Abstract
Pectinodontid limpets are important members of deep-sea hot vents and cold seeps as can be seen by their conspicuous presence in both extant and extinct systems. They have traditionally been classified into different genera and species based on shell and radula characteristics; the reliability of these characters has been questioned but not tested thoroughly. Here, for the first time in taxa endemic to deep-sea chemosynthetic ecosystems, we combine substrate translocation with molecular data to assess the plasticity and variability of key phenotypic characters. Molecular data revealed that several ‘species’ of extant vent/seep pectinodontids actually represent intergrading morphotypes of a single, highly plastic, evolutionary lineage, with each morphological trait being possibly influenced differently by environmental and genetic factors. Our results challenge previous interpretations of paleoecology at fossil chemosynthetic ecosystems and highlight the importance of modern analogues in understanding fossil systems.
Keywords: chemosynthetic ecosystems, pectinodontidae, phenotypic variability, population genetics
1. Introduction
To understand the evolutionary processes and history of extant biodiversity, it is crucial to compare data from living and extinct taxa [1]. A key limitation in the interpretation of paleoecology is the reliance on morphology and extrapolation from the knowledge of modern analogues; and that only some groups have well-preserved fossil records. Molluscs produce hard armour, and consequently, the phylum has one of the most complete fossil records. However, extreme flexibility and adaptability in modifying their body plan [2], combined with a high degree of phenotypic plasticity [3], means the usefulness of taxonomic characters cannot always be shared across taxa. Without the knowledge of relationships between morphological characters and genetic/environmental factors shaping them in living relatives, it is difficult to confidently reconstruct paleoecology and it is often especially challenging for deep-sea groups.
Modern benthic taxa inhabiting deep-sea hot vents and cold seeps mostly evolved recently in the Late Mesozoic or Cenozoic, when brachiopods were replaced by molluscs [4]. Pectinodontid limpets in the genera Bathyacmaea Okutani, Tsuchida & Fujikura, 1992 [5] and Serradonta Okutani, Tsuchida & Fujikura, 1992 [5] are thought to be restricted to vents and seeps and are known from both extant and fossil systems dating back to the Upper Cretaceous [6], making them an ideal group to study. Both genera are found in the western Pacific and are especially diverse around Japan, but species from this region have been described based on morphology alone [7,8]. Taxonomy has historically relied on shell shape, shell sculpture and radula characteristics: Bathyacmaea has a flat, broad shell and smooth-edged radulae, grazing on flat surfaces, whereas Serradonta has a curved, narrow shell and serrated radula thought to be specialist inhabiting siboglinid worm tubes [6]. Translocation experiments show that many shallow-water patellogastropod limpets with distinct ecophenotypes [3,9,10] are presented by a single molecular operational taxonomic unit (MOTU, [11]) [12]. The reliability of these characters has been questioned in pectinodontids [13] but has yet to be thoroughly tested. Here, we used substrate translocation combined with molecular data to tease apart factors influencing morphological characters in deep-sea pectinodontid limpets.
2. Material and methods
(a). Acquisition of limpets
A total of 96 pectinodontid limpets were collected from vents in the Okinawa Trough and seeps in Sagami Bay and the South China Sea using slurp guns mounted on remotely operated vehicles (ROVs) Hyper-Dolphin and KAIKO, or the deep-submergence vehicle (DSV) Jiaolong (electronic supplementary material, table S1). Limpets recovered were preserved in 99% ethanol or kept alive in 4°C seawater. Limpets were identified as Bathyacmaea or Serradonta based on shell morphology. Bathyacmaea were collected from Bathymodiolus mussels and Serradonta from tubes of Lamellibrachia worms.
(b). Live-rearing experiment
To investigate the effect of substrate on shell and radula morphology, both Bathyacmaea and Serradonta obtained from Sagami Bay seep were reared in aquaria under atmospheric pressure using 4°C artificial seawater, with an air pump. Upon recovery, responsive individuals were placed on their original substrates, and only those that survived for a week were used in the experiment. This resulted in 16 Bathyacmaea and 8 Serradonta. Half of the Bathyacmaea were kept on mussel shells, while the other half were transferred to Lamellibrachia tubes; vice versa for Serradonta. Limpets were reared for three months and grazed on naturally growing bacterial film.
(c). Shell and radula morphology
Limpets were dissected to isolate their shell and radula. Shells were subjected to X-ray micro-CT using a ScanXmate-D160TSS105 (Comscantecno, Japan) to visualize their morphology; three-dimensional reconstruction was carried out using Amira v. 5.3. Radula was imaged using a scanning electron microscopy (SEM), after cleaning in half-strength commercial bleach and washed in MilliQ water. Specimens were observed uncoated at 15 kV using a Hitachi TM-3000SEM.
(d). Molecular methods
Of all limpets, 93 were successfully sequenced for the mitochondrial COI gene, including 81 Bathyacmaea and 12 Serradonta (electronic supplementary material, table S2). The nuclear H3 gene was sequenced from 23 Bathyacmaea and 10 Serradonta (electronic supplementary material, table S3) and the nuclear 18S rRNA gene was sequenced from nine Bathyacmaea and six Serradonta (electronic supplementary material, table S4). Published methods were followed for DNA extraction, amplification and sequencing [14]. Primer pairs included LCO1490/HCO2198 for the COI region [15], H3aF/H3aR for the H3 region [16] and 18e/18dh for the 18S region [17]. Sequences were aligned using Clustal W in MEGA6 [18].
Resulting COI (517 bp), H3 (249 bp) and 18S (695 bp) alignments were used for downstream analyses (electronic supplementary material, tables S2–S4). For phylogenetic reconstruction, the most suitable substitution model, determined using Model Selection in MEGA6, was Tamura 3-parameter (gamma) for COI, Kimura 2-parameter for H3 and Jukes-Cantor for 18S. The maximum-likelihood (ML) tree was generated using MEGA6 (1000 bootstraps) and Bayesian tree using MrBayes v. 3.2.6 (5 million generations, topologies sampled every 1000 generations, 25% ‘burn-in’) [19]. Pectinodontid sequences were obtained from GenBank, and the vetigastropod Lepetodrilus elevatus was used as an outgroup. Parsimonious haplotype networks were estimated using TCS v. 1.21 [20]. Arlequin v. 3.5.2.2 [21] was used to calculate the haplotype number, haplotype diversity, nucleotide diversity and carry out neutrality test with Fu's Fs for COI and H3 genes. For the COI gene, the same software was used for population comparisons based on pairwise Fst.
3. Results
(a). Natural phenotypic variations
Traditional shell and radular characters of specimens collected (figure 1), combined with their geographical localities and environments (figure 2; electronic supplementary material, table S1), indicated that the following described species were collected: Bathyacmaea nipponica Okutani, Tsuchida & Fujikura, 1992 [5] (Sagami Bay, seep), B. secunda Okutani, Fujikura & Sasaki, 1993 [22] (Okinawa Trough, vent), B. tertia Sasaki, Okutani & Fujikura, 2003 [7] (Okinawa Trough, vent) and Serradonta vestimentifericola Okutani, Tsuchida & Fujikura, 1992 [5] (Sagami Bay, seep). For these species, our collecting sites included their type localities. Additionally, B. cf. tertia (South China Sea, seep) and S. cf. vestimentifericola (Okinawa Trough, vent) were also collected.
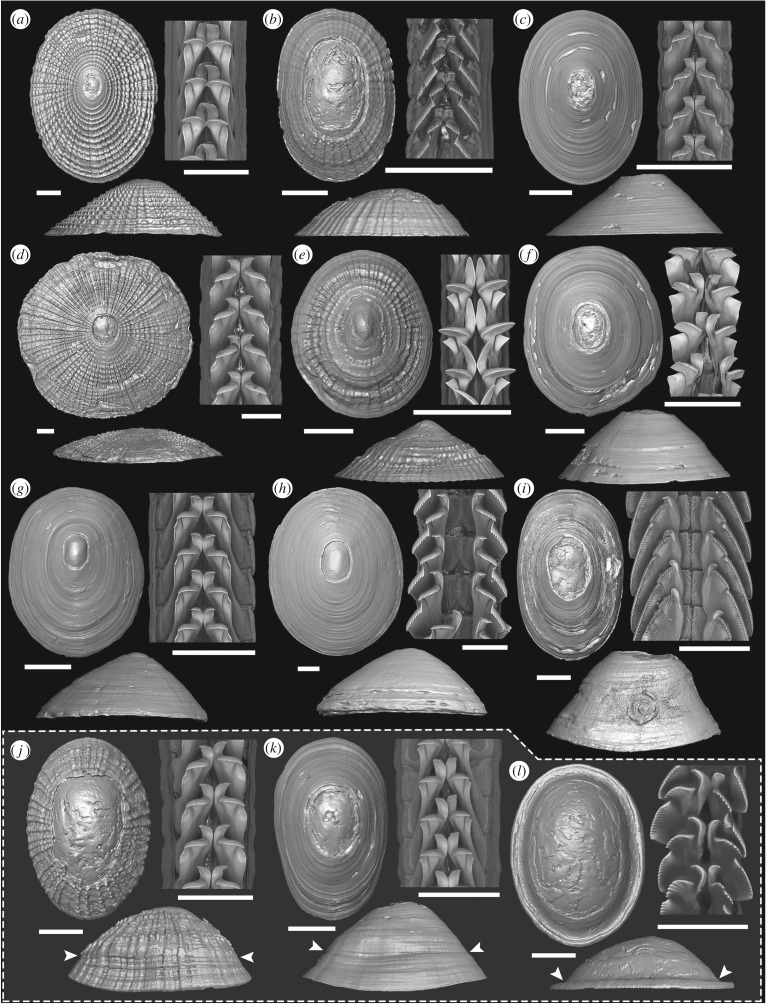
Figure 1.
Representative shell and radula morphology of vent/seep pectinodontids. (a–c) Bathyacmaea nipponica, Off Hatsushima seep. (d,e) Bathyacmaea secunda, Iheya North Knoll vent. (f) Bathyacmaea tertia, Iheya North Knoll vent. (g,h) Bathyacmaea cf. tertia, Jiaolong Ridge seep. (i) Serradonta vestimentifericola, Off Hatsushima seep. (j,k) Bathyacmaea nipponica translocated from mussels to worm tubes. (l) Serradonta vestimentifericola translocated from worm tubes to mussels. Scale bars, left (shell) = 2 mm; right (radula) = 100 µm. Arrowheads indicate the start of shell deposition during translocation.
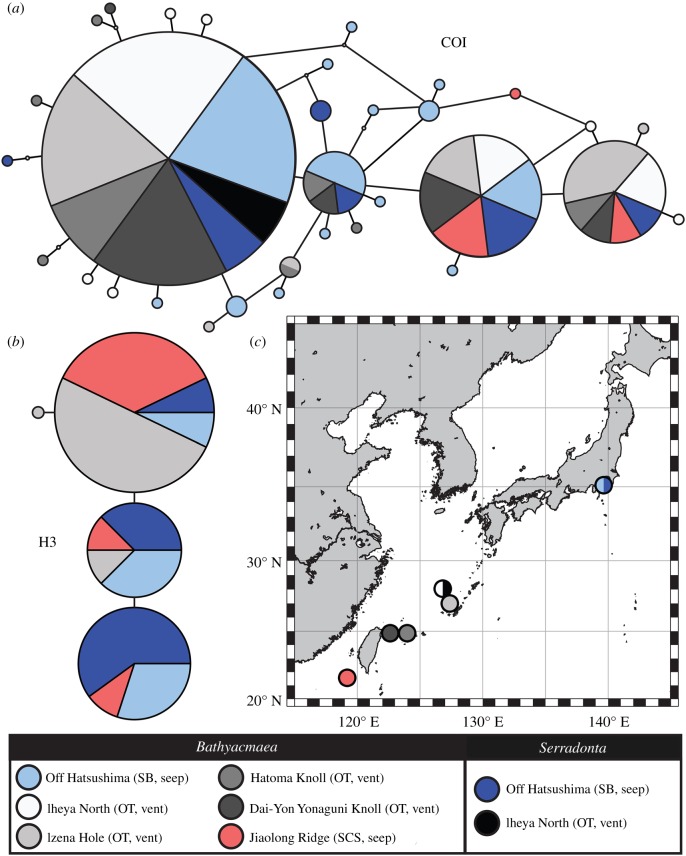
Figure 2.
Haplotype networks and localities of vent/seep pectinodontids. (a) COI fragment, 517 bp. (b) H3 fragment, 249 bp. (c) Map of localities sampled in the present study. The colour legend applies to the entire figure. Each node in haplotype networks represents one base difference, and small open circles indicate unsampled haplotypes. OT, Okinawa Trough; SB, Sagami Bay; SCS, South China Sea. (Online version in colour.)
A large degree of variation among shell and radula morphology was observed within each ‘species’ and locality (figure 1a–h), and this variation was greater than expected for presumed taxonomically informative features [5,7,22,23]. Although we found shells and radula combinations matching original descriptions (figure 1a,d), there were often discrepancies. In extreme cases, individuals had flat shells characteristic of Bathyacmaea but serrated radula characteristic of Serradonta (figure 1b,h), although the outermost cusp was not broadened to the same extent. All but three specimens of Bathyacmaea, however, exhibited typical smooth cusps, and all specimens of Serradonta examined had serrated cusps. Furthermore, many individuals contained considerable differences in morphology along the radular ribbon (electronic supplementary material, figure S1).
(b). Effect of substrate on the morphology
Of the 24 individuals used in the three-month experiment, 14 Bathyacmaea (6 transplanted and 8 on the native substrate) and 4 Serradonta (2 transplanted and 2 on the native substrate) survived to the end. In the case of Bathyacmaea, three of the surviving transplanted individuals left worm tubes at early stages and lived on the aquarium wall, despite several attempts at reattachment (electronic supplementary material, figure S2). Shells of surviving individuals clearly exhibited a scar absent before the experiment, marking the change of environment and the beginning of new shell deposition (arrowhead, figure 1j–l; electronic supplementary material, figure S2).
Limpets transplanted to non-native substrates underwent an obvious shift in shell morphology conforming to the substrate shape, from flat to curved in Bathyacmaea (figure 1j,k; apical angle measured from side view shifting from 100°–105° to 55°–71°) and vice versa in Serradonta (figure 1l; apical angle shifting from 60°–77° to 96°–100°). Shell sculptures remained unchanged, unlike changes in sculpture observed for patellogastropods [10,24]. The radula did not change across the entire ribbon when examined post-experiment. Individuals reared on native substrates did not deviate in shell shape (electronic supplementary material, figure S2a,c; apical angle 92°–96° before and 91°–98° after experiment in Bathyacmaea, 62°–76° before and 58°–70° after experiment in Serradonta), although Bathyacmaea that left mussel shells and lived on aquarium glass became more flattened (electronic supplementary material, figure S2b; apical angle shifting from 93°–100° to 116°–122°).
(c). Phylogeny and population genetics
Both COI and H3 haplotype networks (figure 2a,b) demonstrated that all individuals of Bathyacmaea and Serradonta from all localities belonged to a single well-mixed population with moderate genetic diversity. To corroborate this, both ML and Bayesian phylogenetic reconstructions using the COI gene or concatenated sequences of COI, H3 and 18S genes (electronic supplementary material, figure S3) clearly placed all individuals into a single MOTU, regardless of morphology or locality. Molecular diversity indices for COI and H3 genes are shown in table 1; the overall negative Fu's Fs that were significant for a number of populations suggest departures from mutation-drift equilibrium. The 18S sequences were identical across all but one individual differing by one base and were not used for population inferences (electronic supplementary material, table S4).
Table 1.
Diversity indices for COI and H3 genes showing sample size (N), haplotype number (Nh), haplotype diversity (h), nucleotide diversity (π), Fu's Fs. OT, Okinawa Trough; SB, Sagami Bay; SCS, South China Sea; Significant differences (p < 0.02) shown in italics.
| genus | locality | COI (517 bp) |
H3 (249 bp) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Nh | h (s.d.) | π (s.d.) | Fu's Fs | N | Nh | h (s.d.) | π | Fu's Fs | ||
| Bathyacmaea | Off Hatsushima (SB, seep) | 23 | 12 | 0.8933 (0.0495) | 0.0039 (0.0025) | −6.8899 | 7 | 3 | 0.7143 (0.1267) | 0.0034 (0.0031) | −0.2373 |
| Iheya North Knoll (OT, vent) | 18 | 9 | 0.8039 (0.0907) | 0.0038 (0.0025) | −3.7540 | — | |||||
| Izena Hole (OT, vent) | 15 | 6 | 0.7905 (0.0785) | 0.0040 (0.0027) | −0.7463 | 9 | 3 | 0.4167 (0.1907) | 0.0018 (0.0020) | −1.0811 | |
| Hatoma Knoll (OT, vent) | 10 | 8 | 0.9333 (0.0773) | 0.0045 (0.0031) | −4.4690 | — | |||||
| Dai-Yon Yonaguni Knoll (OT, vent) | 11 | 5 | 0.7091 (0.1366) | 0.0029 (0.0021) | −1.0844 | — | |||||
| Jiaolong Ridge (SCS, seep) | 4 | 3 | 0.8333 (0.2224) | 0.0019 (0.0019) | −0.8873 | 7 | 3 | 0.5238 (0.2086) | 0.0031 (0.0029) | −0.4377 | |
| Serradonta | Off Hatsushima (SB, seep) | 10 | 7 | 0.9333 (0.0620) | 0.0041 (0.0028) | −3.1329 | 10 | 3 | 0.6000 (0.1305) | 0.0029 (0.0027) | −0.1006 |
| Iheya North Knoll (OT, vent) | 2 | 1 | — | 0.0000 (0.0000) | — | — | |||||
Pairwise Fst estimates among populations using the COI gene (table 2) also indicated a lack of genetic structure between Bathyacmaea and Serradonta morphotypes. Significant genetic differentiation was only detected within Bathyacmaea, between the Izena Hole vent and the Off Hatsushima seep as well as between Jiaolong Ridge seep and four other sites.
Table 2.
Genetic structure analysis of pectinodontids using the COI gene (517 bp), showing the fixation index (Fst). OT, Okinawa Trough; SB, Sagami Bay; SCS, South China Sea. N, sample size. Significant differences (p < 0.05) shown in italics.
| genus | N | locality | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bathyacmaea | 23 | Off Hatsushima (SB, seep) | 1 | — | |||||||
| 18 | Iheya North Knoll (OT, vent) | 2 | 0.0352 | — | |||||||
| 15 | Izena Hole (OT, vent) | 3 | 0.0827 | −0.0038 | — | ||||||
| 10 | Hatoma Knoll (OT, vent) | 4 | −0.0087 | −0.0004 | 0.0426 | — | |||||
| 11 | Dai-Yon Yonaguni Knoll (OT, vent) | 5 | −0.0015 | −0.0433 | 0.0135 | −0.0447 | — | ||||
| 4 | Jiaolong Ridge (SCS, seep) | 6 | 0.3104 | 0.2664 | 0.1391 | 0.3277 | 0.3684 | — | |||
| Serradonta | 10 | Off Hatsushima (SB, seep) | 7 | 0.0279 | 0.0393 | 0.0100 | 0.0216 | 0.0173 | 0.1867 | — | |
| 2 | Iheya North Knoll (OT, vent) | 8 | −0.1236 | −0.1392 | 0.0476 | −0.2209 | −0.1425 | 0.7108 | 0.0486 | — |
Haplotypes generated were deposited under DDBJ/ENA/GenBank accession nos. LC456808–LC456842 and MN130949–MN130950.
4. Discussion
Taken together, our results show that vent/seep pectinodontid limpets sampled between Sagami Bay, Japan and the South China Sea belong to a single species that is highly variable in shell and radula morphology. Unlike suggestions from earlier studies [6,8,25], Bathyacmaea and Serradonta do not represent separate evolutionary lineages that diverged to adapt to different substrate types. Both mitochondrial and nuclear markers indicated genetic homogeneity among Bathyacmaea and Serradonta, supporting them being ecophenotypes of the same taxon. This is also supported by population genetic inferences with similar haplotype and nucleotide diversity between the two for the COI fragment, as well as a lack of genetic structure separating the two (tables 1 and 2). The scarcity of material is a significant limiting factor when studying deep-sea animals, often leading to the underestimation of phenotypic variability, as certainly has been the case for these limpets. The detection of genetic structure between Sagami Bay seep and Izena Hole vents is interesting, although the two still share the main haplotypes (figure 2a). The southernmost Jiaolong Ridge seep was recovered as genetically distinct at a population level compared to four other sites, but it was only represented by four individuals.
The earliest names available among these species are B. nipponica and S. vestimentifericola, published simultaneously in the same work, and are the type species of the two genera [5]. We hereby give precedence to the name B. nipponica as the First Reviser following Art.24.2 of the International Code of Zoological Nomenclature [26]. The names B. secunda, B. tertia and S. vestimentifericola are here formally synonymized with B. nipponica. Accordingly, Serradonta is treated as a junior synonym of Bathyacmaea. Two other named species, B. subnipponica and B. kanesunosensis, from seeps in the Nankai Trough (between Sagami Bay and Okinawa Trough), also likely to belong to the same complex. However, as no samples were collected from type localities, we tentatively treat them as valid species. Genetic sequences of B. lactea described from the South China Sea are very similar to B. becki from the southwest Pacific [23] but differ from B. nipponica (electronic supplementary material, figure S3), indicating that two species of Bathyacmaea inhabit South China Sea seeps.
Shell shape, shell sculpture and radula morphology were clearly determined by separate factors in B. nipponica, and the interplay between development and the environment can result in complex arrangements of morphology in this species. The shell shape is dictated by substrate constraints and easily shifts during life, as has been shown for shallow-water patellogastropod limpets [10]. Shell sculptures ranged between strongly latticed (figure 1a) to smooth (figure 1c) with various intergrades (figure 1b,e); this did not change with the substrate or during rearing. The Oligocene Pectinodonta palaeoxylodia also had smooth shells [13], suggesting either the same range of variation is present in Pectinodonta or P. palaeoxylodia could be a wood-associated Bathyacmaea. Radula morphology was variable but strongly biased according to the substrate across sizes, and therefore, unlikely to be an effect of ontogenetic change or sex [27,28]. Radula types did not shift with substrate translocation, unlike littorinid snails [29,30].
Pectinodontid limpets are well represented in Mesozoic and Cenozoic fossil seeps [8], but their diversity and paleoecology must be re-evaluated considering the results presented herein, especially with regard to habitat selection. It seems that vent/seep pectinodontids originally appeared in Upper Cretaceous as morphologically plastic generalists on various substrate types and remain as such today. Specimens found attached to ataphrid [6] or provannid [8] snails corroborate this and indicate that perhaps in the Upper Cretaceous, these limpets were even less selective in their substrate.
Samples of Bathyacmaea showing clear shifts in shell shape during growth have not been collected from the natural environment. However, one individual of the seep neolepetopsid limpet Paralepetopsis sasakii exhibiting shell shape shift has been reported, preventing the two forms from being described as separate species [31]. Unlike pectinodontids seen in the present study, in P. sasakii the shell sculpture also changed with shell shape. Considering that limpet-form has evolved at least 54 times in gastropods [32], the taxonomic usefulness of each character is unlikely to be transferable across different groups with distinct evolutionary origins. The determination of truly useful taxonomic characters for Bathyacmaea is a subject for future study, but anatomical reconstruction [33] and shell microstructure [34] are promising candidates.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank the captains/crews of R/Vs NATSUSHIMA, KAIYO, KAIREI, SHINSEI-MARU and XIANGYANGHONG 9, pilots of ROVs Hyper-Dolphin and KAIKO, and DSV Jiaolong for sampling. Cruise principal scientists were Ken Takai, KY14-01/NT15-13; Hideaki Machiyama, NT01-05, Jun-ichiro Ishibashi, NT11-20; Shinsuke Kawagucci, KY14-02/KR15-16; Akinori Yabuki, KS-16-04; Feng Liu/Huaiyang Zhou, Dayang-31. Katsunori Kimoto and Yuriko Nakamura assisted with micro-CT, Ryoko Yamazaki helped with sequencing, Nanami Kishigami aided the rearing experiment. Kei Sato and Robert G. Jenkins are thanked for discussions. Comments from David R. Lindberg and two anonymous reviewers helped improve earlier versions.
Data accessibility
The datasets supporting the results of this article are included within the article's electronic supplementary material; DNA sequences generated were deposited under DDBJ/ENA/GenBank accession nos. LC456808–LC456842 and MN130949–MN130950.
Authors' contributions
C.C. conceived and designed the study, collected specimens, carried out experiments and relevant data analyses, and drafted the manuscript. H.K.W. participated in the design of the study, collected specimens, carried out molecular laboratory work and data analyses. Y.N. and T.T. carried out live-rearing experiments and participated in study design. T.X., J.S. and J.-W.Q. collected specimens and carried out part of the molecular sequencing. T.S. participated in the design of the study and collected specimens. All authors revised the manuscript critically for intellectual content gave their final approval for publication. All authors have agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Competing interests
We declare we have no competing interests.
Funding
C.C. and H.K.W. were supported by a JSPS Grant-in-Aid (grant no. 18K06401). T.X. and J.-W.Q. were supported by a General Research Fund from the Hong Kong government (grant no. 12302917).
References
- 1.Marshall CR. 2017. Five palaeobiological laws needed to understand the evolution of the living biota. Nat. Ecol. Evol. 1, 0165 ( 10.1038/s41559-017-0165) [DOI] [PubMed] [Google Scholar]
- 2.Sigwart JD. 2017. Zoology: molluscs all beneath the Sun, one shell, two shells, more, or none. Curr. Biol. 27, R708–R710. ( 10.1016/j.cub.2017.05.075) [DOI] [PubMed] [Google Scholar]
- 3.McLean JH. 1966. West American prosobranch Gastropoda: superfamilies Patellacea, Pleurotomariacea, Fissurellucea PhD dissertation, Biology Stanford University, Stanford, CA, 255 pp. [Google Scholar]
- 4.Vrijenhoek RC. 2013. On the instability and evolutionary age of deep-sea chemosynthetic communities. Deep Sea Res. Pt. II 92, 189–200. ( 10.1016/j.dsr2.2012.12.004) [DOI] [Google Scholar]
- 5.Okutani T, Tsuchida E, Fujikura K. 1992. Five bathyal gastropods living within or near the Caeyptogena-community of the Hatsushima Islet, Sagami Bay. Venus 51, 137–148. ( 10.18941/venusjjm.51.3_137) [DOI] [Google Scholar]
- 6.Jenkins RG, Kaim A, Hikida Y. 2007. Antiquity of the substrate choice among acmaeid limpets from Late Cretaceous chemosynthesis-based communities. Acta Palaeontol. Pol. 52, 369–373. [Google Scholar]
- 7.Sasaki T, Okutani T, Fujikura K. 2003. New taxa and new records of patelliform gastropods associated with chemoautosynthesis-based communities in Japanese waters. Veliger 46, 189–210. [Google Scholar]
- 8.Saether KP, Little CTS, Marshall BA, Campbell KA. 2012. Systematics and palaeoecology of a new fossil limpet (Patellogastropoda: Pectinodontidae) from Miocene hydrocarbon seep deposits, East Coast Basin, North Island, New Zealand with an overview of known fossil seep pectinodontids. Molluscan Res. 32, 1–15. [Google Scholar]
- 9.Grant AR. 1937. A systematic revision of the genus Acmaea Eschscholtz, including consideration of ecology and speciation PhD thesis, University of California, Berkeley, CA. [Google Scholar]
- 10.Lindberg DR, Pearse JS. 1990. Experimental manipulation of shell color and morphology of the limpets Lottia asmi (Middendorff) and Lottia digitalis (Rathke) (Mollusca: Patellogastropoda). J. Exp. Mar. Biol. Ecol. 140, 173–185. ( 10.1016/0022-0981(90)90125-V) [DOI] [Google Scholar]
- 11.Blaxter M, Mann J, Chapman T, Thomas F, Whitton C, Floyd R, Abebe E. 2005. Defining operational taxonomic units using DNA barcode data. Phil. Trans. R. Soc. B 360, 1935–1943. ( 10.1098/rstb.2005.1725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simison WB, Lindberg DR. 1999. Morphological and molecular resolution of a putative cryptic species complex: a case study of Notoacmea fascicularis (Menke, 1851) (Gastropoda: Patellogastropoda). J. Molluscan Stud. 65, 99–109. ( 10.1093/mollus/65.1.99) [DOI] [Google Scholar]
- 13.Lindberg DR, Hedegaard C. 1996. A deep water patellogastropod from oligocene water-logged wood of Washington State, USA (Acmaeoidea: Pectinodonta). J. Molluscan Stud. 62, 299–314. ( 10.1093/mollus/62.3.299) [DOI] [PubMed] [Google Scholar]
- 14.Ogura T, Watanabe HK, Chen C, Sasaki T, Kojima S, Ishibashi J-I, Fujikura K. 2018. Population history of deep-sea vent and seep Provanna snails (Mollusca: Abyssochrysoidea) in the northwestern Pacific. PeerJ 6, e5673 ( 10.7717/peerj.5673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299. [PubMed] [Google Scholar]
- 16.Colgan DJ, McLauchlan A, Wilson GDF, Livingston SP, Edgecombe GD, Macaranas J, Cassis G, Gray MR. 1999. Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Aust. J. Zool. 46, 419–437. ( 10.1071/ZO98048) [DOI] [Google Scholar]
- 17.Palumbi SR. 1996. PCR and molecular systematics. In Molecular systematics, 2nd edn. (eds Hillis D, Moritz C, Mable BK), pp. 205–248. Sunderland, MA: Sinauer Press. [Google Scholar]
- 18.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. ( 10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- 21.Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. ( 10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 22.Okutani T, Fujikura K, Sasaki T. 1993. New taxa and new distribution records of deepsea gastropods collected from or near the chemosynthetic communities in the Japanese waters. Bull. Nat. Sci. Mus. Tokyo, Ser. A 19, 123–143. [Google Scholar]
- 23.Zhang S-Q, Zhang J-L, Zhang S-P. 2016. A new species of Bathyacmaea (Gastropoda: Pectinodontidae) from a methane seep area in the South China Sea. Nautilus 130, 1–4. [Google Scholar]
- 24.Sorensen FE, Lindberg DR. 1991. Preferential predation by American black oystercatchers on transitional ecophenotypes of the limpet Lottia pelta (Rathke). J. Exp. Mar. Biol. Ecol. 154, 123–136. ( 10.1016/0022-0981(91)90078-B) [DOI] [Google Scholar]
- 25.Sasaki T, Okutani T, Fujikura K. 2006. Anatomy of Bathyacmaea secunda Okutani, Fujikura & Sasaki, 1993 (Patellogastropoda: Acmaeidae). J. Molluscan Stud. 72, 295–309. ( 10.1093/mollus/eyl007) [DOI] [Google Scholar]
- 26.International Commission on Zoological Nomenclature (ICZN). 1999. International code of zoological nomenclature, 4th edn. London, UK: The International Trust for Zoological Nomenclature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.deMaintenon M. 2004. Sexually dimorphic radular morphology in Euplica varians and E. versicolor (Neogastropoda: Columbellidae). Molluscan Res. 24, 179–185. ( 10.1071/MR04011) [DOI] [Google Scholar]
- 28.Martínez-Pita I, Guerra-García JM, Sánchez-España AI, García FJ. 2006. Observations on the ontogenetic and intraspecific changes in the radula of Polycera aurantiomarginata García and Bobo, 1984 (Gastropoda: Opisthobranchia) from Southern Spain. Sci. Mar. 70, 8 ( 10.3989/scimar.2006.70n2227) [DOI] [Google Scholar]
- 29.Andrade SCS, Solferini VN. 2005. Transfer experiment suggests environmental effects on the radula of Littoraria flava (Gastropoda: Littorinidae). J. Molluscan Stud. 72, 111–116. ( 10.1093/mollus/eyi051) [DOI] [Google Scholar]
- 30.Padilla DK. 1998. Inducible phenotypic plasticity of the radula in Lacuna (Gastropoda: Littorinidae). Veliger 4, 201–204. [Google Scholar]
- 31.Warén A, Bouchet P. 2009. New gastropods from deep-sea hydrocarbon seeps off West Africa. Deep Sea Res. Pt. II 56, 2326–2349. ( 10.1016/j.dsr2.2009.04.013) [DOI] [Google Scholar]
- 32.Vermeij GJ. 2016. The limpet form in gastropods: evolution, distribution, and implications for the comparative study of history. Biol. J. Linn. Soc. 120, 22–37. ( 10.1111/bij.12883) [DOI] [Google Scholar]
- 33.Chen C, Linse K, Uematsu K, Sigwart JD. 2018. Cryptic niche switching in a chemosymbiotic gastropod. Proc. R. Soc. B 285, 20181099 ( 10.1098/rspb.2018.1099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuchigami T, Sasaki T. 2005. The shell structure of the recent Patellogastropoda (Mollusca: Gastropoda). Paleontol. Res. 9, 143–168. ( 10.2517/prpsj.9.143) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the results of this article are included within the article's electronic supplementary material; DNA sequences generated were deposited under DDBJ/ENA/GenBank accession nos. LC456808–LC456842 and MN130949–MN130950.