Abstract
Streptococcus pyogenes is an exclusively human pathogen causing a wide range of clinical manifestations from mild superficial infections to severe, life-threatening, invasive diseases. S. pyogenes is consistently susceptible toward penicillin, but therapeutic failure of penicillin treatment has been reported frequently. At the same time, streptococcal resistance to alternative antibiotics, e.g., macrolides, is common. To reduce the application of antibiotics for treatment of S. pyogenes infections, it is mandatory to develop novel therapeutic strategies. Antisense peptide nucleic acids (PNAs) are synthetic DNA derivatives widely applied for hybridization-based microbial diagnostics. They have a high potential as therapeutic agents, because PNA antisense targeting of essential genes was shown to reduce growth of several pathogenic bacterial species. Spontaneous cellular uptake of PNAs is restricted in eukaryotes and in bacteria. To overcome this problem, PNAs can be coupled to cell-penetrating peptides (CPPs) that support PNA translocation over the cell membrane. In bacteria, the efficiency of CPP-mediated PNA uptake is species specific. Previously, HIV-1 transactivator of transcription (HIV-1 TAT) peptide-coupled anti-gyrA PNA was shown to inhibit growth of S. pyogenes. Here, we investigate the effect of 18 CPP-coupled anti-gyrA PNAs on S. pyogenes growth and virulence. HIV-1 TAT, oligolysine (K8), and (RXR)4XB peptide-coupled anti-gyrA PNAs efficiently abolished bacterial growth in vitro. Consistently, treatment with these three CPP-PNAs increased survival of larvae in a Galleria mellonella infection model.
Keywords: Peptide Nucleic Acids, Antisense therapy, Cell Penetrating Peptides, Streptococcus pyogenes
Introduction
Streptococcus pyogenes (group A streptococcus [GAS]) is a Gram-positive, exclusively human pathogen responsible for a variety of diseases ranging from mild self-limiting superficial infections of the throat or skin to life-threatening invasive diseases, including bacteremia and necrotizing fasciitis. The global burden of streptococcal infections is high, with 18 million invasive infections per year and 500,000 deaths.1 The impact of GAS diseases is especially high in resource-limited settings, and a rise of global invasive disease burden caused by GAS has been reported recently.2 Untreated superficial infections often lead to the development of severe invasive infections or autoimmune sequelae.1, 3
To date, penicillin is the standard treatment of streptococcal pharyngitis, because GAS is invariably susceptible toward penicillin. Macrolides are recommended as alternate antibiotics for the treatment of S. pyogenes infections in patients who are allergic to β-lactams or in cases of penicillin failure.4 Resistance rates to macrolides in the United States have remained relatively low.5 In contrast, a rise of macrolide resistance in S. pyogenes has been observed in Europe, followed by a decrease in erythromycin resistance in several European countries.6 Today, a major goal of public health is to limit the application and distribution of antibiotics. One possible strategy is the application of antisense therapeutics targeting essential genes or antibiotic-resistance genes. Desired features of S. pyogenes-specific antimicrobials are a high specificity for the target gene, effective uptake into the bacterial cell, low unspecific toxicity, high stability, and—for the eradication of intracellular bacteria—import into eukaryotic cells. Antisense peptide nucleic acids (PNAs) potentially combine these properties and have been tested as antimicrobial agents in many bacterial species. PNAs are synthetic DNA derivatives, which bind sequence specific to DNA and RNA and are able to form stable duplexes and triplexes.7 The nucleic acid sugar-phosphate backbone is replaced by a pseudo-peptide backbone, resulting in a high chemical stability and resistance to nucleases and proteases.7, 8 Cellular uptake of PNAs is limited by bacterial membranes and cell walls. Coupling of PNAs to cell-penetrating peptides (CPPs) may facilitate PNA translocation into bacteria and thereby enhance antimicrobial efficiency. CPPs are naturally occurring or designed peptides that are able to penetrate cell membranes and have been used for the introduction of different kinds of cargo into eukaryotic cells and bacteria.9, 10 Typical examples of CPPs used in bacteria are the synthetic (KFF)3K and the HIV-1 transactivator of transcription (HIV-1 TAT)-derived peptides. (KFF)3K facilitated uptake of PNAs, among others, in Escherichia coli and Staphylococcus aureus.11, 12 HIV-1 TAT was able to penetrate Listeria monocytogenes, S. aureus, and S. epidermidis.13, 14 We observed previously that HIV-1 TAT-coupled anti-gyrA PNAs were able to inhibit growth in S. pyogenes.15 In this study, we tested anti-gyrA PNAs coupled to 18 different CPPs. We selected CPPs, which have been tested before as carrier molecules in eukaryotic cells and were known to exhibit low toxicity (Table 1). We found that HIV-1 TAT, oligolysine (K8), and (RXR)4XB-coupled anti-gyrA PNAs efficiently abolished growth of S. pyogenes in vitro. In a Galleria mellonella infection model, treatment of infected larvae with these CPP-PNAs increased survival.
Table 1.
CPP-PNA Anti-gyrA Conjugates for Antisense Studies in S. pyogenes
| CPP | CPP Sequence | CPP-PNA Designation | Reference |
|---|---|---|---|
| Antennapedia homeodomain (Penetratin) | RQIKIWFQNRRMKWKK | Ant-anti-gyrA PNA | 16 |
| Ant-anti-gyrA scPNA | |||
| ELA | Dansyl-G-C-ELALE LALEALEAALELA | ELA-anti-gyrA PNA | 17 |
| ELA-anti-gyrA scPNA | |||
| HIV-1 TAT (48–57) | GRKKRRQRRRYK | TAT-anti-gyrA PNA | 18 |
| TAT-anti-gyrA scPNA | |||
| Human calcitonin | LGTYQDFNKFHTFPQTAIGVGAP | hCalcitonin-anti-gyrA PNA | 19 |
| hCalcitonin-anti-gyrA scPNA | |||
| (KFF)3K | KFFKFFKFFK | (KFF)3K-anti-gyrA PNA | 20 |
| (KFF)3K-anti-gyrA scPNA | |||
| MAP | KLALKLALKALKAALKLA | MAP-anti-gyrA PNA | 21, 22 |
| MAP-anti-gyrA scPNA | |||
| mVE-cadherin (pVEC) | LLIILRRRIRKQAHAHSK | mVE-cadherin-anti-gyrA PNA | 21, 22 |
| mVE-cadherin-anti-gyrA scPNA | |||
| M918 | MVTVLFRRLRIRRACGPPRVRV | M918-anti-gyrA PNA | 23 |
| M918-anti-gyrA scPNA | |||
| Oligoarginin (R6) | RRRRRR | R6-anti-gyrA PNA | 21, 24, 25 |
| R6-anti-gyrA scPNA | |||
| Oligoarginin (R10) | RRRRRRRRRR | R10-anti-gyrA PNA | |
| R10-anti-gyrA scPNA | |||
| Oligoleucine (L6) | LLLLLL | L6-anti-gyrA PNA | this work |
| L6-anti-gyrA scPNA | |||
| Oligolysin (K8) | KKKKKKKK | K8-anti-gyrA PNA | 26 |
| K8-anti-gyrA scPNA | |||
| PDESTK | PDESTK | PDESTK-anti-gyrA PNA | 27 |
| PDESTK-anti-gyrA scPNA | |||
| TLM | PLSSIFSRIGDP | TLM-anti-gyrA PNA | 28 |
| TLM-anti-gyrA scPNA | |||
| TP10 | AGYLLGKINLKALAALAKKIL | TP10-anti-gyrA PNA | 29 |
| TP10-anti-gyrA scPNA | |||
| Transportan | GWTLNSAGYLLGKINLKALAALAKKIL | Transportan-anti-gyrA PNA | 30 |
| Transportan-anti-gyrA scPNA | |||
| VT5 | DPKGDPKGVTVTVTVTVTGKGDPKPD | VT5-anti-gyrA PNA | 31 |
| VT5-anti-gyrA scPNA | |||
| (RXR)4XB | RXRRXRRXRRXRXB | (RXR)4XB-anti-gyrA PNA | 32 |
| (RXR)4XB-anti-gyrA scPNA |
Results
Design of CPP-Coupled Anti-gyrA PNAs Specific for S. pyogenes
In a previous study, we observed antimicrobial effects of peptide-coupled anti-gyrA antisense PNAs specific for S. pyogenes.15 Growth inhibition by this construct was caused by antisense targeting of the essential gene gyrA. Its gene product represents the subunit A of the DNA topoisomerase gyrase, which is involved in replication and is thus required for bacterial growth. Since carrier molecules show a species-specific influence on cargo uptake, we wanted to explore the effect of a variety of CPPs coupled to anti-gyrA antisense PNAs on S. pyogenes (Table 1). Peptides were coupled to PNAs via a flexible ethyleneglycol linker (8-amino-3, 6-dioxaoctanoic acid). The sequence of anti-gyrA antisense PNAs was tgcatttaag-NH2, covering gyrA −5 to 5. The sequence of the corresponding control PNAs (scrambled PNAs [scPNAs]) was attagactgt-NH2. scPNAs were composed of the same base pairs as the antisense PNAs in a randomized order.
Antimicrobial Effect of CPP-Coupled Anti-gyrA PNAs on S. pyogenes
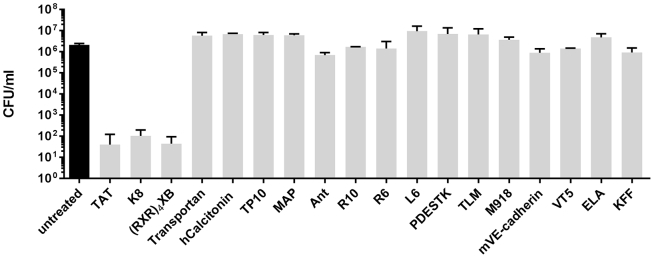
To determine the impact of different CPPs on the efficacy of antisense PNAs targeting S. pyogenes, a prescreening approach was performed. S. pyogenes M49 strain 591 was incubated for 6 h with 10 μM CPP-anti-gyrA PNA conjugates. Reduction of bacterial counts caused by different CPP-coupled antisense PNAs compared with an untreated control was determined. From 18 CPP-antisense PNA conjugates, three showed an antimicrobial effect in this assay: TAT-anti-gyrA PNA, K8-anti-gyrA PNA, and (RXR)4XB-anti-gyrA PNA (Figure 1). Similar results were obtained with S. pyogenes M1 strain AP1, with the exception of K8-anti-gyrA PNAs, which did not show any antimicrobial effect in AP1 (data not shown; Figure 3B).
Figure 1.
Reduction of the Bacterial Count following Treatment of S. pyogenes 591 (M49) with 10 μM CPP-Anti-gyrA PNAs for 6 h
PNA conjugates are indicated by the name of the respective CPP. Data are presented as mean values and SD. The experiment was performed twice.
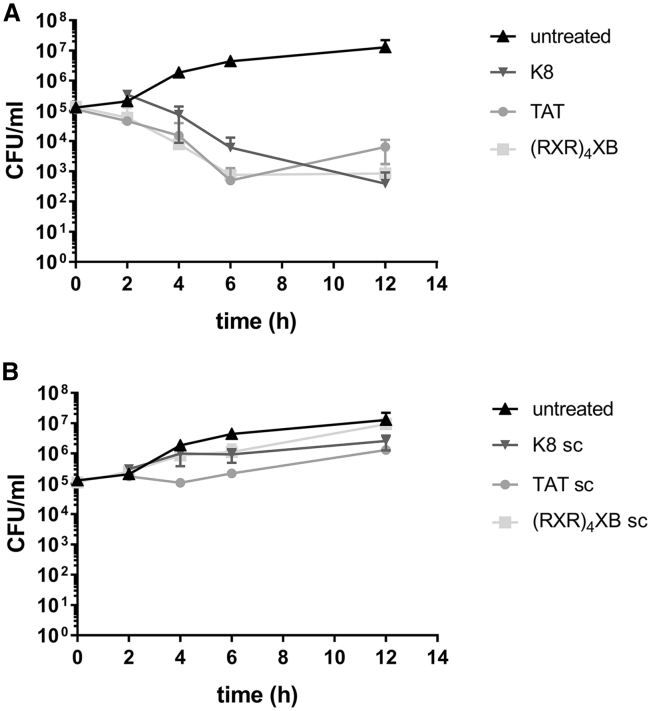
Figure 3.
Killing Kinetics of CPP-Anti-gyrA PNA Treatment
(A) Bacterial counts following treatment of S. pyogenes 591 (M49) with CPP-anti-gyrA PNAs. (B) Bacterial counts following treatment of S. pyogenes 591 (M49) with CPP-anti-gyrA scPNAs. PNA conjugates are indicated by the name of the respective CPP. Scrambled PNA controls are indicated by sc. Data are presented as mean values and SD. Sample size: n ≥ 3.
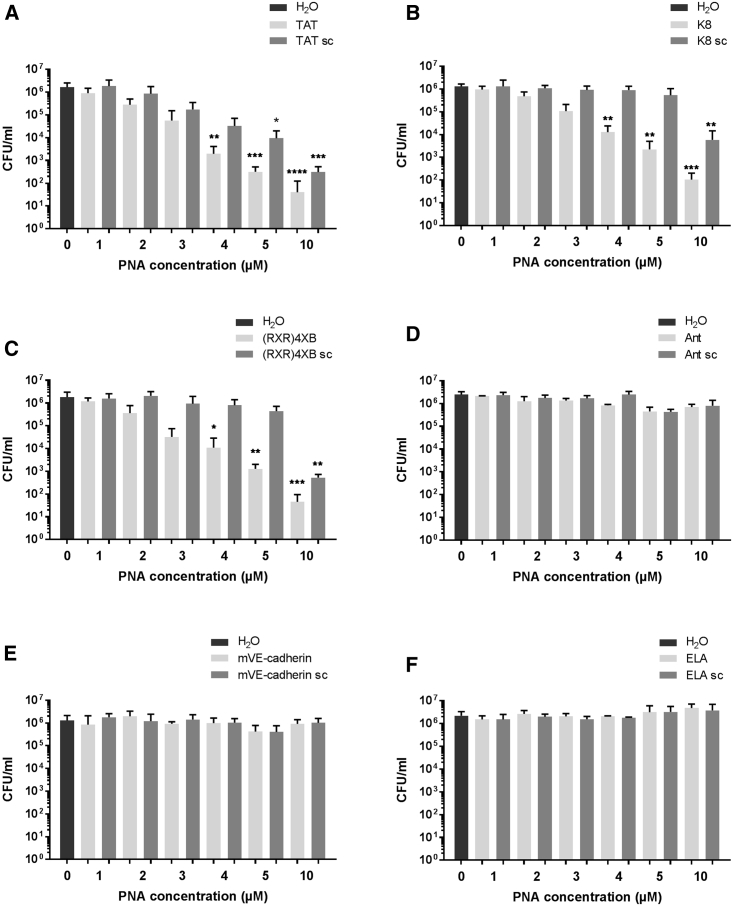
Six CPP-anti-gyrA PNA conjugates were selected for further analyses: three constructs that showed antimicrobial activity in the pilot experiment and three constructs that did not show any effect. Concentration-dependent bactericidal activity was investigated by treatment of S. pyogenes in a CPP-anti-gyrA PNA conjugate concentration range from 1 to 10 μM (Figure 2). Reduction of bacterial counts was observed following incubation of S. pyogenes with TAT-anti-gyrA PNA (Figure 2A), K8-anti-gyrA PNA (Figure 2B), and (RXR)4XB-anti-gyrA PNA (Figure 2C), respectively. Colony-forming units (CFU) per milliliter in treated samples were significantly reduced compared with the untreated control sample in a concentration range from 4 to 10 μM PNA. TAT-anti-gyrA scPNA caused a significant reduction of CFU per milliliter following treatment with 5 and 10 μM scPNA, hinting toward a toxic effect of TAT CPP at higher concentrations (Figure 2A). K8-anti-gyrA scPNA and (RXR)4XB-anti-gyrA scPNA showed a significant reduction of bacterial counts following treatment with 10 μM scPNA (Figures 2B and 2C). In contrast, no reduction of CFU per milliliter was observed following incubation with an Antennapedia homeodomain-derived CPP (Ant)-anti-gyrA PNA, ELA-anti-gyrA PNA, mVE-cadherin-anti-gyrA PNA, and the corresponding scrambled control CPP-PNAs (Figures 2D–2F).
Figure 2.
Concentration-Dependent Reduction of the Bacterial Counts following Treatment of S. pyogenes 591 (M49) with CPP-Anti-gyrA PNAs for 6 h
PNA conjugates are indicated by the name of the respective CPP: TAT (A), K8 (B), (RXR)4XB (C), Ant (D), mVE-cadherin (E), and ELA (F). Scrambled PNA controls are indicated by sc. Data are presented as mean values and SD. Statistical significance was determined using the Kruskal-Wallis test. Differences between PNA conjugate samples and the mock control (H2O) were expressed as *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. Sample size: n = 5 (A–C); n = 3 (D–F).
Minimum Inhibitory Concentration
We determined the minimum inhibitory concentration (MIC) of the CPP-antisense PNAs that showed antimicrobial activity in the kill assay (Table 2). K8-anti-gyrA PNA and TAT-anti-gyrA PNA showed the lowest MIC at 15.6 μM. (RXR)4XB-anti-gyrA PNA was less effective with a MIC of 62.5 μM. All scPNA controls showed a lower antimicrobial activity than the corresponding antisense constructs.
Table 2.
MIC of CPP-PNA Anti-gyrA Conjugates
| CPP-PNA | MIC (μM) |
|---|---|
| K8-anti-gyrA PNA | 15.6 |
| K8-anti-gyrA scPNA | 62.5 |
| TAT-anti-gyrA PNA | 15.6 |
| TAT-anti-gyrA scPNA | 62.5 |
| (RXR)4XB-anti-gyrA PNA | 62.5 |
| (RXR)4XB-anti-gyrA scPNA | 125 |
Bactericidal Kinetics of CPP-Coupled Anti-gyrA PNAs
To monitor reduction of bacterial counts over the course of the experiment, S. pyogenes was treated with 5 μM CPP-antisense PNAs (Figure 3A) or scrambled control PNAs (Figure 3B). Samples were taken at time points 2, 4, 6, and 12 h following antisense treatment. CFUs per milliliter were determined by plating of serial dilutions. During the course of the experiment, no complete clearance was achieved.
Susceptibility of Different S. pyogenes Isolates to CPP-Anti-gyrA PNAs
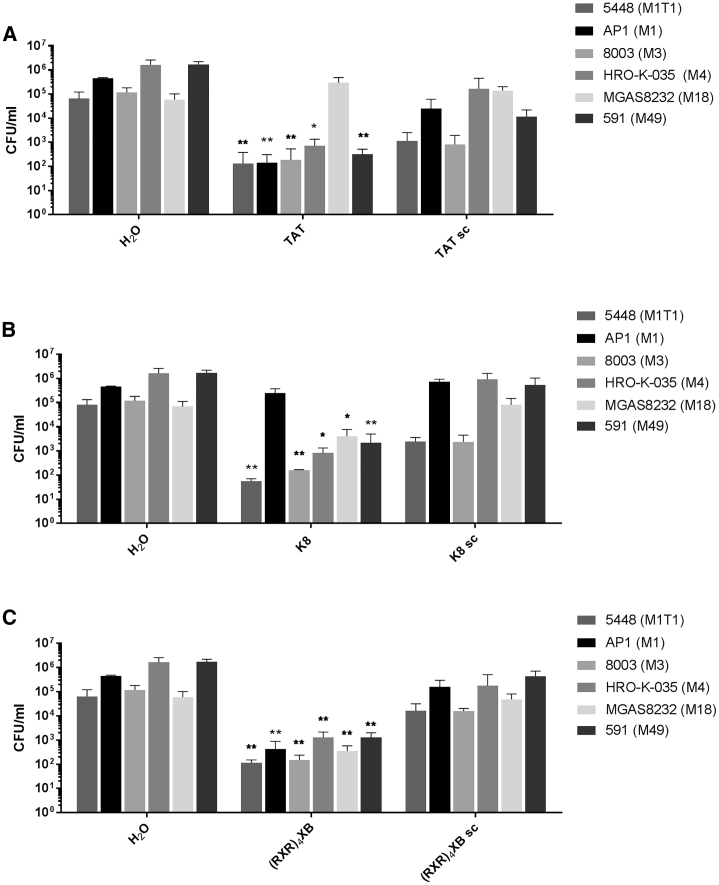
We determined the antimicrobial effect of CPP-PNA conjugates on different S. pyogenes isolates representing distinct M serotypes of epidemiological relevance. Bacterial strains were treated with 5 μM CPP-PNA constructs.
Samples were collected after 6 h, and bacterial counts were analyzed (Figure 4). TAT-anti-gyrA PNA exhibited antimicrobial activity against all strains except MGAS8232 (M18). K8-anti-gyrA PNA was effective toward all strains with the exception of AP1 (M1). In contrast, (RXR)4XB-anti-gyrA PNA was able to reduce bacterial counts of all strains tested in this experiment.
Figure 4.
Susceptibility of Different S. pyogenes Isolates to CPP-Anti-gyrA PNA Treatment
Reduction of the bacterial count following treatment of different S. pyogenes strains with 5 μM CPP-anti-gyrA PNAs. PNA conjugates are indicated by the name of the respective CPP. Scrambled PNA controls are indicated by sc. (A) TAT-anti-gyrA PNA and TAT-anti-gyrA scPNA. (B) K8-anti-gyrA PNA and K8-anti-gyrA scPNA. (C) (RXR)4XB-anti-gyrA PNA and (RXR)4XB-anti-gyrA scPNA. Data are presented as mean values and SD. Statistical significance was determined using the Kruskal-Wallis test. Differences between PNA conjugate samples and the corresponding mock control (H2O) were expressed as *p ≤ 0.05; **p ≤ 0.01. Sample size: n = 5.
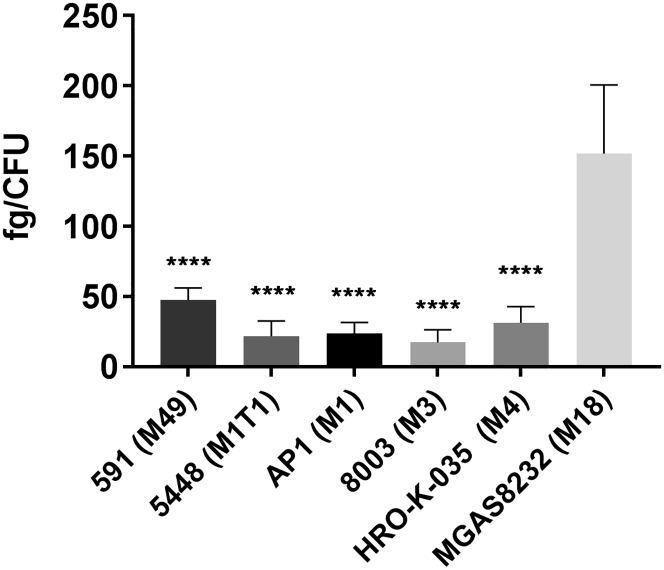
Hyaluronic Acid Content in Different S. pyogenes Isolates
To determine whether differential capsule production was correlated to sensitivity toward CPP-PNA conjugates, hyaluronic acid (HA) was extracted from S. pyogenes strains. HA content of MGAS8232 (M18) was significantly higher than in all other isolates tested (Figure 5). Since TAT-anti-gyrA PNAs were not effective in MGAS8232 (M18) (Figure 4A), this result indicates that HA represents a barrier for TAT-antisense PNAs but neither for K8-antisense PNAs nor for (RXR)4XB-antisense PNAs.
Figure 5.
Hyaluronic Acid Content of Different S. pyogenes Serotypes
Statistical significance was determined using one-way ANOVA/Tukey’s multiple comparisons test. Data are presented as mean values and SD. Differences between S. pyogenes serotypes and S. pyogenes MGAS8232 were expressed as ****p ≤ 0.0001. Sample size: n = 5.
CPP-Anti-gyrA PNAs Affect the Abundance of Target Gene Transcripts in S. pyogenes
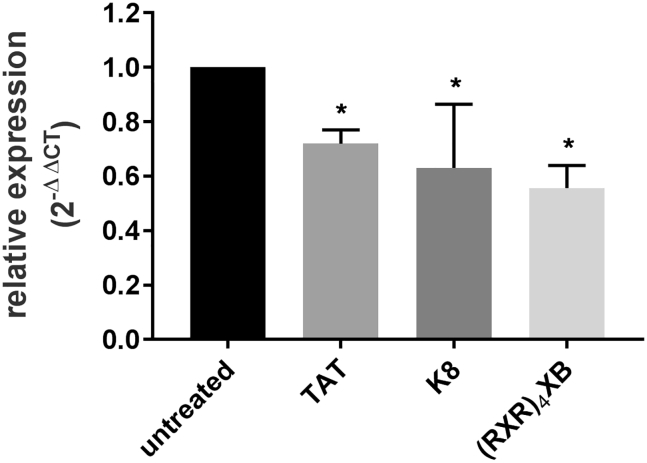
The influence of S. pyogenes treatment with CPP-anti-gyrA PNAs on the amount of gyrA mRNA was investigated by reverse transcription, followed by quantitative real-time PCR (Figure 6). Bacteria were treated with a sublethal dose of CPP-PNA conjugates.
Figure 6.
Relative Expression of gyrA following Treatment with CPP-Antisense PNAs
Reduction of the transcript level was observed following treatment of S. pyogenes strains with 2 μM CPP-anti-gyrA PNAs. PNA conjugates are indicated by the name of the respective CPP. Data are presented as mean values and SD. Statistical significance was determined using the Mann-Whitney U-test. Differences between PNA conjugate samples and the untreated control were expressed as *p ≤ 0.05. Sample size: n = 3.
Following incubation, total RNA was extracted, and qRT-PCR was performed. Transcript abundance of the 5S RNA gene was used for normalization. The gyrA mRNA level in mock-treated S. pyogenes samples served as control. Treatment with 2 μM TAT-anti-gyrA PNA, K8-anti-gyrA PNA, and (RXR)4XB-anti-gyrA PNA led to a significant reduction of gyrA transcript compared with the untreated control sample (Figure 6). The gyrA mRNA level decreased to 70%, 60%, and 56%, respectively, of the amount detected in the mock-treated bacteria.
Evaluation of CPP-Antisense PNA Conjugates in a G. mellonella Infection Model
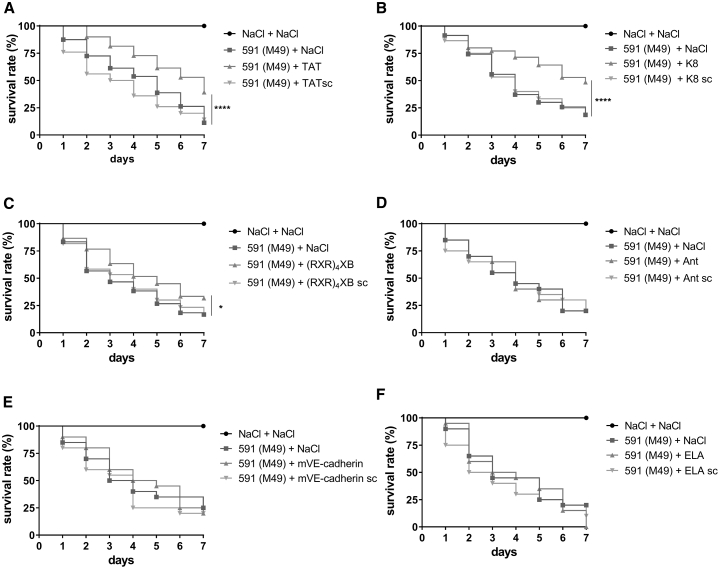
Antimicrobial efficiency of CPP-antisense PNA conjugates was evaluated in vivo, using a G. mellonella infection model. Larvae were infected with S. pyogenes strain 591 (M49) and treated with 4 nmol CPP-PNAs. Survival of larvae was observed over 7 days. We compared survival of larvae treated with CPP-anti-gyrA PNAs with mock-treated larvae. Larvae treated with TAT-anti-gyrA PNA, K8-anti-gyrA PNA, or (RXR)4XB-anti-gyrA PNA showed increased survival compared with mock-treated larvae (Figures 7A–7C). Ant-, mVE-cadherin-, and ELA-anti-gyrA PNAs that did not show antimicrobial effects in vitro did not affect survival of infected larvae (Figures 7D–7F).
Figure 7.
Survival of Galleria mellonella Larvae Treated with 4 nmol CPP-PNAs following Infection with S. pyogenes
PNA conjugates are indicated by the name of the respective CPP: TAT (A), K8 (B), (RXR)4XB (C), Ant (D), mVE-cadherin (E), and ELA (F). Scrambled PNA controls are indicated by sc. Statistical significance was determined using the log-rank test. Differences between curves were expressed as *p ≤ 0.05; ****p ≤ 0.0001. Sample size: n = 60 larvae per group (A–C); n = 20 larvae per group (D–F).
One promising therapeutic strategy is a combination of antisense agents with conventional antibiotics to reduce the concentration needed for efficient antibiotics treatment.
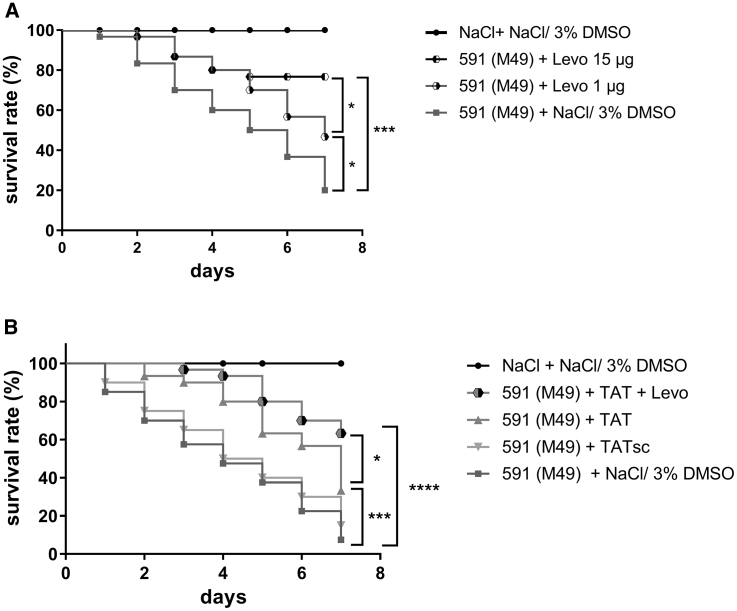
Previously, we observed that a combination of TAT-anti-gyrA PNA with antibiotics targeting gyrase subunits resulted in synergistic or additive antimicrobial effects on S. pyogenes in vitro.15 To test whether TAT-anti-gyrA PNA treatment could enhance antibiotics efficiency in vivo, we first treated infected G. mellonella larvae with 1 μg levofloxacin, which increased survival of infected larvae from 20% to 46% (Figure 8A). In combination with 4 nmol TAT-anti-gyrA PNA, survival was increased to 63% (Figure 8B). A comparable survival of infected larvae was achieved by application of 15 μg levofloxacin (Figure 8A). Combination of PNA and levofloxacin also increased survival of the infected larvae compared with TAT-anti-gyrA PNA treatment alone (33%) (Figure 8B).
Figure 8.
Survival of Galleria mellonella Larvae Treated with Levofloxacin Alone or in Combination with TAT-Anti-gyrA PNA following Infection with S. pyogenes
(A) Levo, levofloxacin. (B) TAT, 4 nmol TAT-anti-gyrA PNA. Scrambled PNA controls are indicated by sc. Statistical significance was determined using the log-rank test. Differences between curves were expressed as *p ≤ 0.05; ***p ≤ 0.001; ****p ≤ 0.0001. Sample size: n = 30 larvae per group.
Discussion
PNAs are nucleic acid derivatives with a variety of properties rendering them suitable as antisense molecules, including chemical and thermal stability, strong binding to DNA and RNA, reasonable solubility, a lack of immunogenicity, and low intracellular toxicity.33
However, poor delivery into target cells hampers application of PNAs as antisense therapeutics. In bacteria, the cell membrane, the bacterial cell wall, and extracellular surface structures, such as the lipopolysaccharide layer or the capsule, represent barriers limiting cellular uptake of PNAs. Treatment of intracellular pathogens poses an additional challenge, because PNAs have to be delivered into the host cell, escape the endosomal pathway, and finally penetrate the bacteria. One possible strategy to improve cellular uptake is PNA coupling to CPPs. The efficiency of a given CPP to enhance delivery is species specific. In Gram-negative bacteria, among others, (KFF)3K and HIV-1-TAT have been identified as useful carriers.34, 35 Growth of intracellular Salmonella enterica Serovar Typhimurium could be inhibited by (RXR)4XB-conjugated antisense peptide-phosphorodiamidate morpholino oligomers.36 In Gram-positive bacteria, (KFF)3K was efficient in S. aureus.12 Intracellular L. monocytogenes could be targeted with HIV-1-TAT- and (RXR)4XB-conjugated PNAs.14 In our previous study, we found that HIV-1-TAT-coupled anti-gyrA PNAs showed antimicrobial activity in S. pyogenes.15
The aim of this study was to compare the efficiency of potential carrier molecules delivering antisense PNAs in S. pyogenes. We tested 18 CPPs belonging to different classes. Three CPPs were shown to support uptake of anti-gyrA PNAs: the cationic CPPs K8 and HIV-1-TAT and the arginine-rich, amphipathic peptide (RXR)4XB. In general, basic residues support internalization of CPPs into cells, because their positive charge initiates interaction with the negatively charged surface. Specifically, it has been shown that arginine residues were more effective than lysines and that the replacement of lysine residues with arginine improved cellular uptake.26, 37 Here, we show that oligoarginine-coupled antisense PNAs were not able to inhibit S. pyogenes growth, whereas K8-conjugated anti-gyrA PNA showed an antimicrobial effect. In eukaryotic cells, insertion of 6-aminohexanoic acid (X) or β-alanine (B) residues into oligoarginine R8 decreased the cellular uptake but increased the splice-correction activity of the resulting compound.38 We observed that in contrast to R8, which did not function as a carrier in S. pyogenes, (RXR)4XB was able to mediate uptake of antisense PNAs. Penetratin (Ant) did not support antisense PNA uptake into S. pyogenes. This result is in accordance with an observation in L. monocytogenes. PNA uptake into L. monocytogenes was mediated by HIV-1-TAT and (RXR)4XB but not by Ant.14 PDSTK, a peptide derived from a PEST-like sequence from yeast, was able to support PNA antisense effects in S. aureus.12 We did not detect antimicrobial activity of PDSTK-conjugated antisense PNA in S. pyogenes.
We further analyzed the bactericidal effect of TAT-, K8-, and (RXR)4XB-anti-gyrA PNAs. All three CPP-anti-gyrA PNAs showed a dose-dependent antimicrobial effect in kill assays. Incubation of S. pyogenes with up to 5 μM K8- and (RXR)4XB-conjugated scPNAs did not lead to the reduction of bacterial counts. In contrast, TAT-anti-gyrA scPNA showed a sequence-independent antimicrobial effect in this assay, indicating a toxic influence of the TAT peptide on S. pyogenes under these conditions. In a previous study, we tested the effect of the TAT peptide alone on S. pyogenes and did not observe any antibacterial activity up to 20 μM.15 Six hours following treatment with 5 μM or 10 μM TAT-, K8-, and (RXR)4XB-anti-gyrA PNAs, a log CFU reduction of three or four, respectively, was observed, but no clearance was achieved. In contrast, L. monocytogenes could be cleared after 20 min incubation with 8 μM TAT- and (RXR)4XB-antisense PNAs specific for the gene of RNA polymerase α subunit (rpoA).14
MIC determination revealed that TAT-anti-gyrA PNA and K8-anti-gyrA PNA were effective at the same MIC of 15.6 μM, whereas the respective scrambled controls showed a MIC of 62.5 μM. Here, no toxic effect of the TAT peptide was observed. The MICs of (RXR)4XB-anti-gyrA PNA and its corresponding scrambled control were 62.5 μM and 125 μM, respectively. Compared with L. monocytogenes, these MIC values are rather high. TAT- and (RXR)4XB-anti-rpoA PNAs exhibited a MIC of 1–4 μM, depending on the L. monocytogenes isolate tested.14
To assess whether the bactericidal effect of the CPP-antisense PNAs is sufficient for treatment of a S. pyogenes infection in vivo, a G. mellonella infection model was used.39 TAT-, K8-, and (RXR)4XB-anti-gyrA PNAs increased survival of infected G. mellonella larvae. In contrast, treatment of infected larvae with Ant-, mVE-cadherin-, and ELA-anti-gyrA PNAs, which did not show bactericidal activity in vitro, did not affect survival. A combination of anti-gyrA PNAs with antibiotics targeting gyrase subunit A was shown to result in synergistic or additive antimicrobial effects on S. aureus and S. pyogenes in vitro.15, 40 Here, we demonstrate that combination therapy of infected larvae with TAT-anti-gyrA PNAs and levofloxacin led to increased survival rates compared with each treatment alone, supporting the idea that a combination of antisense PNAs with conventional antibiotics is a potent strategy to decrease the concentration of antibiotics during treatment of S. pyogenes infections.
We were surprised that from 18 CPP-PNA conjugates tested in this study, only three showed an efficient antimicrobial effect. For future experiments, different types of carriers should be investigated. One possible alternative to peptide carriers is vitamin B12, which has been successfully used in E. coli and Salmonella Typhimurium.41 The authors showed that vitamin B12 worked more efficiently in E. coli than (KFF)3K, which is widely used in this organism.
Furthermore, we will aim at the identification of additional antisense target genes specific for S. pyogenes. Beside other essential genes, antisense targeting of virulence factor genes is a promising strategy. For instance, antisense PNAs directed against ska, the gene coding for streptokinase, could potentially diminish S. pyogenes virulence. Streptokinase is involved in the lysis of fibrin clots and thereby supports bacterial spreading. It has been shown that a small compound inhibiting ska expression was able to improve survival in a murine infection model.42
In this study, we were able to confirm that gyrA is a suitable target for PNA-mediated antisense inhibition of gene expression in S. pyogenes. We found that TAT-, K8-, and (RXR)4XB-anti-gyrA PNAs showed antibacterial activity in vitro and in vivo with comparable characteristics. TAT-conjugated scPNAs showed in vitro an unspecific effect, probably caused by TAT toxicity, which was not apparent in vivo. K8-coupled anti-gyrA PNA showed high antimicrobial efficiency in vitro and was effective on all S. pyogenes serotypes tested except AP1. In contrast, (RXR)4XB-coupled anti-gyrA PNA showed high bactericidal efficiency in the kill assay but exhibited higher MICs than TAT and K8-PNA conjugates. Additionally, the effect of (RXR)4XB-coupled anti-gyrA PNA on larvae survival in the G. mellonella infection model was lower compared with the other conjugates. Overall, our results underline the importance of suitable vectors for PNA delivery to achieve optimal antimicrobial function and identified efficient CPPs for testing of additional S. pyogenes target genes.
Materials and Methods
PNA Synthesis
CPP-PNAs were synthesized and purified by high-performance liquid chromatography (HPLC) (Peps4LS, Heidelberg, Germany). Sequences of all CPP-PNAs used in this work are listed in Table 1.
Bacterial Strains and Culture Conditions
S. pyogenes strains were cultured in Todd-Hewitt broth, supplemented with 0.5% yeast extract (THY; Oxoid, Thermo Fisher Scientific, Darmstadt, Germany), at 37°C under a 5% CO2/20% O2 atmosphere. All strains used in this study are listed in Table 3.
Table 3.
S. pyogenes Strains
| Strain | M Type | Isolation | Reference |
|---|---|---|---|
| 5448 | M1T1 | STSS | Dr. Nikolai Siemens, Karolinska Institut, Lund, Sweden |
| AP1 | M1 | sepsis | Centre for Reference and Research on Streptococci, Prague, Czech Republic |
| M3 8003 | M3 | necrotizing fasciitis | Dr. Nikolai Siemens, Karolinska Institut, Lund, Sweden |
| HRO-K-035 | M4 | throat infection | clinical isolate, University Medicine Rostock, Germany |
| MGAS8232 | M18 | ARF | 43 |
| 591 | M49 | skin | R. Lütticken, Aachen, Germany |
ARF, acute rheumatic fever; STSS, streptococcal toxic-shock syndrome.
Bacterial Kill Assay
Overnight cultures of the respective S. pyogenes strain were diluted in PBS/brain heart infusion (BHI) (7/2) to approximately 2 × 105 CFU/mL. 450 μl bacterial suspension, containing ∼105 CFU, was transferred to a 2-mL reaction tube. 50 μL PNA was added to a final PNA concentration of 1–10 μM or as indicated. 50 μL H2O served as mock control. The reaction tubes were incubated for 6 h at 37°C and 7 rpm (Rotor SB3; Stuart, Staffordshire, UK). Viable cell counts were determined by plating appropriate dilutions on THY agar plates. CFUs were determined by visual inspection following overnight incubation at 37°C under a 5% CO2/20% O2 atmosphere. At time point 0, the viable cell count corresponded to 1–3 × 105 CFU/mL. The initial CPP screen has been performed in two biological replicates. Each subsequent experiment has been performed in at least three independent biological replicates, as indicated in the figure legends.
Extraction of Total RNA
For RNA isolation, 450 μL bacterial suspension (107 CFU/mL in THY) was prepared for each experimental condition, treated with 2 μM CPP-PNA conjugates, and incubated in a 2-mL reaction tube for 6 h at 37°C and 7 rpm (Rotor SB3; Stuart, Staffordshire, UK). Subsequently, five samples per condition were pooled. Bacteria were pelleted immediately, shock frozen in liquid nitrogen, and stored at −80°C until use. Bacterial cells were disrupted in a homogenizer (Peqlab Biotechnologie, Erlangen, Germany). Total RNA was extracted, according to the protocol, supplied with the Direct-zol RNA MiniPrep Kit (Zymo Research, Irvine, CA). After extraction, RNA was treated with acid phenol:chloroform:isoamyl alcohol (125:24:1; pH 4.5; Thermo Fisher Scientific), and TURBO DNase (Thermo Fisher Scientific), according to the manufacturer’s instructions. RNA was stored at −80° C until further use.
Reverse Transcription Followed by quantitative real-time PCR
cDNA synthesis was performed using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, Thermo Fisher Scientific). Quantitative real-time PCR amplification was conducted with SYBR Green (Thermo Fisher Scientific) using the ViiA 7 Real-Time PCR System (Applied Biosystems, Darmstadt, Germany). The 5S rRNA gene served as internal control. Relative expression was calculated using the 2−ΔΔct method.44 Primers were designed based on the full genome sequence of S. pyogenes M49 strain NZ131 (NCBI: NC011375): gyrA-specific primers: 5′-TGAGTGTCATTGTGGCAAGAGC-3′ and 5′-AGAGAATACGACGATGCACAGG-3′; 5S-specific primers: 5′-AGCGACTACCTTATCTCACAG-3′ and 5′-GAGATACACCTGTACCCATG-3′.
Determination of the MIC
MIC determination was performed following the protocol of the Clinical and Laboratory Standards Institute (CLSI).45 In MIC assays containing CPP-PNAs, lysed horse blood (LHB)/cation-adjusted Mueller-Hinton broth (CAMHB) medium was supplemented with 0.02% acetic acid and 0.4% BSA. MICs were recorded as the lowest concentration where no turbidity was observed in the wells.
G. mellonella Infection Model
Larvae of the greater wax moth G. mellonella were obtained from Reptilienkosmos (Niederkrüchten, Germany). For infection experiments, S. pyogenes strains were grown overnight in THY, washed twice in a 0.9% NaCl solution, and suspended in 0.9% NaCl to a final concentration of 1–3 × 108 CFU/mL. Larvae with a weight of 150–200 mg were infected with 1–3 × 106 CFU/larva. Bacteria were injected into the hemocoel of the larvae between the last two pairs of pro-legs using a microapplicator (World Precisions Instruments, Sarasota, FL) and a fine dosage syringe (Omnican F; B. Braun, Melsungen, Germany; 0.01–1 mL, 0.30 × 12 mm). As mock control, 0.9% NaCl was injected. For CPP-PNA treatment, larvae were injected 30 min postinfection with 4 nmol CPP-PNA/larva. Larvae were incubated for 7 days, and survival was monitored daily.46
Statistical Analyses
All experiments were performed at least three times or as indicated by sample size (n). Statistical significance was determined using the tests indicated in the respective figure legends. Statistical analyses were performed using GraphPad Prism 7 software.
Author Contributions
N.P. designed this study. G.B., A.-L.L., R.P., S.K., and A.S. conducted the experiments. N.P., G.B., A.J., and B.K. contributed to the interpretation of data and manuscript writing.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
Support for the work of G.B. and B.K. was provided by a grant from the Ministerium für Bildung, Wissenschaft und Kultur, Mecklenburg-Vorpommern (ESF/14-BM-A55-0010/16). Support for the work of N.P. was provided by the University Medicine Rostock (Forun 889008).
References
- 1.Carapetis J.R., Steer A.C., Mulholland E.K., Weber M. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Sims Sanyahumbi A., Colquhoun S., Wyber R., Carapetis J.R. Global disease burden of group A streptococcus. In: Ferretti J.J., Stevens D.L., Fischetti V.A., editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. University of Oklahoma Health Sciences Center; 2016. [PubMed] [Google Scholar]
- 3.Cunningham M.W. Pathogenesis of group A streptococcal infections and their sequelae. Adv. Exp. Med. Biol. 2008;609:29–42. doi: 10.1007/978-0-387-73960-1_3. [DOI] [PubMed] [Google Scholar]
- 4.Bisno A.L., Gerber M.A., Gwaltney J.M., Jr., Kaplan E.L., Schwartz R.H., Infectious Diseases Society of America Practice guidelines for the diagnosis and management of group A streptococcal pharyngitis. Clin. Infect. Dis. 2002;35:113–125. doi: 10.1086/340949. [DOI] [PubMed] [Google Scholar]
- 5.Richter S.S., Heilmann K.P., Beekmann S.E., Miller N.J., Miller A.L., Rice C.L., Doern C.D., Reid S.D., Doern G.V. Macrolide-resistant Streptococcus pyogenes in the United States, 2002-2003. Clin. Infect. Dis. 2005;41:599–608. doi: 10.1086/432473. [DOI] [PubMed] [Google Scholar]
- 6.Cattoir V. Mechanisms of antibiotic resistance. In: Ferretti J.J., Stevens D.L., Fischetti V.A., editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. University of Oklahoma Health Sciences Center; 2016. [PubMed] [Google Scholar]
- 7.Nielsen P.E., Egholm M. An introduction to peptide nucleic acid. Curr. Issues Mol. Biol. 1999;1:89–104. [PubMed] [Google Scholar]
- 8.Demidov V.V., Potaman V.N., Frank-Kamenetskii M.D., Egholm M., Buchard O., Sönnichsen S.H., Nielsen P.E. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 9.Gupta B., Levchenko T.S., Torchilin V.P. Intracellular delivery of large molecules and small particles by cell-penetrating proteins and peptides. Adv. Drug Deliv. Rev. 2005;57:637–651. doi: 10.1016/j.addr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Lehto T., Ezzat K., Wood M.J.A., El Andaloussi S. Peptides for nucleic acid delivery. Adv. Drug Deliv. Rev. 2016;106(Pt A):172–182. doi: 10.1016/j.addr.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Eriksson M., Nielsen P.E., Good L. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J. Biol. Chem. 2002;277:7144–7147. doi: 10.1074/jbc.M106624200. [DOI] [PubMed] [Google Scholar]
- 12.Nekhotiaeva N., Awasthi S.K., Nielsen P.E., Good L. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 2004;10:652–659. doi: 10.1016/j.ymthe.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Abushahba M.F., Mohammad H., Seleem M.N. Targeting multidrug-resistant Staphylococci with an anti-rpoA peptide nucleic acid conjugated to the HIV-1 TAT cell penetrating peptide. Mol. Ther. Nucleic Acids. 2016;5:e339. doi: 10.1038/mtna.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abushahba M.F., Mohammad H., Thangamani S., Hussein A.A., Seleem M.N. Impact of different cell penetrating peptides on the efficacy of antisense therapeutics for targeting intracellular pathogens. Sci. Rep. 2016;6:20832. doi: 10.1038/srep20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patenge N., Pappesch R., Krawack F., Walda C., Mraheil M.A., Jacob A., Hain T., Kreikemeyer B. Inhibition of growth and gene expression by PNA-peptide conjugates in Streptococcus pyogenes. Mol. Ther. Nucleic Acids. 2013;2:e132. doi: 10.1038/mtna.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derossi D., Joliot A.H., Chassaing G., Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 17.Turner Y., Wallukat G., Säälik P., Wiesner B., Pritz S., Oehlke J. Cellular uptake and biological activity of peptide nucleic acids conjugated with peptides with and without cell-penetrating ability. J. Pept. Sci. 2010;16:71–80. doi: 10.1002/psc.1198. [DOI] [PubMed] [Google Scholar]
- 18.Vivès E., Brodin P., Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 19.Tréhin R., Krauss U., Beck-Sickinger A.G., Merkle H.P., Nielsen H.M. Cellular uptake but low permeation of human calcitonin-derived cell penetrating peptides and Tat(47-57) through well-differentiated epithelial models. Pharm. Res. 2004;21:1248–1256. doi: 10.1023/b:pham.0000033013.45204.c3. [DOI] [PubMed] [Google Scholar]
- 20.Vaara M., Porro M. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 1996;40:1801–1805. doi: 10.1128/aac.40.8.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller J., Kretzschmar I., Volkmer R., Boisguerin P. Comparison of cellular uptake using 22 CPPs in 4 different cell lines. Bioconjug. Chem. 2008;19:2363–2374. doi: 10.1021/bc800194e. [DOI] [PubMed] [Google Scholar]
- 22.Palm C., Netzereab S., Hällbrink M. Quantitatively determined uptake of cell-penetrating peptides in non-mammalian cells with an evaluation of degradation and antimicrobial effects. Peptides. 2006;27:1710–1716. doi: 10.1016/j.peptides.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 23.El-Andaloussi S., Johansson H.J., Holm T., Langel U. A novel cell-penetrating peptide, M918, for efficient delivery of proteins and peptide nucleic acids. Mol. Ther. 2007;15:1820–1826. doi: 10.1038/sj.mt.6300255. [DOI] [PubMed] [Google Scholar]
- 24.Maiolo J.R., Ferrer M., Ottinger E.A. Effects of cargo molecules on the cellular uptake of arginine-rich cell-penetrating peptides. Biochim. Biophys. Acta. 2005;1712:161–172. doi: 10.1016/j.bbamem.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Tünnemann G., Ter-Avetisyan G., Martin R.M., Stöckl M., Herrmann A., Cardoso M.C. Live-cell analysis of cell penetration ability and toxicity of oligo-arginines. J. Pept. Sci. 2008;14:469–476. doi: 10.1002/psc.968. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell D.J., Kim D.T., Steinman L., Fathman C.G., Rothbard J.B. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Pept. Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 27.Roth A.F., Sullivan D.M., Davis N.G. A large PEST-like sequence directs the ubiquitination, endocytosis, and vacuolar degradation of the yeast a-factor receptor. J. Cell Biol. 1998;142:949–961. doi: 10.1083/jcb.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manceur A., Wu A., Audet J. Flow cytometric screening of cell-penetrating peptides for their uptake into embryonic and adult stem cells. Anal. Biochem. 2007;364:51–59. doi: 10.1016/j.ab.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 29.El-Andaloussi S., Järver P., Johansson H.J., Langel U. Cargo-dependent cytotoxicity and delivery efficacy of cell-penetrating peptides: a comparative study. Biochem. J. 2007;407:285–292. doi: 10.1042/BJ20070507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindgren M., Gallet X., Soomets U., Hällbrink M., Bråkenhielm E., Pooga M., Brasseur R., Langel U. Translocation properties of novel cell penetrating transportan and penetratin analogues. Bioconjug. Chem. 2000;11:619–626. doi: 10.1021/bc990156s. [DOI] [PubMed] [Google Scholar]
- 31.Oehlke J., Krause E., Wiesner B., Beyermann M., Bienert M. Extensive cellular uptake into endothelial cells of an amphipathic beta-sheet forming peptide. FEBS Lett. 1997;415:196–199. doi: 10.1016/s0014-5793(97)01123-x. [DOI] [PubMed] [Google Scholar]
- 32.Abes R., Moulton H.M., Clair P., Yang S.T., Abes S., Melikov K., Prevot P., Youngblood D.S., Iversen P.L., Chernomordik L.V., Lebleu B. Delivery of steric block morpholino oligomers by (R-X-R)4 peptides: structure-activity studies. Nucleic Acids Res. 2008;36:6343–6354. doi: 10.1093/nar/gkn541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma C., Awasthi S.K. Versatility of peptide nucleic acids (PNAs): role in chemical biology, drug discovery, and origins of life. Chem. Biol. Drug Des. 2017;89:16–37. doi: 10.1111/cbdd.12833. [DOI] [PubMed] [Google Scholar]
- 34.Bai H., You Y., Yan H., Meng J., Xue X., Hou Z., Zhou Y., Ma X., Sang G., Luo X. Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials. 2012;33:659–667. doi: 10.1016/j.biomaterials.2011.09.075. [DOI] [PubMed] [Google Scholar]
- 35.Ghosal A., Nielsen P.E. Potent antibacterial antisense peptide-peptide nucleic acid conjugates against Pseudomonas aeruginosa. Nucleic Acid Ther. 2012;22:323–334. doi: 10.1089/nat.2012.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitev G.M., Mellbye B.L., Iversen P.L., Geller B.L. Inhibition of intracellular growth of Salmonella enterica serovar Typhimurium in tissue culture by antisense peptide-phosphorodiamidate morpholino oligomer. Antimicrob. Agents Chemother. 2009;53:3700–3704. doi: 10.1128/AAC.00099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wender P.A., Mitchell D.J., Pattabiraman K., Pelkey E.T., Steinman L., Rothbard J.B. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters. Proc. Natl. Acad. Sci. USA. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu R.P., Youngblood D.S., Hassinger J.N., Lovejoy C.E., Nelson M.H., Iversen P.L., Moulton H.M. Cell-penetrating peptides as transporters for morpholino oligomers: effects of amino acid composition on intracellular delivery and cytotoxicity. Nucleic Acids Res. 2007;35:5182–5191. doi: 10.1093/nar/gkm478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai C.J., Loh J.M., Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7:214–229. doi: 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dryselius R., Nekhotiaeva N., Good L. Antimicrobial synergy between mRNA- and protein-level inhibitors. J. Antimicrob. Chemother. 2005;56:97–103. doi: 10.1093/jac/dki173. [DOI] [PubMed] [Google Scholar]
- 41.Równicki M., Wojciechowska M., Wierzba A.J., Czarnecki J., Bartosik D., Gryko D., Trylska J. Vitamin B12 as a carrier of peptide nucleic acid (PNA) into bacterial cells. Sci. Rep. 2017;7:7644. doi: 10.1038/s41598-017-08032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun H., Xu Y., Sitkiewicz I., Ma Y., Wang X., Yestrepsky B.D., Huang Y., Lapadatescu M.C., Larsen M.J., Larsen S.D. Inhibitor of streptokinase gene expression improves survival after group A streptococcus infection in mice. Proc. Natl. Acad. Sci. USA. 2012;109:3469–3474. doi: 10.1073/pnas.1201031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smoot J.C., Barbian K.D., Van Gompel J.J., Smoot L.M., Chaussee M.S., Sylva G.L., Sturdevant D.E., Ricklefs S.M., Porcella S.F., Parkins L.D. Genome sequence and comparative microarray analysis of serotype M18 group A streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA. 2002;99:4668–4673. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Weinstein M.P., Patel J.B., Burnham C.-A., Campeau S., Conville P.S., Doern C., Eliopoulos G.M., Galas M.F., Humphries R.M., Jenkins S.G. 11th Edition. Clinical and Laboratory Standards Institute; 2018. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. [Google Scholar]
- 46.Mukherjee K., Altincicek B., Hain T., Domann E., Vilcinskas A., Chakraborty T. Galleria mellonella as a model system for studying Listeria pathogenesis. Appl. Environ. Microbiol. 2010;76:310–317. doi: 10.1128/AEM.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]