Abstract
To perceive pathogens, plants employ pattern recognition receptor (PRR) complexes, which then transmit these signals via the receptor‐like cytoplasmic kinase BIK1 to induce defense responses. How BIK1 activity and stability are controlled is still not completely understood. Here, we show that the Hippo/STE20 homolog MAP4K4 regulates BIK1‐mediated immune responses. MAP4K4 associates and phosphorylates BIK1 at Ser233, Ser236, and Thr242 to ensure BIK1 stability and activity. Furthermore, MAP4K4 phosphorylates PP2C38 at Ser77 to enable flg22‐induced BIK1 activation. Our results uncover that a Hippo/STE20 homolog, MAP4K4, maintains the homeostasis of the central immune component BIK1.
Keywords: BIK1, Flg22‐triggered immunity, MAP4K4, PP2C38
Subject Categories: Immunology, Plant Biology, Signal Transduction
Introduction
Plants employ a multi‐layered immune system to combat pathogens 1. The first layer in plant innate immunity is the recognition of pathogen‐associated molecular patterns (PAMPs) such as bacterial flagellin or fungal chitin by cell‐surface pattern recognition receptors (PRRs), which ultimately activate a signaling cascade resulting in the establishment of PAMP‐triggered immunity (PTI), including calcium influx, oxidative burst, MAPK activation, and defense gene reprogramming 2, 3, 4, 5. Successful pathogens evolved effectors that are delivered into a host cell to inhibit PTI, but in an evolutionary arms race, plants also developed R proteins to recognize the invading effectors. R proteins detect effectors either by direct interaction or indirectly sensing the status of a host protein (“guardee”) to trigger a strong resistance response including a hypersensitive cell death (HR) which is termed effector‐triggered immunity (ETI) 6.
In Arabidopsis, flg22, a synthetic 22‐amino acid flagellin peptide, is perceived by the PRR complex FLAGELLIN‐SENSING 2 (FLS2) and BRI1‐ASSOCIATED RECEPTOR KINASE 1 (BAK1) 7, 8, 9. In the absence of flg22 10, FLS2 forms homodimers 11, 12, 13, heterodimerizes rapidly with BAK1 upon flg22 perception 7, 14. FLS2/BAK1 then phosphorylates BIK1 and activated BIK1 and then phosphorylates and activates RBOHD which is responsible for PRR‐triggered ROS production 11, 12, 15, 16, 17.
To regulate the optimal duration and amplitude of immune responses, the abundance, activity, and subcellular localization of PRRs are tightly controlled via their synthesis, phosphorylation, and ubiquitination 3, 18, 19, 20, 21. Regarding FLS2, two U‐box E3 ubiquitin ligases, PUB12 and PUB13, directly polyubiquitinate FLS2 and lead to flagellin‐induced FLS2 degradation. Furthermore, BAK1 promotes PUB13‐mediated FLS2 ubiquitination by phosphorylation of PUB13 22. PUB25 and PUB26 target non‐activated BIK1 for degradation, and CPK28 phosphorylates PUB25/26 to boost their activity and promote BIK1 degradation, whereas heterotrimeric G proteins inhibit PUB25/26 activity to stabilize BIK1 18, 23, 24, 25. The protein phosphatase PP2C38 dephosphorylates BIK1 to avoid premature extra‐activation in the resting state, but upon PAMP treatment, PP2C38 is phosphorylated at Ser77 and enables BIK1 activation by its dissociation from BIK1 26. BIK1 also localizes to the nucleus and interacts with WRKY transcription factors to negatively regulate JA/SA responses. EFR phosphorylates BIK1 to regulate its interaction with WRKY TFs 27.
MAPK cascades are evolutionarily highly conserved signaling modules found in all eukaryotes composed of MAPKKK‐MAPKK‐MAPK modules. In some cases, MAPKKKKs act as additional upstream regulators. Arabidopsis has 10 putative MAP4Ks 28. In this study, we show that MAP4K4 associates and regulates BIK1‐mediated plant immunity. map4k4 mutants resemble bik1 mutants in having compromised flg22‐induced immune responses. We show that MAP4K4 associates with BIK1. Phosphorylation of BIK1 by MAP4K4 stabilizes BIK1 protein levels. Moreover, we found that MAP4K4 phosphorylates PP2C38, revealing flg22‐induced BIK1 activation is mediated by MAP4K4 regulated phosphorylation of PP2C38. Our findings uncover another phosphorylation cascade regulating the central immune component BIK1.
Results
Flg22‐induced responses are compromised in map4k4 mutants
To investigate the function of MAP4K4 in plant immunity processes, two independent T‐DNA insertion mutants, map4k4‐1 (SALK_086087C) and map4k4‐2 (SALK_065417C), were obtained from NASC (Figs EV1A and 1B). Semiquantitative RT–PCR analysis indicated that both map4k4‐1 and map4k4‐2 are loss‐of‐function mutants with no detectable full‐length transcript (Fig EV1C). We analyzed the flg22‐triggered responses including reactive oxygen species (ROS) induction and gene expression reprogramming in the homozygous map4k4 knockout plants. The flg22‐induced ROS production was slightly but significantly reduced in map4k4 plants (Fig 1A). We further analyzed if MAP4K4 affects the PTI marker gene FRK1 induction by flg22 by quantitative RT–PCR (qRT–PCR). The results show that the flg22‐induced expression of FRK1 was also compromised in map4k4 plants (Fig 1B).
Figure EV1. Analysis of MAP4K4 T‐DNA insertion lines.
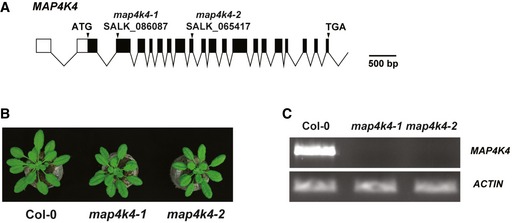
- T‐DNA location for the lines SALK_086087 (map4k4‐1) and SALK_065417 (map4k4‐2). The gene structure was generated by Exon‐Intron Graphic Maker (http://wormweb.org/exonintron).
- Morphological phenotype of 4‐week‐old Col‐0, map4k4‐1, and map4k4‐2.
- qRT–PCR showed that expression of MAP4K4 was not detectable in the map4k4 mutants. Actin was used as the control.
Figure 1. MAP4K4 regulates flg22 responses.

- ROS burst response in wild‐type (WT) and map4k4 lines in response to 1 μM flg22. Values are means ± SEM, n = 16 (biological replicates). Results are representative of three independent experiments. Statistical analysis was performed using a one‐way ANOVA with Tukey post‐test, P < 0.01.
- qRT–PCR analysis of the expression of PTI marker gene FRK1 in wild‐type and map4k4‐1 seedlings 1 h after treatment with 1 μM flg22. Bars represent means ± SD based on three biological replicates, P < 0.01.
- Venn diagram of the overlap between the flg22‐induced genes (fold change ≥ 2 and P‐value ≤ 0.05) in wild type and map4k4‐1.
- Heat map of gene expression response to flg22 in wild type and map4k4‐1; red and blue indicate high and low expression levels.
- GO enrichment analysis of MAP4K4‐dependent flg22‐induced genes.
- qRT–PCR analysis of 14‐day‐old seedlings treated with H2O or 1 μM flg22 for 1 h. Values are the means of three biological replicates ± SD, P < 0.01.
To understand the MAP4K4‐mediated transcriptional regulation in response to flg22, we performed global transcriptome analysis by RNA sequencing (RNA‐seq). Fourteen‐day‐old seedlings of Col‐0 and map4k4‐1 were treated with 1 μM flg22 or H2O for 1 h. Compared with mock (H2O) treatment, 2,273 and 1,953 genes were induced more than 2‐fold (P value < 0.05) upon flg22 treatment in Col‐0 and map4k4‐1, respectively, with 1,844 genes induced in both genotypes (Fig 1C). Strikingly, the overall response to flg22 was decreased in map4k4‐1 (Fig 1D). We further assessed the contribution of MAP4K4 to the flg22 response by analyzing the MAP4K4‐dependent flg22‐induced genes. A total of 429 genes were induced in wild type but not in map4k4‐1 (Fig 1C and Dataset EV1). These genes are enriched in the Gene Ontology (GO) terms defense, stimulus, and stress response (Fig 1E). Thus, MAP4K4 positively regulates a proportion of flg22‐induced genes. These RNA‐seq results were confirmed by qRT–PCR analysis of relevant genes (Fig 1F). We also checked whether MAP4K4 regulated flg22‐induced MAPK activation, indicating that MAPK activation was not significantly changed in map4k4‐1 mutants (Fig EV2). Collectively, these data show that MAP4K4 positively regulates a subset of PTI responses.
Figure EV2. flg22‐induced MAPK activation.

Two‐week‐old seedlings of Col‐0 and map4k4‐1 were treated with 100 nM flg22, and samples were collected at 0, 10, and 20 min after treatment. Activated MPKs were detected by Western blot using pTEpY antibodies.Source data are available online for this figure.
map4k4 mutants show enhanced resistance to infection by Pst DC3000
We assessed the role of MAP4K4 in plant immunity by spray inoculation of homozygous map4k4 knockout plants with virulent Pseudomonas syringae pv. tomato strain DC3000 (Pst DC3000). Bacterial titers at 2 h (day 0) after inoculation were similar in wild‐type Col‐0 and map4k4 leaves. However, at day 3 after infection, map4k4 showed significantly lower bacterial titers than wild type (Figs 2A and EV3A). To confirm whether the map4k4 mutant phenotype is caused by the T‐DNA insertion in MAP4K4, we generated transgenic complementation lines expressing MAP4K4 under its native promoter in the map4k4‐1 mutant background. Of five independent T3 single copy homozygous plants generated, two lines (c867 and c872), which showed comparable levels of expression of MAP4K4 to Col‐0, were further assessed (Fig EV3B). The disease resistance phenotype of map4k4‐1 was rescued by expression of MAP4K4 in the two complementation lines (Fig 2B).
Figure 2. MAP4K4 regulates immunity to DC3000.

- Left panel, map4k4‐1 shows enhanced resistance to Pseudomonas syringae pv. tomato DC3000. The growth of Pst DC3000 after inoculation in the indicated genotypes (Col‐0, white bars, map4k4‐1, green bars). Bacterial titers were determined 2 h (day 0) and 3 days after inoculation. Right panel, disease symptoms of wild type and map4k4‐1 at 3 days after inoculation. Values are the means ± SD, n = 6 (biological replicates), P < 0.01.
- map4k4‐1 disease resistance phenotype was recovered to wild‐type levels in two independent complementation lines, c867 and c872. Bacterial titers were determined as described above. Values are the means ± SD, n = 6 (biological replicates), P < 0.01.
- qRT–PCR analysis of PR1 expression levels in Col‐0, map4k4‐1, and c867 after mock (MgCl2) or Pst DC3000 inoculation, values are the means ± SD, n = 3 (biological replicates), P < 0.01.
- Flg22‐protection assay. Col‐0, map4k4‐1, and c867 were pretreated with H2O or flg22 1 day before Pst DC3000 inoculation. The bacterial population was measured 3 days after infection. Values are the means ± SD, n = 6. **P < 0.01. Results are representative of three independent experiments.
Figure EV3. Reduced growth of Pst DC3000 in map4k4 and rescue in the complementation lines.

- map4k4‐2 show enhanced resistance to Pseudomonas syringae pv. tomato DC3000, growth of Pst DC3000 after inoculation in the indicated genotypes (Col‐0, white bars; map4k4, black bars). Bacterial titers were determined at 2 h (day 0) and 3 d after inoculation. Bars represent the average of six replicates, and error bars show SD. Results are representative of three independent experiments. **P < 0.01 (Student's t‐test). Photographs were taken at 3 days after inoculation.
- qRT–PCR analysis of the expression of MAP4K4 in Col‐0, map4k4‐1, complementation line C867, and C872. Values are means ± SD, n = 3 (biological replicates). Different letters indicate statistical significance based on one‐way ANOVA with Tukey post‐test, and samples sharing letters are not significantly different, P < 0.01. Right panel, morphological phenotype of 4‐week‐old Col‐0, map4k4‐1, C867, and C872.
Pathogenesis‐related protein 1 (PR1) was strongly induced in response to pathogens 29, 30. To determine whether MAP4K4 affects PR1 induction, the Col‐0, map4k4‐1, and complementation line c867 were inoculated with Pst DC3000 or MgCl2 (mock), respectively. Total RNA was isolated from the inoculated leaves after 48 h, and expression levels of PR1 were measured by qRT–PCR. As shown in Fig 2C, in mock conditions, PR1 transcripts were comparable between Col‐0, map4k4‐1, and c867. Upon Pst DC3000 infection, PR1 was induced in all genotypes, but map4k4‐1 exhibited significantly higher upregulation of PR1 compared to Col‐0 and the complementation line c867.
MAP4K4 is required for flg22‐induced resistance to Pseudomonas syringae
To determine whether MAP4K4 is required for PAMP‐induced resistance to Pst DC3000, a flg22‐protection assay was carried out 31. One day before Pst DC3000 inoculation, Col‐0, map4k4‐1, and complementation line c867 were pretreated with water or 1 μM flg22, and the bacterial population was determined 3 days after pathogen inoculation. The flg22‐induced resistance to Pst DC3000 was significantly enhanced in Col‐0 and complementation line c867, but not in map4k4‐1 (Fig 2D). Collectively, these results indicate that MAP4K4 positively regulates flg22‐triggered immunity responses.
MAP4K4 is localized to the plasma membrane and the cytosol
To reveal the molecular mechanism underlying the immunity phenotype of map4k4, we first determined subcellular localization of MAP4K4. Searching PubMed and the Arabidopsis subcellular database SUBA 32, which integrate prediction and experimental data, mass spectrometry showed that MAP4K4 (AT5G14720) is localized to plasma membranes and the cytosol 33, 34, 35, 36, 37. To further confirm its localization, MAP4K4‐GFP was transiently expressed in Arabidopsis mesophyll protoplasts. Using the plasma membrane dye FM4‐64, MAP4K4‐GFP partially co‐localized with FM4‐64 (yellow part), indicating that MAP4K4 partially localized to the cytosol and partially to the plasma membrane (Fig 3A).
Figure 3. MAP4K4 localizes to the plasma membrane and cytosol and associates with BIK1.

- Subcellular localization of MAP4K4‐GFP in Arabidopsis mesophyll protoplasts. MAP4K4‐GFP (green) partially co‐localized with the plasma membrane dye FM4‐64 (red). Fourteen hours after transformation, photographs were taken when the plasma membrane of mesophyll protoplasts was stained by FM4‐64 (< 3 min). Scale bars = 10 μm.
- Autophosphorylation of recombinant HisMBP‐MAP4K4 wild type, no autophosphorylation in the kinase‐inactive HisMBP‐MAP4K4 D139A protein. The phosphorylation level was determined by Pro‐Q Diamond phosphoprotein gel stain (upper panel), and protein loading amounts were visualized by Coomassie G‐250 staining (lower panel). The predicted molecular weight of HisMBP‐MAP4K4 is 118.4 kDa.
- BiFC visualization of interaction between MAP4K4 and BIK1 in Arabidopsis mesophyll protoplasts. BIK1‐nYFP and MAP4K4‐cYFP were co‐expressed in protoplasts to show fluorescence of complemented YFP signal (green), and 14 h after transformation, the results were photographed by confocal laser scanning microscopy, scale bars = 10 μm. Chl indicates Chloroplast, and BF indicates Bright field. nYFP and cYFP constructs were used as empty controls. MAP3K5‐K375M and MKK4 were used as control.
- MAP4K4 associates with BIK1 in vivo by Co‐IP. BIK1‐HA and MAP4K4‐GFP were co‐expressed in plants by crossing of 35S::BIK1‐HA and 35S::MAP4K4‐GFP transgenic lines. Fourteen‐day‐old plants were treated (+) with 1 μM flg22 or H2O (−) for 10 min. GFP trap beads were used for immunoprecipitation, and target proteins were detected by Western blots using anti‐GFP or anti‐HA antibodies. The predicted molecular weight of BIK1‐HA, MAP4K4‐GFP is 49.3 and 103.8 kDa, respectively.
Source data are available online for this figure.
MAP4K4 is a conserved active protein kinase in plants
In the Arabidopsis genome, 10 putative MAP4Ks are annotated 28. MAP4K4 is a typical kinase containing the conserved 11 major protein kinase subdomains 38, which are highly conserved in diverse flowering plants (Fig EV4). To determine whether MAP4K4 has kinase activity, MAP4K4 and a kinase‐dead version MAP4K4 D139A (mutated in the ATP‐binding pocket) were fused to His‐MBP and subsequently expressed and purified from Escherichia coli. MAP4K4 showed strong autophosphorylation, whereas the kinase‐dead MAP4K D139A only displayed a weak background signal, indicating that MAP4K4 is an active protein kinase (Fig 3B).
Figure EV4. Conserved MAP4K4 kinase domain in diverse plant species.

Protein sequence alignments of MAP4K4 kinase domain from different plants species. The multiple alignment was generated by CLUSTALW. The identity of conserved residues is represented with a white font on black background, whereby similar residues are represented with a black font on gray background.
MAP4K4 associates with BIK1
Reminiscent of map4k4, bik1 mutants exhibit less flg22‐induced immune responses but more resistance to Pst DC3000 11, 12. We thus studied whether MAP4K4 associates with BIK1 in planta. We first performed BiFC assays in protoplasts. BIK1‐nYFP and MAP4K4‐cYFP were transiently co‐expressed in Arabidopsis mesophyll protoplasts, and YFP fluorescence could only be detected in cells co‐expressing BIK1‐nYFP with MAP4K4‐cYFP. Similarly, YFP signals were observed in cells co‐expressed with MAP3K5K375M‐nYFP and MKK4‐cYFP. However, no fluorescence was detected in negative controls in which BIK1‐nYFP was co‐expressed with MKK4‐cYFP or MAP3K5K375M‐nYFP was co‐expressed with MAP4K4‐cYFP (Fig 3C). YFP fluorescence was observed mainly at the plasma membrane of Arabidopsis mesophyll protoplasts, suggesting that MAP4K4 associates with BIK1 at the plasma membrane. The in vivo association between MAP4K4 and BIK1 was further tested by Co‐IP. MAP4K4‐GFP and BIK1‐HA fusion proteins were stably co‐expressed in Arabidopsis by crossing MAP4K4‐GFP with BIK1‐HA transgenic lines. The MAP4K4‐GFP protein was immunoprecipitated with GFP trap agarose beads, and co‐immunoprecipitation (co‐IP) was then detected by anti‐HA antibodies. As shown in Fig 3D, BIK1‐HA was detected upon co‐IP using extracts from BIK1‐HA/MAP4K4‐GFP transgenic plants, but not from BIK1‐HA plants, indicating that MAP4K4 associates with BIK1 in vivo. Interestingly, the association between BIK1‐HA and MAP4K4‐GFP was not altered upon flg22 treatment (Fig 3D). Notably, MAP4K4 and BIK1 do not associate with MAP3K5 and MKK4, respectively (Fig EV5).
Figure EV5. Negative controls for MAP4K4 Co‐IP .

- MAP4K4‐GFP does not associate with MAP3K5K375M‐HA.
- BIK1‐HA does not associate with MKK4‐GFP.
- MAP3K5K375M‐HA associates with MKK4‐GFP.
- MAP4K4‐HA does not associate with GFP.
- BIK1‐HA does not associate with GFP.
MAP4K4 regulates BIK1 stability by attenuating proteasome‐dependent degradation of BIK1
To investigate whether MAP4K4 regulates BIK1 abundance, we generated single copy BIK1‐HA transgenic overexpression lines under control of the 35S promoter by Agrobacterium‐mediated transformation. map4k4‐1 was crossed with 35S::BIK1‐HA, and sibling lines carrying either map4k4‐1 or MAP4K4 genotypes were isolated from the F3 generation. BIK1‐HA protein abundances were determined by Western blotting using anti‐HA antibodies. The results showed that BIK1‐HA proteins accumulated to a significantly lower level in map4k4 compared to WT (Fig 4A), indicating that MAP4K4 is required for BIK1 accumulation. To determine whether BIK1‐HA proteins are regulated by protein degradation, we treated 35S::BIK1‐HA plants with the proteasome inhibitor MG132. Addition of MG132 increased BIK1‐HA proteins levels in map4k4‐1 (Fig 4B), indicating that MAP4K4 regulates BIK1 stability through the 26S proteasome degradation pathway.
Figure 4. MAP4K4 regulates BIK1 stability and activation.

-
ADecreased BIK1 abundance in map4k4‐1. 35S::BIK1‐HA was introduced into map4k4‐1 by crossing, and sibling lines carrying either map4k4‐1 or MAP4K4 genotype were isolated from F3 generation. BIK1 levels were detected by Western blot analysis using anti‐HA antibodies.
-
BMAP4K4 regulates BIK1 accumulation through the proteasome pathway. Ten‐day‐old 35S::BIK1‐HA seedlings of WT or map4k4‐1 were treated with DMSO (−) or the proteasome inhibitor MG132 (50 μM) for 5 h, and then, the plants were collected for immunoblot analysis.
-
CFlg22‐induced BIK1 phosphorylation assay. Ten‐day‐old 35S::BIK1‐HA seedlings of WT or map4k4‐1 were treated with H2O (−) or 1 μM flg22 for 15 min, and BIK1 phosphorylation levels were detected by mobility‐shift assay using anti‐HA immunoblot.
-
D–FIdentification of phosphorylation sites by LC‐MS/MS analysis. Five residues in GST‐BIK1‐K105E were phosphorylated in vitro by HisMBP‐MAP4K4. The peptide sequences are shown above the spectrum with the PH designating phosphorylated residue. In the spectrum, the matched fragment ions are shown in red line for terminal series ions and in orange for immonium ions and modified ions.
-
GS233 and T242 were required for BIK1 hyper‐phosphorylation. BIK1‐HA, BIK1S233‐HA, and BIK1S242‐HA mutants were expressed in Col‐0 protoplasts and treated with 100 nM flg22 for 15 min, and BIK1 phosphorylation levels were detected by mobility‐shift assay using anti‐HA immunoblot. Each experiment was performed at least three times with similar results.
Source data are available online for this figure.
Flg22‐induced BIK1 phosphorylation depends on MAP4K4
BIK1 phosphorylation levels are increased upon flg22 treatment and can be measured by mobility‐shift Western blot assays 11, 12. Since MAP4K4 associates with BIK1 in vivo, we thus tested whether MAP4K4 regulates flg22‐induced BIK1 hyper‐phosphorylation 18. Ten‐day‐old 35S::BIK1‐HA map4k4‐1 and 35S::BIK1‐HA MAP4K4 plants were treated with 1 μM flg22 or H2O (mock), and phosphorylation levels were detected after a 10‐min treatment. In the absence of flg22, BIK1 abundance was significantly reduced in the map4k4‐1 mutant (Fig 4C). BIK1 phosphorylation was significantly increased upon flg22 treatment in 35S::BIK1‐HA MAP4K4, whereas the hyper‐phosphorylated BIK1 could be hardly detected in 35S::BIK1‐HA map4k4‐1 (Fig 4C). These data indicate that MAP4K4 regulates flg22‐induced BIK1 phosphorylation. Notably, BIK1 abundance was decreased in map4k4‐1 before and after flg22 treatment (Fig 4C), revealing that MAP4K4 regulates not only the under‐phosphorylated BIK1 before and after immune activation but also the proportion of activated BIK1 protein upon flg22 perception.
MAP4K4 directly phosphorylates BIK1
We further examined whether MAP4K4 directly phosphorylates BIK1 by in vitro kinase assays. Kinase‐dead GST‐BIK1‐K105E was incubated with MBP‐MAP4K4. BIK1 was then subjected to LC‐MS/MS analysis to identify the MAP4K4‐phosphorylated residues. The protein sequence coverage was comparable between GST‐BIK1‐K105E alone and GST‐BIK1‐K105E incubated with MBP‐MAP4K4 (89% vs. 87%). No phosphorylated residues were detected in GST‐BIK1‐K105E alone. Five amino acid residues, S19, S26, S233, S236, and T242, were identified as residues phosphorylated by MAP4K4 (Fig 4D–F). Whereas S19 and S26 are in the N terminus, S233, S236, and T242 lie within the activation loop of the BIK1 kinase domain 17. Among these phosphorylation sites, S236 and T242 are essential for BIK1 kinase activity and immune function and Ala substitution at S236 and T242 leads to compromised flg22‐induced responses, whereas BIK1T242D enhances flg22‐induced PTI 11, 39. S233 also affects the basal resistance to Pst DC3000 and BIK1 function in integrating flagellin and SA responses 39. Recently, S236 was reported to affect BIK1 stability through the proteasome system 18, indicating that phosphorylation of S236 by MAP4K4 stabilizes BIK1. Since S236 affects flg22‐induced hyper‐phosphorylation of BIK1 11, we assessed whether S233 and T242 are required for flg22‐induced hyper‐phosphorylation of BIK1 in protoplasts. In line with previous reports 11, 26, BIK1 shows a significant mobility shift in Col‐0 protoplasts upon flg22 treatment (Fig 4G), and S233A and T242A mutations abolished the band shift, indicating that S233 and T242 are major phosphorylation sites of BIK1 in response to flg22 (Fig 4G).
MAP4K4 associates with and phosphorylates PP2C38 at S77
PAMP‐induced hyper‐phosphorylation of BIK1 is regulated by the protein phosphatase PP2C38, which is determined by the phosphorylation status of S77 in PP2C38 26. The compromised flg22‐induced BIK1 hyper‐phosphorylation in map4k4 prompted us to determine whether MAP4K4 regulates BIK1 activation through phosphorylation of PP2C38. To assess whether MAP4K4 associates with PP2C38 in vivo, we performed Co‐IP using the protoplast system. As shown in Fig 5A, MAP4K4 associated with PP2C38. To test whether MAP4K4 phosphorylates PP2C38, we performed mass spectrometric analysis. Serine 77 was identified as the only phosphorylated residue when PP2C38 was incubated with MAP4K4, but not in PP2C38 alone (Fig 5B). Consistent with published results 26, the S77A mutation abolished the band shift in response to flg22 in protoplasts (Fig 5C), indicating that S77 is the major residue that is phosphorylated upon flg22 treatment. We further assessed whether MAP4K4 regulates PP2C38 phosphorylation levels in vivo, and PP2C38 showed a strong reduction in the phospho‐band in map4k4 when compared to wild type (Fig 5D). Our in vitro and in vivo data together indicate that MAP4K4 regulates PP2C38 S77 phosphorylation upon flg22 perception.
Figure 5. MAP4K4 associates and phosphorylates PP2C38.

- MAP4K4 associates with PP2C38 in vivo by Co‐IP. MAP4K4‐GFP and PP2C38‐HA were co‐expressed in protoplasts. GFP trap beads were used for immunoprecipitation, and target proteins were detected by Western blots using anti‐GFP or anti‐HA antibodies. The predicted molecular weight of MAP4K4‐GFP and PP2C38‐HA is 103.8 and 48.1 kDa, respectively.
- Identification of phosphorylation sites by LC‐MS/MS analysis. In vitro phosphorylation site of HisMBP‐PP2C38 by HisMBP‐MAP4K4. The peptide sequence is shown above the spectrum with the PH designating phosphorylated residue. In the spectrum, the matched fragment ions are shown in red line for terminal series ions and in orange for immonium ions and modified ions.
- S77 is required for flg22‐induced PP2C38 band shift. PP2C38‐HA and PP2C38‐S77A‐HA were expressed in Col‐0 and map4k4 protoplasts and treated with 1 μM flg22 for 30 min. The phosphorylated PP2C38 was detected by Western blot mobility‐shift assay.
- flg22‐induced PP2C38 band shift was compromised in map4k4 mutants. PP2C38‐HA were expressed in Col‐0 and map4k4 protoplasts and treated with 1 μM flg22 for 30 min.
- PP2C38 dephosphorylates MAP4K4 by its phosphatase activity. HisMBP‐MAP4K4 was incubated with HisMBP‐PP2C38 or HisMBP‐PP2C38D88N D289N (phosphatase‐inactive) or alone, and the phosphorylation level of MAP4K4 was determined by Pro‐Q Diamond phosphoprotein gel stain. All the experiments were performed three times with similar results.
Source data are available online for this figure.
Given the association between PP2C38 and MAP4K4, we tested whether PP2C38 affects MAP4K4 phosphorylation levels in vitro. HisMBP‐MAP4K4 was incubated with HisMBP‐PP2C38 or HisMBP‐PP2C38D88N D289N (phosphatase‐inactive PP2C38) or alone. HisMBP‐MAP4K4 phosphorylation levels were not affected by HisMBP‐PP2C38D88N D289N, but HisMBP‐PP2C38 reduced the phosphorylation levels of HisMBP‐MAP4K4 (Fig 5E).
MAP4K4 regulates BIK1‐mediated immunity
Consistent with previous reports 23, 25, overexpression of BIK1 in Col‐0 leads to an enhanced PAMP‐triggered ROS burst (Fig 6A). To determine whether the reduced BIK1 accumulation contributes to the impaired immunity phenotype in map4k4 plants, we generated map4k4‐1 35S::BIK1‐HA lines. We observed that the decreased ROS burst after flg22 treatment in the map4k4‐1 mutant was completely rescued by 35S::BIK1‐HA (Fig 6A), indicating that the compromised immune responses in the map4k4 line were indeed caused by reduced BIK1 accumulation. We further generated map4k4‐1 bik1 double mutants (Fig 6B) and observed a reduced ROS burst than in the single map4k4‐1 and bik1 mutants (Fig 6C). Since bik1 shows an autoimmune response in terms of upregulation of PR1 expression, we compared PR1 expression levels between Col‐0, map4k4, bik1, and map4k4‐1 bik1 double mutants. qRT–PCR analysis revealed that map4k4‐1 bik1 double mutants show higher upregulation of PR1 (Fig 6D), which is consistent with the stronger autoimmune response of map4k4‐1 bik1 than bik1 (Fig 6B).
Figure 6. MAP4K4 regulates BIK1‐mediated immunity.

- BIK1 transgene restores flg22‐induced ROS burst in map4k4‐1. 35S::BIK1‐HA WT or 35S::BIK1‐HA map4k4‐1 were generated by crossing as described before. ROS burst response in the indicated genotypes in response to 1 μM flg22. Values are means ± SEM, n = 16. Results are representative of three independent experiments.
- Morphological phenotype of 6‐week‐old Col‐0, map4k4‐1, bik1, and map4k4‐1 bik1 mutants.
- ROS burst response in Col‐0, map4k4‐1, bik1, and map4k4‐1 bik1 mutants. Values are means ± SEM, n = 16. Results are representative of three independent experiments.
- Relative expression level of PR1 in Col‐0, map4k4‐1, bik1, and map4k4‐1 bik1 mutants by qRT–PCR analysis. Values are means ± SD, n = 3 (biological replicates).
- Morphological phenotype of 6‐week‐old Col‐0, map4k4‐1, map4k3, and map4k4‐1 map4k3 mutants.
- Relative expression level of PR1 in Col‐0, map4k4‐1, map4k3, and map4k4‐1 map4k3 mutants by qRT–PCR analysis. Values are means ± SD, n = 3 (biological replicates).
- ROS burst response in Col‐0, map4k4‐1, map4k3, and map4k4‐1 map4k3 mutants. Values are means ± SEM, n = 16 (biological replicates).
MAP4K3/SIK1 functions with MAP4K4 in regulating BIK1‐mediated immunity
Similar to MAP4K4, MAP4K3/SIK1 was recently reported to positively regulate flg22‐induced immunity responses by stabilizing BIK1 40. To determine the genetic redundancy between MAP4K3 and MAP4K4, we generated map4k3 map4k4 double mutants (Fig 6E). map4k3 map4k4 double mutants exhibit a more severe dwarfism than single mutants (Fig 6E), and consistently, PR1 expression was much higher in map4k3 map4k4 double than single mutants (Fig 6F). We further tested the flg22‐induced ROS burst showing more reduced H2O2 accumulation in map4k3 map4k4 double than in single mutants (Fig 6G). Together, these results indicate that MAP4K3 acts additively with MAP4K4.
Discussion
Plants employ cell‐surface‐localized receptors consisting of receptor‐like kinases (RLKs) and receptor‐like proteins (RLPs) to perceive pathogens. The receptor‐like cytoplasmic kinases (RLCKs) act in concert with RLKs and RLPs and regulate PRR‐mediated signaling cascades by phosphorylation 20. Notably, the activity and stability of PRRs and RLCKs are fine‐tuned at multiple levels via sophisticated regulatory mechanisms 17, 25, 41. In yeast and mammalian cells, a fourth level of kinases, termed STE20/MAP4Ks, not only phosphorylate and regulate downstream MAP3Ks but also regulate upstream signaling components 42, 43. Here, we report a MAP3K kinase in Arabidopsis, MAP4K4, whose kinase domain is highly conserved across plant species. MAP4K4 associates and phosphorylates BIK1. The MAP4K4‐phosphorylated S236 and T242 residues lie within the activation loop of BIK1. Activation loop phosphorylation is one of the most common mechanisms for regulating protein kinase activity, and S236 and T242 play essential roles for BIK1 kinase activity and function. Notably, S236 was required for BIK1 stability through the ubiquitin‐proteasome pathway 18. BIK1 abundance is decreased in map4k4 mutants, and BIK1 abundance can be partially rescued by the proteasome inhibitor MG132, indicating that MAP4K4 regulates BIK1 stability through phosphorylation of S236 (Fig 7). Interestingly, BIK1 associates with MAP4K4 independently of flg22 treatment, which is consistent with the results that BIK1 abundance was decreased in map4k4 before and after flg22 treatment.
Figure 7. Model: MAP4K4 regulates BIK1 stability and activation by phosphorylation.

In the absence of flg22, BIK1 interacts with FLS2, BAK1, SIK1, MAP4K4, and PP2C38. SIK1 and MAP4K4 stabilize BIK1 by phosphorylation. PP2C38 maintains BIK1 activity at minimum levels. Upon flg22 treatment, BIK1 is released from FLS2 and BAK1. MAP4K4 phosphorylates PP2C38 at Ser77, thereby enabling BIK1 activation. In parallel or alternatively, SIK1 and MAP4K4 phosphorylate and stabilize BIK1. SIK1 and BIK1 phosphorylate and activate RBOHD producing H2O2.
The appropriate amplitude of signaling cascades is often regulated by phosphorylation, and phosphorylation homeostasis is controlled by kinases and phosphatases in response to a variety of signals. The protein phosphatase type 2C PP2C38 directly dephosphorylates BIK1 and keeps its phosphorylation status at a minimum level in the absence of elicitation. Upon PAMP perception, PP2C38 is phosphorylated at S77 by BIK1. Phosphorylated PP2C38 is released from BIK1 and enables BIK1 activation 26. In vitro kinase assays and mass spectrometry analysis showed that MAP4K4 phosphorylates PP2C38 at S77, and flg22‐induced PP2C38 mobility shift was decreased in map4k4, indicating that MAP4K4 phosphorylates PP2C38 at S77 in vivo. Furthermore, flg22‐induced BIK1 activation was compromised in map4k4. Collectively, we propose that MAP4K4 regulates flg22‐induced BIK1 activation through phosphorylation of PP2C38 (Fig 7). Notably, MAP4K4 directly phosphorylates BIK1 at S233, S236, and T242, so the alternative model is that MAP4K4 regulates flg22‐induced BIK1 activation via phosphorylation of BIK1 (Fig 7).
Recently, a member of the MAP4K family, MAP4K3/SIK1 was reported to stabilize BIK1 40. The authors have analyzed all the 10 MAP4K4 family members, but only found map4k3/sik1 showing a phenotype in terms of plant growth and immunity. The authors also found that map4k3/sik1 mutants showed an autoimmune phenotype only under specific conditions, indicating that growth conditions affect MAP4K3 function. Consistent with these results (Zhang et al, Fig 1B) 40, both map4k3 and map4k4 mutants exhibit smaller sizes than wild‐type plants (Fig 6E), and, likely due to different laboratory conditions, we found that MAP4K4 also plays a role in immunity processes. Comparing map4k3 and map4k4 mutants, they show some common phenotypes. Both mutants exhibit enhanced resistance to Pst DC3000, but a reduced flg22‐induced ROS burst and reduced flg22‐induced Pst DC3000 resistance. Furthermore, BIK1 abundance is decreased in both map4k3 and map4k4 mutants. MAP4K3 and MAP4K4 are both active protein kinases and phosphorylate BIK1 at S236 which is required for BIK1 stability. The reduced flg22‐induced ROS burst in map4k3 map4k4 double mutants further indicates additive roles between MAP4K3 and MAP4K4. However, MAP4K4 displays unique features. MAP4K4 phosphorylates BIK1 at T242 which is required for BIK1 kinase activity and flg22‐induced Pst DC3000 resistance 39. Furthermore, flg22‐induced BIK1 activation is decreased in map4k4, but not map4k3 40, indicating that BIK1 hyper‐phosphorylation was dependent on MAP4K4 but not MAP4K3. Therefore, although these MAP4Ks share certain functions, they also show distinct features that collectively add to the robustness of immune signaling in Arabidopsis. The discovery of the functions of other MAP4K proteins should be of major interest to further our understanding of the conserved and diverse mechanisms of signal transduction in plants.
Materials and Methods
Plant materials
The Arabidopsis ecotype Columbia‐0 (Col‐0) was used as wild‐type control. map4k4‐1 (SALK_086087), map4k4‐2 (SALK_065417), bik1 (SALK_005291), and map4k3/sik1‐1 (SALK_046158) T‐DNA insertion mutants were ordered from the Nottingham Arabidopsis Stock Centre (NASC), and homozygous T‐DNA insertion mutants were isolated by allele‐specific primers (Dataset EV2). Plants were grown either on soil or on plates [half‐strength Murashige and Skoog (MS, Sigma M6899), 0.5% sucrose (sigma S5016), 1% agar (sigma A1296), and 0.5% MES (sigma M8250), pH adjusted to 5.7 with KOH] at 23°C in 16‐h day‐length conditions.
Pseudomonas assays
Plants were grown in Jiffy‐7 pellet in a growth chamber (Percival) under 12 h light at 23°C. Pseudomonas syringae pv. tomato (Pst) DC3000 strains were grown for 24 h at 28°C on King Agar B (Sigma 60786) with 50 mg/l rifampicin. Four‐ to 5‐week‐old plants were spray inoculated with bacterial suspensions at 1 × 108 cfu/ml in 10 mM MgCl2 containing 0.04% Silwet L‐77 (LEHLE SEEDS). Bacterial titers were determined as previously described 44. For flg22 protection assay 31, 1 μM flg22 (Genscript) was infiltrated into leaves 1 day before Pst inoculation.
ROS burst assay
Plants were grown on Jiffy‐7 pellets with short photoperiod in Percival growth chamber (12 h light at 23°C). After 4–5 weeks, leaf disks (17.8 mm2) were used for ROS assay as described previously 45. Luminescence was measured using a TECAN Infinite 200 PRO microplate readers, and signal integration time was 0.5 s.
Constructs
Phusion High‐Fidelity DNA Polymerase (New England Biolabs, NEB) was used in PCR cloning and mutagenesis. MAP4K4, BIK1, and PP2C38 coding sequences were amplified from Col‐0 cDNA, MAP4K4 locus were amplified from Col‐0 genomic DNA, and the entry clones were generated by pDONR207 (Invitrogen, Gateway BP recombination reactions). Site‐directed mutagenesis was created by PCR with complementary primers carrying a mutation (s) and followed by Dpn I (NEB) digestion. For recombinant protein constructs, HisMBP‐MAP4K4, HisMBP‐MAP4K4 D139A and HisMBP‐PP2C38 and GST‐BIK1 were generated by pDEST‐HisMBP 46 and pDEST 15 (Invitrogen, Gateway LR recombination reactions), respectively. pGWnY and pGWcY 47 were used to generate BiFC constructs. p2GWF7 48 was used to generate a transient expression construct 35S::MAP4K4‐GFP. All the constructs were verified by sequencing.
Recombinant protein purification
HisMBP‐MAP4K4, HisMBP‐MAP4K4 D139A, HisMBP‐PP2C38, and HisMBP‐PP2C38 D88N D289N were expressed in Rosetta (Novagen), and GST‐BIK1‐K105E were expressed in BL21‐AI (Invitrogen). HisMBP fusion protein expression was induced overnight at 20°C with 1 mM isopropyl b‐D‐1‐thiogalactopyranoside (IPTG) and purified by Ni‐NTA Agarose (Qiagen, 30230). Eluted protein was desalted by PD‐10 desalting columns (GE healthcare, 17‐0851‐01). GST fusion protein expression was induced overnight at 20°C by 1 mM IPTG and 0.2% l‐arabinose and purified by Glutathione Sepharose 4B (GE healthcare, 17‐0756‐01). The purification procedures were according to the manufacturer's instructions.
In vitro phosphorylation and mass spectrometry
In vitro kinase assay was performed in a mixture containing 20 mM Tris–HCl, pH 7.5, 5 mM EGTA, 1 mM DTT, 50 μM ATP, and 10 mM MgCl2 at 28°C for 1 h. The kinase reactions were terminated by adding SDS sample buffer and boiling at 70°C for 10 min. The protein phosphorylation level was determined by Pro‐Q Diamond phosphoprotein gel stain (Molecular Probes, P33300) according to the manufacturer's instructions; images were captured by Biorad ChemiDoc Imagers.
Phosphopeptides were analyzed according to the methods described previously 49. Briefly, protein samples were separated by SDS–PAGE and stained by Coomassie (SimplyBlue SafeStain, Invitrogen), and the target protein bands were excised and digested with Trypsin (Promega, V5113). After ZipTip purification, samples were proceeded to LTQ Orbitrap mass‐spectrometer (Thermo Scientific). The LC‐MS/MS spectra were analyzed using Mascot against the Arabidopsis database (TAIR10), and the identified phosphopeptides were confirmed manually.
Protein localization and BiFC in Arabidopsis mesophyll protoplasts
Arabidopsis mesophyll protoplast isolation and transformation were performed according to the method described by 50. For protein localization, 100 μl protoplasts (around 2 × 104 cells) were transformed with 5 μg plasmid. For BiFC, 100 μl protoplasts were transformed with 10 μg total plasmid (5 μg of each construct). Protoplast plasma membrane was stained with 15 μM FM4‐64 dye (Molecular Probes, T13320). Fluorescent signals were visualized 16 h after transfection with Zeiss LSM 710 confocal microscope.
MG132 treatment
MG132 (selleckchem) was dissolved in DMSO. Plants were grown on MS plates (1/2 MS, 0.5% sucrose, 1% agar, and 0.05% MES, pH 5.7) at 23°C in 16‐h day‐length conditions. Five‐day‐old plants were transferred to 24‐well plates with liquid 1/2 MS. After 5 days, old 1/2 MS medium was replaced by fresh 1/2 MS with 1% DMSO or 100 μM MG132, and samples were collected 10 h after treatment.
Co‐immunoprecipitation (Co‐IP) assay
The co‐immunoprecipitation in protoplasts was performed as previously described with minor modification 12. Briefly, protoplasts or tissues were harvested and lysed with extraction buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X‐100, Roche cOmplete ULTRA EDTA‐free protease inhibitor cocktail, PhosSTOP phosphatase inhibitor cocktail). Extracts were incubated with GFP‐Trap (ChromoTek) at 4°C for 4 h and washed four times with 500 μl washing buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Triton X‐100). The dry beads were mixed with SDS sample buffer, heated at 95°C for 5 min, and then filtered by Pierce spin columns. Samples were separated on 10–12% SDS–PAGE and analyzed by Western blot with the anti‐GFP antibody (1:10,000, Molecular Probes, A‐11122) and anti‐HA antibody (1:10,000, Roche, 11867423001), the PVDF membrane was developed by ECL (Biorad), and images were captured by Biorad ChemiDoc Imagers.
RNA extraction and RT–qPCR
RNA was extracted using NucleoSpin RNA Plant kit (Macherey Nagel). 1 μg total RNA was reverse‐transcribed using SuperScript III First‐Strand Synthesis SuperMix (Invitrogen, 18080400) according to the manufacturer's instructions. RT–qPCR was performed using StepOnePlus Real‐Time PCR System (Applied Biosystems) with Power SYBR Green PCR Master Mix (Applied Biosystems). The PCR reaction program was as follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Gene expression values were normalized to At4g05320 (UBQ10). The primers for real‐time PCR are listed in Dataset EV2.
RNA‐seq and transcriptome analysis
Seeds were surface sterilized and stratified on MS plates (1/2 MS, 0.5% sucrose, 1% agar, and 0.05% MES, pH 5.7) for 3 days at 4°C. Then, plants were grown in a growth chamber at 23°C with a 16‐h photoperiod. Fourteen‐day‐old seedlings were treated with 1 μM flg22 or H2O for 1 h, and three biological repeat samples were proceeded to total RNA extraction. mRNA sequencing libraries were constructed in KAUST Bioscience Core Lab using TruSeq Stranded mRNA LT Sample Prep Kit and manufacturer's instructions (Illumina). Sequencing was performed on each library to generate 151‐bp paired‐end reads for transcriptome sequencing on Illumina Genome Analyzer platform (HiSeq 4000). Low‐quality reads were trimmed using a Trimmomatic 51. After pre‐processing the Illumina reads, the transcript structures were reconstructed using TopHat for aligning with the genome (TAIR10) and Cufflinks for gene structure predictions 52, 53. Genes with expression fold change ≥ 2 and P‐value < 0.05 have been chosen to define the differential expression. Gene Ontology analysis was performed by AgriGO 54 toolkit and database using singular enrichment analysis tool and default parameters; graphical results were presented.
MAPK activation
Two‐week‐old plants were grown on half‐strength MS medium and were treated with 100 nM flg22. Protein was lysed with extraction buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X‐100, Roche cOmplete ULTRA EDTA‐free protease inhibitor cocktail, PhosSTOP phosphatase inhibitor cocktail). Samples were separated on 10% SDS–PAGE and analyzed by Western blot with the anti‐pTEpY antibody (1:3,000, Cell Signaling Technology, #4370) and anti‐rabbit antibody (1:10,000, Sigma, A6154).
Author contributions
YJ and HH designed the project, YJ, BH, and HZ conducted the experiments, YJ and KGM analyzed the RNA‐seq data, JB and JC contributed unpublished constructs and plant materials, and YJ and HH wrote the article with input from all co‐authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Source Data for Expanded View
Review Process File
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
This work was supported by the King Abdullah University of Science and Technology projects BAS/1/1062‐01‐01 and URF/1/2965‐01‐01 and the LabEx Saclay Plant Sciences‐SPS ANR‐10‐LABX‐0040‐SPS.
EMBO Reports (2019) 20: e47965
Contributor Information
Yunhe Jiang, Email: yunhe.jiang@kaust.edu.sa.
Heribert Hirt, Email: heribert.hirt@kaust.edu.sa.
Data availability
The datasets produced in this study are available in the following databases: RNA‐Seq data: Sequence Read Archive PRJNA548363 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA548363).
References
- 1. Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- 2. Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern‐triggered immunity (PTI). Mol Plant 8: 521–539 [DOI] [PubMed] [Google Scholar]
- 3. Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- 4. Latrasse D, Jégu T, Li H, de Zelicourt A, Raynaud C, Legras S, Gust A, Samajova O, Veluchamy A, Rayapuram N et al (2017) MAPK‐triggered chromatin reprogramming by histone deacetylase in plant innate immunity. Genome Biol 18: 131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang D, Wang G, Zhou J‐M (2017) Receptor kinases in plant‐pathogen interactions: more than pattern recognition. Plant Cell 29: 618–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cui H, Tsuda K, Parker JE (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- 7. Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JDG, Felix G, Boller T (2007) A flagellin‐induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- 8. Gómez‐Gómez L, Boller T (2000) FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis . Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- 9. Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J (2013) Structural basis for flg22‐induced activation of the Arabidopsis FLS2‐BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- 10. Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- 11. Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S et al (2010) Receptor‐like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- 12. Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor‐like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun W, Cao Y, Jansen Labby K, Bittel P, Boller T, Bent AF (2012) Probing the Arabidopsis flagellin receptor: FLS2‐FLS2 association and the contributions of specific domains to signaling function. Plant Cell 24: 1096–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heese A, Hann DR, Gimenez‐Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor‐like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y et al (2014) The FLS2‐associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338 [DOI] [PubMed] [Google Scholar]
- 16. Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones Jonathan D, Shirasu K, Menke F, Jones A et al (2014) Direct regulation of the NADPH Oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- 17. Lin W, Li B, Lu D, Chen S, Zhu N, He P, Shan L (2014) Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc Natl Acad Sci 111: 3632–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang J, Grubb LE, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li M, Li L et al (2018) A regulatory module controlling homeostasis of a plant immune kinase. Mol Cell 69: 493–504.e6 [DOI] [PubMed] [Google Scholar]
- 19. Mithoe SC, Menke FLH (2018) Regulation of pattern recognition receptor signalling by phosphorylation and ubiquitination. Curr Opin Plant Biol 45: 162–170 [DOI] [PubMed] [Google Scholar]
- 20. Liang X, Zhou J‐M (2018) Receptor‐like cytoplasmic kinases: central players in plant receptor kinase‐mediated signaling. Annu Rev Plant Biol 69: 267–299 [DOI] [PubMed] [Google Scholar]
- 21. Yu X, Feng B, He P, Shan L (2017) From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol 55: 109–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky Frederikke G, Rathjen John P, MacLean D, Romeis T et al (2014) The calcium‐dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16: 605–615 [DOI] [PubMed] [Google Scholar]
- 24. Liang X, Ma M, Zhou Z, Wang J, Yang X, Rao S, Bi G, Li L, Zhang X, Chai J et al (2018) Ligand‐triggered de‐repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Res 28: 529–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S et al (2016) Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife 5: e13568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Couto D, Niebergall R, Liang X, Bücherl CA, Sklenar J, Macho AP, Ntoukakis V, Derbyshire P, Altenbach D, Maclean D et al (2016) The Arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLoS Pathog 12: e1005811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lal NK, Nagalakshmi U, Hurlburt NK, Flores R, Bak A, Sone P, Ma X, Song G, Walley J, Shan L et al (2018) The receptor‐like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe 23: 485–497.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Champion A, Picaud A, Henry Y (2004) Reassessing the MAP3K and MAP4K relationships. Trends Plant Sci 9: 123–129 [DOI] [PubMed] [Google Scholar]
- 29. Uknes S, Mauch‐Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis . Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Breen S, Williams SJ, Outram M, Kobe B, Solomon PS (2017) Emerging insights into the functions of pathogenesis‐related protein 1. Trends Plant Sci 22: 871–879 [DOI] [PubMed] [Google Scholar]
- 31. Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
- 32. Hooper CM, Castleden IR, Tanz SK, Aryamanesh N, Millar AH (2017) SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res 45: D1064–D1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nuhse TS, Stensballe A, Jensen ON, Peck SC (2003) Large‐scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics 2: 1234–1243 [DOI] [PubMed] [Google Scholar]
- 34. Benschop JJ, Mohammed S, O'Flaherty M, Heck AJ, Slijper M, Menke FL (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis . Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- 35. Zhang ZJ, Peck SC (2011) Simplified enrichment of plasma membrane proteins for proteomic analyses in Arabidopsis thaliana . Proteomics 11: 1780–1788 [DOI] [PubMed] [Google Scholar]
- 36. Elmore JM, Liu J, Smith B, Phinney B, Coaker G (2012) Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol Cell Proteomics 11: M111.014555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rayapuram N, Bonhomme L, Bigeard J, Haddadou K, Przybylski C, Hirt H, Pflieger D (2014) Identification of novel PAMP‐triggered phosphorylation and dephosphorylation events in Arabidopsis thaliana by quantitative phosphoproteomic analysis. J Proteome Res 13: 2137–2151 [DOI] [PubMed] [Google Scholar]
- 38. Hanks S, Quinn A, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241: 42–52 [DOI] [PubMed] [Google Scholar]
- 39. Laluk K, Luo H, Chai M, Dhawan R, Lai Z, Mengiste T (2011) Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS‐INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP‐triggered immunity in Arabidopsis . Plant Cell 23: 2831–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang M, Chiang Y‐H, Toruño TY, Lee D, Ma M, Liang X, Lal NK, Lemos M, Lu Y‐J, Ma S et al (2018) The MAP4 kinase SIK1 ensures robust extracellular ROS burst and antibacterial immunity in plants. Cell Host Microbe 24: 379–391.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zou Y, Wang S, Zhou Y, Bai J, Huang G, Liu X, Zhang Y, Tang D, Lu D (2018) Transcriptional regulation of the immune receptor FLS2 controls the ontogeny of plant innate immunity. Plant Cell 30: 2779–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dan I, Watanabe NM, Kusumi A (2001) The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol 11: 220–230 [DOI] [PubMed] [Google Scholar]
- 43. Machida N, Umikawa M, Takei K, Sakima N, Myagmar B‐E, Taira K, Uezato H, Ogawa Y, Kariya K‐i (2004) Mitogen‐activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c‐Jun N‐terminal kinase. J Biol Chem 279: 15711–15714 [DOI] [PubMed] [Google Scholar]
- 44. García AV, Blanvillain‐Baufumé S, Huibers RP, Wiermer M, Li G, Gobbato E, Rietz S, Parker JE (2010) Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog 6: e1000970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith JM, Heese A (2014) Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leave tissue in response to living Pseudomonas syringae . Plant Methods 10: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nallamsetty S, Austin BP, Penrose KJ, Waugh DS (2005) Gateway vectors for the production of combinatorially‐tagged His6‐MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli . Protein Sci 14: 2964–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hino T, Tanaka Y, Kawamukai M, Nishimura K, Mano S, Nakagawa T (2011) Two Sec13p homologs, AtSec13A and AtSec13B, redundantly contribute to the formation of COPII transport vesicles in Arabidopsis thaliana . Biosci Biotechnol Biochem 75: 1848–1852 [DOI] [PubMed] [Google Scholar]
- 48. Karimi M, Depicker A, Hilson P (2007) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2007) In‐gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- 50. Yoo S‐D, Cho Y‐H, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- 51. Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA‐Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 45: W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Source Data for Expanded View
Review Process File
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Data Availability Statement
The datasets produced in this study are available in the following databases: RNA‐Seq data: Sequence Read Archive PRJNA548363 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA548363).


