SUMMARY
Both the structure and the amount of sleep are important for brain function. Entry into deep, restorative stages of sleep is time-dependent; short sleep bouts selectively eliminate these states. Fragmentation-induced cognitive dysfunction is a feature of many common human sleep pathologies. Whether sleep structure is normally regulated independent of the amount of sleep is unknown. Here we show that, in Drosophila melanogaster, activation of a subset of serotonergic neurons fragments sleep without major changes in the total amount of sleep, dramatically reducing long episodes that may correspond to deep sleep states. Disruption of sleep structure results in learning deficits which can be rescued by pharmacologically or genetically consolidating sleep. We identify two reciprocally connected sets of ellipsoid body neurons which form the heart of a serotonin-modulated circuit that controls sleep architecture. Taken together, these findings define a circuit essential for controlling the structure of sleep independent of its amount.
eTOC Blurb
Liu et al. image, perturb, and analyze a serotonin-modulated circuit in the Drosophila brain which regulates sleep architecture without affecting the amount of sleep. Dysregulation of this circuit impairs cognitive function.
INTRODUCTION
Sleep has a regular daily structure. In both humans and Drosophila, nighttime sleep occurs in long consolidated bouts, while daytime sleep is much more fragmented [1, 2]. For diurnal animals, long periods of consolidated sleep at night facilitate progression into the deep sleep stages that are associated with positive health and cognitive benefits. Conversely, fragmentation of sleep during the daytime active period may reflect maintaining a higher arousal state. Sleep fragmentation is also a hallmark of sleep-maintenance insomnia, but can occur without changes in the amount of sleep in a number of neurological conditions. In humans, normal aging and disorders such as obstructive sleep apnea, restless leg syndrome, narcolepsy and chronic pain [3] are associated with altered sleep architecture. Fragmentation decreases sleep quality and is associated with learning and memory deficits, attention deficit, hypertension and diabetes [4], even when total sleep time is not significantly decreased [3, 5]. Sleep structure therefore has importance that is independent of the total amount of sleep, and understanding its regulation will be critical to our understanding of the functions of sleep.
One candidate modulator of human sleep structure is serotonin (5-phydroxytryptamine, 5HT). Drugs that increase serotonergic signaling are widely used to treat a variety of conditions, including mood disorders, and their use is closely associated with sleep fragmentation [6, 7]. 5HT has been known to be involved in regulation of sleep for many years, but how and where it acts is complex. In vertebrates, some evidence suggests that 5HT functions in sleep induction and maintenance [8, 9] but other evidence suggests a role in promotion of wakefulness [10]. Most recent work recognizes the complexity and context-dependence of 5HT action [11] but an understanding of how it regulates the structure of sleep remains elusive.
Drosophila melanogaster has been used to study sleep for almost two decades [12, 13]. Strong conservation of molecular processes across phyla, coupled with the more genetically tractable and smaller central nervous system of Drosophila, make it a powerful system for uncovering basic mechanisms of brain function. A role for 5HT in promoting sleep has been demonstrated using mutants lacking tryptophan hydroxylase (Trh; the rate-limiting synthetic enzyme) and mutants for the 5HT1a and 5HT2b receptors [14, 15]. 5HT has also been implicated in other forms of quiescence and behavioral suppression and appears to achieve this suite of effects via independent modulation multiple brain loci [16].
In the present study, we show that sleep/wake structure is regulated independently of the amount of sleep by a 5HT-modulated neural circuit in the ellipsoid body (EB), a prominent brain structure of the central complex, which mediates sensory integration and motor coordination [for review see 17]. Activation of two anatomically distinct sets of EB neurons results in a similar fragmentation pattern with no change in total sleep. Functional connectivity between these two sets of EB neurons suggests they form an interactive loop that responds to external inputs to sculpt sleep structure. Inappropriate activation of this circuit by increasing the activity of serotonergic inputs generates sleep fragmentation and a learning deficit. Together, our data suggest that sleep structure is regulated separately from the amount of sleep and that this regulation is required for normal cognitive function.
RESULTS
Serotonergic neurons control sleep structure
To examine the role of 5HT in regulation of sleep structure in Drosophila, we targeted expression of warmth-activated dTrpA1 [18] and light-gated Chrimson (Hoopfer et al., 2015) channels to a subset of 5HT+ neurons using Trh-GAL4 [Trh-GAL4(III), 19]. Activation of these neurons for 24 h by either increasing ambient temperature from 21 °Cto 27 °C or pulsing light (5 sec on/5 sec off ) (Figure 1A-B) dramatically increased the number of sleep episodes, indicating that increased serotonin release fragments sleep. This can be seen in the raw data (Figures 1C and E), but is more clearly demonstrated by calculating the change from baseline (Figures 1D and F) during the activation day. This method compares each genotype to itself (thus controlling for small differences in genetic background) and also allows the general, genotype-independent, effects of neuronal activation conditions [c.f. 20] to be directly measured. While both heat and light affected baseline sleep, 5HT+ neuron activation did not have a uniform effect on total sleep across different manipulations compared to controls. Activation with heat had no effect on total sleep compared to controls (Figure 1G-H), while light activation increased nighttime sleep compared to controls (Figure 1I-J). This total sleep effect with light was seen in Trh lines (see Figures 1E-F and S1L-M), but not in downstream circuit element lines (Figure S5F and S6F), suggesting that it may be a dominant light-interaction phenotype from upstream of the fragmentation circuit. Interestingly, there was a consistent and effector-independent increase in the amount of sleep at the beginning of the day after cessation of both dTrpA1 and Chrimson activation compared to controls (Figure 1H and J), suggesting that homeostatic mechanisms may be engaged by “low quality” sleep even when total sleep is unchanged (dTrpA1) or increased (Chrimson). A second, independent Trh-GAL4 line produced the same fragmentation and rebound sleep phenotypes with both dTrpA1 and Chrimson, (Trh-GAL4 (II); Figure S1).
Figure 1. Activation of Trh-GAL4+ neurons fragments sleep but has no consistent effect on total amount.
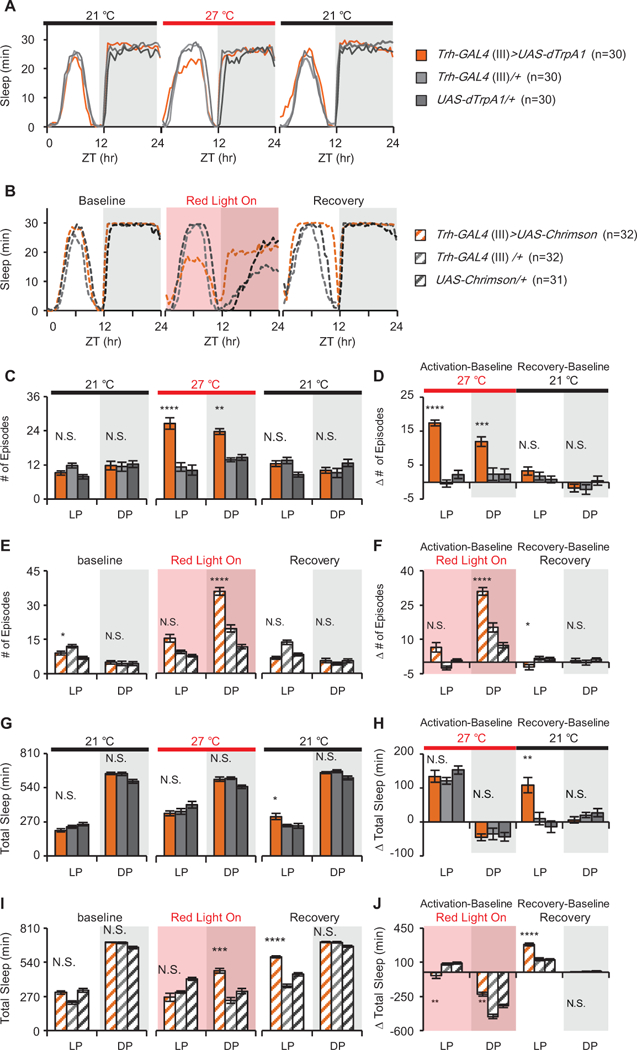
Movement data captured in 1 min bins were used to extract sleep using the standard definition of a sleep bout as 5 min of inactivity [13]. Sleep profiles of baseline, activation and recovery days. The experimental genotypes, Trh>dTrpA1 (A) and Trh>Chrimson (B), are shown in orange, UAS/+ and GAL4/+ controls are in gray. Activation of Trh-GAL4 cells increases the number of sleep episodes (C and E). The change from baseline day (in minutes) is shown (D and F) for the day of activation and the recovery day. Heat activation has no significant effect on the amount of sleep compared to controls (G), while light activation causes an increase in sleep during the night (I). The change in total sleep time (minutes) compared to the baseline day for activation and recovery days shows that both heat (H) and light (J) have significant effects on total sleep in all genotypes compared to the baseline day. Throughout all figures unless specified: LP: light period; DP: dark period. ZT: Zeitgeber Time. Asterisks in the figure indicate significant differences between the experimental genotype and both GAL4 and UAS controls. *p<0.05; **p<0.01; *** p<0.001; **** p<0.0001; N.S.: not significant. Red bar and black bar indicate data collected at 27°C and 21°C, respectively. Red shade indicates data collected when red light was on. Complete statistical methods and all individual comparisons are shown in STables 2–7. See also Figure S1.
To explore the effects of activation of this subset of serotonergic neurons on sleep structure, we examined the distribution of sleep episode durations over the course of the day (Figure 2). dTrpA1 activation of serotonergic neurons robustly decreased episode length, which is visualized by the shift of the cumulative distribution of the experimental population to the left (middle panel of Figure 2A for daytime, Figure S2 for night and Chrimson activation). Long sleep episodes, which occur almost exclusively during the nighttime, may reflect a behaviorally important deep sleep state [21]. Since an episode length of 200 min reflected the midpoint of the distribution, we looked at changes using this cutoff as a threshold for “long” episodes. Activation significantly reduced the ratio of long episodes in both the day and night (Figure 2D). Importantly, this was the result of fewer flies exhibiting long episodes (Figure 2E), suggesting the population data reflects the state of the majority of the animals rather than arising from the behavior of a few outliers. Similar results were obtained using thresholds <200 min (Table S1 and S2).
Figure 2. Activation of Trh-GAL4+ neurons alters sleep structure by changing the probability of state transitions.
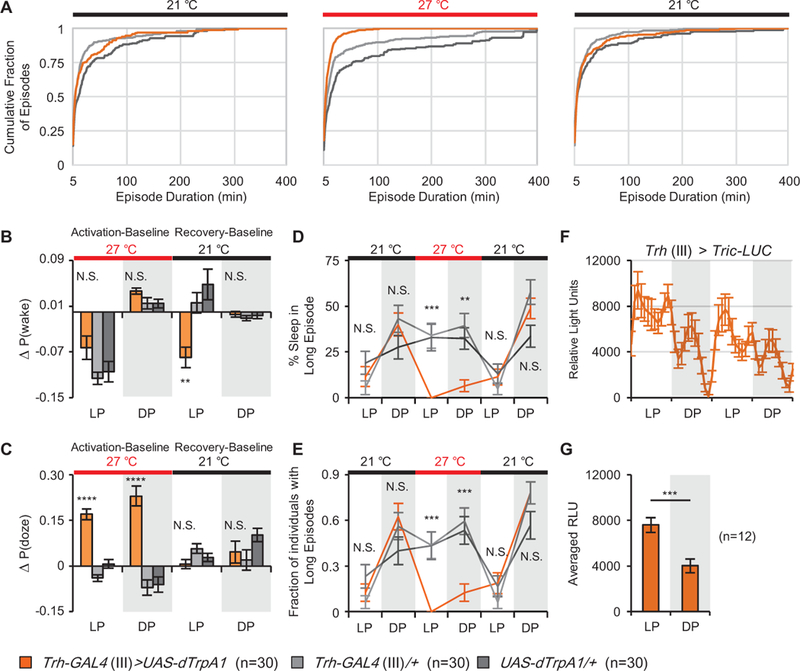
(A) The cumulative distribution of daytime sleep episode durations for baseline (left), activation (middle) and recovery (right) days is shown. (B) Probability of transition from a sleep to an awake state, P(wake), is significantly decreased compared to controls during rebound sleep. (C) Probability of transition from an awake to a sleep state, P(doze), significantly increased upon activation of Trh-GAL4+ neurons. Data are presented as the absolute change from baseline day. (D) The percent of total sleep occurring in episodes >200 min was significantly reduced when Trh-GAL4+ neurons were activated. (E) The number of individuals with long episodes was decreased. Data in D and E are presented as mean + binomial standard error. (F-G) Trh-GAL4+ neurons exhibit a rhythmic daily activity pattern as visualized with a Tric-LUC reporter in two days. Data are presented as mean + SEM. See also Table S1–2, Figures S1–2.
The changes in sleep structure we see with activation of this subset of serotonergic neurons reflect changes in the probabilistic processes underlying sleep. The probability of transitioning from a sleep to an awake state, P(wake), and the probability of falling asleep from a wake state, P(doze), can be approximated using power law distributions, suggesting that they are history-dependent and consistent with sleep being a multi-stage process [2, 22]. P(wake) can be thought of as arousal and P(doze) as sleep drive. In wild type flies, P(wake) is the major determinant of total sleep time and is increased by elevated dopaminergic tone [2]. Calculation of these metrics during and after activation of serotonergic neurons shows that during activation there are significant increases in P(doze), leading to more transitions between wake and sleep, but no alteration in P(wake) (Figures 2BC, S1–2). After release from activation, there is a significant decrease in arousal, or P(wake), during the recovery day that is associated with an increase in total sleep (Figure 1H and J and S1). These analyses show that activation of these neurons alters the state-dependent transitions that define the structure of sleep and wake and imply that a decrease in arousal is a homeostatic response to fragmentation.
While artificial activation of a neuron can reveal potential functions, it is important to know if, and in what context, activation normally occurs. To determine the normal activity pattern of Trh-GAL4+ neurons in freely behaving flies, we employed Tric-LUC, a calcium sensor which drives luciferase expression in response to neuronal activity [23]. Luciferase levels peaked after lights on and then again before lights off, exhibiting generally higher activity during the day than the night (Figure 2F-G and S1O); this may reflect synchronization with circadian- driven locomotor activity [24] and is similar to the behavior of vertebrate 5HT neurons [9]. The function of the nighttime peak is unclear, but given the many functions of 5HT may not be sleep-related. Our data are consistent with the observation that sleep is normally more fragmented during the day than during the night [2] and with the fact that 5HT+ neurons are wake-active in mammals [10].
Sleep fragmentation disrupts learning
Sleep fragmentation results in impairment of daytime function and cognitive processing in humans, mammals and flies [25–28]. In mammals, these effects can occur in the absence of sleep loss and are believed to be the result of a failure to enter deep sleep stages required for the cognitive benefits of sleep [29]. To determine whether fragmented sleep induced by increasing the activity of 5HT neurons leads to cognitive deficits, we measured associative learning in an odor-shock aversive paradigm. Activation of the Trh-GAL4+ subset of serotonergic neurons for 24 h prior to training resulted in a significant impairment in aversive learning measured immediately after training (Figure 3A). No learning deficit was observed when flies were trained and tested at permissive temperature (Figure S3A).
Figure 3. Sleep fragmentation disrupts learning.
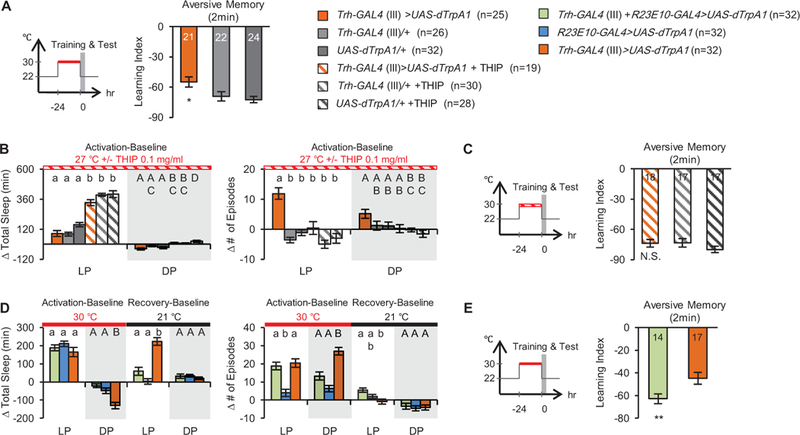
(A) Activation of Trh-GAL4+ neurons for 24 h (red bar) before training and testing disrupts aversive olfactory learning. Schematic of experiment is shown at left, data for 2 min memory at right. (B) Effects of 0.1 mg/ml THIP on the change in total sleep and episode number relative to baseline day. Statistically similar LP (lower case) and DP (upper case) groups are marked by the same letter; different letters indicate significant differences (p<0.05) between groups. (C) Application of THIP while Trh-GAL4+ neurons were activated (red striped bar) rescues learning. Schematic of experiment is shown at left. (D) Effects of co-activation of dorsal fan-shaped body and Trh-GAL4+ neurons on sleep. (E) Co-activation of dorsal fan-shaped body with Trh-GAL4+ neurons (red bar) rescues learning. For panels A, C and E, the number of independent reciprocal experiments is indicated on the bars. See also Figure S3.
To ask whether this learning deficit was due to changes in sleep structure, we consolidated sleep with gaboxadol (THIP), a GABAA agonist that promotes sleep in Drosophila [30] and specifically increases deep non-REM sleep in humans and rats [31, 32]. Feeding of 0.1 mg/ml THIP during activation of 5HT neurons across all genotypes significantly increased daytime sleep but did not increase nighttime sleep, likely due to a ceiling effect (Figure 3B). Locomotor activity during wake periods was not decreased (data not shown), indicating enhancement of sleep by the drug, rather than sedation. The number of sleep episodes was rescued to the level of control flies by the drug (Figure 3B, compare orange-striped bars to gray- striped bars) indicating that this drug has effects on sleep structure as well as amount. Treatment of flies with 0.1 mg/ml THIP during activation of Trh-GAL4+ neurons completely rescued post-activation learning deficit (Figure 3C). Importantly, control flies receiving THIP the day before training did not show any apparent enhancement of learning, implying that the improvement in learning resulted from rescue of fragmented sleep rather than a direct effect of the drug or the change in total sleep. Similar results were obtained with genetic rescue of 5HT-induced fragmentation. Co-activation of the sleep-promoting dorsal fan-shaped body [FB, 33] in the central complex significantly rescued both sleep fragmentation (Figure 3D) induced by activation of 5HT+ neurons and learning (Figure 3E). Activation of this area by itself before training did not enhance learning (Figure S3B). Together these rescue experiments implicate serotonergic regulation of sleep structure as a significant determinant of behavioral plasticity.
Serotonergic signaling to the ellipsoid body (EB) regulates sleep structure via 5HT7
To determine the cellular mechanisms controlling sleep structure, we needed to determine the targets of Trh-GAL4+ neurons and the 5HT receptor subtype they contained. Previous work using 5HT receptor mutants had shown that signaling via 5HT2b in a subset of FB neurons and 5HT1a in the mushroom bodies (MB, a learning center that also regulates sleep) is important for normal amounts of sleep (Yuan et al., 2006; Qian et al., 2017). Serotonergic innervation of mushroom bodies comes from dorsal paired medial neurons [34] which, when activated with dTrpA1, strongly increase total sleep [35]. Likewise, FB neurons promote sleep [15, 33, 36]. Because we saw no change in the amount of sleep compared to controls when we activated the Trh-GAL4+ neurons using dTrpA1, we suspected that 5HT regulation of sleep architecture involved a different 5HTR and likely a different brain region. To identify candidate regions and receptors, we expressed mCD8::GFP under control of Trh-GAL4 and co-stained with anti-5HT. Figure 4A shows that serotonergic neurons in this GAL4 line project to FB and ellipsoid body (EB), a region known to regulate locomotor activity in response to sensory inputs [17], but do not innervate the MB.
Figure 4. Serotonergic signaling to the EB regulates sleep structure via 5HT7Rs.
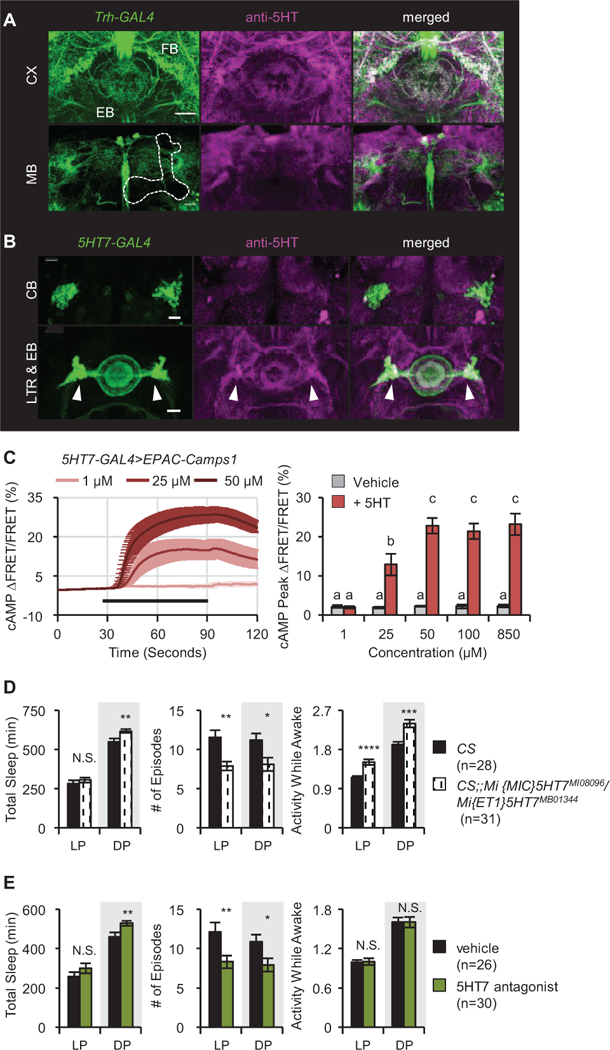
(A) Expression pattern of TrH(III)>GFP (green) costained with anti-5HT (magenta). Upper panels show sections at the level of the central complex (CX); lower panels the level of the mushroom body (MB, outlined). FB, fan-shaped body; EB, ellipsoid body. (B) Expression pattern of 5HT7>GFP (green) with anti-5HT (magenta). Upper panels, EB cell bodies (CB), lower panels, EB processes and lateral triangle region (LTR, white arrowheads) where serotonergic inputs enter. Scale bar = 20 μm for all images. (C) 5HT7-GAL4+ neurons in the EB respond to 5HT. Left: cAMP levels after application of 5HT (black bar) in the presence of 1 μM TTX. n = 8–9 for all groups. Right: quantified data compared to vehicle. (D) 5HT7R mutants exhibit more consolidated sleep in both the day (LP) and the night (DP) as well as increased nighttime sleep and mild hyperactivity. (E) Feeding of the 5HT7 antagonist (SB258719) to wild type flies consolidates sleep. Activity while awake is not affected. Data are shown for Day 4 of drug application. See also Figure S7.
The EB has been shown to express several 5HT receptors including 5HT7 [37, 38], a receptor subtype implicated in mammalian sleep [11]. Expressing CD8::GFP under control of 5HT7-GAL4 and co-staining with anti-5HT revealed a colocalization of serotonergic inputs and 5HT7-GAL4+ processes [Fig 4B and 37]. To determine if these neurons responded to 5HT, we expressed EPAC, a cyclic nucleotide sensor, in the cells and bath applied 5HT in the presence of TTX. There was a dose-dependent increase in cAMP levels consistent with expression of 5HT7 in that subset of EB cells (Figure 4C). 5HT was also able to increase calcium levels in 5HT7-GAL4+ neurons, but only in the absence of TTX, implying that 5HT modulates the efficacy of basally active excitatory inputs to these cells rather than directly depolarizing them (Figure S4A).
To determine the role of 5HT7 in regulation of sleep fragmentation, we assessed the effects of loss of receptor function with mutants and a specific inhibitor. Figure 4D shows data from transheterozygote Mi{MIC}5HT7MI08096/ Mi{ET1}5HT7MB01344 animals which have reduced 5HT7 mRNA levels (Figure S4D). These flies have much more consolidated sleep in both the day and the night, with only a small increase in total sleep at night. To confirm the acute role of 5HT7 receptors in regulation of sleep structure, the selective 5HT7 receptor antagonist SB258719 [37] was fed to wildtype flies. Sleep became more consolidated, with again a mild effect on total sleep that was limited to the nighttime (Figure 4E). The consolidation effects of reduced 5HT7R activity could not be explained by locomotor dysfunction since animals were either normal (drug) or mildly hyperlocomotor (mutant) rather than less active when awake (Figure 4E). In aggregate, these results suggest that 5HT7 in the central complex regulates sleep and sleep structure in a manner opposite to that of 5HT1a and 5HT2b [14, 15].
Multiple EB neuron populations control sleep structure
Because the FB has been shown to strongly regulate the amount of sleep [15, 33, 36] and because we had implicated 5HT7, which is expressed in EB, in fragmentation, we focused our attention on EB to identify the circuitry involved in the amount-independent regulation of sleep structure. Early anatomical studies of EB described 5 subtypes of neurons, grouped according to their projections to the characteristic “rings” of the EB: R1, R2, R3, R4 m and R4d [39, 40]. More recent functional and anatomical studies suggest that there is substantial diversity within these subgroups [41, 42]. To begin to determine the elements involved in regulation of sleep architecture, we screened 41 EB-expressing GAL4 lines from Janelia, VDRC and other published collections for sleep fragmentation using dTrpA1 [43] and identified two lines that, when expressing dTrpA1, generated activation-dependent sleep fragmentation without changing total sleep.
The first was 5HT7-GAL4 (Figure 5A; Video S1) which we have shown receives serotonergic inputs and responds directly to 5HT (Figure 4). This line projects to R2, R3 and R4d and expresses in about 100 cells. Activation of 5HT7-GAL4+ neurons with either heat or light had no significant effect on total sleep in the experimental genotype compared to controls, but increased the number of sleep episodes during the day by over 4-fold (Figure 5B, S5). Similar to the effects of stimulating Trh-GAL4+ neurons, there was a rebound increase in total sleep after activation ceased (Figure 5B, S5). The second line identified in the screen was VT038828-GAL4 (Figure 5D; Video S2). This line, which we abbreviate as “VT-GAL4”, labels a smaller population of 16–18 EB neurons that project only to R2. Activation of VT-GAL4+ neurons had no effect on total sleep compared to controls but significantly increased the number of daytime sleep episodes (Figure 5E, S6). Unlike Trh-GAL4+ and 5HT7-GAL4+ neurons, however, there was no sleep rebound following release from activation. These R2 neurons are distinct from the group that was recently shown to influence sleep homeostasis [44, data not shown].
Figure 5. Two populations of EB neurons control sleep structure without affecting the amount of sleep.
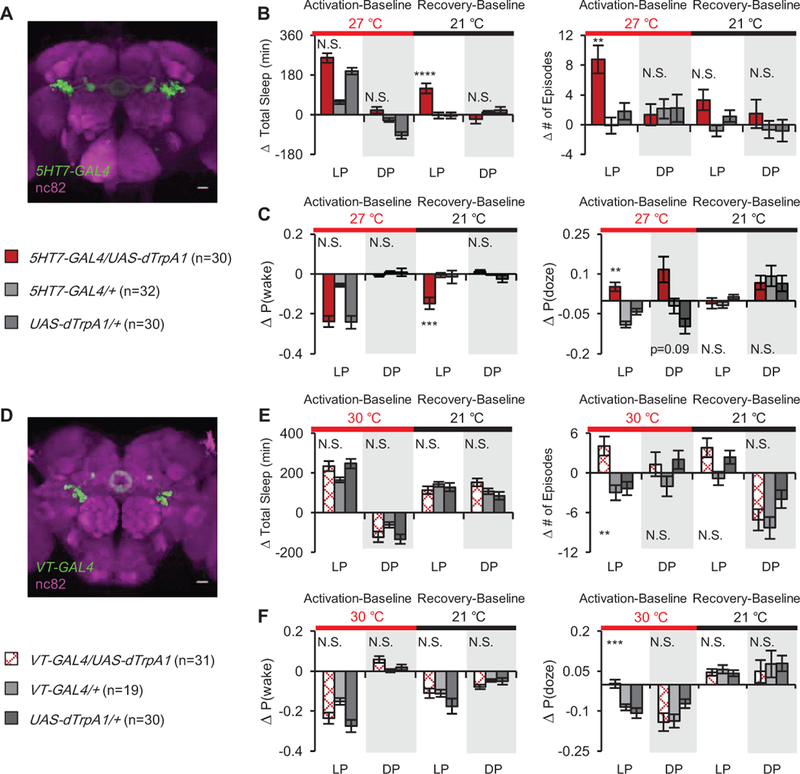
(A) Expression of 5HT7-GAL4. (B) Activation of 5HT7-GAL4+ neurons increases the number of daytime sleep episodes without changing total sleep compared to control, inducing rebound sleep after activation ends. Data are presented as the change (in min) from baseline day. (C) The probability of state transition from sleep to wake, P(wake), significantly decreased during rebound sleep, while probability of transition from wake to sleep, P(doze), increased during activation. Data are expressed as change from baseline day. (D) Expression of VT038828-GAL4 (VT-GAL4). (E) Activation of VT-GAL4+ neurons significantly increased the number of daytime episodes, with no effect on total sleep compared to controls. (F) P(doze) is increased compared to controls upon activation but there were no changes in P(wake). See also Figures S5–6, Videos S1-2.
The underlying parameter that drives fragmentation in both VT-GAL4+ and 5HT7-GAL4+ neurons is P(doze), or sleep drive (Figure 5, S5–6). As was seen with stimulation of Trh-GAL4+ neurons, activation of either group of neurons elevates the probability of transition from the wake to sleep state with almost no change in P(wake). The sleep rebound seen after activation of 5HT7-GAL4+ neurons is associated with a decrease in arousal, or P(wake), mirroring the effects of Trh-GAL4+ neurons. These analyses suggest that these three sets of neurons are affecting the same process to increase sleep fragmentation during their activation and that neither directly affects the arousal system.
An intra-EB circuit controls sleep structure
Given that 5HT7-GAL4+ neurons project to R2, R3 and R4d, we asked whether VT-GAL4+ R2 neurons were a subset of the 5HT7-GAL4+ neurons. To do this, we generated a LexA line (VT-LexA) using the promoter fragment present in VT038828-GAL4. This driver largely recapitulates the expression pattern of the VT-GAL4 line, staining all but two of the 16–18 VT-GAL4+ neurons (Figure S7A). To our surprise, VT-LexA did not label any of the 5HT7-GAL4+ neurons (Figure 6A, Video S3). Consistent with this, the 5HT responsiveness of the VT-GAL4+ cells was much lower than that of the 5HT7-GAL+ cells (Figure S7B). These data indicate that 5HT7-GAL4+ and VT-GAL4+ cells represent two distinct populations of neurons, both of which regulate sleep structure without affecting the amount of sleep.
Figure 6. An intra-EB circuit controls sleep structure.
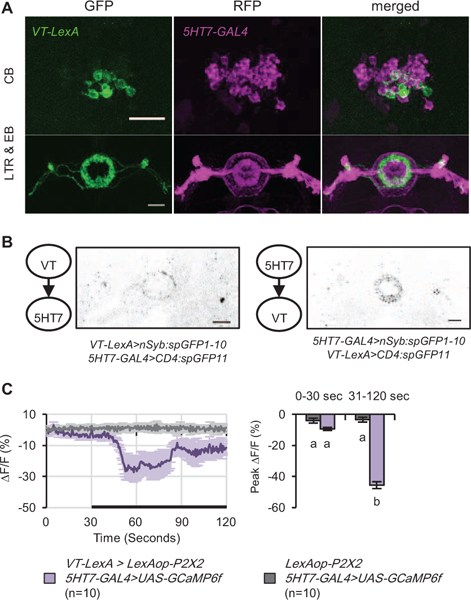
(A) 5HT7>RFP and VT>GFP cells do not overlap. (B) Reciprocal activity-dependent synaptic GRASP signals were detected between VT-LexA+ and 5HT7-GAL4+ neurons using a mAb which detects only holoGFP [54]. (C) Activation of VT-LexA+ neurons expressing P2X2 by application of ATP significantly decreased calcium levels in GCaMP6-expressing 5HT7-GAL4+ neurons. Black bar indicates ATP application. Statistically similar groups are marked by the same letter, with different letters indicating significant differences (p<0.05) between groups. See also Figure S7A-C, Videos S3.
To test the hypothesis that VT- and 5HT7-GAL4+ neurons are part of the same circuit, we employed active GRASP [45], a technique that uses reconstitution of holoGFP to determine if one cell population releases vesicles at synapses that contact a second cell type. Figure 6B shows that when the presynaptic fragment of GFP is expressed in VT-LexA+ neurons (Figure S7C), synapses with postsynaptic 5HT7-GAL4+ neurons can be visualized. GRASP is present both in the R2 ring and in the lateral triangle. Active GRASP experiments performed with 5HT7-GAL4 as the presynaptic cell and VT-LexA as the postsynaptic partner demonstrate that the connectivity between these two groups is reciprocal (Figure 6B, S7C).
To probe the functionality of these connections, we expressed P2X2, an ATP-gated cation channel, in VT-LexA+ cells and GCaMP in 5HT7-GAL4+ cells [46]. Application of ATP led to a large decrease in calcium levels in 5HT7-GAL4+ cells compared to controls (Figure 6C), suggesting an inhibitory connection. GCaMP signals from VT-LexA+ neurons were too dim to reliably assess the ability of activation of 5HT7-GAL4+ neurons to alter their calcium levels.
The EB fragmentation circuit sends outputs to multiple areas
Testing the connectivity of VT-LexA+ and 5HT7-GAL4+ neurons with active GRASP and P2X2 indicated that they are part of the same circuit, but these experiments did not provide insight into how they transfer information to the rest of the brain. To determine the outputs of these cell groups, we utilized trans-Tango [47]. This technique allows the identification of all neurons that receive synaptic inputs from a GAL4 population. Figure 7 shows the downstream connections made by VT-GAL4+ and 5HT7-GAL4+ respectively (Videos S4 and S5).
Figure 7. The EB fragmentation circuit has high interconnectivity but limited output.
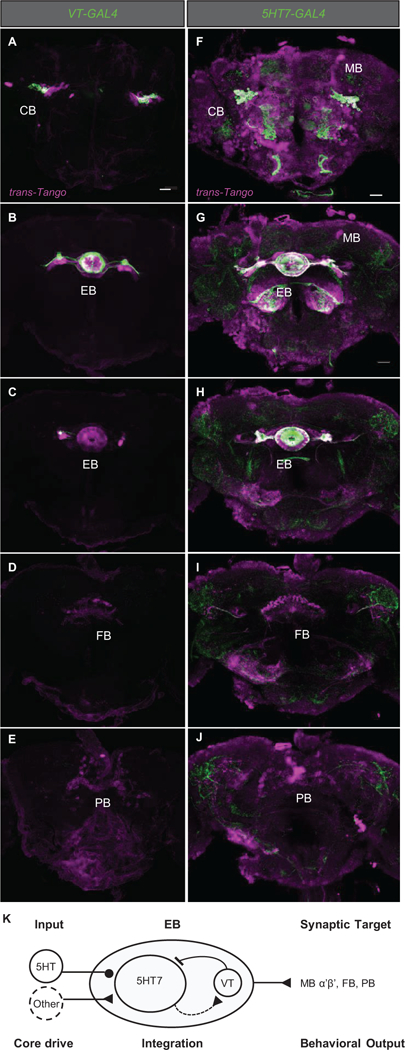
5 confocal sections at different levels of the same brain from anterior to posterior are shown for trans-Tango targeted brain regions with VT-GAL4 (A-E) and 5HT7-GAL4 (F-J). Expression of the GAL4 marked with GFP is shown in green, trans-Tango expressing mtdTomato in magenta and white indicates GAL4+ neurons making synapses on themselves. VT-GAL4+ neurons target locally in the central complex: ca. 2–3 GAL4+ neurons and a population of ca. 60 other EB neurons in the region of 5HT7-GAL4. Fan-shaped body (FB) and protocerebral bridge (PB) also receive synapses. 5HT7-GAL4+ neurons target themselves within the EB, and output to mushroom body (MB) α’β’ lobes, FB and PB. (K) Proposed model for the circuit controlling sleep structure. See also Figure S7D, Videos S4-5.
VT-GAL4+ neurons have a very limited connectivity (Figure 7 A-E). Within the EB, VT-GAL4+ neurons make strong synapses with ca. 60 other EB neurons whose morphology suggests the 5HT7-GAL4+ group. This would indicate that the VT-GAL4+ group is connected to over 50% of the 5HT7-GAL4+ neurons. Two to three of the cells in each hemisphere express both markers (Figure S7D), indicating a small amount of recurrent connectivity. Outside the EB, VT-GAL4+ cells connect to both FB and protocerebral bridge (PB) cells, but do not connect to neurons outside the central complex.
In contrast to VT-GAL4+, 5HT7-GAL4+ neurons make strong connections within their population (about 60% have both markers, Figure 7F-J). They also connect to ca. 25 other EB cells, presumably including most or all of the VT-GAL4+ cells. Like VT-GAL4, 5HT7-GAL4 connects to subsets of FB and PB within the central complex. Outside the central complex, there are connections to mushroom body prime lobes.
DISCUSSION
A dedicated circuit for regulation of sleep structure
In this study, we describe a circuit which regulates of sleep structure without affecting the total amount of sleep. Figure 7K shows a cartoon of the EB sleep structure circuit and its postulated connectivity. 5HT acts to enhance the response of 5HT7-GAL4+ neurons to basally active excitatory inputs. 5HT-dependent calcium signals are blocked by TTX, while its ability to increase cAMP is not (Figure 4C, S4A-B), supporting the existence of these active excitatory inputs to 5HT7-GAL4+ cells. In contrast, VT-GAL4+ cells do not have basally-active excitatory inputs (Figure S4C). 5HT modulation of the circuit likely occurs primarily via inputs to 5HT7-GAL4+ neurons since the response of VT-GAL4+ neurons is weaker and lower affinity (Figure S7B). Whether there are other, perhaps situationally-active, inputs to this circuit is currently unknown.
Within the EB the circuit is complex. VT-GAL4+ neurons are functionally connected with the 5HT7-GAL4+ group (Figure 6B). VT-GAL4+ neurons provide feedback inhibition to a subset of 5HT7-GAL4+ neurons (Figures 6C) which enhances fragmentation, likely via output to noncentral complex regions. How inhibition of a subset of the 5HT7-GAL4+ cells acts to modulate the behavioral output of the rest of the population is not yet clear, but we note that many of the 5HT7-GAL4+ cells are GABAergic (data not shown). While all the details of the circuit’s complex dynamics remain to be discovered, it is clear that this circuit has a profound and selective effect on sleep architecture.
5HT is important for integration of sleep with other functions in both vertebrates and invertebrates
The circuit we describe is modulated by 5HT, a neurochemical known to be important for regulation of behavioral states in many species. While 5HT in mammals is important in a wide variety of contexts [48], it was controversial for nearly half a century whether it promoted sleep or wakefulness [10]. In Drosophila, 5HT has only been thought to promote sleep [14–16, 35]. Our data show that upregulation of serotonergic signaling can also induce sleep fragmentation, suggesting that 5HT’s role in sleep in flies exhibits a complexity similar to that of its roles in mammals. The genesis of this apparently conserved complexity may be the extensive involvement of 5HT in non-sleep processes. For an animal in the wild, sleep has inherent risks: predation and loss of opportunities for mating or feeding are just a few. Sleep/wake systems in the brain must control arousal state in collaboration with systems that assess competing needs. 5HT, because it is central to so many critical behavioral circuits, is ideally poised to be an integration point for sleep and the general state of the animal. The diverse, circuit-specific, roles in sleep that 5HT exhibits across phyla may be a result of its ubiquity.
The role we have uncovered for 5HT as a regulator of sleep architecture, aligns well with this idea. The daily neuronal activity profile reported by Tric-LUC [23] in sleep fragmentationgenerating neurons maps to dawn and dusk, when crepuscular organisms such as fruit flies are most active. Fragmentation of sleep at these times would presumably be beneficial since flies would not enter into deep sleep states at times when they should be feeding and mating. Interestingly, the circuit we describe accomplishes this feat by increasing P(doze), leaving the scaling of P(wake), a parameter associated with dopamine and arousal [2], free to be modulated by other factors (e.g. danger from predation, appearance of potential mates). The fact that long sleep bouts can be prevented without putting the animal into a hyperaroused state is advantageous, allowing flexible responsiveness to changing conditions. The involvement of P(Doze), a parameter associated with sleep drive, is also congruent with the sleep-promoting role of 5HT in other brain regions.
Sleep consolidation has a critical function
While controlled sleep fragmentation appears to assist in active period behavior, there is also a need for consolidated sleep. In both mammals and Drosophila, sleep has electrophysiologically distinct substrates with progressively higher arousal thresholds that appear in an ordered fashion during a sleep episode [2, 21, 49, 50]. The deeper sleep stages in mammals, REM and slow wave sleep, are strongly associated with maintenance of cognitive function [for review see 51]. Fragmentation of sleep, because it truncates sleep episodes before deeper stages are reached, can result in a selective deprivation of deep sleep stages even when total sleep is not changed. In this study, we demonstrate that decreasing sleep consolidation, without changing the amount of sleep, can disrupt associative learning. These results suggest that in Drosophila, like mammals, there are time-dependent changes in the depth of sleep that are important for its beneficial effects. This idea is also supported by modeling and analysis of the structure of fly sleep which indicate that there are time-dependent changes in the probability of sleep-wake transitions consistent with the existence of deep sleep stages that are only accessed in long sleep episodes [2].
Fragmentation of sleep induced by activation of 5HT inputs to the EB also produced an increase in sleep after the activation was terminated. Excess sleep in the recovery day after a perturbation is a hallmark of a homeostatic process. Homeostatic regulation of total sleep time has been previously demonstrated in Drosophila [12, 13], but our data suggest that there is also homeostatic regulation of sleep quality. In mammals, individual sleep substates have been demonstrated to be homeostatically regulated-selective deprivation of REM or slow wave sleep, in the absence of loss of total sleep time, drive rebound increases of the deprived stage [52, 53] and mechanical sleep fragmentation has been shown to lead to an increase in total sleep [29]. The ability of the EB circuit we describe in Drosophila to selectively modulate sleep structure, without changing the total amount of sleep, has allowed us for the first time to selectively probe the cognitive importance of long sleep bouts and deep sleep stages in the fly. The fact that fragmentation triggers rebound sleep implies that these long sleep bouts may also be important for the general health benefits of sleep.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Leslie C. Griffith (griffith@brandeis.edu). All unique and stable reagents generated in this study are available from the Lead Contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Flies were raised in a 12 h:12h light/dark cycle on modified Brent and Oster cornmeal-dextrose- yeast agar food [55] Per batch: 60 l H2O, 600 g Agar, 1,950 g flaked yeast, 1,451 g cornmeal, 6,300 g dextrose, 480 g NaKT, 60 g CaCl2, 169 g Lexgard dissolved in ethanol. All flies for experiments were raised at 25°C in an incubator after eclosion except for animals carrying UAS-dTrpA1 which were raised at 18°C. Mated females were employed for all sleep experiments unless specified. The genetic background in the w;;5HT7 or yw;;5HT7 mutants with a genetic insertion in the 5HT7 gene was replaced with the Canton-S background.
The following lines were previously described: UAS-dTrpA1 [18], UAS-Kir2.1 [56], UAS-PKA-mC* [57], UAS-BDK33(PKAr) [58], UAS-mCD8::GFP [59], UAS-mCD8::RFP, LexAop2- mCD8::GFP [60], UAS-Chrimson-tdTomato [61]. The following lines were obtained from the Bloomington Drosophila Stock Center at Indiana University (Bloomington, IN, USA): Trh-GAL4 (II) (stock #38388), Trh-GAL4 (III) (stock #38389), Mi{ET1}5HT7MB01344 (stock#23066), Mi{MIC}5HT7MI08096(stock#44745), UAS-GCaMP6f (stock#42747), UAS-sSyb:spGFP1 – 10,LexAop-CD4:spGFP11 (stock#64314), LexcAop-nSyb:spGFP1–10, UAS-CD4:spGFP11 (stock#64315), LexAop-P2X2 (stock#76030). The lines below were obtained from Vienna Drosophila Resource Center (VDRC, Vienna, Austria): VT038828-GAL4 (stock #201975). And UAS-Epac1-camps(55A) [62] was kindly provided by Dr. Orie Shafer. 5HT7Dro-GAL4 [37] flies were a gift from Dr. Charles Nichols.
The VT038828-LexA transgenic line was based on VT038828-GAL4. The 2285 bp promoter fragment was amplified from genomic DNA of wild-type flies using the same primers as used for VT038828-GAL4 (forward primer 5 ‘-AGTTTTTCCCATTTCCCATCAACAAAA-3 ‘ and reverse primer 5’-CCGGAGGACCCCAGGACTATGTCTAC-3’). This fragment was then cloned into plasmid pBPnlsLexA::p65Uw (Addgene Cat. #26230) using FseI and AatII restriction sites. The sequence was confirmed by sequencing using primers as follow: forward 5’- CAGGGTTATTGTCTCATGAGCGGATAC-3’ and reverse 5’-ACGTCTGCTCGGCTCGAACATTCATTC-3’. The plasmid was inserted into a VK00027 site using PhiC31 mediated recombination (Rainbow Inc.).
METHOD DETAILS
Behavioral analysis
Sleep assay
Newly enclosed flies were raised in standard vials, and mating was allowed to happen freely in vials. Flies were 2–5 day old at the start of each experiment. Individuals were placed into 65 mm x 5 mm glass. All sleep tubes contained 2% agarose with 5% sucrose food. Flies were entrained at 25°C in 12 h: 12 h light/dark (LD) conditions for 2–3 days. Activity was then recorded for 2 days in LD then switched to constant darkness (DD) for another 2 days (data not shown). For experiments with dTrpAl-activation, after the entrainment at 21°C for 2–3 days, we collected the following 1 day data as baseline, and elevated the temperature to 27°C (or 30°C for EB lines) for 1 day to activate the neurons expressing the dTrpA1, then shifted the temperature back to 21°C for another day to test if the effects are reversible. For experiments with Chrimson-activation, after entrainment at 25°C for 2–3 days, we collected one day as “Baseline”, then applied a red LED stimulus with 5 seconds on and 5 seconds off for 24 h to activate the neurons expressing the mtdTomato-tagged Chrimson, labelled as “Red Light On”. After we turned off the red LED stimulus, data for the next day is labelled as “Recovery Day”.
For drug administration in dTrpA1-activation experiments with gaboxadol (THIP) (Sigma Cat. No. T101), a direct-acting GABAA agonist which has been shown to be effective for the treatment of insomnia, 1mg/ml or 0.1 mg/ml was applied on the day when serotonergic neurons were activated from ZT0. For drug administration experiments with SB258719 (Tocris bioscience Cat. No. 2726, Bristol, UK), a highly selective antagonist at mammalian 5HT7 receptors, 1 mM drug in the sleep food was applied. CS wildtype flies were employed. Four days of sleep data in LD conditions after the loading day were obtained and presented to assess the drug effect.
Learning assay
The conditioning protocol was as described previously (Aso et al., 2010). A group of ~50 flies in a training tube alternately received 10% octan-3-ol (OCT; Merck) and 10% 4-methylcyclohexanol (MCH; Sigma-Aldrich) for 1 min in a constant air stream for immediate memory. For electric-shock induced memory, flies were loaded into shock tubes where one odor was presented with shock (1.25 sec of 90 V shock every 5 sec, 12 shocks in a minute) and the other odor was presented without shock. After a given retention time (2 min), the trained flies were allowed to choose between OCT and MCH for 2 min in a T-maze. A performance index was calculated by subtracting the number of flies choosing the unpaired odor from the number of flies choosing the conditioned odor divided by the total number of flies who chose an odor. A learning index was then calculated by taking the mean performance indices of the two reciprocally trained groups. Half of the trained groups received reinforcement together with the first presented odor and the other half with the second odor to cancel the effect of the order of reinforcement. To examine the fragmented effect on learning ability, flies were moved to 30°C incubator from 18°C incubator for 24 h prior to the conditioning or flies were trained and tested at 22°C with or without drug. Training and testing sessions were performed in a 65% relative humidity behavioral room at 22°C.
Calculation of relative sleep changes
The behavioral patterns of individual flies were monitored using the DAM (Drosophila Activity Monitor) system (Trikinetics, Waltham). Sleep parameters were analyzed using an in-house MATLAB program described preciously [63] from averages of 2 days of LD data in most experiments. Sleep manipulation (activation of neurons with dTrpA1) was carried out for 1 day. Total sleep, number of sleep episodes, max episode length, and activity while awake were analyzed for 12 h light and dark periods (LP and DP). In addition to analyzing the raw daily data, to control for the effect on sleep from elevated temperature, the sleep changes (ΔSleep) during and/or after manipulation were calculated for analysis where sleep was normalized to its baseline day sleep (ΔSleep = sleep on manipulation/recovery day – sleep on baseline day). Sleep episode data was pooled within experimental groups to calculate the empirical cumulative distribution function of episode duration. Based on a pilot analysis of baseline sleep cumulative distribution functions, sleep episodes were separated into “short” and “long” using a 200 minute threshold. The percentage of flies that at least one long episode, and the total duration of all long episodes performed by each fly was calculated. The binomial standard error is reported for the percentage of flies with a long sleep episode, defined as (square root(, where is the proportion of flies with a long episode and n is the number of flies.
Immunohistochemistry
Immunolabelling was done with a standard protocol. Briefly, brains were dissected in ice-cold PBS and fixed in 4% paraformaldehyde (vol/vol) on ice for 1 hr. Brains were incubated in PBS containing 0.5% Triton X-100, 10% NGS, primary antibodies over two nights and secondary antibodies for one night each with 3 × 5 min washes between each incubation. Frontal optical sections of whole-mount brains using Vectashield (Vector Laboratories, Inc. Cat. No. H-1000) were visualized by a Leica TCS SP5 confocal microscope with a 40x oil objective lens. Primary antibodies were used as follows: mouse anti-GFP (1:200, Roche Applied Science Cat. No. 11814460001), rabbit anti-GFP (1:1000, Invitrogen Cat. No. A-11122), rabbit anti-serotonin (1:1000, Sigma Cat. No. S5545), rabbit anti Ds-Red (1:200, Clontech Cat. No. 632496), chicken anti-GFP (1:1000, Abcam Cat. No. 13970), rabbit anti-hCD4 (1:500, Novus Cat. No. NBP1–86143). Alexa Fluor secondary antibodies (1:200, Invitrogen, 488 goat anti-mouse Cat. No. A11001, 488 goat anti-rabbit Cat. No. A-11008, 633 goat anti-mouse Cat. No. A-21052, and 635 goat anti-rabbit Cat. No. A-31576) and Alexa Fluor 488 (1:200, goat anti-chicken Invitrogen Cat. No. A-11039)) were used to visualize staining patterns. Confocal stacks were analyzed with the freely available FIJI (Image-J) software and plugins [64].
Real-time quantitative PCR
~30 brains of each genotype were used for RNA extraction. cDNA was synthesized using SuperScript III First-Strand Synthesis System (Invitrogen Cat. No. 18080) and used as a template for q-PCR using primers for 5HT7 and RPL32 (reference gene). Primers for 5HT7 were designed with the following sequences: forward primer 5’-GCATGGTGCGGAAATTGAGG-3’; reverse primer 5’-ACGGATATGGCACACAGA-3’; and for RPL32: forward primer 5’-GGACAGTATCTGATGCCCAAC-3’; reverse primer 5 ‘-ATCTCGCCGCAGTAAAGGC-3 ‘. The mRNA levels of 5HT7 were measured using QuantiFast SYBR Green PCR kit (Qiagen Cat. No. 204054) on a Mastercycler realplex2 instrument (Eppendorf). The relative mRNA levels were calculated using the comparative delta CT method.
Ex vivo functional fluorescence imaging
Adult hemolymph-like saline (AHL) consists of (in mM) 108 NaCl, 5 KCl, 2 CaCl2, 8.2 MgCl2, 4 NaHCO3, 1 NaH2PO4-H2O, 5 trehalose, 10 sucrose, 5 HEPES; pH 7.5 [65]. Serotonin hydrochloride (5HT) was purchased from Tocris Bioscience (Cat. No. 3547, Minneapolis, MN). 5HT was dissolved directly in AHL immediately prior to the experiment, and kept in light- shielded containers to prevent degradation. Tetrodotoxin (TTX) was purchased from TOCRIS Bioscience (Cat. No. 1078) was diluted to 1 uM. L-Glutamic acid was purchased from Sigma (Cat. No. G8415) and diluted to 5 mM for the experiment. Adenosine 5’-triphosphate magnesium salt (ATP) was purchased from Sigma (Cat. No. A9187), diluted to 2.5 mM and re-pH to 7.5 for all experiments.
Female flies of genotypes w1118;5HT7-GAL4/Sp;UAS-EPAC-Camps1/+, w1118;5HT7-GAL4/LexAop-P2X2; VT038828-LexAUAS-GCaMP6f, w118;5HT7-GAL4/LexAop-P2X2; UAS-GCaMP6f/+ and male flies of w1118;LexAop-P2X2/+;VT038828-GAL4/UAS-GCaMP6f aged 310 days post-eclosion were employed. Flies were anesthetized on ice, and brains were dissected into AHL at room temperature. Dissected brains were then pinned to a custom-made chamber with AHL. Brains expressing the FRET sensor EPACcamps1 were exposed to fluorescent light for 5 min to minimize differences in photobleaching rates between the CFP and YFP fluorophores, as YFP has been described to photobleach more slowly than CFP [62, 66]. Perfusion flow was established over the brain with a gravity-fed ValveLink perfusion system (Automate Scientific, Berkeley, CA). Solution containing 5HT, ATP or Glutamate was delivered by switching perfusion flow from the vehicle channel to drug channel. To control for the effects of switching channels, a vehicle control trial was performed by switching to a third channel with the same vehicle solution for the same duration as the experimental trial.
Imaging was performed using an Olympus BX51WI fluorescence microscope (Olympus, Center Valley, PA) under an Olympus x60 (0.90W, LUMPlanFI) water-immersion objective, and all recordings were captured using a charge-coupled device camera (Hamamatsu ORCAC472–80- 12AG). All images were captured using μManager acquisition software [67]. Regions of interests (ROIs) were selected in the EB.
For EPAC, ROIs were analyzed using custom software developed in MATLAB (The MathWorks, Natick, MA). This analysis script was modified as previously described [35]. Briefly, identical ROIs were selected from both the CFP and YFP emissions channels, and the fluorescence resonance energy transfer (FRET) signal (YFP/CFP ratio) was calculated for each time point and normalized to the ratio of the first 20 frames of baseline recording. The relative cAMP changes were determined by plotting the normalized CFP/YFP ratio (percentage) over time. The peak percent change values were determined as the mean maximum values for each group during drug perfusion period.
For GCaMP6f, ROIs were analyzed using ImageJ [64, and National Institutes of Health, Bethesda, MD]. Briefly, the percent change in fluorescence over time was calculated using ΔF/F = (Fn – F0)/F0 × 100%, where Fn is the fluorescence at time point n, and F0 is the fluorescence at time 0. Peak fluorescence change values were determined as the maximum (or minimum) percentage change observed for each trace before/during drug perfusion period.
CaLexA bioluminescence recording
The recording was done as described in Guo et al., 2017. Basically, food containing 1% agar and 5% sucrose was heated to melt and supplemented with 20 mM of D-luciferin potassium salt (GOLDBIO), 250 μl of which was then distributed into every other well in a 96-well plate. Plates were allowed to cool down completely before use. Individual flies were loaded into each well and the plate was then sealed with a transparent adhesive (TopSeal-A PLUS, Perkin Elmer). Every well was punctured with two to three small holes to allow air circulation, and recording was with a TopCount NXT luminescence counter (Perkin Elmer) in an incubator under LD cycles at 25°C. Data were then analysed with MATLAB.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis
Statistical analyses for behavioral data were performed using GraphPad Prism 7. Datasets that did not have a normal distribution, nonparametric statistics (Kruskal-Wallis, followed by Dunn’s multiple comparisons test) was applied. Otherwise, a one-way ANOVA, followed by Holm-Sidak’s test was applied when datasets passed normality test. For experiments that had multiple variables, a two-way ANOVA was performed. Bonferroni multiple comparisons after two-way ANOVA were used for each analysis period (LP and DP), and were performed to determine which pairs were significantly different and if major effects are significantly different. Figs are all presented as mean ± SEM in a uniform style for clarity. Asterisk (*) always indicates a significant difference between the experimental group and both the control groups. The significance level of statistical tests was set to 0.05. All statistical analyses are available at Mendeley Data: see the link below.
Statistical analyses for functional imaging were performed using GraphPad Prism 7. In EPAC measurements, a two-way ANOVA was used to determine statistical significance among different dosages of 5HT or between the experimental groups and the control groups. In P2X2-GCamP6f experiments, a two-way ANOVA was used to determine statistical significance for treatments (before and after ATP perfusion) and subjects (experimental and control groups). Tukey’s multiple comparisons test was followed where a significance takes place in the ANOVA. Results are expressed as mean ± SEM. Datasets are marked with letters (a, b or c) for statistical equivalence groups; i.e., data that are significantly different are indicated by different letters. Significance level was set at 0.05. In GCaMP6f experiments with application of 5HT, a paired t-test was performed to determine the drug perfusion effects due to the fact that these datasets passed normality tests.
DATA AND CODE AVAILABILITY
The MATLAB scripts for analysis of P(Wake)/P(Doze) have been deposited in GitHub and can be accessed at: https://github.com/Griffith-Lab/Fly_Sleep_Probability
Statistical analyses reported in this paper are archived at Mendeley Data: DOI: 10.17632/3tf592t65v.1
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-GFP | Roche Applied Science | Cat#11814460001 RRID:AB_390913 |
| Rabbit polyclonal anti-GFP | Invitrogen | Cat# A-11122 RRID: AB_221569 |
| Rabbit anti-serotonin | Sigma | Cat# S5545 RRID: AB_477522 |
| Rabbit polyclonal anti-Ds-Red | Clontech | Cat# 632496 RRID: AB_10013483 |
| Chicken anti-GFP | Abcam | Cat# 13970 RRID: AB_300798 |
| Rabbit anti-hCD4 | Novus | Cat# NBP1–86143 RRID: 11037580 |
| Alexa Fluor 488 (goat anti-mouse) | Invitrogen | Cat# A-11001 RRID: AB_2534069 |
| Alexa Fluor 488 (goat anti-rabbit) | Invitrogen | Cat# A-11008 RRID: AB_143165 |
| Alexa Fluor 488 (goat anti-chicken) | Invitrogen | Cat# A-11039 RRID: AB_142924 |
| Alexa Fluor 633 (goat anti-mouse) | Invitrogen | Cat# A-21052 RRID:AB_2535719 |
| Alexa Fluor 635 (goat anti-rabbit) | Invitrogen | Cat# A-31576 RRID: AB_2536186 |
| Vectashield | Vector Laboratories, Inc. | Cat# H-1000 RRID: AB_2336789 |
| Bacterial and Virus Strains | ||
| pBPnlsLexA::p65Uw | [55] | Addgene Cat#26230 |
| NEB Stable Competent E. coli (High Effiency) | NEB | Cat# C3040H |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tetrodotoxin (TTX) | Tocris Bioscience | Cat# 1078 |
| Adenosine 5’-triphosphate magnesium salt (ATP) | Sigma | Cat# A9187 |
| Serotonin hydrochloride (5-HT) | Sigma | Cat# H9523 |
| Gaboxadol Hydrochloride (THIP) | Sigma | Cat# T101 |
| SB258719 hydrochloride | Tocris Bioscience | Cat# 2726 |
| D-Luciferin Firefly, potassium salt | BIOSYNTH AG | Cat# L-8220 |
| Critical Commercial Assays | ||
| SuperScript III First-Stand Synthesis System | Invitrogen | Cat# 18080 |
| QuantiFast SYBR Green PCR kit | Qiagen | Cat# 204054 |
| Experimental Models: Organisms/Strains | ||
| Drosophila: UAS-dTrpA1 | [18] | RRID: BDSC_26263 |
| Drosophila: UAS-Kir2.1 | [56] | FlyBase: FBtp0039831 |
| Drosophila: UAS-Chrimson-tdTomato | [57] | |
| Drosophila: UAS-mCD8::GFP | [58] | RRID: BDSC_5136 |
| Drosophila: UAS-mCD8::RFP,LexAop2-mCD8::GFP | [55] | RRID: BDSC_32229 |
| Drosophila: Trh-GAL4 (II) | Bloomington Drosophila Stock Center | RRID: BDSC_38388 |
| Drosophila: Trh-GAL4 (III) | Bloomington Drosophila Stock Center | RRID: BDSC_38389 |
| Drosophila: PBac{WH}5-HT7f05214 | Bloomington Drosophila Stock Center | RRID: BDSC_18848 |
| Drosophila: Mi{MIC}5-HT7MI00215 | Bloomington Drosophila Stock Center | RRID: BDSC_30667 |
| Drosophila: Mi{ET1}5-HT7MB01344 | Bloomington Drosophila Stock Center | RRID: BDSC_23066 |
| Drosophila: Mi{ET1}5-HT7MB04445 CG31008MB04445 | Bloomington Drosophila Stock Center | RRID: BDSC_24705 |
| Drosophila: Mi{MIC}5-HT7MI08096 | Bloomington Drosophila Stock Center | RRID: BDSC_44745 |
| Drosophila: UAS-GCaMP6f | Bloomington Drosophila Stock Center | RRID: BDSC_42747 |
| Drosophila: UAS-nSyb:spGFP1–10, LexAop-CD4:spGFP11 | [45] | RRID: BDSC_64314 |
| Drosophila: LexAop-nSyb:spGFP1–10, UAS-CD4:spGFP11 | Bloomington Drosophila Stock Center | RRID: BDSC_64315 |
| Drosophila: LexAop-P2X2 | [46] | RRID: BDSC_76030 |
| Drosophila: VT038828-GAL4 | Vienna Drosophila Resource Center (VDRC) | RRID: BDSC_v201975 |
| Drosophila: UAS-Epac1-camps(55A) | [59] | Dr. Orie Shafer |
| Drosophila: 5-HT7Dro-GAL4 | [37] | Dr. Charles Nichols |
| Drosophila: UAS-myrGFP,QUAS-mtdTomato-3xHA;trans-Tango | [47] | RRID: BDSC_77124 |
| Drosophila: Tric-LUC | [23] | Dr. Michael Rosbash |
| Drosophila: R23E10-GAL4 | [60] | RRID:BDSC_49032 |
| Oligonucleotides | ||
| RT forward primer for 5-HT7: GCATGGTGCGGAAATTGAGG | This paper | N/A |
| RT reverse primer for 5-HT7: CCACGGATATGGCACACAGA | This paper | N/A |
| RT forward primer for RPL32: GGACAGTATCTGATGCCCAAC | This paper | N/A |
| RT reverse primer for RPL32: ATCTCGCCGCAGTAAAGGC | This paper | N/A |
| Amplification for VT038828 promoter forward primer: AGTTTTTCCCATTTCCCATCAACAAAA | http://stockcenter.vdrc.at/control/product/~VIEW_INDEX=0/~VIEW_SIZE=100/~product_id=201975 | N/A |
| Amplification for VT038828 promoter reverse primer: CCGGAGGACCCCAGGACTATGTCTAC | http://stockcenter.vdrc.at/control/product/~VIEW_INDEX=0/~VIEW_SIZE=100/~product_id=201975 | N/A |
| Sequencing forward primer for VT038828-LexA: CAGGGTTATTGTCTCATGAGCGGATAC | This paper | N/A |
| Sequencing reverse primer for VT038828-LexA: ACGTCTGCTCGGCTCGAACATTCATTC | This paper | N/A |
| Software and Algorithms | ||
| Fiji | http://fiji.sc | RRID: SCR_002285 |
| GraphPad Prism 7 | GraphPad Software | RRID: SCR_002798 |
| MATLAB R2012b | MathWorks | RRID: SCR_001622 |
| Leica TCS SP5 confocal microscope | Leica | RRID: SCR_002140 |
Highlights.
Serotonin plays a role in regulation of sleep architecture independent of total sleep
Serotonergic signaling to the ellipsoid body via 5HT7 receptors fragments sleep
Learning deficits resulting from fragmentation can be rescued by consolidating sleep
Two reciprocally connected sets of EB neurons control sleep architecture
ACKNOWLEDGEMENTS
This work was supported by R01 MH67284 to LCG and F32 NS098624 to TDW. We also thank Elena Kuklin for help with the trans-Tango experiments and Johanna Flyer-Adams for early work on learning.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Faraut B, Andrillon T, Vecchierini MF, and Leger D. (2017). Napping: A public health issue. From epidemiological to laboratory studies. Sleep medicine reviews 35, 85100. [DOI] [PubMed] [Google Scholar]
- 2.Wiggin TD, Goodwin PR., Donelson NC, Liu C, Trinh K, Sanyal S, and Griffith LC. (2019). Sleep structure and quantity are determined by behavioral transition probability in Drosophila melanogaster. bioRxiv 263301; doi: 10.1101/263301 [Google Scholar]
- 3.Bonnet MH, and Arand DL (2003). Clinical effects of sleep fragmentation versus sleep deprivation. Sleep medicine reviews 7, 297–310. [DOI] [PubMed] [Google Scholar]
- 4.Van Someren EJ, Cirelli C, Dijk DJ, Van Cauter E, Schwartz S, and Chee MW (2015). Disrupted Sleep: From Molecules to Cognition. J Neurosci 35, 13889–13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antunes BM, Campos EZ, Parmezzani SS, Santos RV, Franchini E, and Lira FS (2017). Sleep quality and duration are associated with performance in maximal incremental test. Physiology & behavior 177, 252–256. [DOI] [PubMed] [Google Scholar]
- 6.Biard K, Douglass AB, and De Koninck J. (2015). The effects of galantamine and buspirone on sleep structure: Implications for understanding sleep abnormalities in major depression. J Psychopharmacol 29, 1106–1111. [DOI] [PubMed] [Google Scholar]
- 7.Ensrud KE, Blackwell TL, Redline S, Ancoli-Israel S, Paudel ML, Cawthon PM, Dam TT, Barrett-Connor E, Leung PC., Stone KL, et al. (2009). Sleep disturbances and frailty status in older community-dwelling men. Journal of the American Geriatrics Society 57, 2085–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solarewicz JZ, Angoa-Perez M, Kuhn DM, and Mateika JH (2015). The sleep-wake cycle and motor activity, but not temperature, are disrupted over the light-dark cycle in mice genetically depleted of serotonin. American journal of physiology 308, R10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oikonomou G, Altermatt M, Zhang RW, Coughlin GM, Montz C, Gradinaru V, and Prober DA (2019). The Serotonergic Raphe Promote Sleep in Zebrafish and Mice. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portas CM, Bjorvatn B, and Ursin R. (2000). Serotonin and the sleep/wake cycle: special emphasis on microdialysis studies. Prog Neurobiol 60, 13–35. [DOI] [PubMed] [Google Scholar]
- 11.Monti JM (2011). Serotonin control of sleep-wake behavior. Sleep medicine reviews 15, 269–281. [DOI] [PubMed] [Google Scholar]
- 12.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, and Pack AI (2000). Rest in Drosophila is a sleep-like state. Neuron 25, 129–138. [DOI] [PubMed] [Google Scholar]
- 13.Shaw PJ, Cirelli C, Greenspan RJ, and Tononi G. (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Q, Joiner WJ, and Sehgal A. (2006). A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol 16, 1051–1062. [DOI] [PubMed] [Google Scholar]
- 15.Qian Y, Cao Y, Deng B, Yang G, Li J, Xu R, Zhang D, Huang J, and Rao Y. (2017). Sleep homeostasis regulated by 5HT2b receptor in a small subset of neurons in the dorsal fan-shaped body of drosophila. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pooryasin A, and Fiala A. (2015). Identified Serotonin-Releasing Neurons Induce Behavioral Quiescence and Suppress Mating in Drosophila. J Neurosci 35, 12792–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strausfeld NJ, and Hirth F. (2013). Deep homology of arthropod central complex and vertebrate basal ganglia. Science 340, 157–161. [DOI] [PubMed] [Google Scholar]
- 18.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, and Garrity PA. (2008). An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alekseyenko OV, Lee C, and Kravitz EA (2010). Targeted manipulation of serotonergic neurotransmission affects the escalation of aggression in adult male Drosophila melanogaster. PLoS ONE 5, e10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parisky KM, Agosto Rivera JL, Donelson NC, Kotecha S, and Griffith LC (2016). Reorganization of Sleep by Temperature in Drosophila Requires Light, the Homeostat, and the Circadian Clock. Curr Biol 26, 882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Alphen B, Yap MH, Kirszenblat L, Kottler B, and van Swinderen B. (2013). A dynamic deep sleep stage in Drosophila. J Neurosci 33, 6917–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida K, Shi S, Ukai-Tadenuma M, Fujishima H, Ohno RI, and Ueda HR (2018). Leak potassium channels regulate sleep duration. Proc Natl Acad Sci U S A 115, E9459–E9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo F, Chen X, and Rosbash M. (2017). Temporal calcium profiling of specific circadian neurons in freely moving flies. Proc Natl Acad Sci U S A 114, E8780–E8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo F, Holla M, Diaz MM, and Rosbash M. (2018). A Circadian Output Circuit Controls Sleep-Wake Arousal in Drosophila. Neuron 100, 624–635 e624. [DOI] [PubMed] [Google Scholar]
- 25.Cipolli C, Mazzetti M, and Plazzi G. (2013). Sleep-dependent memory consolidation in patients with sleep disorders. Sleep medicine reviews 17, 91–103. [DOI] [PubMed] [Google Scholar]
- 26.Rolls A, Colas D, Adamantidis A, Carter M, Lanre-Amos T, Heller HC, and de Lecea L. (2011). Optogenetic disruption of sleep continuity impairs memory consolidation. Proc Natl Acad Sci U S A 108, 13305–13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stepanski EJ (2002). The effect of sleep fragmentation on daytime function. Sleep 25, 268–276. [DOI] [PubMed] [Google Scholar]
- 28.Seugnet L, Suzuki Y, Vine L, Gottschalk L, and Shaw PJ. (2008). D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr Biol 18, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baud MO, Magistretti PJ., and Petit JM (2015). Sustained sleep fragmentation induces sleep homeostasis in mice. Sleep 38, 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dissel S, Angadi V, Kirszenblat L, Suzuki Y, Donlea J, Klose M, Koch Z, English D, Winsky-Sommerer R, van Swinderen B, et al. (2015). Sleep restores behavioral plasticity to Drosophila mutants. Curr Biol 25, 1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faulhaber J, Steiger A, and Lancel M. (1997). The GABAA agonist THIP produces slow wave sleep and reduces spindling activity in NREM sleep in humans. Psychopharmacology (Berl) 130, 285–291. [DOI] [PubMed] [Google Scholar]
- 32.Lancel M. (1997). The GABA(A) agonist THIP increases non-REM sleep and enhances non-REM sleep-specific delta activity in the rat during the dark period. Sleep 20, 10991104. [DOI] [PubMed] [Google Scholar]
- 33.Donlea JM, Pimentel D, and Miesenbock G. (2014). Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81, 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee PT., Lin HW, Chang YH, Fu TF, Dubnau J, Hirsh J, Lee T, and Chiang AS (2011). Serotonin-mushroom body circuit modulating the formation of anesthesia-resistant memory in Drosophila. Proc Natl Acad Sci U S A 108, 13794–13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haynes PR., Christmann BL, and Griffith LC (2015). A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, and Kume K. (2012). Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci 15, 1516–1523. [DOI] [PubMed] [Google Scholar]
- 37.Becnel J, Johnson O, Luo J, Nassel DR, and Nichols CD (2011). The serotonin 5-HT7Dro receptor is expressed in the brain of Drosophila, and is essential for normal courtship and mating. PLoS ONE 6, e20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gnerer JP, Venken KJ, and Dierick HA (2015). Gene-specific cell labeling using MiMIC transposons. Nucleic Acids Res 43, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Pena A, Acebes A, Rodriguez JR, Chevalier V, Casas-Tinto S, Triphan T, Strauss R, and Ferrus A. (2014). Cell types and coincident synapses in the ellipsoid body of Drosophila. Eur J Neurosci 39, 1586–1601. [DOI] [PubMed] [Google Scholar]
- 40.Hanesch U, Fischbach KF, Heisenberg M. (1989). Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Research 257, 343–366. [Google Scholar]
- 41.Wolff T, Iyer NA, and Rubin GM (2015). Neuroarchitecture and neuroanatomy of the Drosophila central complex: A GAL4-based dissection of protocerebral bridge neurons and circuits. J Comp Neurol 523, 997–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omoto JJ, Nguyen BM, Kandimalla P, Lovick JK, Donlea JM, and Hartenstein V (2018). Neuronal Constituents and Putative Interactions Within the Drosophila Ellipsoid Body Neuropil. Frontiers in neural circuits 12, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu JY, Liu C, and Griffith LC (2019). EB screen paper. In preparation. [Google Scholar]
- 44.Liu S, Liu Q, Tabuchi M, and Wu MN (2016). Sleep Drive Is Encoded by Neural Plastic Changes in a Dedicated Circuit. Cell 165, 1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macpherson LJ, Zaharieva EE, Kearney PJ., Alpert MH, Lin TY, Turan Z, Lee CH, and Gallio M. (2015). Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nature communications 6, 10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao Z, Macara AM, Lelito KR, Minosyan TY, and Shafer OT (2012). Analysis of functional neuronal connectivity in the Drosophila brain. J Neurophysiol 108, 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talay M, Richman EB, Snell NJ, Hartmann GG, Fisher JD, Sorkac A, Santoyo JF, Chou-Freed C, Nair N, Johnson M, et al. (2017). Transsynaptic Mapping of Second-Order Taste Neurons in Flies by trans-Tango. Neuron 96, 783–795 e784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo M, Li Y, and Zhong W. (2016). Do dorsal raphe 5-HT neurons encode “beneficialness”? Neurobiol Learn Mem 135, 40–49. [DOI] [PubMed] [Google Scholar]
- 49.Yap MHW, Grabowska MJ, Rohrscheib C, Jeans R, Troup M, Paulk AC, van Alphen B, Shaw PJ., and B. van Swinderen (2017). Oscillatory brain activity in spontaneous and induced sleep stages in flies. Nature communications 8, 1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colten HR, and Altevogt BM, eds. (2006). Sleep Disorders and Sleep Deprivation: An Unmet Public Health Problem, (Washington (DC)). [PubMed] [Google Scholar]
- 51.Stickgold R. (2013). Parsing the role of sleep in memory processing. Curr Opin Neurobiol 23, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ocampo-Garces A, Molina E, Rodriguez A, and Vivaldi EA (2000). Homeostasis of REM sleep after total and selective sleep deprivation in the rat. J Neurophysiol 84, 26992702. [DOI] [PubMed] [Google Scholar]
- 53.Ferrara M, De Gennaro L, and Bertini M. (1999). Selective slow-wave sleep (SWS) deprivation and SWS rebound: do we need a fixed SWS amount per night? Sleep Res Online 2, 15–19. [PubMed] [Google Scholar]
- 54.Gorostiza EA, Depetris-Chauvin A, Frenkel L, Pirez N, and Ceriani MF (2014). Circadian pacemaker neurons change synaptic contacts across the day. Curr Biol 24, 2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brent MM, and Oster II (1974). Nutritional Substitution- a new approach to microbial control for Drosophila cultures. Volume 51 (Dro. Inf. Ser.), pp. 155–157. [Google Scholar]
- 56.Baines RA, Uhler JP, Thompson A, Sweeney ST, and Bate M. (2001). Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci 21, 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li W, Ohlmeyer JT, Lane ME, and Kalderon D. (1995). Function of protein kinase A in hedgehog signal transduction and Drosophila imaginal disc development. Cell 80, 553–562. [DOI] [PubMed] [Google Scholar]
- 58.Rodan AR, Kiger JA Jr., and Heberlein U. (2002). Functional dissection of neuroanatomical loci regulating ethanol sensitivity in Drosophila. J Neurosci 22, 94909501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee T, and Luo L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461. [DOI] [PubMed] [Google Scholar]
- 60.Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, and Rubin GM (2010). Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoopfer ED, Jung Y, Inagaki HK, Rubin GM, and Anderson DJ (2015). P1 interneurons promote a persistent internal state that enhances inter-male aggression in Drosophila. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO., Lohse MJ, and Taghert PH (2008). Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron 58, 223237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donelson NC, Donelson N, Kim EZ, Slawson JB, Vecsey CG, Huber R, and Griffith LC (2012). High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS ONE 7, e37250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang JW, Wong AM, Flores J, Vosshall LB, and Axel R. (2003). Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell 112, 271282. [DOI] [PubMed] [Google Scholar]
- 66.Pirez N, Christmann BL, and Griffith LC (2013). Daily rhythms in locomotor circuits in Drosophila involve PDF. J Neurophysiol 110, 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edelstein A, Amodaj N, Hoover K, Vale R, and Stuurman N. (2010). Computer control of microscopes using uManager. Curr Protocols Mol Biol 92, 14.20.11–14.20.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MATLAB scripts for analysis of P(Wake)/P(Doze) have been deposited in GitHub and can be accessed at: https://github.com/Griffith-Lab/Fly_Sleep_Probability
Statistical analyses reported in this paper are archived at Mendeley Data: DOI: 10.17632/3tf592t65v.1


