Abstract
Revegetation plantings are a key management tool for ecological restoration. Revegetation success is usually measured using ecological traits, however, genetic diversity should also be considered as it can influence fitness, adaptive capacity and long-term viability of revegetation plantings and ecosystem functioning. Here we review the global literature comparing genetic diversity in revegetation plantings to natural stands. Findings from 48 studies suggest variable genetic outcomes of revegetation, with 46% demonstrating higher genetic diversity in revegetation than natural stands and 52% demonstrating lower diversity. Levels of genetic diversity were most strongly associated with the number of source sites used—where information was available, 69% of studies showing higher genetic diversity in revegetation reported using multiple provenances, compared with only 33% for those with lower diversity. However, with a few exceptions, it was unclear whether differences in genetic diversity between revegetation and natural stands were statistically significant. This reflected insufficient reporting of statistical error and metadata within the published studies, which limited conclusions about factors contributing to patterns. Nonetheless, our findings indicate that mixed seed sourcing can contribute to higher genetic diversity in revegetation. Finally, we emphasize the type of metadata needed to determine factors influencing genetic diversity in revegetation and inform restoration efforts.
Keywords: ecological restoration, genetic diversity, heterozygosity, industry practices, provenancing, seed sourcing
1. Introduction
Effective restoration of degraded ecosystems is essential for improving the status of native biodiversity and ecosystem services [1,2]. Revegetation—the seeding or planting of native species—is commonly used to restore degraded ecosystems and habitats affected by anthropogenic changes such as agriculture, forestry, urbanization, mining [3] and altered water quality [4]. Annually, an estimated $US2 trillion are spent worldwide on restoration [5], with continued and growing investment in restoration facilitated by large global efforts such as The Bonn Challenge, aiming to restore 350 million hectares of degraded land by 2030 (http://www.bonnchallenge.org/content/challenge; accessed 5 March 2019). Understanding the factors that influence the long-term viability of restoration efforts will be essential for maximizing outcomes of these investments, especially under climate change and other environmental changes.
To date, there has been little emphasis on assessing genetic outcomes of revegetation plantings [6,7], despite the importance of genetic variation in revegetation being recognized for some time [7,8]. Traditionally, restoration success has been measured by three ecosystem attributes: species and functional diversity, vegetation structure and ecological processes [9,10]. Evaluating the genetic diversity of plantings used in revegetation (rather than forestry plantations) compared with natural stands provides an evolutionary perspective for determining restoration success. Overall genetic diversity is positively associated with population fitness in sexually reproducing species [11,12], suggesting that standing genetic variation within populations is closely tied to adaptive capacity and resilience to environmental change (e.g. [13]). Both empirical and theoretical evidence suggest that the long-term viability of populations will depend upon capturing genetic variation to enable efficient selection and facilitate adaptation to environmental change (e.g. [14,15]), as well as maximizing population sizes to avoid risks associated with inbreeding, genetic load and drift [12,16,17]. Maintaining genetic diversity to support adaptability is particularly pertinent given projections of rapid climate change, compounded by additional stresses such as habitat fragmentation, ecosystem degradation and the unprecedented spread and proliferation of invasive species [17–20]. Furthermore, genetic diversity can influence ecosystem function [21], for example enhancing habitat and nutrient retention [22] as well as recovery following disturbance [13]. Consequently, broadening the genetic basis of revegetation plantings may be critical for enhancing their long-term viability and adaptive potential across generations under environmental change [18,23,24].
While the importance of capturing genetic diversity from natural populations for revegetation purposes is widely appreciated, a multitude of factors can compromise the revegetation process, leading to suboptimal genetic outcomes. For example, in fragmented landscapes, traditional seed sourcing strategies that favour collecting material from the local geographical area—‘local provenancing’—can result in sourcing seed from small isolated remnant populations lacking adequate genetic diversity [25]. Logistics of seed collection and industry demands for cost-effective seedlings can limit the number of parent plants targeted for seed and thus the genetic diversity that is introduced into revegetation plantings [8,26,27]. Selecting source material from distant populations—‘non-local provenancing’—may result in maladapted germplasm or outbreeding depression in revegetation plantings, leading to poor plant performance, especially in cases of strong local adaptation [20,25]. However, risks associated with mixing gene pools are often overstated [28] and sourcing strategies advocating the broader genetic sampling, including non-local genotypes, are gaining momentum [25,29–31]. Specifically, the inclusion of climate-adapted genotypes—genotypes from regions of predicted future climates—is being widely encouraged based on the assumption that local seed sources may be maladaptive under future environmental conditions [29,30], while recognizing these predictions remain largely untested. Consequently, the long-term success of revegetation plantings is dependent on understanding factors that influence the capture of genetic diversity and the establishment of industry guidelines that promote optimal genetic outcomes.
The aim of this study was to review restoration outcomes from a genetic perspective, through a synthesis of studies that provide estimates of genetic diversity for both revegetation plantings and natural stands. In particular, we asked how well revegetation plantings capture genetic diversity compared with natural stands, and what restoration practices influence success in capturing this diversity. We hypothesized that genetic outcomes of revegetation plantings would be variable and dependent on seed sourcing strategies. We discuss trends identified in this review, highlight data required to improve future assessments and provide recommendations to address knowledge gaps to guide ongoing restoration efforts.
2. Literature review and analysis
We ran a Web of Science literature search on 22 January 2018 using the following search terms: TS = ((restor* OR reveg*) AND (genetic* OR genom*) AND (diversity OR allel* OR heterozygo* OR distance) AND (plant* OR tree* OR shrub* OR grass* OR herb* OR annual OR perennial)). The resulting 1381 publications were filtered to only those studies comparing genetic diversity in both revegetation planting(s) (hereafter referred to as revegetation) and natural stand(s). Studies matching this criterion were included irrespective of whether the natural stand was the seed source for the revegetation or simply a reference natural stand, and irrespective of the cohort sampled. We excluded studies where plantings were not part of ecosystem restoration plantings (e.g. botanic garden plantings), used bred cultivars, involved natural colonization events (e.g. following site restoration or regeneration from natural soil seed banks) or were non-restoration experimental plantings (e.g. provenance trials). Three studies with restoration experimental plantings were retained as these formed part of wider restoration projects and remained focused on restoration methods and outcomes [32–34]. Conference abstracts with unavailable datasets were also excluded.
Genetic diversity data were mined from studies, including the genetic metric used and associated error (where available), sample size, type and number of molecular markers. Data in graphs were extracted [35] using DataThief (III v 1.7; [36])—a program shown to have both high validity and reliability [37]. Estimates of H [38], Hs [39] and Nei's h [32,40] were included under expected heterozygosity (He). To increase sample sizes, estimates of unbiased expected heterozygosity were converted back to He using the equations of Nei [41] (see Supplementary Methods in the electronic supplementary material). Where present, 95% confidence intervals (CI) were converted to standard errors (s.e.; see Supplementary Methods in the electronic supplementary material). Where necessary, reported numbers of polymorphic loci were converted to a percentage to match the majority of data for this metric [33,42]. For Li et al., 2005 [43], P99 estimates were used for percentage polymorphic loci.
Where available in the published study, we also collected information on the seed sourcing approach and restoration practices uses (‘metadata’, electronic supplementary material, appendix A1). This metadata included the number of source sites and age of planting. Additional metadata, such as number of sampled plants from source sites, were not collected as they were either not available or not reported frequently enough for analysis. Based on best assessment from the information provided in a study, the cohort sampled was assigned as ‘adult’ (adult in natural stands or original plants in revegetation), ‘seed’ (seed collected from ‘adult’ plants) or ‘progeny’ (juvenile plants in natural stands or F1 or higher generation in revegetation). Latest plant family names were used as per ‘The Plant List’ (http://www.theplantlist.org; accessed June 2018).
Data from specific publications were further filtered and processed to partition multiple, independent datasets within a publication into separate studies [33,44–48]; to exclude natural colonization sites or revegetation sites where natural individuals were still present [38,47,49]; to include individual site estimates rather than across site averages [39,50]; to include ‘large’ rather than ‘small’ natural remnants as natural site comparisons [51]; to ensure a single estimate per genetic metric per site [52]; and in one case to include natural stand data from a complementary study [53,54] (see Supplementary Methods in the electronic supplementary material for more detail).
Where data for multiple cohorts were available [35,39,55–60], the adult cohort was retained and seed or juvenile data excluded from the primary analysis, in line with the majority of studies reporting adult data (seed and juvenile data were used later to explore the potential effect of cohort sampled; see below). Similarly, only the most recent data—i.e. data collected at the longest time since planting—were included for studies with multiple time points [32,34].
Too few studies reported errors around metrics to allow for formal meta-analysis (17/48; 35%). We assessed correlations between the error and actual estimates of expected heterozygosity (He) data (the metric with the largest dataset) to see if error could be approximated based on available data (see Supplementary Methods in the electronic supplementary material). Correlations were generally too weak to allow for estimates of error (|r| < 0.55; see Supplementary Methods in the electronic supplementary material; electronic supplementary material, figure S1). Consequently, genetic diversity estimates without error were used for further analyses.
Patterns of genetic diversity between natural stands and revegetation were explored using metrics with sufficient representation across studies (n ≥ 17). Estimates of expected heterozygosity (He, n = 41), number of alleles (A, n = 24), allelic richness (Ar, n = 17) and percentage polymorphic loci (P, n = 27) were contrasted. Expected heterozygosity was used in favour of observed heterozygosity owing to a greater reporting of this metric (41 versus 24 studies, respectively) and a high correlation between the two metrics (adjusted r2 = 0.70 and 0.91 for natural stands and revegetation, respectively, from studies reporting both metrics, n = 23). In addition to genetic diversity, estimates of inbreeding (fixation) were compared (F, n = 18), including F, FIS, f and fAFLP estimates.
To compare natural stands and revegetation, average genetic diversity and inbreeding estimates for each site type within each study was calculated in R v. 3.5.0 [61], with range data recorded to allow for variation among sites in lieu of error estimates. Studies were subsequently binned in the following hierarchy:
Q1 Is the average genetic diversity in revegetation higher or lower than natural stands?
Q2 Is the average genetic diversity in revegetation within the range of estimates seen in natural stands? That is, despite average genetic diversity being higher or lower, does the value fall within the bounds of variation seen between natural stands (for cases where more than 1 natural stand was included)?
Q3 Do the estimated ranges for genetic diversity of revegetation and natural stands overlap, despite the average for revegetation not falling within the range of natural stands? That is, does between-site variation observed in natural stands and revegetation plantings overlap (for cases where genetic diversity estimates for more than 1 stand for both natural and revegetation sites is given)?
In lieu of a formal meta-analysis, a single average s.e. for each site type—revegetation plantings and natural stands—was calculated for the small subset of studies with error data (n = 17 studies in total; He = 12, A = 8, Ar = 6, P = 1, F = 4). The average error was used as a proxy for assessing the influence of statistical error on results, with studies binned as having higher or lower average genetic diversity in revegetation plantings compared to natural stands based on average diversity ±1 average s.e. Where appropriate, s.d. was converted to s.e. (see Supplementary Methods in the electronic supplementary material).
We explored whether the number of planted seed sources influenced the genetic diversity captured in revegetation by comparing results for Q1 to the effect of using single or multiple (more than 1 for at least one revegetation planting within the study) sources, where data were available. We assessed the influence of cohort sampled on the Q1 results, as the level of genetic diversity in revegetation plantings may change over generations, with differences between natural stands and revegetation plantings potentially more prevalent in subsequent generations (primarily owing to drift or gene flow from external sources). For this analysis only, we included all cohort data for those studies that sampled multiple cohorts [35,39,55–60]. Here, we calculated average genetic diversity for each cohort within natural stands and revegetation plantings, allowing us to contrast diversity estimates between ‘adults’ and ‘F1’ (‘seed’ or ‘progeny’). We assessed the number of seed sources and cohort effects on observed differences in genetic diversity between natural stands and revegetation plantings (Q1) using chi-squared tests.
We compared average genetic diversity in revegetation plantings and their natural seed source(s) using a subset of studies that cited the natural stand source site(s) used for the revegetation planting. We calculated average genetic diversity for natural source stands and revegetation plantings within a study for the four diversity metrics—He, A, Ar and P—and scored as higher or lower in revegetation plantings as per Q1.
Finally, we explored publication bias by investigating year of publication versus overall results of average genetic diversity in revegetation being higher, lower or the same compared to natural stands (Q1). We used this approach because traditional meta-analysis tests for publication bias (e.g. funnel plots) were not possible using the data available, as there was a lack of reporting of statistical error. Instead we used chi-squared tests of year of publication versus revegetation category, using 5 year windows beginning at 1995, to assess these differences. While this approach is not directly testing for publication bias, it does at least test for potential temporal changes in revegetation strategies that may, or may not, influence the genetic outcomes considered here.
3. Genetic outcomes in revegetation plantings
We identified 48 studies that compared genetic diversity in natural stands and revegetation plantings, from 41 published journal articles, representing 41 different plant species (table 1; electronic supplementary material, appendix A1; appendix A).
Table 1.
Counts of studies comparing genetic diversity in revegetation plantings to natural stands by marker, geographical location, habitat and plant form. Numbers in brackets indicate numbers of different species.
| markers |
location |
habitat |
forma |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| microsatellite | 21 | (19) | North America | 20 | (15) | terrestrial | 39 | (36) | tree | 21 | (19) |
| dominant marker | 18 | (17) | Australia/New Zealand | 9 | (7) | aquatic | 9 | (5) | forb | 17 | (16) |
| codominant enzyme | 8 | (6) | Asia | 8 | (8) | grass | 10b | (6) | |||
| SNPc | 1 | (1) | Europe | 7 | (7) | ||||||
| South America | 4 | (4) |
aApproximate form.
bThree terrestrial and seven seagrass studies; three terrestrial species and three aquatic species.
cSingle nucleotide polymorphism.
The four genetic diversity metrics (He, A, Ar and P) demonstrated consistent outcomes with average genetic diversity in revegetation plantings consistently higher, lower or equal to natural stands across all metrics. Three exceptions to this were Broadhurst, 2011 [62] and Fant et al., 2013 [50] (A lower in revegetation plantings while He and Ar were higher) and Pakkad et al., 2008 [58] (He lower in revegetation plantings while A was higher). To enable a single binning for the three questions across all metrics, these three studies were scored as per results for He (most frequently reported metric; [50,62] = higher; [58] = lower).
(a). Is average genetic diversity in revegetation higher or lower than natural stands (Q1)?
Overall, the results showed a near even split in the number of studies finding average genetic diversity in revegetation to be higher than natural stands (n = 22, 46% of studies) compared to studies finding lower genetic diversity in revegetation (n = 25, 52% of studies; figure 1, electronic supplementary material, figure S2). Only a single study found equal diversity between natural stands and revegetation across all tested metrics ([44] ‘a’). Analyses suggested no evidence of publication bias, based on year of publication, with studies reporting both higher and lower genetic diversity captured in revegetation irrespective of time (electronic supplementary material, figure S3; χ2 = 2.659, d.f. = 3, p = 0.447, using 5 year bins and excluding two categories with only a single record—year = 1995–1999 and comparative diversity = ‘same’; electronic supplementary material, table S1a).
Figure 1.
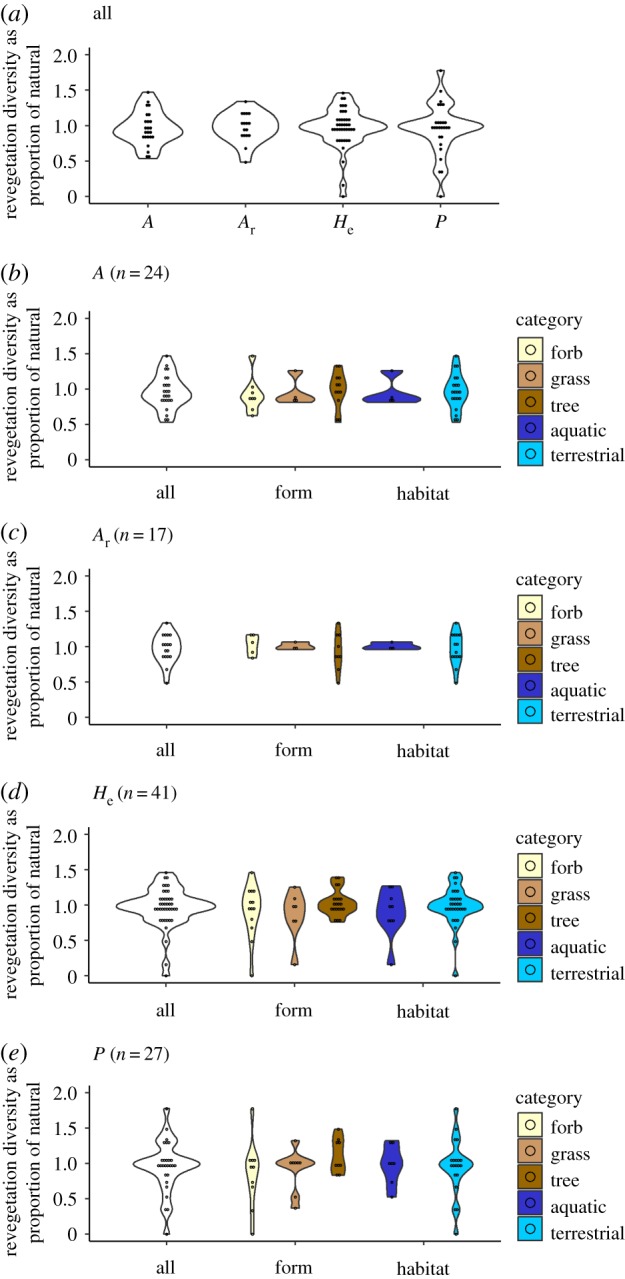
Average genetic diversity in revegetation as a proportion of average genetic diversity in natural stands. (a) All studies by genetic diversity metric. (b–e) Proportion diversity by metric (white) and by ‘form’ (browns) or ‘habitat’ (blues) within metric. A, number of alleles; Ar, allelic richness; He, expected heterozygosity; P, percentage polymorphic loci.
(b). Is average diversity in revegetation within the range of natural stands (Q2)?
Estimates for more than one natural stand were reported in 35 studies, providing the range data required to address Q2. Of these 35 studies, data were split approximately into thirds between revegetation having average genetic diversity within, above or below natural variation (electronic supplementary material, appendix A1). Average genetic diversity of revegetation was within the range of natural stand estimates for all metrics reported in 14 studies (40%), irrespective of which site type had the higher genetic diversity. Nine studies (26%) had average genetic diversity of revegetation both higher than average natural diversity and above the range of natural stand estimates for at least one metric. Similarly, 12 (34%) studies had average genetic diversity in revegetation both lower than average natural diversity and below the range of natural stand estimates for at least one metric.
(c). Is there overlap in the range of diversity estimates for natural stands and revegetation (Q3)?
Variation in diversity estimates overlapped between revegetation and natural stands in the majority of cases (electronic supplementary material, appendix A1). Of those studies where average genetic diversity in revegetation was above or below the range of natural stand estimates for at least one metric, 15 studies had more than one revegetation estimate reported and thus range data available for both site types. Overlapping variation in estimates was found for nine studies (60%), with no overlap for at least one metric in the remaining six studies (40%). For these latter six studies, average genetic diversity in revegetation was higher than natural stands in two studies and lower in four studies.
(d). Additional comparisons—inbreeding, source sites, cohort and reported error
Comparisons of inbreeding between site types showed similar patterns to that of genetic diversity. For half the studies reporting inbreeding, revegetation was more inbred—higher positive F estimate—than natural stands (9/18; electronic supplementary material, appendix A1 and figure S4). Despite being more inbred, revegetation estimates in these studies were still generally within the range of natural stand estimates (Q2, 5/7 studies with range data) or had overlapping variation with natural stand estimates (Q3, 6/6 studies with available data). Revegetation was less inbred than natural stands in 5 of the 18 studies, having either lower positive F values than natural stands (n = 3) or negative F estimates compared to positive natural stand F estimates (n = 2). As well as being less inbred, revegetation in these studies was below the natural range (4/4 studies with range data, Q2), though all studies except one had overlapping ranges (3/4, Q3). For the remaining four studies, inbreeding estimates were negative for both natural stands and revegetation, with revegetation being the same as natural stands (one study) or having values closer to 0 (three studies). For these latter three studies, one was within the range of natural estimates, another was above the range of natural estimates with no overlap in variation between site types and the third lacked range data.
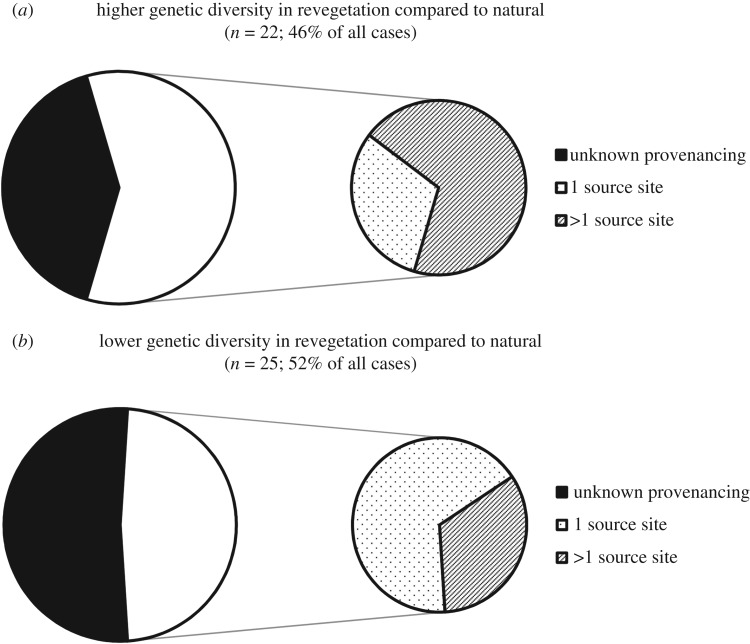
Based on 26 studies reporting the number of source sites, revegetation plantings with higher average genetic diversity compared to natural stands had more frequently been established from multiple source sites than was the case for revegetation with lower diversity (69% and 33% respectively; figure 2). Conversely, a single source site was more common for revegetation with lower average genetic diversity than natural stands (67%; figure 2). This difference was nearly significant (χ2 = 3.222, d.f. = 1, p = 0.073; n = 25, excludes ‘same’ genetic diversity category with only a single record; electronic supplementary material, table S1b).
Figure 2.
Proportion of studies citing single (1) or multiple (more than 1 for at least one revegetation planting) sites used for revegetation seed sourcing. Data divided into studies where revegetation had higher (a) or lower (b) genetic diversity than natural stands. Left hand pie charts = proportion of studies with known (white) and unknown (black) number of source sites. Right hand pie charts = proportion of studies with known number of source sites that used single (dots) or multiple (lines) seed source sites.
The cohort sampled in either revegetation or natural stands did not influence the overall results. The number of records for exploring the effect of cohort sampled increased to 66 when including all cohort estimates for studies sampling multiple cohorts (n = 8 studies), and all pairwise cohort comparisons between site types within these studies. Average genetic diversity in revegetation was lower than natural stands more often when comparing estimates from revegetation ‘F1’ (‘seed’ or ‘progeny’) cohorts with natural ‘adult’ cohorts (85%; 11/13 comparisons), than when comparing estimates from the same cohort in both site types (54%, 19/35 comparisons for ‘adult to adult’ or 64%, 7/11 comparisons for ‘F1 to F1’ comparisons; electronic supplementary material, table S1c). This difference, however, was not significant (excluding ‘same’ genetic diversity category with only a single record; n = 65, χ2 = 4.437, d.f. = 3, p = 0.218; electronic supplementary material, table S1c). Within the eight studies that sampled across multiple cohorts, three had the same result—average genetic diversity higher or lower (Q1)—irrespective of cohort sampled [35,39,59]. Results for the remaining five studies differed depending on cohort sampled and metric assessed, however there was no consistent trend (electronic supplementary material, table S2).
For comparisons of cited source site versus revegetation, results were again generally consistent across the four diversity metrics, with revegetation having either the same and higher (considered ‘higher’), or the same and lower (considered ‘lower’) average genetic diversity across measured metrics within a study. Average genetic diversity was lower in revegetation compared to source sites for half the source-revegetation comparisons (14/28; 50%). Approximately one third had higher diversity in the revegetation plantings (35.7%; 10/28) and one study reported consistent estimates between revegetation and source sites across all measured metrics. The three remaining studies had mixed results—higher He though lower A and Ar in revegetation [63]; higher A though lower He [58]; higher Ar though lower He and A (one pairing in [50]).
For the small subset of studies reporting error estimates (He, n = 12; A, n = 8; Ar, n = 6; P, n = 1, F, n = 4), average genetic diversity in revegetation and natural stands mostly did not differ by more than 1 s.e. (averaged). Exceptions involved two He comparisons, one where average genetic diversity in revegetation was higher and one where it was lower than in natural stands.
4. Discussion
Our review provides a genetic assessment of revegetation outcomes from around the world. Our analysis of 48 suitable studies indicated varied genetic outcomes in revegetation plantings, with approximately half the studies showing higher average diversity in revegetation plantings compared to natural stands, and half showing lower diversity. Our analyses also suggested that genetic diversity in revegetation plantings typically does not differ greatly from natural variability, with a few exceptions. These findings indicate that revegetation plantings do have the capacity to restore natural levels of genetic diversity to sites. However, this can vary greatly and those factors influencing this variability remain unclear. Here we discuss the trends found in this review, highlight key information and knowledge gaps and conclude with recommendations to improve future assessments of factors influencing genetic diversity in revegetation plantings.
Our analysis suggests that higher diversity in revegetation plantings compared to natural stands may be more likely when seed from multiple source sites is used, although this result was only nearly significant. This provides support for ‘mixed provenancing’—the use of seed from multiple source sites across the landscape—as a tool for broadening the genetic base of revegetation plantings, with potential benefits for enhancing fitness, adaptability and resilience to environmental change of replanted populations [25,29]. Evolutionary and ecological benefits of higher genetic diversity sourced from multiple populations still require further characterization, but evidence from empirical studies of plant re-introductions suggests improved population ecological outcomes in plantings established using multiple seed sources [64]. Furthermore, high genetic diversity generally may enhance adaptability, especially where there is uncertainty around the fitness of different genetic variants or provenances under future environmental change [18]. The lower diversity in revegetation plantings attributed to using a single seed source is likely owing to lower total diversity in single versus multiple natural populations, but could also be owing to a lower total number of sampled parent plants—a likely outcome of collecting from one rather than multiple populations, or the use of seed from genetically depauperate sources, such as small, isolated remnants in highly modified landscapes [25]. Sampling from a large number of parents from large, genetically diverse sites could explain the higher genetic diversity of revegetation plantings in some studies despite only a single source site being used.
Though not significant, the possible effect found in this review of cohort sampled on differences in genetic diversity in revegetation plantings compared to natural stands highlights the need to consider change over time, especially under environmental change. For example, some studies indicated that potential gene flow from external sources may be enhancing genetic diversity within revegetation plantings (e.g. [52,65]). Gene flow could assist in enhancing effective population size and adaptive genetic diversity, and thus the potential fitness and adaptability of revegetation plantings aiming to maintain and restore ecosystem services [66–68]. Understanding changes to genetic diversity over time, including the potential influences of gene flow, will help meet the overall objective of local revegetation efforts. By undertaking genetic comparisons between stands across a longer time interval, it also becomes possible to encompass the impact of other anthropogenic processes on genetic diversity, such as clearing of old growth forests (e.g. [69]).
Determining the statistical and biological significance of differences observed in this review was hindered by two key information gaps—(i) the lack of error estimates for genetic diversity metrics and (ii) the absence of information (metadata) relating to restoration practices used for the revegetation plantings. Whether differences observed represent true increases or decreases in diversity compared with natural stands, or simply non-significant random variation is difficult to assess in the absence of error estimates. Similarly, in the absence of error estimates it is difficult to determine the effect size of factors that might influence genetic diversity. Diversity estimates should become more accurate as genetic measures of diversity shift from being based on microsatellites to being based on thousands of SNPs, with only one study included in this review using SNPs (although another study using SNPs has been published subsequent to our review period [70]).
While our findings suggest patterns of genetic diversity in revegetation plantings are likely to be influenced by seed sourcing strategies, the majority of variation found in this review could not be explained owing to a lack of information on restoration practices. Moving forward, especially as alternative seed sourcing strategies for revegetation emerge [29,30], it will be important to understand the influence of revegetation practices, in particular different seed sourcing strategies, on capturing genetic diversity in revegetation and enhancing evolutionary and ecosystem-level outcomes. Furthermore, it may be important to consider not only overall genetic diversity, but the types of genetic variants included within revegetation plantings. For instance, climate-adjusted provenancing for revegetation is gaining increasing interest internationally [30,66]. This approach aims to improve adaptive potential of plantings by including seed from climatically different areas, and thus to capture both genetic diversity and pre-adapted genotypes, into seed mixes. However, the effectiveness of this approach remains untested from both genetic and ecological perspectives.
5. Conclusion and recommendations
We conclude that revegetation plantings have the potential to capture levels of genetic diversity similar to those of natural stands, and that this may be facilitated by using multiple seed sources. However, current genetic outcomes in revegetation appear variable and the reasons for this variation remain unclear. Improvements to revegetation practices are dependent on adequate metadata allowing for meta-analyses to identify factors that influence restoration success. With globally increasing investments in ecological restoration, such assessments will be essential for guiding future revegetation strategies, especially in the face of ongoing environmental change [2]. In light of these findings, we propose the following recommendations to improve our understanding of the links between revegetation practices, genetic diversity, and the long-term viability and resilience of restoration investments.
-
1.
Record and report restoration practices used in revegetation plantings to enable assessments of different approaches on genetic, and ecological, outcomes. This should include the number and location of source sites, year of establishment and size of the original planting (number of plants, size of planting area). Additional useful information includes the number of plants sampled at each source site, size of source population (number of individuals and geographical area) and type of material planted (seed versus seedlings).
-
2.
Monitor and evaluate changes in genetic diversity over time, especially where comparisons can encompass multiple generations. This type of information can help assess the potential long-term effects of any loss of genetic diversity in revegetation or whether this can be restored through ongoing gene flow.
-
3.
Report standard errors and sample sizes associated with genetic metrics to enable robust statistical comparisons within and across studies. This type of information is essential for any future meta-analyses and can help in identifying factors influencing genetic diversity across studies.
Supplementary Material
Supplementary Material
Acknowledgements
Thank-you to Karen Bell, Peter Harrison and five anonymous reviewers whose constructive feedback greatly improved this manuscript.
Appendix A. List of 41 publications used in the analysis of genetic diversity in revegetation plantings compared to natural stands
Aavik T, Edwards PJ, Holderegger R, Graf R, Billeter R. 2012 Genetic consequences of using seed mixtures in restoration: a case study of a wetland plant Lychnis flos-cuculi. Biol. Conserv. 145, 195–204. (doi:10.1016/j.biocon.2011.11.004)
Alonso MA, Guillo A, Perez-Botella J, Crespo MB, Juan A. 2014 Genetic assessment of population restorations of the critically endangered Silene hifacensis in the Iberian Peninsula. J. Nature Conserv. 22, 532–538. (doi:10.1016/j.jnc.2014.08.007)
Álvares-Carvalho SV Silva-Mann R, Gois IB, Melo MFV, Oliveira AS, Ferreira RA, Gomes LJ. 2017 Restoration over time and sustainability of Schinus terebinthifolius Raddi. Genet. Mol. Res. 16, gmr16029669. (doi:10.4238/gmr16029669)
Broadhurst LM. 2013 A genetic analysis of scattered Yellow Box trees (Eucalyptus melliodora A.Cunn. ex Schauer Myrtaceae) and their restored cohorts. Biol. Conserv. 161, 48–57. (doi:10.1016/j.biocon.2013.02.016)
Broadhurst LM. 2011 Genetic diversity and population genetic structure in fragmented Allocasuarina verticillata (Allocasuarinaceae)—implications for restoration. Austral. J. Botany 59, 770–780. (doi:10.1071/BT11253)
Burgarella C, Navascues M, Soto A, Lora A, Fici S. 2007 Narrow genetic base in forest restoration with holm oak (Quercus ilex L.) in Sicily. Annals Forest Sci. 64, 757–763. (doi:10.1051/forest:2007055)
Campanella JJ, Bologna PAX, Smalley JV, Avila DN, Lee KN, Areche EC, Slavin LJ. 2013 An analysis of the population genetics of restored Zostera marina plantings in Barnegat Bay New Jersey. Popul. Ecol. 55, 121–133. (doi:10.1007/s10144-012-0351-4)
Czarnecki DM, Rao MN, Norcini JG, Gmitter Jr FG, Deng Z. 2008 Genetic diversity and differentiation among natural, production, and introduced populations of the narrowly endemic species Coreopsis leavenworthii (Asteraceae). J. Am. Soc. Hortic. Sci. 133, 234–241. (doi:10.21273/JASHS.133.2.234)
Dolan RW, Marr DL, Schnabel A. 2008 Capturing genetic variation during ecological restorations: an example from Kankakee Sands in Indiana. Restor. Ecol. 16, 386–396. (doi:10.1111/j.1526-100X.2007.00318.x)
Fant JB, Holmstrom RM, Sirkin E, Etterson JR, Masi S. 2008 Genetic structure of threatened native populations and propagules used for restoration in a clonal species American beachgrass (Ammophila breviligulata Fern.). Restor. Ecol. 16, 594–603. (doi:10.1111/j.1526-100X.2007.00348.x)
Fant JB, Kramer A, Sirkin E, Havens K. 2013 Genetics of reintroduced populations of the narrowly endemic thistle, Cirsium pitcheri (Asteraceae). Botany 91, 301–308. (doi:10.1139/cjb-2012-0232)
Fotinos TD, Namoff S, Lewis C, Maschinski J, Griffith MP, von Wettberg EJB. 2015 Genetic evaluation of a reintroduction of Sargent's Cherry Palm, Pseudophoenix sargentii. J. Torrey Botanical Soc. 142, 51–62. (doi:10.3159/TORREY-D-14-00004.1)
Frick KM, Ritchie AL, Krauss SL. 2014 Field of dreams: restitution of pollinator services in restored bird-pollinated plant populations. Restor. Ecol. 22, 832–840. (doi:10.1111/rec.12152)
Gustafson DJ, Gibson DJ, Nickrent DL. 2002 Genetic diversity and competitive abilities of Dalea purpurea (Fabaceae) from remnant and restored grasslands. Int. J. Plant Sci. 163, 979–990. (doi:10.1086/342709)
Gustafson DJ, Gibson DJ, Nickrent DL. 2004 Competitive relationships of Andropogon gerardii (Big Bluestem) from remnant and restored native populations and select cultivated varieties. Funct. Ecol. 18, 451–457. (doi:10.1111/j.0269-8463.2004.00850.x)
Jordan R, Dillon SK, Prober SM, Hoffmann AA. 2016 Landscape genomics reveals altered genome wide diversity within revegetated stands of Eucalyptus microcarpa (Grey Box). New Phytol. 212, 992–1006. (doi:10.1111/nph.14084)
Li YY, Chen XY, Zhang X, Wu TY, Lu HP, Cai YW. 2005 Genetic differences between wild and artificial populations of Metasequoia glyptostroboides: implications for species recovery. Conserv. Biol. 19, 224–231. (doi:10.1111/j.1523-1739.2005.00025.x)
Li H, Chen G. 2009 Genetic variation within the endangered mangrove species Sonneratia paracaseolaris (Sonneratiaceae) in China detected by inter-simple sequence repeats analysis. Biochem. Syst. Ecol. 37, 260–265. (doi:10.1016/j.bse.2009.04.005)
Liu M-H, Chen X-Y, Zhang X, Shen D-W. 2008 A population genetic evaluation of ecological restoration with the case study on Cyclobalanopsis myrsinaefolia (Fagaceae). Plant Ecol. 197, 31–41. (doi:10.1007/s11258-007-9357-y)
Lloyd MW, Burnett Jr RK, Engelhardt KAM, Neel MC. 2012 Does genetic diversity of restored sites differ from natural sites? A comparison of Vallisneria americana (Hydrocharitaceae) populations within the Chesapeake Bay. Conserv. Genet. 13, 753–765. (doi:10.1007/s10592-012-0324-3)
Neto OC, Aguiar AV, Twyford AD, Neaves LE, Pennington RT, Lopes AV. 2014 Genetic and ecological outcomes of Inga vera subsp affinis (Leguminosae) tree plantations in a fragmented tropical landscape. PLoS ONE 9, e99903. (doi:10.1371/journal.pone.0099903)
Pakkad G, Al Mazrooei S, Blakesley D, James C, Elliott S, Luoma-Aho T, Koskela J. 2008 Genetic variation and gene flow among Prunus cerasoides DDn populations in northern Thailand: analysis of a rehabilitated site and adjacent intact forest. New For. 35, 33–43. (doi:10.1007/s11056-007-9059-2)
Ramp JM, Collinge SK, Ranker TA. 2006 Restor. genetics of the vernal pool endemic Lasthenia conjugens (Asteraceae). Conserv. Genet. 7, 631–649. (doi:10.1007/s10592-005-9052-2)
Reiker J, Schulz B, Wissemann V, Gemeinholzer B. 2015 Does origin always matter? Evaluating the influence of nonlocal seed provenances for ecological restoration purposes in a widespread and outcrossing plant species. Ecol. Evol. 5, 5642–5651. (doi:10.1002/ece3.1817)
Reynolds LK, Waycott M, McGlathery KJ, Orth RJ, Zieman JC. 2012 Eelgrass restoration by seed maintains genetic diversity: case study from a coastal bay system. Marine Ecol. Progr. Ser. 448, 223–233. (doi:10.3354/meps09386)
Ritchie AL, Krauss SL. 2012 A genetic assessment of ecological restoration success in Banksia attenuata. Restor. Ecol. 20, 441–449. (doi:10.1111/j.1526-100X.2011.00791.x)
Ritchie AL, Nevill PG, Sinclair EA, Krauss SL. 2017 Does restored plant diversity play a role in the reproductive functionality of Banksia populations? Restor. Ecol. 25, 414–423. (doi:10.1111/rec.12456)
Severns PM. 2013 Genetic differentiation in an artificial population of the threatened plant Lupinus oreganus (Fabaceae). Botany 91, 319–322. (doi:10.1139/cjb-2012-0278)
Smulders MJM, van der Schoot J, Geerts R, Antonisse-de Jong AG, Korevaar H, van der Werf A, Vosman B. 2000 Genetic diversity and the reintroduction of meadow species. Plant Biol. 2, 447–454. (doi:10.1055/s-2000-6780)
Souza EMS, Pereira GS, Silva-Mann R, Alvares-Carvalho SV, Ferreira RA. 2016 A comparative framework of the Erythrina velutina tree species in reforested land and native populations. Genet. Mol. Res. 15, gmr15028534. (doi:10.4238/gmr.15028534)
Stevens MI, Clarke AC, Clarkson FM, Goshorn M, Gemmill CEC. 2015 Are current ecological restoration practices capturing natural levels of genetic diversity? A New Zealand case study using AFLP and ISSR data from mahoe (Melicytus ramiflorus). N. Z. J. Ecol. 39, 190–197.
Sujii PS, Schwarcz KD, Grando C, Silvestre E de A, Mori GM, Brancalion PHS, Zucchi MI. 2017 Recovery of genetic diversity levels of a neotropical tree in Atlantic forest restoration plantations. Biol. Conserv. 211, 110–116. (doi:10.1016/j.biocon.2017.05.006)
Travis SE, Sheridan P. 2006 Genetic structure of natural and restored shoalgrass Halodule wrightii populations in the NW Gulf of Mexico. Mar. Ecol. Progr. Ser. 322, 117–127. (doi:10.3354/meps322117)
Wang Z-S, Liu H, Wei N, Xu W-X, An S-Q. 2010 Contribution of progeny cohorts to the restoration of genetic diversity in the post-harvest dragon spruce (Picea asperata) stands. Forestry 83, 307–314. (doi:10.1093/forestry/cpq013)
Williams SL. 2001 Reduced genetic diversity in eelgrass transplantations affects both population growth and individual fitness. Ecol. Appl. 11, 1472–1488. (doi:10.1890/1051-0761(2001)011[1472:RGDIET]2.0.CO;2)
Williams SL, Davis CA. 1996 Population genetic analyses of transplanted eelgrass (Zostera marina) beds reveal reduced genetic diversity in southern California. Restor. Ecol. 4, 163–180. (doi:10.1111/j.1526-100X.1996.tb00117.x)
Yokogawa M, Kaneko S, Takahashi Y, Isagi Y. 2013 Genetic consequences of rapid population decline and restoration of the critically endangered herb Polemonium kiushianum. Biol. Conserv. 157, 401–408. (doi:10.1016/j.biocon.2012.09.010)
Young AG, Murray BG. 2000 Genetic bottlenecks and dysgenic gene flow into re-established populations of the grassland daisy, Rutidosis leptorrhynchoides. Austr. J. Botany 48, 409–416. (doi:10.1071/BT98083)
Zavodna M, Abdelkrim J, Pellissier V, Machon N. 2015 A long-term genetic study reveals complex population dynamics of multiple-source plant reintroductions. Biol. Conserv. 192, 1–9. (doi:10.1016/j.biocon.2015.08.025)
Zhang X, Chen X-Y, Zhang D. 2006 Effect of regeneration method on RAPD-based genetic variation of Cyclobalanopsis glauca (Fagaceae). New For. 32, 347–356. (doi:10.1007/s11056-006-9008-5)
Zhang ZY, Wang H, Chen W, Pang XM, Li YY. 2016 Genetic diversity and structure of native and non-native populations of the endangered plant Pinus dabeshanensis. Genet. Mol. Res. 15, gmr15027937. (doi:10.4238/gmr.15027937)
Data Accessibility
Appendix A lists the publications used in this analysis, with appendix A1 (see electronic supplementary material) containing the results of analyses.
Authors' contributions
R.J. and M.F.B. conceived the study. R.J. performed the primary search and analysis. All authors contributed to the study design, data-mining, interpretation of results, and writing and revision of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Scholes RJ, et al. 2018. IPBES (2018): Summary for policymakers of the assessment report on land degradation and restoration of the Intergovernmental Science- Policy Platform on Biodiversity and Ecosystem Services. Bonn, Germany: IPBES Secretariat. [Google Scholar]
- 2.Miller BP, et al. 2017. A framework for the practical science necessary to restore sustainable, resilient, and biodiverse ecosystems. Restor. Ecol. 25, 605–617. ( 10.1111/rec.12475) [DOI] [Google Scholar]
- 3.Hobbs R, Norton D. 1996. Towards a conceptual framework for restoration ecology. Restor. Ecol. 4, 93–110. ( 10.1111/j.1526-100X.1996.tb00112.x) [DOI] [Google Scholar]
- 4.Bayraktarov E, Saunders MI, Abdullah S, Mills M, Beher J, Possingham HP, Mumby PJ, Lovelock CE. 2016. The cost and feasibility of marine coastal restoration. Ecol. Appl. 26, 1055–1074. ( 10.1890/15-1077) [DOI] [PubMed] [Google Scholar]
- 5.Cunningham S. 2008. Rewealth!: stake your claim in the $2 trillion development trend that's renewing the world. New York, NY: McGraw-Hill Professional. [Google Scholar]
- 6.Mijangos JL, Pacioni C, Spencer PBS, Craig MD. 2015. Contribution of genetics to ecological restoration. Mol. Ecol. 24, 22–37. ( 10.1111/mec.12995) [DOI] [PubMed] [Google Scholar]
- 7.Thomas E, Jalonen R, Loo J, Boshier D, Gallo L, Cavers S, Bordács S, Smith P, Bozzano M. 2014. Genetic considerations in ecosystem restoration using native tree species. For. Ecol. Manage. 333, 66–75. ( 10.1016/j.foreco.2014.07.015) [DOI] [Google Scholar]
- 8.Campbell RK, Sorensen FC. 1984. Genetic implications of nursery practices. In Forest nursery manual: production of bareroot seedlings (eds Duryea ML, Landis TD), pp. 183–191. The Hague/Boston/Lancaster: Martinus Nijhoff/Dr W Junk Publishers. [Google Scholar]
- 9.Ruiz-Jaen MC, Aide T. 2005. Restoration success: how is it being measured? Restor. Ecol. 13, 569–577. ( 10.1111/j.1526-100X.2005.00072.x) [DOI] [Google Scholar]
- 10.Wortley L, Hero JM, Howes M. 2013. Evaluating ecological restoration success: a review of the literature. Restor. Ecol. 21, 537–543. ( 10.1111/rec.12028) [DOI] [Google Scholar]
- 11.Reed DH, Frankham R. 2003. Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237. ( 10.1046/j.1523-1739.2003.01236.x) [DOI] [Google Scholar]
- 12.Leimu R, Mutikainen P, Koricheva J, Fischer M. 2006. How general are positive relationships between plant population size, fitness and genetic variation? J. Ecol. 94, 942–952. ( 10.1111/j.1365-2745.2006.01150.x) [DOI] [Google Scholar]
- 13.Reusch TBH, Ehlers A, Hammerli A, Worm B. 2005. Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proc. Natl Acad. Sci. USA 102, 2826–2831. ( 10.1073/pnas.0500008102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch M, Lande R.. 1993. Evolution and extinction in response to environmental change. In Biotic interactions and global change (eds Mareiva PM, Kingsolver JG, Huey RB), pp. 234–250. Sunderland MA: Sinauer Associates. [Google Scholar]
- 15.Markert JA, Champlin DM, Gutjahr-Gobell R, Grear JS, Kuhn A, McGreevy TJ, Roth A, Bagley MJ, Nacci DE. 2010. Population genetic diversity and fitness in multiple environments. BMC Evol. Biol. 10, 5–9. ( 10.1186/1471-2148-10-205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann AA, Sgrò CM, Kristensen TN. 2017. Revisiting adaptive potential, population size, and conservation. Trends Ecol. Evol. 32, 506–517. ( 10.1016/j.tree.2017.03.012) [DOI] [PubMed] [Google Scholar]
- 17.Breed MF, Ottewell KM, Gardner MG, Marklund MHK, Stead MG, Harris JBC, Lowe HAJ. 2015. Mating system and early viability resistance to habitat fragmentation in a bird-pollinated eucalypt. Heredity 115, 100–107. ( 10.1038/hdy.2012.72) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sgrò CM, Lowe AJ, Hoffmann AA. 2011. Building evolutionary resilience for conserving biodiversity under climate change. Evol. Appl. 4, 326–337. ( 10.1111/j.1752-4571.2010.00157.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prober SM, Williams KJ, Broadhurst LM, Doerr VAJ. 2017. Nature conservation and ecological restoration in a changing climate: what are we aiming for? Rangel. J. 39, 477–486. ( 10.1071/RJ17069) [DOI] [Google Scholar]
- 20.Breed MF, et al. 2018. Priority actions to improve provenance decision-making. Bioscience 68, 510–516. ( 10.1093/biosci/biy050) [DOI] [Google Scholar]
- 21.Hughes AR, Inouye BD, Johnson MTJ, Underwood N, Vellend M. 2008. Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623. ( 10.1111/j.1461-0248.2008.01179.x) [DOI] [PubMed] [Google Scholar]
- 22.Reynolds LK, McGlathery KJ, Waycott M. 2012. Genetic diversity enhances restoration success by augmenting ecosystem services. PLoS ONE 7, e38397. ( 10.1371/journal.pone.0038397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vander Mijnsbrugge K, Bischoff A, Smith B. 2010. A question of origin: where and how to collect seed for ecological restoration. Basic Appl. Ecol. 11, 300–311. ( 10.1016/j.baae.2009.09.002) [DOI] [Google Scholar]
- 24.Prober SM, Doerr VAJ, Broadhurst LM, Williams KJ, Dickson F. 2019. Shifting the conservation paradigm: a synthesis of options for renovating nature under climate change. Ecol. Monogr. 89, 1–23. ( 10.1002/ecm.1333) [DOI] [Google Scholar]
- 25.Broadhurst LM, Lowe A, Coates DJ, Cunningham SA, McDonald M, Vesk PA, Yates C. 2008. Seed supply for broadscale restoration: maximizing evolutionary potential. Evol. Appl. 1, 587–597. ( 10.1111/j.1752-4571.2008.00045.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guarino L, Ramanatha Rao V, Reid R (eds). 1995. Collecting plant genetic diversity: technical guidelines. Rome Italy: CAB International. [Google Scholar]
- 27.Kettle CJ, Ennos RA, Jaffre T, McCoy S, Le Borgne T, Gardner M, Hollingsworth PM. 2012. Importance of demography and dispersal for the resilience and restoration of a critically endangered tropical conifer Araucaria nemorosa. Divers. Distrib. 18, 248–259. ( 10.1111/j.1472-4642.2011.00835.x) [DOI] [Google Scholar]
- 28.Weeks AR, et al. 2011. Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evol. Appl. 4, 709–725. ( 10.1111/j.1752-4571.2011.00192.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breed MF, Stead MG, Ottewell KM, Gardner MG, Lowe AJ. 2013. Which provenance and where? Seed sourcing strategies for revegetation in a changing environment. Conserv. Genet. 14, 1–10. ( 10.1007/s10592-012-0425-z) [DOI] [Google Scholar]
- 30.Prober SM, Byrne M, McLean EH, Steane DA, Potts BM, Vaillancourt RE, Stock WD. 2015. Climate-adjusted provenancing: a strategy for climate-resilient ecological restoration. Front. Ecol. Evol. 3, 65 ( 10.3389/fevo.2015.00065) [DOI] [Google Scholar]
- 31.Standards Reference Group SERA. 2017. National standards for the practice of ecological restoration in Australia, 2nd edn Society for Ecological Restoration Australasia; See www.seraustralasia.com. [Google Scholar]
- 32.Ramp JM, Collinge SK, Ranker TA. 2006. Restoration genetics of the vernal pool endemic Lasthenia conjugens (Asteraceae). Conserv. Genet. 7, 631–649. ( 10.1007/s10592-005-9052-2) [DOI] [Google Scholar]
- 33.Smulders MJM, van der Schoot J, Geerts R, Antonisse-de Jong AG, Korevaar H, van der Werf A, Vosman B. 2000. Genetic diversity and the reintroduction of meadow species. Plant Biol. 2, 447–454. ( 10.1055/s-2000-6780) [DOI] [Google Scholar]
- 34.Zavodna M, Abdelkrim J, Pellissier V, Machon N. 2015. A long-term genetic study reveals complex population dynamics of multiple-source plant reintroductions. Biol. Conserv. 192, 1–9. ( 10.1016/j.biocon.2015.08.025) [DOI] [Google Scholar]
- 35.Sujii PS, Schwarcz KD, Grando C, Silvestre EdA, Mori GM, Brancalion PHS, Zucchi MI. 2017. Recovery of genetic diversity levels of a neotropical tree in Atlantic forest restoration plantations. Biol. Conserv. 211, 110–116. ( 10.1016/j.biocon.2017.05.006) [DOI] [Google Scholar]
- 36.Tummer B. 2006. DataThief III. See http://datathief.org/.
- 37.Flower A, McKenna JW, Upreti G. 2016. Validity and reliability of GraphClick and DataThief III for data extraction. Behav. Modif. 40, 396–413. ( 10.1177/0145445515616105) [DOI] [PubMed] [Google Scholar]
- 38.Alonso MA, Guillo A, Perez-Botella J, Crespo MB, Juan A. 2014. Genetic assessment of population restorations of the critically endangered Silene hifacensis in the Iberian Peninsula. J. Nat. Conserv. 22, 532–538. ( 10.1016/j.jnc.2014.08.007) [DOI] [Google Scholar]
- 39.Neto OC, Aguiar AV, Twyford AD, Neaves LE, Pennington RT, Lopes AV. 2014. Genetic and ecological outcomes of Inga vera subsp affinis (Leguminosae) tree plantations in a fragmented tropical landscape. PLoS ONE 9, e99903 ( 10.1371/journal.pone.0099903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Souza EMS, Pereira GS, Silva-Mann R, Alvares-Carvalho SV, Ferreira RA. 2016. A comparative framework of the Erythrina velutina tree species in reforested land and native populations. Genet. Mol. Res. 15, gmr15028534 ( 10.4238/gmr.15028534) [DOI] [PubMed] [Google Scholar]
- 41.Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fant JB, Holmstrom RM, Sirkin E, Etterson JR, Masi S. 2008. Genetic structure of threatened native populations and propagules used for restoration in a clonal species American beachgrass (Ammophila breviligulata Fern.). Restor. Ecol. 16, 594–603. ( 10.1111/j.1526-100X.2007.00348.x) [DOI] [Google Scholar]
- 43.Li YY, Chen XY, Zhang X, Wu TY, Lu HP, Cai YW. 2005. Genetic differences between wild and artificial populations of Metasequoia glyptostroboides: implications for species recovery. Conserv. Biol. 19, 224–231. ( 10.1111/j.1523-1739.2005.00025.x) [DOI] [Google Scholar]
- 44.Williams SL. 2001. Reduced genetic diversity in eelgrass transplantations affects both population growth and individual fitness. Ecol. Appl. 11, 1472–1488. ( 10.1890/1051-0761(2001)011[1472:RGDIET]2.0.CO;2) [DOI] [Google Scholar]
- 45.Stevens MI, Clarke AC, Clarkson FM, Goshorn M, Gemmill CEC. 2015. Are current ecological restoration practices capturing natural levels of genetic diversity? A New Zealand case study using AFLP and ISSR data from mahoe (Melicytus ramiflorus). NZ J. Ecol. 39, 190–197. [Google Scholar]
- 46.Gustafson DJ, Gibson DJ, Nickrent DL. 2004. Competitive relationships of Andropogon gerardii (Big Bluestem) from remnant and restored native populations and select cultivated varieties. Funct. Ecol. 18, 451–457. ( 10.1111/j.0269-8463.2004.00850.x) [DOI] [Google Scholar]
- 47.Dolan RW, Marr DL, Schnabel A. 2008. Capturing genetic variation during ecological restorations: an example from Kankakee Sands in Indiana. Restor. Ecol. 16, 386–396. ( 10.1111/j.1526-100X.2007.00318.x) [DOI] [Google Scholar]
- 48.Gustafson DJ, Gibson DJ, Nickrent DL. 2002. Genetic diversity and competitive abilities of Dalea purpurea (Fabaceae) from remnant and restored grasslands. Int. JPant Sci. 163, 979–990. ( 10.1086/342709) [DOI] [Google Scholar]
- 49.Williams SL, Davis CA. 1996. Population genetic analyses of transplanted eelgrass (Zostera marina) beds reveal reduced genetic diversity in southern California. Restor. Ecol. 4, 163–180. ( 10.1111/j.1526-100X.1996.tb00117.x) [DOI] [Google Scholar]
- 50.Fant JB, Kramer A, Sirkin E, Havens K. 2013. Genetics of reintroduced populations of the narrowly endemic thistle, Cirsium pitcheri (Asteraceae). Botany 91, 301–308. ( 10.1139/cjb-2012-0232) [DOI] [Google Scholar]
- 51.Jordan R, Dillon SK, Prober SM, Hoffmann AA. 2016. Landscape genomics reveals altered genome wide diversity within revegetated stands of Eucalyptus microcarpa (Grey Box). New Phytol. 212, 992–1006. ( 10.1111/nph.14084) [DOI] [PubMed] [Google Scholar]
- 52.Czarnecki DM, Rao MN, Norcini JG, Gmitter FG Jr, Deng Z. 2008. Genetic diversity and differentiation among natural, production, and introduced populations of the narrowly endemic species Coreopsis leavenworthii (Asteraceae). J. Am. Soc. Hortic. Sci. 133, 234–241. ( 10.21273/JASHS.133.2.234) [DOI] [Google Scholar]
- 53.Campanella JJ, Bologna PAX, Smith SM, Rosenzweig EB, Smalley JV. 2010. Zostera marina population genetics in Barnegat Bay New Jersey, and implications for grass bed restoration. Popul. Ecol. 52, 181–190. ( 10.1007/s10144-009-0170-4) [DOI] [Google Scholar]
- 54.Campanella JJ, Bologna PAX, Smalley JV, Avila DN, Lee KN, Areche EC, Slavin LJ. 2013. An analysis of the population genetics of restored Zostera marina plantings in Barnegat Bay New Jersey. Popul. Ecol. 55, 121–133. ( 10.1007/s10144-012-0351-4) [DOI] [Google Scholar]
- 55.Ritchie AL, Krauss SL. 2012. A genetic assessment of ecological restoration success in Banksia attenuata. Restor. Ecol. 20, 441–449. ( 10.1111/j.1526-100X.2011.00791.x) [DOI] [Google Scholar]
- 56.Ritchie AL, Nevill PG, Sinclair EA, Krauss SL. 2017. Does restored plant diversity play a role in the reproductive functionality of Banksia populations? Restor. Ecol. 25, 414–423. ( 10.1111/rec.12456) [DOI] [Google Scholar]
- 57.Broadhurst LM. 2013. A genetic analysis of scattered Yellow Box trees (Eucalyptus melliodora A.Cunn. ex Schauer Myrtaceae) and their restored cohorts. Biol. Conserv. 161, 48–57. ( 10.1016/j.biocon.2013.02.016) [DOI] [Google Scholar]
- 58.Pakkad G, Al Mazrooei S, Blakesley D, James C, Elliott S, Luoma-Aho T, Koskela J. 2008. Genetic variation and gene flow among Prunus cerasoides DDn populations in northern Thailand: analysis of a rehabilitated site and adjacent intact forest. New For. 35, 33–43. ( 10.1007/s11056-007-9059-2) [DOI] [Google Scholar]
- 59.Severns PM. 2013. Genetic differentiation in an artificial population of the threatened plant Lupinus oreganus (Fabaceae). Botany 91, 319–322. ( 10.1139/cjb-2012-0278) [DOI] [Google Scholar]
- 60.Fotinos TD, Namoff S, Lewis C, Maschinski J, Griffith MP, von Wettberg EJB. 2015. Genetic evaluation of a reintroduction of Sargent's Cherry Palm, Pseudophoenix sargentii. J. Torrey Bot. Soc. 142, 51–62. ( 10.3159/TORREY-D-14-00004.1) [DOI] [Google Scholar]
- 61.R Core Team. 2018. R: a language and environment for statistical computing. Vienna Austria: R Foundation for Statistical Computing; See Https://www.r-project.org/ [Google Scholar]
- 62.Broadhurst LM. 2011. Genetic diversity and population genetic structure in fragmented Allocasuarina verticillata (Allocasuarinaceae)—implications for restoration. Aust. JBt. 59, 770–780. ( 10.1071/BT11253) [DOI] [Google Scholar]
- 63.Liu M-H, Chen X-Y, Zhang X, Shen D-W. 2008. A population genetic evaluation of ecological restoration with the case study on Cyclobalanopsis myrsinaefolia (Fagaceae). Plant Ecol. 197, 31–41. ( 10.1007/s11258-007-9357-y) [DOI] [Google Scholar]
- 64.Godefroid S, et al. 2011. How successful are plant species reintroductions? Biol. Conserv. 144, 672–682. ( 10.1016/j.biocon.2010.10.003) [DOI] [Google Scholar]
- 65.Frick KM, Ritchie AL, Krauss SL. 2014. Field of dreams: restitution of pollinator services in restored bird-pollinated plant populations. Restor. Ecol. 22, 832–840. ( 10.1111/rec.12152) [DOI] [Google Scholar]
- 66.Aitken SN, Bemmels JB. 2016. Time to get moving: assisted gene flow of forest trees. Evol. Appl. 9, 271–290. ( 10.1111/eva.12293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jordan R, Hoffmann AA, Dillon SK, Prober SM. 2017. Evidence of genomic adaptation to climate in Eucalyptus microcarpa: implications for adaptive potential to projected climate change. Mol. Ecol. 26, 6002–6020. ( 10.1111/mec.14341) [DOI] [PubMed] [Google Scholar]
- 68.Pina-Martins F, Baptista J, Pappas G, Paulo OS. 2019. New insights into adaptation and population structure of cork oak using genotyping by sequencing. Glob. Chang. Biol. 25, 337–350. ( 10.1111/gcb.14497) [DOI] [PubMed] [Google Scholar]
- 69.Gerwein AJB, Kesseli RV. 2006. Genetic diversity and population structure of Quercus rubra (Fagaceae) in old-growth and secondary forests in southern New England. Rhodora 108, 1–18. ( 10.3119/05-9.1) [DOI] [Google Scholar]
- 70.Viana JPG, et al. 2018. Genomic diversity is similar between Atlantic Forest restorations and natural remnants for the native tree Casearia sylvestris Sw. PLoS ONE 13, e0192165 ( 10.1371/journal.pone.0192165) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Appendix A lists the publications used in this analysis, with appendix A1 (see electronic supplementary material) containing the results of analyses.