Abstract
Natural products have drastically improved our lives by providing an excellent source of molecules to fight cancer, pathogens, and cardiovascular diseases that have revolutionized medicine. Fungi are prolific producers of diverse natural products and several recent advances in synthetic biology, genetics, bioinformatics and natural product chemistry have greatly enhanced our ability to efficiently mine their genomes for the discovery of novel drugs. In this article, we provide an overview of improved heterologous expression platforms for targeted production of fungal secondary metabolites, of advances in chemical and bioinformatics dereplication, and of novel bioinformatic platforms to discover biosynthetic genes involved in the production of metabolites with specific bioactivities. These advances, coupled with the presence of vast numbers of biosynthetic gene clusters in fungal genomes whose natural products remain unknown, have revitalized efforts to mine the fungal treasure chest and renewed the promise of discovering new drugs.
Graphical abstract
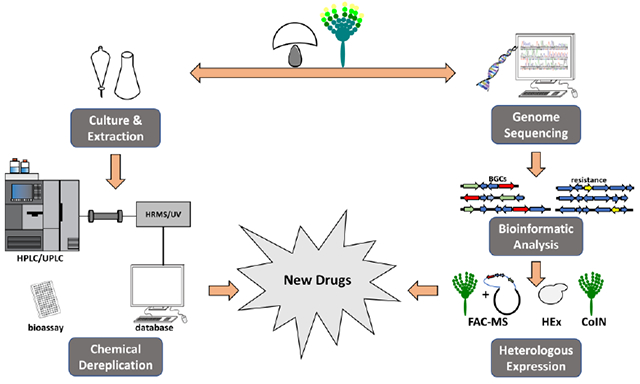
Introduction
The rampant rise of antimicrobial resistance to known drugs is a major challenge for medicine and agriculture [1,2]. Antibiotics, immunosuppressants, anticancers, cholesterol-lowering and other drugs are largely derived from natural products, which are compounds that are naturally synthesized by an organism [3]. Many natural products are also called secondary or specialized metabolites (SMs), because, unlike primary metabolites, they are not essential for survival under laboratory conditions, they are generally uniquely produced by one or a small group of closely related organisms, and often have low molecular weight and potent bioactivities [4,5].
Fungi are an rich source of SMs and several important drugs have been isolated from diverse fungal organisms [6]. But with only a small slice of SM pathways functionally characterized [7], we are just starting to tap into fungal chemodiversity and appreciate its full potential. Usually the genes involved in the synthesis of a specific SM are clustered together in the genome, forming a biosynthetic gene cluster (BGC) [8]. There is an estimated 5.1 million extant species of fungi, and genome sequencing and bioinformatics analysis have shown that the genomes of species in ascomycetes, arguably the largest of the fungal phyla with an estimated 500,000–3 million species, can encode for up to 80 BGCs each [9]. Furthermore, population genomic studies examining within-species variation in BGCs have shown that the genomes of individual isolates do not always capture the entire spectrum of BGCs present in a species [7,10], further increasing the genomic potential of fungal chemodiversity.
This review offers an overview of different approaches to fungal drug discovery, focusing on three areas with exciting recent advances: building efficient platforms for heterologous expression of BGCs whose products remain unknown, developing novel techniques to target chemical and bioinformatic identification of known SMs to address dereplication issues of ‘re-discovery’, and developing bioinformatic tools for efficiently identifying BGCs involved in the production of SMs with specific bioactivities.
Drug discovery using new heterologous expression platforms
In recent years, fungal genome sequencing and the development of specialized software for the prediction of fungal BGCs such as antiSMASH, SMURF and FunGeneClusterS [11–13], have revealed that most filamentous fungi contain dozens of BGCs in their genomes and have the potential to produce many SMs [7]. However, the major challenge hindering efforts to discover novel SMs is that the products of most of these BGCs remain unknown and are difficult to activate under laboratory conditions [3]. One common approach for tackling this challenge is BGC activation using gene overexpression, deletion/overexpression of chromatin modifying enzymes [14]. But the techniques involved are not always efficient and can only be performed on genetically amenable fungi, making their adoption across the Fungal Kingdom difficult. To overcome this issue, several different strategies have been developed, such as growth under diverse nutritional media, co-culture with other organisms, and use of epigenetic modifiers [15–18]. A common characteristic of all these strategies is that they require study of BGCs in their native organisms. An alternative approach is to study BGCs outside their native genomes. This can be achieved through genetic engineering and heterologous expression of BGCs in well-developed platforms in a variety of organism hosts, such as filamentous fungi in the genus Aspergillus (subphylum Pezizomycotina, phylum Ascomycota) [19–22], or budding yeasts in the subphylum Saccharomycotina (phylum Ascomycota), such as the baker’s yeast Saccharomyces cerevisiae [23] or the methylotrophic yeast Komagataella (Pichia) pastoris [24].
One recently developed and highly promising platform is the co-inducible nitrate (CoIN) expression system for SMs in Aspergillus nidulans [14]. This platform takes advantage of the sterigmatocystin (ST) BGC, in which the transcription factor (afIR) and its cofactor (afIS) can effectively regulate the other ST biosynthetic genes. The afIR and afIS promoters were substituted with a bidirectional nitrate-inducible promoter (niaD/niiA) which in turn can regulate the expression of the promoter regions within the ST BGC. In addition, the genes of interest are flanked with small DNA regions to direct homologous recombination at the genomic loci wA and yA associated with spore pigmentation, which enables for facile selection of the transformants. An A. nidulans strain devoid of ST was designed and as proof-of-concept, the three genes involved in β-carotene biosynthesis were constructed with specific ST promoters. The strain produced β-carotene with an excellent yield at the desired induction. This is a promising platform for the study of BGCs whose SM products remain unknown, especially when these products may be toxic for the host because it utilizes an inducible promoter (Figure 1).
Figure 1.
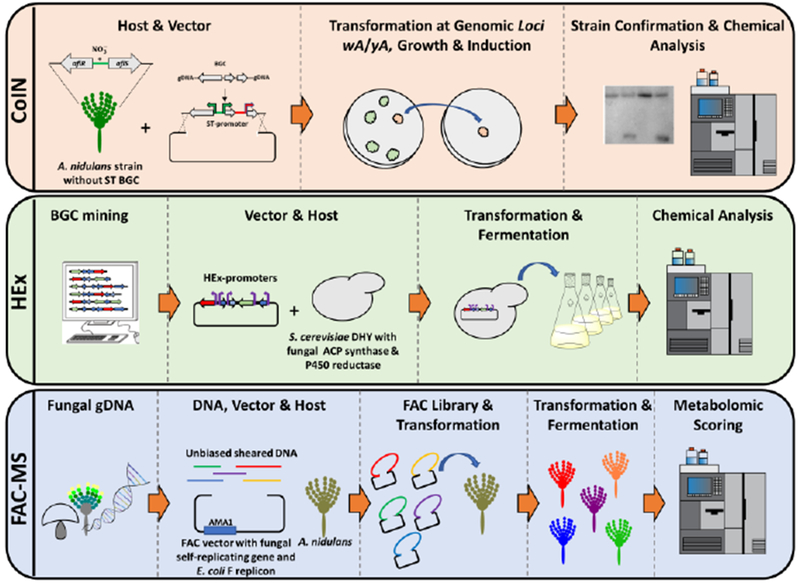
Schematic of three heterologous platforms recently developed to characterize novel fungal secondary metabolites [14,25,26].
Another robust platform called Heterologous Expression (HEx) was recently developed in S. cerevisiae (Figure 1) [25]. For this platform 30 auto-inducible, coordinated and sufficiently unique HEx promoters have been designed. A new S. cerevisiae strain (DHY) was engineered with increased mitochondrial stability and high sporulation. In addition, this strain features the A. nidulans npgA gene encoding for a 4′-phosphopantetheinyl transferase, and a P450 reductase from Aspergillus terreus to facilitate the biosynthesis of SMs derived from non-ribosomal synthetases (NRPSs) and polyketide synthases (PKSs). To prove the versatility of this system, the authors computationally predicted BGCs containing PKSs or UbiA-type sesquiterepene cyclases from every ascomycete and basidiomycete genome available in GenBank and used phylogenetic analyses to select 41 unique BGCs. They used yeast homologous recombination to build vectors containing the 41 BGCs and transformed them in the DHY strain. Excitingly, more than half (22/41) of these BGCs produced detectable levels of different compounds.
Another new heterologous platform also uses A. nidulans but this time with a fungal artificial chromosome (FAC) self-replicating vector [26,27]. This system works by shearing the DNA in an unbiased fashion in large DNA fragments (30–100 kb), which can contain entire BGCs. The DNA fragments are then assembled together with the FAC vector that contains the E. coli F replicon and the Aspergillus autonomously replicating sequence (AMA1). The group was able to produce a library of FACs containing 156 BGCs from three Aspergillus species, which were confirmed by insert end-sequencing and alignment to the reference genome. Of these FACs, 56 contained uncharacterized BGCs; these were transformed in A. nidulans and the strains were cultured, extracted and analyzed by liquid chromatography (LC) coupled with high-resolution mass spectrometry (HRMS). Metabolomic scoring was used to identify novel SMs produced by the transformant strains. To confirm that the BGC was involved in the biosynthesis of the SM, for each strain that produced novel SMs, A. nidulans was also transformed with the FAC vector containing the BGC with the synthase gene disrupted. In this work, 17 metabolites were produced by 15 different FACs (Figure 1).
Dereplication strategies for drug discovery
Recent advances in purification methods, extremely sensitive analytical techniques and cheaper sequencing are flooding databases with ever increasing amounts of data, rendering systematic analysis and prioritization challenging. In the context of drug discovery, two key questions are how one can take advantage of these sophisticated analytical techniques to prioritize promising novel SM candidates with interesting bioactivity from a pool of (known and unknown) metabolites (chemical dereplication) and how one can navigate large genomic databases to identify BGCs involved in producing new SMs (bioinformatic dereplication) (Figure 2).
Figure 2.
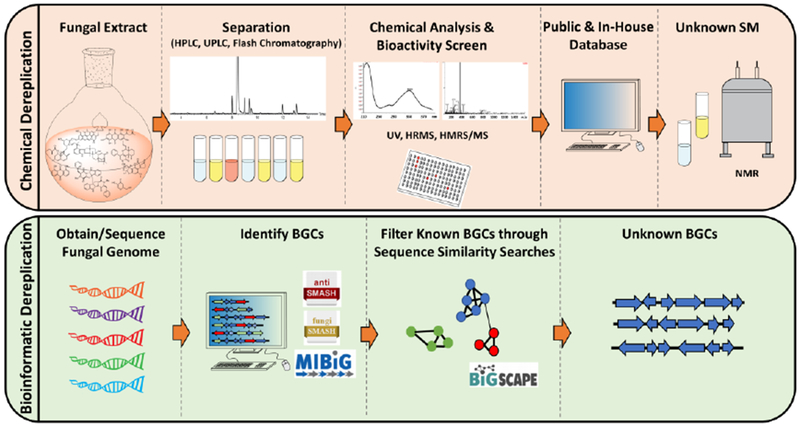
Chemical and bioinformatic dereplication work-flow for the discovery of unknown secondary metabolites from fungi.
Chemical dereplication
One of the main challenges in discovering novel SMs is in distinguishing unknown from known compounds. Chemical dereplication is essential for avoiding re-characterization of already known SMs. In recent years, great advances in chemical separation by liquid chromatography and detection by HRMS and diode array in combination with the use of databases have facilitated a faster chemical dereplication [28]. El-Elimat et al. have optimized a protocol that utilizes a short chromatographic method (10 minutes) with a deconvoluting program to analyze complex mixtures (ACD/IntelliXtracts) and an in-house database containing HRMS, MS/MS and UV data for each SM. This method allowed the group to identify that, among all the analyzed fungal crude extracts with activity against cancer cells, only 50% contained novel SMs [29]. The importance of a good database was highlighted by the work of Kildergaard et al., who were able to identify bioactive compounds from marine-derived fungi and show how fractionation of crude extracts and a comprehensive library that contains HRMS, MS/HRMS, and UV data facilitated the purification of novel SMs and derivatives of known SMs with activities against cancer cells, fungi and bacteria [30,31] (Figure 2). They also implemented their dereplication strategy by using an unbiased peak picking algorithm which further improved the identification of unknown SMs. Zani and Carroll have developed an open access nuclear magnetic resonance (NMR) and MS database for SMs called DEREP-NP [32]. To be able to use this database, the crude extracts need to be fractionated or purified, but it allows for quick identification of known SMs.
Open-access databases of compounds exist (MetLin, MassBank) [33,34], but they feature a limited number of microbial SMs and should be further implemented with HRMS, MS fragmentation and UV data to allow characterization of known SMs. Recently, a new database called Global Natural Products Social molecular networking (GNPS) was developed where researchers can upload raw or processed MS/MS data [35]. This database offers various features, such as molecular explorer, which provides all the datasets available for a specific molecule and its analogues [35]. Similarly, the molecular networking feature allows users to correlate and visualize a set of spectra for similar molecules. Naman et al. have applied this to cyanobacteria and shown that this tool, in combination with bioactivity screening, was extremely useful for the identification of SMs with novel chemical features and led to the characterization of a novel cyclic peptide active against a range of cancer cells [36]. Another novel strategy to induce production of cryptic SMs was developed by Oakley and co-workers. Their idea was to minimize the production of known SMs by gene deletion of the main BGCs, and it was called ‘genetic dereplication’ [37]. In this case, eight BGCs were deleted in A. nidulans, and HPLC-UV/MS analysis showed a very simple metabolite profile. The strain was now able to produce a novel metabolite derived from NRPS called aspercryptin. In addition, heterologous expression of BGCs in this A. nidulans strain could lead to an easier SM identification as the background from the host metabolites is considerably reduced. Together with advances in genetic modifications in fungi, this strategy offers novel way to discover new secondary metabolites.
Bioinformatic dereplication
As more fungal genomes become available and software for the mining of BGCs improve their accuracy of detection, the possibility of distinguishing unknown from known compounds can also take place at the level of BGCs. For example, sequence similarity searches and evolutionary analyses harbor tremendous potential for mining genomes to identify BGCs that uniquely appear in one or a few species or to identify all the organisms (including ones that are potentially experimentally more tractable) that contain a BGC of interest.
One key requirement for this bioinformatic dereplication approach are databases that contain functionally characterized BGCs and their products. Toward this end, Medema et al. recently established a community standard for the information necessary to create a high-quality repository of BGCs and the SMs they encode [38]. Furthermore, they implemented this community standard as part of the MIBiG repository, on an online database that allows individual users to upload new data as well as to obtain currently known BGCs [39]. As of the end of 2018, the MIBiG repository contains more than 1,800 complete and partial entries for BGCs and their products from diverse fungi, bacteria and plants (Figure 2).
But how does one analyze genomic data to identify BGCs that are unlike those already in MIBiG? One solution to the challenge is offered by a newly developed computational tool named Biosynthetic Gene Similarity Clustering and Prospecting Engine (BiG-SCAPE) [40]. BiG-SCAPE is a similarity-based algorithm aimed at constructing the network of BGC sequence similarities; examination of such a network can reveal “families” of BGCs that are far apart from or do not contain BGCs already present in the MIBiG repository.
Drug discovery with a target in mind
One highly interesting feature of BGCs is that they sometimes contain a self-resistance gene for self-protection against the SM produced; in recent years, several such self-resistance genes have been identified in fungal BGCs, including transporter genes that accelerate SM secretion, genes that detoxify the SM by changing its chemical structure, or additional copies of the target protein that carry mutations that render them insensitive to the SM’s effects [41]. Great examples of BGCs with extra copy of the target protein are the lovastatin BGC, which contains an extra copy of the (3S)-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase [42], the mycophenolic acid BGC, which contains an additional IMP dehydrogenase [43], the fumagillin BGC which contains two copies of the methionine aminopeptidase [44,45], and the sporiofungin A BGC, which contains an additional copy of the β-1,3-glucan synthase FKS1 [46] (Table 1).
Table 1.
Natural products from fungi with a homolog of their target protein as the self-resistance gene within their BGC
| Compound | Organism | Self-resistance gene | Activity | Ref. |
|---|---|---|---|---|
| Lovastatin | A. terreus | IvrA (HMG-CoA reductase) | Treats hypercholesterolemia by inhibiting HMG-CoA reductase | [42,48] |
| Mycophenolic acid | Penicillium brevicompactum | mpaF(IMP dehydrogenase) | Immunosuppressive | [43] |
| Fumagillin | A. fumigatus | AfuA_8g00460 (methionine amino peptidase I) AfuA_8g00410 (methionine amino peptidase II) |
anti-angiogenesis | [44,45] |
| Sporiofungin | Pezicula radicicola | prfks1a (glucan synthase FKS1) | Antifungal | [46] |
| Fellutamide | A. nidulans | inpE (β6 proteasome subunit) | Proteasome inhibitor | [47] |
| Aspterric acid | A. terreus | aspD (dihydroxyacid dehydratase) | Herbicide – Inhibition of the DHAD enzyme involved in the branched-chain amino acid biosynthetic pathway in plants | [48] |
Efforts to identify BGCs containing such self-resistance genes have increased because their functional characterization allows for the identification of not only a novel SM but also its specific gene target (as well as clues about the molecular processes or pathways that the SM likely affects). Recently, an exciting pipeline was developed for Actinobacteria that identified resistance genes in the context of a BGC, called Antibiotic Resistance Target Seeker (ARTS) [47]. This platform identifies resistance genes by looking for a copy of a housekeeping gene within a BGC and evidence of horizontal gene transfer. The platform was shown to be able to identify several BGCs containing resistance genes, including already discovered examples.
More recently, the Fungal Resistance Gene-directed Genome mining (FRIGG) pipeline was created to identify resistance gene within BGC in fungi [48]. This pipeline does not require any knowledge of the resistance gene, but it is designed to identify BGCs that contain a homolog to a housekeeping gene present elsewhere in the genome through a strict search aimed at limiting false-positive hits for resistance genes. Testing of FRIGG on the genomes of 51 Aspergillus and Penicillium species resulted in the identification of 72 unique resistance genes within BGCs, one of which was the already characterized fellutamide B BGC, demonstrating the applicability of this platform [49,50].
Self-resistance genes can also be employed to guide discovery of drugs that affect them. Yan et al. were interested in identifying novel herbicide leads that could inhibit dihydroxyacid dehydratase (DHAD), an enzyme that is part of the branched chain amino acid (BCAA) pathway [51]. Since the BCAA pathway is also found in fungi, the authors searched fungal genomes for BGCs containing a DHAD homolog; strikingly, they identified DHAD homologs within four different BGCs across different fungi, one of which was A. terreus. Expression of the A. terreus BGC in S. cerevisiae showed that it produced aspterric acid, which was effective at inhibiting the growth of Arabidopsis thaliana. The work was able to establish the mode of action of this compound and showed by in vivo and in vitro experiments that it is a competitive inhibitor of DHAD.
Beyond the cluster paradigm
Although most fungal SM pathways are encoded by BGCs, examination of fungal genomes suggests that there is substantial variation in the genomic arrangement of fungal SM pathways [7]. On one hand, SM pathways can be split into two or more BGCs, such as in the case of dothistromin [52], cephalosporin [53], and the meroterpenoids austinol and dehydroaustinol [54]. On the other hand, SM pathways can be intertwined, as in the case of the genes involved in the biosynthesis for the SMs pseurotin and fumagillin, which are found as part of a single supercluster [45]. Irrespective of their genomic arrangement, a hallmark characteristic of genes in SM pathways is that they are all part of the same regulatory network [55]. Thus, an alternative approach is to use global co-expression data sets to identify sets of genes, including encoding for enzymes such as PKSs and NRPSs, that are co-expressed across a range of different conditions or experiments [55], irrespective of whether these genes are part of a BGC or not.
This gene co-expression network approach has been recently successfully employed in both plants [55], which are not always organized into BGCs [7], and fungi [56,57]. For example, a recent study used 155 transcriptomic experiments to construct the gene co-expression network of Aspergillus niger [56]. Examination of co-expression subnetworks that contain BGCs revealed two transcription factors that are not parts of BGCs but whose over-expression led to the up-regulation of several BGCs in the A. niger genome [56], suggesting that they are involved in the global regulation of SM in this species.
Conclusion and outlook
Fungi have for a long time been considered as a great source of drugs, but only with the sequencing of many fungal genomes and the discovery that they contain many uncharacterized BGCs, we realized that their true potential in producing bioactive molecules is still largely untapped. In order to exploit this great resource, new platforms for heterologous expression have been designed in different organisms with success in expressing natural products from traditionally hard to mine fungi, such as Basidiomycetes [21]. These different platforms have shown to be very robust in expressing BGCs from a range of different fungi, which allowed the identification of novel metabolites. These platforms have great potential for drug discovery and they can provide examples to further improve existing platforms. Progress has been made in the development of pipelines that allow efficient identification of unknown molecules with bioactivities. Chemical dereplication can offer a systematic way to process crude extracts. Efforts in developing comprehensive and publicly available chemical databases with detailed data about metabolites (e.g. HRMS, HRMS/MS, and UV spectra) need to be made to facilitate drug discovery. New bioinformatic tools have been developed to identify novel strategies to mine the growing number of fungal genomes available. These novel bioinformatic tools allow to better identify BGCs and provide some information about their products. This is an exciting time to discover new compounds from fungi and this review has highlighted some of the recent advances that will facilitate this process.
Highlights.
Fungi are an excellent source of drugs and their full potential in producing different metabolites is still largely untapped.
Several heterologous platforms have been developed to produced novel metabolites.
Chemical and bioinformatic pipelines can successfully identified unknown metabolites.
Bioinformatic tools can identify fungal biosynthetic gene clusters involved in the biosynthesis of novel bioactive compounds.
Acknowledgements
Research in the Rokas laboratory has been supported by the National Science Foundation, the Searle Scholars Program, the Guggenheim Foundation, the Burroughs Wellcome Trust, the National Institutes of Health, the Beckman Scholars Program and the March of Dimes. The Keller laboratory acknowledges support of NIH R01GM112739–01 for this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
special interest (●) or outstanding interest (●●)
- 1.Demain AL, Vaishnav P: Natural products for cancer chemotherapy. Microb Biotechnol 2011, 4:687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Revie NM, Iyer KR, Robbins N, Cowen LE: Antifungal drug resistance: evolution, mechanisms and impact. Curr Opin Microbiol 2018, 45:70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skellam E: Strategies for Engineering Natural Product Biosynthesis in Fungi. Trends Biotechnol 2018, in press. [DOI] [PubMed] [Google Scholar]
- 4.Newman DJ, Cragg GM: Natural Products as Sources of New Drugs from 1981 to 2014. J Nat Prod 2016, 79:629–661. [DOI] [PubMed] [Google Scholar]
- 5.Keller NP, Turner G, Bennett JW: Fungal Secondary Metabolism - from Biochemistry to Genomics. Nat Rev Microbiol 2005, 3:937–947. [DOI] [PubMed] [Google Scholar]
- 6.Aly AH, Debbab A, Proksch P: Fifty years of drug discovery from fungi. fungal Divers 2011, 50:3–19. [Google Scholar]
- 7.Rokas A, Wisecaver JH, Lind AL: The birth, evolution and death of metabolic gene clusters in fungi. Nat Rev Microbiol 2018, 16:731–744. [DOI] [PubMed] [Google Scholar]
- 8.Keller NP: Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol 2018, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackwell M: The Fungi: 1, 2, 3 – 5.1 Million Species? Am J Bot 2011, 98:426–438. [DOI] [PubMed] [Google Scholar]
- 10.Lind AL, Wisecaver JH, Lameiras C, Wiemann P, Palmer JM, Keller NP, Rodrigues F, Goldman GH, Rokas A: Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol 2017, 15:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, de los Santos ELC, Kim HU, Nave M, et al. : antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 2017, 45:W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND: SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 2010, 47:736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vesth TC, Brandi J, Andersen MR: FunGeneClusterS: Predicting fungal gene clusters from genome and transcriptome data. Synth Syst Biotechnol 2016, 1:122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiemann P, Soukup AA, Folz JS, Wang PM, Noack A, Keller NP: CoIN : co - inducible nitrate expression system for secondary metabolites in Aspergillus nidulans. Fungal Biol Biotechnol 2018, 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frisvad JC: Media and Growth Conditions for Induction of Secondary Metabolite Production In Fungal Secondary Metabolism. . Humana Press; 2012:47–58. [DOI] [PubMed] [Google Scholar]
- 16.Strauss J, Reyes-Dominguez Y: Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet Biol 2011, 48:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfannenstiel BT, Greco C, Sukowaty AT, Keller NP: The epigenetic reader SntB regulates secondary metabolism, development and global histone modifications in Aspergillus flavus. Fungal Genet Biol 2018, 120:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Netzker T, Fischer J, Weber J, Mattern DJ, Konig CC, Valiante V, Schroeckh V, Brakhage AA: Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol 2015, 6:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang Y, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang S, Sung CT, Wang CCC, Oakley BR: An Efficient System for Heterologous Expression of Secondary Metabolite Genes in Aspergillus nidulans. J Am Chem Soc 2013, 135:7720–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams K, Szwalbe AJ, Mulholland NP, Vincent JL, Bailey AM, Willis CL, Simpson TJ, Cox RJ: Heterologous Production of Fungal Maleidrides Reveals the Cryptic Cyclization Involved in their Biosynthesis. Angew Chemie Int Ed 2016, 55:6784–6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberti F, Khairudin K, Venegas ER, Davies JA, Hayes PM, Willis CL, Bailey AM, Foster GD: Heterologous expression reveals the biosynthesis of the antibiotic pleuromutilin and generates bioactive semi-synthetic derivatives. Nat Commun 2017, 8:1831. [DOI] [PMC free article] [PubMed] [Google Scholar]; ● This work described the heterologous production of the antibiotic pleuromutilin in A. oryzae as well as several analogues with increased bioactivities.
- 22.Hoefgen S, Lin J, Fricke J, Stroe MC, Mattern DJ, Kufs JE, Hortschansky P, Brakhage AA, Hoffmeister D, Valiante V: Facile assembly and fluorescence-based screening method for heterologous expression of biosynthetic pathways in fungi. Metab Eng 2018, 48:44–51. [DOI] [PubMed] [Google Scholar]
- 23.Billingsley JM, Denicola AB, Tang Y: Technology development for natural product biosynthesis in Saccharomyces cerevisiae. Curr Opin Biotechnol 2016, 42:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao L, Cai M, Shen W, Xiao S, Zhou X, Zhang Y: Engineered fungal polyketide biosynthesis in Pichia pastoris: a potential excellent host for polyketide production. Microb Cell Fact 2013, 12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harvey CJB, Tang M, Schlecht U, Horecka J, Fischer CR, Lin H, Li J, Naughton B, Cherry J, Miranda M, et al. : HEx: A heterologous expression platform for the discovery of fungal natural products. Sci Adv 2018, 4:eaar5459. [DOI] [PMC free article] [PubMed] [Google Scholar]; ● This study describes the development of a highly optimized S. cerevisiae strain that can be used to efficiently expressed fungal biosynthetic genes with high efficiency and control.
- 26.Bok JW, Ye R, Clevenger KD, Mead D, Wagner M, Krerowicz A, Albright JC, Goering AW, Thomas PM, Kelleher NL, et al. : Fungal artificial chromosomes for mining of the fungal secondary metabolome. BMC Genomics 2015, 16:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clevenger KD, Bok JW, Ye R, Miley GP, Verdan MH, Velk T, Chen C, Yang K, Robey MT, Gao P, et al. : A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat Chem Biol 2017, 13:895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]; ● This paper describes a novel system for the efficient expression of whole BGC in A. nidulans using a self-replicating vector. A metabolomic scoring platform was developed to precisely identified novel secondary metabolites.
- 28.Nielsen KF, Larsen TO: The importance of mass spectrometric dereplication in fungal secondary metabolite analysis. Front Microbiol Microbiol 2015, 6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-elimat T, Figueroa M, Ehrmann BM, Cech NB, Pearce CJ, Oberlies NH: High-Resolution MS, MS/MS, and UV Database of Fungal Secondary Metabolites as a Dereplication Protocol for Bioactive Natural Products. J Nat Prod 2013, 76:1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]; ● This work highlighted how the combination of bioassay screening together with LC-MS/MS molecular networking can facilitate drug discovery.
- 30.Kildgaard S, Mansson M, Dosen I, Klitgaard A, Frisvad JC, Larsen TO, Nielsen KF: Accurate Dereplication of Bioactive Secondary Metabolites from Marine-Derived Fungi by UHPLC-DAD-QTOFMS and a MS/HRMS Library. Mar Drugs 2014, 12:3681–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kildgaard S, Subko K, Phillips E, Goidts V, Cruz M De, Caridad D, Gotfredsen CH, Id BA, Frisvad JC, Nielsen KF, et al. : A Dereplication and Bioguided Discovery Approach to Reveal New Compounds from a Marine-Derived Fungus Stilbella fimetaria. Mar Drugs 2017, 15:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zani CL, Carroll AR: Database for Rapid Dereplication of Known Natural Products Using Data from MS and Fast NMR Experiments. J Nat Prod 2017, 80:1758–1766. [DOI] [PubMed] [Google Scholar]
- 33.Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, Koellensperger G, Huan T, Uritboonthai W, Aisporna AE, et al. : METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal Chem 2018, 90:3156–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, et al. : MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom 2010, 45:703–714. [DOI] [PubMed] [Google Scholar]
- 35.Wang M., Carver J J., Phelan W., Sanchez LM, Garg N, Peng Y, Nguyen DD., Watrous J., Kapono CA., Luzzatto-Knaan T., et al. : Sharing and community curation of mass spectrometry data with GNPS. Nat Biotechnol 2017, 34:828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naman CB, Rattan R, Nikoulina SE, Lee J, Miller BW, Moss NA, Armstrong L, Boudreau PD, Debonsi HM, Valeriote FA, et al. : Integrating Molecular Networking and Biological Assays To Target the Isolation of a Cytotoxic Cyclic Octapeptide, Samoamide A, from an American Samoan Marine Cyanobacterium. J Nat Prod 2017, 80:625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang Y-M, Ahuja M, Oakley CE, Entwistle R, Asokan A, Zutz C, Wang CCC, Oakley BR: Development of Genetic Dereplication Strains in Aspergillus nidulans Results in the Discovery of Aspercryptin. Angew Chemie Int Ed 2016, 55:1662–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Epstein SC, Charkoudian LK, Medema MH: A standardized workflow for submitting data to the Minimum Information about a Biosynthetic Gene cluster (MIBiG) repository: prospects for research-based educational experiences. Stand Genomic Sci 2018, 13:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, et al. : Minimum Information about a Biosynthetic Gene cluster. Nat Chem Biol 2015, 11:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro-Munoz J, Selem-Mojica N, Mullowney M, Kautsar S, Tryon J, Parkinson E, Santos EDL, Yeong M, Cruz-Morales P, Abubucker S, et al. : A computational framework for systematic exploration of biosynthetic diversity from large-scale genomic data. bioRxiv 2018, doi: 10.110/45270. [DOI] [Google Scholar]
- 41.Keller NP: Translating biosynthetic gene clusters into fungal armor and weaponry. Nat Chem Biol 2015, 11:671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR: Modulation of Polyketide Synthase Activity by Accessory Proteins During Lovastatin Biosynthesis. Science 1999, 284:1368–1373. [DOI] [PubMed] [Google Scholar]
- 43.Regueira TB, Kildegaard KR, Hansen BG, Mortensen UH, Hertweck C, Nielsen J: Molecular Basis for Mycophenolic Acid Biosynthesis in Penicillium brevicompactum. Appl Environ Microbiol 2011, 77:3035–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin H, Chooi Y, Dhingra S, Xu W, Calvo AM, Tang Y: The Fumagillin Biosynthetic Gene Cluster in Aspergillus fumigatus Encodes a Cryptic Terpene Cyclase Involved in the Formation of β-trans-Bergamotene. J Am Chem Soc 2013, 2:2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiemann P, Guo C, Palmer JM, Sekonyela R, Wang CCC: Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci 2013, 110:17065–17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yue Q, Li Y, Chen L, Zhang X, Liu X, An Z, Bills GF: Genomics-driven discovery of a novel self-resistance mechanism in the echinocandin-producing fungus Pezicula radicicola. Environ Microbiol 2018, 20:3154–3167. [DOI] [PubMed] [Google Scholar]
- 47.Alanjary M, Kronmiller B, Adamek M, Blin K, Weber T, Huson D, Philmus B, Ziemert N: The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids Res 2017, 45:42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kjærbølling I, Vesth T, Andersen MR: Resistance Gene-Directed Genome Mining of 50 Aspergillus species. bioRxiv 2018, doi: 10.1101/457903 [DOI] [PMC free article] [PubMed] [Google Scholar]; ●● This paper describes the development of a new bioinformatic platform for fungi that allows the identification of BGCs with self-resistance genes.
- 49.Kjærbølling I, Vesth TC, Frisvad JC, Nybo JL, Theobald S, Kuo A, Bowyer P, Matsuda Y, Mondo S, Lyhne EK, et al. : Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proc Natl Acad Sci 2018, 115:E753–E761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeh HH, Ahuja M, Chiang YM, Oakley CE, Moore S, Yoon O, Hajovsky H, Bok JW, Keller NP, Wang CCC, et al. : Resistance Gene-Guided Genome Mining: Serial Promoter Exchanges in Aspergillus nidulans Reveal the Biosynthetic Pathway for Fellutamide B, a Proteasome Inhibitor. ACS Chem Biol 2016, 11:2275–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Y, Liu Q, Zang X, Yuan S, Bat-erdene U, Nguyen C, Gan J, Zhou J, Jacobsen SE, Tang Y: Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature 2018, 559:415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]; ●● This work shows how the identification of BGCs with self-resistance genes allowed for a fast identification of the mode of action of the herbicide aspterric acid.
- 52.Bradshaw RE, Slot JC, Moore GG, Chettri P, de Wit PJGM, Ehrlich KC, Ganley ARD, Olson MA, Rokas A, Carbone I, et al. : Fragmentation of an aflatoxin-like gene cluster in a forest pathogen. New Phytol 2013, 198:525–535. [DOI] [PubMed] [Google Scholar]
- 53.Gutiérrez S, Fierro F, Casqueiro J, Martin JF: Gene organization and plasticity of the beta-lactam genes in different filamentous fungi. Antonie Van Leeuwenhoek 1999, 75:81–94. [DOI] [PubMed] [Google Scholar]
- 54.Lo H-C, Entwistle R, Guo C-J, Ahuja M, Szewczyk E, Hung J-H, Chiang Y-M, Oakley BR, Wang CCC: Two Separate Gene Clusters Encode the Biosynthetic Pathway for the Meroterpenoids Austinol and Dehydroaustinol in Aspergillus nidulans J Am Chem Soc 2012, 134:4709–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wisecaver JH, Borowsky AT, Tzin V, Jander G, Kliebenstein DJ, Rokas A: A Global Coexpression Network Approach for Connecting Genes to Specialized Metabolic Pathways in Plants. Plant Cell 2017, 29:944–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schäpe P, Kwon MJ, Baumann B, Gutschmann B, Jung S, Lenz S, Nitsche B, Paege N, Schütze T, Cairns TC, et al. : Updating genome annotation for the microbial cell factory Aspergillus niger using gene co-expression networks. Nucleic Acids Res 2019, 47:559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersen MR, Nielsen JB, Klitgaard A, Petersen LM, Zachariasen M, Hansen TJ, Blicher LH, Gotfredsen CH, Larsen TO, Nielsen KF, et al. : Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc Natl Acad Sci 2013, 110:E99–E107. [DOI] [PMC free article] [PubMed] [Google Scholar]


