma, and CF. Likewise, other groups of both pro-fibrotic and anti-fibrotic miRNA have been identified to play a role in interstitial pulmonary fibrosis. Lung diseases are the leading cause of morbidity and mortality worldwide.71 According to the World Health Organization (WHO), COPD is the fourth leading cause of death worldwide and predicted to become the third leading cause by 2030.72 On the other hand, asthma, a complex, heritable disease, affects more than 300 million people globally, and idiopathic pulmonary fibrosis (IPF), a chronic fibrotic lung disease, affects approximately 3 million people worldwide, with the incidence increasing with age.73, 74 The GLOBOCAN 2018 database reports 2.09 million new cases and 1.76 million deaths from lung cancers.75 Hence identifying the molecular mechanisms involved in the development and progression of these diseases is important to public health. Many reports are now investigating microRNA-mediated post-transcriptional gene silencing in lung diseases. Much attention and research remain to be conducted to explore the function and pathological role of miRNA in respiratory diseases. In the following sections, we will be focusing on aberrant miRNA expression, their target sites, and findings in the five most common lung diseases (Table 1).
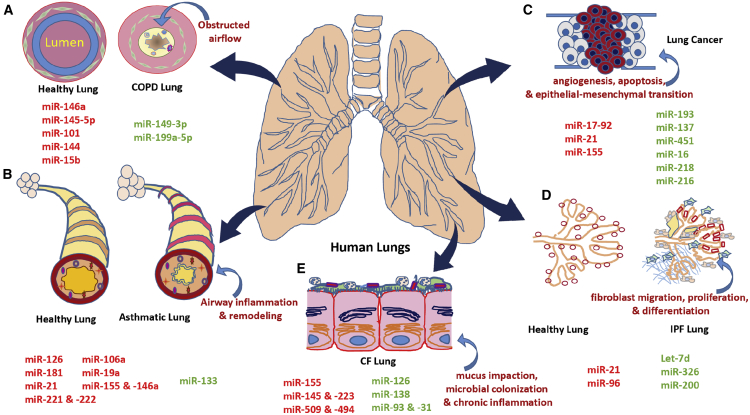
Figure 2.
Schematic of MicroRNAs Implicated in Lung Pathophysiology in Different Lung Diseases
(A) Chronic obstructive pulmonary disease (COPD) is a chronic inflammatory lung disease that causes obstructed airflow from the lungs and attenuates mucociliary clearance (MCC), leading to mucous obstruction, and provides a nutrient-rich environment for bacterial reproduction, leading to pulmonary infections and chronic inflammation. (B) Asthma, characterized by the hallmarks of airway inflammation, airway remodeling, airway hyperresponsiveness, and reversible airway obstruction. (C) Lung cancer is associated with excessive pulmonary cell proliferation, apoptosis, angiogenesis, and epithelial-mesenchymal transition. (D) In idiopathic pulmonary fibrosis, the normal lung tissue is replaced by more heavily scarred lung tissue, which makes it difficult for the patient to breathe and deliver needed oxygen to the body. This causes the aberrantly activated lung epithelium to produce mediators of fibroblast migration, proliferation, and differentiation into active myofibroblasts. (E) In cystic fibrosis, aberrant or nonexistent CFTR function compromises the airway surface liquid, leading to mucous impaction and sub-optimal ciliary beating. This results in microbial colonization and chronic inflammation, which further compromise mucociliary clearance. microRNAs in red and green indicate whether the miRNA is elevated or reduced in lung-associated diseases, respectively. CF, cystic fibrosis; COPD, chronic obstructive pulmonary disease; IPF, idiopathic pulmonary fibrosis.
Table 1.
List of miRNAs that Are Differentially Expressed in Lung Diseases (COPD, asthma, lung cancer, IPF, and CF), with Their Respective Target Sites and Findings Listed Above
| Lung Diseases | Specific miRNA | Expression Level | Target Site/Host Gene | Findings | References |
|---|---|---|---|---|---|
| COPD | miR-146a | high | COX-2 | targets 3′ UTR of the Cox2 mRNA and suppresses the expression | 1 |
| miR-149-3p | low | TLR-4, MyD88 | reduced expression causes overexpression of TLR-4 and MyD88 | 2 | |
| miR-145-5p | high | SMAD3, CFTR, SLC26A9 | involved in Th2 response activation, blocks chloride ion channel | 3, 4 | |
| miR-199a-5p | low | Unfolded protein responses | intensification of the UPR | 5 | |
| miR-101 and miR-144 | high | MKP-1, TGF-β signaling | induce inflammatory responses | 6 | |
| miR-15b | high | SMAD7 | induces TGF-β signaling | 7 | |
| Asthma | miR-126 | high | TLRs | activation of inflammatory pathways | 8 |
| miR-21 | high | IL-12p35 | modulates IL-12 expression and polarizes Th cells toward Th2 response | 9 | |
| miR-155 and miR-146a | high | transcription factor PU.1 and IL-4 | contributes to immediate inflammation and allergic reactions | 10 | |
| miR-133a | low | RhoA | excessive bronchial smooth muscle (BSM) contraction | 11 | |
| miR-221 and miR-222 | high | p21WAF1 and p27kip1 | involved in mast cell activation and release several growth factors | 12 | |
| miR-106a | high | IL-10 | increases pro-inflammatory cytokines release | 13, 14 | |
| miR-181 | high | NF-κB | induces increased TCR sensitivity | 15 | |
| miR-19a | high | PI3K, JAK-STAT, NF-κB signaling | promotes allergic inflammatory phenotype | 16, 17 | |
| Lung cancer | miR-193 | low | KRAS | promotes cellular proliferation, differentiation, and migration | 18 |
| miR-17-92 | high | myc | promotes hyper-proliferation of lung epithelial cells | 19 | |
| miR-21 | high | PTEN, PDCD4 | promotes growth and invasion in NSCLC | 20 | |
| miR-137 | low | SLC22A18 | promotes aggressive tumor progression | 21 | |
| miR-451 | low | RAB14 | induces tumor differentiation and shorter survival | 22 | |
| miR-16 | low | p27, Bcl-2, Bax, and caspase-3 | induces cell proliferation and apoptosis | 23 | |
| miR-218 | low | HMGB1 | leads to aggressive cell proliferation, migration, and invasion | 24 | |
| miR-155 | high | Apaf-1 | resistance to therapy and associated with shorter survival | 25 | |
| miR-216 | low | eIF4B, ZEB1 | tumor growth, proliferation, metastasis, and chemoresistance | 26 | |
| IPF | Let-7d | low | HMGA2 | increases mesenchymal markers (ACTA2, VIM) and decreases epithelial markers (cytokeratin and TJP1) | 27 |
| miR-21 | high | SMAD7 | promotes excessive extracellular matrix (ECM) gene transcription | 28 | |
| miR-96 | high | FoxO3a | increases PI3K-Akt activity, thereby promoting IPF fibroblasts | 29 | |
| miR-326 | low | 3′ UTR of TGF-β | upregulates profibrotic genes | 30 | |
| miR-200 | low | TGF-β signaling | induces epithelial-mesenchymal transition and tumor metastasis | 31 | |
| CF | miR-126 | low | TOM1 | causes excessive inflammatory response and airway obstruction | 32, 33 |
| miR-138 | low | SIN3A | resuscitates the CFTR expression | ||
| miR-155 | high | MAPK and PI3K/Akt signaling | activates proinflammatory cytokine IL-8 to attract neutrophils | 34 | |
| miR-145 and miR-223 | high | 3′ UTR of CFTR | decrease CFTR expression and cause inflammation | 35 | |
| miR-509 and miR-494 | high | NF-κB signaling | repress CFTR expression and induce pro-inflammatory cytokines | 36 | |
| miR-93 and miR-31 | low | 3′ UTR of IL-8, IRF-1 | promote increased production of cathepsin S | 37 |
High and low indicate whether the miRNA is elevated or reduced in lung-associated diseases, respectively. CF, cystic fibrosis; COPD, chronic obstructive pulmonary disease; IPF, idiopathic pulmonary fibrosis.
miRNA in COPD
COPD is a common airway complication that comprises chronic obstructive bronchitis and lung emphysema.76 COPD is a multifactorial disease that represents the leading cause of higher morbidity and mortality globally, and it is also expected that COPD will become the third leading cause of death worldwide by 2030 because of increased prevalence with older age, environmental risk factors, excessive cigarette smoking, and noxious gases.77, 78 The hallmarks of COPD are characterized by chronic inflammation in the lungs, a shorter interval between breathing, severe cough, and repetitive impediment across the tracheal wall during inhalation (Figure 2A).79 Several miRNA have been implicated in the pathobiology of COPD.80, 81, 82, 83, 84
In COPD patients, increased secretion of prostaglandin E2 (PGE2) results in collagen overproduction, ultimately reducing lung capacity and accelerating COPD.69 COPD patients demonstrate decreased miR-146a expression and increased expression of its target, Cox-2, with a consequent increase in PGE2 levels.80 Matrix metalloproteases (MMPs) play a major role in respiratory inflammation and structural remodeling in COPD patients. During the early stage of COPD, cigarette smoke induces macrophages, lymphocytes, and neutrophils to be deposited in the walls of bronchioles, alveolar ducts, and alveoli.85, 86 Macrophage-derived MMPs including MMP-2, MMP-9, and MMP-12 degrade and solubilize extracellular matrix proteins, collagen, and elastin.87, 88. MMP-12 is overexpressed in the lungs of COPD patients.89 Graff et al.90 demonstrated that miR-452, an MMP-12-targeting microRNA, is significantly downregulated in COPD patients, resulting in overexpression of MMP-12.
Shen at al.91 have shown that levels of miR-149-3p play a protective role in COPD by suppressing the Toll-like receptor 4/nuclear factor κB (TLR4/NF-κB) pathway by targeting two distinct signaling intermediates, namely, TLR4 and MyD88.91, 92, 93 miR-149-3p levels are progressively suppressed in non-COPD smokers, followed by stable COPD smokers, with maximal suppression observed in smokers with acute exacerbation COPD.91 Dysregulation of TLR4 expression has multiple downstream effects by increasing the expression of proinflammatory cytokines interleukin-1β (IL-1β), IL-6, IL-8, IL-10, tumor necrosis factor-alpha (TNF-α), and interferon-γ (IFN-γ).94, 95, 96 Persistent activation of the TLR-4 signaling and MyD88 dysregulation also induces matrix metalloproteinase 1 (MMP-1) via MyD88 and IRAK1 pathways, which plays an important role in COPD.
Another microRNA, miR-145-5p, is significantly upregulated in patients with COPD and smokers, and can serve as a promising biomarker of COPD.97 Tobacco smoking is the principal risk factor for COPD. Cigarette smokers and COPD patients demonstrate chronic induction of TGF-β signaling.98, 99, 100, 101, 102 We have demonstrated that TGF-β upregulates miR-145-5p in bronchial epithelial cells.100 miR-145-5p dysregulation can have multiple downstream effects, which can lead to a progressive decline in lung function. For instance, miR-145-5p is involved in Th2 response activation, macrophage differentiation, and recruitment of eosinophils.103, 104 Likewise, TGF-β-mediated miR-145-5p induction plays an important role in the regulation of airway smooth muscle (ASM) function in COPD patients by targeting SMAD3 that negatively regulates the release of pro-inflammatory cytokines.105 Cystic fibrosis transmembrane conductance regulator (CFTR) functions as a chloride channel involved in maintaining airway fluid homeostasis, as well as regulating innate immune responses in the airway. Smokers and COPD patients demonstrate an acquired CFTR dysfunction even though they have regular copies of the CFTR gene.106, 107 Acquired CFTR dysfunction results in impaired mucociliary clearance and dysfunctional airway innate immune responses, which result in chronic microbial infection and lung inflammation.108, 109, 110, 111, 112, 113, 114, 115 In COPD patients, expression of CFTR-targeting miRNA miR-101 and miR-144 is upregulated with consequent CFTR suppression.116 We have recently demonstrated that TGF-β signaling and cigarette smoke (via TGF-β signaling) upregulate miR-145-5p to suppress CFTR, as well as an important CFTR modifier SLC26A9, which also functions as a backup Cl− channel.100
miR-144 and miR-15b are potential mediators of the TGF-β signaling cascade and genes that are functionally associated with the TGF-β superfamily involved in the development and progression of COPD, inflammatory response, and airway epithelial repair after injury.99, 117 The miR-15b expression is higher in COPD patients with a concomitant decrease in the inhibitory SMAD7.118 Expression of another microRNA, miR-199a-5p, is diminished in COPD patients because of hypermethylation of CpG sites in the miR-199a-5p promoter.119 Decreased expression of miR-199a-5p leads to an intensification of the unfolded protein responses (UPRs) and contributes to lung cell apoptosis and lung inflammation.119
miRNA in Asthma
Asthma is a chronic inflammatory disease of the airway system characterized by fatal airway obstruction, tissue remodeling, bronchial epithelial hyperresponsiveness, and chronic inflammation (Figure 2B). Asthmatic patients also experience intermittent periods of wheezing, heavy tightness in the chest, and shortness of periodic breathing.120, 121, 122 According to the WHO, asthma caused 225,000 deaths all over the world in 2005, and with this current trend, the number will reach 430,000 by 2030.123 Both genetic and environmental factors are considered important triggers to the pathogenesis of asthma. Chronic inflammation in asthmatic patients is associated with persistent deposition of mast cells and eosinophils that promotes increased cytokine production by Th2 cells, resulting in mucous hypersecretion, bronchial hyperactivity, elevated immunoglobulin E (IgE) levels, and eosinophils infiltration.124, 125
Several miRNA play important roles in the pathology of asthma. These can be broadly categorized into pro-inflammatory miRNA and anti-inflammatory miRNA. MicroRNA-126 promotes inflammation by inducing the overexpression of Toll-like receptors (TLRs) present on T helper 2 cells (Th2).121 miR-126 antagonism activates the PU.1 transcription factor, which modulates Th2 cell function via negative regulation of GATA3 expression.126 Another study confirmed this role for miR-126 using antagomiRs to miR-126.127 They showed that miR-126 antagonism suppresses Th2-driven bronchial inflammation, mucous hypersecretion, and airway hyperresponsiveness (AHR) in mice.
IL-13, a pleiotropic Th2 cell-derived effector cytokine, plays a central role in the pathogenesis of asthma.128 Lu et al.122 showed that IL-13 induces the overexpression of miR-21 and underexpression of miR-1 in transgenic mice compared with the control mice. They also demonstrated that IL-13-induced miR-21 overexpression is IL-13Rα1 dependent, whereas allergen-induced miR-21 overexpression is IL-13Rα1 and STAT6 independent, meaning that miR-21 induction is associated with leukocytes recruitment/activation in IL-13Rα1−/− mice. By using bioinformatics tools and target site validation approaches, Lu et al.122 found that IL-12p35 is the putative target of miR-21. On the other hand, increased levels of miR-21 negatively modulate the expression of IL-12, which is a pro-inflammatory cytokine that induces the production of IFN-γ, involved in adaptive immune responses including Th1 cell polarization.129 IL-13 also upregulates RhoA expression, which is responsible for bronchial smooth muscle (BSM) contraction that contributes to airway narrowing in people with asthma.130 IL-13-induced RhoA upregulation was found to be a consequence of miR-133a suppression and STAT6 dependent.131 IL-13 secretion itself is subject to miRNA-mediated regulation. miR-145 plays a vital role in the onset and pathogenesis of allergic airways disease by inducing Th2 cells to release IL-5 and IL-13.103
Altered expression of miR-155 and miR-146a affects the local immune response in allergic asthma.132, 133, 134, 135 Increased expression of miR-155 and miR-146a is associated with Th2-mediated increased cytokine IL-4 release that induces B cells to undergo class switching to secrete more IgE and contributes to immediate inflammation and allergic reactions.136 Intranasal administration of miR-155 in mice activates the expression of chemokine eotaxin-1/CCL11 and eotaxin-2/CCL24, as well as an eotaxin-1/2/CCR3 pathway, which are essential for eosinophil recruitment.134, 137 miR-155 knockout mice demonstrated elevated levels of PU.1, transcription factor suggesting that PU.1 is a direct target of miR-155. PU.1 negatively regulates Th2 cytokines (IL-4, IL-5, IL-9, and IL-13), which play a vital role in the pathophysiology of asthma.138 Antagonism with miR-155 suppresses the inflammation in asthmatic patients and can be considered a novel lead to asthma therapy.139
In severe asthmatic patients, TGF-β increases expression of miR-221 and miR-222 with consequent ASM hyper-proliferation and IL-6 secretion. IL-6 is involved in mast cell activation and release of several growth factors.140 Mast cells are responsible for the early-phase reaction of allergic inflammation and involved in the secretion of preformed and lipid-derived mediators.141 Perry et al.142 discovered that increased secretion of IL-6 in ASMCs was associated with reduced expression of cyclin-dependent kinase inhibitor p21WAF1 and tumor suppressor p27kip1. They also found that miR-221 and miR-222 regulate the level of p21WAF1 and p27kip1 expression in mast cells and induce the abnormal inflammatory and proliferative responses with severe asthma.143
miR-181a and miR-19a are also considered inflammatory miRNA in asthma. miR-181a upregulation induces increased TCR sensitivity and lowers the T cell activation threshold.144, 145 Likewise, upregulation of miR-19a stimulates Th2 cytokine production by augmenting the phosphatidylinositol 3-kinase (PI3K), JAK-STAT, and NF-κB signaling pathways, and drives asthma pathogenesis.146, 147 Decreased secretion of anti-inflammatory cytokines such as IL-10 plays an important role in asthma.148 miR-106a significantly decreases the synthesis of anti-inflammatory cytokine IL-10 with a concurrent augmentation of proinflammatory cytokines release.149, 150 miR-106a antagonism promotes IL-10 secretion and helps mitigate asthmatic conditions by increasing Th2 response in mouse models of asthma. Other miRNA are also reported to regulate IL-10 synthesis as well. Likewise, IL-10 induces expression of other miRNA such as miR-146a, miR-146b, miR-155, miR-132, miR-21, and miR-125a in asthmatic patients by regulating TLR and NF-κB-mediated signaling pathways.151, 152
miRNA in Lung Cancer
Lung cancer is the leading cause of cancer morbidity and mortality worldwide.153 Impairment in proper microRNA processing, frequent epigenetic changes in cellular regulatory elements, activation of oncogenes, suppression of tumor suppressor genes, impairment with Drosha, DGCR8, and Dicer activity, and potential effects of cigarette smoke or allergens have all been found as possible mechanisms for microRNA dysfunction in lung cancer.1 Figure 2C summarises the dysregulation of miRNA in lung cancer. Although most reports have demonstrated suppression of miRNA targeting oncogenes with a concomitant increase in their target gene expression, some miRNA such as miR-17-92 cluster and miR-155 are overexpressed in small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC), respectively. Hayashita et al.154 showed that the overexpression of miR-17-92 promotes the hyper-proliferation of lung epithelial cells and leads to lung cancer. Upregulation of miR-17-92 cluster by Myc plays an important role in the oncogenesis of the more aggressive SCLC.155 miR-155 upregulation in NSCLC has been associated with resistance to chemotherapy.156 miR-155 targets the apoptotic protease-activating factor 1 (Apaf-1), which increases the sensitivity of lung cancer cells to cisplatin-induced DNA damage and apoptosis.157 Roosbroeck et al.156 have shown that overexpression of miR-155 also suppresses the expression of tumor protein (TP53) in lung cancers. They reported that the miR-155/TP53-negative regulatory feedback loop is involved in resistance to therapy and decreased survival in lung cancer.
miR-21 is one of the most extensively identified and studied miRNA in different types of cancer, including NSCLC.158 miR-21 functions as an anti-apoptotic and pro-survival factor, and overexpression of miR-21 is a prognostic and diagnostic biomarker for lung cancer.159 Shen et al.160 have reported that miR-21 post-transcriptionally silences the expression of the tumor suppressor PTEN and promotes growth and invasion in NSCLC.
miR-193a (miR-193a-3p and miR-193a-5p) functions as a tumor suppressor in lung cancer.161 Fan et al.162 first reported the detailed association between miR-193a-3p expression and lung cancer, and also identified the unique target genes for miR-193a-3p by using a xenograft mouse model. Their study indicated that overexpression of miR-193a-3p in NSCLC tissues suppressed cell viability, cellular migration, and proliferation by targeting the KRAS oncogene that regulates cellular proliferation, differentiation, and migration in lung cancer.163
SLC22A18, an organic cation transporter, plays an important role in lung cancer.164 Using a large cohort of NSCLC patients, the study demonstrated that overexpression of miR-137 drastically suppresses the proliferation and migration in NSCLC patients. Conversely, decreased expression of miR-137 directly suppresses SLC22A18 expression and promotes aggressive tumor progression.165 Zhang et al.165 demonstrated that overexpression of SLC22A18 was associated with tumor progression and patients’ prognosis. They also showed that miRNA-137 serves as a tumor suppressor by directly targeting SLC22A18 and inhibiting NSCLC cell proliferation, invasion, and migration. Moreover, in NSCLC, miR-137 also targets paxillin, Cdc42, and Cdk6, and inhibits the proliferation and migration of NSCLC cells.166, 167
miR-218 is a putative tumor suppressor in NSCLC.168 Expression of a mature miR-218 is depleted in NSCLC, and overexpression of miR-218 negatively regulates cell proliferation and invasiveness by reducing the expression of JAK3, IL-6R, and phosphorylated STAT3 in lung cancer. miR-218 also targets high mobility group box-1 (HMGB1) that binds to chromatin and facilitates access of transcriptional factors.169, 170 Overexpression of HMGB1 leads to aggressive cell tumorigenesis and tumor metastasis, and this can be diminished by miR-218.171, 172 Likewise, miR-451 and miR-216 are suppressed in NSCLC. Wang et al.173 demonstrated that reduced expression of miR-451 in NSCLC tissues was significantly associated with tumor differentiation, pathological stage, and shorter overall survival in NSCLC patients. miR-451 targets the oncogene Ras-related protein (RAB14), a member of the RAS oncogene family, and its dysfunction has been reported in various types of lung cancer.174
mir-216 functions as a tumor suppressor by regulating eukaryotic initiation factor 4B (eIF4B) and zinc-finger E-box-binding homeobox 1 (ZEB1), and downregulation of miR-216a expression causes tumor growth, proliferation, metastasis, and chemoresistance contributing to NSCLC progression.175 The mir-216 expression can also serve as a biomarker for NSCLC progression. Both eIF4B and ZEB1 act as oncogenes and induce several proto-oncogenic signaling pathways such as Ras-mitogen-activated protein kinase (MAPK) and PI3K/mammalian target of rapamycin (mTOR).176, 177 Zinc-finger E-box miR-145-5p also plays an overall important role in NSCLC. Recently, Hu et al.178 demonstrated that expression of miR-203 and miR-145 is downregulated in NSCLC and suggested that both function as tumor suppressors. It has been shown that TGF-β signaling plays an important role in epithelial-mesenchymal transition (EMT).179 EMT is considered an important step in tumor progression. miR-145 and miR-203 suppress the TGF-β-induced EMT and invasion by repressing SMAD3 in NSCLC cells where SMAD3 has an important role in the EMT and tumor metastasis.178
miR-34a is suppressed in human NSCLC tissues, and restoration of miR-34a expression inhibits cell growth and tumor formation.180 miR-34 loci were frequently hypermethylated and downregulated in human NSCLCs.181 miR-34a regulates epidermal growth factor receptor (EGFR) directly or EGFR signaling and function as a vital tumor suppressor in NSCLC with EGFR as a novel target.182 Because EGFR is involved in oncogenesis such as excessive DNA synthesis, dysregulated cell cycle, uncontrolled cell proliferation, cell invasion, and metastasis, miR-34a-mediated EGFR downregulation inhibits cellular proliferation, promotes cellular apoptosis, and induces cell-cycle progression in NSCLC cell lines.183
miRNA in IPF
IPF is a chronic, progressive, and fibrotic lung disease.184 The disease is characterized by fibroblast proliferation, extracellular matrix remodeling, epithelial scarring, and excessive accumulation of collagen in parenchymal tissue (Figure 2D).185 Several studies have identified cigarette smoking, exposure to commonly prescribed drugs, environmental factors, and genetic predisposition as the potential causes for IPF.186
Pulmonary fibrosis in IPF is characterized by excessive synthesis and secretion of cytokines such as TGF-β, TNF-α, FGFs, interleukin-1 (IL-1), and monocyte chemoattractant protein-1 (MCP-1) from activated inflammatory cells, such as macrophages and eosinophils.187 Other studies reported that several miRNA affect the networks of cytokines and exacerbate the disease.188, 189, 190 TGF-β expression is tightly regulated at different stages such as transcription, post-transcriptional mRNA stability, and processing and posttranslational processing.191 TGF-β promotes fibroblast differentiation into more fibrogenic myofibroblasts and acts as the primary regulator of fibrotic lung diseases.192 TGF-β signaling can lead to transcription activating SMAD3, or the inhibitory SMAD7.193 TGF-β induces SMAD3, which has been shown to suppress the expression of let-7d by binding to the upstream region of let-7.194 TGF-β induces high mobility group A2 (HMGA2) by inhibiting let-7 expression. TGF-β-induced EMT is associated with smad-dependent overexpression of HMGA2 which results in transcription of multiple factors involved in EMT.195 Downregulation of let-7 expression and consequent overexpression of HMGA2 increased expression of mesenchymal markers ACTA2 and VIM and decreased expression of epithelial markers cytokeratin and TJP1. Liu et al.184 demonstrated that TGF-β signaling leads to significant overexpression of miR-21 in the bleomycin-induced lungs of mice, and this functions as an amplifying circuit to increase the fibrogenic activity of TGF-β, thereby promoting lung fibrosis. miR-21 overexpression suppresses the inhibitory SMAD7, as well as leading to enhanced phosphorylation of SMAD2 with consequent fibrogenic effects.
Das et al.196 showed that miR-326 plays a protective role in lung fibrosis by downregulating TGF-β expression and attenuating fibrotic response. Downregulation of miR-326 expression is a crucial mediator of IPF by acting on different components of TGF-β signaling pathways. In bleomycin-induced lung fibrosis, the miR-326 expression is suppressed. miR-326 mimics decreased TGF-β expression and consequently attenuated the bleomycin-induced fibrotic response. miR-326 was also implicated in the downregulation of other profibrotic genes, such as Ets1, Smad3, and MMP-9, and upregulation of antifibrotic genes, such as Smad7, involved in the TGF-β signaling pathway. Nho et al.197 demonstrated that increased expression of miR-96 correlates with decreased expression of the FoxO3a transcription factor in most IPF fibroblasts. FoxO3a is ubiquitously expressed in cells and regulates cell proliferation and survival, and coordinates responses to DNA damages.198 FoxO3a regulates functions as a checkpoint in the cell cycle, triggers the repair of DNA damage, and protects cells from oxidative stress.199 By suppressing FoxO3a, miR-96 suppresses p27, p21, and Bim-1 expression, which leads to increased cell proliferation.200
Yang et al.201 showed that miR-200 family members (200a, 200b, 200c) suppress EMT and reverse the fibrogenetic function of pulmonary fibroblasts. miR-200 family members target GATA3, ZEB1, and ZEB2 genes implicated in EMT and tumor metastasis.202, 203 miR-200 mimics suppressed the overexpression of SMA-α and Fn, the marker of the myofibroblasts in lung fibroblasts of mice with pulmonary fibrosis.204 Yang et al.201 also showed that members of the miR-200 family act as negative regulators of TGF-β-mediated lung fibrosis and attenuate the TGF-β-mediated expression of mesenchymal markers, and could serve as candidate therapeutics to treat lung fibrosis.
miRNA in CF
CF is one of the monogenic, lethal genetic (autosomal recessive) lung disorders common in Caucasian populations. CF is also reported in African and Asian populations with a lower incidence.79, 205 Several studies demonstrated that the underlying reason for CF is a dysfunctional CFTR as a consequence of mutation in the CFTR gene. CFTR localizes to the mucosal side of the airway epithelium and is involved in Cl− efflux and Na+ absorption. The net effect of CFTR action is a mild osmotic gradient that drives paracellular water flow, maintaining the airway surface liquid, which is critical for ciliary beating and mucous clearance.111 CF, as a consequence of CFTR dysfunction, is best characterized by altered chloride ion transport, depletion of airway surface liquid, airway obstruction, and an excessive inflammatory response (Figure 2E).206 Mucociliary dysfunction facilitates both chronic and acute bacterial infection by several opportunistic microorganisms named Pseudomonas aeruginosa and Staphylococcus aureus, and causes excessive inflammation.207, 208
miRNA in CF pathophysiology can be broadly categorized into two types, those directly regulating CFTR and those that modulate the consequent inflammation and remodeling because of CFTR dysfunction. miR-138 belongs to the first category in that it regulates SIN3A expression.209 SIN13A is a transcriptional repressor that is mobilized to the promoter region of CFTR repressing its expression.210 ΔF508 is the most predominant mutation in the CFTR mutation in CF and promotes the misfolding and degradation of CFTR.211, 212 However, a small proportion of ΔF508 does make it to the surface, and once on the surface is active; hence suppression of CFTR expression will further exacerbate CF. miR-138-mediated SIN13A suppression resuscitates CFTR expression and, consequently, activity.
miR-509-3p and miR-494 are upregulated in CF lungs compared with non-CF healthy controls.213 The NF-κB signaling pathway regulates the expression of both miRNA. Both these miRNA are known to directly regulate CFTR, suggesting that under inflammatory stimuli predisposing to NF- κB signaling, miR-509-3p and miR-494 repress CFTR expression and consequently its function cooperatively by binding to its 3′ UTR. Likewise, increased expression of miR-145, miR-223, and miR-494 in CF individuals who are carrying homozygous or heterozygous ΔF508 CFTR mutation leads to decreased CFTR expression.214 Oglesby et al.214 have shown that microbial colonization in CF alters miRNA expression, which can directly modulate CFTR expression or indirectly affect ΔF508 CFTR by promoting inflammation. They showed that of the 255 miRNA with potential seed target sites in CFTR mRNA, miRNA miR-145, -223, and -494, with targeting a highly conserved region of 3′ UTR of CFTR, are upregulated. Interestingly, they showed that Pseudomonas-conditioned media, including lipopeptides, lipopolysaccharide (LPS), and CpG DNA, induce the overexpression of these miRNA. This study supports the previous reports by Gillen et al.215 and Megiorni et al.216 demonstrating the role of these miRNA in CFTR mRNA suppression.
miRNA that promote or suppress inflammation can indirectly alter CFTR expression. As discussed above, inflammatory stimuli can lead to the expression of miR-509-3p and miR-494 repressing CFTR mRNA. Oglesby et al.217 reported that the expression of miR-126 is significantly suppressed in CF lungs with a concomitant increase in its target TOM1. TOM1 has also been considered a negative regulator of TLR2, TLR4, and IL-1β and the TNF-α-induced signaling pathway, and inhibits the activity of transcription factors NF-κB and AP-1.218, 219 Overexpression of TOM1 decreases NF-κB activity even upon LPS stimulation. Likewise, knocking down TOM1 in LPS-stimulated cells increases NF-κB mediated IL-8 expression. Their studies indicate that increased expression of TOM1 via miR-126 downregulation may act in an anti-inflammatory role and counter the effects of other proinflammatory regulators in CF lungs.217 Fabbri et al.220 analyzed the microRNA profile in CF bronchial epithelial cells infected with Pseudomonas aeruginosa. In that study, they showed that P. aeruginosa infection decreases the expression of miR-93 in CF, in parallel with overexpression of pro-inflammatory IL-8. They identified a potential target site in the 3′ UTR region of IL-8 mRNA. Downregulation of miR-31 in CF airway epithelial cells promotes increased production of cathepsin S.221 Cathepsin S is a potent elastase that promotes remodeling of the extracellular matrix via its proteolytic activity and is reported in CF lungs, along with cancer and heart disease.222 Weldon et al. showed that transcription factor IRF-1 is the target for miR-31, and increased levels of IRF-1 due to the downregulation of miR-31 results in overexpression and secretion of cathepsin B by CF airway epithelial cells.
miRNA-Targeted Interventions as Therapy in Respiratory Diseases
Identifying clinically relevant miRNA is important for exploiting their therapeutic potential. Given that miRNA expression profiles are similar for both human and mouse lung, in most cases mouse models can be used to study the effects of aberrant microRNAomes in lung diseases while also identifying therapeutic leads to reverse the downstream effects of the dysregulated miRNA (Figure 3).59 By using nucleic acid-based inhibitors such as small interfering RNAs (siRNAs), miRNA mimics, and miRNA inhibitors, researchers are trying to restore the normal microRNAome and improve clinical outcomes. The mechanism of cellular uptake of antisense oligonucleotide (ASO) depends on the structure of ASO and the cell type.223 Various energy-dependent and non-energy-dependent entry pathways are believed to be involved in oligonucleotide internalization.224 However, effective delivery of the oligonucleotides to their intracellular site of action remains a major challenge, and therapeutic applications can be limited because of problems associated with in vivo delivery of these therapeutic oligonucleotides and possible off-target effects.225 The airway system uniquely consists of pulmonary surfactants, which are zwitterionic lipids that possess cationic properties at the pH of the respiratory tract.226 Moschos et al.227 demonstrated that anionic oligonucleotides are designed in a way to be absorbed by the respiratory surfactant and efficiently taken up by the cells. Moreover, the miRNA mimics, siRNAs, or antagomiRs can stimulate the immune system or saturate the post-transcriptional gene silencing mechanism.228 Several strategies such as SNPs in the miRNA gene, miRNA 3′ tailing, editing, and methylation are being designed to minimize off-target effects, enhance uptake, and increase their stability.229
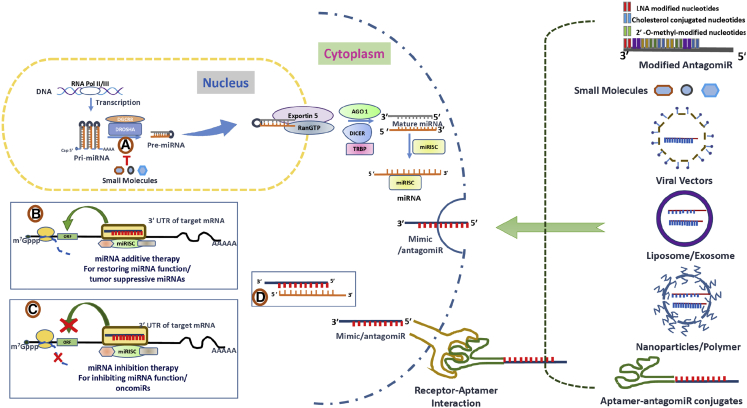
Figure 3.
Therapeutic Approaches to Rescue miRNA Dysfunction
Exosome/liposome, viral vectors (lentivirus [LV], adeno-associated virus [AAV], adeno, and plasmid), nanoparticles/polymers, aptamer-mediated antagomiR, and miRNA mimic delivery into the pulmonary cells. (A) Small molecules bind to Drosha and Dicer processing sites of human miRNAs that are disease associated and inhibit their biogenesis. (B) miRNA mimics function like endogenous miRNAs restoring the activity of a miRNA. (C and D) Binding of single-stranded antagomiRs having complementary sequences to the target endogenous miRNA genome sequence and inhibiting the synthesis of disease-causing miRNAs (C), and antagomiRs having seed sequence sequesters the endogenous free miRNA target inhibiting the activity (D). AGO, Argonaute proteins; DGCR8, DiGeorge syndrome critical region gene 8; m7G cap, 7-methylguanosine; miRISC, miRNA-induced silencing complex; miRNA, microRNA; pre-miRNA, miRNA precursor; pri-miRNA, primary miRNA; RAN-GTP, Ras-related nuclear protein coupled with guanosine-5′-triphosphate; T, inhibitory effect; TRBP, transactivating response RNA-binding protein.
Therapies Using Mimics to Restore MicroRNA Levels
Earlier efforts for delivery of mimics focused on direct intratumoral injections (in case of cancers) or by viral vectors. Unfortunately, using modified viral vectors as therapeutic vehicles has some limitations and is considered controversial due to the risk of integration of viral DNA into transcriptionally active sites in host genome possibly dysregulating the expression of oncogenes or imparting excessive immunogenicity.230 Lately, liposome and nanoparticle-based drugs have been used to facilitate the delivery and uptake of miRNA mimics or inhibitors and siRNAs. Trang et al.231 explored therapeutic delivery of lipid-based let-7 and miR-34 formulations to show tumor-suppressive effects in a KRAS mouse model for lung cancer, and Rai et al.232 showed that a miR-7 expressing plasmid has anti-proliferative effects against EGFR oncogene addicted lung cancer cells using liposomal delivery. Also, Chen et al.233 found that GC4 single-chain variable fragment (scFv)-targeted nanoparticles containing miR-34a actively reduce the tumor size as well as survivin expression, an inhibitor-of-apoptosis protein, by targeting the MAPK pathway in lung metastasis. MRX34 is the first microRNA (miRNA) mimic encapsulated in a liposomal nanoparticle system to facilitate target cellular uptake to be tested in a clinical setting.234 However, researchers are trying to overcome the liposome-based therapies due to charged molecules in liposomes and low pH sensitivity.235 On the other hand, Xiao et al.236 identified one small molecule activator of miR-34a called Rubone, which can upregulate the miR-34a expression in hepatocellular carcinoma. Young et al.237 reported that small-molecule activator induces the expression of miR-122 in liver cancer cells and promotes the apoptosis through caspase activation. Chen et al.238 identified a small-molecule activator derived from the photoreaction of naphthalene-1,4-dione and acetylenes and demonstrated its application is rescuing levels of miR-1 and miR-122 miRNA which are involved in tumorigenesis.239 For the treatment of pulmonary diseases, miRNA-based therapeutics can be formulated as aerosols and delivered through inhalation that might decrease systemic exposure and reduce the possible toxicity and off-target effects.240
Therapies Targeting miRNA
Anti-sense oligonucleotide-based techniques (antagomirs, locked nucleic acid [LNA], and miRNA sponges) have also been designed to inhibit onco-miRs in lung cancer.240 Chemical modifications like 2′-O-methyl group in antagomir gives the required stability against nucleases, and insertion of cholesterol moiety into the passenger strand facilitates cellular uptake. Antagomirs, also known as anti-miRs, are chemically synthesized oligonucleotides complementary to the miRNA and designed to bind to and interfere with their function (Figure 3D).241 We have shown that CFTR and SLC26A9 suppression in primary human bronchial epithelium redifferentiated ex vivo can be rescued by miR-145 antagonism with the consequent restoration of chloride efflux.100 An antagomir targeting miR-9 rescues protein phosphatase 2A (PP2A) activity with the consequent restoration of dexamethasone (DEX)-induced GR nuclear translocation and restores steroid sensitivity in AHR.242 Use of LNA-based anti-miRs in which the ribose sugar ring in each nucleotide is “locked” with a methylene bridge between 2′-O and the 4′-C groups confers high affinity to target the miRNA sequence and improves resistance to nucleases. “Miravirsen,” an LNA-based drug, effectively inhibits miR-122, which plays a crucial role in hepatitis C virus (HCV) replication.243 Of note, multiple miRNA “sponges,” considered as transgenes, have been suggested that encode RNA transcripts consisting of several tandem repeats of the miRNA target sequence, serving as decoys to compete with native mRNA targets for miRISC binding, thereby lowering sequestering of the miRNA to prevent it from binding to its cellular target sites.244
On the other hand, high-throughput screening and reporter based assays have identified several small molecules from a small-molecule drug library that act by either inhibiting the formation of active RNA-induced silencing complex (RISC) or preventing the processing of pri-miRNA to mature miRNA (Figure 3A).245, 246 Gumireddy et al.247 reported that azobenzene inhibits the expression of miR-21, an anti-apoptotic factor that is elevated in various cancers such as breast, ovarian, and lung cancer, as well as glioblastomas.248 Later on, several studies subsequently identified other diverse small-molecule modifiers that can act as activators or inhibitors of miRNA-mediated post-transcriptional gene silencing. Shi et al.249 reported that AC1MMYR2, a potent and selective inhibitor of miR-21, reverses EMT and suppresses tumor growth and progression. Young et al.237 also discovered one small-molecule inhibitor that suppresses the HCV replication in the liver cells by targeting miR-122, and thereby functions as a novel treatment approach in HCV infection.
Aptamers, an emerging class of therapeutics, are high-affinity single-stranded nucleic acid ligands that exhibit specificity and avidity comparable with or exceeding that of antibodies, and can be generated against most targets.250, 251, 252 Unlike antibodies, aptamers can be synthesized chemically and hence offer significant advantages in terms of production cost, more straightforward regulatory approval, and lower immunogenicity when administered in preclinical doses 1,000-fold higher than those used for animal and human therapeutic application.253, 254 Aptamers are highly specific and can discriminate between related proteins that share common sets of structural domains.255, 256 Nucleic acid aptamers are already approved for use in humans (e.g., Macugen).257, 258 Different strategies have been employed to develop cell-specific aptamers for the delivery of oligonucleotide-based therapies.259, 260 Upon receptor-mediated uptake, miRNA cargo is processed by DICER and incorporated in the RISC, and finally binds to the target of interest (Figures 3A and 3B).235 MUC1 aptamer functionalized as nanoparticles and coupled with miR-29b has demonstrated selective delivery of miRNA-29b to lung tumor cells and tissues.261 Likewise, aptamers conjugated to miR-34c and miR-212 have been shown to suppress proliferation of NSCLC or promote susceptibility of NSCLC cells to TNF-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis.262, 263 Also, Esposito et al.264 characterized a selective RNA-based aptamer (GL21.T) that is conjugated with tumor suppressor let-7g miRNccA sequence and binds with high affinity to the oncogenic tyrosine kinase receptor, Axl. They found that specific delivery of this multifunctional conjugate complex to the Axl-expressing cancer cells and suppression of the let-7g targeting gene expression resulted in the inhibition of cancer cell progression and invasion, as well as reduction of tumor growth, in a xenograft model of lung adenocarcinoma.265 Hence aptamer-miRNA conjugates can function as novel tools with the therapeutic potential to inhibit cancer cell survival and migration in vitro and in vivo in lung cancer.
Conclusions
The field of miRNA in lung health and disease is ever evolving. Indeed, the lung is continuously exposed to different stresses such as chemical irritants, free radicals, and air pollutants, so it is likely that miRNA play a permanent role in the host defense and cellular responses against/under these external stresses. Even though significant studies have been made in determining the pathological (or therapeutic) role of miRNA in lung diseases, much remains to be done. Rising evidence supports the hypothesis that deregulation of protein expression because of abnormal unique miRNA expression signature is directly or indirectly linked to the pathogenesis of pulmonary disorders. The major challenge for researchers is in identifying a defined molecular pathway involving a particular miRNA because each mRNA can regulate multiple genes, and multiple miRNA can regulate a single gene. Although the study of the microRNAome itself can identify molecular pathways in lung health and disease, characterization of genes involved in post-transcriptional gene silencing, such as DICER1, Argonaute, TRBP, and so forth, can provide additional information in the pathophysiology of an aberrant microRNAome. Of note, the peripheral lung clock has been implicated in several lung pathologies, and several reports have mentioned the role of miRNA in regulating the molecular clock.266, 267, 268, 269 Several miRNA have been known to modulate genes involved in the lung peripheral molecular clock.270, 271, 272 Although the lung can provide a unique inhalation-based delivery route for these therapeutics, epithelial barrier functions coupled with the mucociliary escalator can result in decreased bioavailability of these therapeutics. Hence efforts to improve therapeutic formulations that can increase residence time and release, for instance, mucoadhesive nanoparticles, can open new avenues for various lung diseases and improve the therapeutic outcomes in patients.
Author Contributions
R.K.D.: manuscript outline, preparation of the draft manuscript, and preparation of figures and the table. S.C.: critical reading and editing of the draft manuscript. H.U.: critical reading and editing of the draft manuscript and writing of the introduction section. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported in part by Flight Attendant Medical Research Institute (FAMRI; grant CIA150086) and 1R01HL147715-01 from the National Institutes of Health to HU.
References
- 1.Brown D., Rahman M., Nana-Sinkam S.P. miRNA in respiratory disease. A clinician’s overview. Ann. Am. Thorac. Soc. 2014;11:1277–1285. doi: 10.1513/AnnalsATS.201404-179FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pritchard C.C., Cheng H.H., Tewari M. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The miRNA of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Ameis D., Khoshgoo N., Iwasiow B.M., Snarr P., Keijzer R. miRNA in Lung Development and Disease. Paediatr. Respir. Rev. 2017;22:38–43. doi: 10.1016/j.prrv.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Sessa R., Hata A. Role of miRNA in lung development and pulmonary diseases. Pulm. Circ. 2013;3:315–328. doi: 10.4103/2045-8932.114758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catalanotto C., Cogoni C., Zardo G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016;17:E1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iqbal M.A., Arora S., Prakasam G., Calin G.A., Syed M.A. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol. Aspects Med. 2018 doi: 10.1016/j.mam.2018.07.003. Published online August 18, 2018. [DOI] [PubMed] [Google Scholar]
- 9.Li H., Zhao X., Shan H., Liang H. miRNA in idiopathic pulmonary fibrosis: involvement in pathogenesis and potential use in diagnosis and therapeutics. Acta Pharm. Sin. B. 2016;6:531–539. doi: 10.1016/j.apsb.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salimian J., Mirzaei H., Moridikia A., Harchegani A.B., Sahebkar A., Salehi H. Chronic obstructive pulmonary disease: miRNA and exosomes as new diagnostic and therapeutic biomarkers. J. Res. Med. Sci. 2018;23:27. doi: 10.4103/jrms.JRMS_1054_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pua H.H., Ansel K.M. MicroRNA regulation of allergic inflammation and asthma. Curr. Opin. Immunol. 2015;36:101–108. doi: 10.1016/j.coi.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boucherat O., Potus F., Bonnet S. microRNA and Pulmonary Hypertension. Adv. Exp. Med. Biol. 2015;888:237–252. doi: 10.1007/978-3-319-22671-2_12. [DOI] [PubMed] [Google Scholar]
- 13.Szymczak I., Wieczfinska J., Pawliczak R. Molecular Background of miRNA Role in Asthma and COPD: An Updated Insight. Biomed Res. Int. 2016;2016:7802521. doi: 10.1155/2016/7802521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alipoor S.D., Adcock I.M., Garssen J., Mortaz E., Varahram M., Mirsaeidi M., Velayati A. The roles of miRNA as potential biomarkers in lung diseases. Eur. J. Pharmacol. 2016;791:395–404. doi: 10.1016/j.ejphar.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Britten R.J., Davidson E.H. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 16.Steiman-Shimony A., Shtrikman O., Margalit H. Assessing the functional association of intronic miRNA with their host genes. RNA. 2018;24:991–1004. doi: 10.1261/rna.064386.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smalheiser N.R., Torvik V.I. Mammalian miRNA derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Joshi P., Middleton J., Jeon Y.-J., Garofalo M. miRNA in lung cancer. World J. Methodol. 2014;4:59–72. doi: 10.5662/wjm.v4.i2.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebrahimi A., Sadroddiny E. miRNA in lung diseases: Recent findings and their pathophysiological implications. Pulm. Pharmacol. Ther. 2015;34:55–63. doi: 10.1016/j.pupt.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borchert G.M., Lanier W., Davidson B.L. RNA polymerase III transcribes human miRNA. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 22.Macfarlane L.A., Murphy P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Y., Zhang X., Graves P., Zeng Y. A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA. 2012;18:2083–2092. doi: 10.1261/rna.033688.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters L., Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Maltby S., Plank M., Tay H.L., Collison A., Foster P.S. Targeting MicroRNA Function in Respiratory Diseases: Mini-Review. Front. Physiol. 2016;7:21. doi: 10.3389/fphys.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J. Control of protein synthesis and mRNA degradation by miRNA. Curr. Opin. Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Lytle J.R., Yario T.A., Steitz J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valinezhad Orang A., Safaralizadeh R., Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brodersen P., Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 30.Carroll A.P., Goodall G.J., Liu B. Understanding principles of miRNA target recognition and function through integrated biological and bioinformatics approaches. Wiley Interdiscip. Rev. RNA. 2014;5:361–379. doi: 10.1002/wrna.1217. [DOI] [PubMed] [Google Scholar]
- 31.Wang X. Composition of seed sequence is a major determinant of microRNA targeting patterns. Bioinformatics. 2014;30:1377–1383. doi: 10.1093/bioinformatics/btu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNA results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 34.Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNA and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 35.Wakiyama M., Takimoto K., Ohara O., Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiriakidou M., Tan G.S., Lamprinaki S., De Planell-Saguer M., Nelson P.T., Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 37.Chendrimada T.P., Finn K.J., Ji X., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 39.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 40.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 41.Saito Y., Liang G., Egger G., Friedman J.M., Chuang J.C., Coetzee G.A., Jones P.A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 42.Chen J., Wang M., Guo M., Xie Y., Cong Y.S. miR-127 regulates cell proliferation and senescence by targeting BCL6. PLoS ONE. 2013;8:e80266. doi: 10.1371/journal.pone.0080266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhai L., Wu R., Han W., Zhang Y., Zhu D. miR-127 enhances myogenic cell differentiation by targeting S1PR3. Cell Death Dis. 2017;8:e2707. doi: 10.1038/cddis.2017.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott G.K., Mattie M.D., Berger C.E., Benz S.C., Benz C.C. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 45.Chuang J.C., Jones P.A. Epigenetics and miRNA. Pediatr. Res. 2007;61:24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 46.Lin S.L., Miller J.D., Ying S.Y. Intronic microRNA (miRNA) J. Biomed. Biotechnol. 2006;2006:26818. doi: 10.1155/JBB/2006/26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang H., Hata A. Control of Drosha-Mediated MicroRNA Maturation by Smad Proteins. Enzymes. 2012;32:123–136. [Google Scholar]
- 48.Guil S., Cáceres J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 49.Tsang W.P., Kwok T.T. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30:953–959. doi: 10.1093/carcin/bgp094. [DOI] [PubMed] [Google Scholar]
- 50.Trabucchi M., Briata P., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M.G. KSRP promotes the maturation of a group of miRNA precursors. In: Großhans H., editor. Regulation of miRNA. Springer; 2010. pp. 36–42. [Google Scholar]
- 51.Ruggiero T., Trabucchi M., Ponassi M., Corte G., Chen C.Y., al-Haj L., Khabar K.S., Briata P., Gherzi R. Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling. BMC Mol. Biol. 2007;8:28. doi: 10.1186/1471-2199-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou X., Benson K.F., Ashar H.R., Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 53.De Martino I., Visone R., Fedele M., Petrocca F., Palmieri D., Martinez Hoyos J., Forzati F., Croce C.M., Fusco A. Regulation of microRNA expression by HMGA1 proteins. Oncogene. 2009;28:1432–1442. doi: 10.1038/onc.2008.495. [DOI] [PubMed] [Google Scholar]
- 54.Weng T., Chen Z., Jin N., Gao L., Liu L. Gene expression profiling identifies regulatory pathways involved in the late stage of rat fetal lung development. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006;291:L1027–L1037. doi: 10.1152/ajplung.00435.2005. [DOI] [PubMed] [Google Scholar]
- 55.Schittny J.C. Development of the lung. Cell Tissue Res. 2017;367:427–444. doi: 10.1007/s00441-016-2545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang M., Shi J., Huang Y., Lai L. Expression of canonical WNT/β-CATENIN signaling components in the developing human lung. BMC Dev. Biol. 2012;12:21. doi: 10.1186/1471-213X-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartram U., Speer C.P. The role of transforming growth factor beta in lung development and disease. Chest. 2004;125:754–765. doi: 10.1378/chest.125.2.754. [DOI] [PubMed] [Google Scholar]
- 58.Lebeche D., Malpel S., Cardoso W.V. Fibroblast growth factor interactions in the developing lung. Mech. Dev. 1999;86:125–136. doi: 10.1016/s0925-4773(99)00124-0. [DOI] [PubMed] [Google Scholar]
- 59.Williams A.E., Moschos S.A., Perry M.M., Barnes P.J., Lindsay M.A. Maternally imprinted miRNA are differentially expressed during mouse and human lung development. Dev. Dyn. 2007;236:572–580. doi: 10.1002/dvdy.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu Y., Thomson J.M., Wong H.Y., Hammond S.M., Hogan B.L. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev. Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu Y., Okubo T., Rawlins E., Hogan B.L. Epithelial progenitor cells of the embryonic lung and the role of miRNA in their proliferation. Proc. Am. Thorac. Soc. 2008;5:300–304. doi: 10.1513/pats.200710-162DR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hayashi T., Koyama N., Azuma Y., Kashimata M. Mesenchymal miR-21 regulates branching morphogenesis in murine submandibular gland in vitro. Dev. Biol. 2011;352:299–307. doi: 10.1016/j.ydbio.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 63.Carraro G., Shrestha A., Rostkovius J., Contreras A., Chao C.M., El Agha E., Mackenzie B., Dilai S., Guidolin D., Taketo M.M. miR-142-3p balances proliferation and differentiation of mesenchymal cells during lung development. Development. 2014;141:1272–1281. doi: 10.1242/dev.105908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang Z., Cushing L., Ai X., Lü J. miR-326 is downstream of Sonic hedgehog signaling and regulates the expression of Gli2 and smoothened. Am. J. Respir. Cell Mol. Biol. 2014;51:273–283. doi: 10.1165/rcmb.2013-0127OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cushing L., Jiang Z., Kuang P., Lü J. The roles of miRNA and protein components of the microRNA pathway in lung development and diseases. Am. J. Respir. Cell Mol. Biol. 2015;52:397–408. doi: 10.1165/rcmb.2014-0232RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harris K.S., Zhang Z., McManus M.T., Harfe B.D., Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. USA. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kataoka Y., Takeichi M., Uemura T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells. 2001;6:313–325. doi: 10.1046/j.1365-2443.2001.00427.x. [DOI] [PubMed] [Google Scholar]
- 68.Lü J., Qian J., Chen F., Tang X., Li C., Cardoso W.V. Differential expression of components of the microRNA machinery during mouse organogenesis. Biochem. Biophys. Res. Commun. 2005;334:319–323. doi: 10.1016/j.bbrc.2005.05.206. [DOI] [PubMed] [Google Scholar]
- 69.Angulo M., Lecuona E., Sznajder J.I. Role of miRNA in lung disease. Arch. Bronconeumol. 2012;48:325–330. doi: 10.1016/j.arbres.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nana-Sinkam S.P., Hunter M.G., Nuovo G.J., Schmittgen T.D., Gelinas R., Galas D., Marsh C.B. Integrating the MicroRNome into the study of lung disease. Am. J. Respir. Crit. Care Med. 2009;179:4–10. doi: 10.1164/rccm.200807-1042PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garantziotis S., Schwartz D.A. Ecogenomics of respiratory diseases of public health significance. Annu. Rev. Public Health. 2010;31:37–51. doi: 10.1146/annurev.publhealth.012809.103633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adeloye D., Chua S., Lee C., Basquill C., Papana A., Theodoratou E., Nair H., Gasevic D., Sridhar D., Campbell H., Global Health Epidemiology Reference Group (GHERG) Global and regional estimates of COPD prevalence: Systematic review and meta-analysis. J. Glob. Health. 2015;5:020415. doi: 10.7189/jogh.05-020415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomsen S.F. Exploring the origins of asthma: Lessons from twin studies. Eur. Clin. Respir. J. 2014;1(Suppl 1) doi: 10.3402/ecrj.v1.25535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martinez F.J., Collard H.R., Pardo A., Raghu G., Richeldi L., Selman M., Swigris J.J., Taniguchi H., Wells A.U. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Primers. 2017;3:17074. doi: 10.1038/nrdp.2017.74. [DOI] [PubMed] [Google Scholar]
- 75.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Piñeros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 76.Pahal P, Avula A, Sharma S. Emphysema. [Updated 2019 Aug 14]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2019 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK482217/.
- 77.Mahboub B.H., Vats M.G., Al Zaabi A., Iqbal M.N., Safwat T., Al-Hurish F., Miravitlles M., Singh D., Asad K., Zeineldine S., Al-Hajjaj M.S. Joint statement for the diagnosis, management, and prevention of chronic obstructive pulmonary disease for Gulf Cooperation Council countries and Middle East-North Africa region, 2017. Int. J. Chron. Obstruct. Pulmon. Dis. 2017;12:2869–2890. doi: 10.2147/COPD.S136245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ford E.S. Trends in mortality from COPD among adults in the United States. Chest. 2015;148:962–970. doi: 10.1378/chest.14-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oglesby I.K., McElvaney N.G., Greene C.M. miRNA in inflammatory lung disease—master regulators or target practice? Respir. Res. 2010;11:148. doi: 10.1186/1465-9921-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sato T., Liu X., Nelson A., Nakanishi M., Kanaji N., Wang X., Kim M., Li Y., Sun J., Michalski J. Reduced miR-146a increases prostaglandin E2in chronic obstructive pulmonary disease fibroblasts. Am. J. Respir. Crit. Care Med. 2010;182:1020–1029. doi: 10.1164/rccm.201001-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Perry M.M., Moschos S.A., Williams A.E., Shepherd N.J., Larner-Svensson H.M., Lindsay M.A. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J. Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strillacci A., Griffoni C., Sansone P., Paterini P., Piazzi G., Lazzarini G., Spisni E., Pantaleo M.A., Biasco G., Tomasi V. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp. Cell Res. 2009;315:1439–1447. doi: 10.1016/j.yexcr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 83.Chakrabarty A., Tranguch S., Daikoku T., Jensen K., Furneaux H., Dey S.K. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc. Natl. Acad. Sci. USA. 2007;104:15144–15149. doi: 10.1073/pnas.0705917104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dhami R., Gilks B., Xie C., Zay K., Wright J.L., Churg A. Acute cigarette smoke-induced connective tissue breakdown is mediated by neutrophils and prevented by alpha1-antitrypsin. Am. J. Respir. Cell Mol. Biol. 2000;22:244–252. doi: 10.1165/ajrcmb.22.2.3809. [DOI] [PubMed] [Google Scholar]
- 86.Chinnapaiyan S., Unwalla H.J. Mucociliary dysfunction in HIV and smoked substance abuse. Front. Microbiol. 2015;6:1052. doi: 10.3389/fmicb.2015.01052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Churg A., Wang R.D., Tai H., Wang X., Xie C., Dai J., Shapiro S.D., Wright J.L. Macrophage metalloelastase mediates acute cigarette smoke-induced inflammation via tumor necrosis factor-alpha release. Am. J. Respir. Crit. Care Med. 2003;167:1083–1089. doi: 10.1164/rccm.200212-1396OC. [DOI] [PubMed] [Google Scholar]
- 88.Hautamaki R.D., Kobayashi D.K., Senior R.M., Shapiro S.D. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 89.Qu P., Du H., Wang X., Yan C. Matrix metalloproteinase 12 overexpression in lung epithelial cells plays a key role in emphysema to lung bronchioalveolar adenocarcinoma transition. Cancer Res. 2009;69:7252–7261. doi: 10.1158/0008-5472.CAN-09-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Graff J.W., Powers L.S., Dickson A.M., Kim J., Reisetter A.C., Hassan I.H., Kremens K., Gross T.J., Wilson M.E., Monick M.M. Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PLoS ONE. 2012;7:e44066. doi: 10.1371/journal.pone.0044066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shen W., Liu J., Zhao G., Fan M., Song G., Zhang Y., Weng Z., Zhang Y. Repression of Toll-like receptor-4 by microRNA-149-3p is associated with smoking-related COPD. Int. J. Chron. Obstruct. Pulmon. Dis. 2017;12:705–715. doi: 10.2147/COPD.S128031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu G., Zhang Z., Xing Y., Wei J., Ge Z., Liu X., Zhang Y., Huang X. MicroRNA-149 negatively regulates TLR-triggered inflammatory response in macrophages by targeting MyD88. J. Cell. Biochem. 2014;115:919–927. doi: 10.1002/jcb.24734. [DOI] [PubMed] [Google Scholar]
- 93.Doz E., Noulin N., Boichot E., Guénon I., Fick L., Le Bert M., Lagente V., Ryffel B., Schnyder B., Quesniaux V.F., Couillin I. Cigarette smoke-induced pulmonary inflammation is TLR4/MyD88 and IL-1R1/MyD88 signaling dependent. J. Immunol. 2008;180:1169–1178. doi: 10.4049/jimmunol.180.2.1169. [DOI] [PubMed] [Google Scholar]
- 94.Pons J., Sauleda J., Regueiro V., Santos C., López M., Ferrer J., Agustí A.G., Bengoechea J.A. Expression of Toll-like receptor 2 is up-regulated in monocytes from patients with chronic obstructive pulmonary disease. Respir. Res. 2006;7:64. doi: 10.1186/1465-9921-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.MacRedmond R.E., Greene C.M., Dorscheid D.R., McElvaney N.G., O’Neill S.J. Epithelial expression of TLR4 is modulated in COPD and by steroids, salmeterol and cigarette smoke. Respir. Res. 2007;8:84. doi: 10.1186/1465-9921-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nadigel J., Préfontaine D., Baglole C.J., Maltais F., Bourbeau J., Eidelman D.H., Hamid Q. Cigarette smoke increases TLR4 and TLR9 expression and induces cytokine production from CD8(+) T cells in chronic obstructive pulmonary disease. Respir. Res. 2011;12:149. doi: 10.1186/1465-9921-12-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang M., Huang Y., Liang Z., Liu D., Lu Y., Dai Y., Feng G., Wang C. Plasma miRNA might be promising biomarkers of chronic obstructive pulmonary disease. Clin. Respir. J. 2016;10:104–111. doi: 10.1111/crj.12194. [DOI] [PubMed] [Google Scholar]
- 98.Celedón J.C., Lange C., Raby B.A., Litonjua A.A., Palmer L.J., DeMeo D.L., Reilly J.J., Kwiatkowski D.J., Chapman H.A., Laird N. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD) Hum. Mol. Genet. 2004;13:1649–1656. doi: 10.1093/hmg/ddh171. [DOI] [PubMed] [Google Scholar]
- 99.Königshoff M., Kneidinger N., Eickelberg O. TGF-beta signaling in COPD: deciphering genetic and cellular susceptibilities for future therapeutic regimen. Swiss Med. Wkly. 2009;139:554–563. doi: 10.4414/smw.2009.12528. [DOI] [PubMed] [Google Scholar]
- 100.Dutta R.K., Chinnapaiyan S., Rasmussen L., Raju S.V., Unwalla H.J. A Neutralizing Aptamer to TGFBR2 and miR-145 Antagonism Rescue Cigarette Smoke- and TGF-β-Mediated CFTR Expression. Mol. Ther. 2019;27:442–455. doi: 10.1016/j.ymthe.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Unwalla H.J., Ivonnet P., Dennis J.S., Conner G.E., Salathe M. Transforming growth factor-β1 and cigarette smoke inhibit the ability of β2-agonists to enhance epithelial permeability. Am. J. Respir. Cell Mol. Biol. 2015;52:65–74. doi: 10.1165/rcmb.2013-0538OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chinnapaiyan S., Dutta R.K., Nair M., Chand H.S., Rahman I., Unwalla H.J. TGF-β1 increases viral burden and promotes HIV-1 latency in primary differentiated human bronchial epithelial cells. Sci. Rep. 2019;9:12552. doi: 10.1038/s41598-019-49056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Collison A., Mattes J., Plank M., Foster P.S. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J. Allergy Clin. Immunol. 2011;128:160–167.e4. doi: 10.1016/j.jaci.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Y., Zhang M., Zhong M., Suo Q., Lv K. Expression profiles of miRNA in polarized macrophages. Int. J. Mol. Med. 2013;31:797–802. doi: 10.3892/ijmm.2013.1260. [DOI] [PubMed] [Google Scholar]
- 105.O’Leary L., Sevinç K., Papazoglou I.M., Tildy B., Detillieux K., Halayko A.J., Chung K.F., Perry M.M. Airway smooth muscle inflammation is regulated by microRNA-145 in COPD. FEBS Lett. 2016;590:1324–1334. doi: 10.1002/1873-3468.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sloane P.A., Shastry S., Wilhelm A., Courville C., Tang L.P., Backer K., Levin E., Raju S.V., Li Y., Mazur M. A pharmacologic approach to acquired cystic fibrosis transmembrane conductance regulator dysfunction in smoking related lung disease. PLoS ONE. 2012;7:e39809. doi: 10.1371/journal.pone.0039809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dransfield M.T., Wilhelm A.M., Flanagan B., Courville C., Tidwell S.L., Raju S.V., Gaggar A., Steele C., Tang L.P., Liu B., Rowe S.M. Acquired cystic fibrosis transmembrane conductance regulator dysfunction in the lower airways in COPD. Chest. 2013;144:498–506. doi: 10.1378/chest.13-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kulshreshtha A., Ahmad T., Agrawal A., Ghosh B. Proinflammatory role of epithelial cell-derived exosomes in allergic airway inflammation. J. Allergy Clin. Immunol. 2013;131 doi: 10.1016/j.jaci.2012.12.1565. 1194–1203, 1203.e1–e14. [DOI] [PubMed] [Google Scholar]
- 109.Dawson D.C., Smith S.S., Mansoura M.K. CFTR: mechanism of anion conduction. Physiol. Rev. 1999;79(Suppl 1):S47–S75. doi: 10.1152/physrev.1999.79.1.S47. [DOI] [PubMed] [Google Scholar]
- 110.Conner G.E., Wijkstrom-Frei C., Randell S.H., Fernandez V.E., Salathe M. The lactoperoxidase system links anion transport to host defense in cystic fibrosis. FEBS Lett. 2007;581:271–278. doi: 10.1016/j.febslet.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boucher R.C. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 2004;23:146–158. doi: 10.1183/09031936.03.00057003. [DOI] [PubMed] [Google Scholar]
- 112.Hogg J.C., Chu F., Utokaparch S., Woods R., Elliott W.M., Buzatu L., Cherniack R.M., Rogers R.M., Sciurba F.C., Coxson H.O., Paré P.D. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N. Engl. J. Med. 2004;350:2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 113.Noone P.G., Leigh M.W., Sannuti A., Minnix S.L., Carson J.L., Hazucha M., Zariwala M.A., Knowles M.R. Primary ciliary dyskinesia: diagnostic and phenotypic features. Am. J. Respir. Crit. Care Med. 2004;169:459–467. doi: 10.1164/rccm.200303-365OC. [DOI] [PubMed] [Google Scholar]
- 114.Chinnapaiyan S., Parira T., Dutta R., Agudelo M., Morris A., Nair M., Unwalla H.J. HIV Infects Bronchial Epithelium and Suppresses Components of the Mucociliary Clearance Apparatus. PLoS ONE. 2017;12:e0169161. doi: 10.1371/journal.pone.0169161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chinnapaiyan S., Dutta R., Bala J., Parira T., Agudelo M., Nair M., Unwalla H.J. Cigarette smoke promotes HIV infection of primary bronchial epithelium and additively suppresses CFTR function. Sci. Rep. 2018;8:7984. doi: 10.1038/s41598-018-26095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hassan F., Nuovo G.J., Crawford M., Boyaka P.N., Kirkby S., Nana-Sinkam S.P., Cormet-Boyaka E. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS ONE. 2012;7:e50837. doi: 10.1371/journal.pone.0050837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen Y., Thomas P.S., Kumar R.K., Herbert C. The role of noncoding RNAs in regulating epithelial responses in COPD. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018;315:L184–L192. doi: 10.1152/ajplung.00063.2018. [DOI] [PubMed] [Google Scholar]
- 118.Ezzie M.E., Crawford M., Cho J.H., Orellana R., Zhang S., Gelinas R., Batte K., Yu L., Nuovo G., Galas D. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67:122–131. doi: 10.1136/thoraxjnl-2011-200089. [DOI] [PubMed] [Google Scholar]
- 119.Hassan T., Carroll T.P., Buckley P.G., Cummins R., O’Neill S.J., McElvaney N.G., Greene C.M. miR-199a-5p silencing regulates the unfolded protein response in chronic obstructive pulmonary disease and α1-antitrypsin deficiency. Am. J. Respir. Crit. Care Med. 2014;189:263–273. doi: 10.1164/rccm.201306-1151OC. [DOI] [PubMed] [Google Scholar]
- 120.Williams A.E., Larner-Svensson H., Perry M.M., Campbell G.A., Herrick S.E., Adcock I.M., Erjefalt J.S., Chung K.F., Lindsay M.A. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLoS ONE. 2009;4:e5889. doi: 10.1371/journal.pone.0005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mattes J., Collison A., Plank M., Phipps S., Foster P.S. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. USA. 2009;106:18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lu T.X., Munitz A., Rothenberg M.E. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vianello A., Caminati M., Crivellaro M., El Mazloum R., Snenghi R., Schiappoli M., Dama A., Rossi A., Festi G., Marchi M.R. Fatal asthma; is it still an epidemic? World Allergy Organ. J. 2016;9:42. doi: 10.1186/s40413-016-0129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McBrien C.N., Menzies-Gow A. The Biology of Eosinophils and Their Role in Asthma. Front. Med. (Lausanne) 2017;4:93. doi: 10.3389/fmed.2017.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bradding P. Asthma: eosinophil disease, mast cell disease, or both? Allergy Asthma Clin. Immunol. 2008;4:84–90. doi: 10.1186/1710-1492-4-2-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chang H.C., Han L., Jabeen R., Carotta S., Nutt S.L., Kaplan M.H. PU.1 regulates TCR expression by modulating GATA-3 activity. J. Immunol. 2009;183:4887–4894. doi: 10.4049/jimmunol.0900363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Collison A., Herbert C., Siegle J.S., Mattes J., Foster P.S., Kumar R.K. Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulm. Med. 2011;11:29. doi: 10.1186/1471-2466-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Corren J. Role of interleukin-13 in asthma. Curr. Allergy Asthma Rep. 2013;13:415–420. doi: 10.1007/s11882-013-0373-9. [DOI] [PubMed] [Google Scholar]
- 129.Hammad Mahmoud Hammad R., Hamed D.H.E.D., Eldosoky M.A.E.R., Ahmad A.A.E.S., Osman H.M., Abd Elgalil H.M., Mahmoud Hassan M.M. Plasma microRNA-21, microRNA-146a and IL-13 expression in asthmatic children. Innate Immun. 2018;24:171–179. doi: 10.1177/1753425918763521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chiba Y., Matsusue K., Misawa M. RhoA, a possible target for treatment of airway hyperresponsiveness in bronchial asthma. J. Pharmacol. Sci. 2010;114:239–247. doi: 10.1254/jphs.10r03cr. [DOI] [PubMed] [Google Scholar]
- 131.Chiba Y., Tanabe M., Goto K., Sakai H., Misawa M. Down-regulation of miR-133a contributes to up-regulation of Rhoa in bronchial smooth muscle cells. Am. J. Respir. Crit. Care Med. 2009;180:713–719. doi: 10.1164/rccm.200903-0325OC. [DOI] [PubMed] [Google Scholar]
- 132.Malmhäll C., Johansson K., Winkler C., Alawieh S., Ekerljung L., Rådinger M. Altered miR-155 Expression in Allergic Asthmatic Airways. Scand. J. Immunol. 2017;85:300–307. doi: 10.1111/sji.12535. [DOI] [PubMed] [Google Scholar]
- 133.Plank M.W., Maltby S., Tay H.L., Stewart J., Eyers F., Hansbro P.M., Foster P.S. MicroRNA Expression Is Altered in an Ovalbumin-Induced Asthma Model and Targeting miR-155 with Antagomirs Reveals Cellular Specificity. PLoS ONE. 2015;10:e0144810. doi: 10.1371/journal.pone.0144810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Malmhall C., Alawieh S., Lu Y., Sjöstrand M., Bossios A., Eldh M., Rådinger M. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J. Allergy Clin. Immunol. 2014;133 doi: 10.1016/j.jaci.2013.11.008. 1429–1438, 1438.e1421–e1427. [DOI] [PubMed] [Google Scholar]
- 135.Zhang Y., Xue Y., Liu Y., Song G., Lv G., Wang Y., Wang Y., Li X., Yang L. MicroRNA-146a expression inhibits the proliferation and promotes the apoptosis of bronchial smooth muscle cells in asthma by directly targeting the epidermal growth factor receptor. Exp. Ther. Med. 2016;12:854–858. doi: 10.3892/etm.2016.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Steinke J.W., Borish L. Th2 cytokines and asthma. Interleukin-4: its role in the pathogenesis of asthma, and targeting it for asthma treatment with interleukin-4 receptor antagonists. Respir. Res. 2001;2:66–70. doi: 10.1186/rr40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pope S.M., Zimmermann N., Stringer K.F., Karow M.L., Rothenberg M.E. The eotaxin chemokines and CCR3 are fundamental regulators of allergen-induced pulmonary eosinophilia. J. Immunol. 2005;175:5341–5350. doi: 10.4049/jimmunol.175.8.5341. [DOI] [PubMed] [Google Scholar]
- 138.Vigorito E., Perks K.L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P.P., Miska E.A., Rodriguez A., Bradley A. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou H., Li J., Gao P., Wang Q., Zhang J. miR-155: A Novel Target in Allergic Asthma. Int. J. Mol. Sci. 2016;17:1773. doi: 10.3390/ijms17101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Perry M., Baker J., Chung K.F. miR-221 and miR-222 target p21 and p27 in airway smooth muscle to elicit hyper proliferation in severe asthmatics. Eur. Respir. J. 2011;38(Suppl 55):749. [Google Scholar]
- 141.Galli S.J., Tsai M., Piliponsky A.M. The development of allergic inflammation. Nature. 2008;454:445–454. doi: 10.1038/nature07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Perry M.M., Baker J.E., Gibeon D.S., Adcock I.M., Chung K.F. Airway smooth muscle hyperproliferation is regulated by microRNA-221 in severe asthma. Am. J. Respir. Cell Mol. Biol. 2014;50:7–17. doi: 10.1165/rcmb.2013-0067OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Qin H.B., Xu B., Mei J.J., Li D., Liu J.J., Zhao D.Y., Liu F. Inhibition of miRNA-221 suppresses the airway inflammation in asthma. Inflammation. 2012;35:1595–1599. doi: 10.1007/s10753-012-9474-1. [DOI] [PubMed] [Google Scholar]
- 144.Li Q.J., Chau J., Ebert P.J., Sylvester G., Min H., Liu G., Braich R., Manoharan M., Soutschek J., Skare P. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 145.Wang J.-W., Li K., Hellermann G., Lockey R.F., Mohapatra S., Mohapatra S. Regulating the Regulators: microRNA and Asthma. World Allergy Organ. J. 2011;4:94–103. doi: 10.1186/1939-4551-4-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Simpson L.J., Patel S., Bhakta N.R., Choy D.F., Brightbill H.D., Ren X., Wang Y., Pua H.H., Baumjohann D., Montoya M.M. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat. Immunol. 2014;15:1162–1170. doi: 10.1038/ni.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hou C., Chen Y., Huang X., Huang Q., Li M., Tan X. miR-19 targets PTEN and mediates high mobility group protein B1(HMGB1)-induced proliferation and migration of human airway smooth muscle cells. PLoS ONE. 2019;14:e0219081. doi: 10.1371/journal.pone.0219081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Borish L., Aarons A., Rumbyrt J., Cvietusa P., Negri J., Wenzel S. Interleukin-10 regulation in normal subjects and patients with asthma. J. Allergy Clin. Immunol. 1996;97:1288–1296. doi: 10.1016/s0091-6749(96)70197-5. [DOI] [PubMed] [Google Scholar]
- 149.Sharma A., Kumar M., Aich J., Hariharan M., Brahmachari S.K., Agrawal A., Ghosh B. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc. Natl. Acad. Sci. USA. 2009;106:5761–5766. doi: 10.1073/pnas.0808743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sharma A., Kumar M., Ahmad T., Mabalirajan U., Aich J., Agrawal A., Ghosh B. Antagonism of mmu-mir-106a attenuates asthma features in allergic murine model. J. Appl. Physiol. (1985) 2012;113:459–464. doi: 10.1152/japplphysiol.00001.2012. [DOI] [PubMed] [Google Scholar]
- 151.He X., Jing Z., Cheng G. miRNA: new regulators of Toll-like receptor signalling pathways. BioMed Res. Int. 2014;2014:945169. doi: 10.1155/2014/945169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhou R., Hu G., Liu J., Gong A.Y., Drescher K.M., Chen X.M. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 2009;5:e1000681. doi: 10.1371/journal.ppat.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Travis W.D., Brambilla E., Nicholson A.G., Yatabe Y., Austin J.H.M., Beasley M.B., Chirieac L.R., Dacic S., Duhig E., Flieder D.B., WHO Panel The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 154.Hayashita Y., Osada H., Tatematsu Y., Yamada H., Yanagisawa K., Tomida S., Yatabe Y., Kawahara K., Sekido Y., Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 155.Takahashi T., Obata Y., Sekido Y., Hida T., Ueda R., Watanabe H., Ariyoshi Y., Sugiura T., Takahashi T. Expression and amplification of myc gene family in small cell lung cancer and its relation to biological characteristics. Cancer Res. 1989;49:2683–2688. [PubMed] [Google Scholar]
- 156.Van Roosbroeck K., Fanini F., Setoyama T., Ivan C., Rodriguez-Aguayo C., Fuentes-Mattei E., Xiao L., Vannini I., Redis R.S., D’Abundo L. Combining Anti-Mir-155 with Chemotherapy for the Treatment of Lung Cancers. Clin. Cancer Res. 2017;23:2891–2904. doi: 10.1158/1078-0432.CCR-16-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zang Y.S., Zhong Y.F., Fang Z., Li B., An J. MiR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf-1 expression. Cancer Gene Ther. 2012;19:773–778. doi: 10.1038/cgt.2012.60. [DOI] [PubMed] [Google Scholar]
- 158.Bica-Pop C., Cojocneanu-Petric R., Magdo L., Raduly L., Gulei D., Berindan-Neagoe I. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell. Mol. Life Sci. 2018;75:3539–3551. doi: 10.1007/s00018-018-2877-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Markou A., Zavridou M., Lianidou E.S. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer (Auckl.) 2016;7:19–27. doi: 10.2147/LCTT.S60341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shen H., Zhu F., Liu J., Xu T., Pei D., Wang R., Qian Y., Li Q., Wang L., Shi Z. Alteration in Mir-21/PTEN expression modulates gefitinib resistance in non-small cell lung cancer. PLoS ONE. 2014;9:e103305. doi: 10.1371/journal.pone.0103305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liang H., Liu M., Yan X., Zhou Y., Wang W., Wang X., Fu Z., Wang N., Zhang S., Wang Y. miR-193a-3p functions as a tumor suppressor in lung cancer by down-regulating ERBB4. J. Biol. Chem. 2015;290:926–940. doi: 10.1074/jbc.M114.621409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fan Q., Hu X., Zhang H., Wang S., Zhang H., You C., Zhang C.Y., Liang H., Chen X., Ba Y. MiR-193a-3p is an Important Tumour Suppressor in Lung Cancer and Directly Targets KRAS. Cell. Physiol. Biochem. 2017;44:1311–1324. doi: 10.1159/000485491. [DOI] [PubMed] [Google Scholar]
- 163.Jancík S., Drábek J., Radzioch D., Hajdúch M. Clinical relevance of KRAS in human cancers. J. Biomed. Biotechnol. 2010;2010:150960. doi: 10.1155/2010/150960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lei M., Cheng Q., Zhao Y., Liu T., Wang X., Deng Y., Yang J., Zhang Z. [Expression and its clinical significance of SLC22A18 in non-small cell lung cancer] Zhongguo Fei Ai Za Zhi. 2012;15:17–20. doi: 10.3779/j.issn.1009-3419.2012.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang B., Liu T., Wu T., Wang Z., Rao Z., Gao J. microRNA-137 functions as a tumor suppressor in human non-small cell lung cancer by targeting SLC22A18. Int. J. Biol. Macromol. 2015;74:111–118. doi: 10.1016/j.ijbiomac.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 166.Bi Y., Han Y., Bi H., Gao F., Wang X. miR-137 impairs the proliferative and migratory capacity of human non-small cell lung cancer cells by targeting paxillin. Hum. Cell. 2014;27:95–102. doi: 10.1007/s13577-013-0085-4. [DOI] [PubMed] [Google Scholar]
- 167.Zhu X., Li Y., Shen H., Li H., Long L., Hui L., Xu W. miR-137 inhibits the proliferation of lung cancer cells by targeting Cdc42 and Cdk6. FEBS Lett. 2013;587:73–81. doi: 10.1016/j.febslet.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 168.Yang Y., Ding L., Hu Q., Xia J., Sun J., Wang X., Xiong H., Gurbani D., Li L., Liu Y., Liu A. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol. Cancer. 2017;16:141. doi: 10.1186/s12943-017-0710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lotze M.T., Tracey K.J. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 170.Dumitriu I.E., Baruah P., Manfredi A.A., Bianchi M.E., Rovere-Querini P. HMGB1: guiding immunity from within. Trends Immunol. 2005;26:381–387. doi: 10.1016/j.it.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 171.Shang G.H., Jia C.Q., Tian H., Xiao W., Li Y., Wang A.H., Dong L., Lin D.J. Serum high mobility group box protein 1 as a clinical marker for non-small cell lung cancer. Respir. Med. 2009;103:1949–1953. doi: 10.1016/j.rmed.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 172.Zhang C., Ge S., Hu C., Yang N., Zhang J. MiRNA-218, a new regulator of HMGB1, suppresses cell migration and invasion in non-small cell lung cancer. Acta Biochim. Biophys. Sin. (Shanghai) 2013;45:1055–1061. doi: 10.1093/abbs/gmt109. [DOI] [PubMed] [Google Scholar]
- 173.Wang R., Wang Z.X., Yang J.S., Pan X., De W., Chen L.B. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14) Oncogene. 2011;30:2644–2658. doi: 10.1038/onc.2010.642. [DOI] [PubMed] [Google Scholar]
- 174.Guo B., Wang W., Zhao Z., Li Q., Zhou K., Zhao L., Wang L., Yang J., Huang C. Rab14 Act as Oncogene and Induce Proliferation of Gastric Cancer Cells via AKT Signaling Pathway. PLoS ONE. 2017;12:e0170620. doi: 10.1371/journal.pone.0170620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang R.T., Xu M., Xu C.X., Song Z.G., Jin H. Decreased expression of miR216a contributes to non-small-cell lung cancer progression. Clin. Cancer Res. 2014;20:4705–4716. doi: 10.1158/1078-0432.CCR-14-0517. [DOI] [PubMed] [Google Scholar]
- 176.Shahbazian D., Parsyan A., Petroulakis E., Hershey J., Sonenberg N. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle. 2010;9:4106–4109. doi: 10.4161/cc.9.20.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ahn Y.H., Gibbons D.L., Chakravarti D., Creighton C.J., Rizvi Z.H., Adams H.P., Pertsemlidis A., Gregory P.A., Wright J.A., Goodall G.J. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J. Clin. Invest. 2012;122:3170–3183. doi: 10.1172/JCI63608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hu H., Xu Z., Li C., Xu C., Lei Z., Zhang H.T., Zhao J. MiR-145 and miR-203 represses TGF-β-induced epithelial-mesenchymal transition and invasion by inhibiting SMAD3 in non-small cell lung cancer cells. Lung Cancer. 2016;97:87–94. doi: 10.1016/j.lungcan.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 179.Roberts A.B., Tian F., Byfield S.D., Stuelten C., Ooshima A., Saika S., Flanders K.C. Smad3 is key to TGF-beta-mediated epithelial-to-mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine Growth Factor Rev. 2006;17:19–27. doi: 10.1016/j.cytogfr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 180.Wiggins J.F., Ruffino L., Kelnar K., Omotola M., Patrawala L., Brown D., Bader A.G. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tanaka N., Toyooka S., Soh J., Kubo T., Yamamoto H., Maki Y., Muraoka T., Shien K., Furukawa M., Ueno T. Frequent methylation and oncogenic role of microRNA-34b/c in small-cell lung cancer. Lung Cancer. 2012;76:32–38. doi: 10.1016/j.lungcan.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 182.Li Y.L., Liu X.M., Zhang C.Y., Zhou J.B., Shao Y., Liang C., Wang H.M., Hua Z.Y., Lu S.D., Ma Z.L. MicroRNA-34a/EGFR axis plays pivotal roles in lung tumorigenesis. Oncogenesis. 2017;6:e372. doi: 10.1038/oncsis.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lui V.W., Grandis J.R. EGFR-mediated cell cycle regulation. Anticancer Res. 2002;22(1A):1–11. [PubMed] [Google Scholar]
- 184.Liu G., Friggeri A., Yang Y., Milosevic J., Ding Q., Thannickal V.J., Kaminski N., Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wynn T.A., Ramalingam T.R. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat. Med. 2012;18:1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.American Thoracic Society American Thoracic Society. Idiopathic pulmonary fibrosis: diagnosis and treatment. International consensus statement. American Thoracic Society (ATS), and the European Respiratory Society (ERS) Am. J. Respir. Crit. Care Med. 2000;161:646–664. doi: 10.1164/ajrccm.161.2.ats3-00. [DOI] [PubMed] [Google Scholar]
- 187.Pottier N., Maurin T., Chevalier B., Puisségur M.P., Lebrigand K., Robbe-Sermesant K., Bertero T., Lino Cardenas C.L., Courcot E., Rios G. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS ONE. 2009;4:e6718. doi: 10.1371/journal.pone.0006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kang H. Role of miRNA in TGF-β Signaling Pathway-Mediated Pulmonary Fibrosis. Int. J. Mol. Sci. 2017;18:E2527. doi: 10.3390/ijms18122527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xie T., Liang J., Guo R., Liu N., Noble P.W., Jiang D. Comprehensive microRNA analysis in bleomycin-induced pulmonary fibrosis identifies multiple sites of molecular regulation. Physiol. Genomics. 2011;43:479–487. doi: 10.1152/physiolgenomics.00222.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Huang C., Xiao X., Yang Y., Mishra A., Liang Y., Zeng X., Yang X., Xu D., Blackburn M.R., Henke C.A., Liu L. MicroRNA-101 attenuates pulmonary fibrosis by inhibiting fibroblast proliferation and activation. J. Biol. Chem. 2017;292:16420–16439. doi: 10.1074/jbc.M117.805747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shen Z.J., Esnault S., Rosenthal L.A., Szakaly R.J., Sorkness R.L., Westmark P.R., Sandor M., Malter J.S. Pin1 regulates TGF-beta1 production by activated human and murine eosinophils and contributes to allergic lung fibrosis. J. Clin. Invest. 2008;118:479–490. doi: 10.1172/JCI32789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hinz B., Phan S.H., Thannickal V.J., Galli A., Bochaton-Piallat M.L., Gabbiani G. The myofibroblast: one function, multiple origins. Am. J. Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J.L., Heuchel R., Itoh S., Kawabata M., Heldin N.E., Heldin C.H., ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 194.Pandit K.V., Corcoran D., Yousef H., Yarlagadda M., Tzouvelekis A., Gibson K.F., Konishi K., Yousem S.A., Singh M., Handley D. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Thuault S., Valcourt U., Petersen M., Manfioletti G., Heldin C.H., Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J. Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Das S., Kumar M., Negi V., Pattnaik B., Prakash Y.S., Agrawal A., Ghosh B. MicroRNA-326 regulates profibrotic functions of transforming growth factor-β in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2014;50:882–892. doi: 10.1165/rcmb.2013-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nho R.S., Im J., Ho Y.Y., Hergert P. MicroRNA-96 inhibits FoxO3a function in IPF fibroblasts on type I collagen matrix. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014;307:L632–L642. doi: 10.1152/ajplung.00127.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Peck B., Ferber E.C., Schulze A. Antagonism between FOXO and MYC Regulates Cellular Powerhouse. Front. Oncol. 2013;3:96. doi: 10.3389/fonc.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gu T.L., Tothova Z., Scheijen B., Griffin J.D., Gilliland D.G., Sternberg D.W. NPM-ALK fusion kinase of anaplastic large-cell lymphoma regulates survival and proliferative signaling through modulation of FOXO3a. Blood. 2004;103:4622–4629. doi: 10.1182/blood-2003-03-0820. [DOI] [PubMed] [Google Scholar]
- 200.Chen J., Gomes A.R., Monteiro L.J., Wong S.Y., Wu L.H., Ng T.T., Karadedou C.T., Millour J., Ip Y.C., Cheung Y.N. Constitutively nuclear FOXO3a localization predicts poor survival and promotes Akt phosphorylation in breast cancer. PLoS ONE. 2010;5:e12293. doi: 10.1371/journal.pone.0012293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yang S., Banerjee S., de Freitas A., Sanders Y.Y., Ding Q., Matalon S., Thannickal V.J., Abraham E., Liu G. Participation of miR-200 in pulmonary fibrosis. Am. J. Pathol. 2012;180:484–493. doi: 10.1016/j.ajpath.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yan W., Cao Q.J., Arenas R.B., Bentley B., Shao R. GATA3 inhibits breast cancer metastasis through the reversal of epithelial-mesenchymal transition. J. Biol. Chem. 2010;285:14042–14051. doi: 10.1074/jbc.M110.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 204.Rajasekaran S., Rajaguru P., Sudhakar Gandhi P.S. miRNA as potential targets for progressive pulmonary fibrosis. Front. Pharmacol. 2015;6:254. doi: 10.3389/fphar.2015.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Scotet V., Duguépéroux I., Saliou P., Rault G., Roussey M., Audrézet M.P., Férec C. Evidence for decline in the incidence of cystic fibrosis: a 35-year observational study in Brittany, France. Orphanet J. Rare Dis. 2012;7:14. doi: 10.1186/1750-1172-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chmiel J.F., Davis P.B. State of the art: why do the lungs of patients with cystic fibrosis become infected and why can’t they clear the infection? Respir. Res. 2003;4:8. doi: 10.1186/1465-9921-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Worlitzsch D., Tarran R., Ulrich M., Schwab U., Cekici A., Meyer K.C., Birrer P., Bellon G., Berger J., Weiss T. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 2002;109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tognon M., Köhler T., Gdaniec B.G., Hao Y., Lam J.S., Beaume M., Luscher A., Buckling A., van Delden C. Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J. 2017;11:2233–2243. doi: 10.1038/ismej.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ramachandran S., Karp P.H., Jiang P., Ostedgaard L.S., Walz A.E., Fisher J.T., Keshavjee S., Lennox K.A., Jacobi A.M., Rose S.D. A microRNA network regulates expression and biosynthesis of wild-type and DeltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. USA. 2012;109:13362–13367. doi: 10.1073/pnas.1210906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lutz M., Burke L.J., Barreto G., Goeman F., Greb H., Arnold R., Schultheiss H., Brehm A., Kouzarides T., Lobanenkov V., Renkawitz R. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 2000;28:1707–1713. doi: 10.1093/nar/28.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rowe S.M., Miller S., Sorscher E.J. Cystic fibrosis. N. Engl. J. Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 212.Farinha C.M., Matos P., Amaral M.D. Control of cystic fibrosis transmembrane conductance regulator membrane trafficking: not just from the endoplasmic reticulum to the Golgi. FEBS J. 2013;280:4396–4406. doi: 10.1111/febs.12392. [DOI] [PubMed] [Google Scholar]
- 213.Ramachandran S., Karp P.H., Osterhaus S.R., Jiang P., Wohlford-Lenane C., Lennox K.A., Jacobi A.M., Praekh K., Rose S.D., Behlke M.A. Post-transcriptional regulation of cystic fibrosis transmembrane conductance regulator expression and function by miRNA. Am. J. Respir. Cell Mol. Biol. 2013;49:544–551. doi: 10.1165/rcmb.2012-0430OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Oglesby I.K., Chotirmall S.H., McElvaney N.G., Greene C.M. Regulation of cystic fibrosis transmembrane conductance regulator by microRNA-145, -223, and -494 is altered in ΔF508 cystic fibrosis airway epithelium. J. Immunol. 2013;190:3354–3362. doi: 10.4049/jimmunol.1202960. [DOI] [PubMed] [Google Scholar]
- 215.Gillen A.E., Gosalia N., Leir S.H., Harris A. MicroRNA regulation of expression of the cystic fibrosis transmembrane conductance regulator gene. Biochem. J. 2011;438:25–32. doi: 10.1042/BJ20110672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Megiorni F., Cialfi S., Dominici C., Quattrucci S., Pizzuti A. Synergistic post-transcriptional regulation of the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) by miR-101 and miR-494 specific binding. PLoS ONE. 2011;6:e26601. doi: 10.1371/journal.pone.0026601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Oglesby I.K., Bray I.M., Chotirmall S.H., Stallings R.L., O’Neill S.J., McElvaney N.G., Greene C.M. miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. J. Immunol. 2010;184:1702–1709. doi: 10.4049/jimmunol.0902669. [DOI] [PubMed] [Google Scholar]
- 218.Katoh Y., Shiba Y., Mitsuhashi H., Yanagida Y., Takatsu H., Nakayama K. Tollip and Tom1 form a complex and recruit ubiquitin-conjugated proteins onto early endosomes. J. Biol. Chem. 2004;279:24435–24443. doi: 10.1074/jbc.M400059200. [DOI] [PubMed] [Google Scholar]
- 219.Yamakami M., Yokosawa H. Tom1 (target of Myb 1) is a novel negative regulator of interleukin-1- and tumor necrosis factor-induced signaling pathways. Biol. Pharm. Bull. 2004;27:564–566. doi: 10.1248/bpb.27.564. [DOI] [PubMed] [Google Scholar]
- 220.Fabbri E., Borgatti M., Montagner G., Bianchi N., Finotti A., Lampronti I., Bezzerri V., Dechecchi M.C., Cabrini G., Gambari R. Expression of microRNA-93 and Interleukin-8 during Pseudomonas aeruginosa-mediated induction of proinflammatory responses. Am. J. Respir. Cell Mol. Biol. 2014;50:1144–1155. doi: 10.1165/rcmb.2013-0160OC. [DOI] [PubMed] [Google Scholar]
- 221.Weldon S., McNally P., McAuley D.F., Oglesby I.K., Wohlford-Lenane C.L., Bartlett J.A., Scott C.J., McElvaney N.G., Greene C.M., McCray P.B., Jr., Taggart C.C. miR-31 dysregulation in cystic fibrosis airways contributes to increased pulmonary cathepsin S production. Am. J. Respir. Crit. Care Med. 2014;190:165–174. doi: 10.1164/rccm.201311-1986OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Cheng X.W., Shi G.P., Kuzuya M., Sasaki T., Okumura K., Murohara T. Role for cysteine protease cathepsins in heart disease: focus on biology and mechanisms with clinical implication. Circulation. 2012;125:1551–1562. doi: 10.1161/CIRCULATIONAHA.111.066712. [DOI] [PubMed] [Google Scholar]
- 223.Liao W., Dong J., Peh H.Y., Tan L.H., Lim K.S., Li L., Wong W.F. Oligonucleotide Therapy for Obstructive and Restrictive Respiratory Diseases. Molecules. 2017;22:E139. doi: 10.3390/molecules22010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ahmed M.S., Dutta R.K., Manandhar P., Li X., Torabi H., Barrios A., Wang P., Chinnapaiyan S., Unwalla H.J., Moon J.H. A guanylurea-functionalized conjugated polymer enables RNA interference in ex vivo human airway epithelium. Chem. Commun. (Camb.) 2019;55:7804–7807. doi: 10.1039/c9cc02856k. [DOI] [PubMed] [Google Scholar]
- 225.Järver P., Coursindel T., Andaloussi S.E., Godfrey C., Wood M.J., Gait M.J. Peptide-mediated Cell and In Vivo Delivery of Antisense Oligonucleotides and siRNA. Mol. Ther. Nucleic Acids. 2012;1:e27. doi: 10.1038/mtna.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tanaka M., Nyce J.W. Respirable antisense oligonucleotides: a new drug class for respiratory disease. Respir. Res. 2001;2:5–9. doi: 10.1186/rr32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Moschos S.A., Frick M., Taylor B., Turnpenny P., Graves H., Spink K.G., Brady K., Lamb D., Collins D., Rockel T.D. Uptake, efficacy, and systemic distribution of naked, inhaled short interfering RNA (siRNA) and locked nucleic acid (LNA) antisense. Mol. Ther. 2011;19:2163–2168. doi: 10.1038/mt.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Grijalvo S., Alagia A., Jorge A.F., Eritja R. Covalent Strategies for Targeting Messenger and Non-Coding RNAs: An Updated Review on siRNA, miRNA and antimiR Conjugates. Genes (Basel) 2018;9:E74. doi: 10.3390/genes9020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 230.Lundstrom K. Viral Vectors in Gene Therapy. Diseases. 2018;6:42. doi: 10.3390/diseases6020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Trang P., Wiggins J.F., Daige C.L., Cho C., Omotola M., Brown D., Weidhaas J.B., Bader A.G., Slack F.J. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Rai K., Takigawa N., Ito S., Kashihara H., Ichihara E., Yasuda T., Shimizu K., Tanimoto M., Kiura K. Liposomal delivery of MicroRNA-7-expressing plasmid overcomes epidermal growth factor receptor tyrosine kinase inhibitor-resistance in lung cancer cells. Mol. Cancer Ther. 2011;10:1720–1727. doi: 10.1158/1535-7163.MCT-11-0220. [DOI] [PubMed] [Google Scholar]
- 233.Chen Y., Zhu X., Zhang X., Liu B., Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol. Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Bouchie A. First microRNA mimic enters clinic. Nat. Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 235.Kim D.-H., Behlke M.A., Rose S.D., Chang M.S., Choi S., Rossi J.J. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 236.Xiao Z., Li C.H., Chan S.L., Xu F., Feng L., Wang Y., Jiang J.D., Sung J.J., Cheng C.H., Chen Y. A small-molecule modulator of the tumor-suppressor miR34a inhibits the growth of hepatocellular carcinoma. Cancer Res. 2014;74:6236–6247. doi: 10.1158/0008-5472.CAN-14-0855. [DOI] [PubMed] [Google Scholar]
- 237.Young D.D., Connelly C.M., Grohmann C., Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J. Am. Chem. Soc. 2010;132:7976–7981. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- 238.Chen X., Huang C., Zhang W., Wu Y., Chen X., Zhang C.Y., Zhang Y. A universal activator of miRNA identified from photoreaction products. Chem. Commun. (Camb.) 2012;48:6432–6434. doi: 10.1039/c2cc32157b. [DOI] [PubMed] [Google Scholar]
- 239.Han C., Yu Z., Duan Z., Kan Q. Role of microRNA-1 in human cancer and its therapeutic potentials. Biomed Res. Int. 2014;2014:428371. doi: 10.1155/2014/428371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Fujita Y., Takeshita F., Kuwano K., Ochiya T. RNAi Therapeutic Platforms for Lung Diseases. Pharmaceuticals (Basel) 2013;6:223–250. doi: 10.3390/ph6020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of miRNA in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 242.Li J.J., Tay H.L., Maltby S., Xiang Y., Eyers F., Hatchwell L., Zhou H., Toop H.D., Morris J.C., Nair P. MicroRNA-9 regulates steroid-resistant airway hyperresponsiveness by reducing protein phosphatase 2A activity. J. Allergy Clin. Immunol. 2015;136:462–473. doi: 10.1016/j.jaci.2014.11.044. [DOI] [PubMed] [Google Scholar]
- 243.Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 244.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Tan G.S., Chiu C.H., Garchow B.G., Metzler D., Diamond S.L., Kiriakidou M. Small molecule inhibition of RISC loading. ACS Chem. Biol. 2012;7:403–410. doi: 10.1021/cb200253h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Chen Y., Tang H. High-throughput screening assays to identify small molecules preventing photoreceptor degeneration caused by the rhodopsin P23H mutation. Methods Mol. Biol. 2015;1271:369–390. doi: 10.1007/978-1-4939-2330-4_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Gumireddy K., Young D.D., Xiong X., Hogenesch J.B., Huang Q., Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew. Chem. Int. Ed. Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Chan J.A., Krichevsky A.M., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 249.Shi Z., Zhang J., Qian X., Han L., Zhang K., Chen L., Liu J., Ren Y., Yang M., Zhang A. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res. 2013;73:5519–5531. doi: 10.1158/0008-5472.CAN-13-0280. [DOI] [PubMed] [Google Scholar]
- 250.Nimjee S.M., Rusconi C.P., Sullenger B.A. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 251.Que-Gewirth N.S., Sullenger B.A. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14:283–291. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- 252.Bala J., Chinnapaiyan S., Dutta R.K., Unwalla H. Aptamers in HIV research diagnosis and therapy. RNA Biol. 2018;15:327–337. doi: 10.1080/15476286.2017.1414131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Eyetech Study Group Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina. 2002;22:143–152. doi: 10.1097/00006982-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 254.Eyetech Study Group Anti-vascular endothelial growth factor therapy for subfoveal choroidal neovascularization secondary to age-related macular degeneration: phase II study results. Ophthalmology. 2003;110:979–986. doi: 10.1016/S0161-6420(03)00085-X. [DOI] [PubMed] [Google Scholar]
- 255.Doudna J.A., Cech T.R., Sullenger B.A. Selection of an RNA molecule that mimics a major autoantigenic epitope of human insulin receptor. Proc. Natl. Acad. Sci. USA. 1995;92:2355–2359. doi: 10.1073/pnas.92.6.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Rusconi C.P., Scardino E., Layzer J., Pitoc G.A., Ortel T.L., Monroe D., Sullenger B.A. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 257.Calvo-González C., Reche-Frutos J., Donate-López J., García-Feijoó J., Leila M., Fernández-Pérez C., Garcia-Sánchez J. Combined Pegaptanib sodium (Macugen) and photodynamic therapy in predominantly classic juxtafoveal choroidal neovascularisation in age related macular degeneration. Br. J. Ophthalmol. 2008;92:74–75. doi: 10.1136/bjo.2007.128942. [DOI] [PubMed] [Google Scholar]
- 258.Lee J.H., Canny M.D., De Erkenez A., Krilleke D., Ng Y.S., Shima D.T., Pardi A., Jucker F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc. Natl. Acad. Sci. USA. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhou J., Rossi J.J. Cell-type-specific, Aptamer-functionalized Agents for Targeted Disease Therapy. Mol. Ther. Nucleic Acids. 2014;3:e169. doi: 10.1038/mtna.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Catuogno S., Esposito C.L., de Franciscis V. Aptamer-Mediated Targeted Delivery of Therapeutics: An Update. Pharmaceuticals (Basel) 2016;9:E69. doi: 10.3390/ph9040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Perepelyuk M., Sacko K., Thangavel K., Shoyele S.A. Evaluation of MUC1-Aptamer Functionalized Hybrid Nanoparticles for Targeted Delivery of miRNA-29b to Nonsmall Cell Lung Cancer. Mol. Pharm. 2018;15:985–993. doi: 10.1021/acs.molpharmaceut.7b00900. [DOI] [PubMed] [Google Scholar]
- 262.Russo V., Paciocco A., Affinito A., Roscigno G., Fiore D., Palma F., Galasso M., Volinia S., Fiorelli A., Esposito C.L. Aptamer-miR-34c Conjugate Affects Cell Proliferation of Non-Small-Cell Lung Cancer Cells. Mol. Ther. Nucleic Acids. 2018;13:334–346. doi: 10.1016/j.omtn.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Iaboni M., Russo V., Fontanella R., Roscigno G., Fiore D., Donnarumma E., Esposito C.L., Quintavalle C., Giangrande P.H., de Franciscis V., Condorelli G. Aptamer-miRNA-212 Conjugate Sensitizes NSCLC Cells to TRAIL. Mol. Ther. Nucleic Acids. 2016;5:e289. doi: 10.1038/mtna.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Esposito C.L., Cerchia L., Catuogno S., De Vita G., Dassie J.P., Santamaria G., Swiderski P., Condorelli G., Giangrande P.H., de Franciscis V. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol. Ther. 2014;22:1151–1163. doi: 10.1038/mt.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Cerchia L., Esposito C.L., Camorani S., Rienzo A., Stasio L., Insabato L., Affuso A., de Franciscis V. Targeting Axl with an high-affinity inhibitory aptamer. Mol. Ther. 2012;20:2291–2303. doi: 10.1038/mt.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sundar I.K., Yao H., Sellix M.T., Rahman I. Circadian molecular clock in lung pathophysiology. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015;309:L1056–L1075. doi: 10.1152/ajplung.00152.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Krakowiak K., Durrington H.J. The Role of the Body Clock in Asthma and COPD: Implication for Treatment. Pulm. Ther. 2018;4:29–43. doi: 10.1007/s41030-018-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hansen K.F., Sakamoto K., Obrietan K. miRNA: a potential interface between the circadian clock and human health. Genome Med. 2011;3:10. doi: 10.1186/gm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Mehta N., Cheng H.Y. Micro-managing the circadian clock: The role of miRNA in biological timekeeping. J. Mol. Biol. 2013;425:3609–3624. doi: 10.1016/j.jmb.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 270.Malmhäll C., Alawieh S., Lötvall J., Rådinger M. MicroRNA-146a and microRNA-155 expression in induced sputum of allergic asthmatics. Eur. Respir. J. 2015;46(Suppl 59) PA2552. [Google Scholar]
- 271.Wang W., Chen J., Dai J., Zhang B., Wang F., Sun Y. MicroRNA-16-1 Inhibits Tumor Cell Proliferation and Induces Apoptosis in A549 Non-Small Cell Lung Carcinoma Cells. Oncol. Res. 2016;24:345–351. doi: 10.3727/096504016X14685034103194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Bhattacharyya S., Balakathiresan N.S., Dalgard C., Gutti U., Armistead D., Jozwik C., Srivastava M., Pollard H.B., Biswas R. Elevated miR-155 promotes inflammation in cystic fibrosis by driving hyperexpression of interleukin-8. J. Biol. Chem. 2011;286:11604–11615. doi: 10.1074/jbc.M110.198390. [DOI] [PMC free article] [PubMed] [Google Scholar]