Abstract
Objective:
To investigate the effect of long noncoding RNA GM16343 on interleukin 36β promotion of CD8+T cells in tumor microenvironment regulation.
Methods:
The differentially expressed long noncoding RNA in interleukin 36β-stimulated mouse CD8+T cells was screened by gene chip technology, and the significant differentially expressed long noncoding RNAs were verified by real-time polymerase chain reaction. The lentiviral vector that overexpresses or knockdown GM16343 was constructed, transfected into CD8+T cells, and stimulated with interleukin 36β, and the amount of interferon γ secreted was detected by enzyme-linked immunosorbent assay. A mouse subcutaneous xenograft model that stably express interleukin 36β was established, and the tumor size and mouse survival time were observed by stimulation with CD8+T cells overexpression or knockdown of GM16343.
Results:
A total of 12 long noncoding RNAs with significant differences were screened by gene chip analysis. Real-time polymerase chain reaction showed that the difference in GM16343 was larger, and the difference between the groups was observed to be the most significant. Compared to control group, CD8+T cells overexpressing GM16343 increased the secretion of interferon γ, and the tumor diameter of the mice after stimulation showed significant reduction, and the survival time showed significant prolongation. Compared to control group, the CD8+T cells after GM16343 were knocked down. The interferon γ secretion was decreased, and no significant change in tumor diameter and survival time was observed.
Conclusion:
Interleukin 36β may enhance antitumor immune response of CD8+T cells by regulating GM16343.
Keywords: lncRNA, GM16343, IL-36β, colorectal cancer, tumor microenvironment, tumor immunity
Introduction
Cancer is the second leading cause of death in the world. According to the statistics put forwarded by World Health Organization, one-sixth of the world’s deaths is caused by cancer. In China, the incidence of cancer accounted for 22% of the world’s total prevalence, and it is still on the rise.1 There into, colorectal cancer is the third largest cancer in the world, with increasing morbidity and mortality in Asian countries, and the age of onset in individuals with colorectal cancer tends to be younger.2 Therefore, treating cancer has always been a difficult problem worldwide. In the recent years, tumor microenvironment has become the hot research topic of many researchers. This is because it provides a rich nutritional environment for growth, proliferation, and metastasis of cancer cells. Also, the role of cytokines in cancer immunotherapy has become increasingly prominent.3 The advent of various tumor targets and the emergence of their antibodies and inhibitors made cancer treatment more targeted, significantly reduced the side effects, and greatly improved the prognosis as well as quality of life of patients. Recent studies have shown that cytokine interleukin 36 (IL-36) can promote the secretion of interferon γ (IFN-γ) by type 1 lymphocytes, inducing strong antitumor effects both in vivo and in vitro.4 In this study, we further explored the mechanism of IL-36β activating CD8+T cells in antitumor immune response to find new targets for cancer immunotherapy.
According to the human genome project, only 1.2% of all genome encodes proteins, most of the remaining are transcribed into noncoding RNA (ncRNA).5 The ncRNA plays an important role in tumorigenesis, development, metastasis, cell proliferation, and apoptosis, affecting multiple functions associated with digestive, respiratory, and circulatory systems.6,7 Long noncoding RNA (lncRNA) is an ncRNA with length of greater than 200 nt.8 Long ncRNA plays an important role in regulating life activities such as epigenesis,9,10 cell cycle,11 and cell differentiation.12 At present, little is known about lncRNAs, and complex studies have reported that lncRNA can regulate Toll-like receptor signaling pathways, playing an important role in the immune response and in turn promoting or inhibiting the inflammatory signal transduction.13 This suggested that lncRNA might be involved in IL-36 activation of CD8+T cells antitumor immune response, which in turn plays an important role in tumor immunotherapy.
Materials and Methods
Cell Culture and Stimulation
From the spleens of C57BL/6J mice, CD8+T cells were isolated by immunomagnetic beads sorting, and MyD88 (MyD88 is a signaling molecule of IL-36β signal transduction pathway) was isolated from MyD88 knockout (KO) mice. The flow cytometer detects cell purity above 95%. The cell density was adjusted to 1.0 × 106/mL and seeded in a 96-well plate, and CD8+T cells were stimulated according to the use of IL-36β (100 ng/mL) or not. The experimental mice were divided into 4 groups: wild type (WT) + IL-36β, MyD88 KO + IL-36β, WT control, and MyD88 KO control. The cells were incubated with 1640 medium containing 10% fetal calf serum in an incubator containing 5% CO2 at 37°C for 72 hours.
Long Noncoding RNAs Screening
The lncRNA that was differentially expressed by CD8+T cells was stimulated by IL-36β according to the former described method and was screened using the Human LncRNA Microarray V3.0 chip of Kangcheng Technology Co., Ltd (Shanghai, China) and records up or down 2- or 5-fold lncRNAs.
Bioinformatics Analysis
Functional annotation and classification of differential lncRNAs from cellular components, molecular functions, and biological processes to obtain differential lncRNA potential target genes, namely, Gene ontology (GO) analysis and differential messenger RNA-related genomes and metabolism were acquired using Kyoto encyclopedia of genes and genomes database pathway for performing pathway enrichment analysis.
Real-Time Polymerase Chain Reaction
Total RNA was extracted using a TRIZOL kit (Shanghai Thermo Fisher Scientific, Shanghai, China), and then the complementary DNA (cDNA) was synthesized according to a reverse transcription kit (purchased from QIAGEN, Germany). According to the results of gene chip, the differential lncRNA (5 upregulated, 7 downregulated, P < .05) was verified by polymerase chain reaction (PCR). The lncRNA with the largest difference fold was upregulated or downregulated for subsequent experiments. β-actin was used as the internal reference gene, and the relative expression was expressed as 2−ΔΔCt. The primer sequences are listed in Table 1.
Table 1.
The Primer Sequence.
| Gene Symbol | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| AK029357 | TCCCTGGGCACAGTAATA | CCAAGCCTCACTTCCTAA |
| AK034605 | CGGCTCCTTAGGCTTGTT | TTAGTGGGCAGGGCAGAT |
| AK043298 | CGCATCAGCCTCAACAG | TTGCCATTAAGGTAGAGTTTC |
| AK052878 | AGGGCTGGACATGAAAGT | GTGGGATGGACAGAAGGT |
| AK150191 | ACGGATGTACTTGCTTGG | GGAGGTATGGGATGAGGA |
| D930032P07Rik | ACCTGGAAATCTCAACTC | GATGGTGCCTCTTCTTAC |
| Gm12522 | ACCGACATACTGTGGTACTGA | CCCGAAGCCATTGATTTA |
| Gm16343 | GCATCTTTGTGACGCTCTA | TTCCCTTCTACTTTCCTAACT |
| Gm20632 | TTTTATAGACGCAGACCC | GAGAAACAGCGAGCAA |
| Gm25445 | AGAGGAGGACTGCTGTTG | TCATAGAGGGATAGGCTTG |
| Gm26763 | ATGAGAATTGACGAGGGA | CATTTGTGACTTTGGTGC |
| uc.213 | GTCGTTTGTGCGTCTA | GTCCCAGTCATTTCCA |
| β-actin | ATGCATGACGATATGGCT | ATGAGGTACTCTGTGAGGT |
ShRNA Synthesis and Lentivirus Packaging
Three short hairpin RNA (shRNAs) and negative control shRNA (NC-shRNA) (Table 2) designed and synthesized by Shanghai Novobio Co, Ltd (Shanghai, China) were packed into lentiviral vectors for inducing different target gene fragments. After transfection of CD8+T cells, the transfection efficiency and subsequent functional tests were tested.
Table 2.
Sequences of shRNA.
| shRNA-1 | 5′-CCGGGCATCTTTGTGACGCTCTAGTTTCAA GAGAACTAGAGCGTCACAAAGATGCTTTT TT GGTACC-3′ |
| 5′-AATTGGTACCAAAAAAGCATCTTTGTGAC GCTCTAGTTCTCTTGAAACTAGAGCGTCA CAAAGATGA-3′ | |
| shRNA-2 | 5′-CCGGGGTCAGTGATTGCATCTTTGTTTCA AGAGAACAAAGATGCAATCACTGACC TT TTTT GGTACC-3′ |
| 5′-AATTGGTACCAAAAAAGGTCAGTGATTGC ATCTTTGTTCTCTTGAAACAAAGATGCAA TCACTGACC-3′ | |
| shRNA-3 | 5′-CCGG GGCTGCTTACTGAGATGATCTTTCA AGAGAAGATCATCTCAGTAAGCAGCC TT TTTT GGTACC-3′ |
| 5′-AATTGGTACCAAAAAAGGCTGCTTACTGA GATGATCTTCTCTTGAAAGATCATCTCAG TAAGCAGCC-3′ | |
| NC-shRNA | 5′-CCGGGTTCTCCGAACGTGTCACGTTTCAA GAGAACGTGACACGTTCGGAGAAC TTTT TT GGTACC-3′ |
| 5′-AATTGGTACCAAAAAAGTTCTCCGAACG TGTCACGTTCTCTTGAAACGTGACACGTT CGGAGAAC-3′ |
Abbreviations: NC-shRNA, negative control shRNA; shRNA, short hairpin RNA.
Construction of Lentiviral Vector Overexpressing LncRNA
The lentiviral vector pcDNA-3.1-GM16343(LV-pcDNA-3.1-GM16343) was constructed by Shanghai Novobio Co, Ltd. After transfection of CD8+T cells, the transfection efficiency was detected by flow cytometry, and subsequent overexpression and knockdown efficiencies were detected by real-time PCR (RT-PCR).
Enzyme-Linked Immunosorbent Assay
The level of IFN-γ in cell culture supernatant was measured by IFN-γ enzyme-linked immunosorbent assay kit (Biolegend, San Diego, CA). According to the kit procedure, the optical density value was detected and recorded at the end of the test, the concentration standard curve was drawn, and the unknown serum IFN-γ value was calculated.
Establishment of CT-26 Cell Line of Colon Cancer in Mice With Stable Overexpression of IL-36
The WT and stable expression of IL-36β in mouse colon cancer CT-26 cell line was donated by Professor Xueguang Zhang, Soochow University, China. CT-26 cell line that stably expresses IL-36β was named as CT-26-IL-36β. Meanwhile, the WT CT-26 cells without IL-36β stimulation were named as CT-26-WT.
Animal Model
CT-26-WT and CT-26-IL-36β cells in logarithmic growth phase at a concentration of 1 × 107 cells/mL were selected, and 0.1 mL of tumor cells were implanted in the inguinal region of mice, that is, about 1 × 106 cancer cells. To study the effect of GM16343 on tumor growth and prognosis, on third day of tumor formation, CD8+T cells overexpressed or knocked down GM16343, followed by injection of 100 μL of total of 6 × 106 cells into the tumor tissue. Multiple injections were used in multiple sites, and the control group was injected with phosphate-buffered saline. Seven days after the first injection, the injection was again performed according to the original protocol. Tumor size was measured every 2 days, and the time of death of the mice was recorded. The experiment was divided into 5 groups: CT-26-WT, CT-26-IL-36β, CT-26-IL-36β+CD8+T-WT, CT-26-IL-36β+CD8+T-LV-pcDNA3.1-GM16343, and CT-26- IL-36β+CD8+T-shRNA-1. Five mice were present in each group, and the diameter of tumors was expressed as mean ± standard error of mean.
Statistical Analysis
Intergroup data were analyzed by t test. Survival analysis was performed by log-rank test. SPSS 19.0 was used for data processing, and P values <.05 were considered to be statistically significant.
Results
Screening of LncRNA–RNA Pair
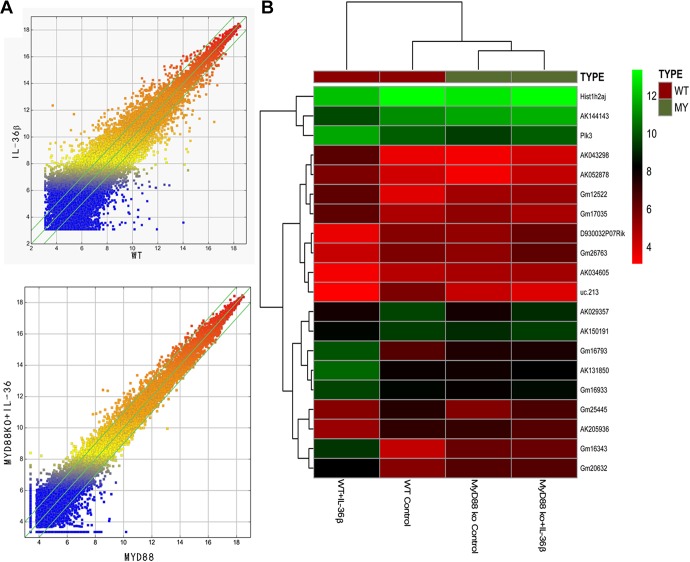
There are 395 lncRNAs with a difference of more than 2-fold in the WT + IL-36β group when compared to the WT control group. Among them, 178 were raised and 217 were lowered. There were 57 lncRNAs with a difference of more than 5 times, and 25 of these were upregulated and 32 were downregulated (Figure 1). In view of the above results, a total of 12 lncRNAs with strong original signals, small intragroup differences, and significant differences between groups (up to 5 and down to 7) were screened (Table 3).
Figure 1.
Differential long noncoding RNA (lncRNA) expression was screened by gene chip. A, Scatter map of lncRNA gene chip: wild type (WT) versus interleukin 36β (IL-36β), MyD88 knockout (KO) versus MyD88 KO + IL-36β. B, Cluster map of lncRNA genes. The expression of GM16343 was significantly higher in the WT + IL-36β group than in other groups.
Table 3.
Screening of 12 Pairs of LncRNA–mRNA Pairs by Microarray.
| Gene Symbol | Fold Change— LncRNAs | Regulation— LncRNAs | Near by Gene | Near by Gene Symbol | Fold Change—mRNAs |
|---|---|---|---|---|---|
| AK029357 | 2.5008092 | Down | NM_001162950 | Hif3a | 2.9988887 |
| AK034605 | 3.1426615 | Down | NM_170671 | Mycbpap | 2.0812213 |
| AK043298 | 5.2223806 | Up | NM_007897 | Ebf1 | 2.9260796 |
| AK052878 | 2.5520272 | Up | NM_175303 | Sall4 | 2.2033957 |
| AK150191 | 2.1601552 | Down | NM_001198825 | App | 2.5293029 |
| D930032P07Rik | 6.1818344 | Down | NM_175459 | Glis3 | 2.4360102 |
| Gm12522 | 2.2462635 | Up | NM_026831 | Mybphl | 6.1898953 |
| Gm16343 | 8.7983439 | Up | NM_178020 | Hyal3 | 2.9652731 |
| Gm20632 | 3.4684051 | Up | ENSMUST00000078756 | Hist2h2aa2 | 2.4074269 |
| Gm25445 | 2.3226365 | Down | NM_139309 | Fktn | 2.1246144 |
| Gm26763 | 4.0120147 | Down | NM_001205101 | Gm13212 | 2.0957883 |
| uc.213 | 2.4441461 | Down | NM_010453 | Hoxa5 | 2.3571229 |
Abbreviations: lncRNA, long noncoding RNA; mRNA, messenger RNA.
As shown in Figure 1B, Gm16343 in WT + IL-36β group was highly expressed than that in WT control group. Compared to MyD88 KO + IL-36β group, the MyD88 KO control group showed no obvious change in GM16343 expression. MyD88 is an essential molecule for IL-36 receptor intracellular signal transduction. After MyD88 is KO, IL-36 signal cannot be transmitted downstream. As mentioned above, GM16343 was chosen for further research.
Bioinformatics Analysis Results of Differential LncRNAs
Gene ontology and pathway analysis of differential lncRNAs showed that these lncRNAs were not only involved in cytokine regulation, translation regulation, immunity, proliferation, cell adhesion, and cell differentiation but were also involved in apoptosis, autophagy, and activation of related signaling pathways, such as tumor necrosis factor, NOD-like receptor, p53, and Rap1 (Figure 2).
Figure 2.
Differential long noncoding RNAs (lncRNAs) potential target genes gene ontology (GO) and pathway analysis.
Real-Time PCR Results
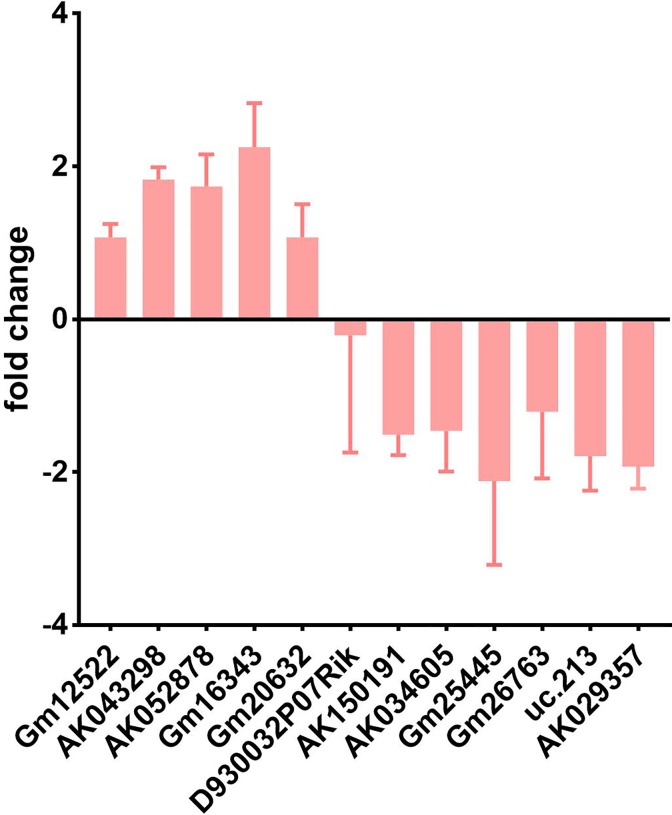
CD8+T cells of group WT + IL-36β and WT control were cultured for 72 hours, and then RT-PCR was performed. The results of RT-PCR analysis showed that after 3 repeated experiments, the changes in AK043298, AK052878, and Gm16343 were consistent with that of chip expression. Among them, the upregulated lncRNA GM16343 showed a larger difference, and the difference between the groups remained the most significant (Figure 3).
Figure 3.
Validation of differential long noncoding RNAs (lncRNA) real-time polymerase chain reaction (PCR). Among the 12 pairs of lncRNAs screened by gene chip, the upregulated lncRNA GM16343 showed the most significant difference.
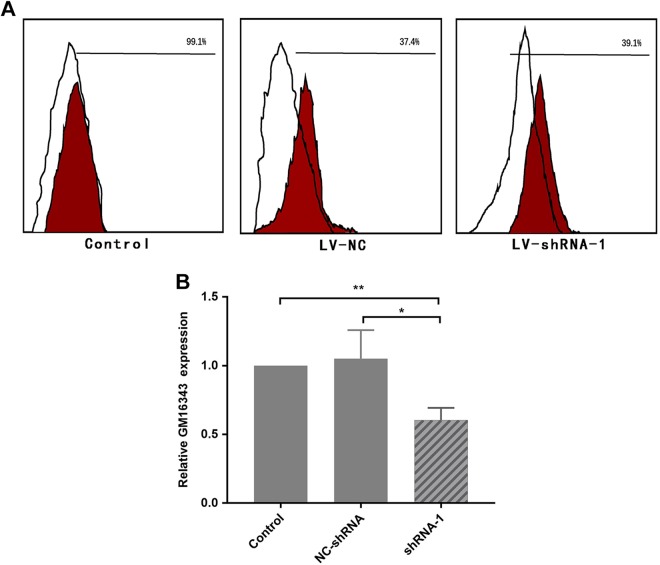
ShRNA-GM16343 Screening
The 3 shRNAs and NC-shRNAs constructed in the previous stage were transfected into CD8+T cells, respectively, and then stimulated with IL-36β at a concentration of 100 ng/mL. The transfection efficiency was detected by flow cytometry after 6 hours, and the expression of GM16343 was detected by RT-PCR by 48 hours. The results showed that the transfection efficiency of CD8+T cells was 39.1%; shRNA-1 demonstrated the most significant inhibitory effect on GM16343 (P < .05), and shRNA-1 interference efficiency was 39.75%. Therefore, shRNA-1 was used for follow-up functional test (Figure 4).
Figure 4.
Screening of short hairpin RNA (shRNA)-GM16343. The rate of shRNA transfected CD8+T cells by flow cytometry was 39.1% (A). The shRNA interference efficiency was about 39.75% (B).
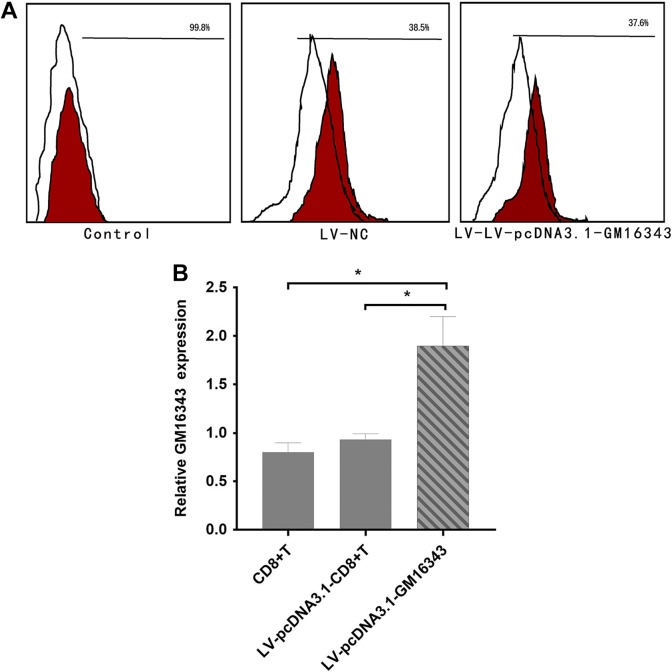
Verification of Lentivirus Vector pcDNA3.1-GM16343
The transfection efficiency of lentiviral vector pcDNA3.1-GM16343 transfected with CD8+T cells and whether it can significantly promote the expression of GM16343 were further investigated. After transfection, it was stimulated with IL-36β with a concentration of 100 ng/mL. After 6 hours, the transfection efficiency was detected by flow cytometry, and after 48 hours, the expression level of GM16343 was detected by PCR. The results showed that the transfection efficiency of CD8+T cells was 37.6%, LV-pcDNA3.1-GM16343 significantly promoted the expression of GM16343 (P < .05), and the overexpression ratio of LV-pcDNA3.1-GM16343 was 1.9 times (Figure 5).
Figure 5.
Verification of the overexpressing vector LV-pcDNA3.1-GM16343. The rate of LV-pcDNA3.1-GM16343 transfected with CD8+T cells by flow cytometry was 37.6% (A). The overexpression ratio of LV-pcDNA3.1-GM16343 was 1.9 times (B).
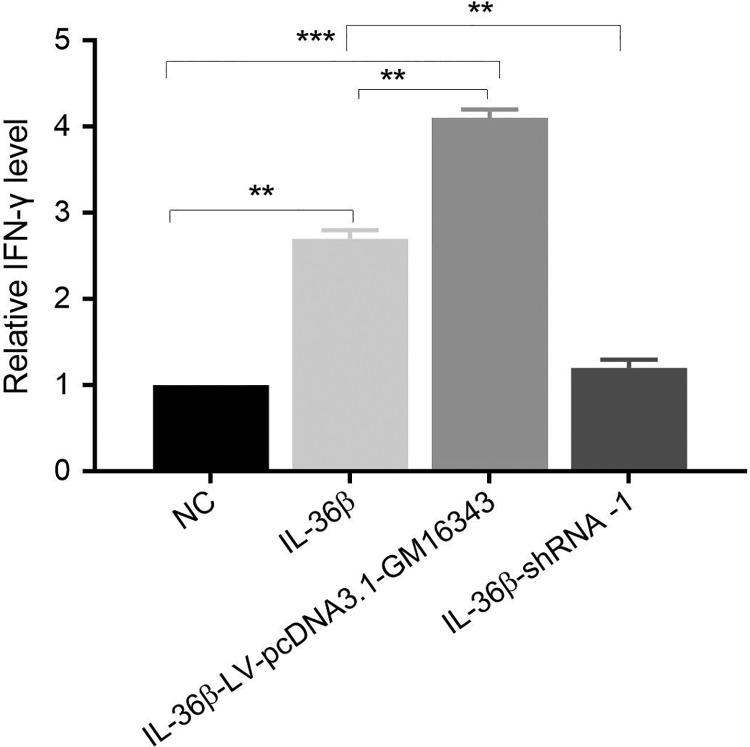
Interferon-γ Secretion in CD8+T cells in Which GM16343 Was Overexpressed or Knocked Down
After overexpression of GM16343 or knockdown of GM16343, CD8+T cells were stimulated with or without IL-36β, respectively. The results showed that the secretion of IFN-γ in CD8+T cells stimulated by IL-36β (IL-36β group) was significantly higher than that of the control group (NC group). The IFN-γ secretion by IL-36β-stimulated CD8+T cells which overexpressed GM16343 was significantly increased (IL-36β-LV-pcDNA3.1-GM16343 group). The amount of IFN-γ secreted by IL-36β-stimulated CD8+T cells that knocked down GM16343 (IL-36β-shRNA-1 group) was significantly lowered than that of the above 2 groups, but no significant changes were observed when compared to the NC group. These results suggested that IL-36β might promote the secretion of IFN-γ by CD8+T cells via GM16343 (Figure 6).
Figure 6.
Interferon γ (IFN-γ) secretion in CD8+T cells overexpressed or knocked down.
GM16343 Significantly Inhibits the Growth of Tumor by Influencing Antitumor Immune Effect of CD8+T Cells
CD8+T cells transfected with LV-pcDNA3.1-GM16343 or shRNA-GM16343 were injected into tumor tissues, and the tumor diameter was measured every 2 days. The results showed that IL-36β significantly inhibited tumor growth, and CD8+T-LV-pcDNA3.1-GM16343 enhanced tumor suppressive effects (P < .01). The CT-26-IL-36β+CD8+T-shRNA-1 group showed significant inhibition of tumor growth when compared to CT-26-WT group (P < .01), but no significant difference was observed when compared to CT-26-IL-36β (P > .05; Figure 7A and B).
Figure 7.
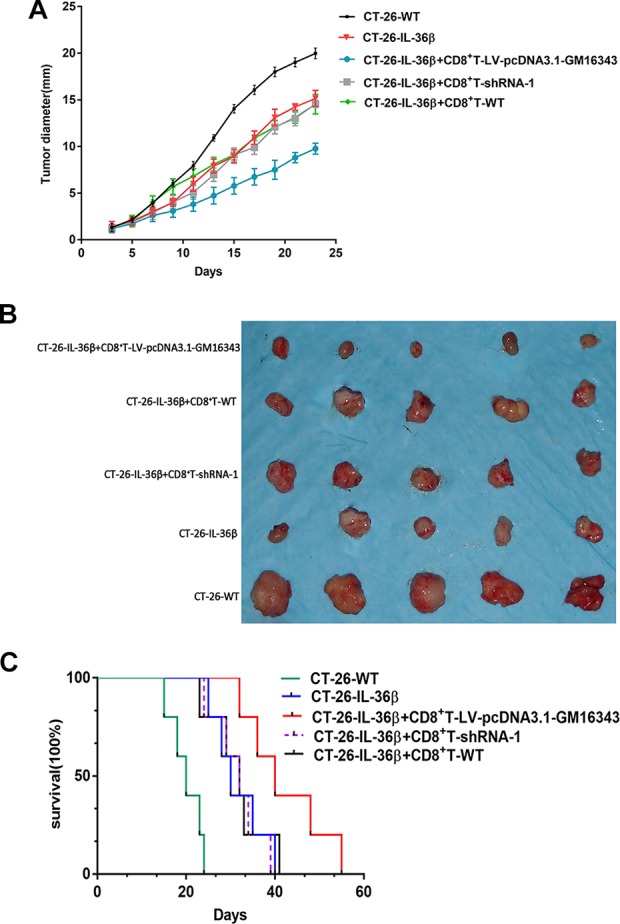
(A) Tumor diameter after overexpression or knockdown of GM16343. The vectors were injected into the tumor tissues. Seven days after the first injection, the injection was again performed according to the original protocol. The tumor diameter was measured every 2 days. Five mice in each group and the diameter of tumors were expressed as mean ± standard error of mean (SEM). *P < .05 (CT-26-IL-36β+CD8+T-LV-pcDNA3.1-GM16343 group was compared with CT-26-IL-36β group). (B) The picture showing the tumor nodules in each group. (C) Survival of mice was monitored. Five mice in each group. CT-26-IL-36β+CD8+T-LV-pcDNA3.1-GM16343 group was compared with CT-26-IL-36β group (P < .01) and determined by log-rank test. IL-36β indicates interleukin 36β.
GM16343 Affects Mouse Prognosis by Influencing the Antitumor Immune Effects of CD8+T Cells
CD8+T cells transfected with pcDNA3.1-GM16343 or shRNA-GM16343 were injected into the tumor tissues, and the survival of the mice was observed daily. The results showed that the survival time of CT-26-IL-36β+CD8+T-LV-pcDNA3.1-GM16343 group was significantly longer than that of CT-26-WT group (P < .01). The survival time of CT-26-IL-36β+CD8+T-shRNA-1 group was significantly shorter than that of the CT-26-WT group, but no significant differences were observed when compared to CT-26-IL-36β group (Figure 7B).
Discussion
In this study, CT-26 colon cancer cells were injected into mice to induce tumorigenesis. This study aimed to explore the biological mechanism of IL-36β promoting CD8+T cells to regulate tumor microenvironment. Interleukin 36 plays an important role in the pathogenesis of several diseases such as psoriasis,14 pneumonia,15 chronic kidney disease,16 and rheumatoid arthritis.17 During skin inflammation, IL-36 promotes inflammation by activating keratinocytes, antigen-presenting cells, and T cells. In addition, IL-36 also promoted dendritic cell maturation and T cell proliferation.18 Studies have shown that IL-36 promoted the activation of NLRP3 inflammation in renal innate cells and immune cells, and also the activation of IL-23/IL17 axis, thereby promoting renal fibrosis and accelerating tubulointerstitial lesions.16 Recently, more and more studies have been conducted on the role of IL-36 in tumor microenvironment. Weinstein et al found that IL-36γ can be detected in colorectal cancer tissues, and IL-36γ produced by macrophages and vascular endothelial cells promoted and maintained tumor immune response.19 Moreover, IL-36α showed close relation with the size of colorectal tumors and tumor–node–metastasis (TNM) staging, and IL-36α also acts as an independent prognostic factor.20 In vivo experiments showed that the tumor volume of fibrosarcoma mice treated with IL-36 gene was significantly reduced, the expression of tumor marker Ki-67 was decreased, and the expression of IFN-γ was enhanced.21 Previous studies have confirmed that IL-36γ can stimulate CD8+T cells to secrete IFN-γ and promoted the proliferation of CD8+T cells, NK cells, and γδT cells, which thus promoted immune response of the tumors. Therefore, this experiment verified the tumor immune effect by detecting the amount of IFN-γ. In vivo experiments have proved that CD8+T cells and NK cells are necessary for IL-36γ to exert its antitumor effects.4 MyD88 is an important transduction protein in the Toll-like receptor signaling pathway, and IL-36 receptor is a Toll-like receptor. Therefore, when IL-36 binds to its receptor, signal transduction into the cell depends on MyD88, and so MyD88 is considered to be an essential molecule for IL-36 intracellular signal transduction. While KO of MyD88 cannot transmit IL-36 signals downstream.22,23
As a regulator of gene expression in the immune system, lncRNAs play a key role in immune system,24,25 especially in cancer immunotherapy. The lncRNAs are used as prognostic markers and in gene targeting therapy.26,27 Pang et al analyzed more than 100 lncRNAs that are differentially expressed in CD8+T cells, and the results showed to have the characteristics of regulating cell differentiation, evolutionary protection, and promoter regulation.28 In conclusion, this study aimed to explore the mechanism of lncRNA in enhancing antitumor response of CD8+T cells by IL-36β.
Here, the gene chip and bioinformatics analysis were performed to show the involvement of differentially altered lncRNAs in a variety of biological functions and signaling pathways. Among them, the GM16343 demonstrated the biggest difference. Real-time PCR results showed that the changing trend of GM16343 was consistent with that of the chip results. Overexpression or knockdown of GM16343 gene in CD8+T cells stimulated these CD8+T cells with IL-36β, and the expression of IFN-γ was in turn affected by the expression of GM16343. To verify the antitumor effects of GM16343 on IL-36β, CD8+T cells overexpressing or silencing GM16343 was injected into the tumor tissues of mice. These tumors stably expressed IL-36β. The results showed that the expression of GM16343 significantly inhibited the growth of tumors and improved the survival time of mice. Therefore, we determined that lncRNA GM16343 participated in the process of IL-36β promotion in the secretion of IFN-γ by CD8+T cells, providing a new understanding on the biological process of IL-36 in the direction of tumor immunity. How IL-36β regulates the expression of lncRNA GM16343 may be related to transcriptional regulation, histone modification, and lncRNA stability regulation. Xie et al found that in squamous cell carcinomas, TP63 binds to super enhancers and activates LINC01503 transcription.29 Long ncRNA CCAT1 is activated by H3K27 acetylation and affects cell proliferation and migration.30 In addition, the recruitment of CCR4–NOT complex by the adaptor protein IGF2BP1 leads to a decrease in the stability of lncRNA HULC and promotes degradation.31 Hence, IL-36β may promote the expression of lncRNA GM16343 through the above regulatory mechanisms.
In conclusion, IL-36β may promote the secretion of IFN-γ by CD8+T cells through lncRNA GM16343, thereby exerting an antitumor effects. We believed that the results of this study provide new insights into the understanding of the role of IL-36 in tumor immunity, but its specific regulatory mechanisms should be further studied.
Abbreviations
- GO
gene ontology
- IL
interleukin
- IFN- γ
interferon γ
- KO
knockout
- lncRNA
long noncoding RNA
- ncRNA
noncoding RNA
- NC-shRNA
negative control shRNA
- RT-PCR
real-time polymerase chain reaction
- shRNA
short hairpin RNA
- WT
wild type
Footnotes
Authors’ Note: Deli Mao, Chenrui Hu and Jianglei Zhang have contributed equally to this work. Xin Zhao and Xiaoqiang Dong proposed the research and are guarantors. Deli Mao wrote the manuscript. Deli Mao, Chenrui Hu and Jianglei Zhang contributed equally to the current work. All authors contributed to the study design and data interpretation and have reviewed the final version of the manuscript. This study was approved by the First Affiliated Hospital of Soochow University Ethical Committee (approval no. 2012634). The Soochow University Medical College Animal Care and Use Committee approved the experimental procedures (approval no. 2018265). All animal housing and experiments were conducted in strict accordance with the Institutional Guidelines for Care and Use of Laboratory Animals.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by grants from the National Science Foundation of China (NSFC, no. 31770985), Social Development Project of Jiangsu Province, China (no. BE2019665), Postdoctoral Science Foundation Grant of China (no. 2016M591913), Jiangsu Provincial Medical Youth Talent, China (no. QNRC2016732), Jiangsu Provincial “ Six Peaks Talent ” Program, China (no. 2016-WSW-043). Suzhou Municipal Project of Gusu Health Talent, Young Top Talent, China (no. 2018-057), Gusu Health Talents Cultivation Program, China (no. GSWS2019028), Scientific Research Program of Jiangsu Provincial “333 Projects”, China (no. BRA2019327), Science and Technology Program of Suzhou City, China (no. SYS2019053, no. SLC201906), Provincial Natural Science Foundation of Jiangsu Province, China (no. BK20161225), Scientific Research Program of Jiangsu Provincial Commission of Health and Family Planning, China (no. H201620) and The Natural Science Foundation of the Jiangsu Higher Education Institution of China (grant no. 18KJB320015).
ORCID iDs: Deli Mao  https://orcid.org/0000-0001-5430-0883
https://orcid.org/0000-0001-5430-0883
References
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5): E359–E386. [DOI] [PubMed] [Google Scholar]
- 2. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. [DOI] [PubMed] [Google Scholar]
- 3. Lalani AA, McGregor BA, Albiges L, et al. Systemic treatment of metastatic clear cell renal cell carcinoma in 2018: current paradigms, use of immunotherapy, and future directions. Eur Urol. 2019;75(1):100–110. [DOI] [PubMed] [Google Scholar]
- 4. Wang X, Zhao X, Feng C, et al. IL-36γ transforms the tumor microenvironment and promotes type 1 lymphocyte-mediated antitumor immune responses. Cancer Cell. 2015;28(3):296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang C, Shen C, Feng T, Li H. Noncoding RNA in NK cells. J Leukoc Biol. 2019;105(1):63–71. [DOI] [PubMed] [Google Scholar]
- 7. Zhan F, Shen J, Wang R, et al. Role of exosomal small RNA in prostate cancer metastasis. Cancer Manag Res. 2018;10:4029–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(1):R17–R29. [DOI] [PubMed] [Google Scholar]
- 9. Xu Y, Ge Z, Zhang E, et al. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis. 2017;8(10):e3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlson HL, Quinn JJ, Yang YW, Thornburg CK, Chang HY, Stadler HS. LncRNA-HIT functions as an epigenetic regulator of chondrogenesis through Its recruitment of p100/CBP complexes. PLoS Genet. 2015;11(12):e1005680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu YW, Kang CM, Zhao JJ, et al. LncRNA PLAC2 down-regulates RPL36 expression and blocks cell cycle progression in glioma through a mechanism involving STAT1. J Cell Mol Med. 2018;22(1):497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hou J, Long H, Zhou C, et al. Long noncoding RNA Braveheart promotes cardiogenic differentiation of mesenchymal stem cells in vitro. Stem Cell Res Ther. 2017;8(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo J, Tu J, Hu Y, Song G, Yin Z. Cathepsin G cleaves and activates IL-36γ and promotes the inflammation of psoriasis. Drug Des Devel Ther. 2019;13:581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nanjo Y, Newstead MW, Aoyagi T, et al. Overlapping roles for interleukin-36 cytokines in protective host defense against murine legionella pneumophila pneumonia. Infect Immun. 2019;87(1). doi:10.1128/IAI.00583-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chi HH, Hua KF, Lin YC, et al. IL-36 signaling facilitates activation of the NLRP3 inflammasome and IL-23/IL-17 axis in renal inflammation and fibrosis. J Am Soc Nephrol. 2017;28(7):2022–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frey S, Derer A, Messbacher ME, et al. The novel cytokine interleukin-36α is expressed in psoriatic and rheumatoid arthritis synovium. Ann Rheum Dis. 2013;72(9):1569–1574. [DOI] [PubMed] [Google Scholar]
- 18. Foster AM, Baliwag J, Chen CS, et al. IL-36 promotes myeloid cell infiltration, activation, and inflammatory activity in skin. J Immunol. 2014;192(12):6053–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weinstein AM, Giraldo NA, Petitprez F, et al. Association of IL-36γ with tertiary lymphoid structures and inflammatory immune infiltrates in human colorectal cancer. Cancer Immunol Immunother. 2019;68(1):109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang ZS, Cong ZJ, Luo Y, et al. Decreased expression of interleukin-36α predicts poor prognosis in colorectal cancer patients. Int J Clin Exp Pathol. 2014;7(11):8077–8081. [PMC free article] [PubMed] [Google Scholar]
- 21. Solahaye-Kahnamouii S, Farhadi F, Rahkare-Farshi M, et al. The effect of interleukin 36 gene therapy in the regression of tumor. Iran J Cancer Prev. 2014;7(4):197–203. [PMC free article] [PubMed] [Google Scholar]
- 22. Nishida A, Inatomi O, Fujimoto T, Imaeda H, Tani M, Andoh A. Interleukin-36α induces inflammatory mediators from human pancreatic myofibroblasts via a MyD88 dependent pathway. Pancreas. 2017;46(4):539–548. [DOI] [PubMed] [Google Scholar]
- 23. Li C, Maillet I, Mackowiak C, et al. Experimental atopic dermatitis depends on IL-33 R signaling via MyD88 in dendritic cells. Cell Death Dis. 2017;8(4):e2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morchikh M, Cribier A, Raffel R, et al. HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol Cell. 2017;67(3):387–399. e385. [DOI] [PubMed] [Google Scholar]
- 25. Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18(9):962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang R, Tang J, Chen Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017;8:15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smolle MA, Calin HN, Pichler M, Calin GA. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. FEBS J. 2017;284(13):1952–1966. [DOI] [PubMed] [Google Scholar]
- 28. Pang KC, Dinger ME, Mercer TR, et al. Genome-wide identification of long noncoding RNAs in CD8+T cells. J Immunol. 2009;182(12):7738–7748. [DOI] [PubMed] [Google Scholar]
- 29. Xie JJ, Jiang YY, Jiang Y, et al. Super-enhancer-driven long non-coding RNA LINC01503, regulated by TP63, is over-expressed and oncogenic in squamous cell carcinoma. Gastroenterol. 2018;154(8):2137–2151. e2131. [DOI] [PubMed] [Google Scholar]
- 30. Zhang E, Han L, Yin D, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45(6):3086–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hammerle M, Gutschner T, Uckelmann H, et al. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatol. 2013;58(5):1703–1712. [DOI] [PubMed] [Google Scholar]