The intracellular pathogen Coxiella burnetii employs a type 4B secretion system (T4BSS) that promotes growth by translocating effectors of eukaryotic pathways into host cells. T4BSS regulation modeled in Legionella pneumophila indicates IcmS facilitates effector translocation. Here, we characterized type 4B secretion by a Coxiella ΔicmS mutant that exhibits intracellular growth defects. T4BSS substrates demonstrated increased, equivalent, or decreased secretion by the ΔicmS mutant relative to wild-type Coxiella. Similar to the Legionella T4BSS, IcmS dependency in Coxiella was determined by C-terminal and/or internal secretion signals. However, IcmS inhibited secretion of some effectors by Coxiella that were previously shown to be translocated by Legionella. Thus, Coxiella has a unique IcmS regulatory mechanism that both positively and negatively regulates T4BSS export.
KEYWORDS: Coxiella, Dot/Icm, IcmS, Q fever, chaperone, effector proteins, regulation, type 4B secretion
ABSTRACT
Macrophage parasitism by Coxiella burnetii, the cause of human Q fever, requires the translocation of proteins with effector functions directly into the host cell cytosol via a Dot/Icm type 4B secretion system (T4BSS). Secretion by the analogous Legionella pneumophila T4BSS involves signal sequences within the C-terminal and internal domains of effector proteins. The cytoplasmic chaperone pair IcmSW promotes secretion and binds internal sites distinct from signal sequences. In the present study, we investigated requirements of C. burnetii IcmS for host cell parasitism and effector translocation. A C. burnetii icmS deletion mutant (ΔicmS) exhibited impaired replication in Vero epithelial cells, deficient formation of the Coxiella-containing vacuole, and aberrant T4BSS secretion. Three secretion phenotypes were identified from a screen of 50 Dot/Icm substrates: IcmS dependent (secreted by only wild-type bacteria), IcmS independent (secreted by both wild-type and ΔicmS bacteria), or IcmS inhibited (secreted by only ΔicmS bacteria). Secretion was assessed for N-terminal or C-terminal truncated forms of CBU0794 and CBU1525. IcmS-inhibited secretion of CBU1525 required a C-terminal secretion signal whereas IcmS-dependent secretion of CBU0794 was directed by C-terminal and internal signals. Interchange of the C-terminal 50 amino acids of CBU0794 and CBU1525 revealed that sites within the C terminus regulate IcmS dependency. Glutathione S-transferase-tagged IcmSW bound internal sequences of IcmS-dependent and -inhibited substrates. Thus, the growth defect of the C. burnetii ΔicmS strain is associated with a loss of T4BSS chaperone activity that both positively and negatively regulates effector translocation.
IMPORTANCE The intracellular pathogen Coxiella burnetii employs a type 4B secretion system (T4BSS) that promotes growth by translocating effectors of eukaryotic pathways into host cells. T4BSS regulation modeled in Legionella pneumophila indicates IcmS facilitates effector translocation. Here, we characterized type 4B secretion by a Coxiella ΔicmS mutant that exhibits intracellular growth defects. T4BSS substrates demonstrated increased, equivalent, or decreased secretion by the ΔicmS mutant relative to wild-type Coxiella. Similar to the Legionella T4BSS, IcmS dependency in Coxiella was determined by C-terminal and/or internal secretion signals. However, IcmS inhibited secretion of some effectors by Coxiella that were previously shown to be translocated by Legionella. Thus, Coxiella has a unique IcmS regulatory mechanism that both positively and negatively regulates T4BSS export.
INTRODUCTION
Central to disease pathogenesis by the human Q fever bacterium Coxiella burnetii are invasion and replication within mononuclear phagocytes, such as alveolar macrophages (1, 2). C. burnetii has a unique intracellular lifestyle that involves acid activation of metabolism within a phagolysosome-like compartment called the Coxiella-containing vacuole (CCV). An important virulence determinant of C. burnetii is the ability to resist degradation in the CCV. Another critical factor for pathogen growth is the manipulation of host cell functions by proteins with effector functions translocated directly into the host cell cytoplasm by the C. burnetii Dot/Icm type 4B secretion system (T4BSS) (3–9). Genetic tools have aided functional analysis of C. burnetii Dot/Icm substrates with ∼140 identified to date (10). Characterized C. burnetii effectors are known to modulate apoptotic and pyroptotic cell death pathways (11–14), mitogen-activated protein kinase signaling (15), endosomal trafficking through manipulation of clathrin machinery (16, 17), and autophagy/phosphoinositide metabolism (18, 19).
Although progress has been made in characterizing C. burnetii effectors, considerable gaps in knowledge remain regarding regulation of T4BSS export. For example, how is effector translocation controlled to temporally orchestrate events specific to different stages of Coxiella’s lengthy infectious cycle? A general physiological requirement for effector delivery is metabolic activation of C. burnetii, a process that only occurs after CCV acidification (20, 21). Consequently, translocation of effector-reporter chimeras is not detected until 4 to 8 h postinfection when the nascent CCV has matured to an endolysosomal state (22). Early inhibition of transcription impedes effector translocation, but it is unclear whether this effect is directly related to Dot/Icm function or due to a general decline in metabolic fitness (22). Nonetheless, these data indicate the Dot/Icm T4BSS is not required for early infection events, such as internalization or CCV acidification (4).
A C-terminal secretion signal of ∼30 amino acids controls the export of some proteins by the Dot/Icm system (23–31). C-terminal secretion signals exhibit an E block of negatively charged amino acids located between positions −20 to −10 that is flanked by a regular distribution of serine/threonine residues and amino acids with bulky hydrophobic side chains (26, 27). Efficient export of L. pneumophila Dot/Icm substrates can also require the chaperone complex IcmSW, which binds to substrates on sites exclusive from signal sequences (28, 32–37). IcmSW, along with DotL, DotM, and DotN, forms a T4BSS coupling protein complex (T4CP) on the cytoplasmic surface of the inner membrane that promotes export of T4BSS effectors by an unknown mechanism (32, 37–41).
L. pneumophila icmS and/or icmW mutants exhibit moderate growth defects in human macrophage-like cells (36). Similarly, C. burnetii icmS transposon mutants have a moderate growth defect, whereas mutagenesis of icmW results in moderate to severe defects (7, 8, 42). Functional redundancy within the effector pool and IcmSW-independent translocation of some substrates likely mitigate impaired T4BSS function (24, 28, 43). C. burnetii icmS and icmW complement L. pneumophila mutants with corresponding gene deletions (44, 45). These results suggest that IcmSW functions are similar between C. burnetii and L. pneumophila. However, there are discrepancies in substrate translocation between the two pathogens (46, 47), and T4BSS activity and IcmS function have not been directly investigated in C. burnetii. Here, characterization of a C. burnetii ΔicmS mutant reveals that intracellular growth defects correspond with both positive and negative regulatory roles for IcmS in T4BSS export. These data suggest that IcmS regulation of T4BSS activity in C. burnetii is distinct from that of L. pneumophila.
RESULTS
C. burnetii ΔicmS mutant has an intracellular growth defect.
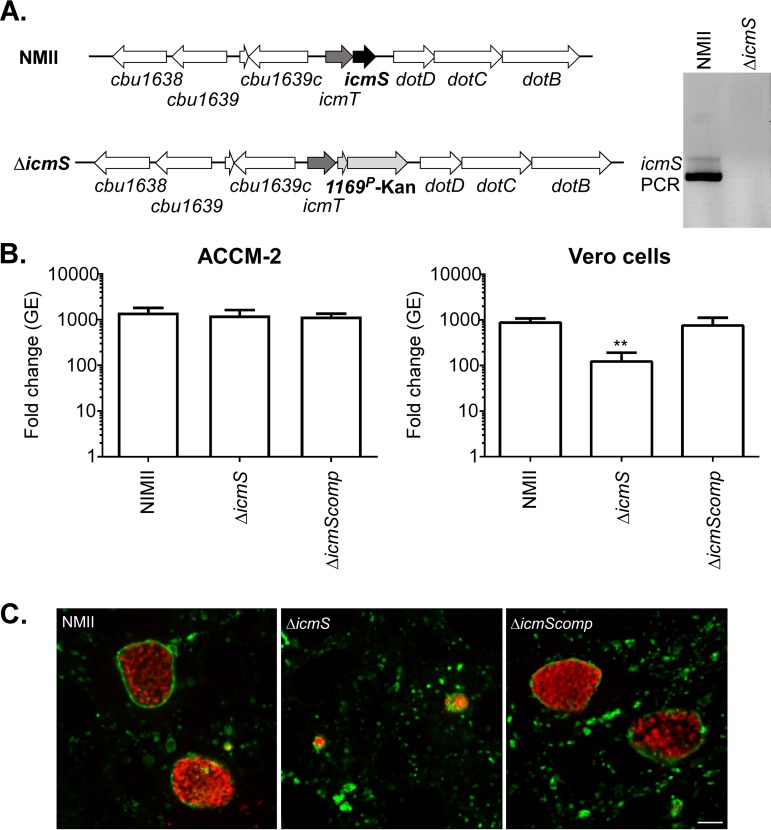
C. burnetii icmS is downstream of icmT in a predicted two-gene operon within the ∼23-kbp dot/icm locus (46) (Fig. 1A). To evaluate the role of icmS in C. burnetii effector translocation and host cell colonization, we constructed an icmS deletion mutant by allelic exchange (5). Deletion of icmS was confirmed by PCR (Fig. 1A). Growth of wild-type C. burnetii, the ΔicmS mutant, and the ΔicmS mutant complemented with a single copy of icmS under the control of its native promoter was measured in the synthetic medium ACCM-2 or Vero cells. Six-day genome equivalent (GE) increases for the three strains were indistinguishable in ACCM-2. However, in Vero cells, the ΔicmS mutant had a statistically significant (P < 0.001) growth defect, with 10-fold less GE than wild-type C. burnetii or the complemented ΔicmS mutant (Fig. 1B). Deficient replication by the ΔicmS mutant correlated with defective CCV development. By immunofluorescence, mutant bacteria occupied aberrantly small CD63-positive vacuoles, whereas both wild-type bacteria and the complemented mutant resided in single large and spacious CD63+ vacuoles (Fig. 1C).
FIG 1.
C. burnetii ΔicmS has a moderate intracellular growth defect. (A) Schematic depicting the arrangement of icmS and adjacent genes in wild-type C. burnetii (NMII) and the ΔicmS mutant. Replacement of icmS with a Kanr cassette by allelic exchange resulted in the ΔicmS mutant. Deletion of icmS was confirmed by PCR using oligonucleotides specific to icmS. (B) Replication of wild-type C. burnetii, the ΔicmS mutant, and the complemented ΔicmS mutant (ΔicmScomp) grown in ACCM-2 or Vero cells. Histograms show fold change in bacterial genome equivalents (GE) at 6 days postinfection relative to the day of infection. ACCM-2 results are expressed as the means from three independent experiments. Vero cell results are expressed as the means of two biological replicates from three independent experiments. Error bars indicate the standard deviations from the means, and asterisks indicate a statistically significant difference (P < 0.001) from cells infected with wild-type C. burnetii. (C) Fluorescence micrographs of Vero cells infected for 4 days with wild-type C. burnetii, ΔicmS, or ΔicmScomp strains. Cells were immunostained for C. burnetii (red) and the lysosomal marker CD63 (green). Scale bar, 10 μm.
C. burnetii Dot/Icm substrates display three profiles of IcmS dependency.
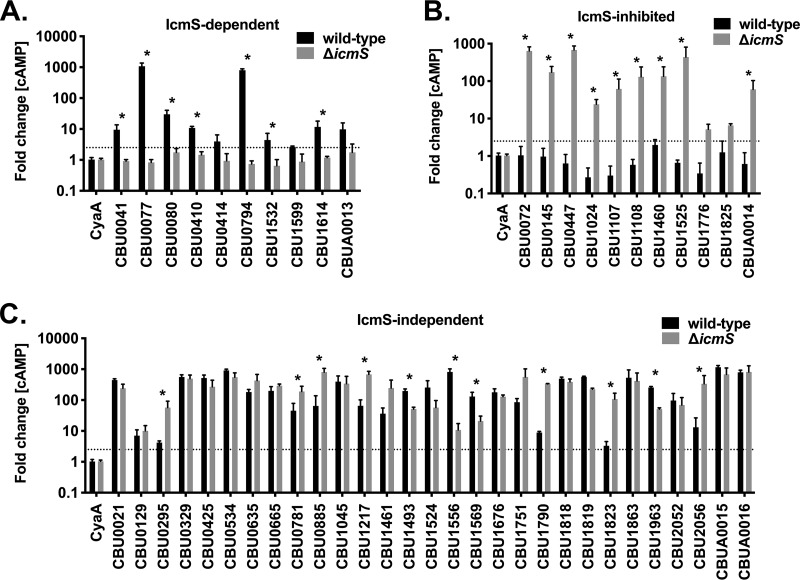
IcmS control over T4BSS effector translocation was assessed for 50 C. burnetii proteins previously reported as Dot/Icm substrates (Table 1). Due in part to past limitations of C. burnetii genetics, 44 of the Dot/Icm substrates selected were originally identified using L. pneumophila to produce candidate effectors fused to the reporter tags CyaA or BlaM. Fourteen substrates were subsequently validated in C. burnetii (3, 6, 8, 15, 16, 28, 30, 31, 46–49). Substrates with defined host cell effector functions included CBU0021 (CvpB, Cig2), CBU0041 (CirA), CBU0072 (AnkA), CBU0077 (MceA) CBU0665 (CvpA), CBU0885, CBU1524 (CaeA), CBU1532 (CaeB), CBU1679 (Cem9), CBU1751 (Cig57), and CBU1823 (IcaA) (3, 6, 16, 28, 31, 46–51). THP-1 macrophages were infected for 48 h with wild-type C. burnetii or the ΔicmS mutant producing effectors fused at their N termini with CyaA, a reporter tag that, when delivered to the host cell cytoplasm, catalyzes the production of cyclic AMP (cAMP). Tested substrates were deemed secreted by C. burnetii if they produced ≥2.5-fold more cAMP versus the negative control (CyaA only) (4, 16, 46, 47). Secretion results from wild-type and ΔicmS bacteria were separated into three groups (Fig. 2). Ten substrates secreted by only wild-type C. burnetii were classified as IcmS dependent (Fig. 2A), 29 substrates secreted by both the wild type and the ΔicmS mutant were classified as IcmS independent (Fig. 2C), and 11 substrates secreted by only the ΔicmS mutant were classified as IcmS inhibited (Fig. 2B). Within the IcmS-independent group the cAMP levels were significantly greater for four and seven substrates secreted by wild-type bacteria and the ΔicmS mutant, respectively. Here and elsewhere in this study, immunoblots demonstrated that differences in cAMP production between strains were not due to inconsistent expression of CyaA fusion proteins (see Fig. S1 and S2 in the supplemental material). Translocation assays were also conducted with Vero cells infected for 48 h with C. burnetii expressing CBU0794 or CBU1525 fused to the β-lactamase (BlaM) fluorescent reporter tag. Measurement of BlaM translocation within individual cells by fluorescence confocal imaging coupled with computational image analysis confirmed the IcmS-dependent and IcmS-inhibited secretion phenotypes of CBU0794 and CBU1525, respectively (Fig. S3). Together, CyaA and BlaM translocation assays indicate that expression of IcmS by wild-type C. burnetii promotes the secretion of CBU0794 but inhibits the export of CBU1525, a substrate that was previously shown to be translocated by the wild-type L. pneumophila T4BSS (3, 30, 48, 49).
TABLE 1.
Dot/Icm substrates tested in this studya
| ORF | Name(s) | Size (kDa) | Translocation results (reference[s])b |
|---|---|---|---|
| CBU0021 | CvpB, Cig2 | 93.1 | Lp CyaA (28), Cb CyaA (47) |
| CBU0041 | CirA | 82.7 | Lp BlaM [80] (48) |
| CBU0072 | AnkA | 45.5 | Lp CyaA (WT < IcmS) (31), Lp CyaA (49) |
| CBU0077 | MceA | 29.6 | Lp CyaA, Cb BlaM [29] (3), Cb BlaM 38% (8) |
| CBU0080 | 18.7 | Lp CyaA (3), Cb BlaM [1] (6) | |
| CBU0129 | 11.1 | Lp BlaM 70% (48) | |
| CBU0145 | AnkB | 38.5 | Lp CyaA (WT > IcmS) (31), Lp CyaA (49) |
| CBU0295 | 56.8 | Lp CyaA (3), Lp BlaM [2] (6) | |
| CBU0329 | 23.3 | Lp CyaA (3) | |
| CBU0410 | 63.3 | Lp BlaM [45] (48) | |
| CBU0414 | 30.4 | Lp BlaM [45] (48) | |
| CBU0425 | CirB | 51.9 | Lp CyaA (3) |
| CBU0447 | AnkF | 21.3 | Lp CyaA (WT = IcmS) (31), Lp CyaA (49) |
| CBU0534 | 46.1 | Cb CyaA (47) | |
| CBU0635 | 55.3 | Lp CyaA, Cb BlaM [7.7] (3) | |
| CBU0665 | CvpA | 38.0 | Cb CyaA (47) |
| CBU0781 | AnkG | 38.6 | Lp CyaA (WT = IcmS) (31), Lp CyaA (49) |
| CBU0794 | 53.1 | Lp BlaM [80] (48) | |
| CBU0885 | 44.9 | Lp CyaA (28), Lp BlaM (6), Cb CyaA (47) | |
| CBU1024 | AnkH | 88.1 | Lp CyaA (WT > IcmS) (31), Lp CyaA (49) |
| CBU1045 | 40.0 | Lp BlaM [40] (48) | |
| CBU1107 | 35.5 | Lp CyaA (3) | |
| CBU1108 | 21.8 | Lp CyaA (3) | |
| CBU1217 | 57.8 | Lp BlaM [10] (48), Cb CyaA (47) | |
| CBU1460 | 29.6 | Lp BlaM [40] (48) | |
| CBU1461 | 67.1 | Lp BlaM [2] (48) | |
| CBU1493 | 64.2 | Cb CyaA (47) | |
| CBU1524 | CaeA | 25.1 | Lp CyaA (3), Cb BlaM [10], Cb BlaM [38] (8) |
| CBU1525 | 43.9 | Lp CyaA (3) | |
| CBU1532 | CaeB | 16.5 | Lp CyaA (3) |
| CBU1556 | CvpC | 64.6 | Lp BlaM [50] (48), Cb CyaA (47) |
| CBU1569 | 63.1 | Lp BlaM [90] (48) | |
| CBU1599 | 12.5 | Lp BlaM [10] (48) | |
| CBU1614 | 16.2 | Cb CyaA (46) | |
| CBU1676 | 41.8 | Lp CyaA (15), Cb CyaA (47) | |
| CBU1751 | Cig57 | 48.8 | Lp BlaM [60] (48) |
| CBU1776 | 24.0 | Lp CyaA (3) | |
| CBU1790 | 73.8 | Cb CyaA (46) | |
| CBU1818 | CvpD | 53.9 | Lp CyaA (15), Cb CyaA (47) |
| CBU1819 | 42.3 | Cb CyaA (47) | |
| CBU1823 | IcaA | 40.3 | Lp CyaA, Cb BlaM [10] (3), Cb BlaM [13] (8) |
| CBU1825 | 13.3 | Lp CyaA (3) | |
| CBU1863 | CvpE | 66.2 | Cb CyaA (47) |
| CBU1963 | 72.9 | Lp CyaA (3) | |
| CBU2052 | CirD | 34.0 | Lp CyaA (3) |
| CBU2056 | 25.4 | Lp CyaA (3) | |
| CBUA0013 | CpeB | 28.0 | Lp CyaA (30), Cb CyaA and BlaM (30) |
| CBUA0014 | CpeC | 8.7 | Lp CyaA (30), Cb CyaA and BlaM (30) |
| CBUA0015 | CpeD | 26.3 | Lp CyaA (30), Cb CyaA and BlaM (30) |
| CBUA0016 | CpeE | 39.5 | Lp CyaA (30), Cb CyaA and BlaM (30) |
C. burnetii Nine Mile (RSA 493) open reading frames (ORFs) of proteins tested for secretion by the C. burnetii Dot/Icm T4BSS are indicated, along with names and predicted molecular weights.
Translocation results with beta-lactamase (BlaM) and adenylate cyclase (CyaA) reporter assays in L. pneumophila (Lp) or C. burnetii (Cb) are indicated for the referenced studies. Where reported, the percentage of BlaM-positive cells is indicated in brackets, as well as the relative secretion efficiency in wild-type L. pneumophila (WT) versus the icmS mutant (IcmS).
FIG 2.
C. burnetii Dot/Icm substrates display three profiles of IcmS dependency. THP-l cells were infected for 48 h with wild-type C. burnetii or the icmS mutant (ΔicmS) producing the indicated Dot/Icm substrates fused to CyaA. Histograms depict the fold change in cytosolic [cAMP] relative to cells infected with C. burnetii producing CyaA alone (CyaA). (A) Ten substrates were secreted only by wild-type C. burnetii (IcmS dependent); (B) 29 substrates were secreted by both wild-type C. burnetii and the ΔicmS mutant (IcmS independent); (C) 11 substrates were secreted only by the ΔicmS mutant (IcmS inhibited). The cutoff for positive secretion is indicated by a dotted line at 2.5-fold change [cAMP]. Error bars indicate the standard deviations from the means for two independent experiments conducted in duplicate. Asterisks indicate substrates with significantly different translocation efficiencies by wild-type versus ΔicmS bacteria (P < 0.01, Student t test).
IcmS dependency is consistent throughout infection.
To examine whether C. burnetii IcmS dependency changes from early to late infection, cAMP assays were conducted at 8, 24, 48, 72, and 120 h postinfection with substrates that were IcmS dependent (CBU0794 and CBUA0013), IcmS independent (CBUA0016 and CBU1863), or IcmS inhibited (CBU0072 and CBU1525) (Fig. S4). Translocation signals at 8 h postinfection were consistently lower than signals at the later time points, a result that likely reflects C. burnetii in the early stages of metabolic activation (22). Interestingly, the greatest fold changes in cAMP concentration ([cAMP]) occurred at 24 h postinfection, which is earlier than the maximal growth of C. burnetii observed at 48 to 72 h postinfection. Overall, the translocation phenotypes of IcmS-dependent, -independent, and -inhibited substrates remained consistent from 24 to 120 h postinfection as C. burnetii is transitioning from the exponential growth to the early stationary phase (4). This behavior suggested that IcmS control of T4BSS translocation is not temporally regulated for substrates constitutively expressed during C. burnetii growth in cultured macrophages.
Differences in C-terminal amino acid composition distinguish IcmS-inhibited and IcmS-dependent substrates.
A subset of L. pneumophila effectors that harbor C-terminal sequences enriched in acidic residues (D and E) at E-block positions −20 to −10 exhibit less dependence on IcmS for export (28). To determine whether specific physiochemical properties in the C-terminal secretion signals of C. burnetii substrates correlated with IcmS-related secretion phenotypes, the probability of specific amino acids at positions −35 to −1 was calculated (Fig. 3 and Table S3). For this analysis, substrates were grouped according to statistically different or similar levels of secretion by C. burnetii wild type versus the ΔicmS mutant (P < 0.01). Based on these criteria, several substrates secreted by both C. burnetii wild-type and the ΔicmS mutant strains, but at different levels (Fig. 2B), were removed from the IcmS-independent group and included with the IcmS-dependent or IcmS-inhibited substrates. Overall, IcmS-inhibited substrates exhibited a typical E block of negatively charged amino acids within the −20 to −10 region of the C terminus. These sequences were flanked immediately upstream by amino acids with hydrophobic side chains and downstream by serine/threonine and hydrophobic residues. By comparison, E-block residues were not prominent in C-terminal 35 amino acids of IcmS-dependent substrates that instead exhibited greater enrichment of serine and proline residues. IcmS-independent secretion signals were a hybrid of IcmS-inhibited and -dependent signals, with both E-block residues and C-terminal prolines. C-terminal sequences of CBU0080 and CBU1024 were clear outliers among IcmS-dependent and -inhibited secretion signals, respectively, suggesting that factors other than C-terminal amino acid composition can also influence IcmS dependency (Table S3). However, overall trends indicated that, similar to L. pneumophila, C. burnetii effectors with strong E blocks exhibit less dependence on IcmS for export (28, 31, 32). Conversely, deficient export of substrates harboring strong E blocks by wild-type bacteria is unique to C. burnetii.
FIG 3.
Effectors translocated by the C. burnetii ΔicmS mutant exhibit C-terminal signal sequences enriched in acidic residues. The probability of a particular amino acid at each position in the 35 C-terminal residues of IcmS-dependent, IcmS-independent, and IcmS-inhibited substrates. Amino acids are colored according to their physiochemical and structural properties: negatively charged E and D (red); positively charged K and R (orange); bulky hydrophobic F, I, L, and V (blue); helix breaker P (green); small polar S and T (magenta); and other residues (black).
IcmS dependency is regulated by sites within the C-terminal 50 amino acids.
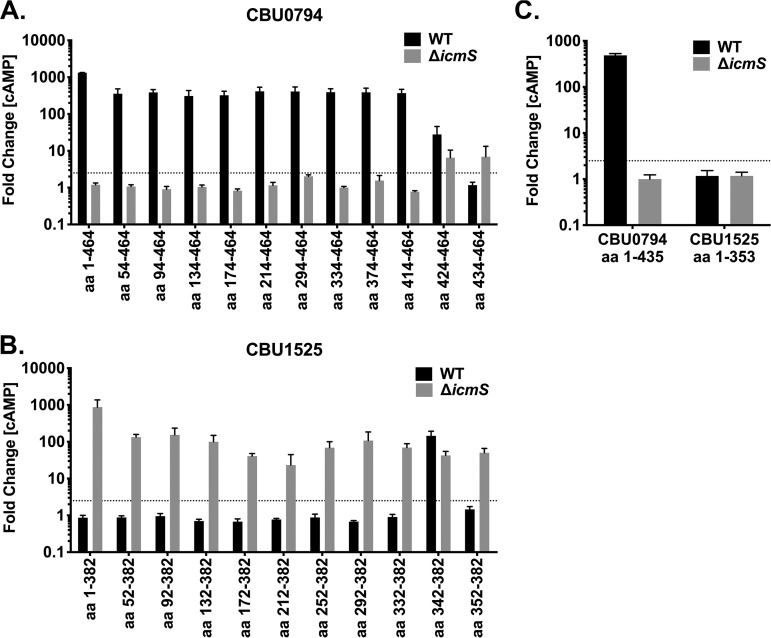
Direct binding of IcmS to L. pneumophila effectors has been demonstrated with the binding sequence mapped to sites internal to the C terminus of SidG (32, 34, 35, 37, 40). To map the region mediating C. burnetii IcmS-dependent and -inhibited translocation, export was tested for deletion derivatives of CBU0794 (IcmS dependent) and CBU1525 (IcmS inhibited) by wild-type C. burnetii or the ΔicmS mutant. CyaA fusions of these proteins were made with progressive N-terminal deletions. The smallest truncated forms of CBU0794 and CBU1525 contained amino acids 434 to 464 (CBU0794434-464) and 353 to 382 (CBU1525353-382), respectively. N-terminal deletion derivatives containing at least 50 C-terminal amino acids were secreted similar to full-length protein, but larger N-terminal truncations perturbed the IcmS dependency of substrate secretion (Fig. 4A and B). CBU0794424–464 and CBU1525343–382 were secreted by both wild-type C. burnetii and the ΔicmS mutant, whereas CBU0794434–464 and CBU1525353–382 were only secreted by the ΔicmS mutant. These data suggest that residues encompassed within the region from positions −50 to −40 control IcmS dependency of CBU0794 and CBU1525 secretion. Secretion of CBU0794434–464 and CBU1525353–382 by only the ΔicmS mutant indicated that both constructs harbor a secretion signal that is inhibited by IcmS. To further investigate how the C-terminal 30 amino acids contribute to IcmS dependency, translocation was assessed for CBU07941–435 and CBU15251–353 (Fig. 4C). Similar to the full-length protein, CBU07941–435 was secreted by wild-type bacteria but not by the icmS mutant, which, together with the N-terminal truncation data, suggests that IcmS-dependent export of CBU0794 by wild-type C. burnetii can be directed by a C-terminal and/or an internal secretion signal. In contrast, CBU15251–352 was not secreted by the ΔicmS mutant, thereby confirming that its C-terminal secretion signal is required for IcmS-inhibited export. These data suggest that CBU1525 has a C-terminal secretion signal whereas CBU0794 has at least two signal sequences, a C-terminal signal and an internal signal, similar to SidJ (37).
FIG 4.
IcmS dependency is regulated by sites within the C-terminal 50 amino acids. THP-l cells were infected for 48 h with wild-type C. burnetii or the icmS mutant producing CyaA fused to the indicated amino acids. Histograms depict the fold change in cytosolic [cAMP] for cells infected with C. burnetii producing N-terminal truncated forms of CBU0794 (IcmS dependent) (A) or CBU1525 (IcmS inhibited) (B) or C-terminal truncated forms of both proteins (C). The cutoff for positive secretion is indicated by a dotted line at 2.5-fold change [cAMP]. Error bars indicate the standard deviations from the means for two independent experiments conducted in duplicate.
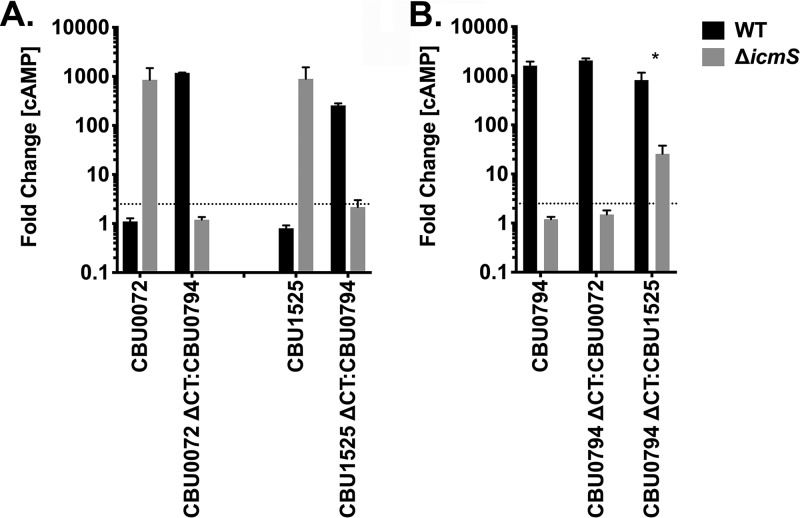
Exchange of C-terminal secretion signals changes IcmS dependency.
To further explore how Dot/Icm secretion signals affect IcmS regulation, CyaA translocation assays were conducted with C-terminal domain swap constructs. The C-terminal 50 amino acids from the IcmS-inhibited substrates CBU0072 and CBU1525 were removed and replaced with the C-terminal 50 amino acids from the IcmS-dependent substrate CBU0794. Conversely, two different CBU0794 constructs were generated fused to the C-terminal 50 amino acids of CBU0072 or CBU1525. THP-1 cells were infected for 48 h with C. burnetii producing the CyaA fusions of the native substrate or domain swap constructs, and cytosolic [cAMP] was measured (Fig. 5). The CBU0072 and CBU1525 chimeras were exported by wild-type C. burnetii, which was in opposition to the IcmS-inhibited export of the native substrates. This result indicated C-terminal secretion signals contribute to export of CBU0072 and CBU1525 by the ΔicmS mutant. The CBU0794 ΔCT:CBU0072 chimera exhibited IcmS-dependent secretion similar to native CBU0794. Interestingly, the CBU0794 ΔCT:CBU1525 chimera was secreted by both C. burnetii strains, but at significantly greater levels by wild-type bacteria. Thus, only a subset of secretion signals within IcmS-inhibited substrates are sufficient for protein translocation by the ΔicmS mutant. These results illustrate that the C-terminal secretion signal is one of several factors that can influence IcmS dependency.
FIG 5.
Exchange of C-terminal secretion signals alters IcmS dependency. Histograms depict the fold change in cytosolic [cAMP] for THP-l cells infected for 48 h with wild-type C. burnetii or the icmS mutant producing the indicated CyaA fusion proteins. (A) Secretion of CBU0072 and CBU1525 fused to the C-terminal 50 amino acids of CBU0794 (CBU0072 ΔCT:CBU0794 or CBU1525 ΔCT:CBU0794, respectively). (B) Secretion of CBU0794 fused to the C-terminal 50 amino acids of CBU0072 or CBU1525 (CBU0794 ΔCT:CBU0072 or CBU0794 ΔCT:CBU1525, respectively). The cutoff for positive secretion is indicated by a dotted line at 2.5-fold change [cAMP]. Error bars indicate the standard deviations from the means for two independent experiments conducted in duplicate. The asterisk indicates a statistically significant difference (P < 0.01).
IcmS-dependent and -inhibited effectors bind IcmSW.
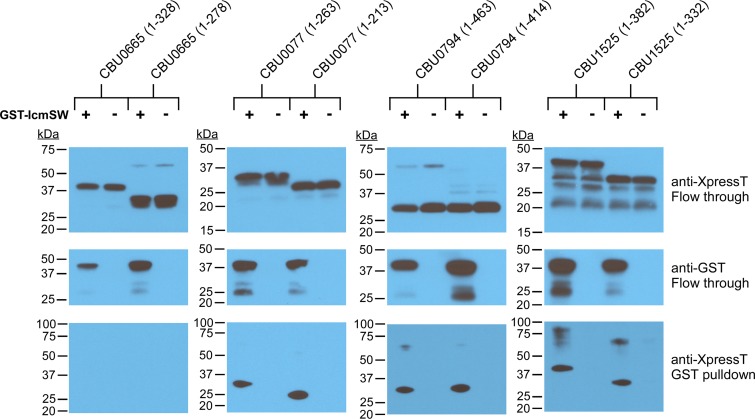
Affinity tag fusion to IcmS disrupts interactions with IcmW required for chaperone recognition of synthesized effectors (39). Therefore, pulldowns with IcmW N-terminally tagged with glutathione S-transferase (GST) were conducted to investigate effector engagement by the IcmS-containing chaperone complex. In vitro transcription and translation (IVTT) was used to simultaneously produce GST-IcmW, IcmS, and selected IcmS-dependent, -independent, or -inhibited substrates N-terminally tagged with XpressT (52). IVTT extracts were incubated with GST-affinity beads to collect proteins complexed with GST-IcmSW. Immunoblots of GST pulldown samples demonstrated IcmSW binding to CBU0794 (IcmS dependent) and CBU1525 (IcmS inhibited) but not CBU0665 (IcmS independent) (Fig. 6). For reasons that remain unclear, only an ∼30-kDa N-terminal fragment of CBU0794 was produced by IVTT. Therefore, we performed pulldowns with CBU0077, a second IcmS-dependent substrate that also bound GST-IcmSW. Deletion of substrate C-terminal 50 amino acids did not alter binding to GST-IcmSW, suggesting that both substrates have internal IcmS binding sites. Together, these data indicate that IcmSW recognition of internal sites within IcmS-dependent or -inhibited effectors can positively or negatively regulate T4BSS translocation.
FIG 6.
IcmSW binds IcmS-dependent and -inhibited substrates. Immunoblots of GST-tagged IcmW and XpressT-tagged CBU0077, CBU0794, CBU066, or CBU1525 with or without their C-terminal 50 amino acids. GST-IcmW, IcmS, and individual substrates were produced together by IVTT. After incubation of cell extracts with GST-affinity beads, unbound (Flow through) and bead-bound (GST pulldown) proteins were immunoblotted with antibodies against the GST or XpressT tags.
DISCUSSION
C. burnetii encodes a virulence-associated T4BSS that exhibits conserved functions with the L. pneumophila Dot/Icm secretion apparatus. Historical limitations of C. burnetii genetics hindered direct investigations of T4BSS function. Thus, L. pneumophila was largely used to gain an understanding of the C. burnetii T4BSS and to identify the effector cohort. In this study, we used current techniques for C. burnetii genetic manipulation to investigate the role of the T4BSS chaperone protein IcmS in intracellular growth of C. burnetii and effector secretion. Gene deletion by allelic exchange produces a C. burnetii ΔicmS mutant with a partial intracellular growth defect that correlates with production of a small CCV. The ∼10-fold replication defect of the ΔicmS deletion mutant in Vero cells is consistent with published reports of growth by icmS transposon mutants in different cell lines (7, 8, 42). Overall, the moderate growth defects observed for icmS mutants here and elsewhere (7, 8, 42) may reflect a low number of IcmS-dependent effectors (20% in this study) and/or functional redundancy in the effector repertoire. One poorly growing transposon mutant occupied multiple, small CCVs, indicating defective homotypic fusion of Coxiella phagosomes. However, the transposon insertion eliminated only two amino acids from the C-terminal end of IcmS; thus, it is unclear how this small deletion results in the multivacuolar phenotype (19).
Studies of L. pneumophila T4BSS function have employed gene deletion or point mutations to identify IcmSW-binding and C-terminal signal sequences required for secretion. These reports indicate that L. pneumophila T4BSS effector translocation can be directed by a C-terminal secretion signal, by both C-terminal and internal secretion signals, or by an internal secretion signal (24, 27, 28, 32, 34, 37, 40, 53). Of the 50 C. burnetii proteins tested in this study, 36 are secreted by the ΔicmS mutant at levels significantly greater than or equal to wild-type C. burnetii. For this group of effectors, C-terminal sequence alignments show enrichment of acidic E-block residues in the region from −20 to −10. Strong E-block motifs containing ≥3 acidic residues are most prominent within the set of IcmS-inhibited substrates, including CBU1525 and CBU0072. CBU1525 encodes a C-terminal secretion signal typical of L. pneumophila Dot/Icm substrates exported in the absence of IcmS. Replacement of the C-terminal 50-amino-acid peptide of IcmS-dependent CBU0794 with that of CBU1525 results in a hybrid protein that is secreted by the C. burnetii ΔicmS mutant. CBU0072 encodes an atypical C-terminal secretion signal with acidic resides (ELEEE, amino acids −25 to −21) located outside the normal E-block location (28). The CBU0794 ΔCT:CBU0072 chimera is not secreted by the ΔicmS mutant, indicating that C-terminal sequences of IcmS-inhibited effectors are functionally diverse. Indeed, eight substrates that exhibit positive or equivalent secretion by the ΔicmS mutant relative to wild-type C. burnetii do not exhibit prominent E-block motifs (CBU0072, CBU0129, CBU0534, CBU0665 [CvpA], CBU0781, CBU1024, CBU1818 [CvpD], and CBU1819).
Early secretion of L. pneumophila SidJ is mediated by a conventional C-terminal signal sequence, while late secretion is orchestrated by an internal signal (37). N-terminal truncation constructs of CBU0794 and CBU1525 with ≥50 C-terminal amino acids exhibit the same IcmS-dependent and -inhibited secretions, respectively, consistent with the IcmS dependency of full-length proteins. However, CBU0794 and CBU1525 constructs that encode fewer C-terminal residues exhibit altered secretion. Specifically, the C-terminal 40 amino acids of CBU0794 and CBU1525 are secreted by both wild-type bacteria and the ΔicmS mutant, whereas 30-amino-acid constructs of both substrates are only secreted by the ΔicmS mutant. This indicates that the −50 to −40 sequences of CBU0794 and CBU1525 contain sites that regulate IcmS dependency. Additionally, both substrates harbor a secretion signal within the C-terminal 30 amino acids that is inhibited by IcmS. The L. pneumophila effector SidJ contains multiple IcmS binding sites distinct from the C-terminal 40-amino-acid peptide that encodes a secretion signal (37). Thus, it is unclear how IcmS represses the secretion of CBU0794 and CBU1525 C-terminal 30-amino-acid peptides by C. burnetii. Interestingly, the CBU07941–435 construct that lacks only the C-terminal 30 amino acids is translocated by wild-type C. burnetii at levels comparable to the full-length protein. Reminiscent of the L. pneumophila effectors SidG and SidJ (32, 37), these characteristics of CBU0794 are suggestive of both C-terminal and internal T4BSS secretion signals. In contrast, CBU1525 and CBU0072 are not translocated by wild-type C. burnetii, and their C-terminal 30-amino-acid peptides are required for export by the ΔicmS mutant. We were unable to identify infection conditions that derepressed secretion of full-length CBU1525 or CBU0072 by wild-type C. burnetii. However, swapping out the endogenous C-terminal 50-amino-acid sequences from CBU1525 or CBU0072 with the C-terminal 50 amino acids of CBU0794 produces chimeric proteins that are secreted by wild-type C. burnetii but not by the ΔicmS mutant. Together, the domain swap experiments indicate that sites within the C-terminal 50 amino acids promote secretion of CBU0072 and CBU1525 by the ΔicmS mutant while inhibiting secretion by wild-type bacteria. However, C-terminal signals from CBU0072 and CBU1525 do not abrogate IcmS-mediated secretion of CBU0794 by wild-type C. burnetii. Thus, IcmS-dependent secretion of CBU0794 that is mediated by an internal secretion signal(s) exerts greater control over substrate translocation than the IcmS-inhibited C-terminal signals from CBU0072 or CBU1525. Along with pulldown experiments that show that IcmSW binds both IcmS-dependent and -inhibited substrates, these data indicate a regulatory hierarchy involving IcmS controls export of Dot/Icm substrates in C. burnetii. Indeed, IcmSW chaperone activity both positively and negatively regulates substrate translocation in C. burnetii. To our knowledge, inhibition of substrate translocation in the presence of IcmS is unique to the C. burnetii T4BSS.
All of the 11 IcmS-inhibited substrates described in this study were initially identified in screens that used L. pneumophila as a surrogate for validating Dot/Icm substrate translocation (3, 30, 48, 49), including five ankyrin-repeat (Ank) family effectors. AnkA, -B, -F, -G, and -H are secreted at greater levels by the ΔicmS mutant than by wild-type C. burnetii. However, when expressed in L. pneumophila, AnkB, -F, -G, and -H are secreted more efficiently by wild-type L. pneumophila than by an ΔicmS mutant, whereas AnkA secretion increases in an ΔicmS mutant (31). We interpret these differences in substrate translocation to mean that a subset of C. burnetii effectors are controlled by IcmS regulatory mechanisms that are distinct from the L. pneumophila Dot/Icm system.
In pilot immunoblot experiments we were unable to detect IcmS or IcmW in C. burnetii protein lysates with monospecific sera against L. pneumophila IcmS or IcmW. However, a previous microarray study indicates stable expression of icmS and icmW throughout the C. burnetii infection cycle (54). C. burnetii IcmS and IcmW proteins exhibit 56 and 60% identities to the respective L. pneumophila proteins. Impaired growth of L. pneumophila ΔicmS or ΔicmW mutants is restored by genetic complementation with respective C. burnetii homologs (45), indicating the chaperone complex functions similarly in the two pathogens. Consequently, discrepancies in substrate translocation between C. burnetii and L. pneumophila observed here and previously (46, 47) likely result from species-specific factors that modulate IcmSW activity. Several recent L. pneumophila studies indicate IcmS binding is regulated, in part, by its interactions with other components of the T4CP, including IcmW, DotL, and LvgA (38–40). Notably, C. burnetii lacks a homolog of LvgA, an adaptor protein unique to L. pneumophila that binds hydrophobic residues on IcmS (39). C. burnetii encodes a known homolog of DotL, but an adaptor protein that binds IcmS has not been identified.
Our data suggest that IcmS dependency in C. burnetii is regulated according to specific sites within Dot/Icm substrates that function as secretion signals or IcmS binding sites. Overall, substrates secreted in the absence of IcmS, including both IcmS-inhibited and -independent substrates, often exhibit C-terminal signals containing an acidic E-block motif, whereas IcmS-dependent substrates typically lack E-block secretion signals. Exceptions to this trend, exemplified by CBU0080 and CBU1024, indicate that the E-block motif is not the only factor controlling IcmS dependency. Indeed, the observation that IcmS-inhibited secretion of CBU1525 is abrogated by deletion of the −50 to −40 region suggests that IcmS dependency can be regulated by sites distinct from the C-terminal 30 amino acids that contains the secretion signal. Additionally, interactions between IcmS and other proteins within the C. burnetii T4CP, for example, a functional analog to LvgA, could alter IcmS activity. In this model, interactions between IcmS and different T4CP adaptor proteins modulate which substrate sites are bound by IcmS and whether IcmS positively or negatively regulates specific types of secretion signals. Detailed characterization of the C. burnetii T4CP is critical for determining the molecular basis of Dot/Icm substrate translocation by the C. burnetii and how it differs from the L. pneumophila T4BSS.
In summary, this study shows a complex interplay between the chaperone IcmS and the C. burnetii T4BSS effector pool. Three export behaviors based on IcmS dependency were identified in addition to an internal IcmS binding site. Several intriguing questions remain unanswered. For example, what are the precise physiochemical properties of internal sites engaged by IcmSW? Further, does IcmSW target effector subsets with common functions and/or temporal constraints that modify host cell functions at specific stages of C. burnetii’s lengthy infectious cycle? A better understanding of regulation of C. burnetii T4BSS secretion will provide important insight into the pathogen-host cell relationship.
MATERIALS AND METHODS
C. burnetii and mammalian cells.
The C. burnetii Nine Mile, phase II (NMII; clone 4, RSA439) strain was utilized throughout this work. Wild-type C. burnetii and genetic transformants were grown microaerobically in ACCM-2, as previously described (55). For storage, bacteria were pelleted after 6 days of growth, washed three times in phosphate-buffered saline (PBS; 1 mM KH2PO4, 155 mM NaCl, 3 mM Na2HPO4 [pH 7.4]), suspended in cell freezing medium (RPMI 1640 medium containing 10% dimethyl sulfoxide and 10% fetal bovine serum [FBS]; Invitrogen), and then frozen at –80°C. Escherichia coli Stellar (BD Clontech) and PIR1 cells (Invitrogen) were used for recombinant DNA procedures and cultivated in Luria-Bertani (LB) broth. E. coli transformants were selected on LB agar plates containing 50 μg/ml of kanamycin or 10 μg/ml of chloramphenicol. African green monkey kidney (Vero) cells (CCL-81; ATCC) were maintained in RPMI 1640 medium containing 10% FBS at 37°C and 5% CO2. THP-1 monocytes (TIB-202; ATCC) were differentiated into macrophage-like cells by incubation overnight in RPMI 1640 medium containing 10% FBS and 200 nM phorbol myristate acetate (Sigma-Aldrich). C. burnetii replication in host cells or ACCM-2 was measured by quantitative PCR of genome equivalents (GE) as previously described using a probe specific to dotA (56, 57).
Generation and complementation of an icmS mutant.
The plasmids and oligonucleotide primers used in this study are listed in Tables S1 and S2, respectively, in the supplemental material. Restriction enzymes were obtained from New England Biolabs. PCR was performed using Accuprime Pfx or Taq polymerase (Thermo Fisher Scientific). PCR primers were obtained from Integrated DNA Technologies. All cloning procedures were conducted using an In-Fusion PCR cloning system (BD Clontech), and DNA was transformed into either E. coli Stellar or PIR1 competent cells. For targeted inactivation of C. burnetii icmS (cbu1642), the 5′ and 3′ flanking regions of the gene were amplified from NMII genomic DNA by PCR using the upstream and downstream oligonucleotide pairs CBU1642-5′-F/CBU1642-5′-R and CBU1642-3′-F/CBU1642-3′-R, respectively. The 5′ and 3′ fragments were cloned into BamHI/SalI-digested pJC-CAT (5) by In-Fusion, resulting in the formation of an internal AgeI site between the 5′ and 3′ regions and the creation of pJC-CAT::CBU1642-5′3′. The 1169P-Kan cassette was amplified from pJB-Kan (55) by PCR with P1169-Kan-AgeI-KO-rev-F and P1169-Kan-AgeI-KO-rev-R oligonucleotides and cloned into AgeI-digested pJC-CAT::CBU1642-5′3′ to create pJC-CAT::CBU1642-5′3′-Kan. For complementation of the C. burnetii ΔicmS mutant, the cbu1642 promoter and gene region were amplified from NMII genomic DNA by PCR using the oligonucleotides CBU1642comp-F and CBU1642comp-R. The cbu1642comp fragment was cloned into EcoRI-digested pMiniTn7T-CAT (5) by In-Fusion to create pMiniTn7T-CAT::cbu1642comp.
Electroporation of C. burnetii and selection of allelic-exchange mutants were conducted as previously described (5, 55). The C. burnetii ΔicmS strain was subsequently expanded by culture in ACCM-2 containing kanamycin, and clones were isolated by limiting dilution in ACCM-2. Deletion of icmS and clonality were verified by PCR. PCR validation was accomplished by amplifying an internal icmS gene fragment with specific primer pairs, using wild-type or mutant genomic DNA as the template. Generation and verification of a Tn7-based genetic complement of the C. burnetii ΔicmS strain were performed as previously described (5).
Indirect immunofluorescence.
Vero cells infected at a multiplicity of infection (MOI) of ∼25 were fixed for 20 min in 4% paraformaldehyde plus PBS, followed by permeabilization for 5 min in 0.1% saponin in PBS. Cells were stained for indirect immunofluorescence as previously described (52, 58). Rabbit anti-C. burnetii serum and a mouse monoclonal antibody directed against the lysosomal marker CD63 (LAMP3) (clone H5C6, BD Biosciences) were used as primary antibodies. Alexa Fluor 488- and 594- IgG (Invitrogen) were used as secondary antibodies. Coverslips were mounted using ProLong Gold (Thermo Fischer Scientific). Microscopy was conducted using a modified Perkin-Elmer UltraView spinning-disk confocal system connected to a Nikon Eclipse Ti-E inverted microscope. Images were obtained using Metamorph software (Molecular Devices, Inc.) and processed with ImageJ software (National Institutes of Health).
Translocation assays.
Full-length, partial, and chimeric genes encoding published Dot/Icm substrates were amplified by PCR from NMII genomic DNA using the oligonucleotide primers listed in Table S2. PCR products were cloned by In-Fusion into the unique SalI site of pJB-CAT-CyaA or pJB-CAT-BlaM to create the translocation reporter plasmids listed in Table S1. CyaA and BlaM translocation assays were performed using THP-1 macrophage-like cells (5 × 105 per well) in 24-well plates. Cells were washed once with RPMI medium plus 10% FBS, infected with C. burnetii transformants expressing CyaA or BlaM fusion proteins at an MOI ∼ 50, and incubated in RPMI medium plus 10% FBS for 48 h. For CyaA translocation assays, the concentration of cAMP in lysates from infected cells was determined using the cAMP enzyme immunoassay (GE Healthcare) as previously described (4). Positive secretion of CyaA fusion proteins by wild-type C. burnetii or the ΔicmS mutant was scored as ≥2.5-fold more cytosolic cAMP than the negative control (wild-type C. burnetii expressing CyaA alone) in accordance with the previously validated threshold for secretion (4, 16, 30, 46, 47). For BlaM translocation assays, infected cells in glass bottom plates were loaded for 1 h with CCF4-AM (LiveBLAzer-FRET B/G loading kit; Invitrogen) in solution with 15 mM probenecid (30). Cells were replenished with fresh medium and immediately imaged on a LSM710 confocal laser-scanning microscope (Carl Zeiss Micro Imaging) with a 405-nm diode laser and detectors at 520 to 630 nm for CCF4-AM substrate (green) and 410 to 488 nm for CCF4 product (blue), respectively. Image segmentation and signal quantitation were performed using CellProfiler as previously described (59). Positive translocation of BlaM fusion proteins was scored as a ratio of blue to green signal ≥2.5-fold more than the mean of uninfected cells.
Protein binding assays.
Full-length C. burnetii icmS, IcmW, cbu0077, cbu0665, cbu0794, and cbu1525, and respective gene fragments encoding proteins with 50-amino-acid C-terminal deletions were amplified by PCR using the oligonucleotide primers listed in Table S2. PCR products were cloned by Infusion into pEXP1-DEST (Invitrogen) digested with BsrGI/HindIII (New England Biolabs) to generate plasmids encoding proteins with N-terminal Xpress tags. For production of the IcmSW fusion tagged with GST on the N terminus of IcmW, icmS was amplified by PCR and fused downstream of an amplicon encoding icmW. The resulting gene fusion was inserted into pDEST15 (Invitrogen) digested with BsrGI/BamHI. Effector-encoding plasmids were mixed with the IcmSW-encoding plasmid in equal amounts for in IVTT reactions using the RTS 100 E. coli HY kit (Roche) to achieve cell-free production of protein. IVTT reaction mixtures were incubated at room temperature for 1 h with glutathione-Sepharose 4B (GE Healthcare). Glutathione-Sepharose was pelleted in Pierce spin cups (Thermo Fisher) by centrifugation at 1,000 × g for 1 min and washed five times in Tris-buffered saline (pH 7.2) containing 1% Triton X-100. Glutathione-Sepharose was resuspended in SDS-PAGE loading buffer and boiled for 10 min. Proteins were separated by SDS-PAGE on a 10% gel and then immunoblotted as described above using mouse monoclonal antibodies directed against the GST tag (EMD Millipore) to detect IcmW or the Xpress tag (Invitrogen) to detect effector proteins.
Statistical analysis.
Statistical analyses were performed using a one-way analysis of variance or a Student t test and Prism software (GraphPad Software, Inc., La Jolla, CA).
Supplementary Material
ACKNOWLEDGMENT
This study was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Disease.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00431-19.
REFERENCES
- 1.Stein A, Louveau C, Lepidi H, Ricci F, Baylac P, Davoust B, Raoult D. 2005. Q fever pneumonia: virulence of Coxiella burnetii pathovars in a murine model of aerosol infection. Infect Immun 73:2469–2477. doi: 10.1128/IAI.73.4.2469-2477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham JG, MacDonald LJ, Hussain SK, Sharma UM, Kurten RC, Voth DE. 2013. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell Microbiol 15:1012–1025. doi: 10.1111/cmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey KL, Newton HJ, Luhrmann A, Roy CR. 2011. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2:e00175. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beare PA, Larson CL, Gilk SD, Heinzen RA. 2012. Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol 78:4580–4589. doi: 10.1128/AEM.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, Luo ZQ, Samuel JE. 2013. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol 195:3914–3924. doi: 10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez E, Cantet F, Fava L, Norville I, Bonazzi M. 2014. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog 10:e1004013. doi: 10.1371/journal.ppat.1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, Roy CR. 2014. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog 10:e1004286. doi: 10.1371/journal.ppat.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crabill E, Schofield WB, Newton HJ, Goodman AL, Roy CR. 2018. Dot/Icm-translocated proteins important for biogenesis of the Coxiella burnetii-containing vacuole Identified by screening of an effector mutant sublibrary. Infect Immun 86. doi: 10.1128/IAI.00758-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larson CL, Martinez E, Beare PA, Jeffrey B, Heinzen RA, Bonazzi M. 2016. Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions. Future Microbiol 11:919–939. doi: 10.2217/fmb-2016-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luhrmann A, Nogueira CV, Carey KL, Roy CR. 2010. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci U S A 107:18997–19001. doi: 10.1073/pnas.1004380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckart RA, Bisle S, Schulze-Luehrmann J, Wittmann I, Jantsch J, Schmid B, Berens C, Lührmann A. 2014. Antiapoptotic activity of Coxiella burnetii effector protein AnkG is controlled by p32-dependent trafficking. Infect Immun 82:2763–2771. doi: 10.1128/IAI.01204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klingenbeck L, Eckart RA, Berens C, Luhrmann A. 2013. The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell Microbiol 15:675–687. doi: 10.1111/cmi.12066. [DOI] [PubMed] [Google Scholar]
- 14.Cunha LD, Ribeiro JM, Fernandes TD, Massis LM, Khoo CA, Moffatt JH, Newton HJ, Roy CR, Zamboni DS. 2015. Inhibition of inflammasome activation by Coxiella burnetii type IV secretion system effector IcaA. Nat Commun 6:10205. doi: 10.1038/ncomms10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lifshitz Z, Burstein D, Schwartz K, Shuman HA, Pupko T, Segal G. 2014. Identification of novel Coxiella burnetii Icm/Dot effectors and genetic analysis of their involvement in modulating a mitogen-activated protein kinase pathway. Infect Immun 82:3740–3752. doi: 10.1128/IAI.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larson CL, Beare PA, Howe D, Heinzen RA. 2013. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc Natl Acad Sci U S A 110:E4770–E4779. doi: 10.1073/pnas.1309195110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latomanski EA, Newton P, Khoo CA, Newton HJ. 2016. The effector Cig57 hijacks FCHO-mediated vesicular trafficking to facilitate intracellular replication of Coxiella burnetii. PLoS Pathog 12:e1006101. doi: 10.1371/journal.ppat.1006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler LJ, Reed SR, Sarraf SA, Arteaga DD, Newton HJ, Roy CR. 2016. Effector protein Cig2 decreases host tolerance of infection by directing constitutive fusion of autophagosomes with the Coxiella-containing vacuole. mBio 7:e01127-16. doi: 10.1128/mBio.01327-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez E, Allombert J, Cantet F, Lakhani A, Yandrapalli N, Neyret A, Norville IH, Favard C, Muriaux D, Bonazzi M. 2016. Coxiella burnetii effector CvpB modulates phosphoinositide metabolism for optimal vacuole development. Proc Natl Acad Sci U S A 113:E3260–E3269. doi: 10.1073/pnas.1522811113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackstadt T, Williams JC. 1981. Incorporation of macromolecular precursors by Coxiella burnetii in an axenic medium, p 431–440. In Burgdorfer W, Anacker RL (ed), Rickettsiae and rickettsial diseases. Academic Press, Inc, New York, NY. [Google Scholar]
- 21.Omsland A, Cockrell DC, Fischer ER, Heinzen RA. 2008. Sustained axenic metabolic activity by the obligate intracellular bacterium Coxiella burnetii. J Bacteriol 190:3203–3212. doi: 10.1128/JB.01911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton HJ, McDonough JA, Roy CR. 2013. Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One 8:e54566. doi: 10.1371/journal.pone.0054566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubori T, Hyakutake A, Nagai H. 2008. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol 67:1307–1319. doi: 10.1111/j.1365-2958.2008.06124.x. [DOI] [PubMed] [Google Scholar]
- 24.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A 102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog 5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubori T, Nagai H. 2016. The Type IVB secretion system: an enigmatic chimera. Curr Opin Microbiol 29:22–29. doi: 10.1016/j.mib.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Huang L, Boyd D, Amyot WM, Hempstead AD, Luo Z-Q, O’Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. 2011. The E block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol 13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. 2013. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A 110:707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham JG, Winchell CG, Sharma UM, Voth DE. 2015. Identification of ElpA, a Coxiella burnetii pathotype-specific Dot/Icm type IV secretion system substrate. Infect Immun 83:1190–1198. doi: 10.1128/IAI.02855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, Omsland A, Heinzen RA. 2011. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol 193:1493–1503. doi: 10.1128/JB.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, Heinzen RA. 2009. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol 191:4232–4242. doi: 10.1128/JB.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cambronne ED, Roy CR. 2007. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog 3:e188. doi: 10.1371/journal.ppat.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coers J, Monahan JJ, Roy CR. 1999. Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nat Cell Biol 1:451–188. doi: 10.1038/15687. [DOI] [PubMed] [Google Scholar]
- 34.Ninio S, Zuckman-Cholon DM, Cambronne ED, Roy CR. 2005. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol Microbiol 55:912–926. doi: 10.1111/j.1365-2958.2004.04435.x. [DOI] [PubMed] [Google Scholar]
- 35.Bardill JP, Miller JL, Vogel JP. 2005. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol 56:90–103. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- 36.Coers J, Kagan JC, Matthews M, Nagai H, Zuckman DM, Roy CR. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol Microbiol 38:719–736. doi: 10.1046/j.1365-2958.2000.02176.x. [DOI] [PubMed] [Google Scholar]
- 37.Jeong KC, Sutherland MC, Vogel JP. 2015. Novel export control of a Legionella Dot/Icm substrate is mediated by dual, independent signal sequences. Mol Microbiol 96:175–188. doi: 10.1111/mmi.12928. [DOI] [PubMed] [Google Scholar]
- 38.Kwak MJ, Kim JD, Kim H, Kim C, Bowman JW, Kim S, Joo K, Lee J, Jin KS, Kim YG, Lee NK, Jung JU, Oh BH. 2017. Architecture of the type IV coupling protein complex of Legionella pneumophila. Nat Microbiol 2:17114. doi: 10.1038/nmicrobiol.2017.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, Xu D, Wan M, Yin L, Wang X, Wu L, Liu Y, Liu X, Zhou Y, Zhu Y. 2017. Structural insights into the roles of the IcmS-IcmW complex in the type IVb secretion system of Legionella pneumophila. Proc Natl Acad Sci U S A 114:13543–13548. doi: 10.1073/pnas.1706883115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutherland MC, Nguyen TL, Tseng V, Vogel JP. 2012. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog 8:e1002910. doi: 10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vincent CD, Friedman JR, Jeong KC, Sutherland MC, Vogel JP. 2012. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol 85:378–391. doi: 10.1111/j.1365-2958.2012.08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez E, Cantet F, Bonazzi M. 2015. Generation and multi-phenotypic high-content screening of Coxiella burnetii transposon mutants. J Vis Exp 13:e52851. doi: 10.3791/52851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Connor TJ, Adepoju Y, Boyd D, Isberg RR. 2011. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A 108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zusman T, Yerushalmi G, Segal G. 2003. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect Immun 71:3714–3723. doi: 10.1128/iai.71.7.3714-3723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamboni DS, McGrath S, Rabinovitch M, Roy CR. 2003. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol 49:965–976. doi: 10.1046/j.1365-2958.2003.03626.x. [DOI] [PubMed] [Google Scholar]
- 46.Beare PA, Sandoz KM, Larson CL, Howe D, Kronmiller B, Heinzen RA. 2014. Essential role for the response regulator PmrA in Coxiella burnetii type 4B secretion and colonization of mammalian host cells. J Bacteriol 196:1925–1940. doi: 10.1128/JB.01532-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, Valdivia RH, Heinzen RA. 2015. Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication. Infect Immun 83:661–670. doi: 10.1128/IAI.02763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, Banga S, Mertens K, Weber MM, Gorbaslieva I, Tan Y, Luo ZQ, Samuel JE. 2010. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A 107:21755–21760. doi: 10.1073/pnas.1010485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber MM, Faris R, van Schaik EJ, McLachlan JT, Wright WU, Tellez A, Roman VA, Rowin K, Case ED, Luo ZQ, Samuel JE. 2016. The type IV secretion system effector protein CirA stimulates the GTPase activity of RhoA and Is required for virulence in a mouse model of Coxiella burnetii infection. Infect Immun 84:2524–2533. doi: 10.1128/IAI.01554-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fielden LF, Moffatt JH, Kang Y, Baker MJ, Khoo CA, Roy CR, Stojanovski D, Newton HJ. 2017. A farnesylated Coxiella burnetii effector forms a multimeric complex at the mitochondrial outer membrane during Infection. Infect Immun 85:e01046-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA. 2009. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J Bacteriol 191:1369–1381. doi: 10.1128/JB.01580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeong KC, Sexton JA, Vogel JP. 2015. Spatiotemporal regulation of a Legionella pneumophila T4SS substrate by the metaeffector SidJ. PLoS Pathog 11:e1004695. doi: 10.1371/journal.ppat.1004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandoz KM, Popham DL, Beare PA, Sturdevant DE, Hansen B, Nair V, Heinzen RA. 2016. Transcriptional profiling of Coxiella burnetii reveals extensive cell wall remodeling in the small cell variant developmental form. PLoS One 11:e0149957. doi: 10.1371/journal.pone.0149957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, Samuel JE, Heinzen RA. 2011. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol 77:3720–3725. doi: 10.1128/AEM.02826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA. 2009. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci U S A 106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Howe D, Shannon JG, Winfree S, Dorward DW, Heinzen RA. 2010. Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect Immun 78:3465–3474. doi: 10.1128/IAI.00406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howe D, Melnicakova J, Barak I, Heinzen RA. 2003. Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell Microbiol 5:469–480. doi: 10.1046/j.1462-5822.2003.00293.x. [DOI] [PubMed] [Google Scholar]
- 59.Larson CL, Heinzen RA. 2017. High-content imaging reveals expansion of the endosomal compartment during Coxiella burnetii parasitophorous vacuole maturation. Front Cell Infect Microbiol 7:48. doi: 10.3389/fcimb.2017.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.