The heat-stable nucleoid-structuring (H-NS) protein coordinates the expression of over 200 genes in E. coli, with a large number involved in both bacterial virulence and drug resistance. We report on the novel observation of a dynamic compaction of the bacterial chromosome in response to exposure to high levels of salt. This stress response results in the detachment of H-NS proteins and their subsequent expulsion to the periphery of the cells. We found that this behavior is related to mechanical properties of the bacterial chromosome, in particular, to how tightly twisted and coiled is the chromosomal DNA. This behavior might act as a biomechanical response to stress that coordinates the expression of genes involved in adapting bacteria to a salty environment.
KEYWORDS: Escherichia coli, H-NS, chromosome organization, nucleoid-associated proteins, osmotic stress, stress response
ABSTRACT
The heat-stable nucleoid-structuring (H-NS) protein is a global transcriptional regulator implicated in coordinating the expression of over 200 genes in Escherichia coli, including many involved in adaptation to osmotic stress. We have applied superresolved microscopy to quantify the intracellular and spatial reorganization of H-NS in response to a rapid osmotic shift. We found that H-NS showed growth phase-dependent relocalization in response to hyperosmotic shock. In stationary phase, H-NS detached from a tightly compacted bacterial chromosome and was excluded from the nucleoid volume over an extended period of time. This behavior was absent during rapid growth but was induced by exposing the osmotically stressed culture to a DNA gyrase inhibitor, coumermycin. This chromosomal compaction/H-NS exclusion phenomenon occurred in the presence of either potassium or sodium ions and was independent of the presence of stress-responsive sigma factor σS and of the H-NS paralog StpA.
IMPORTANCE The heat-stable nucleoid-structuring (H-NS) protein coordinates the expression of over 200 genes in E. coli, with a large number involved in both bacterial virulence and drug resistance. We report on the novel observation of a dynamic compaction of the bacterial chromosome in response to exposure to high levels of salt. This stress response results in the detachment of H-NS proteins and their subsequent expulsion to the periphery of the cells. We found that this behavior is related to mechanical properties of the bacterial chromosome, in particular, to how tightly twisted and coiled is the chromosomal DNA. This behavior might act as a biomechanical response to stress that coordinates the expression of genes involved in adapting bacteria to a salty environment.
INTRODUCTION
Bacteria are under constant pressure to adapt and flourish within unstable and often unpredictable environments. One of the more effective means by which bacteria adapt to environmental stressors is that of acquiring genetic traits through the mechanism of horizontal gene transfer (HGT). Expression of most horizontally acquired genes, however, simply incurs a metabolic cost or worse for the host, placing the cells at a competitive disadvantage. Bacteria have therefore evolved to mitigate the costs of HGT, with many enteric bacteria, such as Escherichia coli and Salmonella, utilizing the heat-stable nucleoid-structuring (H-NS) protein (1, 2). H-NS acts as a global transcriptional silencer of foreign (or xenogeneic) DNA acquired through HGT, primarily by binding to adenine-rich and thymine-rich regions of the chromosome (1, 3). H-NS has been implicated in coordinating a range of bacterial stress responses such as adaptation to changes in pH, temperature, and osmolarity (4). Because of these observations, for decades H-NS was thought to primarily regulate the stress response of enterobacteria. However, with the later advent of high-throughput sequencing techniques, a crucial role for H-NS in silencing xenogeneic DNA was identified (5). It is now generally appreciated that H-NS, like a eukaryotic histone, is a general factor that has evolved many regulatory roles, including controlling the expression of AT-rich genes that are transcriptionally responsive to a variety of conditions, including stress.
H-NS polymerizes on DNA to both block RNAP access to promoters and likely also to trigger RNAP stalls during its translocation/elongation along the DNA template (6–10). As a consequence of its function, it also acts as a “domainin” (a DNA binding protein) that prevents the diffusion of supercoils generated during replication and transcription along the DNA molecule, instead keeping such supercoils trapped at a local region of the chromosome (10–14). Indeed, the ability of H-NS to trap supercoils was identified in early studies on the biochemical properties of the molecule (15). One reason that H-NS may contribute to RNAP stalling is that of preventing the free diffusion of positive supercoils in front of the translocating polymerase.
H-NS has been implicated in the regulation of the E. coli and Salmonella osmotic responses both globally and specifically by several independent studies. H-NS is a direct negative regulator of the proV locus (where it was also named OsmZ in early studies) (16, 17). Globally, H-NS affects levels of σS (σ38 or RpoS), the RNAP sigma subunit that mediates the enterobacterial general stress response to conditions, including starvation and osmolarity (18, 19).
Supercoiling tension along any stretch of DNA represents a combination of the activities of topoisomerases (stable changes to linking number), DNA binding proteins (domainins), and helicases/polymerases, which transiently cause supercoiling stress via the translocational unwinding of the DNA template (20). The findings that H-NS constrains supercoils, that H-NS negatively regulates the expression of several osmotically induced genes, and that high osmolarity alters the overall average negative supercoiling of several genes and plasmid reporters have led many to speculate that H-NS controls supercoiling in the cell (2, 15, 21). However, it is unclear exactly how H-NS binding is affected by DNA supercoiling, whether supercoiling is affected by H-NS directly or indirectly, and whether external triggers such as osmolarity or temperature can modulate the ability of H-NS to directly affect DNA supercoiling. These problems arise, in part, from the paucity of tools with sufficient resolution to dissect how H-NS responds in vivo to perturbations in environmental conditions.
In this study, we focused on the spatiotemporal distribution of H-NS binding along the E. coli chromosome, in response to osmotic stress, and at different phases of growth (both exponential and early stationary phase). Previously, it was found that several highly expressed nucleoid-associated proteins (NAPs) such as H-NS distribute in a more or less random fashion about the nucleoid volume (22). We should comment, since it is often incorrectly cited, that while H-NS was reported to cluster into a single focus within E. coli (22), this observation proved to represent an artifact caused by weak dimerization of the fluorescent protein mEos2 (23). This weak dimerization of the fused fluorescent label, combined with the fact that H-NS itself forms extended protein filaments (24–26), likely led to mEos2-mediated aggregation of the H-NS nucleoprotein complexes. Changes to the dimerization domain of mEos2, resulting in the highly monomeric mEos3.2 (27), mitigate this effect and reveal that H-NS-mEos3.2 is dispersed in small punctate foci randomly throughout the nucleoid.
We have observed a pronounced, almost immediate spatial redistribution of H-NS following osmotic shock during the stationary phase of growth. A rapid increase in osmolarity causes the bacterial chromosome to tightly condense, at which point H-NS apparently detaches from the chromosome and migrates toward the periphery of the cell. This behavior is not observed in exponential phase under the same stress conditions; rather, H-NS remains distributed throughout the nucleoid despite a slight compaction of the chromosome. If, however, we subject exponentially replicating cells to the DNA gyrase inhibitor coumermycin (28–30) during osmotic shock, we observe both increased compaction of the chromosome and an apparent detachment/exclusion of H-NS from the nucleoid volume, similar to what was observed in stationary phase. Dissociation of H-NS from the chromosome was also measured in chromatin immunoprecipitation (ChIP) assays. Notably, the observed compaction and H-NS redistribution were independent of the general stress response sigma factor σS and of the nucleoid-associated H-NS paralog StpA.
RESULTS
In situ analysis of H-NS and nucleoid localization.
We proceeded to visualize the spatial distribution of H-NS and the nucleoid using superresolved radial fluctuation (SRRF) imaging (31), an approach similar to techniques such as superresolution optical fluctuation imaging (SOFI) (32) that make use of temporal correlations in an image stack to achieve superresolved image reconstructions. SRRF imaging applied to a conventional, wide-field image stack can result in a resolution of approximately 100 to 150 nm, which provided sufficient detail for this study (see Fig. S3 in the supplemental material).
For the imaging, the chromosomal hns gene was engineered with a 3′ in-frame fusion to the gene encoding the monomeric photoactivatable fluorescent protein mEos3.2 (further fused at the 3′ end with a sequence encoding a FLAG epitope tag) to generate a chimeric protein, H-NS-mEos3.2FLAG. While we did not make use of the photoswitchable properties of mEos3.2 here, its proven ability to remain truly monomeric when fused to H-NS made it an ideal choice for this study (23). Our initial pilot studies utilized this chimeric protein expressed from a low-copy-number plasmid using the native hns promoter. However, we found that plasmid-mediated expression of H-NS-mEos3.2FLAG led to the appearance of punctate foci (Fig. S4), which may have been caused by overexpression of the protein or H-NS association with the expression plasmids themselves, as H-NS was previously shown to bind the hns promoter (33).
To circumvent these issues, we constructed an E. coli strain with a chromosomally encoded version of H-NS-mEos3.2FLAG. The sequence was recombined into the hns locus, leaving it under the control of the native hns promoter, to generate strain WN3334. During imaging, this strain did not show the same problematic foci as were observed with the plasmid-encoded chimeric protein. We verified that the chimeric H-NS-mEos3.2FLAG protein was functional and capable of silencing loci at a level similar to that seen with wild-type H-NS (Fig. S5). To observe the nucleoid, we employed DAPI (4′,6-diamidino-2-phenylindole) staining and merged the two images to analyze where H-NS-mEos3.2FLAG localized with respect to the bulk chromosome.
Dynamics of the nucleoid in response to osmotic shock during growth and stationary phase.
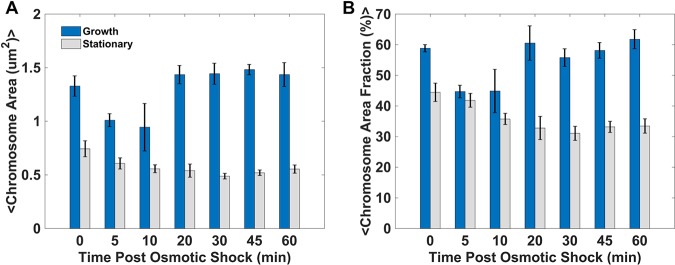
A previous study reported by Cagliero and Jin (34) showed that RNAP was displaced and that the nucleoid condensed dramatically under conditions of osmotic shock when exponentially growing E. coli cells were treated with NaCl. We examined the dynamic response of the nucleoid to osmotic stress by fixing cells at a series of time points following induction of osmotic shock. Panels A and B of Fig. 1 show the absolute area and fractional area (relative to that of the cell) of the nucleoid, respectively, for cells in the exponential-growth phase or the stationary-growth phase. The analysis examined DAPI-stained chromosomal DNA at 0, 5, 10, 20, 30, 45, and 60 min after the addition of 300 mM KCl.
FIG 1.
Dynamics of chromosome compaction following osmotic shock. (A and B) Absolute, projected chromosome area (A) and the area fraction (i.e., normalized to the projected area of the cell) (B), for both exponential-phase and stationary-phase cells, up to 60 min after osmotic shock. Error bars represent standard deviations of the means (see Table S1 in the supplemental material).
In rapidly growing cells, the nucleoid showed significant condensation between 5 and 10 min, but by 20 min the images of the chromosome were difficult to differentiate from those taken before the changes in preinduction. We presume that this was due to rapid adaptation of the cells to osmotic stress, whereby the cells immediately import K+ to prevent dehydration and stabilize turgor pressure and begin to accumulate osmolytes and small molecule osmoprotectants such as proline within minutes. Similar dynamics seen during the exponential phase were previously reported by Cagliero and Jin, although the level of compaction reported in minimal media appears to be greater than what we observed. While we are unsure of the origin of this discrepancy, we note that Cagliero and Jin provided limited data in minimal media and induced shock with roughly twice the level of osmolarity as was used in this study. In the stationary phase, the nucleoid was more compact preshock and condensed less dramatically than the nucleoid of cells in the exponential-growth phrase after the addition of KCl. However, after the addition of KCl, the nucleoid remained condensed and did not return to its preshock size after 60 min.
H-NS is displaced from the stationary-phase chromosome during osmotic shock.
Concurrent with imaging the chromosome, we examined the localization of H-NS-mEos3.2FLAG at various time points after osmotic shock in both rapidly growing cells (Fig. 2) and stationary-phase cells (Fig. 3). In both figures, images are provided for 0, 5, 10, and 45 min postinduction. Images representing additional time points are provided elsewhere (see Fig. S6 in the supplemental material).
FIG 2.

SRRF images of exponential-growth-phase dynamic response to osmotic stress (300 mM KCl). From left to right, t = 0, 5, 10, and 45 min postinduction. (A to D) H-NS (H-NS-mEos3.2FLAG). (E to H) Chromosome. (I to L) Merged images. The merged images were magnified to show details (magnified regions are indicated by boxes in the images above the merged images). All scale bars are 2 μm.
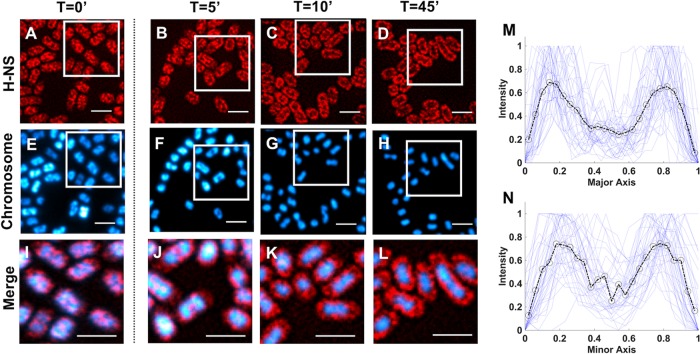
FIG 3.
SRRF images of stationary-phase dynamic response to osmotic stress (300 mM KCl). From left to right, t = 0, 5, 10, and 45 min postinduction. (A to D) H-NS-mEos3.2FLAG. (E to H) Chromosome. (I to L) Merged images. The merged images were magnified to show details (magnified regions are indicated by boxes in the images above the merged images). All scale bars are 2 μm. (M and N) Major-axis (M) and minor-axis (N) normalized intensity cross sections of H-NS distribution at 30 min postinduction. Intensity traces from individual bacteria (40 cells) are shown, with the average given by the dashed line (mean pixel values are indicated by circles).
After induction of osmotic shock, no noticeable response was observed in the spatial distribution of H-NS during the rapid growth phase (Fig. 2). However, during stationary phase (Fig. 3), after 5 min, H-NS appears to have reorganized throughout the cell volume, forming a ringlike pattern that was fully established by 10 min postinduction and maintained for at least 60 min, which is the longest period for which we measured. The chromosome, which was already more condensed in stationary phase than in exponential phase, continued to compact until roughly 10 min postinduction. From these images, the osmotically shocked stationary-phase chromosome appears to have been tightly condensed within the center of each cell, with the bulk of H-NS surrounding it.
Coumermycin inhibition of DNA gyrase alters the nucleoid response to osmotic shock during rapid growth.
We hypothesized that extreme DNA compaction, affected by supercoiling, might play a role in the spatial reorganization of H-NS. Cells were treated with 5 μg/ml coumermycin for 30 min and then imaged at 30 min after osmotic induction. Coumermycin is an antibiotic that acts as a DNA gyrase inhibitor by blocking access of ATP to the enzyme and may also inhibit cell growth and DNA synthesis (28–30). No effect was seen in stationary phase; however, cells within exponential phase displayed a spatial reorganization of H-NS-mEos3.2FLAG similar to what was previously observed only in stationary phase (Fig. 4). Once again, the bacterial chromosome was observed to have been significantly more compact than before osmotic induction and H-NS was again seen to have been excluded from the chromosomal volume and pushed toward the periphery of the bacterial cell. We note, however, that this behavior was not seen in exponentially growing cells treated with coumermycin alone.
FIG 4.
Analysis of cells treated with coumermycin (Table S2). (A) SRRF images of exponential-growth-phase cells subjected to osmotic stress treatment (300 mM KCl) for 30 min without (left column) or with (right column) coumermycin (Coum; 5 μg/ml). From top to bottom, H-NS-mEos3.2FLAG, DAPI-stained chromosome, and merged images. Scale bars are 2 μm. (B and C) Comparison of the projected chromosome area (B) and area fraction (C) between exponential growth phase and stationary phase cells. Shown are results for control cells, cells subjected to osmotic shock (KCl), and cells exposed to KCl plus coumermycin.
Chromatin immunoprecipitation of H-NS during osmotic shock.
A prior study examined the effect of transient K+ accumulation on the organization of the nucleoid and the association of RNAP (34). Using cell fractionation, Cagliero and Jin also noted that H-NS appeared to transiently and partially dissociate from the nucleoid before reassembling after osmoadaptation (34). If H-NS does dissociate from the chromosome, it should manifest as a decrease in DNA binding in chromatin immunoprecipitation assays. The alternative hypothesis is that while H-NS migrates to the periphery of the cell during chromosomal compaction, the H-NS-bound DNA sequences remain associated with the protein and “loop” away from the bulk of the nucleoid, which might not be discernible by microscopy.
To test whether H-NS was indeed displaced from its bound loci during osmotic shock, we performed chromatin immunoprecipitation analysis of a nonfluorescent hemagglutinin (HA)-tagged H-NS protein on four known H-NS-bound loci around the chromosome (proV, xapR, yjcF, and bglF) under both stationary-phase and rapid-growth-phase conditions before and after treatment with sodium chloride and/or coumermycin (Fig. 5). Consistent with what was observed by microscopy, we found that H-NSHA association with its DNA targets (e.g., proV) was diminished in response to a rapid increase in osmolarity and that this dissociation was particularly acute during stationary phase. Coumermycin on its own did not dramatically affect H-NSHA binding in either growth phase but dramatically potentiated the displacement of H-NSHA caused by an increase in osmolarity during exponential growth. The fact that H-NSHA behaved similarly to the fluorescent chimeric H-NS protein suggests that the observed displacement was not an artifactual event caused by fusion with the bulky mEos3.2 protein.
FIG 5.
Chromatin immunoprecipitation of H-NSHA at four H-NS-bound loci (proV [upper left], xapR [upper right], yjcF [lower left], and bglF [lower right]) during osmotic shock (KCl) or gyrase inhibition with coumermycin (Coum) or both. Cells, diluted in M9 medium from overnight cultures, were grown to the indicated growth phase. At 30 min after addition of KCl or coumermycin or both, cells were fixed with formaldehyde and DNA-protein complexes were immunoprecipitated as described in Materials and Methods. Enrichment of bound DNA was quantified by real-time PCR and normalized to the amount of DNA in the sample prior to immunoprecipitation. Values above or adjacent to bars indicate the mean levels of ChIP efficiency.
It might be expected that displacement of H-NS from the chromosome would lead to an increase in gene expression from its repressed loci. To test this, we examined the transcript abundance of H-NS repressed loci in the stationary-phase cells treated 30 min with KCl and/or coumermycin (Fig. 6). We observed that the proV transcript was strongly upregulated under these conditions (20-fold increase in steady-state transcript levels). The bglF and yciF genes show more modest 2-fold increases in expression, and the expression level of the xapR transcript was essentially unchanged. There are several possible reasons why some bound loci did not respond transcriptionally to the displacement of H-NS. Under these conditions, the chromosome was tightly compacted and RNAP was expected to be in complex primarily with the sigma subunit σS, which has been shown to positively regulate the proV P1 promoter (35). proV is also known to respond positively to osmotic stress, suggesting that its promoter is optimized for high levels of expression under these experimental conditions (36). In contrast, RNAP-σS is not known to regulate xapR or yjcF or the bglGFB promoters. Indeed, some evidence suggests that the bglGFB operon may be downregulated under conditions where σS is active (37, 38). Therefore, it is likely that H-NS displacement alone is insufficient to activate transcription at many loci and that transcription occurs only in the context of the appropriate promoter, supercoiling, and DNA binding proteins.
FIG 6.

Transcript levels of H-NS-regulated genes in the stationary phase during osmotic shock. RNA was extracted from stationary-phase cultures 30 min after treatment with 300 mM KCl. RNA was reverse transcribed, and transcript levels were measured by quantitative real-time PCR. Fold change is reported compared to identical cultures to which KCl had not been added. All transcripts levels were normalized to those of gyrB as a control. Values above bars represent mean values corresponding to fold changes in expression.
StpA and σS are not involved in the observed compaction of the nucleoid or expulsion of H-NS.
It is possible that H-NS dissociation from DNA is a biophysical consequence of osmotic shock, or it is possible that cellular factors are specifically involved in displacing H-NS from the chromosome. We entertained the idea that H-NS might transiently be displaced from the bacterial chromosome during osmotic shock through competition from its paralog StpA. Notably, StpA demonstrates higher affinity for DNA in vitro in the presence of salt (9, 39). To assess whether StpA plays any role in displacing H-NS from the nucleoid, we visualized H-NS-mEos3.2FLAG localization in an isogenic strain harboring a deletion in stpA. We observed that H-NS was displaced from the chromosome similarly in both the absence and presence of StpA, indicating that competitive binding was not the root cause of H-NS depletion from the chromosome (Fig. S7).
Given that σS is a central and pleiotropic regulator of the cellular response to general stress, including osmotic shock (40), we also assessed whether a member of the σS regulon might be directly or indirectly involved in displacing H-NS from the chromosome under conditions of osmotic shock. However, we found that the localizations of H-NS-mEos3.2FLAG were similar between strains with or without functional σS (Fig. S7).
These findings, along with data corresponding to the speed at which displacement occurs, suggest that H-NS displacement from the chromosome may be a direct consequence of the physical changes that occur inside the cell as a result of osmotic shock and that it is not caused by specific cell-encoded regulatory factors.
DISCUSSION
On average, the bacterial chromosome of E. coli is negatively supercoiled. However, the level of negative superhelicity varies throughout the growth phase, with the overall level decreasing as the cell moves from exponential to stationary phase (20, 41). This transition is thought to be mediated primarily by changes in DNA gyrase activity and the altered binding of nucleoid-associated proteins (NAPs). DNA gyrase induces negative supercoiling, which facilitates transcription by relieving torsional stress along double stranded DNA, while NAPs are conjectured to stabilize the level of local supercoiling throughout the chromosome (29, 41–45).
We have observed that during stationary phase, under conditions of osmotic stress, the E. coli chromosome tightly condenses and the nucleoid-associated protein H-NS is actively excluded from the DNA-containing volume of the cell, relocating toward the membrane periphery. While this behavior was observed in stationary phase, such gross spatial reorganization was not seen under identical conditions during rapid growth—despite the concurrent observation that the chromosome condenses following osmotic shock in both phases, although more so during stationary phase.
We were then able to reproduce a similar effect in the exponential phase upon addition of coumermycin, an antibiotic compound that inhibits DNA gyrase activity. Following exposure to coumermycin, the chromosomal DNA showed an increased level of compaction and H-NS was again excluded from the nucleoid volume, similarly to what we had previously observed only in stationary phase. When we then repeated the coumermycin experiments in stationary phase, again under conditions of osmotic stress, we detected no observable changes. The chromosome remained significantly condensed, with H-NS excluded from the nucleoid volume.
We provide a few possible explanations for our observations. In one model, osmotic stress induces the rapid condensation of the bacterial chromosome. While the mechanism is unclear, Cagliero and Jin posit that the rapid accumulation of cytoplasmic K+ after osmotic stress may induce RNAP to detach from the chromosome and that the absence of transcriptional procession and sliding by the enzyme, which may enlarge the nucleoid structure, leads to condensation. This occurs in both exponential and stationary phase, though to different extents. In stationary phase, where the DNA already displays a reduced level of negative supercoiling before imposition of osmotic stress conditions, DNA condensation is so extreme that H-NS rapidly dissociates from the DNA. In exponential phase, this response is attenuated by the activity of DNA gyrase, which keeps the overall level of negative supercoiling elevated in preparation for higher levels of transcriptional activity than those that occur in stationary phase. The chromosome condenses, but only to a limited extent, and H-NS is still able to remain bound. However, by interfering with DNA gyrase activity by the use of coumermycin, the level of negative helicity is reduced and the chromosome is then able to tightly compact, removing H-NS from the DNA.
Yet another model posits that high osmolarity in the cytoplasm reduces the affinity of H-NS for DNA and that large-scale H-NS dissociation releases previously trapped supercoils, which causes the compaction of DNA (once again, the effect would be greater in stationary-phase cells). This model suggests that the disruption of H-NS binding is the “proximal” event that occurs prior to chromosome compaction. The bacterial cell would then adapt via the production and accumulation of compatible solutes, which would ultimately restore homeostasis. Therefore, the order of events, i.e., whether nucleoid compaction directly causes H-NS to dissociate or whether dissociation of H-NS leads to chromosomal compaction, remains unclear.
It seems perhaps counterintuitive that relaxing of negative supercoils, instead of causing an expansion, leads to a tighter compaction of the nucleoid, and yet this phenomenon has been observed before by other groups (34, 46). Since supercoiling is known to affect the binding affinity of many proteins, it could be that a reduction in negative superhelicity causes other proteins to bind to and further condense the chromosomal DNA. Beyond simply being counterintuitive, a real complication for this model is that several groups have found osmotic stress to increase, not decrease, the level of negative supercoiling within bacterial cells (16, 47, 48). We note that those previous studies examined DNA topology using linkage assays or cruciform formation in plasmid reporters in actively growing cells or after extended periods of growth at high osmolarity, all of which represent conditions that differ from the specific conditions under which we observed chromosomal compaction. But if the extreme compaction of the chromosome and subsequent expulsion of H-NS represent results of increased negative supercoiling, then why do we observe this behavior in stationary phase and not exponential phase unless we inhibit DNA gyrase? While we are unable to answer this question at present, the effect is quite apparent in our data and it suggests a simple biophysical mechanism for responding to osmotic stress by preventing H-NS-mediated gene silencing of particular stress response genes.
Given that σS had no effect on the displacement of H-NS, it does not appear that the general stress response plays an early role in condensing the chromosome or expelling H-NS from the nucleoid volume following osmotic shock. Instead, it is more likely that H-NS displacement is an upstream event that would help augment the osmotic stress response by affecting levels of σS (49). Both σS itself and many σS-regulated genes are induced under conditions of osmotic stress (50, 51), and loss of H-NS has been demonstrated to stabilize σS via derepression of the σS antiadaptors IraM and IraP (19). Dissociation of H-NS from the chromosome shortly after osmotic shock would presumably potentiate activation of the σS regulon.
Finally, we note that during our review of the literature there remained a number of statements and views regarding supercoiling, osmolarity, and gene regulation that were apparently contradictory. It is clear from our study and those performed by others that growth phase, source of osmotic stress, and length of exposure all play a role in what has been observed, which makes comparisons between studies challenging. Furthermore, it remains unclear how well plasmid linking numbers can serve as surrogate metrics for supercoiling-mediated phenomena that may be local. It also remains difficult to dissect which effects represent bona fide regulatory responses to changes in supercoiling and which represent adventitious secondary consequences caused by perturbations in nucleoid structure.
MATERIALS AND METHODS
Strains and plasmids.
These studies employed E. coli strain BW25113 (genotype: F− lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78), previously used as the wild-type strain for the generation of the Keio single-gene-knockout collection (52). Isogenic hns deletion strain JW1225-2 (BW25113; Δhns-746::kan) was used for experiments with plasmid-encoded H-NS-mEos3.2FLAG.
Labeling H-NS with photoactivatable mEos3.2.
The gene encoding the monomeric, photoactivatable fluorescent protein mEos3.2 in plasmid pmEos3.2C (27) was amplified and inserted via Gibson assembly to generate an intermediate construct, producing a chimeric protein where the full-length E. coli H-NS protein from E. coli BW25113 is linked by its C terminus to a linker (sequence: GSAGSAAGSGEF), mEos3.2, and a C-terminal FLAG epitope tag. The entire chimeric construct was subsequently cloned upstream of the chloramphenicol resistance gene (cat [transcribed separately from a downstream promoter]) of plasmid pXG10 (53). This plasmid was then used as a PCR template to generate the fragment hns-mEos3.2-FLAG-cat by PCR using Phusion high-fidelity DNA polymerase (Thermo Fisher) and primers SB039 (5′-GCCGCTGGCGGGATTTTAAGCAAGTGCAATCTACAAAAGAGCTCTTTTTTGTGCGGTGCC) and SB40 (5′-CCTCAACAAACCACCCCAATATAAGTTTGAGATTACTACACAACAGGAGTCCAAGCGAGC).
The purified PCR fragment was subsequently used to replace the chromosomally carried hns gene in strain BW25113, at its native chromosomal location and under the control of its native promoter, using the lambda red recombinase method described previously by Datsenko and Wanner (54) to generate strain WN3334. The tagged chromosomal copy of hns was verified by Sanger sequencing at the TCAG Sequencing Facility (Centre for Applied Genomics, Hospital for Sick Children) using primers EMC003 (5′-GGTGTTATCCACGAAACGG) and EMC004 (5′-CGTTAAATCTGGCACCAAAG).
Derivatives of strain WN3334 lacking rpoS (encoding σS) and stpA were made by performing P1vir-mediated transduction (55) of strain WN3334 by the use of phage P1vir lysates derived from strains JW2644-3 (ΔstpA750::kan) and JW5437-1 (ΔrpoS746::kan) from the Keio E. coli single-gene-knockout collection (52). These strains, selected for growth on both chloramphenicol and kanamycin, were verified by PCR and sequencing of the relevant chromosomal loci.
Cell fixation and sample preparation.
We initially grew bacteria in M9 medium (BioShop) supplemented with MgSO4 (2 mM), CaCl2 (0.1 mM), thiamine (0.01%), Casamino Acids (0.1%), and glucose (1%). Cells were shaken at 220 rpm at 37°C until they reached exponential (∼3 h, optical density at 600 nm [OD600] of ∼0.3) or stationary (∼6 h, OD600 of ∼2) phase. We then exposed the cells to osmotic stress by adding KCl to the culture to reach a final concentration of 300 mM (osmolarity, ∼0.6 Osm/liter). Substituting KCl for NaCl yielded no observable differences in cellular responses (see Fig. S1 in the supplemental material). We concede that the addition of salts can also result in ionic stress upon the cells, but this possibility was not further explored in the present work. After exposure of the cells to the stress, we fixed the bacterial cells with 10× diluted formalin solution in H2O (Sigma-Aldrich) (∼37% formaldehyde stabilized with 10% to 15% methanol). In control experiments, we found that fixation with methanol-free formaldehyde (Thermo Fisher) did not have an effect on the observed results. The exposure time before fixation ranged from 5 to 60 min for the time course experiments, but the exposure time was set at 30 min for all other experiments. Cultures were incubated with formaldehyde and with shaking at room temperature for 30 min. This was followed by 15 min of centrifugation at 1,000 × g, discarding of the supernatant, and washing three times with 1 ml of phosphate-buffered saline (PBS). For each washing step, the pellet was first mixed with PBS for 5 min in an orbital shaker and then centrifuged for 10 min at 1,000 × g at room temperature. After the final wash, the sample was ready to be imaged or was stored at 4°C to be imaged within 1 week.
Samples were mounted by placing 3 μl of fixed cells atop a small 1.5% agarose pad. The agarose pad was then flipped over onto a coverslip that had been precleaned with an initial 30 min of sonication in 3 M KOH, rinsed in distilled water (dH2O), and then again sonicated for 30 min in ethanol (99%). Cells remained immobilized between the coverslip and the agarose pad. The coverslip was then affixed to the microscope slide with a 2.5-mm-diameter CoverWell spacer (Sigma-Aldrich) inserted between the two glass surfaces.
Superresolved microscopy.
Microscopy was performed using an inverted Olympus IX-71 microscope equipped with a 60× oil immersion total internal reflection fluorescence (TIRF) objective (Olympus; APON 60XOTIRF) (numerical aperture [N.A.], 1.49). Images were additionally magnified by a telescope inserted into the collection path of the microscope such that the effective pixel size was ∼73 nm and collected by an electron multiplying charge-coupled-device (EMCCD) camera (Andor, iXon3). All data acquisition was controlled by the open source microscopy program μManager (56).
To obtain superresolution radial fluctuation (SRRF) images, we conducted widefield fluorescence microscopy by imaging mEos3.2 in the green channel. We imaged the fluorescent protein by excitation performed with a low-intensity (∼ 8 W/cm2) 488-nm-wavelength laser (OBIS; Coherent) and transmission of the emission fluorescence using a 520/35-nm band-pass filter (Chroma). We acquired 100 frames for each region of interest (ROI) at a frame rate of 20 Hz. Subsequently, we applied SRRF to the image stack through an open source, graphics processing unit (GPU)-enabled ImageJ plugin (31). SRRF requires the user to define a ring radius for the underlying radiality analysis. A related technique (superresolution quantitative image rating and reporting of error locations [SQUIRREL]), which is also available as an ImageJ plugin, can be used to evaluate the quality of SRRF images (57). Critical to achieving accurate images is the appropriate choice of the ring radius (31). Therefore, we generated SRRF images from our raw image stack for various values of the ring radius, evaluated the resulting images with SQUIRREL, and chose the input value for the ring radius such that the level of error in the SRRF image was minimized (Fig. S2).
Nucleoid imaging.
Fixed cells were stained with DAPI (3′,6-diamidino-2-phynylindole), which enabled visualization of cellular DNA. We added this stain to fixed cultures to reach a final concentration of 0.1 μg/ml and incubated the cultures at room temperature for 5 min. Samples were then centrifuged at 5,000 × g at room temperature for 1 min, the supernatant was discarded, and the pellets were washed by three subsequent resuspensions and centrifugations in 1 ml of fresh PBS. To image the DAPI-stained nucleoid of the bacterial cells, we employed an arc lamp (X-cite Series 120 Q), with a 325/50 excitation band-pass filter and a 447/60 emission band-pass filter inside the filter cube. For each image, a stack of 100 frames was acquired at a frame rate of 20 Hz.
Image analysis and analytics.
To quantify the spatial distribution of H-NS, cells were first manually segregated in ImageJ and then analyzed with custom MATLAB software. Cells that appeared to be dividing or that were larger than roughly 3 μm were removed from the statistical analysis. The principal axes of each cell were identified, and the cells were then rotated through a linear transformation so that their axes aligned. Cross sections were acquired by normalizing the axes of the cell from 0 to 1 and plotting the intensity of a single pixel along the major principal axes.
Chromosome compaction was then quantified from fluorescence images by measuring the projected area occupied by the chromosome within the cell. Closed regions outlining the chromosome were automatically detected within ImageJ and the resulting areas calculated. Note that some cells displayed two distinct globular regions that we took to represent two copies of the chromosome. For those cells, the area of each region was determined separately and then the measurements were added together. Finally, cell boundaries were acquired from bright-field images of the cells using ImageJ and used to normalize the area of the DNA containing nucleoid regions.
Chromatin immunoprecipitation (ChIP).
Chromatin immunoprecipitation assays were carried out as described previously (58). Briefly, E. coli BW25113 Δhns was complemented with pWN426 (pHNSHA), which produces a functional variant of H-NS that carries a C-terminal hemagglutinin (HA) epitope tag that enables the protein to be immunoprecipitated with anti-HA antibodies (5). This strain was grown in M9 medium supplemented with MgSO4 (2 mM), CaCl2 (0.1 mM), thiamine (0.01%), Casamino Acids (0.1%), glucose (1%), and chloramphenicol (10 μg/ml). Overnight cultures were subcultured 1:50 in fresh M9 medium and grown to the exponential (OD600 of ∼0.3) and stationary (OD600 of ∼2) phases (matching the imaging conditions). Cells were then exposed to osmotic stress by adding KCl (300 mM) to the culture for 30 min. Coumermycin (final concentration, 5 μg/ml) was also added to a set of samples 30 min prior to imposition of the osmotic stress.
To cross-link protein to DNAs from the different samples (50 ml each), formaldehyde was added to reach a final concentration of 1% for 15 min at room temperature before quenching with 1.25 mM glycine for 10 min was performed. Cells were washed twice with cold PBS and sonicated to generate chromosomal fragments with an average size of ∼500 bp. Lysates were cleared by centrifugation and precipitated with anti-HA antibody (clone HA-7; Bioshop) using agarose-protein G beads (Calbiochem) as previously described (58). The samples were incubated at 65°C for 5 h to break the DNA-protein cross-links.
DNA fragments that coprecipitated with H-NSHA were quantified by real-time quantitative PCR using SsoFast EvaGreen Supermix (Bio-Rad) according to the manufacturer’s instructions with the following gene-specific primers: for bglF, 5′-ACACTGTTAACCGCCAGGAAGACA-3′ (forward) and 5′-GGATGAAAGCAAAGCGCAAGCAGA-3′ (reverse); for yjcF, 5′-AGTTCCGTGCAGGAAGAGAACCTT-3′ (forward) and 5′-TGGTTACGTCGCTTTCGGCTTACT-3′ (reverse); for proV, 5′-AATATTTGGCGAGCATCCACAGCG-3′ (forward) and 5′-TTTACCCGAGCCGGATAATCCCAT-3′ (reverse); and for xapR, 5′-GCAATCGACGCGATTCTTCCATCAAG-3′ (forward) and 5′-GCAGCGCGTTTAAATATGTCTCAGCC-3′ (reverse).
Reverse transcriptase quantitative PCR (qPCR).
Total RNA was isolated from relevant bacterial strains 30 min after treatment with KCl or coumermycin as described for the chromatin immunoprecipitation experiment. A 0.5-ml volume of culture was combined with 1 ml of RNAprotect reagent (Qiagen). The samples were incubated at room temperature for 30 min, and the cells were harvested by centrifugation at 4,600 × g for 10 min. Total RNA was purified using an Aurum total RNA minikit (Bio-Rad) followed by reverse transcription using an iScript cDNA synthesis kit (Bio-Rad). The resulting cDNA was analyzed by real-time quantitative PCR with gene-specific primers identical to those used in the chromatin immunoprecipitation experiment and with SsoFast Evagreen Supermix (Bio-Rad) according to the manufacturer’s instructions. Transcript levels were normalized to the gyrB gene, amplified with primers 5′-CACTTTCACGGAAACGACCGCAAT-3′ (forward) and 5′-TTACCAACAACATTCCGCAGCGTG-3′ (reverse).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Natural Sciences and Engineering Research Council of Canada (J.N.M. and N.R.) (RG 418251), by an Early Researcher Award from the Ontario Ministry of Research, Innovation and Science (J.N.M. and N.R.), and by the Canadian Institutes of Health Research (W.W.N.) (MOP-86683).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00469-19.
REFERENCES
- 1.Singh K, Milstein JN, Navarre WW. 2016. Xenogeneic silencing and its impact on bacterial genomes. Annu Rev Microbiol 70:199–213. doi: 10.1146/annurev-micro-102215-095301. [DOI] [PubMed] [Google Scholar]
- 2.Dorman CJ. 2007. H-NS, the genome sentinel. Nat Rev Microbiol 5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 3.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog 2:746–752. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer J-P, Danchin A, Bertin P. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol 40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 5.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 6.Kotlajich MV, Hron DR, Boudreau BA, Sun Z, Lyubchenko YL, Landick R. 2015. Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria. Elife 4:e0497. doi: 10.7554/eLife.04970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landick R, Wade JT, Grainger DC. 2015. H-NS and RNA polymerase: a love-hate relationship? Curr Opin Microbiol 24:53–59. doi: 10.1016/j.mib.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Navarre WW. 2016. The impact of gene silencing on horizontal gene transfer and bacterial evolution. Adv Microb Physiol 69:157–186. doi: 10.1016/bs.ampbs.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Boudreau BA, Hron DR, Qin L, van der Valk RA, Kotlajich MV, Dame RT, Landick R. 2018. StpA and Hha stimulate pausing by RNA polymerase by promoting DNA-DNA bridging of H-NS filaments. Nucleic Acids Res 46:5525–5546. doi: 10.1093/nar/gky265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen BA, Landick R. 31 May 2019, posting date Transcription of bacterial chromatin. J Mol Biol doi: 10.1016/j.jmb.2019.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy CD, Cozzarelli NR. 2005. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol 57:1636–1652. doi: 10.1111/j.1365-2958.2005.04799.x. [DOI] [PubMed] [Google Scholar]
- 12.Noom MC, Navarre WW, Oshima T, Wuite GJ, Dame RT. 2007. H-NS promotes looped domain formation in the bacterial chromosome. Curr Biol 17:R913–R914. doi: 10.1016/j.cub.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Leng F, Chen B, Dunlap DD. 2011. Dividing a supercoiled DNA molecule into two independent topological domains. Proc Natl Acad Sci U S A 108:19973–19978. doi: 10.1073/pnas.1109854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 15.Owen-Hughes TA, Pavitt GD, Santos DS, Sidebotham JM, Hulton CSJ, Hinton JCD, Higgins CF. 1992. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 71:255–265. doi: 10.1016/0092-8674(92)90354-F. [DOI] [PubMed] [Google Scholar]
- 16.Higgins CF, Dorman CJ, Stirling DA, Waddell L, Booth IR, May G, Bremer E. 1988. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 17.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. 2007. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat Struct Mol Biol 14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Gottesman S. 2006. Modes of regulation of RpoS by H-NS. J Bacteriol 188:7022–7025. doi: 10.1128/JB.00687-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battesti A, Tsegaye YM, Packer DG, Majdalani N, Gottesman S. 2012. H-NS regulation of IraD and IraM antiadaptors for control of RpoS degradation. J Bacteriol 194:2470–2478. doi: 10.1128/JB.00132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorman CJ, Dorman MJ. 2016. DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression. Biophys Rev 8:89–100. doi: 10.1007/s12551-016-0238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinton JC, Santos DS, Seirafi A, Hulton CS, Pavitt GD, Higgins CF. 1992. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol Microbiol 6:2327–2337. doi: 10.1111/j.1365-2958.1992.tb01408.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Li G-W, Chen C, Xie XS, Zhuang X. 2011. Chromosome organization by a nucleoid-associated protein in live bacteria. Science 333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. 2014. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc Natl Acad Sci U S A 111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arold ST, Leonard PG, Parkinson GN, Ladbury JE. 2010. H-NS forms a superhelical protein scaffold for DNA condensation. Proc Natl Acad Sci U S A 107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vreede J, Dame RT. 2012. Predicting the effect of ions on the conformation of the H-NS dimerization domain. Biophys J 103:89–98. doi: 10.1016/j.bpj.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rimsky S. 2004. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr Opin Microbiol 7:109–114. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Zhang M, Chang H, Zhang Y, Yu J, Wu L, Ji W, Chen J, Liu B, Lu J, Liu Y, Zhang J, Xu P, Xu T. 2012. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods 9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 28.Drlica K, Snyder M. 1978. Superhelical Escherichia coli DNA: relaxation by coumermycin. J Mol Biol 120:145–154. doi: 10.1016/0022-2836(78)90061-x. [DOI] [PubMed] [Google Scholar]
- 29.Hardy CD, Cozzarelli NR. 2003. Alteration of Escherichia coli topoisomerase IV to novobiocin resistance. Antimicrob Agents Chemother 47:941–947. doi: 10.1128/aac.47.3.941-947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gellert M, O'Dea MH, Itoh T, Tomizawa J. 1976. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A 73:4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gustafsson N, Culley S, Ashdown G, Owen DM, Pereira PM, Henriques R. 2016. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat Commun 7:12471. doi: 10.1038/ncomms12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dertinger T, Colyer R, Iyer G, Weiss S, Enderlein J. 2009. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI). Proc Natl Acad Sci U S A 106:22287–22292. doi: 10.1073/pnas.0907866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dersch P, Schmidt K, Bremer E. 1993. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol Microbiol 8:875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 34.Cagliero C, Jin DJ. 2013. Dissociation and re-association of RNA polymerase with DNA during osmotic stress response in Escherichia coli. Nucleic Acids Res 41:315–326. doi: 10.1093/nar/gks988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajkumari K, Kusano S, Ishihama A, Mizuno T, Gowrishankar J. 1996. Effects of H-NS and potassium glutamate on σ(S)- and σ70-directed transcription in vitro from osmotically regulated P1 and P2 promoters of proU in Escherichia coli. J Bacteriol 178:4176–4181. doi: 10.1128/jb.178.14.4176-4181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sleator RD, Hill C. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 37.Madan R, Kolter R, Mahadevan S. 2005. Mutations that activate the silent bgl operon of Escherichia coli confer a growth advantage in stationary phase. J Bacteriol 187:7912–7917. doi: 10.1128/JB.187.23.7912-7917.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madan R, Moorthy S, Mahadevan S. 2008. Enhanced expression of the bgl operon of Escherichia coli in the stationary phase. FEMS Microbiol Lett 288:131–139. doi: 10.1111/j.1574-6968.2008.01346.x. [DOI] [PubMed] [Google Scholar]
- 39.Ali SS, Soo J, Rao C, Leung AS, Ngai D-M, Ensminger AW, Navarre WW. 2014. Silencing by H-NS potentiated the evolution of Salmonella. PLoS Pathog 10:e1004500. doi: 10.1371/journal.ppat.1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lal A, Dhar A, Trostel A, Kouzine F, Seshasayee ASN, Adhya S. 2016. Genome scale patterns of supercoiling in a bacterial chromosome. Nat Commun 7:11055. doi: 10.1038/ncomms11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorman C. 1996. Flexible response: DNA supercoiling, transcription and bacterial adaptation to environmental stress. Trends Microbiol 4:214–216. doi: 10.1016/0966-842X(96)30015-2. [DOI] [PubMed] [Google Scholar]
- 43.Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. 2004. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol 5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouafa ZA, Reverchon S, Lautier T, Muskhelishvili G, Nasser W. 2012. The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii. Nucleic Acids Res 40:4306–4319. doi: 10.1093/nar/gks014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rochman M, Blot N, Dyachenko M, Glaser G, Travers A, Muskhelishvili G. 2004. Buffering of stable RNA promoter activity against DNA relaxation requires a far upstream sequence. Mol Microbiol 53:143–152. doi: 10.1111/j.1365-2958.2004.04126.x. [DOI] [PubMed] [Google Scholar]
- 46.Stuger R, Woldringh CL, van der Weijden CC, Vischer NOE, Bakker BM, van Spanning RJM, Snoep JL, Westerhoff HV. 2002. DNA supercoiling by gyrase is linked to nucleoid compaction. Mol Biol Rep 29:79–82. doi: 10.1023/A:1020318705894. [DOI] [PubMed] [Google Scholar]
- 47.Cheung KJ, Badarinarayana V, Selinger DW, Janse D, Church GM. 2003. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res 13:206–215. doi: 10.1101/gr.401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McClellan JA, Boublíková P, Palecek E, Lilley DM. 1990. Superhelical torsion in cellular DNA responds directly to environmental and genetic factors. Proc Natl Acad Sci U S A 87:8373–8377. doi: 10.1073/pnas.87.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gottesman S. 2019. Trouble is coming: signaling pathways that regulate general stress responses in bacteria. J Biol Chem 294:11685–11700. doi: 10.1074/jbc.REV119.005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronis R. 1995. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of sigma S and many sigma S-dependent genes in Escherichia coli. J Bacteriol 177:3455–3464. doi: 10.1128/jb.177.12.3455-3464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamashino T, Ueguchi C, Mizuno T. 1995. Quantitative control of the stationary phase-specific sigma factor, sigma S, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J 14:594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urban JH, Vogel J. 2007. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res 35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomason LC, Costantino N, Court DL. 1 May 2014, posting date E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol doi: 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- 56.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. 1 October 2010, posting date Computer control of microscopes using μManager. Curr Protoc Mol Biol doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Culley S, Albrecht D, Jacobs C, Pereira PM, Leterrier C, Mercer J, Henriques R. 2018. Quantitative mapping and minimization of super-resolution optical imaging artifacts. Nat Methods 15:263–266. doi: 10.1038/nmeth.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ali SS, Whitney JC, Stevenson J, Robinson H, Howell PL, Navarre WW. 2013. Structural insights into the regulation of foreign genes in Salmonella by the Hha/H-NS complex. J Biol Chem 288:13356–13369. doi: 10.1074/jbc.M113.455378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.