Abstract
The calcineurin pathway is an important signaling cascade for growth, sexual development, stress response, and pathogenicity in fungi. In this study, we investigated the function of CrzA, a key transcription factor of the calcineurin pathway, in an aflatoxin-producing fungus Aspergillus flavus (A. flavus). To examine the role of the crzA gene, crzA deletion mutant strains in A. flavus were constructed and their phenotypes, including fungal growth, spore formation, and sclerotial formation, were examined. Absence of crzA results in decreased colony growth, the number of conidia, and sclerocia production. The crzA-deficient mutant strains were more susceptible to osmotic pressure and cell wall stress than control or complemented strains. Moreover, deletion of crzA results in a reduction in aflatoxin production. Taken together, these results demonstrate that CrzA is important for differentiation and mycotoxin production in A. flavus.
Keywords: CrzA, Calcineurin, aflatoxin, Aspergillus flavus, Asexual development
1. Introduction
Aspergillus flavus is widespread in our environment and contaminates crops such as maize, corn, and peanut [1,2,3]. A. flavus spores germinate in crops and produce detrimental secondary metabolite mycotoxins, including aflatoxins [4,5]. Aflatoxins are harmful fungal mycotoxins that cause carcinogenesis in animals and humans [6,7]. A. flavus is also an opportunistic pathogen in humans and is the second leading pathogen of invasive aspergillosis [8,9,10]. Because of the harmful effects of this fungus, A. flavus causes economic loss to humankind, and developing a better understanding of the biology of this fungus is vital to reduce the adverse effects of A. flavus on humanity.
A. flavus is a heterothallic fungus that can reproduce both sexually and asexually [11,12,13]. Sclerotia are sexual reproductive structures that can be produced by a single strain of one mating type or a cross between opposite mating type strains [14]. Sclerotia can be widespread in soil or crops and survive environmental stresses [14]. A. flavus also produces asexual spores, called conidia, that act as propagates and are infectious [1]. Asexual development is the primary reproductive mode in most Aspergillus species [15]. A. flavus produces the asexual-specific structure, conidiophores, during asexual development. The production of conidiophores is tightly controlled by a variety of positive and negative regulators [16]. Both asexual and sexual development is regulated by several signaling pathways in Aspergillus species [17,18].
The Calcium-calcineurin signaling pathway is highly conserved and plays key roles in fungi, including adaptation to hosts or environments, fungal growth, pathogenesis, and sexual reproduction [19,20]. Calcium is a key ion for signal transduction in response to various environmental stresses [21,22]. During stress, intracellular calcium concentration increases, and then calcium ions in cytoplasm bind to calmodulin. The calcium-calmodulin complex activates the calmodulin-dependent phosphatase calcineurin [23]. Calcineurin dephosphorylates certain targets that function to control the stress response, growth, fungal virulence, and pathogenesis in fungi [24,25]. In fungi, one of the key target proteins of calcineurin is a C2H2 transcription factor Crz1 (or CrzA, calcineurin responsive zinc finger 1) [26]. Activated Crz1 regulates certain genes involved in cell wall integrity and virulence [26]. A variety of studies found that Crz1 plays vital roles in ion homeostasis, cell wall biogenesis, fungal development, stress response, and pathogenesis [26]. In Saccharomyces cerevisiae, loss of CRZ1 results in alteration of mRNA expression of genes involved in cell wall biogenesis, ion homeostasis, and vesicle transport, suggesting that Crz1 is crucial for cell wall biogenesis and ion homeostasis [27,28]. In human pathogenic fungi such as Cryptococcus neoformans and Candida albicans, the absence of CRZ1 attenuates virulence in mouse models [29,30,31]. Crz1 is also involved in fungal pathogenesis in plant pathogenic fungi Magnaporthe oryzae and Botrytis cinerea [32,33]. In Aspergillus species, CrzA is also required for fungal growth, asexual development, and septation [19,34,35]. Absence of crzA results in reduced hyphal growth and asexual development in Aspergillus nidulans [36,37,38]. CrzA is also required for aflatoxin production in A. parasiticus [39]. In A. fumigatus, ΔcrzA mutant strains exhibit abnormal hyphae morphology and septation, reduced asexual sporulation, attenuated virulence, and increased susceptibility to ionic stresses [34,35]. Genome-wide analyses of crzA in A. fumigatus demonstrated that CrzA regulates gene expression associated with ion transport, secondary metabolism, lipid metabolism, and vesicle trafficking [40,41]. These results strongly suggest that CrzA plays diverse roles in Aspergillus species [19,26]. Although CrzA plays important roles in Aspergillus species, these roles have not been demonstrated in A. flavus. In this study, we characterized CrzA in the aflatoxin-producing fungi A. flavus. To examine the role of CrzA, crzA deletion mutant (ΔcrzA) strains were generated, and their phenotypes were analyzed. Similar to the CrzA role in other Aspergillus species, our results demonstrate that CrzA is essential for fungal growth, asexual spore formation, stress response, and aflatoxin production in A. flavus.
2. Results and Discussion
2.1. Summary of CrzA in A. flavus
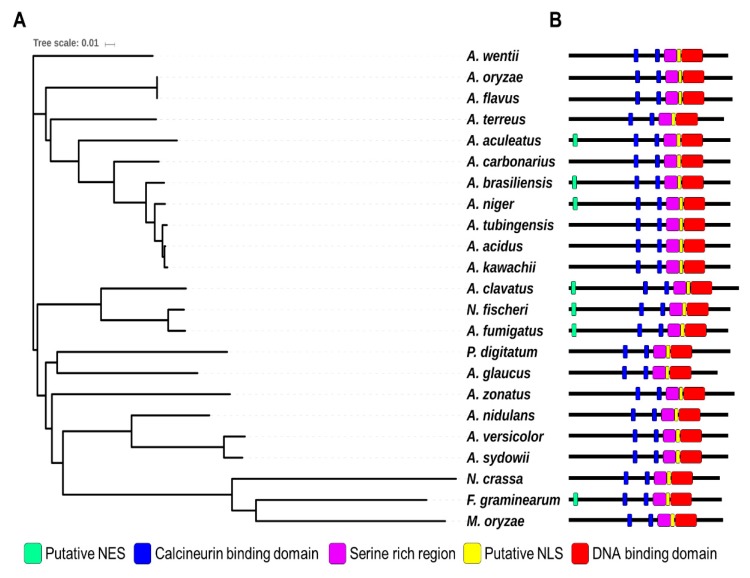
Crz1 is a C2H2-type transcription factor that is a key target for calcineurin in yeast and filamentous fungi [26]. Recent studies analyzed the function and domains of CrzA in two Aspergillus species A. fumigatus and A. nidulans [42,43]. CrzA from both species contains putative a calcineurin-binding domain (CBD), DNA-binding domain (DBD, C2H2 domain), nuclear localization signal (NLS), and serine-rich region. Although A. fumigatus CrzA contains a nuclear export signal (NES) at its N-terminus, A. nidulans CrzA has no NES at its N-terminus. To identify CrzA in A. flavus, we screened the A. flavus NRRL 3357 genome [44] using the protein sequence of A. fumigatus CrzA (XP_750439) and A. nidulans CrzA (XP_663330.1). XP_002381985.1 was identified by protein homology (69% and 67% identity with A. fumigatus CrzA and A. nidulans CrzA, respectively) (Figure 1). AFLA_127920, a putative crzA gene, encodes a 773-amino acid protein that contains a DNA binding domain with two C2H2 Zn-finger domains at the C-terminus. CrzA also has a calcineurin binding domain with calcineurin docking sequence (PxIxT) [45]. The A. flavus CrzA protein contains a putative NLS element (nls-mapper.iab.keio.ac.jp) [45]. However, A. flavus CrzA does not contain an NES at the N-terminus, unlike the A. fumigatus CrzA. The A. flavus CrzA protein also contains CBD and serine-rich regions that are essential for regulating the activity of CrzA [42,43]. The DBD, CBD, and NLS elements of the CrzA homologs in most Aspergillus spp are highly conserved, but the NES element is diverse in 19 Aspergillus species (Figure 1). These results suggest that AFLA_127920 encodes the CrzA homolog of A. flavus.
Figure 1.
Summary of CrzA in Aspergillus species. (A) A phylogenetic tree of the CrzA homologs identified in 23 fungal species including A. flavus AFL3357 (AFL2T_09134), A. oryzae RIB40 (AO090001000491), A. fumigatus Af293 (Afu1g06900), A. nidulans FGSC4 (AN5726), A. niger CBS 513.88 (An18g05920), A. clavatus NRRL 1 (ACLA_027670), A. sydowii (Aspsy1_0149570), A. tubingensis CBS 134.48 (Asptu1_0114323), A. acidus CBS 106 47 (Aspfo1_0139906), A. brasiliensis CBS 101740 (Aspbr1_0197410), A. versicolor CBS 583.65 (Aspve1_0132792), A. kawachii (Aspka1_0180272), A. carbonarius ITEM 5010 (Acar5010_128721), A. aculeatus ATCC16872 (Aacu16872_028665), A. terreus NIH2624 (ATET_02928), A. wentii DTO 134E9 (Aspwe1_0050154), A. zonatus (Aspzo1_0014943), A. glaucus CBS 516 65 (Aspgl1_0031266), Neosartorya fischeri NRRL 181 (NFIA_017790), Penicillium digitatum Pd1 (XP_014531047.1), Neurospora crassa OR74A (XP_962085), Magnaporthe oryzae 70-15 (MGG_05133), and Fusarium graminearum PH-1 (FGSG_01341). A phylogenetic tree of CrzA homologs (or orthologs) was generated by the Clustal Omega package (https://www.ebi.ac.uk/Tools/msa/clustalo/). The tree result was submitted to iTOL (http://itol.embl.de/) to generate the figure. (B) Domains of the CrzA homologs in fungal species. NLS (Nuclear Localization Sequence) and NES (Nuclear Export Signal)
2.2. Roles of CrzA in Asexual Development
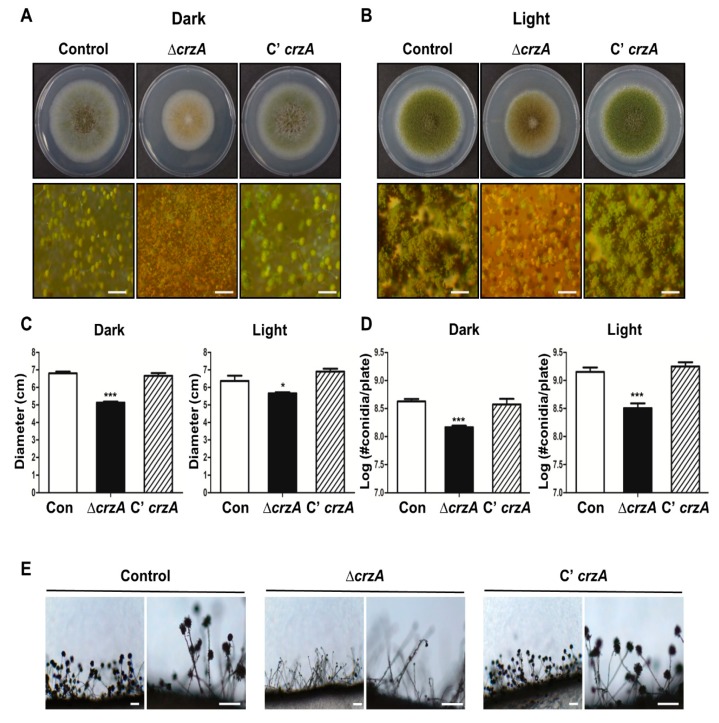
To test the functions of CrzA in A. flavus, the crzA deletion (∆crzA) mutants and complemented strains (C’ crzA) were generated and compared the phenotype of the ∆crzA mutant with the control strain and the complemented strain. Growth and conidiation under light and dark conditions for wild type (WT) and ∆crzA mutant strains were evaluated (Figure 2A,B). The colony diameter of the ΔcrzA strains was significantly decreased compared with control and the complemented strains (Figure 2C). The number of asexual spores in ΔcrzA strains was lower in both light and dark conditions (Figure 2D). The ΔcrzA strains produce light green or brown conidiophores, while control and complemented strains produce green conidiophores (Figure 2A). Microscopic analysis exhibited that the ΔcrzA strains produce abnormal conidiophores in the light condition (Figure 2E). Overall, these results suggest that CrzA is not only necessary for fungal growth but also play a critical role in the formation of conidiophores in A. flavus.
Figure 2.
Phenotypes of the ΔcrzA mutant. (A,B) Colony phenotypes of control (TTJ6.1), ΔcrzA (TDH1.1), and C’crzA (TTJ9.1) strains point-inoculated on solid glucose minimal medium with 0.1% yeast extract (MMYE) media and grown at 37 °C under dark (A) or light (B) condition for 5 days (Scale bars = 500 μm). Quantitative analysis of colony diameter (C) and the number of conidia per plate (D) of WT and velvet deletion mutant strains are shown in (A,B). Error bars indicates the standard errors from the mean of triplicates. (Control versus ΔcrzA strains, * p ≤ 0.05; *** p ≤ 0.001). (E) Conidia formation of control (TTJ6.1), ΔcrzA (TDH1.1), and C’crzA (TTJ9.1) strains was observed under a light microscope at 48 h after inoculation onto solid MMYE media at 37 °C. (Scale bars = 250 μm)
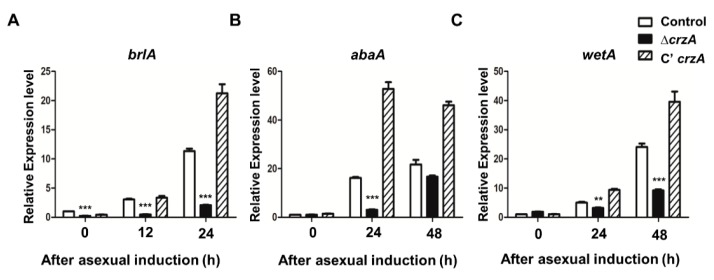
Because CrzA plays an important role in conidiophore production, mRNA expression of genes that regulate asexual development including brlA, abaA, and wetA was assessed in WT and the ΔcrzA mutant strains. We first examined the mRNA expression of brlA, a key initiator of conidiation, during the early phase of conidiation (after asexual induction 12–24 h). The brlA mRNA level was lower in the ΔcrzA mutant compared to control and complemented strains (Figure 3A). Similarly, abaA and wetA mRNA levels in the ΔcrzA mutant were lower compared to those in control and complemented strains (Figure 3B,C). These results demonstrate that CrzA is crucial for the proper expression of key regulators of asexual development.
Figure 3.
mRNA expression of asexual developmental genes in the ΔcrzA mutant. (A–C) mRNA levels of key regulator genes brlA (A), abaA (B), and wetA (C), in control (TTJ6.1), ΔcrzA (TDH1.1), and C’crzA (TTJ9.1) strains after induction of asexual development were assessed by quantitative reverse transcript PCR (qRT-PCR). Error bars indicates the standard errors from the mean of triplicates. (Control versus ΔcrzA strains, ** p ≤ 0.01; *** p ≤ 0.001).
As mention above, CrzA is required for proper the morphology and production of conidiophores. The color of conidiophores of the ΔcrzA mutant strain was changed to brownish in both dark and light conditions (Figure 2A,B). The size of conidiophore heads and the number of conidia in the ΔcrzA mutant strains were each decreased compared to control strains (Figure 2D,E). In addition, the ΔcrzA mutant strains produce less amount of conidia compare to control strains. The phenotypes of ΔcrzA mutant in other Aspergillus species A. nidulans [37] and A. fumigatus [34] are similar to those of the ΔcrzA mutant in A. flavus, implying that CrzA play a conserved role in asexual development in Aspergillus species. Absence of crzA decreased the mRNA expression of brlA, abaA, and wetA (Figure 3). In the model organism A. nidulans and the pathogenic fungus A. fumigatus, brlA expression is dependent on CrzA during asexual development [34]. These results suggest that the altered brlA mRNA expression by deletion of crzA affects morphogenesis of conidiophores in Aspergillus species.
2.3. Roles of CrzA in Sclerocia Formation
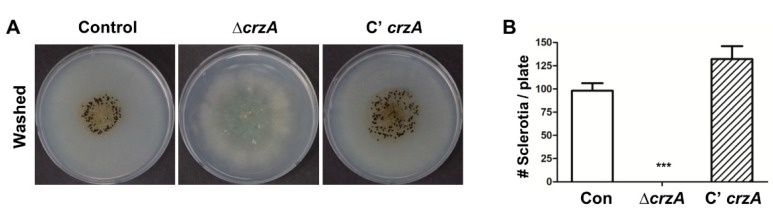
Because CrzA in A. parasiticus is crucial for sclerotial development [39], we examined the role of crzA in sclerocia formation in A. flavus. Control and mutant strains were inoculated into glucose minimal medium with 0.1% yeast extract (MMYE) media and cultured for 7 days under dark conditions. While WT and control and complemented strains produce conidiophores and sclerocia, the ΔcrzA strains do not produce developmental structures under dark conditions, suggesting that CrzA is essential for proper sclerocia production in A. flavus (Figure 4).
Figure 4.
Sclerocia production in the ΔcrzA mutant. Ethanol-washed colony photographs of control (TTJ6.1), ΔcrzA (TDH1.1), and C’crzA (TTJ9.1) strains grown on solid MMYE media for 7 days. (B) Quantitative analysis of sclerocia of strains shown in (A). (Control versus ΔcrzA strains, *** p ≤ 0.001).
2.4. Roles of CrzA in Stress Response
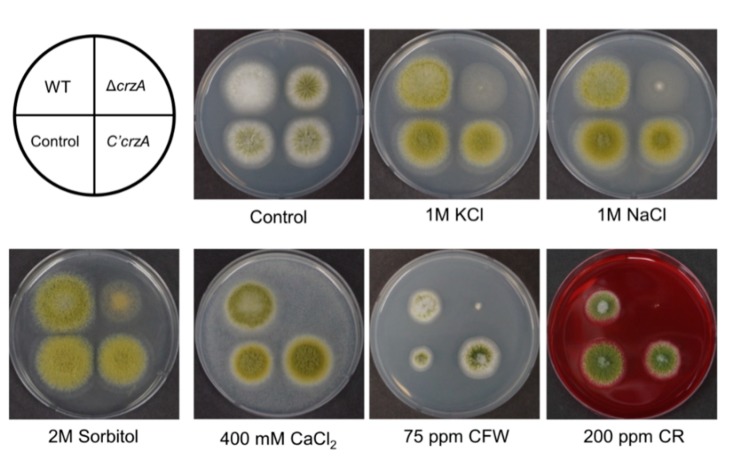
CrzA homologs are involved in cation homeostasis and stress response in A. nidulans and A. fumigatus [34,35,37,46]. Control and mutant strains were inoculated in MMYE with several compounds that are related with osmotic stress (sorbitol, NaCl, and KCl), ion homeostasis (CaCl2), and cell wall stress (calcofluor white (CFW) and Congo red (CR)) (Figure 5). The ΔcrzA mutant strains exhibit reduced colony growth or fungal development under all conditions tested. Importantly, ΔcrzA mutant strains cannot grow at high concentrations of Ca2+ ions. However, ΔcrzA mutant strains are not sensitive to oxidative stresses (1–5 mM H2O2, data not shown). We also examined the thermo-tolerance of ΔcrzA mutant strains, but the thermal sensitivity of ΔcrzA mutant strains was similar to WT. We then checked the contents of trehalose, a protectant for several stresses, in mutant conidia [47]. However, trehalose content of the ΔcrzA mutant conidia was the same as that in WT conidia (Figure S1). This result suggests that CrzA may not be involved in trehalose biosynthesis. Overall, these results demonstrated that the growth of ΔcrzA mutant strains was reduced in various stress conditions in A. flavus (Figure 5), implying that the role of CrzA in cell wall integrity and the stress tolerance is conserved in most filamentous fungi.
Figure 5.
Phenotypes of the ΔcrzA mutant in various stress conditions. WT (NRRL3357), control (TTJ6.1), ΔcrzA (TDH1.1), and C’crzA (TTJ9.1) strains were point inoculated at 37 °C for 3 days on solid MMYE medium containing various compounds including KCl, NaCl, sorbitol, CaCl2, Calcoflour white (CFW), and Congo red (CR).
Previous results from genome-wide analyses found CrzA governs mRNA expression of certain genes associated with calcium metabolism, ion transport, and cell wall integrity [34,36]. Several genes contain CrzA-binding sites (G[A/T]GGC[G/C]) in their promoter regions [41,48,49], thus, we measured the mRNA levels of putative target genes, including rcnA, chsB, pmrA, and pmcA (Figure S2). Under normal conditions, the mRNA expression of these three genes decreased in the ΔcrzA mutant strains in liquid culture. We then added calcium ions to the liquid media and measured mRNA expression. The mRNA levels of rcnA and pmcA, but not chsB were increased in response to high calcium concentration media in WT. However, the mRNA levels of rcnA and pmcA were dramatically decreased in the ΔcrzA mutant strains. While the expression levels of three genes rcnA, chsB, and pmcA were decreased in the ΔcrzA mutant than those in control, pmrA mRNA level was not changed in the ΔcrzA mutant. These results indicate that CrzA supports rcnA and pmcA expression and this activity is conserved in other Aspergillus species [34,43]. Overall, these finding indicate that the roles of CrzA in cell wall biogenesis and stress response are conserved in Aspergillus species, but target genes of CrzA are diverse in Aspergillus species. Additional genome-wide analyses in A. flavus will provide insight into the role of CrzA in Aspergillus species.
2.5. Roles of CrzA in Aflatoxin B1 Production
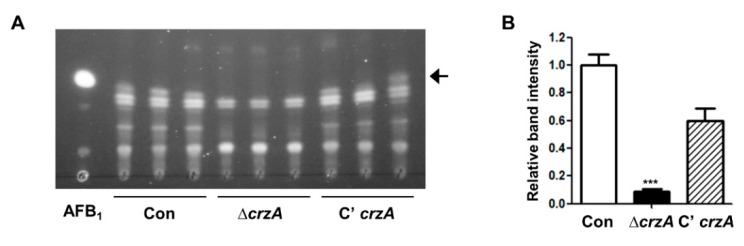
Because CrzA is essential for aflatoxin B1 production in A. parasiticus [39], we examined aflatoxin production in A. flavus. Control, ΔcrzA, and C’crzA strains were inoculated into liquid media and incubated for 7 days under the dark condition. As shown Figure 6, control and complemented strains produce aflatoxin B1, whereas ΔcrzA strains do not produce aflatoxin B1, suggesting that CrzA is required for aflatoxin B1 production. The spot pattern of ΔcrzA mutant on TLC plates was different from that of control strains, speculating that deletion of crzA can alter secondary metabolite biosynthesis (Figure 6). Further high-performance liquid chromatography (HPLC) or other analytic analyses will help to understand the role of CrzA in secondary metabolite biosynthesis.
Figure 6.
Aflatoxin production in the ΔcrzA mutant. (A) Thin-layer chromatography (TLC) of aflatoxin B1 (AFB1) from control (TTJ6.1), ΔcrzA (TDH1.1), and C’crzA (TTJ9.1) strains under dark conditions for 7 days. Black arrow indicates aflatoxin B1. (B) Densitometry of the TLC analysis results is shown in (A). (Control versus ΔcrzA strains, *** p ≤ 0.001).
2.6. Roles of CrzA in Host Colonization
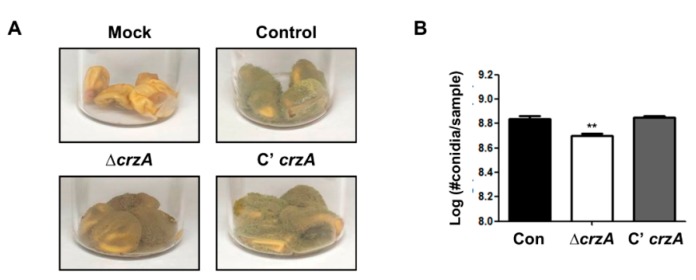
We then examined whether loss of crzA also affects host colonization. Control, ΔcrzA mutant, and complemented strains were inoculated in kernels and incubated for 5 days under light conditions. During conidial production, ΔcrzA mutant strains exhibit a lower sporulation level compared to control and complemented strains (Figure 7).
Figure 7.
Kernel assay in the δcrza mutant. (A) Infected kernels were co-incubated with control (TTJ6.1), ΔcrzA (TDH1.1), and C’crzA (TTJ9.1) strains for 5 days. (B) The number of conidia per samples (A). (Control versus ΔcrzA strains, ** p ≤ 0.01).
3. Conclusions
CrzA is a putative calcineurin target that coordinates fungal growth and asexual or sexual reproduction in A. flavus. The role of CrzA in stress response and cell wall integrity is conserved in Aspergillus species. Importantly, CrzA is crucial for aflatoxin production and host infection. Taken together, we propose that inhibition of calcineurin and CrzA could inhibit survival and aflatoxin production in a variety of environmental conditions and that calcineurin could be an attractive target for anti-fungal agents.
4. Materials and Methods
4.1. Fungal Strains and Culture Conditions
Table 1 shows a list of strains used in this study. To culture A. flavus strains, glucose minimal medium with 0.1% yeast extract (MMYE) was used [50]. A. flavus NRRL 3357.5 (pyrG- auxotrophic mutant strain) was used for transformation and was grown on MMYE with supplements (5 mM uridine (Acros organics, NJ, USA) and 10 mM uracil (Acros organics, NJ, USA)) [51].
Table 1.
Aspergillus flavus strains used in this study.
| Strain Name | Relevant Genotype | References |
|---|---|---|
| NRRL3357 | A. flavus wild type | FGSC 1 |
| NRRL3357.5 | pyrG - | (He et al. 2007) |
| TTJ6.1 | pyrG-; ΔpyrG::AfupyrG+ | This study |
| TDH1.1-3 | pyrG-; ΔcrzA::AfupyrG | This study |
| TTJ9.1-2 | pyrG-; ΔcrzA::AfupyrG; crzA(p)::crzA::FLAG4x::ptrA | This study |
1 Fungal Genetic Stock Center.
4.2. Generation of the CrzA Mutant Strains
The primers used in this study are listed in Table 2. The double-joint PCR (DJ-PCR) method was used for generation of the crzA deletion mutant strain [52]. The 5′ and 3′ flanking regions of the crzA gene were both amplified from A. flavus NRRL 3357 genomic DNA using OHS357:OHS358 and OHS359:OHS360, respectively. The A. fumigatus pyrG+ marker was amplified with the primer pair OHS089:OHS090 from A. fumigatus AF293 genomic DNA. The crzA deletion cassette was amplified with OHS361:OHS362 and introduced into protoplasts of the recipient strain A. flavus NRRL 3357.5. Protoplasts were generated using the VinoTaste® Pro (Novozymes, Denmark) and lysing enzyme (Sigma, St Louis, MO, USA) [53]. Genomic DNA of transformants was isolated and the coding region of crzA was amplified. PCR fragments were digested by several restriction enzyme to verity deletion mutants. Three independent ΔcrzA strains (TDH1.1-3) were isolated.
Table 2.
Oligonucleotides used in this study.
| Name | Sequence (5′ → 3′) a | Purpose |
|---|---|---|
| OHS089 | GCTGAAGTCATGATACAGGCCAAA | AfupyrG Maker_F |
| OHS090 | ATCGTCGGGAGGTATTGTCGTCAC | AfupyrG Maker_R |
| OHS357 | GTTGTCGTCAGGCACCGTCA | 5′ flanking region of crzA |
| OHS358 | GGCTTTGGCCTGTATCATGACTTCA GATACCACGCGAAGCAAGGC | crzA with AfupyrG tail R |
| OHS359 | TTTGGTGACGACAATACCTCCCGAC AGGGTGGAAACCAGATGGCG | crzA with AfupyrG tail F |
| OHS360 | CACCCATTGGTGTTGCGCTC | 3′ flanking region of crzA |
| OHS361 | GGGACGCGGTAGATTGTGCT | crzA nested 5′ NF |
| OHS362 | TCGCACGCTTCCTTCAGGAG | crzA nested 5′ NR |
| OHS367 | AATT GCGGCCGC GGGTTGGGATCCACCGCTTA | 5′ crzA with pro and NotI |
| OHS368 | AATT GCGGCCGC GGCATACCCCGAGTCCCCA | 3′ crzA with NotI |
| OHS407 | GATATGTCGCCACACTGGAC | AflbrlA F_RT |
| OHS408 | CTGTATTCGCGGCTATTCGG | AflbrlA R_RT |
| OHS409 | CTTCCGCACCTTAACAGCAG | AflabaA F_RT |
| OHS410 | GTTTGCCGGAATTGCCAAAG | AflabaA R_RT |
| OHS411 | CCGTCAGATATCCTGCCACA | AflwetA F_RT |
| OHS412 | ATGGATATCGCGGGAGATGG | AflwetA R_RT |
| OHS405 | TATGTCGGTGATGAGGCACA | Aflactin F_RT |
| OHS406 | AACACGGAGCTCGTTGTAGA | Aflactin R_RT |
a Tail sequence is in italic. Restriction enzyme sites are in bold.
For complemented crzA strains, the promoter and ORF regions of crzA was amplified with the primer pair OHS367/OHS368, purified, digested with NotI, and cloned into pMJ4 containing a 4xFLAG tag, the pyrithiamine-resistance gene (ptrA), and the amyB terminator. Then, the resulting plasmid pTJ5 was introduced into the recipient ∆crzA strain TDH1.1 to give rise to TTJ9.1-2. Transformants were verified by qRT-PCR (Figure S3).
4.3. Physiological Studies
To determine the number of conidia, control and mutant strains were point-inoculated onto solid MMYE and incubated at 30 °C for 5 days under dark or light conditions. Colony diameter for each strain was then measured for fungal growth. For conidial production, conidia from each strain were collected from the entire plate and counted using a hemocytometer. For sclerocia production, fungal strains were point-inoculated onto solid MMYE and incubated at 30 °C for 7 days under dark conditions. The cultured plates were then washed with 70% ethanol to facilitate sclerotia counting. All experiments were carried out in triplicate.
4.4. Quantitative Reverse Transcript PCR (qRT-PCR)
Samples were collected as previously described [54,55]. WT and mutants conidia were inoculated into 500 mL of liquid MMYE and incubated at 37 °C and 200 rpm for 16 h. The mycelia were collected by filtering through autoclaved Miracloth (Calbiochem, CA, USA), mono-layered on solid MMYE, and then incubated at 37 °C under light conditions. For RNA extraction, individual samples were collected at a designated time point.
Total RNA isolation was carried out as previously described [56]. Briefly, each sample was mixed with TRIzol Reagent (Invitrogen, USA) and 0.5 mm Zirconia/Silica beads (RPI, IL, USA), and then homogenized using a Mini-Beadbeater (BioSpec Products Inc, OK, USA). After homogenization, the aqueous phase was separated by centrifugation and transferred to new vials. An equal volume of 70% ethanol was added into the aqueous phase and mixed gently. After centrifugation, RNA was collected. To remove genomic contamination, each sample was treated with DNase I (Promega, WI, USA) and pure RNA was isolated using RNeasy mini Kit (Qiagen Inc., Valencia, USA). The RNA concentration was measured by ultraviolet (UV) spectrometry.
To synthesize complementary DNA (cDNA), the GoScript Reverse Transcription system (Promega, WI, USA) was used. Quantitative PCR was performed with iTaq universal SYBR Green supermix (Bio-Rad, CA, USA) and each gene-specific primer set using a CFX96 Touch Real-Time PCR system (Bio-Rad, CA, USA). To calculate the expression levels of target genes, the 2−∆∆CT method was used. For endogenous control, β-actin gene was used. Results shown in this study are representative of two independent experimental replicates. The primers used for quantitative PCR are listed in Table 2.
4.5. Stress Tests
To examine susceptibility to various stresses, 103–104 conidia were inoculated onto MMYE agar media containing the following compounds; Congo red (CR, Sigma, St. Louis, MO, USA) and calcofluor white (CFW, Sigma) for cell wall stress, CaCl2 (Sigma) for ion stress, and NaCl (Fisher, Fair Lawn, NJ, USA), KCl (Fisher), and Sorbitol (Fisher) for osmotic stress. The plates were incubated at 30 °C and photographed between 3 and 5 days post-treatment.
4.6. Aflatoxin Extraction and Thin-Layer Chromatography Analysis
Aflatoxin B1 from each sample was extracted as described by previously [56], with modification. Briefly, approximately 106 spores of each strain were inoculated into 5 mL of liquid complete medium (CM) and cultured for 7 d at 30 °C under dark conditions. To extract Aflatoxin B1, CHCl3 (5 mL) was added to the cultured media and mixed vigorously by a Voltex mixer. The mixed sample was centrifuged and the organic phase was transferred to new vials. Sample was evaporated and resuspended in 50 μL of CHCl3 for TLC (Thin layer chromatography) analysis. Suspended solutions containing Aflatoxin B1 were spotted onto a TLC silica plate (Kiesel gel 60, 0.25 mm; Merck). The plate was putted into a chamber containing chloroform: acetone (9:1, v/v). To capture the image, the TLC plates were exposure to ultra violet (UV) light (366 and 254 nm).
4.7. Kernel Bioassays
Kernel bioassays were carried out as described previously [57]. Corn kernels (Chungnong, Bucheon, South Korea) were washed in 70% ethanol, rinsed with ddH2O, and incubated in bleach. After then, the kernels were washed using ddH2O to remove the bleach, and then dried on paper towels. A hole in the embryo of each kernel was made using a sterile needle. Kernels were put into a new vial and inoculated with a conidial suspension (2 × 105 conidia/mL). After inoculation, vials were placed in a 37 °C incubator under light conditions for 5 days. After the 5-day incubation with light, 2 mL of 0.01% Tween 20 was added to each vial, and then 1 mL of conidial suspension was removed from the vials to count the number of conidia using a hemocytometer.
4.8. Microscopy
Colony photographs were taken with a Pentax MX-1 digital camera. Microscopic images were taken using a Zeiss Lab.A1 microscope equipped with AxioCam 105 and AxioVision (Rel. 4.9) digital imaging software.
4.9. Statistical Analysis
For statistical analysis, GraphPad Prism Version 5.01 software was used. Statistical differences between control and mutant strains were analyzed by the Student’s unpaired t-test. Error bars indicates the standard errors from the mean of triplicates. P values < 0.05 were considered to be significant different between control and ΔcrzA mutant strain. (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001).
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6651/11/10/567/s1, Figure S1: The amount of trehalose in ΔcrzA mutant conidia, Figure S2: Expression of putative calcineurin target genes, Figure S3: Verification of crzA mutants by qRT-PCR, Table S1: Oligonucleotides used for Figure S2.
Author Contributions
S.-Y.L., Y.-E.S., D.-H.L., T.-J.E., and M.-J.K. designed and performed the experiments. S.-Y.L. and H.-S.P. analyzed the date. H.-S.P. wrote the manuscript. S.-Y.L. and Y.-E.S. assisted with writing the manuscript.
Funding
The study at KNU was supported by a National Research Foundation of Korea (NRF) grant, funded by the Korean government (NRF-2019R1F1A1048574).
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
The putative calcineurin target CrzA plays a vital role in fungal growth, differentiation, stress response, and aflatoxin production in an aflatoxin-producing fungus Aspergillus flavus.
References
- 1.Amaike S., Keller N.P. Aspergillus flavus . Annu. Rev. Phytopathol. 2011;49:107–133. doi: 10.1146/annurev-phyto-072910-095221. [DOI] [PubMed] [Google Scholar]
- 2.Fountain J.C., Scully B.T., Ni X., Kemerait R.C., Lee R.D., Chen Z.Y., Guo B. Environmental influences on maize-Aspergillus flavus interactions and aflatoxin production. Front. Microbiol. 2014;5:40. doi: 10.3389/fmicb.2014.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedayati M.T., Pasqualotto A.C., Warn P.A., Bowyer P., Denning D.W. Aspergillus flavus: Human pathogen, allergen and mycotoxin producer. Microbiology. 2007;153:1677–1692. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- 4.Klich M.A. Aspergillus flavus: The major producer of aflatoxin. Mol. Plant Pathol. 2007;8:713–722. doi: 10.1111/j.1364-3703.2007.00436.x. [DOI] [PubMed] [Google Scholar]
- 5.Bennett J.W., Klich M. Mycotoxins. Clin. Microbiol. Rev. 2003;16:497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kensler T.W., Roebuck B.D., Wogan G.N., Groopman J.D. Aflatoxin: A 50-year odyssey of mechanistic and translational toxicology. Toxicol. Sci. 2011;120:S28–S48. doi: 10.1093/toxsci/kfq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar P., Mahato D.K., Kamle M., Mohanta T.K., Kang S.G. Aflatoxins: A Global Concern for Food Safety, Human Health and Their Management. Front. Microbiol. 2016;7:2170. doi: 10.3389/fmicb.2016.02170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan S., Manavathu E.K., Chandrasekar P.H. Aspergillus flavus: An emerging non-fumigatus Aspergillus species of significance. Mycoses. 2009;52:206–222. doi: 10.1111/j.1439-0507.2008.01642.x. [DOI] [PubMed] [Google Scholar]
- 9.Pasqualotto A.C. Differences in pathogenicity and clinical syndromes due to Aspergillus fumigatus and Aspergillus flavus. Med. Mycol. 2009;47:S261–S270. doi: 10.1080/13693780802247702. [DOI] [PubMed] [Google Scholar]
- 10.Rudramurthy S.M., Paul R.A., Chakrabarti A., Mouton J.W., Meis J.F. Invasive Aspergillosis by Aspergillus flavus: Epidemiology, Diagnosis, Antifungal Resistance, and Management. J. Fungi. 2019;5:55. doi: 10.3390/jof5030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horn B.W., Moore G.G., Carbone I. Sexual reproduction in Aspergillus flavus. Mycologia. 2009;101:423–429. doi: 10.3852/09-011. [DOI] [PubMed] [Google Scholar]
- 12.Krijgsheld P., Bleichrodt R., van Veluw G.J., Wang F., Muller W.H., Dijksterhuis J., Wosten H.A. Development in Aspergillus. Stud. Mycol. 2013;74:1–29. doi: 10.3114/sim0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ojeda-Lopez M., Chen W., Eagle C.E., Gutierrez G., Jia W.L., Swilaiman S.S., Huang Z., Park H.S., Yu J.H., Canovas D., et al. Evolution of asexual and sexual reproduction in the aspergilli. Stud. Mycol. 2018;91:37–59. doi: 10.1016/j.simyco.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horn B.W., Gell R.M., Singh R., Sorensen R.B., Carbone I. Sexual Reproduction in Aspergillus flavus Sclerotia: Acquisition of Novel Alleles from Soil Populations and Uniparental Mitochondrial Inheritance. PLoS ONE. 2016;11:e0146169. doi: 10.1371/journal.pone.0146169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes M., Keller N.P., Adams T.H. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 1998;28:1355–1365. doi: 10.1046/j.1365-2958.1998.00907.x. [DOI] [PubMed] [Google Scholar]
- 16.Park H.S., Yu J.H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012;15:669–677. doi: 10.1016/j.mib.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Park H.S., Lee M.K., Han K.H., Kim M.J., Yu J.H. Developmental Decisions in Aspergillus nidulans. In: Hoffmeister D., Gressler M., editors. Biology of the Fungal Cell. 3rd ed. Springer Science & Business Media; Berlin, Germany: 2019. pp. 63–80. [Google Scholar]
- 18.Park H.S., Yu J.H. Molecular Biology of Asexual Sporulation in Filamentous Fungi. In: Hoffmeister D., editor. Biochemistry and Molecular Biology. The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research) Springer; Cham, Germany: 2016. [Google Scholar]
- 19.Juvvadi P.R., Lee S.C., Heitman J., Steinbach W.J. Calcineurin in fungal virulence and drug resistance: Prospects for harnessing targeted inhibition of calcineurin for an antifungal therapeutic approach. Virulence. 2017;8:186–197. doi: 10.1080/21505594.2016.1201250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cyert M.S. Calcineurin signaling in Saccharomyces cerevisiae: How yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 2003;311:1143–1150. doi: 10.1016/S0006-291X(03)01552-3. [DOI] [PubMed] [Google Scholar]
- 21.Clapham D.E. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 22.Liu S., Hou Y., Liu W., Lu C., Wang W., Sun S. Components of the calcium-calcineurin signaling pathway in fungal cells and their potential as antifungal targets. Eukaryot. Cell. 2015;14:324–334. doi: 10.1128/EC.00271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusnak F., Mertz P. Calcineurin: Form and function. Physiol. Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 24.Stie J., Fox D. Calcineurin regulation in fungi and beyond. Eukaryot. Cell. 2008;7:177–186. doi: 10.1128/EC.00326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juvvadi P.R., Lamoth F., Steinbach W.J. Calcineurin as a Multifunctional Regulator: Unraveling Novel Functions in Fungal Stress Responses, Hyphal Growth, Drug Resistance, and Pathogenesis. Fungal Biol. Rev. 2014;28:56–69. doi: 10.1016/j.fbr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thewes S. Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot. Cell. 2014;13:694–705. doi: 10.1128/EC.00038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stathopoulos A.M., Cyert M.S. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matheos D.P., Kingsbury T.J., Ahsan U.S., Cunningham K.W. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 1997;11:3445–3458. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chow E.W., Clancey S.A., Billmyre R.B., Averette A.F., Granek J.A., Mieczkowski P., Cardenas M.E., Heitman J. Elucidation of the calcineurin-Crz1 stress response transcriptional network in the human fungal pathogen Cryptococcus neoformans. PLoS Genet. 2017;13:e1006667. doi: 10.1371/journal.pgen.1006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karababa M., Valentino E., Pardini G., Coste A.T., Bille J., Sanglard D. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol. Microbiol. 2006;59:1429–1451. doi: 10.1111/j.1365-2958.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- 31.Santos M., de Larrinoa I.F. Functional characterization of the Candida albicans CRZ1 gene encoding a calcineurin-regulated transcription factor. Curr. Genet. 2005;48:88–100. doi: 10.1007/s00294-005-0003-8. [DOI] [PubMed] [Google Scholar]
- 32.Choi J., Kim Y., Kim S., Park J., Lee Y.H. MoCRZ1, a gene encoding a calcineurin-responsive transcription factor, regulates fungal growth and pathogenicity of Magnaporthe oryzae. Fungal Genet. Biol. 2009;46:243–254. doi: 10.1016/j.fgb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Schumacher J., de Larrinoa I.F., Tudzynski B. Calcineurin-responsive zinc finger transcription factor CRZ1 of Botrytis cinerea is required for growth, development, and full virulence on bean plants. Eukaryot. Cell. 2008;7:584–601. doi: 10.1128/EC.00426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soriani F.M., Malavazi I., da Silva Ferreira M.E., Savoldi M., Von Zeska Kress M.R., de Souza Goldman M.H., Loss O., Bignell E., Goldman G.H. Functional characterization of the Aspergillus fumigatus CRZ1 homologue, CrzA. Mol. Microbiol. 2008;67:1274–1291. doi: 10.1111/j.1365-2958.2008.06122.x. [DOI] [PubMed] [Google Scholar]
- 35.Cramer R.A., Jr., Perfect B.Z., Pinchai N., Park S., Perlin D.S., Asfaw Y.G., Heitman J., Perfect J.R., Steinbach W.J. Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot. Cell. 2008;7:1085–1097. doi: 10.1128/EC.00086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagiwara D., Kondo A., Fujioka T., Abe K. Functional analysis of C2H2 zinc finger transcription factor CrzA involved in calcium signaling in Aspergillus nidulans. Curr. Genet. 2008;54:325–338. doi: 10.1007/s00294-008-0220-z. [DOI] [PubMed] [Google Scholar]
- 37.Spielvogel A., Findon H., Arst H.N., Araujo-Bazan L., Hernandez-Ortiz P., Stahl U., Meyer V., Espeso E.A. Two zinc finger transcription factors, CrzA and SltA, are involved in cation homoeostasis and detoxification in Aspergillus nidulans. Biochem. J. 2008;414:419–429. doi: 10.1042/BJ20080344. [DOI] [PubMed] [Google Scholar]
- 38.Almeida R.S., Loss O., Colabardini A.C., Brown N.A., Bignell E., Savoldi M., Pantano S., Goldman M.H., Arst H.N., Jr., Goldman G.H. Genetic bypass of Aspergillus nidulans crzA function in calcium homeostasis. G3 Genes Genomes Genet. 2013;3:1129–1141. doi: 10.1534/g3.113.005983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang P.K. Aspergillus parasiticus crzA, which encodes calcineurin response zinc-finger protein, is required for aflatoxin production under calcium stress. Int. J. Mol. Sci. 2008;9:2027–2043. doi: 10.3390/ijms9102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soriani F.M., Malavazi I., Savoldi M., Espeso E., Dinamarco T.M., Bernardes L.A., Ferreira M.E., Goldman M.H., Goldman G.H. Identification of possible targets of the Aspergillus fumigatus CRZ1 homologue, CrzA. BMC Microbiol. 2010;10:12. doi: 10.1186/1471-2180-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Castro P.A., Chen C., de Almeida R.S., Freitas F.Z., Bertolini M.C., Morais E.R., Brown N.A., Ramalho L.N., Hagiwara D., Mitchell T.K., et al. ChIP-seq reveals a role for CrzA in the Aspergillus fumigatus high-osmolarity glycerol response (HOG) signalling pathway. Mol. Microbiol. 2014;94:655–674. doi: 10.1111/mmi.12785. [DOI] [PubMed] [Google Scholar]
- 42.Shwab E.K., Juvvadi P.R., Waitt G., Soderblom E.J., Barrington B.C., Asfaw Y.G., Moseley M.A., Steinbach W.J. Calcineurin-dependent dephosphorylation of the transcription factor CrzA at specific sites controls conidiation, stress tolerance, and virulence of Aspergillus fumigatus. Mol. Microbiol. 2019;112:62–80. doi: 10.1111/mmi.14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manoli M.T., Espeso E.A. Modulation of calcineurin activity in Aspergillus nidulans: The roles of high magnesium concentrations and of transcriptional factor CrzA. Mol. Microbiol. 2019;111:1283–1301. doi: 10.1111/mmi.14221. [DOI] [PubMed] [Google Scholar]
- 44.Nierman W.C., Yu J., Fedorova-Abrams N.D., Losada L., Cleveland T.E., Bhatnagar D., Bennett J.W., Dean R., Payne G.A. Genome Sequence of Aspergillus flavus NRRL 3357, a Strain That Causes Aflatoxin Contamination of Food and Feed. Genome Announc. 2015;3 doi: 10.1128/genomeA.00168-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., Rao A., Hogan P.G. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011;21:91–103. doi: 10.1016/j.tcb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernandez-Ortiz P., Espeso E.A. Phospho-regulation and nucleocytoplasmic trafficking of CrzA in response to calcium and alkaline-pH stress in Aspergillus nidulans. Mol. Microbiol. 2013;89:532–551. doi: 10.1111/mmi.12294. [DOI] [PubMed] [Google Scholar]
- 47.Elbein A.D., Pan Y.T., Pastuszak I., Carroll D. New insights on trehalose: A multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 48.Ries L.N.A., Rocha M.C., de Castro P.A., Silva-Rocha R., Silva R.N., Freitas F.Z., de Assis L.J., Bertolini M.C., Malavazi I., Goldman G.H. The Aspergillus fumigatus CrzA Transcription Factor Activates Chitin Synthase Gene Expression during the Caspofungin Paradoxical Effect. MBio. 2017;8 doi: 10.1128/mBio.00705-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fiedler M.R., Lorenz A., Nitsche B.M., van den Hondel C.A., Ram A.F., Meyer V. The capacity of Aspergillus niger to sense and respond to cell wall stress requires at least three transcription factors: RlmA, MsnA and CrzA. Fungal Biol. Biotechnol. 2014;1:5. doi: 10.1186/s40694-014-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barratt R.W., Johnson G.B., Ogata W.N. Wild-type and mutant stocks of Aspergillus nidulans. Genetics. 1965;52:233–246. doi: 10.1093/genetics/52.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He Z.M., Price M.S., Obrian G.R., Georgianna D.R., Payne G.A. Improved protocols for functional analysis in the pathogenic fungus Aspergillus flavus. BMC Microbiol. 2007;7:104. doi: 10.1186/1471-2180-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu J.H., Hamari Z., Han K.H., Seo J.A., Reyes-Dominguez Y., Scazzocchio C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 53.Lee M.K., Kwon N.J., Lee I.S., Jung S., Kim S.C., Yu J.H. Negative regulation and developmental competence in Aspergillus. Sci. Rep. 2016;6:28874. doi: 10.1038/srep28874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park H.S., Ni M., Jeong K.C., Kim Y.H., Yu J.H. The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans. PLoS ONE. 2012;7:e45935. doi: 10.1371/journal.pone.0045935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park H.S., Yu J.H. Multi-copy genetic screen in Aspergillus nidulans. Methods Mol. Biol. 2012;944:183–190. doi: 10.1007/978-1-62703-122-6_13. [DOI] [PubMed] [Google Scholar]
- 56.Eom T.J., Moon H., Yu J.H., Park H.S. Characterization of the velvet regulators in Aspergillus flavus. J. Microbiol. 2018;56:893–901. doi: 10.1007/s12275-018-8417-4. [DOI] [PubMed] [Google Scholar]
- 57.Christensen S., Borrego E., Shim W.B., Isakeit T., Kolomiets M. Quantification of fungal colonization, sporogenesis, and production of mycotoxins using kernel bioassays. J. Vis. Exp. 2012 doi: 10.3791/3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.