Abstract
Cytogenetically normal acute myeloid leukemia (CN-AML) presents with diverse outcomes in different patients and is categorized as an intermediate prognosis group. It is important to identify prognostic factors for CN-AML risk stratification. In this study, using the TCGA CN-AML dataset, we found that the scavenger receptor stabilin-1 (STAB1) is a prognostic factor for poor outcomes and validated it in three other independent CN-AML datasets. The high STAB1 expression (STAB1high) group had shorter event-free survival compared with the low STAB1 expression (STAB1low) group in both the TCGA dataset (n = 79; p = 0.0478) and GEO: GSE6891 dataset (n = 187; p = 0.0354). Differential expression analysis between the STAB1high and STAB1low groups revealed that upregulated genes in the STAB1high group were enriched in pathways related to cell adhesion and migration and immune responses. We confirmed that STAB1 suppression inhibits cell growth in KG1a and NB4 leukemia cells. Expression correlation analyses between STAB1 and cancer drug targets suggested that patients in the STAB1low group are more sensitive to the BCL2 inhibitor venetoclax, and we confirmed it in cell lines. In conclusion, we identified STAB1 as a prognostic factor for CN-AML in multiple datasets, explored its underlying mechanism, and provided potential therapeutic indications.
Keywords: cytogenetically normal acute myeloid leukemia, STAB1, prognostic factor, therapeutic indications
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease characterized by the expansion of undifferentiated myeloid precursors, resulting in impaired hematopoiesis.1 Recurrent cytogenetic abnormalities are well-established markers for the diagnosis and prognosis of AML.2 Cytogenetically normal AML (CN-AML) accounts for approximately 50% of all AML cases. Currently, CN-AML is considered an intermediate risk disease because some patients respond well to normal treatment and some others do not.3 Therefore, identifying effective prognostic factors for CN-AML is important for providing optimal care to patients.
Recently, various DNA and RNA markers have been proposed as prognostic factors for CN-AML. Mutations in NPM1 and CEBPA have been widely used as prognostic factors for good outcomes in patients with CN-AML;4 in contrast, patients with mutations in FLT3, RUNX1, ASXL1, and TP53 have been associated with adverse outcomes. Furthermore, increased expression of WT1,5 BAALC,6 and ERG7 is reportedly associated with outcomes in patients with CN-AML. Based on gene expression, multiple prognostic factors have been suggested to define CN-AML subgroups. Early in 2008, Metzeler et al.8 developed an 86-probe-set gene expression signature for predicting overall survival (OS) in patients with CN-AML. A stem-cell-associated gene expression signature involving 44 genes was proposed as an indicator of negative prognostic outcomes in patients with primary CN-AML.9 More recently, an integrative prognostic risk score based on molecular markers for gene expression (BAALC, ERG, MN1, and WT1) and mutation (NPM1, FLT3-ITD, CEBPA, and SNP rs16754) was proposed for predicting outcomes in patients with CN-AML.10 However, these prognostic factors lack consistency in different cohorts and have thus not been introduced into clinical practice. In addition, the molecular mechanisms and appropriate therapeutic strategies underlying the groups identified by those markers remain unclear. Therefore, new prognostic factors validated in different CN-AML datasets are still required. Identification of the molecular mechanisms underlying the groups and their potential therapeutic strategies will be of considerable value for academic and clinical studies.
The scavenger receptor stabilin-1 (STAB1), which acts as a scavenging receptor, is induced during chronic inflammation and cancer progression.11 The endocytic ligands of STAB1 include SPARC, a protein that modulates progression of various cancers.12, 13 Meanwhile, the cancer-promoting role of STAB1 has been demonstrated in B16 melanoma and EL-4 lymphoma mouse models.14 Until now, the potential impact of STAB1 expression on the prognosis of CN-AML has not been examined.
In the present study, we identified STAB1 expression as a prognostic factor in multiple CN-AML datasets. We also investigated the potential molecular mechanisms and drug indicators underlying the CN-AML groups identified on the basis of STAB1 stratification. Furthermore, we validated the effect of STAB1 on leukemic cell growth and its sensitivity to the BCL2-targeting drug venetoclax. Our study identified a universal prognostic factor for CN-AML having potential applications in leukemia prognosis and treatment.
Results
STAB1 Is a Potential Prognostic Factor Validated by Multiple CN-AML Datasets
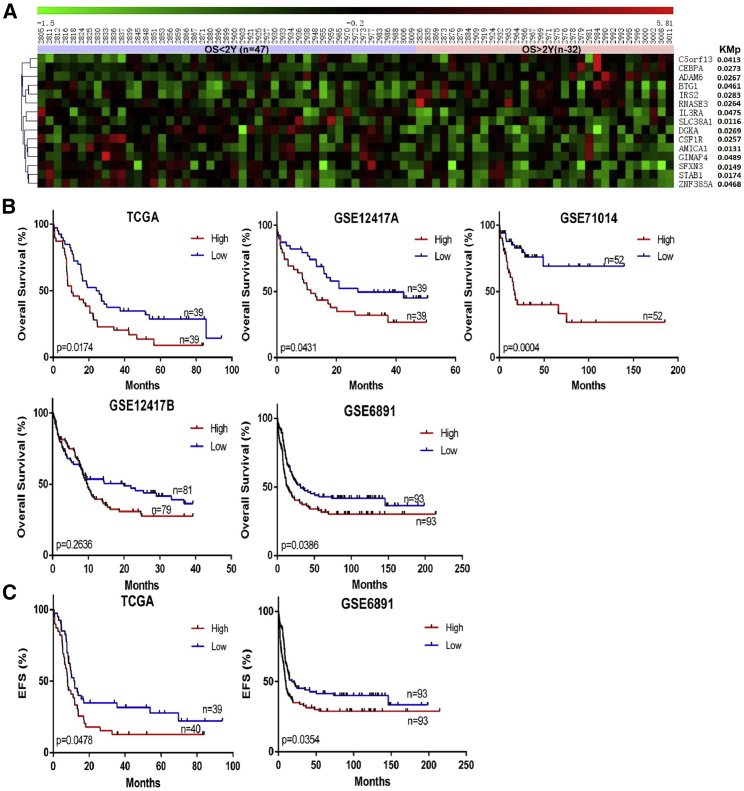
To identify potential prognostic factors for CN-AML, we screened CN-AML samples (n = 79) from the TCGA AML dataset and classified them into two groups based on 2-year OS. Differential expression analysis between the two groups was performed, and 84 differentially expressed genes—34 upregulated and 50 downregulated—were identified. Univariate survival analysis identified 15 genes associated with OS among the differentially expressed genes (Figure 1A). After univariate and multivariate analyses, the expression of six genes (ADAM6, AMICA1, CSF1R, DGKA, IL3RA, and STAB1) was associated with OS and was independent of other clinical factors, including age and mutation of DNMT3A, RUNX1, FLT3-ITD, MT-CYB, WT1, IDH2, NPM1, and IDH1 (p < 0.05) (Tables S1 and S2).
Figure 1.
Selection of STAB1 as a Prognostic Factor for CN-AML
(A) Heatmap of differentially expressed genes associated with OS in the TCGA dataset. (B) OS analysis of STAB1 in five CN-AML datasets. (C) EFS correlation of STAB1 in CN-AML in the TCGA and GSE6891 datasets.
To test whether these six genes are universal prognostic factors for CN-AML, we validated them using four other CN-AML datasets: GEO: GSE12417 test dataset (referred to as GSE12417A in this work; n = 79), GSE12417 training dataset (referred to as GSE12417B in this work; n = 163), GSE71014 (n = 104), and GSE6891 (n = 187). Only STAB1 showed significance in three of the four datasets (Figure 1B; Figure S1). The GSE6891 dataset (n = 187) was selected for further validation because of it having more clinical details. Our results showed that STAB1 expression is a prognostic factor independent of the FLT3, NPM1, and IDH2 mutation status (Table 1). Thus, STAB1 may be a potential prognostic factor in patients with CN-AML.
Table 1.
Multivariable Analysis of STAB1 and Clinical Variables in TCGA and GSE6891 Cohort
| Overall Survival Covariate | TCGA Dataset |
GSE6891 Dataset |
||
|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | |
| STAB1 | 1.98 (1.01−3.03) | 0.0473 | 0.43 (1.05−2.24) | 0.027511 |
| Age | 2.44 (1.15−3.62) | 0.0147 | NA | NA |
| DNMT3A | 2.20 (1.07−3.50) | 0.028 | NA | NA |
| RUNX1 | 2.04 (1.03−5.37) | 0.0417 | NA | NA |
| FLT3-ITD | 1.59 (0.89−3.07) | 0.1124 | 0.57 (1.20−2.60) | 0.004015 |
| MT-CYB | 1.33 (0.66−8.75) | 0.185 | NA | NA |
| WT1 | 1.16 (0.66−5.02) | 0.2461 | NA | NA |
| IDH2 | 0.47 (0.56−2.59) | 0.638 | −0.05 (0.49−1.83) | 0.879925 |
| NPM1 | −0.05 (0.53−1.85) | 0.963 | −0.45 (0.43−0.94) | 0.022836 |
| IDH1 | −1.28 (0.16−1.4) | 0.2016 | NA | NA |
The model was generated from a COX regression model that included age, gene mutation of DNMT3A, and RUNX1, FLT3-ITD, MT-CYB, WT1, IDH2, NPM1, IDH1, and expression level of STAB1. CI, confidence interval; HR, hazard ratio; NA, not available.
Clinical Characteristics of Patients in the STAB1high and STAB1low Groups
To assess the correlation between STAB1 expression and clinical factors in patients with CN-AML, we evaluated the patients’ age, sex, white blood cell (WBC) counts, French-American-British (FAB) classification, mutations, and event-free survival (EFS) and OS status (based on the TCGA and GSE6891 datasets, respectively). In the TCGA cohort, patients in the STAB1low group were more likely to survive for >2 years (p = 0.0392) (Table 2). However, clinical factors such as age, WBC counts, and gene mutation status were not significantly different between the STAB1high and STAB1low groups (Tables S3 and S4). In the GSE6891 dataset, compared with the STAB1high group, the STAB1low group contained fewer female patients (p = 0.0276) and carried fewer FLT3-ITD mutations (p = 0.0255), more NPM1 mutations (p = 0.0004), and more CEBPA mutations (p < 0.0001) (Table 2). Moreover, the STAB1low group had a longer EFS in both the TCGA (p = 0.0478) and GSE6891 (p = 0.0354) datasets (Figure 1C). Thus, lower expression of STAB1 could predict better outcomes in patients with CN-AML.
Table 2.
Significant Clinical Characteristics in the TCGA Cohort and GSE6891 Dataset between the STAB1high and STAB1low Group
| Clinical Factor | STAB1low | STAB1high | p Value |
|---|---|---|---|
| TCGA Dataset | (n = 40) | (n = 39) | |
| OS (≥2 years) | 21 | 11 | 0.0392 |
| OS (<2 years) | 19 | 28 | |
| M5 | 1 | 10 | 0.003 |
| Non-M5 | 39 | 29 | |
| GSE6891 Dataset | (n = 93) | (n = 93) | |
| Female | 38 | 54 | 0.0276 |
| Male | 55 | 39 | |
| EFS (≥1 year) | 53 | 37 | 0.0275 |
| EFS (<1 year) | 40 | 56 | |
| FLT3-ITD negative | 62 | 46 | 0.0255 |
| FLT3-ITD positive | 31 | 47 | |
| NPM1 negative | 53 | 28 | 0.0004 |
| NPM1 positive | 40 | 65 | |
| CEBPA negative | 74 | 91 | <0.0001 |
| CEBPA positive | 19 | 2 | |
| FAB | <0.0001 | ||
| M1 | 41 | 11 | <0.0001 |
| Non-M1 | 46 | 75 | |
| M2 | 24 | 11 | 0.0154 |
| Non-M2 | 63 | 75 | |
| M4 | 6 | 22 | 0.0008 |
| Non-M4 | 81 | 64 | |
| M5 | 11 | 41 | <0.0001 |
| Non-M5 | 76 | 45 | |
Potential Mechanisms Affecting CN-AML Groups Stratified by STAB1 Based on Differential Expression and Regulation
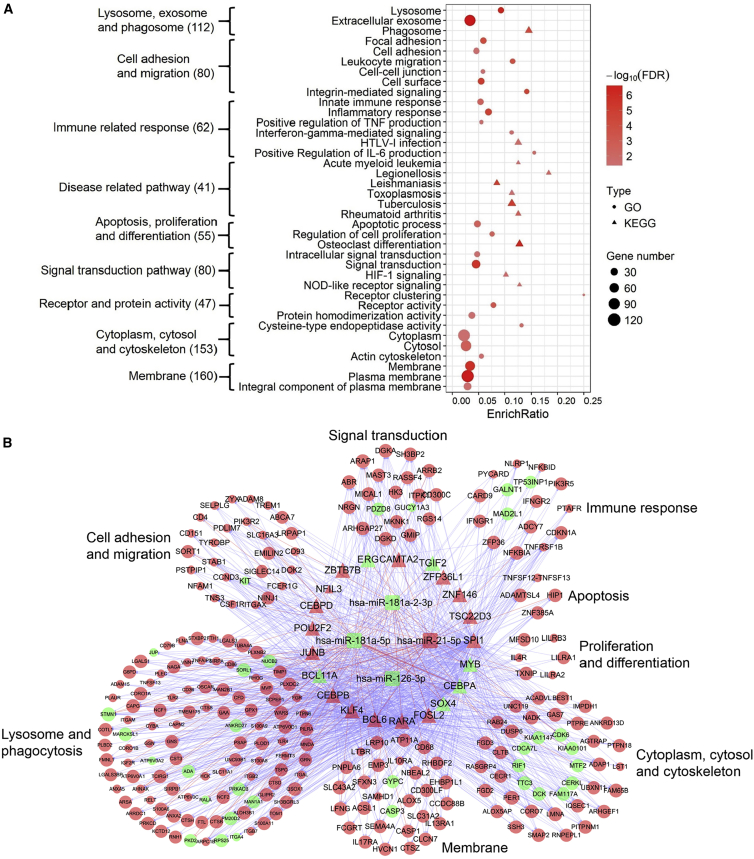
To explore the mechanisms underlying CN-AML groups stratified by STAB1, we identified 353 differentially expressed genes (284 upregulated and 69 downregulated) by comparing the STAB1high and STAB1low groups in the TCGA dataset. Functional analysis of these differentially expressed genes showed enrichment of several gene ontology (GO) functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Functions associated with lysosomes, exosomes, and phagosomes, cell adhesion and migration, and immune-related responses were overrepresented. The enriched GO functions and KEGG pathways corroborate the known functions of STAB1, which are induced in macrophages during inflammation, are involved in cell-cell contact, and may contribute to integrin-mediated leukocyte migration and adhesion (Figure 2A). Several infectious and inflammatory disease-related pathways, such as those involved in legionellosis, toxoplasmosis, and rheumatoid arthritis, were considerably enriched, thereby justifying the reported role of STAB1 as an immune modulator.15 We also identified four microRNAs (miRNAs) that were differentially expressed between the STAB1low and STAB1high groups: hsa-miR-21-5p, hsa-miR-181a-5p, hsa-miR-181-2-3p, and hsa-miR-126-3p. We constructed a miRNA-transcription factor (TF) co-regulatory network combining enriched functional categories to explore the mechanism of STAB1 (Figure 2B). The resulting network included 275 nodes and 2,492 edges comprising four miRNAs and 21 TFs whose abnormal expression contributed to altered gene expression. The downregulation of the cancer suppressor miR-181 family is reportedly associated with good outcomes in AML.16 miR-21-5p is an onco-miRNA and is consistently upregulated in AML.17 Collectively, these TFs and miRNAs may help explain the better outcomes in patients with CN-AML in the STAB1low dataset.
Figure 2.
miRNA-TF Regulation and Its Correlation with STAB1
(A) Enriched GO/KEGG terms of differential genes in the STAB1high versus STAB1l°w groups. Numbers in brackets are the numbers of differential genes in each category. The enrich ratio represents the proportion of enriched genes accounting for the terms. (B) The miRNA-TF co-regulatory network for the enriched terms in (A). Triangles, TFs; circles, target genes; rounded rectangles, miRNAs; red nodes, upregulation in the STAB1high group; green nodes, downregulation.
Potential Drug Indications for Groups Stratified by STAB1
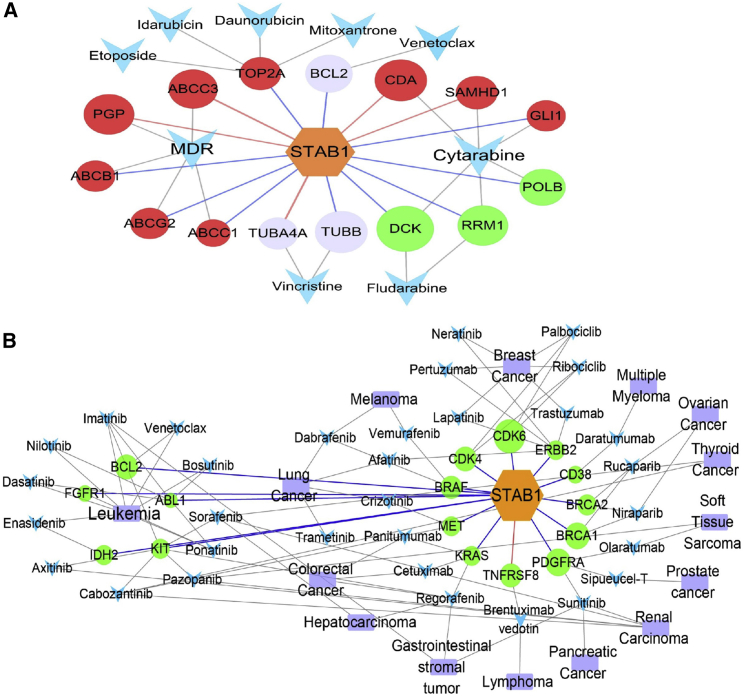
To explore the drug indications for groups stratified by STAB1, we studied the expression correlation between drug target genes and STAB1. First, we identified genes that act as direct targets or transporters of the following 11 clinical leukemia drugs: cytarabine, daunorubicin, idarubicin, mitoxantrone, etoposide, vincristine, dexamethasone, aclarubicin, decitabine, fludarabine, and venetoclax. Expression correlation analyses showed that five (linked with red edges in Figure 3A) and ten genes (linked with blue edges in Figure 3A) were positively and negatively correlated with STAB1, respectively (p < 0.05). The cytarabine-sensitive genes DCK, POLB, and RRM1 were negatively correlated with STAB1, whereas the cytarabine-resistant genes CDA and SAMHD1 were positively correlated with STAB1. These data suggest that patients in the STAB1low group are sensitive to cytarabine. DNA topoisomerase II alpha (TOP2A), a transporter of the drugs etoposide, idarubicin, daunorubicin, and mitoxantrone, was negatively correlated with STAB1. Considering the role of multidrug resistance (MDR)-related proteins in AML cells and the correlation of their expression with resistance to chemotherapy in vitro,18 we investigated the expression correlation between STAB1 and MDR-related genes; PGP and ABCC3 showed a positive correlation, whereas ABCB1, ABCC1, and ABCG2 were negatively correlated with STAB1 expression. This finding indicates the complexity of the interactions of STAB1 with MDR drugs.
Figure 3.
Expression Correlation of STAB1 and Drug Targets in the STAB1high and STAB1low Groups
(A) Correlation of STAB1 expression with gene targets of leukemia drugs. Hexagons, STAB1. (B) Correlation of STAB1 expression with gene targets of FDA-approved cancer drugs. Circles, targeted genes; arrowheads, drugs. The red nodes represent genes resistant to drugs in the STAB1high group; the green nodes represent genes sensitive to drugs in the STAB1high group. The colors of the edges represent different expression correlations (red, positive; blue, negative) between STAB1 and the drug targets. The size of the circle nodes represents the correlation strength between STAB1 and the target genes.
We performed expression correlation analyses between STAB1 and the target genes of 34 US Food and Drug Administration (FDA)-approved cancer drugs (p < 0.05) (Figure 3B). All significant target genes except for TNFRSF8 (also known as CD30), which is the target of brentuximab vedotin used to treat lymphoma, were negatively correlated with STAB1 expression.19 CDK6 (R = −0.52), PDGERA (R = −0.42), BRCA1 (R = −0.41), BCL2 (R = −0.38), and CDK4 (R = −0.38) were the top five genes that were negatively correlated with STAB1 expression.
STAB1 Suppression May Inhibit Leukemia Growth and Induce Cell Apoptosis
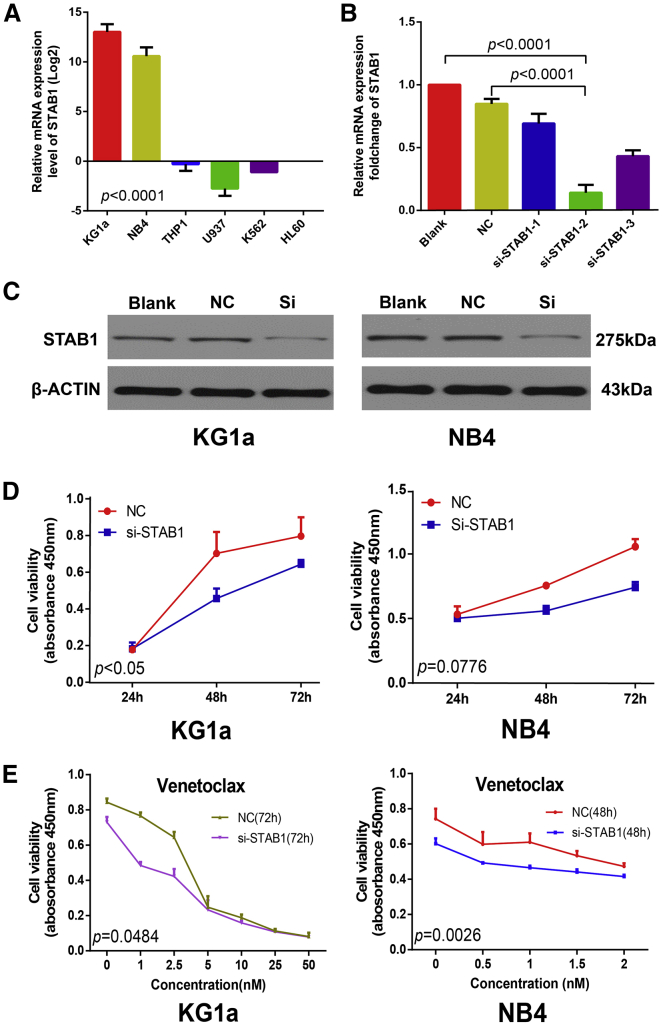
To investigate the role of STAB1 in AML and the mechanism underlying STAB1 stratification, we chose two leukemia cell lines, KG1a and NB4; both of these cell lines have relatively high levels of STAB1 mRNA expression (Figure 4A), and they are suitable in vitro models for loss-of-function experiments. KG1a cells were stably transfected with three si-STAB1s compared with negative control (NC) small interfering RNA (siRNA)-transfected cells and resulted in the most efficient STAB1 downregulation by the si-STAB1-2 (Figure 4B), which was used in the further experiments. Using western blotting, in both the KG1a and NB4 cell lines, the protein level of STAB1 in the si-STAB1 group was found to be downregulated compared with that in the NC group (Figure 4C). CCK8 assays revealed that compared with NC, si-STAB1 led to a marked decrease in KG1a cell proliferation and a decreased tendency toward NB4 cell proliferation (Figure 4D). Fluorescence-activated cell sorting (FACS) analysis showed an increase in the apoptosis of KG1a cells with si-STAB1 transfection compared with NC (Figure S2) (p value not significant). These results suggest that STAB1 suppression inhibits leukemia growth and induces cell apoptosis.
Figure 4.
STAB1 Suppression Inhibits Leukemic Cell Growth and Shows More Sensitivity to Venetoclax in KG1a and NB4 Cells
(A) Relative mRNA expression level (after log2 conversion) of STAB1 in six hematological cell lines analyzed by real-time qPCR. (B) Relative mRNA expression fold change of KG1a cells transfected with NC and three STAB1-specific siRNAs for 48 h. (C) Western blot analysis of KG1a and NB4 cells transfected with si-STAB1-2. (D) Cell proliferation of KG1a and NB4 cells transfected with NC or si-STAB1 for 72 h tested using the CCK8 assay. (E) Cell proliferation graph of KG1a cells and NB4 cells transfected with NC or si-STAB1 treated with venetoclax and tested using the CCK8 assay. Each experiment was repeated at least three times. Error bars represent SD.
The STAB1low Group Showed Greater Sensitivity to Venetoclax in KG1a and NB4 Cells
Because BCL2 is inhibited by venetoclax and BCL2 expression is strongly correlated with sensitivity to venetoclax in hematologic malignancies,20, 21 we inferred that compared with those in the STAB1high group, patients in the STAB1low group may be more sensitive to venetoclax. To assess whether patients in the STAB1low group were more sensitive to venetoclax as well as considering the promising potential of venetoclax, we treated the NC and si-STAB1 cells with venetoclax. KG1a cell viability testing demonstrated that compared with the STAB1high group, the STAB1low group was more sensitive to venetoclax (p = 0.0484) after treatment for 72 h as monotherapy. Assessment of cell viability demonstrated a concentration-dependent decrease up to 5 nmol/mL (Figure 4E). The NB4 cells from the STAB1low group were more sensitive to venetoclax (p = 0.0026) treatment for 48 h, and the assessment of cell viability demonstrated a concentration-dependent decrease of up to 2 nmol/mL (Figure 4E). These data support the prediction that patients in the STAB1low group are sensitive to venetoclax (Figure 3); furthermore, the data corroborate the finding that venetoclax was only modestly effective as monotherapy in AML.21
Discussion
To the best of our knowledge, this is the first report to show that STAB1 expression may be a prognostic factor for CN-AML. Although previous studies have identified promising prognostic factors for CN-AML, none of the identified indicators has been validated using independent datasets.22 Using the TCGA CN-AML data, we identified that compared with higher STAB1 expression, lower STAB1 expression was associated with better survival in patients with CN-AML; furthermore, we validated our findings using three other independent datasets. The GSE71014 dataset mainly includes Asians,23 whereas the other four cohorts (TCGA and the three GEO datasets) mainly include Europeans, indicating the universality of STAB1 as a marker of poor prognosis in patients with CN-AML. Although age, FAB classification, mutations in FLT3, NPM1, and IDH2, and EFS and OS status may affect the survival of patients with CN-AML, we demonstrated STAB1 expression to be an independent prognostic factor in two CN-AML datasets. The potential ease of testing of STAB1 adds to its advantages for clinical applications.
Most previous studies have not explored the potential molecular mechanisms underlying prognostic genes and have not justified the regulatory mechanisms underlying leukemogenesis. In this study, we systematically surveyed genes and miRNAs with differential STAB1 expression. Gene network analysis of STAB1 using the GO and KEGG pathway databases demonstrated that genes in the STAB1high group tended to be associated with leukemogenesis. However, the detailed mechanisms of action of STAB1 in AML leukemogenesis require further studies.
Cancer drugs and inhibitors are being increasingly designed to target specific genes, and these drugs may be used on the basis of the gene expression patterns of cancers.24 On the basis of gene expression, we constructed gene networks combined with drug-specific analyses and investigated the relationships between inhibitor drugs and targets to identify potential drug indicators based on STAB1 expression stratification. STAB1 expression stratification is a potentially valuable strategy for treating patients with CN-AML. Small-molecule inhibitor drugs offer a promising alternative to existing treatment for patients with CN-AML.25 The correlation between STAB1 expression and the gene targets of FDA-approved drugs for various cancers indicates a potential for drug repurposing, which is a promising approach for developing treatment for patients in the STAB1low group. Although many FDA-approved drugs have not yet been applied for AML treatment, they may be clinically beneficial as part of a treatment strategy in patients with CN-AML.
Our study has some limitations. First, the study may be biased due to the presence of confounding factors. However, factors that could influence the results were controlled as much as possible using multivariate analyses; it is therefore unlikely that the survival benefit associated with low STAB1 expression was an artifact. Second, because it is difficult to track patients and obtain clinical samples, we downloaded data from public datasets and validated our findings in cell lines. Third, the mRNA of STAB1 is 7,700-bp long, making constructing plasmids for STAB1 overexpression difficult. Thus, the replenishment experiment was not implemented in this study.
In summary, our study is the first to provide evidence that low STAB1 expression is associated with better outcomes in patients with CN-AML, even after adjusting for known clinical factors. Multiple datasets were used to assess the consistency of our findings. STAB1 overexpression may be a valuable new marker for risk stratification in patients with CN-AML. In addition, drug-specific analyses were applied to identify potential drug indicators based on STAB1 expression stratification. We validated in vitro the suppression of leukemia growth and induction of apoptosis by STAB1 and showed that compared with the STAB1high group, the STAB1low group is more sensitive to venetoclax.
Methods
Clinical Patient Information
We downloaded sequencing data (normalized at level-2 intensity) and clinical information of 79 patients with CN-AML from the TCGA LAML dataset (https://tcga-data.nci.nih.gov/tcga/). We also obtained four CN-AML microarray datasets: GSE12417 test dataset (GSE12417A; n = 79),8 GSE71014 (n = 104),23 GSE12417 training dataset (GSE12417B; n = 163),8 and GSE6891 (n = 187).26 The datasets were normalized as described by Metzeler et al.,8 and the OS data were obtained from their publications and the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The GSE12417 dataset includes a test (referred to as GSE12417A herein) and training (referred to as GSE12417B herein) dataset. Based on clinical experience, the 79 TCGA patients with CN-AML from the TCGA LAML dataset were further classified into two groups on the basis of 2-year OS: OS <2 years (n = 47) and >2 years (n = 32).
Survival Analysis
In the two CN-AML cohorts, TCGA and GSE6891, the association between STAB1 expression and clinical factors, such as age and FLT3 mutations, were analyzed using Fisher’s exact test. High or low STAB1 expression was defined as the median expression level of all CN-AML samples in that cohort. OS was defined as the time from AML diagnosis to death due to any cause or the last clinical follow-up. The EFS was defined as the time from AML diagnosis to an event or the last follow-up. Clinical factors, such as age, sex, WBC counts, mutation status, and FAB classification, were assessed using univariate analyses with the Kaplan-Meier method using GraphPad Prism (https://www.graphpad.com) software. Variables with p < 0.1 were retained for further analysis. After univariate analyses, the significant prognostic factors were combined using multivariate analyses. For multivariate analyses, the multivariable Cox proportional hazards model was used to assess the association between STAB1 expression and OS or EFS in the presence of other known clinical factors such as age, sex, WBC counts, mutation status, and FAB classification. These analyses were performed using the R survival package.27 Hazard ratios with relative 95% confidence intervals were shown in multivariate analyses.
Differential Expression Analysis
Patients with STAB1 expression values greater than the median were classified as the high STAB1 group (STAB1high), whereas the others were considered as the low STAB1 group (STAB1low). The R survival package DESeq28 was used to conduct differential expression analysis with the thresholds false discovery rate (FDR) <0.05 and |fold change| >1.5 between the STAB1high and STAB1low groups. An expression heatmap was constructed and clustering was performed using the MultiExperiment Viewer (MeV) software (http://en.bio-soft.net/chip/MeV.html). Functional enrichment annotations for GO and KEGG pathways were analyzed using the Database for Annotation, Visualization and Integrated Discovery online tool (https://david.ncifcrf.gov/). To further infer the regulatory role of miRNA, DIANA-miRPath v.3.029 and miRNA path30 were used to identify miRNA-regulating pathways.
Construction of the Regulatory Network in CN-AML
We applied the method described in our review31 to merge the predicted and experimentally verified miRNA and TF targets32 (http://bioinfo.life.hust.edu.cn/hTFtarget/). miRNA-gene/TF and TF-gene/miRNA regulatory relations were clearly identified for differentially expressed genes and miRNAs in different CN-AML subgroups. Using in-house-developed scripts, we subsequently identified miRNA-TF-gene feed-forward loops and miRNA-TF feedback loops based on their regulation. These networks were visualized using Cytoscape v.3.5 (https://cytoscape.org/).
Correlation Analysis
We identified the standard AML drugs applied in clinical practice and obtained their drug targets from DrugBank (https://www.drugbank.ca/). Furthermore, we identified genes sensitive or resistant to cytarabine as well as MDR genes from the literature. The target genes of FDA-approved cancer drugs for 23 cancer types were identified using DrugBank.33 Coexpression analysis was performed to identify the expression correlation between drug targets and STAB1. Genes with p < 0.05 were selected for further analysis conducted using the R survival package.
Cell Culture, Transfection with siRNA, and RNA Expression Quantification
KG1a, NB4, THP-1, K562, MOML13, HL60, and U937 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 100 U/mL ampicillin, and incubated in a humidified atmosphere containing 5% CO2 and 95% air at 37°C. For transfection, the cultured cell lines were seeded in six-well plates and transfected with 100 nmol/L STAB1 siRNAs or control oligonucleotides (NC) (RiboBio, Guangzhou, China). The sequences for si-STAB1-1, si-STAB1-2, and si-STAB1-3 were 5′-GGATCGTCTTCTACAACCA-3′, 5′-AGATCACCGTCACCTTTAA-3′, and 5′-GGAACAATGGTCACTTGTA-3′, respectively.
After 48 h of transfection, total RNA was extracted from KG1a cells using RNAiso (Takara) following the manufacturer’s protocol, and 1 μg of the RNA was reverse transcribed using PrimeScript RT Master Mix (Takara). Quantitative real-time PCR (real-time qPCR) of STAB1 was performed using the cDNA as a template in the presence of SYBR Premix Ex Taq (Tli RNaseH Plus; Takara). The levels of mRNA were normalized to the level of actin.
Western Blotting Analysis
The KG1a and NB4 cells were seeded into six-well culture plates, and the cells were then treated with 100 nmol/L STAB1 siRNA and NC and cultured for 48 h. Subsequently, total protein was extracted from the cells, separated using sodium dodecyl sulfate-polyacrylamide-gel electrophoresis, and transferred onto a polyvinylidene fluoride membrane. A gel imaging analysis system (Bio-Rad) was used to detect protein bands. After incubation with secondary antibodies, the membranes were visualized using the software Quantity One (Bio-Rad). β-Actin was used as the standard internal reference. STAB1 antibody was purchased from Bio-Techne.
Assays for Cell Proliferation, Apoptosis, and Drug Treatment
Cells transfected with the siRNA and NC were seeded into 96-well plates (5 × 104 cells/well) in medium containing 10% FBS and cultured for 24, 48, and 72 h. Cell viability testing was performed using the Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan), following the manufacturer’s instructions. For apoptosis assays, the cells were washed with PBS and resuspended in 100 μL of binding buffer containing 5 μL of annexin V and 5 μL of propidium iodide (BD Pharmingen). After incubation for 20 min, fluorescence was quantified by flow cytometry on a FACSCalibur system. Venetoclax was purchased from Selleck (https://www.selleck.cn/). KG1a and NB4 cells were incubated for appropriate durations in RPMI 1640 medium supplemented with 10% FBS and titrated concentrations of venetoclax. Cell viability was assessed at an absorbance of 450 nm using a PerkinElmer EnSpire 2300 multimode plate reader.
Author Contributions
S.-Y.L. analyzed the data and wrote the manuscript. F.-F.H. revised manuscript. F.-F.H., Y.-R.M., H.H., and Q.Z. participated in data analysis. Q.L. offered guidance on laboratory technique. Z.C., Q.L., and H.W. provided specific knowledge on AML. A.-Y.G. and Z.C. designed the study and revised the manuscript. All authors read and approved the final manuscript and submission.
Conflict of interests
The authors declare no competing interests.
Acknowledgments
We thank Dr. Peter J.M. Valk at Erasmus University Medical Center for providing clinical information on the GSE6891 dataset and members working for the TCGA AML group. This work was supported by the National Natural Science Foundation of China (NSFC grants 31822030 and 31771458) and the National Key Research and Development Program of China (grant 2017YFA0700403).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.09.014.
Contributor Information
Zhichao Chen, Email: chenzhichao@hust.edu.cn.
An-Yuan Guo, Email: guoay@hust.edu.cn.
Supplemental Information
References
- 1.Döhner H., Weisdorf D.J., Bloomfield C.D. Acute myeloid leukemia. N. Engl. J. Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr., Laird P.W., Baty J.D., Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Döhner H., Estey E., Grimwade D., Amadori S., Appelbaum F.R., Büchner T., Dombret H., Ebert B.L., Fenaux P., Larson R.A. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel J.P., Gönen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., Van Vlierberghe P., Dolgalev I., Thomas S., Aminova O. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann L., Miething C., Maurer U., Brieger J., Karakas T., Weidmann E., Hoelzer D. High levels of Wilms’ tumor gene (wt1) mRNA in acute myeloid leukemias are associated with a worse long-term outcome. Blood. 1997;90:1217–1225. [PubMed] [Google Scholar]
- 6.Langer C., Radmacher M.D., Ruppert A.S., Whitman S.P., Paschka P., Mrózek K., Baldus C.D., Vukosavljevic T., Liu C.G., Ross M.E., Cancer and Leukemia Group B (CALGB) High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: a Cancer and Leukemia Group B (CALGB) study. Blood. 2008;111:5371–5379. doi: 10.1182/blood-2007-11-124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcucci G., Baldus C.D., Ruppert A.S., Radmacher M.D., Mrózek K., Whitman S.P., Kolitz J.E., Edwards C.G., Vardiman J.W., Powell B.L. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J. Clin. Oncol. 2005;23:9234–9242. doi: 10.1200/JCO.2005.03.6137. [DOI] [PubMed] [Google Scholar]
- 8.Metzeler K.H., Hummel M., Bloomfield C.D., Spiekermann K., Braess J., Sauerland M.C., Heinecke A., Radmacher M., Marcucci G., Whitman S.P., Cancer and Leukemia Group B. German AML Cooperative Group An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzeler K.H., Maharry K., Kohlschmidt J., Volinia S., Mrózek K., Becker H., Nicolet D., Whitman S.P., Mendler J.H., Schwind S. A stem cell-like gene expression signature associates with inferior outcomes and a distinct microRNA expression profile in adults with primary cytogenetically normal acute myeloid leukemia. Leukemia. 2013;27:2023–2031. doi: 10.1038/leu.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damm F., Heuser M., Morgan M., Wagner K., Görlich K., Grosshennig A., Hamwi I., Thol F., Surdziel E., Fiedler W. Integrative prognostic risk score in acute myeloid leukemia with normal karyotype. Blood. 2011;117:4561–4568. doi: 10.1182/blood-2010-08-303479. [DOI] [PubMed] [Google Scholar]
- 11.Kzhyshkowska J. Multifunctional receptor stabilin-1 in homeostasis and disease. ScientificWorldJournal. 2010;10:2039–2053. doi: 10.1100/tsw.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brekken R.A., Puolakkainen P., Graves D.C., Workman G., Lubkin S.R., Sage E.H. Enhanced growth of tumors in SPARC null mice is associated with changes in the ECM. J. Clin. Invest. 2003;111:487–495. doi: 10.1172/JCI16804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chlenski A., Guerrero L.J., Peddinti R., Spitz J.A., Leonhardt P.T., Yang Q., Tian Y., Salwen H.R., Cohn S.L. Anti-angiogenic SPARC peptides inhibit progression of neuroblastoma tumors. Mol. Cancer. 2010;9:138. doi: 10.1186/1476-4598-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karikoski M., Marttila-Ichihara F., Elima K., Rantakari P., Hollmén M., Kelkka T., Gerke H., Huovinen V., Irjala H., Holmdahl R. Clever-1/stabilin-1 controls cancer growth and metastasis. Clin. Cancer Res. 2014;20:6452–6464. doi: 10.1158/1078-0432.CCR-14-1236. [DOI] [PubMed] [Google Scholar]
- 15.Yu X., Guo C., Fisher P.B., Subjeck J.R., Wang X.Y. Scavenger receptors: emerging roles in cancer biology and immunology. Adv. Cancer Res. 2015;128:309–364. doi: 10.1016/bs.acr.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng H., Lal K., Yang F.F., Chen J. The pathological role and prognostic impact of miR-181 in acute myeloid leukemia. Cancer Genet. 2015;208:225–229. doi: 10.1016/j.cancergen.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garzon R., Volinia S., Liu C.G., Fernandez-Cymering C., Palumbo T., Pichiorri F., Fabbri M., Coombes K., Alder H., Nakamura T. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes J.D., Wolf C.R. Molecular mechanisms of drug resistance. Biochem. J. 1990;272:281–295. doi: 10.1042/bj2720281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Younes A., Gopal A.K., Smith S.E., Ansell S.M., Rosenblatt J.D., Savage K.J., Ramchandren R., Bartlett N.L., Cheson B.D., de Vos S. Results of a pivotal phase II study of brentuximab vedotin for patients with relapsed or refractory Hodgkin’s lymphoma. J. Clin. Oncol. 2012;30:2183–2189. doi: 10.1200/JCO.2011.38.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souers A.J., Leverson J.D., Boghaert E.R., Ackler S.L., Catron N.D., Chen J., Dayton B.D., Ding H., Enschede S.H., Fairbrother W.J. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 21.Wei A.H., Tiong I.S. Midostaurin, enasidenib, CPX-351, gemtuzumab ozogamicin, and venetoclax bring new hope to AML. Blood. 2017;130:2469–2474. doi: 10.1182/blood-2017-08-784066. [DOI] [PubMed] [Google Scholar]
- 22.Wang M., Lindberg J., Klevebring D., Nilsson C., Mer A.S., Rantalainen M., Lehmann S., Grönberg H. Validation of risk stratification models in acute myeloid leukemia using sequencing-based molecular profiling. Leukemia. 2017;31:2029–2036. doi: 10.1038/leu.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuang M.K., Chiu Y.C., Chou W.C., Hou H.A., Tseng M.H., Kuo Y.Y., Chen Y., Chuang E.Y., Tien H.F. An mRNA expression signature for prognostication in de novo acute myeloid leukemia patients with normal karyotype. Oncotarget. 2015;6:39098–39110. doi: 10.18632/oncotarget.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Downward J. Cancer biology: signatures guide drug choice. Nature. 2006;439:274–275. doi: 10.1038/439274a. [DOI] [PubMed] [Google Scholar]
- 25.Ramamoorthy K., Ramesh P., Al Bahar S. Primary treatment of acute myeloid leukemia (non M3) in elderly: a review. Gulf J. Oncolog. 2008;4:19–26. [PubMed] [Google Scholar]
- 26.Verhaak R.G., Wouters B.J., Erpelinck C.A., Abbas S., Beverloo H.B., Lugthart S., Löwenberg B., Delwel R., Valk P.J. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin H., Zelterman D. Modeling survival data: extending the Cox model. Technometrics. 2000;44:85–86. [Google Scholar]
- 28.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maragkakis M., Reczko M., Simossis V.A., Alexiou P., Papadopoulos G.L., Dalamagas T., Giannopoulos G., Goumas G., Koukis E., Kourtis K. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Z., Liu T., Huang W., Liu H., Zhang H.M., Li Q., Chen Z., Guo A.Y. MicroRNA regulatory pathway analysis identifies miR-142-5p as a negative regulator of TGF-β pathway via targeting SMAD3. Oncotarget. 2016;7:71504–71513. doi: 10.18632/oncotarget.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H.M., Kuang S., Xiong X., Gao T., Liu C., Guo A.Y. Transcription factor and microRNA co-regulatory loops: important regulatory motifs in biological processes and diseases. Brief. Bioinform. 2015;16:45–58. doi: 10.1093/bib/bbt085. [DOI] [PubMed] [Google Scholar]
- 32.Hu H., Miao Y.R., Jia L.H., Yu Q.Y., Zhang Q., Guo A.Y. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019;47(D1):D33–D38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.