Abstract
Circadian misalignment plays an important role in disease processes and can affect disease severity, treatment outcomes, and even survivorship. In this study, we aim to investigate whether expression and daily oscillation patterns of core circadian clock genes were disturbed in patients with obstructive sleep apnea/hypopnea (OSA) syndrome. We performed real-time quantitative reverse transcriptase-polymerase chain reactions to examine the expression of the nine core circadian clock genes in leukocytes of peripheral blood collected at 12 AM, 6 AM, 12 PM, and 6 PM from 133 patients with OSA and 11 normal controls. Daily expression patterns of the nine circadian clock genes were observed in normal controls, but three of these genes (BMAL1, CLOCK, CRY2) were disrupted in patients with OSA. The expressions of eight circadian clock genes (except PER1) at midnight were significantly downregulated in patients with severe OSA. Binary logistic regression analysis selected CRY1 and PER3 as independent factors for severe OSA and showed that the combined expressions of CRY1 and PER3 enhanced the capability of predicting severe OSA (Odds ratio, 5.800; 95% CI, 1.978 to 17.004; p = 0.001). Our results show that combined expressions of CRY1 and PER3 at midnight could be a potential predictor for severe OSA.
Keywords: obstructive sleep apnea/hypopnea (OSA), circadian rhythm, circadian clock genes
1. Introduction
Obstructive sleep apnea/hypopnea (OSA) syndrome is characterized by repetitive obstruction of the upper airway and causes repetitive hypoxia/reoxygenation. In untreated OSA patients, this ultimately results in an increased risk of hypertension, coronary artery disease, myocardial infarction, stroke, and even sudden death [1,2,3,4,5].
“Circadian clock” means an internal time keeping system that developed from organisms which allows them to anticipate one of the most profound environmental signals, the daily cycle of light/dark. In mammals, circadian rhythm refers to the “body clock” which is an endogenously driven, nearly 24-hour cycle in biochemistry, physiology, or behavior, such as sleep and activity, appetite, hormone levels, metabolism, and gene expression [6]. The circadian master clock resides in the suprachiasmatic nucleus in the hypothalamus. It is now accepted that peripheral blood (PB) cells contain a circadian clock similar to that in the suprachiasmatic nucleus [7]. Until now, at least nine mammalian core circadian clock genes have already been identified: Period1 (PER1), Period2 (PER2), Period3 (PER3), CLOCK, Cryptochrome1 (CRY1), Cryptochrome2 (CRY2), BMAL1, Casein kinase 1ε (CK1ε), and Timeless (TIM). The regulation relies on positive loops (such as CLOCK and BMAL1) and negative loops (such as PER1, PER2, PER3, CRY1, CRY2 and TIM) in the oscillators [8]. Circadian rhythms are ubiquitous phenomena, and are found, for example, in the sleep-wake cycle, body-core temperature, leukocyte count, trace metal concentrations, and levels of many hormones, such as cortisol and melatonin [9,10]. A stable human biological clock helps to regulate sleep patterns, hormone release, and blood pressure, etc. Many aspects of cardiovascular or cerebrovascular physiology are subject to diurnal variation. Serious adverse events including cancers, atherosclerosis, myocardial infarction, and stroke are significantly associated with the disturbance of circadian gene expressions [9].
Until now, there have been very limited reports on the relationships between circadian clock genes and OSA. Sleep is a neurological generated phenomenon who’s timing is under circadian influences. Sleep–wake cycle could modulate circadian clock genes [11] and in turn circadian system could influence apnea occurrence and duration [12,13]. Therefore, in this study, we prospectively investigated the expression and daily oscillation patterns of the nine core circadian clock genes in patients with OSA to further understand the impact of circadian clock regulation on OSA.
2. Materials and Methods
2.1. Patients, Normal Controls, and Samples
The design of this study adhered to the tenets of the Declaration of Helsinki. This study was approved by the Institutional Review Board (IRB) of the Chang Gung Memorial Hospital (CGMH) Ethical Committee (CGMH IRB No. 102-6026B). Written informed consent was obtained from all participants.
We enrolled 144 subjects with the symptoms of sleep-related breathing disorders such as loud snoring, observed apnea or excessive daytime sleepiness, from our sleep clinics. All of the 144 subjects were consecutively admitted for a full-night polysomnography (PSG) at the sleep center of Kaohsiung Chang Gung Memorial Hospital (KCGMH), a 2,600-bed tertiary referral medical center. The criteria for diagnosing OSA are as previously described [14,15,16]. Among the 144 participants, 133 patients were diagnosed with OSA (aged 21–67 years) and 11 subjects were diagnosed as without OSA (aged 26–59 years), and these 11 subjects were categorized as normal controls. The subjects enrolled in this study did not experience shift work or jet lag 1 week before the experiment. Female individuals included in this study were not in their menstrual phases during the experiment. The time schedule and the daily activity of normal controls and patients with OSA were not restricted but they were asked to have breakfast between 7:00 and 8:00, lunch between 11:30 and 13:30, and dinner between 17:30 and 19:30; they were also asked to go to sleep before 24:00 and wake up at around 7:00. Before the PSG, all participants were asked to follow the pre-arranged schedule as described above strictly for 1 week. The collection of PB was carried out four times daily at 12 AM (00:00), 06 AM (06:00), 12 PM (12:00), and 06 PM (18:00), respectively, at the sleep center of KCGMH. For the PB collection at 12 AM, blood-drawing was done under minimal (~20 lux) light density and all individuals fell asleep immediately after blood was drawn.
2.2. Analysis of Expression of Circadian Clock Genes
Isolation of total PB leukocytes, RNA extraction and cDNA synthesis were performed as previously described [17,18]. The expression of the nine circadian clock genes and ACTB gene (as endogenous reference control) was analyzed using real-time quantitative reverse transcriptase- polymerase chain reaction (qRT-PCR) as previously described [17,18]. The relative gene expression was calculated by equalizing differences to the ΔCt of 6 PM in normal controls (i.e., 2−(ΔCt of X -ΔCt of 6PM)); values were normalized so that the expression level of normal controls at 6 PM equaled 1.0.
2.3. Statistical Analysis
Student’s t-test was used to detect the differences of clinical parameters between groups of normal controls and OSA and among groups of normal controls and different severities of OSA. Repeated-measures ANOVA (Analysis of variance) was used to detect the differences among four different time points in the expression of each circadian clock gene. The comparison of daily patterns between different groups of individuals in the expression of each circadian clock gene and the comparison of gene expression of the same gene at the same time point between different groups were evaluated with Univariate analysis of General Linear Model (GLM) with post hoc Bonferroni comparison under GLM. Receiver Operating Characteristic (ROC) curve was plotted and ROC area under the curve (AUC) was calculated to compare the discriminating ability of circadian clock genes. Binary logistic regression modeled the effects of selected independent variables on whether or not the expression of a specific gene could be a predictive marker for severe OSA. The values of ΔCt were used for all statistical analysis. All tests were two-sided with statistical significance set at 0.05 and all computations were made using SPSS 22.0 software (IBM SPSS Statistics for Windows, IBM, Armonk, NY, USA) and Graph Pad Prism 7.04 (GraphPad, San Diego, CA, USA).
3. Results
3.1. Categorization of Patients with OSA
The severities of 133 patients with OSA enrolled in this study were categorized into three major groups according to their apnea/hypopnea index (AHI, /hr.): (1) mild, 5 ≤ AHI < 15, n = 27; (2) moderate, 15 ≤ AHI < 30, n = 27; and (3) severe, AHI ≥ 30 (n = 79); their clinical characteristics are listed in Table 1.
Table 1.
Clinical characteristics of patients with obstructive sleep apnea/hypopnea (OSA) syndrome.
| Characteristic | Normal Controls (n = 11) | OSA (n = 133) | p-Value (1) a | Mild (n = 27) | Moderate (n = 27) | Severe (n = 79) | p-Value (2) b |
|---|---|---|---|---|---|---|---|
| Sex, Male/Female | 5/6 | 111/22 | 0.002 | 19/8 | 24/3 | 68/11 | <0.005 |
| Age, year | 42.55 ± 3.45 c | 42.47 ± 0.91 | 0.981 | 40.89 ± 2.16 | 44.15 ± 2.23 | 42.43 ± 1.12 | 0.736 |
| BMI, kg/m2 | 23.33 ± 0.87 | 26.08 ± 0.28 | 0.007 | 24.39 ± 0.55 | 24.24 ± 0.42 | 27.29 ± 0.35 | <0.001 |
| ESS | 7.6 ± 1.3 | 8.98 ± 0.38 | 0.333 | 8.8 ± 0.9 | 8.7 ± 0.8 | 9.2 ± 0.5 | 0.739 |
| ESS, no. ≤10/>10 | 7/4 | 85/48 | 1.000 | 19/8 | 18/9 | 48/31 | 0.821 |
| AHI, /hour | 2.78 ± 0.44 | 39.94 ± 2.14 | <0.001 | 10.3 ± 0.58 | 20.80 ± 0.79 | 56.59 ± 2.00 | <0.001 |
| AHI in REM, /hour | 4.58 ± 1.14 | 42.71 ± 2.01 | <0.001 | 19.55 ± 3.22 | 34.02 ± 322 | 53.88 ± 2019 | <0.001 |
| % of O2 < 90%, % | 0.22 ± 0.15 | 9.85 ± 1.16 | 0.018 | 1.20 ± 0.31 | 3.09 ± 0.64 | 15.19 ± 1.70 | <0.001 |
| Average O2, % | 96.96 ± 0.20 | 95.02 ± 0.21 | 0.008 | 96.53 ± 0.18 | 96.13 ± 0.23 | 94.12 ± 0.29 | <0.001 |
| LSAT, % | 90.64 ± 0.69 | 75.60 ± 1.13 | <0.001 | 86.11 ± 0.92 | 81.19 ± 1.85 | 70.10 ± 1.45 | <0.001 |
ap value (1) is the p value for comparison between Normal Controls and OSA. The p values indicated were evaluated with Student t-test, except for sex (Chi-square). b p value (2) is the p value for comparison among Normal Controls and different severities of OSA. c Data are presented as mean ± standard error of mean. OSA, obstructive sleep apnea/hypopnea; BMI, body mass index; ESS, Epworth sleepiness scale; AHI, apnea/hypopnea index; REM, rapid eye movement; LSAT, lowest saturation of O2.
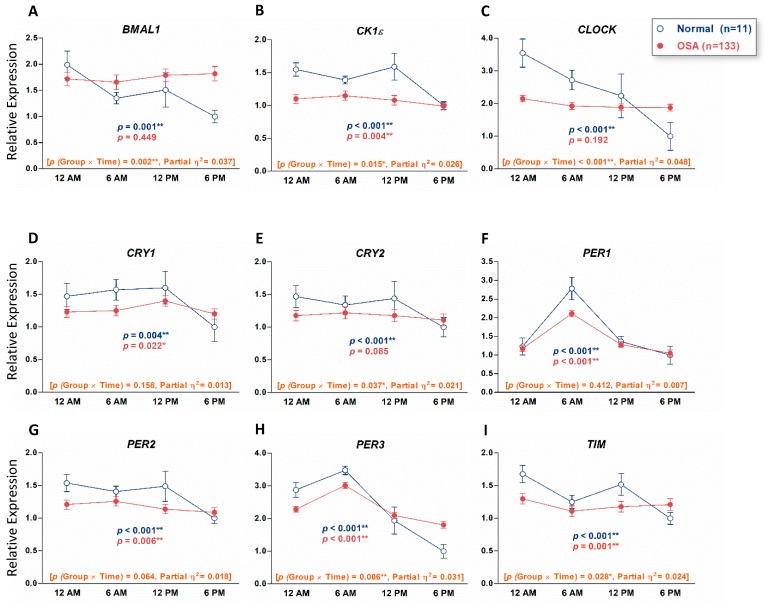
3.2. Circadian Patterns of Expression of Circadian Clock Genes in Healthy Controls and Abolished Daily Patterns in Patients with OSA
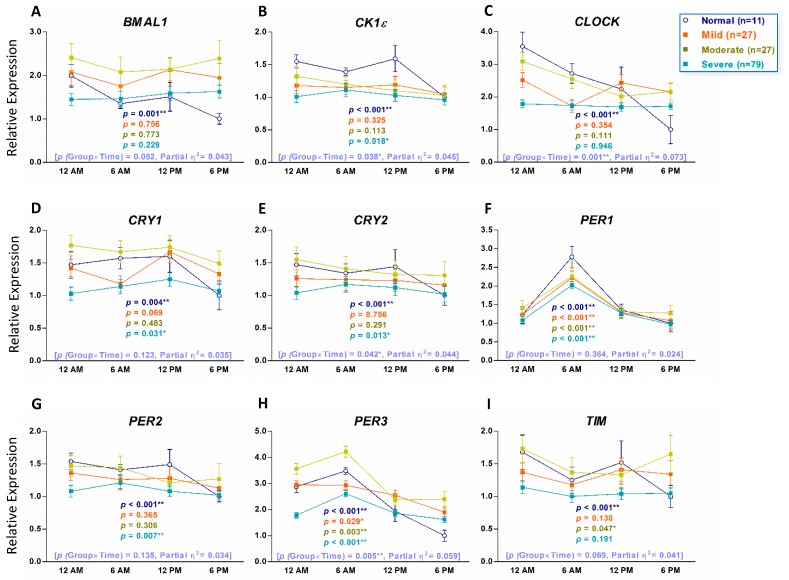
To examine if the expression of circadian clock genes in healthy individuals without OSA showed a time-dependent variation, we used qRT-PCR to analyze PB total leukocytes from 11 healthy individuals who were evaluated by PSG and diagnosed without OSA as normal controls for this study. Repeated-measures ANOVA analyses indicated that in normal controls, all the nine circadian clock genes displayed a time-dependent variation pattern (Figure 1). We also analyzed PB total leukocytes from 133 patients with OSA to examine whether their daily patterns were different from those of normal controls. Repeated-measures ANOVA analyses showed the daily patterns of the BMAL1, CLOCK, and CRY2 genes that changed over time in normal controls and were abolished in patients with OSA (Figure 1A,C,E). We further divided patients with OSA into mild, moderate, and severe groups and analyzed their daily patterns of expression of circadian clock genes. Repeated-measures ANOVA analyses showed that the daily patterns of PER1 and PER3 were consistent in normal controls and all three groups of patients with OSA (Figure 2F,H). Transcripts of BMAL1 and CLOCK showed time variation in normal controls but the daily patterns were abolished in all three groups of patients with OSA (Figure 2A,C). Transcripts of CK1ε, CRY1, CRY2, and PER2 that changed over time in normal controls were abolished in mild and moderate groups of patients with OSA and the time-dependent variation recovered in severe groups of patients with OSA (Figure 2B,D,E,G). The comparison between recovered oscillation patterns of Ck1ε (p = 0.069), CRY1 (p = 0.316), PER1 (p = 0.334), and PER2 (p = 0.122) and their normal control was evaluated by Repeated-measures ANOVA.
Figure 1.
Circadian patterns of the nine circadian clock genes in human peripheral blood (PB) of 133 patients with obstructive sleep apnea (OSA) and 11 normal controls (individuals without OSA). The nine circadian clock genes are BMAL1 (A), CK1ε (B), CLOCK (C), CRY1 (D), CRY2 (E), PER1 (F), PER2 (G), PER3 (H) and TIM (I). The x-axis indicates the time points that PB samples were collected. The y-axis represents the relative mRNA expression level. The value of the mRNA expression at 6 PM in normal controls is designated 1, and the levels of all other mRNA expressions are calibrated to this value. The p values indicated were evaluated with Repeated-measures ANOVA. The numbers in brackets are p values of group × time interaction and partial η2, respectively. ** p < 0.01, * p < 0.05.
Figure 2.
Circadian patterns of the nine circadian clock genes in human peripheral blood (PB) of 133 patients with different severities of obstructive sleep apnea (OSA) and 11 normal controls. The nine circadian clock genes are BMAL1 (A), CK1ε (B), CLOCK (C), CRY1 (D), CRY2 (E), PER1 (F), PER2 (G), PER3 (H) and TIM (I). The x-axis indicates the time points that PB samples were collected. The y-axis represents the relative mRNA expression level. The value of the mRNA expression at 6 PM in normal controls is designated 1, and the levels of all other mRNA expressions are calibrated to this value. The p values indicated were evaluated with Repeated-measures ANOVA. The numbers in brackets are p values of group x time interaction and partial η2, respectively. ** p < 0.01, * p < 0.05.
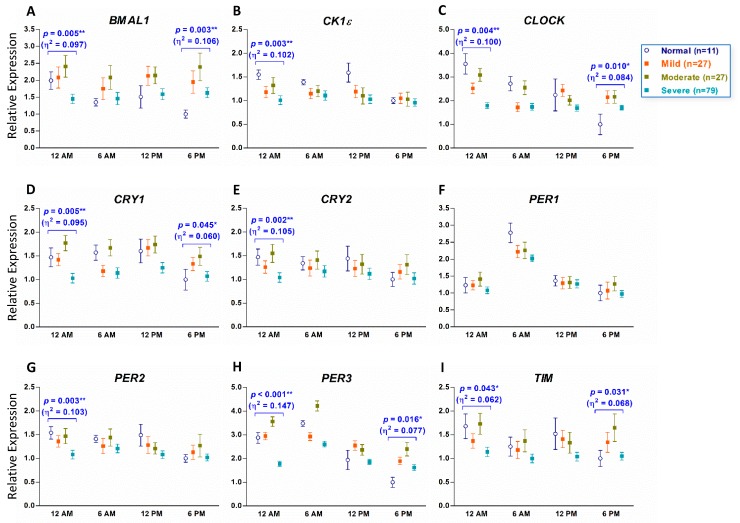
3.3. Downregulation of Circadian Clock Genes in Patients with OSA at Midnight
Transcripts of BMAL1 (p = 0.005, η2 = 0.097), CK1ε (p = 0.003, η2 = 0.102), CLOCK (p = 0.004, η2 = 0.100), CRY1 (p = 0.005, η2 = 0.095), CRY2 (p = 0.002, η2 = 0.105), PER2 (p = 0.003, η2 = 0.103), PER3 (p < 0.001, η2 = 0.147), and TIM (p = 0.043, η2 = 0.062) were significantly downregulated at 12 AM in the severe group of patients with OSA (Figure 3A–E,G–I). PER1 is the only gene that its expression was not different among normal controls and all three groups of patients with OSA at four different time points (Figure 3F). At 6 PM, the expression levels of BMAL1 (p = 0.003, η2 = 0.106), CLOCK (p = 0.010, η2 = 0.084), CRY1 (p = 0.045, η2 = 0.060), PER3 (p = 0.016, η2 = 0.077), and TIM (p = 0.031, η2 = 0.068) were slightly different among the four groups (Figure 3A,C,I).
Figure 3.
Expression of the nine circadian clock genes at different time points in human peripheral blood (PB) of patients with obstructive sleep apnea (OSA) and individuals without OSA. The nine circadian clock genes are BMAL1 (A), CK1ε (B), CLOCK (C), CRY1 (D), CRY2 (E), PER1 (F), PER2 (G), PER3 (H) and TIM (I). The x-axis indicates the time points that PB samples were collected. The y-axis represents the relative mRNA expression level. The value of the mRNA expression at 6 PM in normal controls (individuals without OSA) is designated 1, and the levels of all other mRNA expressions are calibrated to this value. The p values indicated were evaluated with a Univariate analysis of General Linear Model (GLM) with post hoc Bonferroni comparison under GLM. ** p < 0.01, * p < 0.05. The number in the parentheses is the value of partial η2 evaluated with Repeated-measures ANOVA.
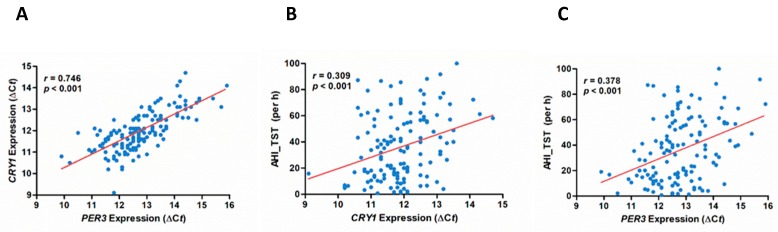
3.4. Correlation and Regression Analysis of Expression of Circadian Clock Genes at Midnight in Patients with OSA
We constructed ROC curves and calculated the AUC to examine the discriminative performance of expression of circadian clock genes at 12 AM between the severe OSA group and non-severe OSA groups. Binary logistic regression analysis selected CRY1 (Odds ratio, 2.963; 95% CI, 1.277 to 6.879; p = 0.011) and PER3 (Odds ratio, 3.746; 95% CI, 1.595 to 8.795; p = 0.002) as independent factors for severe OSA (Table 2). The combined expression of CRY1 and PER3 enhanced the prediction of severe OSA (Odds ratio, 5.800; 95% CI, 1.978 to 17.004; p = 0.001). Pearson correlation analysis showed significant correlations between CRY1 expression and AHI (r = 0.378, p < 0.001) (Figure 4B) and between PER3 expression and AHI (r = 0.309, p < 0.001) (Figure 4C). Our results demonstrated that when the expression of either CRY1 or PER3 is low at midnight, the risk to develop severe OSA is about 3- to 4-fold higher than normal. When the expression of CRY1 and PER3 are both low at midnight, the risk to develop severe OSA is 5.8-fold higher than normal. Thus, the combined expression of CRY1 and PER3 at midnight might have potential to be a predictor for patients with severe OSA.
Table 2.
Binary logistic-regression analysis of predictors of severe obstructive sleep apnea/hypopnea syndrome by expression of CRY1 and PER3.
| Independent Factor | Odds Ratio (95% CI) | p Value |
|---|---|---|
| CRY1 | 2.963 (1.277–6.879) | 0.011 |
| PER3 | 3.746 (1.595–8.795) | 0.002 |
| None | Reference a | |
| CRY1 or PER3 | 2.363 (0.948–5.888) | 0.065 |
| CRY1 and PER3 | 5.800 (1.978–17.004) | 0.001 |
a The reference for the odds ratio is the absence of the corresponding risk factor. CI, confidence interval.
Figure 4.
Correlations of expression of CRY1 and PER3 with apnea/hypopnea index of total sleep time (AHI_TST). Correlation between (A) expression of CRY1 and PER3, (B) expression of CRY1 and AHI_TST, and (C) expression of PER3 and AHI_TST. The values of ΔCt of CRY1 and PER3 obtained from real-time quantitative polymerase chain reaction were used for the expression of PER3 and CRY1 in Pearson’s correlation with AHI_TST (per hour). The r values indicated are the Pearson’s correlation coefficients. Both r and p values indicated were evaluated with Pearson’s correlation test.
4. Discussion
In this current study, we investigated the nine comprehensive circadian clock genes in patients with OSA and found that the transcripts of all nine circadian clock genes displayed daily patterns in PB of normal controls but three of them, BMAL1, CLOCK and CRY2, were arrhythmic in patients with OSA. Burioka et al. examined the PER1 gene only and demonstrated that the expression of PER1 gene peaked at 6AM both in patients with OSA and healthy individuals [19], which coincided with our findings. Our results of BMAL1, CK1ε, CRY1, CRY2, PER1, PER2, and PER3 in normal controls are also consistent with previous reports [18,20,21,22,23,24], demonstrating that the expression of these genes displayed daily oscillation patterns. However, in contrast to our results, previous reports demonstrated that the expression of CLOCK was arrhythmic in human PB [18,22]. Since all the normal controls enrolled in this study are examined by a full-night PSG and diagnosed as without OSA, this may explain the inconsistency between our study and other studies that used normal controls with different inclusion criteria or just simply mentioned the healthy subjects, without confirming via a sleep study.
Consistent with our current study, the expression of clock genes has been shown to be altered in patients with OSA [19,25]. Moreira et al. further revealed that the altered expression of CLOCK gene in seven patients with OSA was not reverted by continuous positive airway pressure (CPAP) treatment [25]. In contrast, Burioka et al. demonstrated that the PER1 expression elevated at 02:00 in patients with OSA was significantly decreased by CPAP treatment [19]. In our study, although we observed that the daily oscillation patterns of circadian clock genes were altered in OSA, the changes of daily oscillation patterns seem not as strongly related to the severities of OSA as the expression levels at midnight. We found that the expression levels of CRY1 and PER3 were significantly decreased at midnight and might potentially predict severe OSA. A recent study also demonstrated a decrease of PER3 in veterans with sleep apnea [26]. PER3 was thought to be less important in the PERIOD gene family but more and more human studies have suggested that PER3 may also be essential in maintaining circadian rhythm [20,27,28,29].
The limitations of this study are the small number of normal controls and only four time points of PB were collected. For better comparison for OSA, we used more strict criteria for selecting normal controls, and only individuals examined by PSG and diagnosed without OSA were enrolled. Therefore, only limited numbers of normal controls were qualified. Our concern is that the normal expression and oscillation patterns of circadian clock genes may not be fully reflected by the limited case numbers as the partial η2 values in Figure 1 and Figure 2 are small (0.01 ≤ η2 ≤ 0.06). Besides, it was very difficult to collect PB from patients more than four times a day although more time points will more precisely reflect the oscillation patterns of circadian clock genes. Although we demonstrated that the expression of CRY1 and PER3 genes at 12AM might be able to predict the OSA disease severity, especially for severe OSA, we only revealed the relationship between circadian clock genes and OSA instead of causal effects. In severe OSA, the expression of CRY1 and PER3 (r = 0.746, p < 0.001) at midnight was highly correlated (Figure 4A) but the correlation between CRY1 and AHI (r = 0.309, p < 0.001) and between PER3 and AHI (r = 0.378, p < 0.001) was not very high. If changes of expression of circadian clock genes could be applied for predicting treatment outcome of OSA, further studies will be needed.
The results of this study may not be sufficient for clinical diagnosis of OSA at present but they do provide a new reference for evaluating the severities of OSA and new directions for future studies. We believe the analysis of expression of circadian clock genes of post-operational OSA patients will strengthen our conclusion. In addition, investigating the mechanisms of downregulated CRY1 and PER3 in severe OSA and the molecular causal effects of circadian clock genes and OSA will also let us take a step forward to the understanding of OSA.
5. Conclusions
This study provides a comprehensive circadian genes survey for patients with OSA. Our results show that the combined expression of PER3 and CRY1 circadian genes at midnight might be a potential predictor for patients with OSA, especially for those with severe OSA. This implementable genetic tool may help clinicians to identify patients with OSA.
Acknowledgments
The authors thank the Biostatistics Center, Kaohsiung Chang Gung Memorial Hospital for statistics work, and Mao-Chang Su, Chien-Hung Chin, Yung-Che Chen, Chean-Chean Yang and Zi-Yin Deng for the assistance in this manuscript.
Author Contributions
Conceptualization, M.-Y.Y. and H.-C.L.; methodology, P.-M.L. and I.-Y.C.; validation, M.-Y.Y. and I.-Y.C.; formal analysis, M.-Y.Y., P.-W.L., I.-Y.C., M.F., C.-F.H., A.M.S. and M.-C.L.; Writing—Original Draft preparation, M.-Y.Y., P.-W.L. and H.-C.L.; Writing—Review and Editing, M.F. and M.-C.L.; project administration, M.-Y.Y. and H.-C.L.; funding acquisition, M.-Y.Y., H.-C.L. and S.-F.L.
Funding
This research was funded by Kaohsiung Chang Gung Memorial Hospital, grant numbers CMRPG8F1281 and CMRPG8G1011 to H.-C.L. and CMRPD8H0011 and CMRPD8J0011 to M.-Y.Y., Ministry of Science and Technology of Taiwan, grant numbers MOST 103-2314-B-182A-060, MOST 104-2314-B-182A-074 and MOST 107-2314-B-182A-086 to H.C.L., MOST 107-2320-B-182-022 to M.-Y.Y., and Research Services Center For Health Information, Chang Gung University, grant number CIRPD1D0031 to H.-C.L.
Conflicts of Interest
H.-C.L. received two research grants from Intuitive Surgical Inc., Sunnyvale, CA. M.F. received a research grant from ImThera Medical Inc., San Diego, CA. However, Intuitive Surgical Inc. and ImThera Medical Inc. had no role in the design or conduct of this study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication. M.-Y.Y., P.-W.L., P.-M.L., I.-Y.C., C.-F.H., A.M.S. and M.-C.L. declare no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Young T., Peppard P.E., Gottlieb D.J. Epidemiology of obstructive sleep apnea. A population health perspective. Am. J. Respir. Crit. Care Med. 2002;165:1217–1239. doi: 10.1164/rccm.2109080. [DOI] [PubMed] [Google Scholar]
- 2.Shamsuzzaman A.S., Gersh B.J., Somers V.K. Obstructive sleep apnea: implications for cardiac and vascular disease. JAMA. 2003;290:1906–1914. doi: 10.1001/jama.290.14.1906. [DOI] [PubMed] [Google Scholar]
- 3.Marin J.M., Carrizo S.J., Vicente E., Agusti A.G. Long-term cardiovascular outcomes in men with obstructive sleep apnoea-hypopnoea with or without treatment with continuous positive airway pressure: an observational study. Lancet. 2005;365:1046–1053. doi: 10.1016/S0140-6736(05)71141-7. [DOI] [PubMed] [Google Scholar]
- 4.Marin J.M., Agusti A., Villar I., Forner M., Nieto D., Carrizo S.J., Barbé F., Vicente E., Wei Y., Nieto F.J., et al. Association between treated and untreated obstructive sleep apnea and risk of hypertension. JAMA. 2012;307:2169–2176. doi: 10.1001/jama.2012.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahlin C., Sandberg O., Gustafson Y., Bucht G., Carlberg B., Stenlund H., Franklin K.A. Obstructive sleep apnea is a risk factor for death in patients with stroke: a 10-year follow-up. Arch. Intern. Med. 2008;168:297–301. doi: 10.1001/archinternmed.2007.70. [DOI] [PubMed] [Google Scholar]
- 6.Becker-Krail D., McClung C. Implications of circadian rhythm and stress in addiction vulnerability. F1000Research. 2016;5:59. doi: 10.12688/f1000research.7608.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silver R., Kriegsfeld L.J. Circadian rhythms have broad implications for understanding brain and behavior. Eur. J. Neurosci. 2014;39:1866–1880. doi: 10.1111/ejn.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albreecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74:246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Bass J., Lazar M.A. Circadian time signatures of fitness and disease. Science. 2016;354:994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckel-Mahan K., Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol. Rev. 2013;93:107–135. doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Chami M., Shaheen D., Ivers B., Syed Z., Badr M.S., Lin H.S., Mateika J.H. Time of day affects the frequency and duration of breathing events and the critical closing pressure during NREM sleep in participants with sleep apnea. J. Appl. Physiol. 2015;119:617–626. doi: 10.1152/japplphysiol.00346.2015. [DOI] [PubMed] [Google Scholar]
- 13.Butler M.P., Smales C., Wu H., Hussain M.V., Mohamed Y.A., Morimoto M., Shea S.A. The circadian system contributes to apnea lengthening across the night in obstructive sleep apnea. Sleep. 2015;38:1793–1801. doi: 10.5665/sleep.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kushida C.A., Littner M.R., Morgenthaler T., Alessi C.A., Bailey D., Coleman J., Jr., Friedman L., Hirshkowitz M., Kapen S., Kramer M., et al. Practice parameters for the indications for polysomnography and related procedures: an update for 2005. Sleep. 2005;28:499–521. doi: 10.1093/sleep/28.4.499. [DOI] [PubMed] [Google Scholar]
- 15.Flemons W.W., Buysse D., Redline S., Pack A., Strohl K., Wheatley J., Young T., Douglas N., Levy P., McNicholas W., et al. Sleep-related breathing disorders in adults: recommendations for syndrome definition and measurement techniques in clinical research. The Report of an American Academy of Sleep Medicine Task Force. Sleep. 1999;22:667–689. [PubMed] [Google Scholar]
- 16.Lin H.C., Friedman M., Chang H.W., Yalamanchali S. Z-palatopharyngoplasty Combined with Endoscopic Coblator Open Tongue Base Resection for Severe Obstructive Sleep Apnea/Hypopnea Syndrome. Otolaryngol. Head Neck Surg. 2014;150:1078–1085. doi: 10.1177/0194599814524722. [DOI] [PubMed] [Google Scholar]
- 17.Yang M.Y., Chang J.G., Lin P.M., Tang K.P., Chen Y.H., Lin H.Y., Liu T.C., Hsiao H.H., Liu Y.C., Lin S.F. Down-regulation of circadian clock genes in chronic myeloid leukemia: Alternative methylation pattern of hPER3. Cancer Sci. 2006;97:1298–1307. doi: 10.1111/j.1349-7006.2006.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang M.Y., Yang W.C., Lin P.M., Hsu J.F., Hsiao H.H., Liu Y.C., Tsai H.J., Chang C.S., Lin S.F. Altered expression of circadian clock genes in human chronic myeloid leukemia. J. Biol. Rhythms. 2011;6:136–148. doi: 10.1177/0748730410395527. [DOI] [PubMed] [Google Scholar]
- 19.Burioka N., Koyanagi S., Endo M., Takata M., Fukuoka Y., Miyata M., Takeda K., Chikumi H., Ohdo S., Shimizu E. Clock gene dysfunction in patients with obstructive sleep apnoea syndrome. Eur. Respir. J. 2008;32:105–112. doi: 10.1183/09031936.00138207. [DOI] [PubMed] [Google Scholar]
- 20.Boivin D.B., James F.O., Wu A., Cho-Park P.F., Xiong H., Sun Z.S. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102:4143–4145. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 21.Fukuya H., Emoto N., Nonaka H., Yagita K., Okamura H., Yokoyama M. Circadian expression of clock genes in human peripheral leukocytes. Biochem. Biophy. Res. Comm. 2007;354:924–928. doi: 10.1016/j.bbrc.2007.01.063. [DOI] [PubMed] [Google Scholar]
- 22.Kusanagi H., Hida A., Satoh K., Echizenya M., Shimizu T., Pendergast J.S., Yamazaki S., Mishima K. Expression profiles of 10 circadian clock genes in human peripheral blood mononuclear cells. Neurosci. Res. 2008;61:136–142. doi: 10.1016/j.neures.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Archer S.N., Viola A.U., Kyriakopoulou V., von Schantz M., Dijk D.J. Inter-individual differences in habitual sleep timing and entrained phase of endogenous circadian rhythms of BMAL1, PER2 and PER3 mRNA in human leukocytes. Sleep. 2008;31:608–617. doi: 10.1093/sleep/31.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hida K., Kusanagi H., Satoh K., Kato T., Matsumoto Y., Echizenya M., Shimizu T., Higuchi S., Mishima K. Expression profiles of PERIOD1, 2, and 3 in peripheral blood mononuclear cells from older subjects. Life Sci. 2009;84:3–7. doi: 10.1016/j.lfs.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Moreira S., Rodrigues R., Barros A.B., Pejanovic N., Neves-Costa A., Pedroso D., Pereira C., Fernandes D., Rodrigues J.V., Barbara C., et al. Changes in Expression of the CLOCK Gene in Obstructive Sleep Apnea Syndrome Patients Are Not Reverted by Continuous Positive Airway Pressure Treatment. Front. Med. 2017;4:187. doi: 10.3389/fmed.2017.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canales M.T., Holzworth M., Bozorgmehri S., Ishani A., Weiner I.D., Berry R.B., Beyth R.J., Gumz M. Clock gene expression is altered in veterans with sleep apnea. Physiol. Genomics. 2019;51:77–82. doi: 10.1152/physiolgenomics.00091.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasan S., van der Veen D.R., Winsky-Sommerer R., Dijk D.J., Archer S.N. Altered sleep and behavioral activity phenotypes inPER3-deficient mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;301:R1821–R1830. doi: 10.1152/ajpregu.00260.2011. [DOI] [PubMed] [Google Scholar]
- 28.Parsons M.J., Lester K.J., Barclay N.L., Archer S.N., Nolan P.M., Eley T.C., Gregory A.M. Polymorphisms in the circadian expressed genes PER3 and ARNTL2 are associated with diurnal preference and GNβ3 with sleep measures. J. Sleep Res. 2014;23:595–604. doi: 10.1111/jsr.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hida A., Kitamura S., Katayose Y., Kato M., Ono H., Kadotani H., Uchiyama M., Ebisawa T., Inoue Y., Kamei Y., et al. Screening of clock gene polymorphisms demonstrates association of a PER3 polymorphism with morningness-eveningness preference and circadian rhythm sleep disorder. Sci. Rep. 2014;4:6309. doi: 10.1038/srep06309. [DOI] [PMC free article] [PubMed] [Google Scholar]