Short abstract
Recapitulating human disease pathophysiology using genetic animal models is a powerful approach to enable mechanistic understanding of genotype–phenotype relationships for drug development. NaV1.7 is a sodium channel expressed in the peripheral nervous system with strong human genetic validation as a pain target. Efforts to identify novel analgesics that are nonaddictive resulted in industry exploration of a class of sulfonamide compounds that bind to the fourth voltage-sensor domain of NaV1.7. Due to sequence differences in this region, sulfonamide blockers generally are potent on human but not rat NaV1.7 channels. To test sulfonamide-based chemical matter in rat models of pain, we generated a humanized NaV1.7 rat expressing a chimeric NaV1.7 protein containing the sulfonamide-binding site of the human gene sequence as a replacement for the equivalent rat sequence. Unexpectedly, upon transcription, the human insert was spliced out, resulting in a premature stop codon. Using a validated antibody, NaV1.7 protein was confirmed to be lost in the brainstem, dorsal root ganglia, sciatic nerve, and gastrointestinal tissue but not in nasal turbinates or olfactory bulb in rats homozygous for the knock-in allele (HOM-KI). HOM-KI rats exhibited normal intraepidermal nerve fiber density with reduced tetrodotoxin-sensitive current density and action potential firing in small diameter dorsal root ganglia neurons. HOM-KI rats did not exhibit nociceptive pain responses in hot plate or capsaicin-induced flinching assays and did not exhibit neuropathic pain responses following spinal nerve ligation. Consistent with expression of chimeric NaV1.7 in olfactory tissue, HOM-KI rats retained olfactory function. This new genetic model highlights the necessity of NaV1.7 for pain behavior in rats and indicates that sufficient inhibition of NaV1.7 in humans may reduce pain in neuropathic conditions. Due to preserved olfactory function, this rat model represents an alternative to global NaV1.7 knockout mice that require time-intensive hand feeding during early postnatal development.
Keywords: Neuropathic pain, SCN9A, NaV1.7 voltage-gated sodium channel, rat knockin, nociception, anti-NaV1.7 antibody
Introduction
To address the ongoing public health crisis driven by opioid abuse and addiction, it is essential to identify new targets and therapies for chronic pain, a critical unmet medical need afflicting over 100 million Americans.1–3 Improved understanding of pain neurobiology has provided opportunities to test novel hypotheses in the clinic in cohorts afflicted by neuropathic pain arising from damage to the somatosensory system that is particularly recalcitrant to treatment.4 However, failure of a large number of these new molecular entities highlights the complexity of drug development in this space.5 By focusing drug development efforts on targets with human genetic validation, the success rate to progress from Phase 1 to drug approval nearly doubles,6–8 pointing to a key strategy for new pain target selection.
The voltage-gated sodium ion channel NaV1.7 regulates action potential (AP) firing in nociceptor neurons within the peripheral nervous system and represents a particularly attractive drug target for the development of novel analgesic therapies due to its exquisite validation via human genetics.9,10 Gain-of-function mutations drive spontaneous or evoked pain in primary erythromelalgia, paroxysmal extreme pain disorder, and small fiber neuropathy,11–13 whereas loss-of-function (LOF) mutations result in congenital insensitivity to pain (CIP) with anosmia.14–16
Attempts to identify antagonists of NaV1.7 for pain has yielded diverse and selective pharmacological tools that bind different sites and block different channel gating states.17–19 Potent and selective antagonists in the sulfonamide chemotype stabilize NaV1.7 in an inactivated conformation via interaction with a site within the fourth voltage-sensor domain.20–25 Lack of robust block of rat NaV1.7 channels by sulfonamides is due to key amino acid sequence differences in this region and has resulted in the utilization of mouse pharmacodynamic and efficacy models to evaluate pre-clinical target engagement in vivo.21,23,24,26,27
Mouse models with various NaV1.7 genetic deficiencies have helped to identify key pain endpoints and behaviors, which are dependent on proper channel function. Global NaV1.7 knockout (KO) in mice recapitulated the CIP phenotype with anosmia.28 These animals require daily hand feeding using an artificial mouse milk formulation for a period of up to three weeks to increase viability; this need is hypothesized to be driven by their anosmic profile in addition to their inbred genetic background. Conditional KO of NaV1.7 in specific dorsal root ganglia (DRG) neuronal sub-populations mitigated pain behavior following both inflammatory and nerve injury insults29–32 and inducible KO of NaV1.7 in adult mice identified NaV1.7-dependent endpoints in the absence of any potential confounding compensatory changes that may arise during development of the nervous system.33 However, the global NaV1.7 KO mouse is difficult to rear, and the conditional NaV1.7 KO mice may not totally recapitulate the human CIP phenotype because of tissue- or age-dependent loss of NaV1.7. There is a need for additional animal models to support research on NaV1.7.
Here, we describe the generation and characterization of the first rat model with a regional deficiency in NaV1.7. We initially set out to generate a humanized NaV1.7 rat expressing a chimeric NaV1.7 protein containing human exon 26 in place of the rat equivalent sequence, encoding for the sulfonamide-binding region, to enable in vivo prosecution of sulfonamide-based chemical matter in rat pain models. Unexpectedly, the inserted human exon 26 was spliced in a tissue-specific manner, resulting in a premature termination codon prior to the fourth voltage-sensor domain. Using a validated selective C-terminal-detecting NaV1.7 monoclonal antibody, we confirmed loss of detectable protein in the nervous system except for olfactory sensory neurons where the intended chimeric NaV1.7 protein was detected, albeit at lower levels. Consistent with these findings, rats homozygous for the knock-in allele (HOM-KI) were deficient in both nociceptive and neuropathic pain behavior yet retained olfactory function. This novel model represents an additional tool to evaluate NaV1.7-dependent pain biology in rats and, due to retention of olfactory function, offers an advantage to the global NaV1.7 KO mouse that requires time-intensive hand feeding during early development.
Materials and methods
Animal model generation
The Scn9atm1(Scn9a*26)rScn9a.AMG/Charles River Laboratories (CRL) Sprague Dawley (SD) rat model was designed by Amgen, adapted and executed by SAGE Labs, Sigma-Aldrich (now a subsidiary of Horizon Discovery, Saint Louis, MO, USA) using zinc finger nuclease (ZFN)-based technology. Briefly, exon 25 of the rat Scn9a gene was replaced with the human SCN9A exon counterpart (exon 26; see Figure 1). Using alternative splicing prediction algorithms, we validated our genetic approach in ensuring that we would not impact the predicted splice donor/acceptor domains in the rat gene. ZFN pairs targeting the rat Scn9a exon 25 were assembled and ZFN mRNAs were transcribed using T7 RNA polymerase-based in vitro transcription method. Each ZFN pair was validated in cultured rat C6 cells by Surveyor nuclease assay. The mRNAs of the active ZFN pair targeting the middle region of the exon (CACCATCATGGTTCTTATAtgcctcAACATGGTA A CCATGATG, ZFN binding sites in uppercase) along with donor DNA plasmid comprising a human SCN9A exon sequence were microinjected into pronuclei of fertilized one-cell embryos from SD rats. In sum, 25 to 30 eggs were transplanted into each pseudo-pregnant female. Resulting live births were screened for correction integration by polymerase chain reaction (PCR) with Cel-1 recombinant nuclease and junction primer pairs, restriction enzyme NdeI digestion, and sequencing of the region flanking the integration site. A total of 57 chimeric animals were screened. Founder 54 displayed the expected integration profile and was selected and backcrossed to a wild-type (WT) SD rat. A large cohort of heterozygous (HET) animals was generated for breeding purposes. HET × HET breeding gave rise to the expected Mendelian ratios [1:2:1; wild type (WT)/ heterozygous (HET) / homozygous (HOM)]. All animal work was performed in accordance with the approved animal protocols overseen by SAGE Labs’ Institutional Animal Care and Use Committee and by the Veterans Administration Connecticut Healthcare System Institutional Animal Care and Use Committee.
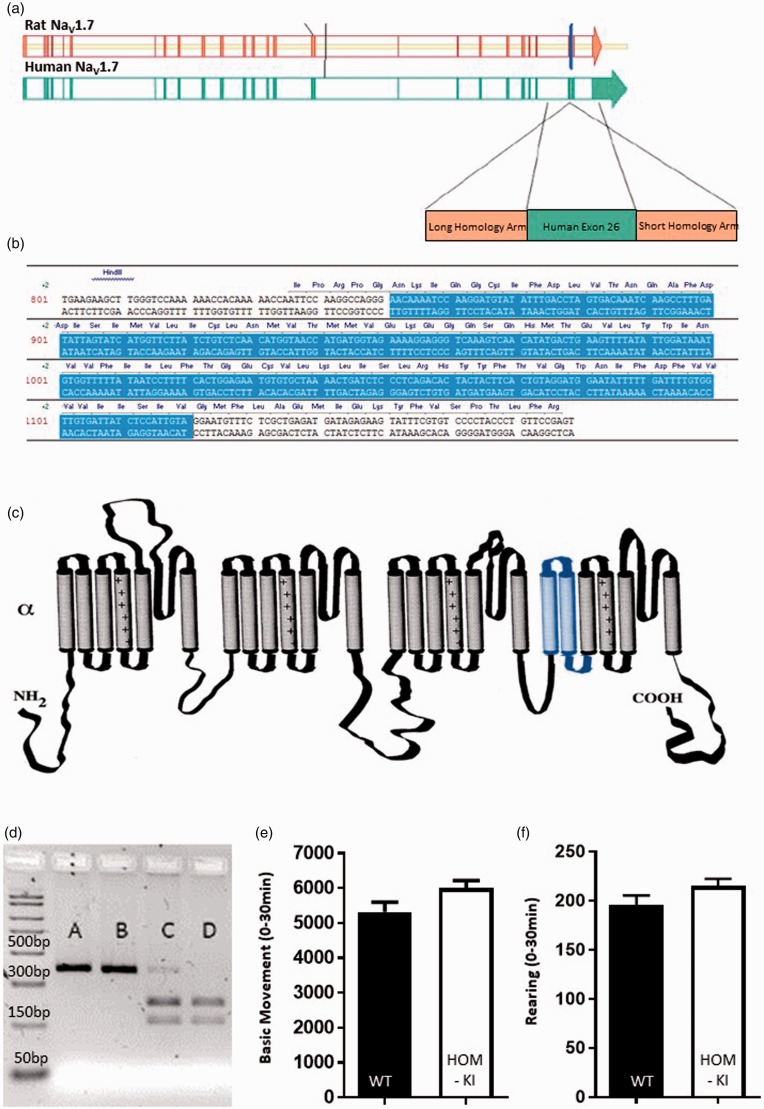
Figure 1.
Strategy to generate humanized chimeric NaV1.7 rat. (a) Rat NaV1.7 exon 25 (blue vertical line) was replaced with the corresponding sequence in human NaV1.7, which is human exon 26. (b) Schematic representation of the Human NaV1.7 exon 26 sequence (blue shading) inserted into rat NaV1.7 sequence (white shading) to generate the resulting chimeric NaV1.7 channel. (c) Predicted topology of rat NaV1.7 protein with insertion of human exon 26 coding sequence (blue shading) comprised of the first two transmembrane domains and S2-S3 intracellular loop of domain IV. (d) Representative restriction digest analysis of PCR products originating from genomic DNA from HOM-KI (A and B), HET (C) and WT (D) rats. (e) Open-field basic movement assessment. HOM-KI (5997 ± 220 counts) and WT (5318 ± 281 counts) rats had similar levels of exploratory movement (t18 = 1.9, p = 0.07, unpaired t-test, n = 10/group). (f) Open-field rearing count assessment. HOM-KI (215 ± 7 counts) and WT (196 ± 10 counts) rats had similar rearing counts (t18 = 1.6, p = 0.13, unpaired t-test, n = 10/group). (Data are mean ± SEM).
Husbandry
The colony was housed and bred at CRL in accordance with the Guide for the Care and Use of Laboratory Animals, 8th Edition (National Research Council Committee for the Update of the Guide for the and Use of Laboratory, 2011). All animals were housed in individual isolator units maintained under positive pressure with a separate conditioned and high-efficiency particulate air-filtered air supply. Within the isolator, animals were housed in solid bottom, polycarbonate cages with wire bar lids. Food was provided ad libitum. Water was provided by water bottles and the bedding in all cages was removed and replenished at least once per week. Feed was a commercial, gamma-irradiated, natural ingredient rodent chow (vacuum-packed) with 18% protein and 5% fat. Bedding was a hard wood shaving (Aspen) that is irradiated and vacuum-packed. At Amgen, adult male and female rats weighing 220 to 400 g were used for experiments and were cared for in accordance to the Guide for the Care and Use of Laboratory Animals, 8th Edition (National Research Council Committee for the Update of the Guide for the and Use of Laboratory, 2011). Animals were group-housed at an Association for Assessment and Accreditation of Laboratory Animal Committee accredited facility in nonsterile ventilated micro-isolator housing on corn cob bedding. All research protocols were approved by Amgen’s Institutional Animal Care and Use Committee. Animals had ad libitum access to pelleted feed (Harlan Teklad 2020X, Indianapolis, IN) and water (on-site generated reverse osmosis) via an automatic watering system. Animals were maintained on a 12:12 h light: dark cycle in rooms at 21 ± 3°C, 50 ± 20% room humidity and had access to enrichment opportunities (nesting materials and plastic domes). All animals were sourced from approved vendors who meet or exceed animal health specifications for the exclusion of specific pathogens. HOM-KI rats exhibited spontaneous scratching and nails were clipped to minimize wounds. Spontaneous scratching behavior was never observed during pain behavioral testing. For this reason, scratching behavior did not confound any behavioral test conducted. Animals with significant scars were excluded from behavioral assays. Although experimenters did not know the genotypes of randomized rat cohorts during testing, it was not possible to use animals without any scars and, for this reason, experiments could not be truly blinded. Animals were allowed at least 1 week acclimation to the facility prior to any procedures. Following completion of behavioral measurements, animals were euthanized with carbon dioxide. All behavioral data were scored by a trained observer blind to the genotype. Global NaV1.7 KO mice were bred and cared for as previously described.28
At the Yale University Center for Neuroscience and Regeneration Research, all animal studies and procedures followed a protocol approved by the Veterans Administration Connecticut Healthcare System Institutional Animal Care and Use Committee. Adult male and female rats weighing 220 to 400 g were used for experiments and were cared for in accordance to the update of Guide for the Care and Use of Laboratory Animals. Animals were group housed at an Association for Assessment and Accreditation of Laboratory Animal Committee accredited facility in nonsterile solid bottom polycarbonate cages with wire bar lids on corn cob bedding; bedding in all cages was removed and replenished at least once per week. Food (Teklad 2018 18% protein rodent diet by Envigo) and water (water bottles) was provided ad libitum. Animals were maintained on a 12:12 h light: dark cycle in rooms at 21 ± 3°C, 50 ± 20% room humidity and had access to enrichment opportunities in the form of nesting materials.
Genotyping
DNA from tail clips or ear samples was extracted using the KAPA Hot Start Mouse Genotyping Kit (KAPA Biosystems). The following primers (IDT DNA, Coralville, IA), originally designed by SAGE, were used to genotype all animals: hScn9a-SAGE-F: 5′-TCTC AACTCCTCCCAGAACC-3′; and hScn9a-SAGE-R: 5′-ACGGCCTTACCTACAATGGA-3′ under a standard 5 step PCR program. Briefly, DNA denaturation was performed at 95°C for 3 min and 96°C for 15 s; annealing at 62°C for 30 s with a 0.5°C drop in temperature per cycle beginning at cycle 2, extension at 72°C for 1.5 min for a total of 37 cycles, including the final extension step at 72°C for 7 min. The resulting PCR products were further processed using Rsal restriction digestion to identify the endogenous rat (WT) allele. The expected calculated product sizes were of 402 base pairs (bp) for the mutant allele and 217 bp + 165 bp for the WT allele (Figure 1(d)). HOM-KI rats are SD-Scn9atm1 (SCN9A*26/SCN9A*26) HET rats are SD-Scn9atm1 (SCN9A*26/+). WT rats are SD-Scn9a+/+.
Initial transcript assessment for characterization
Rat DRGs from all spinal levels of each genotype were dissociated as previously described.28 RNA was isolated using a RNeasy kit (Qiagen Inc.—USA, Germantown, MD) and Covaris S2 Focused Ultrasonicator (Covaris, Woburn, MA) in 400 µl of RLT w/beta-mercaptoethanol (BME) buffer. The concentration of RNA was assessed by NanoDrop (ThermoScientific, Waltham, MA) and 1µg of RNA was converted to cDNA using an Advantage RT-for-PCR kit with random hexamers—(CloneTech/Takara, Mountain View, CA) cDNA fragments, spanning the area of interest were amplified with the following primers: 5′ cDNA primer—5′ATAGATA ATTTCAACCAACAGAAAA3′ and 3′ cDNA primer—5′TGTCCCCTACCCTGTTCC GAGT3′ using Q5 High-Fidelity DNA Polymerase with Q5 High GC Enhancer (NEB, Ipswich, MA). Amplified cDNA fragments were analyzed by 1% TAE agarose gels and sequenced.
Western blots
Samples were isolated and lysed in 1% Triton-X-100, 25mM Hepes, 150 mM NaCl, 1 mM EDTA buffer, 1 Complete EDTA-free Tablet (Sigma Aldrich, St. Louis, MO) using a Covaris S2 Focused Ultrasonicator (Covaris, Woburn, MA). Protein concentration was measured using bicinchoninic acid assay kit (ThermoFisher Scientific, Waltham MA). NaV1.7 was immunoprecipitated with 10 µg NaV1.7 Ab (Neuromab, Cat no. 75–103, the University of California (UC) Davis/National Institutes of Health (NIH) NeuroMab Facility, Davis, CA) or (Millipore Sigma Cat no. AB5390, Burlington, MA) or (Pan-Nav Ab, Alomone Labs, Cat no. ASC-003, Jerusalem, Israel) overnight at 4°C, followed by PeggySue (ProteinSimple, San Jose, CA) blotting with anti-NaV1.7 Ab (Neuromab, Cat no. 75–103, UC Davis/NIH NeuroMab Facility, Davis, CA) followed by Streptavidin (SA)-horse radish peroxidase (HRP) anti-mouse IgG H + L (ProteinSimple, San Jose, CA) detection. Nav stable cells lines were as previously described.26
Immunohistochemistry
All animals used for histology purposes, except for intraepidermal nerve fiber (IENF) analysis (see details below), were transcardially perfused using phosphate-buffered saline (PBS) (Life Technologies, Grand Island, NY) followed by fresh 4% paraformaldehyde/PBS (EMS, Hatfield, PA), until the liver was cleared of all signs of blood (∼50–75 ml of solution). Tissues from WT and HOM-KI adult (10–12-week-old) rats were post-fixed in same fixation solution overnight, rinsed in PBS (3 × 10 min) and embedded in paraffin or cryoprotected in 30% sucrose/PBS for at least 24 h, until the tissue sunk to the bottom of the vessel for embedding in Tissue Freezing Media (ThermoFisher Scientific, Pittsburg, PA). Paraffin blocks were kept at room temperature, while frozen blocks were placed in a −20°C freezer to set and stored therein until ready to use. Long-term storage of samples, when required, was done at −80°C. Tissue blocks were sliced into 40 µM sections on a Leica VT1000S vibratome or 25 µm sections on a Leica CM1850 cryostat (Leica Biosystems, Buffalo Grove, IL), placed onto glass microscope slides (Ink Jet Plus, Thermo Fisher Scientific, Pittsburg, PA), and air dried for 30 min at room temperature. All immunostaining incubations were performed at room temperature, unless otherwise noted. Slides were rehydrated in descending concentrations of ethanol to diH2O (100% ethanol; 100% ethanol; 95% ethanol; 95% ethanol; 80% ethanol; 70% ethanol; running diH2O; 3 min per bath) then processed through a Borg Decloaker for antigen retrieval (Biocare BD1000G1). Samples were then blocked for 30 min in Tris-NaCl-blocking (TNB) buffer (0.1 M Tris-HCL (pH 7.5))/0.15 M NaCl/0.5% (w/v) blocking reagent (PerkinElmer FP1012). A second blocking step was deemed necessary to block nonspecific rat IgG on the surface of the sectioned tissue using rodent block R (BioCARE Cat no. RBR962 G, H, L) for 30 min followed by three 5-min washes in Tris-buffered saline (TBS) buffer. NaV1.7 was detected with a selective anti-NaV1.7 antibody (NeuroMab, Cat no. 75–103 mouse monoclonal IgG1; @ 5 µg/ml in TNB block) overnight at 4°C. An isotype control was also used as an additional control in identical conditions (anti-Mouse IgG1 (BD Bioscience 554121)). Primary antibody incubations were followed by three 5 min washes in TBS and a 1 h incubation in Biotinylated horse anti-mouse solution (Vector BA2001; rat absorbed; 1 µg/ml diluted in TNB block) followed by a 10 min Peroxidazed 1 reaction (Biocare PX968M) and two 5 min washes in 1× TBS. Slides were then incubated for 30 min in a SA-HRP solution (PerkinElmer NEL75000; 1:200 in TNB block) washed twice in 1× TBS and amplified using Tyramide Signal Amplification (TSA) Biotin reagent (PerkinElmer SAT70001EA; 1:50) followed by two washes in 1× TBS. A second identical round of amplification (wash/SA-HRP/wash TSA/wash) was required due to the low affinity of the Nav1.7 antibody against the Nav1.7 rat protein. This second amplification was identical to the first detailed above, with only the TSA kit differing (Perkin Elmer, NEL744001KT, Cyanine 3 system, dilute 1:50 in amplified diluent). Finally, samples were placed under running water for 10 min and mounted in Prolong Gold containing 4', 6-diamidino-2-phenylindole (DAPI, ThermoFisher Cat no.P36935). Imaging was performed using a Zeiss LSM800 equipped with Zen Blue software (Carl Zeiss). Pixel quantification was performed using the Zen Blue analysis module and image assembly performed (with no correction steps) using Adobe Creative Cloud/Photoshop software.
Immunohistochemistry for IENF density in skin
Rats (9–10 weeks of age, both male and female) were deeply anesthetized with ketamine/xylazine (80/5 mg/kg, i.p.) and transcardially perfused with 0.01 M PBS (pH 7.4) followed by ice-cold 4% paraformaldehyde in 0.14 M Sorensen’s phosphate buffer (pH 7.4). Foot pad skin tissues were removed, immersion-fixed in 4% paraformaldehyde (total fixation time 20 min), and cryo-protected with 30% (w/v) sucrose in PBS overnight at 4°C. Tissue sections were cut on a cryostat at 10 µm and mounted on slides (Fisher Scientific, Pittsburgh, PA). Sections were immediately processed for detection of target protein or stored at −20°C for future use.
Sections were incubated in the following solutions: (1) blocking solution (PBS containing 4% normal donkey serum, 2% bovine serum albumin, 0.1% Triton X-100, and 0.02% sodium azide) for 1 h at room temperature; (2) rabbit monoclonal anti-PGP 9.5 antibody (1:300, Abcam, Cat no. ab108986, Batch no. GR3231441-1) in blocking solution at 4°C overnight; (3) PBS, 3 × 10 min each; (4) Alexa Fluor 546-conjugated donkey anti-rabbit IgG (H + L) secondary antibody (Invitrogen) at 1:1000 dilution in blocking solution for 1 h at room temperature; (5) PBS, 3 × 10 min. Tissue sections were mounted in antifade mounting medium with DAPI (Vectashield, Vectorlabs) and were examined with a Nikon Eclipse E800 fluorescence microscope or a Nikon C1 confocal microscope (Nikon USA, Melville, NY).
Light microscopic analysis of axons of sciatic nerve
Rats were deeply anesthetized with ketamine/xylazine (80/5 mg/kg, i.p.) and transcardially perfused with 0.01 M PBS (pH 7.4) followed by ice-cold 4% paraformaldehyde in 0.14 M Sorensen’s phosphate buffer (pH 7.4). Left and right sciatic nerves were dissected carefully from the mid-thigh level (distal to the trifurcation) and transferred to 2% paraformaldehyde plus 2% glutaraldehyde in 0.14 M Sorensen’s phosphate buffer (pH 7.4) at 4°C overnight. Samples were then post-fixed with 1% osmium (Polysciences, Warrington, PA, USA), dehydrated and blocked in 0.5 cm segments and embedded in Epox-812 (Ernest F. Fullam, Latham, NY, USA) using standard plastic embedding protocols. Semithin sections (1 µm) were collected from each tissue block and counterstained with methylene blue and azure II (0.5% each in 0.5% borax) for light microscopy.
Axon diameters were calculated from axonal cross-sectional area measured by ImageJ software. For each animal, 1200 to 1800 axons were measured from four photomicrographs (40×) taken at random areas in tibial and common peroneal nerve cross-sections. Axonal size distributions were presented in 1 µm bins. Data are expressed as the mean ± standard error of the mean (SEM).
DRG RNA isolation for ddPCR
DRGs, pooled from cervical, lumbar, thoracic, and sacral levels, were dissected from four WT and four HOM-KI rats. All DRGs from individual animals were collected in 1.5 mL microcentrifuge tubes containing 500 µL of RNALater buffer (ThermoFisher Cat no. AM7021), stored at 4°C overnight, and transferred to −20°C until processing. DRGs were removed from RNALater solution using forceps cleaned with RNase AWAY™ (ThermoFisher Cat no. 7002) followed by 70% isopropanol, blotted on a fresh Kimwipe (Fisher Scientific Cat no. 06–666), and placed in a new 1.5 mL RNase-free microcentrifuge tube (Kimble Cat no. 749520–0090) containing 350 µl of RLT Plus buffer (Qiagen Cat no. 74134) with 1% BME (Sigma-Aldrich, Cat no. M3148). DRGs were homogenized using an RNase-Free Disposable PELLET PESTLE® (Kimble Cat no. 749520–0090) attached to a PELLET PESTLE® Cordless Motor (Kimble Cat no. 749540–0000) until the lysate was clear and no particulate matter was visible. The homogenate was run through a QIAshredder column (Qiagen Cat no. 79654) for 2 min at full speed in a benchtop centrifuge and the flow-through was retained. RNA was then isolated using the Qiagen RNeasy Mini Plus kit (Qiagen Cat no. 74134) according to manufacturer’s instructions. The resulting RNA was quantified using 1 µl on a NanoDrop Spectrophotometer (Cat no. ND-1000). RNA integrity was assessed with an Agilent RNA 6000 Nano Kit (Agilent Cat no. 5067–1511), and RNA chips run on an Agilent Bioanalyzer 2100 (Agilent Cat no. G2939BA).
cDNA synthesis
Furthermore, 500 ng of total RNA was taken from each DRG sample for synthesis of first-strand cDNA using the SuperScript™ III kit (Invitrogen Cat no. 18080–093). SuperScript reverse transcriptase was replaced by an equal volume of nuclease-free water for the generation of a reverse transcriptase negative control for each sample. Upon completion of the synthesis reaction, each cDNA sample was diluted with 180 µl of IDTE pH 8.0 1× Tris-EDTA (TE) Solution (IDT Cat no. 11–05-01–13) to obtain a final concentration of 5 ng/µl. The diluted cDNA was kept on ice for use in subsequent ddPCR and stored at −20°C.
ddPCR
ddPCR reactions were setup and run according to manufacturer’s protocol using ddPCR™ Supermix for Probes (Bio-Rad Cat no. 1863025). One microliter of cDNA template and 1.25 µl each of 6-fluorescein amidite (FAM)-labeled target gene and hexachrome-fluorescein (HEX)-labeled internal control gene primer/probe assays were added to each reaction. Reactions were emulsified into droplets according to manufacturer’s protocol using QX200™ Droplet Digital™ PCR System (Bio-Rad Cat no. 1864001) and placed in a C1000 thermocycler for PCR amplification (Bio-Rad Cat no. 1851196). Following completion of PCR, the reaction plate was placed in a QX200 reader and droplets were analyzed for presence or absence of amplification. Primers and probes were designed using Primer3 (http://bioinfo.ut.ee/primer3/) or were purchased as pre-designed PrimeTime qPCR Assays (Integrated DNA Technologies [IDT], Coralville, IA). The PrimeTime assays were modified to have a primer/probe ratio of 3.6. Data from the QX200 droplet reader were analyzed on QuantaSoft software V1.5.38.1118 (included in Bio-Rad Cat no. 1864001). The threshold for calling positive droplets was set based on no template control and reverse transcriptase negative control wells. For each sample, the ratio of NaV gene to an internal control gene was determined by dividing the FAM concentration by the HEX concentration. Final ratios were graphed, and unpaired t-tests were calculated using GraphPad Prism v7.02 (GraphPad; La Jolla, CA).
Electrophysiology
Isolation of DRG neurons for whole-cell voltage clamp recordings
DRG neuron isolation and whole-cell patch clamp electrophysiology recordings were conducted using small diameter neurons (20–30 µm) cultured for two to eight days as previously described.26 Neurons were held at −120 mV and stepped to a voltage corresponding to the peak inward sodium current for each individual neuron, which ranged from −30 to 0 mV as determined from current/voltage relationships. Fast-inactivating currents sensitive to tetrodotoxin (TTX-S) currents were defined as peak currents blocked by 0.5 µM TTX, and slow-inactivating currents resistant to tetrodotoxin (TTX-R) currents were defined as residual currents following TTX.
Isolation of DRG neurons for current clamp
DRGs from 4- to 8-week-old WT or HOM-KI rats were harvested and dissociated as described previously34 with minor alterations. In brief, adult rat DRGs were treated in a 20-min incubation at 37°C with 1.5 mg/mL collagenase A (Roche, Indianapolis, IN, USA) and 0.6 mM EDTA, followed by a 17-min incubation at 37°C in 1.5 mg/mL Collagenase D (Roche), 0.6 mM EDTA, and 30 U/mL papain (Worthington Biochemical, Lakewood, NJ, USA). DRGs were then centrifuged and triturated in 0.5 mL of DRG media containing 1.5 mg/mL bovine serum albumin (low endotoxin) and 1.5 mg/mL trypsin inhibitor (Sigma, St. Louis, MO, USA). Cell suspension was seeded onto poly-D-lysine/laminin-coated coverslips (BD), and incubated at 37°C in a 95% air/5% CO2 (vol/vol) incubator.
Whole-cell current-clamp recordings and data analysis
Membrane potentials were recorded from 20 to 30 µm diameter DRG neurons in the current-clamp configuration using an EPC-10 amplifier and the PatchMaster program (HEKA Elektronik, Holliston, MA, USA) at room temperature 3 to 15 h after culture. Patch pipettes were pulled from borosilicate glass (1.65/1.1 outside diameter/inside diameter; World Precision Instruments, Sarasota, FL, USA) using a Sutter Instruments P-97 puller and had a resistance of 0.6 to 1.7 MΩ when filled with internal solution, which contained (in mM) 140 KCl, 3 Mg-ATP, 0.5 EGTA, 5 HEPES at pH 7.3, adjusted to 310 mOsm using dextrose. External bath solution contained (in mM) 140 NaCl, 3 KCl, 2 CaCl2, 2 MgCl2, 10 HEPES at pH 7.3, and was adjusted to 320 mOsm using dextrose. Current-clamp recordings were sampled at 50 KHz and filtered using two Bessel filters at 10 and 2.9 KHz. Small DRG neurons (<30 µm in diameter) with stable (<10% variation) resting membrane potentials (RMPs) more hyperpolarized than −40 mV were included in the analysis. Cells with an input resistance lower than 300 MΩ were excluded from analysis. Input resistance was determined by the slope of a linear fit to hyperpolarizing responses to current steps from −5 pA to −40 pA in −5 pA increments.
Rheobase was defined as the first injection step that resulted in AP firing without subsequent failure and was determined by a series of depolarizing current injections (1 ms) that increased incrementally by 25 pA. APs were defined as rapid increases in membrane potential to > +40 mV with a total amplitude >80 mV. AP frequency was determined by quantifying the number of APs a neuron fired during a 1000 ms current injection. AP amplitudes, slopes, and half-widths were calculated in the FitMaster program (HEKA Elektronik, Holliston, MA, USA).
Current-clamp data are presented as mean ± standard error and significance is assayed via Student’s t-test, unless otherwise noted. All data except firing frequency curves were analyzed in the FitMaster program (HEKA Elektronik, Holliston, MA, USA) and Microsoft Excel. Firing frequency curves were analyzed by two-way analysis of variance (ANOVA) with repeated measures in OriginPro (OriginLab Corporation, Northampton, MA, USA). All data were visualized using IgorPro (WaveMetrics, Lake Oswego, OR, USA) with error bars showing standard error. Spontaneous AP firing was analyzed for significance using z-test.
Behavioral Testing
Open-field activity
Rats were acclimated to the testing room for 1 h and individually placed in an open-field apparatus consisting of a Plexiglas box (41 cm L × 41 cm W × 38 cm H) surrounded by a frame consisting of 48 photocells arranged with 16 in the vertical dimension and 32 in the horizontal dimension (Kinder Scientific, Poway, CA). Photobeam breaks were used as an indication of locomotor activity including basic movement, rearing (counts), and rearing time (sec) over 30 min.
Hot plate
Hot plate paw withdrawal latencies were measured with an Ugo Basile hot/cold plate model 35100 (Columbus Instruments). Animals were habituated to the testing room for 1 h prior to being placed in the center of the hot plate pre-set at 50°C with all four paws touching the surface. The latency for the animal to jump, tap, and/or lick a hind paw was then recorded. Each animal was assessed during three separate trials with at least 10 min between trials. The mean response time was recorded as that animal’s latency to respond to the heat stimulus. The hotplate was wiped down after each trial to remove odor cues. A latency cut-off of 60 s was used to prevent tissue damage.
Olfaction test
To test the animals for olfactory function, a buried food test was used. On three consecutive days, three days prior to test day, one BioServ chocolate Supreme Mini-Treat (Bio-Serv, Frenchtown, NJ) was placed on the floor in the animal’s home cage, along with standard rodent chow, and left overnight. Each morning, the cage was checked to ensure the treat was completely consumed. On the evening before the test, the rodents were food deprived to increase the motivation to eat. On test day, animals were acclimated to the testing room for 1 h in standard polycarbonate clear cages (dimensions: 20″ × 16″ × 8.5″) testing chambers with a wire mesh top. Each cage was filled to 7.62 cm with clean corncob bedding. At the start of the test, the rat was placed in the center of a new cage and allowed to freely explore for 5 min. At the end of 5 min acclimation, the rat was briefly removed from the test cage and one Supreme Mini-Treat was buried 5 cm from a random corner, 1 cm deep. The rat was then reintroduced to the testing cage and the latency to find and begin to eat the Supreme Mini-Treat was recorded as the endpoint. A 15-minute cut-off was used for the assay.
Capsaicin-induced flinching
Rats were acclimated to test box for 30 min. Capsaicin (2 µg/50 µL; Millipore Sigma Cat no. M2028-50mg,) disolved in 10% ETOH (Spectrum Chemical MFG Corp; Cat no. ET108)/1% Tween 80 (Spectrum Chemical MFG Corp; Cat no. PO138)/89% PBS (Invitrogen; Cat no. 10010–031) was injected intra-plantar into the ventral left hind paw. Each rat was observed and flinching behavior was recorded for 1 min.
Spinal nerve ligation
Spinal nerve ligation (SNL) surgery was performed using aseptic surgical techniques and a stereomicroscope.35 Spinal nerve injury was caused by ligating the left L5 and L6 spinal nerves, with special care to avoid damage to the L4 spinal nerve or surrounding area. Under gaseous anesthesia with a mixture of O2 and isoflurane (3% for induction and 2% for maintenance), skin was excised and the longissimus lumborum muscle, part of articular processes (L4-S1), and the fascia above L6 spinal nerve were carefully removed. This procedure provided a clean and spacious working area to enable complete resection of the L6 transverse process, and to separate the L5 spinal nerve from the L4 spinal nerve without damage to L4. The L5 and L6 spinal nerves were each tightly ligated with 6–0 silk thread. The entire surgery procedure beginning from anesthesia and ending with wound clipping of the outside skin lasted 15 min or less.
von Frey behavioral testing
Two weeks postsurgery, mechanical sensitivity was measured by determining the median 50% foot withdrawal threshold for von Frey filaments using the up-down method.36 Rats were placed in a plastic testing box on a metal mesh floor. von Frey filaments (Semmes-Weinstein monofilaments from Stoelting) were applied to the middle glabrous area between the footpads of the plantar surface of the injured hind paw. This plantar area was touched with a series of nine recently calibrated von Frey filaments with approximately exponentially incremental bending forces (von Frey filament numbers: 3.61, 3.8, 4.0, 4.2, 4.41, 4.6, 4.8, 5.0, and 5.2; equivalent to: 0.41, 0.63, 1.0, 1.58, 2.51, 4.07, 6.31, 10 and 15.8 g). The von Frey filament was presented perpendicular to the plantar surface with sufficient force to cause slight bending, and held for approximately 3–4 seconds. Abrupt withdrawal of the foot accompanied by a pain indicative behavior (namely, paw flinching, shaking or licking for more than 2 seconds) was recorded as a response. Any postsurgery rat that displayed a mechanical threshold of more than 3.16 g or less than 0.7 g was eliminated from the study.
Cold plate behavioral testing
Rats were acclimated for 20 min to the test box and then placed onto a 4°C cold plate (IITC Life Science, Woodland Hills, CA). The time for rats to lift or lick a hind paw was recorded with a 5 min cut-off time.
Formalin test
Formalin-induced flinching was measured using the Automated Nociception Analyzer (ANA) (University Anesthesia Research & Development Group, La Jolla, CA). A small metal band was attached to the plantar surface of the test hindpaw with one drop of super glue. Rats where then habituated to the testing chamber for 30 min prior to test onset. At test time, each animal was gently wrapped in a towel with the test paw exposed. Fifty microliters of 2.5% formalin solution (Electron Microscopy Sciences, Hatfield, PA. Cat no. 15742–10) were injected into the dorsal surface of the test paw. Animals were immediately returned to the testing chamber and flinching behavior was scored by the ANA software for 40 min.
Statistical analysis
Data are expressed as mean ± SEM. Behavioral results were generally analyzed using t-tests or a one-way ANOVA with Dunnett’s multiple comparisons post hoc test for significance unless otherwise noted. Statistical calculations and graphs were made using GraphPad Prism 5.01 (GraphPad Software Inc, San Diego, CA).
Results
We set out to design a model to enable in vivo prosecution of sulfonamide-based chemical matter in rat pain assays. Expression of a chimeric NaV1.7 protein would be driven from the endogenous rat NaV1.7 promotor and contain all rat coding exons apart from rat exon 25, which would be replaced by the equivalent human coding sequence in exon 26. These exons encode for the sulfonamide-binding region in the fourth voltage-sensor domain (Figure 1(a) to (c)). Rats were generated using ZFN-based technology and genotyped (Figure 1(d)). HOM-KI rats developed into adults that displayed normal open-field basic movement and rearing behaviors comparable their WT counterparts (Figure 1(e) and (f)).
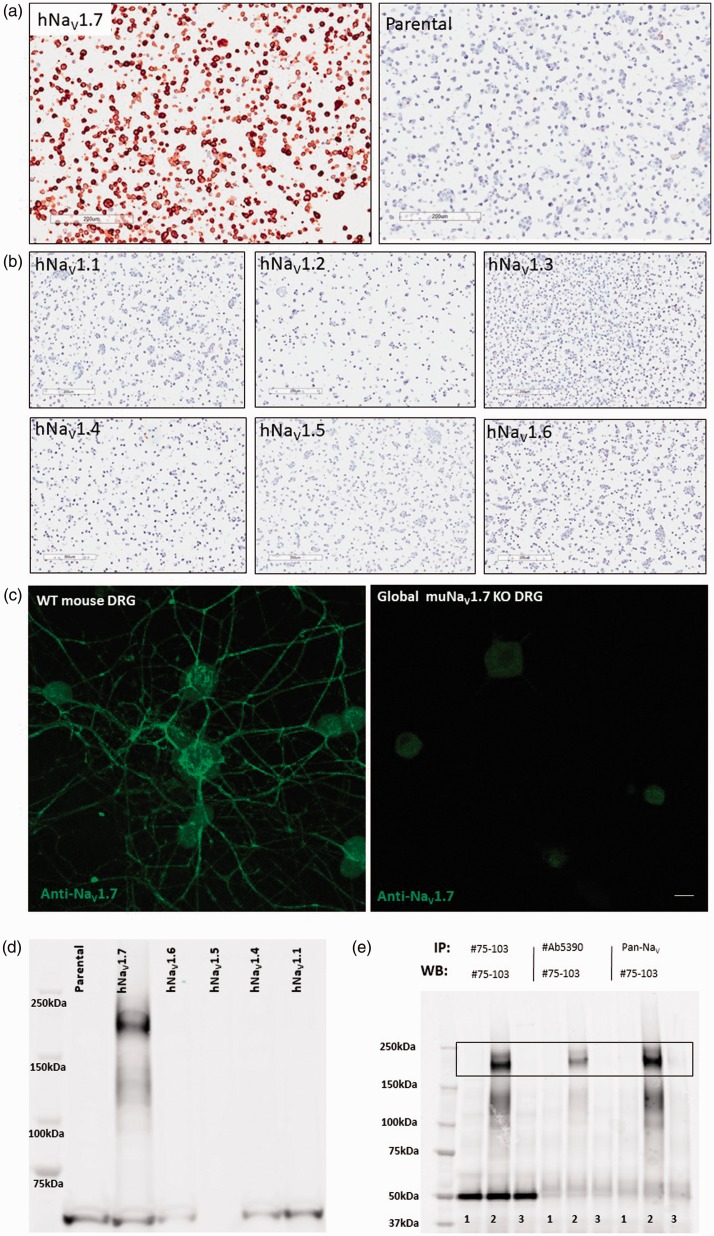
To evaluate the distribution of endogenous and chimeric NaV1.7 rat protein in WT and HOM-KI animals, a NaV1.7 antibody was required. Due to high protein sequence homology between the NaV channel isoforms, utilization of a selective anti-NaV1.7 antibody was mandatory to avoid any confounding signal stemming from other NaV channel family members. We first screened dozens of internal and commercial antibodies to identify an antibody that was selective for NaV1.7 in stable cell lines overexpressing human NaV1.1–1.7 (Figure 2(a) and (b)) and that labeled WT but not global mouse NaV1.7 KO DRG neurons cultured in vitro for five days (Figure 2(c)). The antibody, confirmed for immunocytochemistry and immunofluorescence applications, recognized endogenous human and mouse protein in fixed cultured cells. By aligning the C-terminal, cytoplasmic antigen used to generate the antibody against human, cyno, dog, rat, mouse, and rabbit NaV1.7 sequences (Supplemental Figure 1(A)), we hypothesized that the antibody would cross-react with NaV1.7 from these diverse mammalian tissues. This was confirmed in the species tested by detection of NaV1.7 immunoreactivity in DRG and/or sciatic nerve of human, cyno, rat, and mouse (Supplemental Figure 1B-I). Finally, the antibody specificity was validated for Western blot and immunoprecipitation applications under denaturing conditions in NaV overexpressing HEK293 stable cell lines (Figure 2(d) and (e)).
Figure 2.
Validation of NaV1.7-specific monoclonal antibody on intact and denatured NaV1.7 protein. (a) Positive NaV1.7 immunocytochemistry signal in HEK293 cells overexpressing hNaV1.7 (left panel) but not in corresponding parental HEK293 cells (right panel) used as a negative control. (b) Lack of immunocytochemistry signals in hNaV1.1, hNaV1.2, hNaV1.3, hNaV1.4, hNaV1.5, and hNaV1.6 stable cell lines. (Scale bars = 200 µm for panels A and B). (c) NaV1.7 immunofluorescence signal was observed in adult mouse DRG neurons cultured in vitro for five days from WT (left) but not in NaV1.7 global KO samples (right). Diffuse nonspecific signals from neuronal cell bodies are present in both WT and NaV1.7 KO neurons, while bright neurite labeling is only seen in WT neurons. (Scale bars = 200 µm for panels A and B and 20 µm in panel C). (d) Western blot demonstrating specificity of antibody for human NaV1.7 in HEK293 stable cell lysates (Lane 1: parental HEK293; Lane 2: hNaV1.7; Lane 3: hNaV1.6; Lane 4: hNaV1.5; Lane 5: hNaV1.4; Lane 6: hNaV1.1). (e) NaV1.7 antibody (NeuroMab, Cat no. 75–103) utility for immunoprecipitation (IP) and immunoblotting (IB) hNaV1.7, but not hNaV1.5, from over-expressing HEK293 cell lysates (Lane 1: HEK293 parental (negative control); Lane 2: hNaV1.7; Lane 3: hNaV1.5). WT: wild type; DRG: dorsal root ganglia.
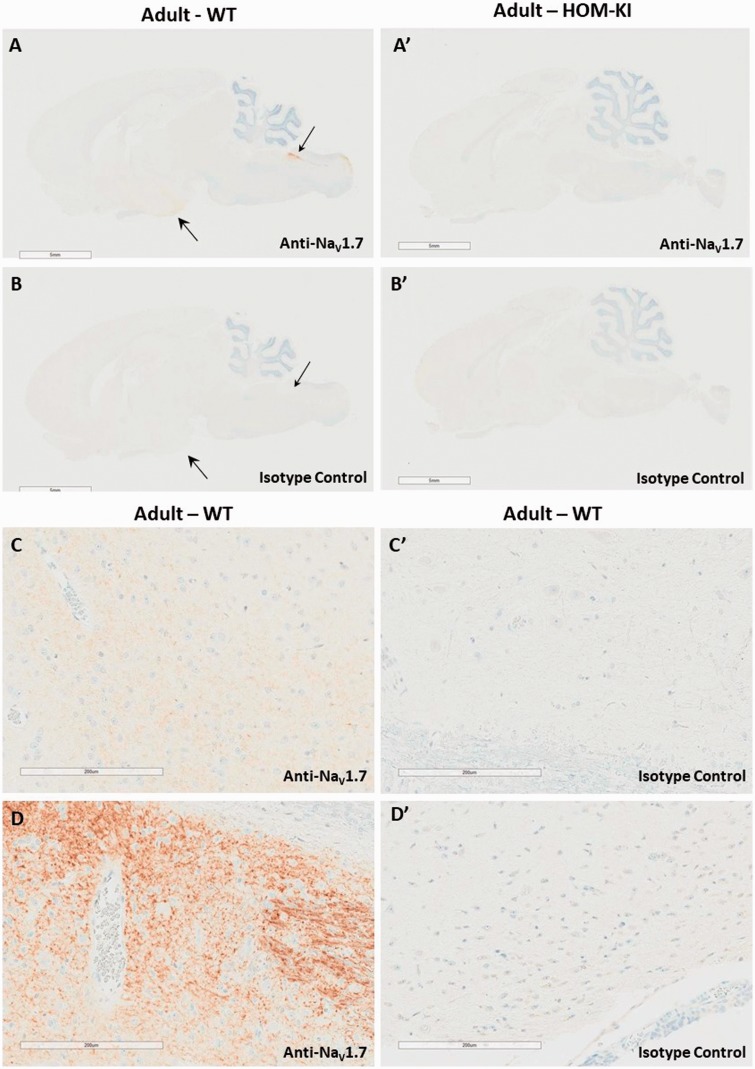
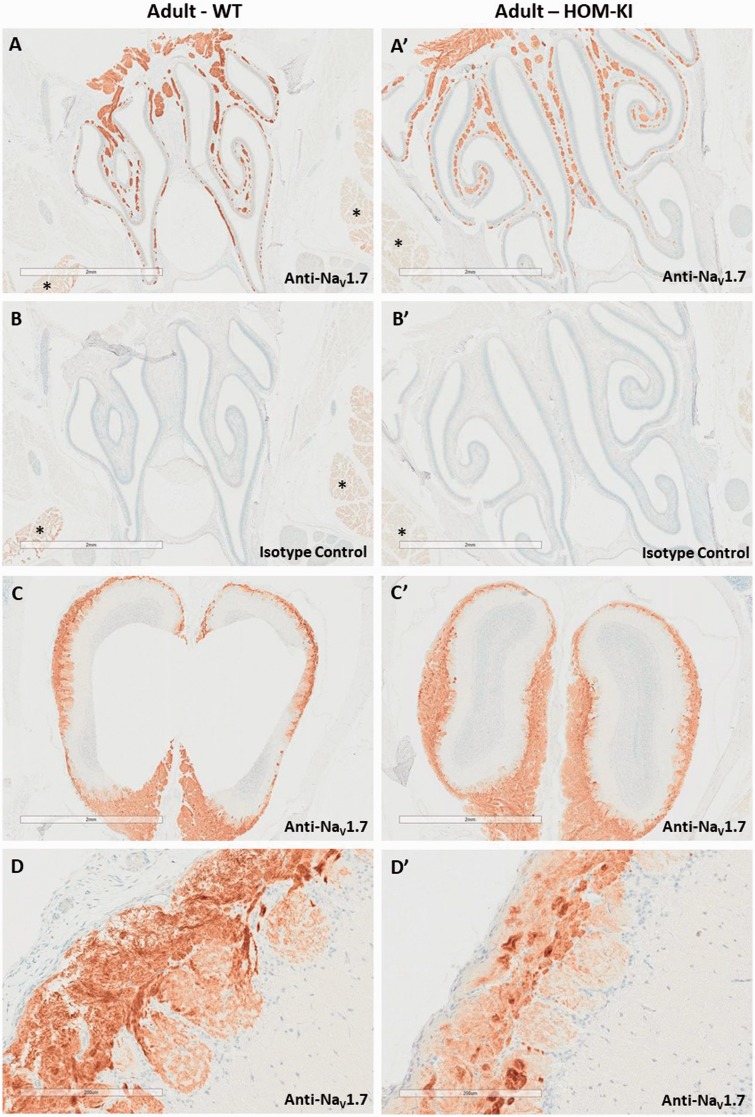
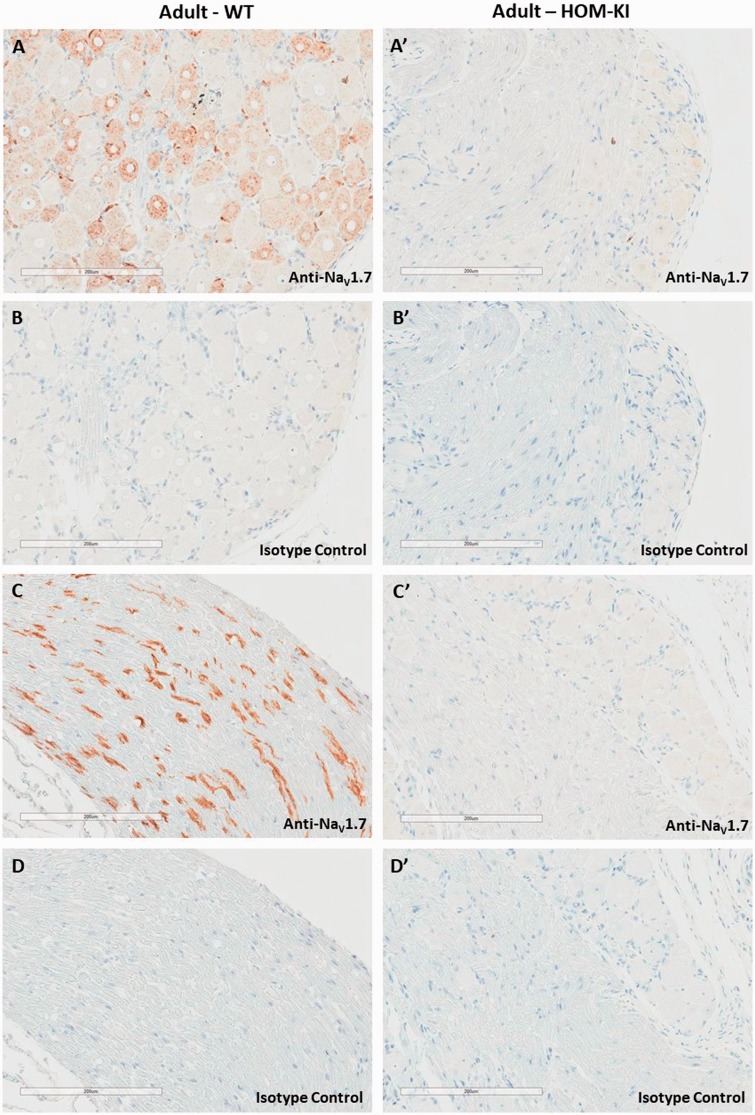
Having identified and validated a selective NaV1.7 antibody, we evaluated the endogenous and chimeric NaV1.7 protein distribution in WT and HOM-KI rat peripheral and central nervous system tissues by immunohistochemistry (IHC). In WT rats, we detected NaV1.7 immunoreactivity in the hypothalamus (large arrow in Figure 3(A) and higher magnification in panel C), solitary tract and area postrema (small arrow in panel A and higher magnification in panel D). To our surprise, the corresponding regions in HOM-KI rats were devoid of NaV1.7 immunoreactivity (Figure 3(A′)) and resembled isotype control patterns (Figure 3(B) for WT and 3B′ for HOM-KI). NaV1.7 immunoreactivity was detected in the nasal turbinates, primarily in the sensory nerve bundles of the lamina propria (Figure 4(A)), olfactory bulb (Figure 4(C)), DRG (Figure 5(A)), and sciatic nerve (Figure 5(C)) in WT rats. Oddly, HOM-KI animals retained NaV1.7 immunoreactivity in olfactory turbinates (Figure 4(A′)) and olfactory bulb (Figure 4(C′)) but were devoid of NaV1.7 immunoreactivity in DRG (Figure 5(A′)) and sciatic nerve (Figure 5(C′)). Regions of the digestive system known to express NaV1.7 were evaluated (jejunum, ileum, cecum and colon) and, unlike WT tissues, HOM-KI tissues were also devoid of NaV1.7 immunoreactivity (Supplemental Figure 2).
Figure 3.
NaV1.7 immunostaining of brain. (A and A′). NaV1.7 immunoreactivity in the hypothalamus (weaker signal, larger arrow head), in the solitary tract and area postrema (shorter arrow head) of WT but not HOM-KI rats. (B and B′) Corresponding isotype control IHC. (C) Higher magnification of the adult WT hypothalamus, showing weaker NaV1.7 immunoreactivity. (C′) Corresponding isotype control. (D) Higher magnification of adult WT dorsal brainstem, consistent with the area postrema (D′) Corresponding isotype control. Scale bars 200 µm. Scale bars 5 mm in A, A′, B, and B′ and 200 µm in C, C′, D, and D′. Three rats of each genotype and gender were evaluated. HOM-KI: rats homozygous for the knock-in allele; WT: wild type.
Figure 4.
NaV1.7 immunostaining in olfactory epithelium and olfactory bulb. (A and A′). NaV1.7 immunoreactivity in nasal turbinates (nerve bundles of the olfactory sensory neurons) from WT and HOM-KI rats. (B and B′). Corresponding isotype control stains. Note the nonspecific reaction product present in salivary glands (*), which is also visible in the isotype controls for each genotype. (C and C′) NaV1.7 immunoreactivity in olfactory bulb from WT and HOM-KI rats. (D and D′) Higher magnification of NaV1.7 immunoreactivity in olfactory bulb from WT and HOM-KI rats. Scale bars = 2 mm for panel A through C′ and 200 µm in panels D and D′. Three rats of each genotype and gender were evaluated.
Figure 5.
NaV1.7 Immunostaining in DRG and sciatic nerve. (A and A′) NaV1.7 immunoreactivity in DRG from WT but not HOM-KI rats. (B and B′). Corresponding isotype control sections. (C and C′) NaV1.7 immunoreactivity in sciatic nerve from WT but not HOM-KI rats. (D and D′) Corresponding isotype control sections. Scale bars 200 µm. Three rats of each genotype and gender were evaluated. HOM-KI: rats homozygous for the knock-in allele; WT: wild type.
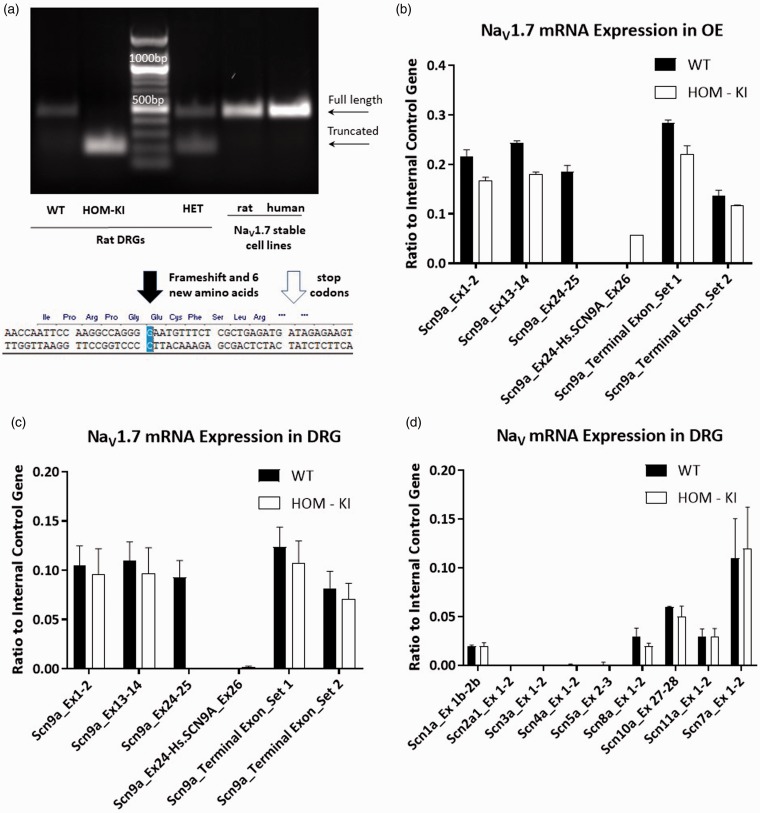
Based on the unexpected differences in NaV1.7 immunoreactivity between WT and HOM-KI rat tissues, we evaluated NaV1.7 transcript levels in olfactory epithelium (OE) and DRG. Using rat and human stable cell lines as positive controls, we compared cDNA products obtained in DRG from each genotype by reverse transcription-polymerase chain reaction (RT-PCR) (Figure 6(a)). WT and HET DRG gave rise to a band of the expected size. However, a smaller band was detected in HET and HOM-KI tissue, indicative of a truncated transcript. Sequencing the truncated band revealed removal of the human exon 26 and retention of an additional nucleotide from the rat sequence that led to a frame-shift, predicted introduction of six novel amino acids and two early stop-codons (Figure 6(a)). Thus, this product lacks the entire sulfonamide-coding region in the fourth voltage-sensor domain. Using an array of primers specific for rat or human exons (Table 1), we evaluated the presence/absence of rat or human transcript regions in WT and HOM-KI NaV1.7 immuno-positive (OE) and immuno-negative (DRG) tissues (Figure 6(b) and (c)). The OE of HOM-KI rats expressed human exon 26 while WT tissue did not, whereas human exon 26 in DRG was nearly undetectable. NaV1.7 amplicons upstream and downstream of the insertion were present in WT and HOM-KI tissues at similar levels. Using primer sets for the nine other members of the voltage-gated sodium channel family, we observed that rat Scn1a (NaV1.1), Scn8a (NaV1.6), Scn10a (NaV1.8), Scn11a (NaV1.9), and NaX (also known as NaG or NaV2.1) transcripts were expressed at similar levels between WT and HOM-KI DRGs (Figure 6(d); p > 0.05 unpaired t-test for each gene). Collectively, these data demonstrate that chimeric NaV1.7 transcript is present in HOM-KI OE and that the transcriptional profile of other NaV isoforms is not impacted in HOM-KI DRGs.
Figure 6.
NaV1.7 transcript analysis in sensory tissues. (a) RT-PCR analysis of adult WT vs. HOM-KI DRGs and reference human and rat NaV1.7 stable HEK293 cell lines. Full-length bands were detected in WT, HET tissue samples as well as in positive controls (stable cell lines) (463 bp) in addition to an unexpected truncated band in HOM-KI and HET samples only (193 bp). Sequence of truncated RT-PCR band revealed splicing out of human exon 26 plus an additional guanine nucleotide from rat exon 26 with a resulting frameshift (black arrow), insertion of six novel amino acids, and pre-mature stop codons (white arrow). (b and c) NaV1.7 amplicons in WT and HOM-KI using primers positioned in various rat or human NaV1.7 exons upstream and downstream of the insertion in olfactory epithelium (OE) (b) and DRG (c). RT-PCR products were detected for rat NaV1.7 amplicons prior to and following human exon 26 in WT and HOM-KI tissue. An amplicon between rat exon 24 and human exon 26 was nominally detected in DRG tissue but was present in OE tissue in HOM-KI rats. (d) RT-PCR of NaV isoforms in DRG tissue. No compensatory expression changes were observed for non-NaV1.7 isoforms in HOM-KI rats. Data are mean ± SEM, four rats of each genotype (two males and two females) were evaluated. HOM-KI: rats homozygous for the knock-in allele; WT: wild type; OE: olfactory epithelium; DRG: dorsal root ganglia.
Table 1.
Primer and probe sequences used for RT-PCR experiments evaluating NaV transcript expression.
| Gene | Location | IDT prime time ID | Right sequence | Probe sequence | LEFT SEQUENCE |
|---|---|---|---|---|---|
| Scn1a | Exon 1b-2b | Rn.PT.58.18149257 | CTCCATCTTGTCATCCTGCAC | /56-FAM/TGCAGTAAG/ZEN/GAACAACATCTCCCGAG/3IABkFQ/ | CAACTGAAGGTGCGGACT |
| Scn2a1 | Exon 1-2 | Rn.PT.58.8446453 | TGCCATCTTTTCATCCTGCT | /56-FAM/TCAGCACTT/ZEN/TCTTACGCGAGGAGC/3IABkFQ/ | CATTCAACACGCACATGGATTC |
| Scn3a | Exon 1-2 | Rn.PT.58.11329215 | CCATCTTTTCATCCTGCACAT | /56-FAM/CCTAAGGCT/ZEN/CTCCATCAGGCAATTCT/3IABkFQ/ | CACCGTCCATTCTAACCATCT |
| Scn4a | Exon 1-2 | Rn.PT.58.13849817 | CAACAGAAAAGTGAACCAGATCTTC | /56-FAM/ACAGCTGAA/ZEN/GTCACGCAGTTCCT/3IABkFQ/ | CCGCTCTTCTCTGCTTCTG |
| Scn5a | Exon 2-3 | Rn.PT.58.8036795 | GCTGAGGACATACAAGGCATT | /56-FAM/TGGCACTGA/ZEN/ACCGGAAGATGGT/3IABkFQ/ | ACCTGGACCCTTTCTATAGTACC |
| Scn7a | Exon 1-2 | Rn.PT.58.12899103 | AGAGCTTAAAGGAGACAACGTG | /56-FAM/AGGATGGAA/ZEN/ACCGCATTGAACCTGA/3IABkFQ/ | GCCCTTGGAAGATGTGGAT |
| Scn8a | Exon 1-2 | Rn.PT.58.6646933 | CCGTAGATGAAAGGCAAACTC | /56-FAM/CGATGAAGA/ZEN/CAGCAAGCCCAAGC/3IABkFQ/ | CAAGCTCAAGAAACCACCAAAG |
| Scn9a | Exon 1-2 | Rn.PT.58.8595593 | TTAGAGGACTGAAGGGAGACA | /56-FAM/AACGCCACC/ZEN/CCTGCTTTGTACA/3IABkFQ/ | ACCTGGACCCATACTATGCT |
| Scn9a | Exon 13-14 | Rn.PT.58.5449652 | CAGTACCATTTCAGCTGCGA | /56-FAM/CCTGTAAAG/ZEN/ATCAAGTTCCCCACTGCA/3IABkFQ/ | GAGCACCACCCAATGACT |
| Scna9a | Exon 24-25 | Custom | ACCAATTCCAAGGCCAG | /56-FAM/CCAAGCTTT/ZEN/TGATATCACCATCATGGTTCTTATATGC/3IABkFQ/ | ATAATCCATGTACTCAGTTTGCC |
| Scn9a-SCN9A | Rn.Exon 24-Hs.Exon 26 | Custom | ACCAATTCCAAGGCCAG | /56-FAM/TCAAGCCTT/ZEN/TGATATTAGTATCATGGTTCTTATCTGTC/3IABkFQ/ | TCATATGTTGACTTTGACCCTC |
| Scn9a | Terminal Exon (Set 1) | Custom | CAGACGGTATCGCCTCAGAC | /56-FAM/ACGTGAACG/ZEN/AGAACTCAAGTCCGGAA/3IABkFQ/ | GAAGGTGGCGAGATGGTTGA |
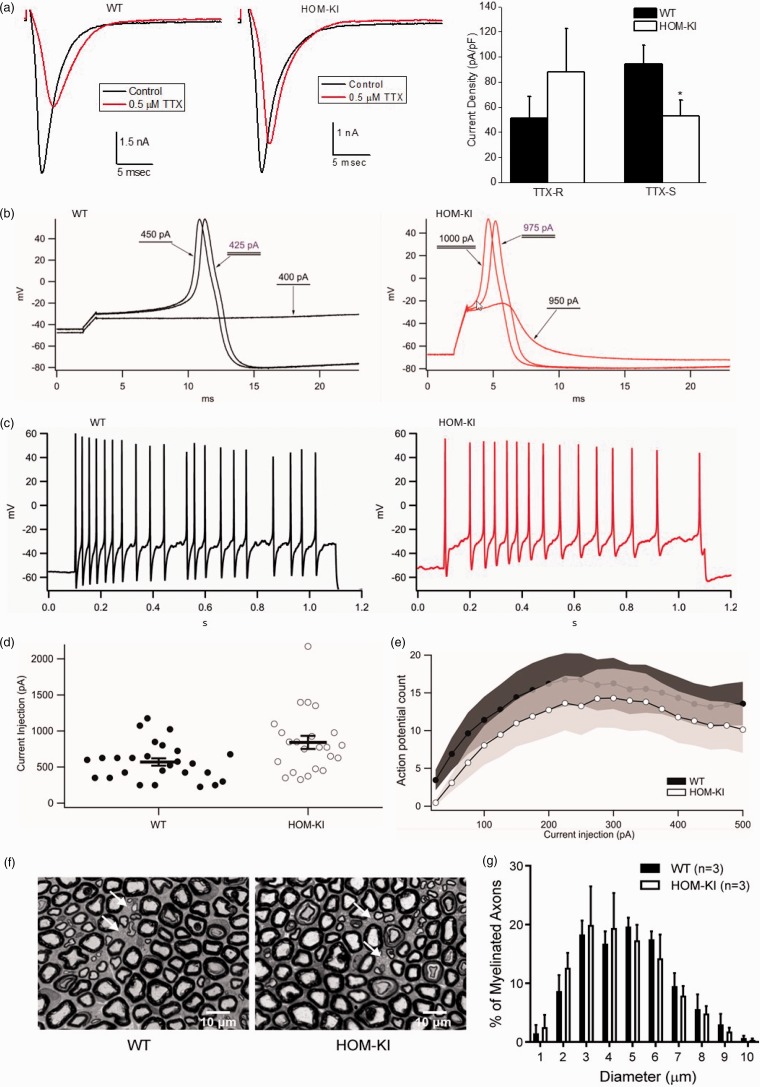
Electrophysiological properties of DRG neurons, including sodium channel current density and AP firing, were evaluated to understand the functional impact of splicing human exon 26 from HOM-KI rats. Whole-cell patch clamp recordings from small diameter cell bodies of acutely dissociated DRG neurons showed TTX-S and TTX-R in both WT and HOM-KI littermates. TTX-S current density was significantly lower in HOM-KI neurons compared to WT neurons, suggesting that some TTX-S current, encoded by NaV1.7, was lost in HOM-KI DRG neurons, whereas the density of TTX-R current was not statistically different between HOM-KI and WT DRG neurons (Figure 7(a)). The RMP of DRG neurons from HOM-KI rats was 3.29 mV more hyperpolarized, a difference that was not significantly different from that of WT littermates (HOM-KI −59.09 ± 1.52 mV, n = 26, compared to WT −55.81 mV ± 1.29, n = 31, p = 0.10, two-tailed unpaired Student’s t-test). Spontaneous AP firing was significantly greater in WT DRG neurons (24.39%; 10/41 neurons) compared to HOM-KI DRG neurons (3.70%; 1/27 neurons, p = 0.023, two-proportion z-test). The threshold current needed to evoke AP firing in HOM-KI DRG neurons was larger, as reflected by the elevated rheobase (Figure 7(b) and (d)). The evoked AP firing frequency in response to a series of depolarizing 1000 ms current injections (25–500 pA) was significantly higher across the range of input stimuli in WT compared to HOM-KI neurons (Figure 7(c) and (e)). Direct comparison of AP characteristics did not show statistically significant differences in the slope of the pre-AP rising phase, time to AP peak, AP overshoot, or AP half-width (Table 2). Collectively, these data indicate that small diameter DRG neurons from HOM-KI rats exhibit lower TTX-S sodium channel current densities, elevated thresholds to fire APs, and reduced AP firing rates reflecting lower overall neuronal excitability. No abnormalities were found during analysis of nerve fibers by light microscopy on epoxy-embedded sciatic nerve samples. Unmyelinated axons could clearly be identified in cross sections of sciatic nerve and the size distribution of myelinated axon diameters was similar between HOM-KI and WT littermates (Figure 7(f) and (g)).
Figure 7.
Reduced HOM-KI DRG neuron excitability. (a) TTX-R and TTX-S current densities in small diameter DRG neurons isolated from WT and HOM-KI rats. Representative traces (left) and summarized data (right). A significant reduction of TTX-S current density, but not TTX-R current density, was observed in HOM-KI rats (*p < 0.05; two-tailed unpaired t-test; n = 14 neurons per group). Two rats of each genotype with mixed gender were evaluated. Data are mean ± SEM. (b) Representative traces illustrating threshold to action potential firing for wild-type (black) and HOM-KI (red) neurons determined by 1 ms current injections. (c) Representative traces of action potential firing in response to 1000 ms of 250 pA current injection in both wild-type (black) and HOM-KI (red) neurons. (d) HOM-KI small diameter DRG neurons exhibited a significant increase in current threshold to action potential firing (HOM-KI 841.30 ± 90.35 pA, n = 23, compared to WT 569.23 ± 50.94 pA, n = 26, p = 0.0096, two-tailed unpaired Student’s t-test). (e) Action potential firing in response to 1000 ms current injections from 25 to 500 pA was reduced in DRG neurons from HOM-KI animals (significant by two-way repeated measures ANOVA with Tukey and Bonferroni tests, p = 0.0054). Shaded regions represent error bars (± 1 SEM) of the action potential firing frequency curves for wild-type (black) and HOM-KI (white) DRG neurons. For (b) to (e), neurons from five rats of each genotype (three males and two females) were evaluated by current clamp. (f) Representative images of tibial nerve cross sections by light microscopy at 100×. There is no apparent difference in gross morphology of myelinated and unmyelinated axons between WT controls and HOM-KI animals. Arrows indicate groups of unmyelinated axons (Scale bar = 10 µm). (g) Quantification of myelinated axon number and size in sciatic nerve from WT controls and HOM-KI rats. Myelinated axon diameter distributions in WT and HOM-KI rats show no statistical difference using multiple unpaired t test. For (f) and (g), three rats of each genotype (two males and one female in WT group; one male and two females in HOM-KI group) were evaluated. HOM-KI: rats homozygous for the knock-in allele; WT: wild type; TTX-S: fast-inactivating currents sensitive to tetrodotoxin; TTX-R: slow-inactivating currents resistant to tetrodotoxin.
Table 2.
Action potential characteristics of DRG neurons from WT and NaV1.7 HOM-KI rats.
| Slope (mV/ms) | Time to peak (ms) | Overshoot (mV) | Half-width (ms) | |
|---|---|---|---|---|
| WT | 3.39 ± 0.90 | 38.15 ± 7.08 | 54.26 ± 1.57 | 2.37 ± 0.23 |
| HOM-KI | 3.55 ± 0.71 | 26.72 ± 5.76 | 51.38 ± 1.57 | 3.00 ± 041 |
| p-value | 0.89 | 0.22 | 0.20 | 0.18 |
HOM-KI: rats homozygous for the knock-in allele; WT: wild type.
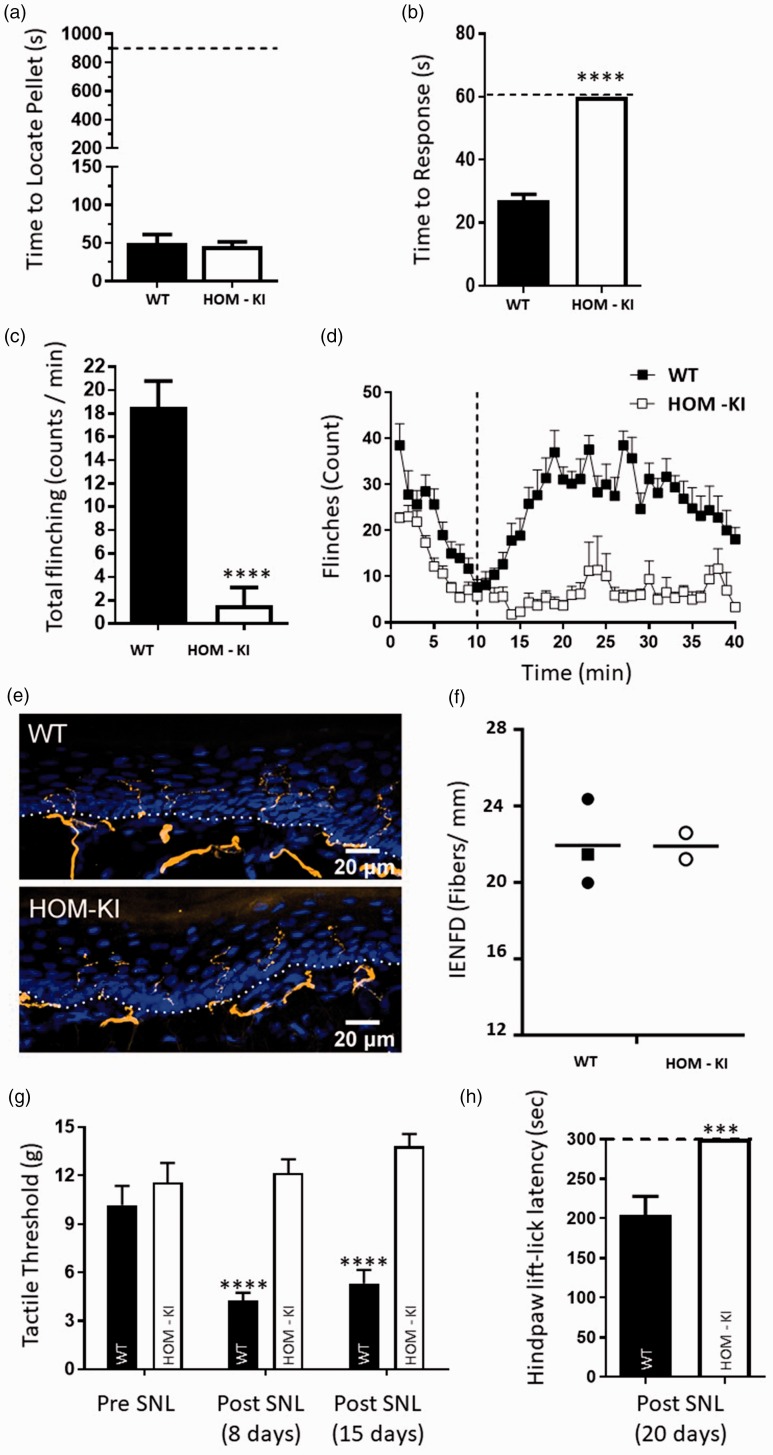
We next evaluated olfactory and peripheral sensory function at the behavioral level in HOM-KI rats. Unlike global NaV1.7 KO mice, HOM-KI rats retained olfactory function and were able to find a buried food pellet like WT rats (Figure 8(a)). HOM-KI rats showed severe sensory deficits to thermal and chemical nociceptive assays, as demonstrated by the lack of response to a 50°C heat stimulus (Figure 8(b)) and nominal response to capsaicin intraplantar injection (Figure 8(c)). HOM-KI rats also had significantly fewer flinches than WT rats in both phases 1 and 2a of the formalin test (Figure 8(d)). Sensory innervation of the epidermis, required for detection and initiation of pain signaling, was evaluated in WT and HOM-KI skin biopsies. The average IENF density in HOM-KI was not different from that of WT samples (Figure 8(e) and (f)). Thus, the lack of pain behavior in HOM-KI rats was not attributable to lack of epidermal nerve fibers. HOM-KI rats were evaluated for their ability to develop neuropathic pain behaviors using the SNL model. WT rats displayed allodynia to tactile and cold stimuli one to three weeks following SNL surgery. HOM-KI rats, however, did not show signs of hypersensitivity to mechanical stimulation following SNL surgery, and displayed similar pre- and post-SNL tactile threshold levels (Figure 8(g)). Moreover, HOM-KI rats were not responsive to a 4°C cold plate following SNL surgery and reached the 5-min assay cut-off time (Figure 8(h)). Taken together, these data suggest that HOM-KI rats do not develop neuropathic pain behavior in the absence of detectable NaV1.7 protein in peripheral sensory neurons.
Figure 8.
Behavioral profiling of WT and HOM-KI rats. (a) Olfactory test. WT (49.8 ± 11.4 s) and HOM-KI (45.8 ± 5.8 s) rats were equally able to detect a buried food pellet (t16 = 0.329, p = 0.75, unpaired t-test; n = 8 WT and n = 10 HOM-KI). (b) Hot plate test. HOM-KI rats (60 s) did not respond to a 50°C heat stimulus, reached the 60 s cut-off time and were significantly different than WT rats (27.1 ± 2 s) (****Mann–Whitney U = 0, p = 0.00001; n = 9 WT and n = 10 HOM-KI). (c) Capsaicin chemical challenge. HOM-KI rats (1.5 ± 1.5 flinches) displayed significantly reduced capsaicin-induced flinching compared to WT rats (18.5 ± 2.2 flinches) (****Mann–Whitney U = 8, p = 0.0001, n = 11/group). (d) Formalin test. HOM-KI rats had significantly less flinching behavior than WT rats in phase 1 (0–10 min, HOM-KI: 133.7 ± 7, WT: 213.6 ± 18, **unpaired t test, t = 0.001) and phase 2a (11–40 min, HOM-KI: 187.6 ± 46, WT: 785.4 ± 46, **** unpaired t test, t = 0.00000006) of the formalin test (n= 10 for WT, n = 9 for HOM-KI). (e) Representative images of IENF in hind paw skin biopsies from WT and HOM-KI rats (9–10 weeks old). IENFs labeled with the pan-axonal marker PGP9.5 (orange) extended from the subdermal plexus into the epidermis (nuclei labelled with DAPI in blue). The dashed line indicates the dermis to epidermis division. HOM-KI rats exhibit normal nerve innervation. Confocal microscopy at 40× (Scale bar = 20 µm) (f) Quantification of intra-epidermal nerve fiber density of hind paw skin biopsies. IENF density in HOM-KI samples (21.90 ± 0.69 fibers/mm) was not statistically different from WT samples (21.94 ± 1.29 fibers/mm) using Mann–Whitney U test. Rectangles represent male samples while circles represent female samples. For (e and f), 3 WT and 2 HOM-KI (one male and two females in WT group; two females in HOM-KI group) rats were evaluated. (g) Spinal nerve ligation—tactile allodynia. HOM-KI rats did not display tactile allodynia as measured with von Frey filaments at eight days (12.2 ± 0.8 g, p = 0.84) or 15 days (13.82 ± 0.76 g, p = 0.084) post-SNL surgery compared to pre-SNL baseline (11.57 ± 1.2 g), indicating lack of development of neuropathic pain whereas WT controls developed tactile allodynia at 8 days (4.3 ± 0.5 g, ****p = 0.000003) and 15 days (5.3 ± 0.8 g, ****p = 0.00009) post-surgery which was significantly different compared to pre-surgery baseline (10.2 ± 1.2 g) (Two-way repeated measures ANOVA followed by Tukey’s multiple comparison test, main effect of time F (2,40) = 6.6762, p = 0.003, main effect of genotype F (1,20) = 36.222, p = 0.00007, n = 11/group). (h) Spinal nerve ligation—cold plate. HOM-KI rats did not respond to a 4°C cold plate and all reached the 300-s cut-off time. This was significantly different from WT controls (mean = 204 ± 24 s) (***Mann–Whitney U = 10, p = 0.0007; n = 10/group) 20 days post-SNL surgery. Data are mean ± SEM unless otherwise stated. HOM-KI: rats homozygous for the knock-in allele; WT: wild type; SNL: spinal nerve ligation.
Discussion
Comparison of the phenotypes of NaV1.7-deficient rats, described herein, NaV1.7 global KO mice and humans with CIP due to LOF NaV1.7 mutations reveal a number of behavioral similarities including lack of acute thermal pain responses, intact motor function, and normal nonnoxious tactile stimuli responses.14,15,28,37,38 These findings support an evolutionarily conserved function of NaV1.7 in nociceptive pain signaling between rodents and humans. However, rats deficient in NaV1.7 retain olfactory function, unlike global KO mice or humans with CIP that both exhibit anosmia.16,28,39 This difference is most likely attributable, at least in part, to the genetic background of the rats in addition to the functional expression of some chimeric NaV1.7 protein in rat olfactory sensory neurons but not in other tissue reported to express NaV1.7, as demonstrated at both the transcript level by PCR and/or the protein level by IHC using a specific antibody directed to the C-terminus of the NaV1.7 protein.
A number of mouse genetic models with tissue-specific or inducible deletion of NaV1.7 have been generated. Tissue-specific deletion of NaV1.7 in NaV1.8-positive neurons, Advillin-positive neurons, or Wnt1-positive neurons impacted nociceptive, inflammatory, and neuropathic pain behavior in various assays.29–32 The specific level of NaV1.7 reduction achieved with these approaches is critically dependent on the quantitative neuronal co-expression profile of NaV1.7 with each of these markers driving Cre-dependent genetic recombination as well as the efficiency of Cre recombination in each of these cell types. Lack of robust co-expression can result in residual NaV1.7 derived from cells not expressing these markers, whereas incomplete efficiency of Cre recombination can result in residual NaV1.7 in cells co-expressing these markers.40 Both scenarios can underestimate the absolute contribution of NaV1.7 to behavioral endpoints.
Inducible deletion of NaV1.7 provides an alternative method to interrogate NaV1.7-dependent pain behavior in adult animals without any confounds of compensatory changes that may occur during nervous system development and maturation as a result of NaV1.7 absence. To this point, NaV1.8 genetic deletion was reported to be associated with an increase in NaV1.7 transcript levels and TTX-S currents in DRG neurons.41 Tamoxifen-inducible deletion of NaV1.7 in adult mice also impacted nociceptive, inflammatory, and neuropathic pain behavior in various assays but, similar to the rat model generated herein, did not impact olfactory function.31,33 It is conceivable that tamoxifen biodistribution to olfactory sensory neurons or efficiency of Cre-mediated gene recombination restricted the level of NaV1.7 knockdown achieved in these sensory neurons. In our rat model, preliminary Western blotting data of NaV1.7 protein level in OE lysates showed reduction of NaV1.7 protein levels by ∼70% (data not shown), whereas in tamoxifen-inducible NaV1.7 KO mice, NaV1.7 transcripts were reduced by ∼50%.33 Collectively, these data indicate that more than 50% to 70% reduction of NaV1.7 mRNA/protein is necessary to impact olfactory function in rodents and that the residual chimeric protein detected in HOM-KI rat olfactory tissue is functional.
Neuropathic pain behavior, as measured by cold allodynia in response to acetone, was partially reversed by tamoxifen following spared nerve injury while mechanical allodynia was partially reversed by tamoxifen after chronic constriction nerve injury31 but not spared nerve injury.33 When tamoxifen was used to delete NaV1.7 prior to spared nerve injury, there was a more robust deficit in the development of cold compared to mechanical allodynia.33 It is not clear why there were different responses to different modalities, but the absolute level of NaV1.7 knockdown achieved in neurons necessary for cold allodynia could be greater than in those for mechanical allodynia. Single cell transcriptional profiling of NaV1.7 expression post-tamoxifen could discern any cell type-specific variation of NaV1.7 knockdown efficiency. By contrast, in our rat genetic model, using SNL, development of tactile allodynia was completely abolished and response times to a 4°C cold plate challenge reached a 5-min cut-off. Although it is difficult to draw generalized conclusions across these studies that utilized different nerve ligation injury models and testing regimens, the herein described NaV1.7 HOM-KI rat model did not display any neuropathic pain behavioral responses, thereby establishing that NaV1.7 is necessary for neuropathic pain in this species. While the involvement of NaV1.7 in human neuropathic pain is still not completely understood, our data suggest that sufficient pharmacologic inhibition of NaV1.7 function in humans could reduce pain in neuropathic conditions stemming from damage to sensory neurons. This contention ultimately requires prosecution of NaV1.7-specific inhibitors in clinical trials using human neuropathic pain cohorts to be properly addressed.
Notably, both HOM-KI rats reported here and global NaV1.7 KO mice exhibited spontaneous scratching behavior in their home cages.28 Spontaneous scratching may be attributable to the interplay between pain and itch with loss of afferent pain input into the spinal cord disinhibiting itch circuity.42 Concomitantly, loss of pain may lead to overgrooming and inability to perceive tissue damage which occurs during this rodent activity. In human NaV1.7 CIP individuals, self-mutilation of the tongue and fingers due to biting has been reported14,43 but spontaneous scratching events have not been noted. Pruritis has been reported in disclosed adverse events in clinical trials with a 1.6 g dose of the NaV1.7 selective sulfonamide inhibitor PF-0508771 from Pfizer in two of five individuals in an erythromelalgia cohort but not with a 150 mg BID dose in a painful diabetic neuropathy cohort.44,45 Pruritis was not listed as an adverse event in healthy volunteers with the NaV1.7 sulfonamide inhibitor GDC-0276 from Genentech.46
Early nerve conduction studies and biopsies in CIP patients with SCN9A mutations reported nerve fibers of normal morphology and size distribution.14,15,47 Skin innervation, as assessed by PGP9.5 immunostaining, has also been reported as normal in a three-year-old male CIP subject with SCN9A mutations.48 These observations suggest that nerve innervation is not impaired in NaV1.7 associated CIP subjects. However, a recent report on SCN9A-related human CIP cases suggests that the pain insensitivity is associated with structural loss of IENFs.43 This suggestion is supported by another recent case report of reduction of IENFs in a 10-year-old female CIP subject.49 These recent case reports raise the important question: does the painless phenotype caused by loss of NaV1.7 function require structural impairment of IENFs? In the NaV1.7 HOM-KI rats, we did not find a deficiency in IENF density, indicating that pain insensitivity in rats does not require structural loss of IENFs. Similarly, in a previously described global NaV1.7 KO mouse model, the skin innervation was reported to be normal.28 We cannot rule out the possibility that there is a species difference between humans and rodents, but certainly in both rats and mice, structural impairment of IENFs is not a prerequisite for NaV1.7-mediated painless phenotypes. Similarly, in small diameter DRG neurons from HOM-KI rats, a reduction, but not absence, of both TTX-S current density and AP firing frequency was observed. These observations echo findings previously described in both global NaV1.7 KO mice28 as well as human iPSC-derived nociceptors expressing a SCN9A LOF mutation,43 and indicate that pain insensitivity does not require complete absence of TTX-S currents or DRG neuron excitability. Absence or loss of function of NaV1.7 as a threshold channel for AP nociceptor firing, by producing ramp currents in response to slow depolarization due to slow onset of closed-state inactivation, and as a channel contributing to the upstroke of APs may account, in part, for the pain insensitivity phenotypes in humans and rodents.50,51 In summary, the rat NaV1.7 genetic model described here represents a powerful tool for pre-clinical experiments in the analgesia space for interrogating NaV1.7-dependent behavioral endpoints as well as assessing on-target vs off-target toxicology findings with selective or non-selective NaV1.7 inhibitors. It will be interesting to determine if visceral pain as well as chemotherapy and cancer-induced bone pain endpoints are impacted in the chimeric rat NaV1.7 model, as these have been reported to be NaV1.7-independent using a tissue-specific NaV1.7 KO approach.31,52,53 Due to the fortuitous finding that rats retained olfactory function, intensive animal husbandry efforts are not required to breed this model, thereby offering an advantage to NaV1.7 global KO mice that demand daily hand feeding during early development for successful generation of experimental cohorts.
Supplemental Material
Supplemental material, MPX881846 Supplemental Material1 for Rat NaV1.7 loss-of-function genetic model: Deficient nociceptive and neuropathic pain behavior with retained olfactory function and intra-epidermal nerve fibers by B Grubinska, L Chen, M Alsaloum, N Rampal, DJ Matson, C Yang, K Taborn, M Zhang, B Youngblood, D Liu, E Galbreath, S Allred, M Lepherd, R Ferrando, TJ Kornecook, SG Lehto, SG Waxman, BD Moyer, S Dib-Hajj and J Gingras in Molecular Pain
Supplemental Material
Supplemental material, MPX881846 Supplemental Material2 for Rat NaV1.7 loss-of-function genetic model: Deficient nociceptive and neuropathic pain behavior with retained olfactory function and intra-epidermal nerve fibers by B Grubinska, L Chen, M Alsaloum, N Rampal, DJ Matson, C Yang, K Taborn, M Zhang, B Youngblood, D Liu, E Galbreath, S Allred, M Lepherd, R Ferrando, TJ Kornecook, SG Lehto, SG Waxman, BD Moyer, S Dib-Hajj and J Gingras in Molecular Pain
Acknowledgments
We would like to thank Danny Chui for feedback and support for the animal model design, Teresa Wegesser, Artem Shkumatov, David Smith, Jim Pretorius, Meiying Qi and Toni Williamson at Amgen for their technical support on the cross species IHC material and Fadia Dib-Hajj and Peng Zhao of Yale Medical School for invaluable technical assistance.
Authors’ Contributions
BG, LC, MA, NR, DJM, CY, KT, MZ, BY, DL, EG, SA, ML, RF, TJK, SGL, SGW, BDM, SD-H, and JG participated in the research design. BG, LC, MA, NR, DJM, CY, KT, MZ, BY, DL, EG, SA, ML, RF, and JG conducted the experiments. BG, LC, MA, NR, DJM, MZ, ML, SGL, SGW, BDM, SD-H, and JG wrote or contributed to the writing of the manuscript.
Declaration of Conflicting Interests
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Stephen Waxman has received remuneration from Amgen for serving on its Neuroscience Advisory Board.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Portions of this work were supported by Center Grant B9253-C from the U.S. Department of Veterans Affairs Rehabilitation Research and Development Service and NIH/NIGMS Medical Scientist Training Program T32GM007205 grant. The Center for Neuroscience and Regeneration Research is a Collaboration of the Paralyzed Veterans of America with Yale University.
Supplemental Material
Supplemental material for this article is available online.
References
- 1.Pizzo Pac NM, Carter-Pokras O, Christopher M, Farrar JT, Follett KA, Heitkemper MM, Inturrisi C, Keefe F, Kerns RD, Lee JS, Loder E, Mackey S, Marinelli R, Payne R, Thernstrom M, Turk DC, Wesselmann U, Zeltzer L. Relieving pain in America, a blueprint for transforming prevention, care, education and research. Washington, DC: Institute of Medicine Report from the Committee on Advancing Pain Research, Care, and Education, 2011. [Google Scholar]
- 2.Holmes D. The pain drain. Nature 2016; 535: S2–S3. [DOI] [PubMed] [Google Scholar]
- 3.Volkow ND, Collins FS. The role of science in addressing the opioid crisis. N Engl J Med 2017; 377: 391–394. [DOI] [PubMed] [Google Scholar]
- 4.Yekkirala AS, Roberson DP, Bean BP, Woolf CJ. Breaking barriers to novel analgesic drug development. Nat Rev Drug Discov 2017; 16: 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knezevic NN, Cicmil N, Knezevic I, Candido KD. Discontinued neuropathic pain therapy between 2009-2015. Expert Opin Investig Drugs 2015; 24: 1631–1646. [DOI] [PubMed] [Google Scholar]
- 6.Mullard A. Calls grow to tap the gold mine of human genetic knockouts. Nat Rev Drug Discov 2017; 16: 515–518. [DOI] [PubMed] [Google Scholar]
- 7.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov 2013; 12: 581–594. [DOI] [PubMed] [Google Scholar]
- 8.Kamb A, Harper S, Stefansson K. Human genetics as a foundation for innovative drug development. Nat Biotechnol 2013; 31: 975–978. [DOI] [PubMed] [Google Scholar]
- 9.Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The Na(V)1.7 sodium channel: from molecule to man. Nat Rev Neurosci 2013; 14: 49–62. [DOI] [PubMed] [Google Scholar]
- 10.Vetter I, Deuis JR, Mueller A, Israel MR, Starobova H, Zhang A, Rash LD, Mobli M. NaV1.7 as a pain target—from gene to pharmacology. Pharmacol Ther 2017; 172: 73–100. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Wang Y, Li S, Xu Z, Li H, Ma L, Fan J, Bu D, Liu B, Fan Z, Wu G, Jin J, Ding B, Zhu X, Shen Y. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J Med Genet 2004; 41: 171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fertleman CR, Baker MD, Parker KA, Moffatt S, Elmslie FV, Abrahamsen B, Ostman J, Klugbauer N, Wood JN, Gardiner RM, Rees M. SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron 2006; 52: 767–774. [DOI] [PubMed] [Google Scholar]
- 13.Faber CG, Hoeijmakers JG, Ahn HS, Cheng X, Han C, Choi JS, Estacion M, Lauria G, Vanhoutte EK, Gerrits MM, Dib-Hajj S, Drenth JP, Waxman SG, Merkies IS. Gain of function Nanu1.7 mutations in idiopathic small fiber neuropathy. Ann Neurol 2012; 71: 26–39. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg YP, MacFarlane J, MacDonald ML, Thompson J, Dube MP, Mattice M, Fraser R, Young C, Hossain S, Pape T, Payne B, Radomski C, Donaldson G, Ives E, Cox J, Younghusband HB, Green R, Duff A, Boltshauser E, Grinspan GA, Dimon JH, Sibley BG, Andria G, Toscano E, Kerdraon J, Bowsher D, Pimstone SN, Samuels ME, Sherrington R, Hayden MR. Loss-of-function mutations in the Nav1.7 gene underlie congenital indifference to pain in multiple human populations. Clin Genet 2007; 71: 311–319. [DOI] [PubMed] [Google Scholar]
- 15.Cox JJ, Reimann F, Nicholas AK, Thornton G, Roberts E, Springell K, Karbani G, Jafri H, Mannan J, Raashid Y, Al-Gazali L, Hamamy H, Valente EM, Gorman S, Williams R, McHale DP, Wood JN, Gribble FM, Woods CG. An SCN9A channelopathy causes congenital inability to experience pain. Nature 2006; 444: 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss J, Pyrski M, Jacobi E, Bufe B, Willnecker V, Schick B, Zizzari P, Gossage SJ, Greer CA, Leinders-Zufall T, Woods CG, Wood JN, Zufall F. Loss-of-function mutations in sodium channel Nav1.7 cause anosmia. Nature 2011; 472: 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payandeh J, Hackos DH. Selective ligands and drug discovery targeting the voltage-gated sodium channel Nav1.7. Handb Exp Pharmacol 2018; 246: 271–306. [DOI] [PubMed] [Google Scholar]
- 18.McKerrall SJ, Sutherlin DP. Nav1.7 inhibitors for the treatment of chronic pain. Bioorg Med Chem Lett 2018; 28: 3141–3149. [DOI] [PubMed] [Google Scholar]
- 19.Mulcahy JV, Pajouhesh H, Beckley J, Delwig A, Du Bois J, Hunter J. Challenges and opportunities for therapeutics targeting the voltage-gated sodium channel isoform NaV1.7. J Med Chem Epub ahead of print 7 May 2019. DOI: 10.1021/acs.jmedchem.8b01906. [DOI] [PMC free article] [PubMed]
- 20.Sun S, Jia Q, Zenova AY, Chafeev M, Zhang Z, Lin S, Kwan R, Grimwood ME, Chowdhury S, Young C, Cohen CJ, Oballa RM. The discovery of benzenesulfonamide-based potent and selective inhibitors of voltage-gated sodium channel Na(v)1.7. Bioorg Med Chem Lett 2014; 24: 4397–4401. [DOI] [PubMed] [Google Scholar]
- 21.Focken T, Liu S, Chahal N, Dauphinais M, Grimwood ME, Chowdhury S, Hemeon I, Bichler P, Bogucki D, Waldbrook M, Bankar G, Sojo LE, Young C, Lin S, Shuart N, Kwan R, Pang J, Chang JH, Safina BS, Sutherlin DP, Johnson JP, Jr., Dehnhardt CM, Mansour TS, Oballa RM, Cohen CJ, Robinette CL. Discovery of aryl sulfonamides as isoform-selective inhibitors of NaV1.7 with efficacy in rodent pain models. ACS Med Chem Lett 2016; 7: 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahuja S, Mukund S, Deng L, Khakh K, Chang E, Ho H, Shriver S, Young C, Lin S, Johnson JP, Jr., Wu P, Li J, Coons M, Tam C, Brillantes B, Sampang H, Mortara K, Bowman KK, Clark KR, Estevez A, Xie Z, Verschoof H, Grimwood M, Dehnhardt C, Andrez JC, Focken T, Sutherlin DP, Safina BS, Starovasnik MA, Ortwine DF, Franke Y, Cohen CJ, Hackos DH, Koth CM, Payandeh J. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science 2015; 350: aac5464. [DOI] [PubMed] [Google Scholar]
- 23.McCormack K, Santos S, Chapman ML, Krafte DS, Marron BE, West CW, Krambis MJ, Antonio BM, Zellmer SG, Printzenhoff D, Padilla KM, Lin Z, Wagoner PK, Swain NA, Stupple PA, de Groot M, Butt RP, Castle NA. Voltage sensor interaction site for selective small molecule inhibitors of voltage-gated sodium channels. Proc Natl Acad Sci U S A 2013; 110: E2724–E2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandrou AJ, Brown AR, Chapman ML, Estacion M, Turner J, Mis MA, Wilbrey A, Payne EC, Gutteridge A, Cox PJ, Doyle R, Printzenhoff D, Lin Z, Marron BE, West C, Swain NA, Storer RI, Stupple PA, Castle NA, Hounshell JA, Rivara M, Randall A, Dib-Hajj SD, Krafte D, Waxman SG, Patel MK, Butt RP, Stevens EB. Subtype-selective small molecule inhibitors reveal a fundamental role for Nav1.7 in nociceptor electrogenesis, axonal conduction and presynaptic release. PLoS One 2016; 11: e0152405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theile JW, Fuller MD, Chapman ML. The selective Nav1.7 inhibitor, PF-05089771, interacts equivalently with fast and slow inactivated Nav1.7 channels. Mol Pharmacol 2016; 90: 540. [DOI] [PubMed] [Google Scholar]
- 26.Kornecook TJ, Yin R, Altmann S, Be X, Berry V, Ilch CP, Jarosh M, Johnson D, Lee JH, Lehto SG, Ligutti J, Liu D, Luther J, Matson D, Ortuno D, Roberts J, Taborn K, Wang J, Weiss MM, Yu V, Zhu DXD, Fremeau RT, Jr, Moyer BD. Pharmacologic characterization of AMG8379, a potent and selective small molecule sulfonamide antagonist of the voltage-gated sodium channel NaV1.7. J Pharmacol Exp Ther 2017; 362: 146–160. [DOI] [PubMed] [Google Scholar]
- 27.Bankar G, Goodchild SJ, Howard S, Nelkenbrecher K, Waldbrook M, Dourado M, Shuart NG, Lin S, Young C, Xie Z, Khakh K, Chang E, Sojo LE, Lindgren A, Chowdhury S, Decker S, Grimwood M, Andrez JC, Dehnhardt CM, Pang J, Chang JH, Safina BS, Sutherlin DP, Johnson JP, Jr., Hackos DH, Robinette CL, Cohen CJ. Selective NaV1.7 antagonists with long residence time show improved efficacy against inflammatory and neuropathic pain. Cell Rep 2018; 24: 3133–3145. [DOI] [PubMed] [Google Scholar]
- 28.Gingras J, Smith S, Matson DJ, Johnson D, Nye K, Couture L, Feric E, Yin R, Moyer BD, Peterson ML, Rottman JB, Beiler RJ, Malmberg AB, McDonough SI. Global Nav1.7 knockout mice recapitulate the phenotype of human congenital indifference to pain. PLoS One 2014; 9: e105895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nassar MA, Stirling LC, Forlani G, Baker MD, Matthews EA, Dickenson AH, Wood JN. Nociceptor-specific gene deletion reveals a major role for Nav1.7 (PN1) in acute and inflammatory pain. Proc Natl Acad Sci U S A 2004; 101: 12706–12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minett MS, Nassar MA, Clark AK, Passmore G, Dickenson AH, Wang F, Malcangio M, Wood JN. Distinct Nav1.7-dependent pain sensations require different sets of sensory and sympathetic neurons. Nat Commun 2012; 3: 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minett MS, Falk S, Santana-Varela S, Bogdanov YD, Nassar MA, Heegaard AM, Wood JN. Pain without nociceptors? Nav1.7-independent pain mechanisms. Cell Rep 2014; 6: 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shields SD, Cheng X, Uceyler N, Sommer C, Dib-Hajj SD, Waxman SG. Sodium channel Na(v)1.7 is essential for lowering heat pain threshold after burn injury. J Neurosci 2012; 32: 10819–10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shields SD, Deng L, Reese RM, Dourado M, Tao J, Foreman O, Chang JH, Hackos DH. Insensitivity to pain upon adult-onset deletion of Nav1.7 or its blockade with selective inhibitors. J Neurosci 2018; 38: 10180–10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dib-Hajj SD, Choi JS, Macala LJ, Tyrrell L, Black JA, Cummins TR, Waxman SG. Transfection of rat or mouse neurons by biolistics or electroporation. Nat Protoc 2009; 4: 1118–1126. [DOI] [PubMed] [Google Scholar]
- 35.Kim SH, Chung JM. An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain 1992; 50: 355–363. [DOI] [PubMed] [Google Scholar]
- 36.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods 1994; 53: 55–63. [DOI] [PubMed] [Google Scholar]
- 37.Ahmad S, Dahllund L, Eriksson AB, Hellgren D, Karlsson U, Lund PE, Meijer IA, Meury L, Mills T, Moody A, Morinville A, Morten J, O'Donnell D, Raynoschek C, Salter H, Rouleau GA, Krupp JJ. A stop codon mutation in SCN9A causes lack of pain sensation. Hum Mol Genet 2007; 16: 2114–2121. [DOI] [PubMed] [Google Scholar]
- 38.Cox JJ, Sheynin J, Shorer Z, Reimann F, Nicholas AK, Zubovic L, Baralle M, Wraige E, Manor E, Levy J, Woods CG, Parvari R. Congenital insensitivity to pain: novel SCN9A missense and in-frame deletion mutations. Hum Mutat 2010; 31: E1670–E1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsen KB, Nicholas AK, Woods CG, Mellgren SI, Nebuchennykh M, Aasly J. Two novel SCN9A mutations causing insensitivity to pain. Pain 2009; 143: 155–158. [DOI] [PubMed] [Google Scholar]
- 40.Lau J, Minett MS, Zhao J, Dennehy U, Wang F, Wood JN, Bogdanov YD. Temporal control of gene deletion in sensory ganglia using a tamoxifen-inducible Advillin-Cre-ERT2 recombinase mouse. Mol Pain 2011; 7: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akopian AN, Souslova V, England S, Okuse K, Ogata N, Ure J, Smith A, Kerr BJ, McMahon SB, Boyce S, Hill R, Stanfa LC, Dickenson AH, Wood JN. The tetrodotoxin-resistant sodium channel SNS has a specialized function in pain pathways. Nat Neurosci 1999; 2: 541–548. [DOI] [PubMed] [Google Scholar]
- 42.Ross SE. Pain and itch: insights into the neural circuits of aversive somatosensation in health and disease. Curr Opin Neurobiol 2011; 21: 880–887. [DOI] [PubMed] [Google Scholar]
- 43.McDermott LA, Weir GA, Themistocleous AC, Segerdahl AR, Blesneac I, Baskozos G, Clark AJ, Millar V, Peck LJ, Ebner D, Tracey I, Serra J, Bennett DL. Defining the functional role of NaV1.7 in human nociception. Neuron 2019; 101: 905.e8–919.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonnell A, Collins S, Ali Z, Iavarone L, Surujbally R, Kirby S, Butt RP. Efficacy of the Nav1.7 blocker PF-05089771 in a randomised, placebo-controlled, double-blind clinical study in subjects with painful diabetic peripheral neuropathy. Pain 2018; 159: 1465–1476. [DOI] [PubMed] [Google Scholar]
- 45.Cao L, McDonnell A, Nitzsche A, Alexandrou A, Saintot PP, Loucif AJ, Brown AR, Young G, Mis M, Randall A, Waxman SG, Stanley P, Kirby S, Tarabar S, Gutteridge A, Butt R, McKernan RM, Whiting P, Ali Z, Bilsland J, Stevens EB. Pharmacological reversal of a pain phenotype in iPSC-derived sensory neurons, patients with inherited erythromelalgia. Sci Transl Med 2016; 8: 335ra56. [DOI] [PubMed] [Google Scholar]
- 46.Rothenberg ME, Tagen M, Chang JH, Boyce-Rustay J, Friesenhahn M, Hackos DH, Hains A, Sutherlin D, Ward M, Cho W. Safety, tolerability, and pharmacokinetics of GDC-0276, a novel NaV1.7 inhibitor, in a first-in-human, single- and multiple-dose study in healthy volunteers. Clin Drug Investig 2019; 39: 873–887. [DOI] [PubMed] [Google Scholar]
- 47.Staud R, Price DD, Janicke D, Andrade E, Hadjipanayis AG, Eaton WT, Kaplan L, Wallace MR. Two novel mutations of SCN9A (Nav1.7) are associated with partial congenital insensitivity to pain. Eur J Pain 2011; 15: 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein CJ, Wu Y, Kilfoyle DH, Sandroni P, Davis MD, Gavrilova RH, Low PA, Dyck PJ. Infrequent SCN9A mutations in congenital insensitivity to pain and erythromelalgia. J Neurol Neurosurg Psychiatry 2013; 84: 386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchi M, Provitera V, Nolano M, Romano M, Maccora S, D'Amato I, Salvi E, Gerrits M, Santoro L, Lauria G. A novel SCN9A splicing mutation in a compound heterozygous girl with congenital insensitivity to pain, hyposmia and hypogeusia. J Peripher Nerv Syst 2018; 23: 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rush AM, Cummins TR, Waxman SG. Multiple sodium channels and their roles in electrogenesis within dorsal root ganglion neurons. J Physiol 2007; 579: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meents JE, Bressan E, Sontag S, Foerster A, Hautvast P, Rosseler C, Hampl M, Schuler H, Goetzke R, Le TKC, Kleggetveit IP, Le Cann K, Kerth C, Rush AM, Rogers M, Kohl Z, Schmelz M, Wagner W, Jorum E, Namer B, Winner B, Zenke M, Lampert A. The role of Nav1.7 in human nociceptors: insights from human induced pluripotent stem cell-derived sensory neurons of erythromelalgia patients. Pain 2019; 160: 1327–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hockley JRF, González-Cano R, McMurray S, Tejada-Giraldez MA, McGuire C, Torres A, Wilbrey AL, Cibert-Goton V, Nieto FR, Pitcher T, Knowles CH, Baeyens JM, Wood JN, Winchester WJ, Bulmer DC, Cendán CM, McMurray G. Visceral and somatic pain modalities reveal NaV 1.7-independent visceral nociceptive pathways. J Physiol 2017; 595: 2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez-Cano R, Tejada MA, Artacho-Cordon A, Nieto FR, Entrena JM, Wood JN, Cendan CM. Effects of tetrodotoxin in mouse models of visceral pain. Mar Drugs 2017; 15(6). pii:E188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, MPX881846 Supplemental Material1 for Rat NaV1.7 loss-of-function genetic model: Deficient nociceptive and neuropathic pain behavior with retained olfactory function and intra-epidermal nerve fibers by B Grubinska, L Chen, M Alsaloum, N Rampal, DJ Matson, C Yang, K Taborn, M Zhang, B Youngblood, D Liu, E Galbreath, S Allred, M Lepherd, R Ferrando, TJ Kornecook, SG Lehto, SG Waxman, BD Moyer, S Dib-Hajj and J Gingras in Molecular Pain
Supplemental material, MPX881846 Supplemental Material2 for Rat NaV1.7 loss-of-function genetic model: Deficient nociceptive and neuropathic pain behavior with retained olfactory function and intra-epidermal nerve fibers by B Grubinska, L Chen, M Alsaloum, N Rampal, DJ Matson, C Yang, K Taborn, M Zhang, B Youngblood, D Liu, E Galbreath, S Allred, M Lepherd, R Ferrando, TJ Kornecook, SG Lehto, SG Waxman, BD Moyer, S Dib-Hajj and J Gingras in Molecular Pain