Abstract
Background
Despite much effort on the treatment of breast cancer over the decades, a great uncertainty regarding the appropriate molecular biomarkers and optimal therapeutic strategy still exists. This research was performed to analyze the association of SPAG5 gene expression with clinicopathological factors and survival outcomes.
Methods
We used a breast cancer database including 5667 patients with a mean follow-up of 69 months. Kaplan-Meier survival analyses for relapse free survival (RFS), overall survival (OS), and distant metastasis-free survival (DMFS) were performed. In addition, ROC analysis was performed to validate SPAG5 as a prognostic candidate gene.
Results
Mean SPAG5 expression value was significantly higher with some clinicopathological factors that resulted in tumor promotion and progression, including poor differentiated type, HER2 positive or TP53 mutated breast cancer. Based on ROC-analysis SPAG 5 is a suitable prognostic marker of poor survival. In patients who received chemotherapy alone, SPAG5 had only a moderate and not significant predictive impact on survival outcomes. However, in hormonal therapy, high SPAG5 expression could strongly predict prognosis with detrimental RFS (HR = 1.57, 95% CI 1.2–2.06, p = 0.001), OS (HR = 2, 95% CI 1.05–3.8, p = 0.03) and DMFS (HR = 2.36, 95% CI 1.57–3.54, p < 0.001), respectively. In addition, SPAG5 could only serve as a survival predictor in ER+, but not ER- breast cancer patients. Patients might also be at an increased risk of relapse despite being diagnosed with a lower grade cancer (well differentiated type).
Conclusions
SPAG5 could be used as an independent prognostic and predictive biomarker that might have clinical utility, especially in ER+ breast cancer patients who received hormonal therapy.
Keywords: SPAG5, Prognosis, Breast cancer, Endocrine therapy, Chemotherapy
Background
Breast cancer is one of the leading types of cancer in women which accounted for about 39,620 deaths among US women in 2013 [1]. Despite much effort on the treatment of breast cancer over the decades, a great uncertainty regarding the optimal therapeutic strategy, especially effective precision medicine for breast cancer still exists [2]. As only those individuals who harbor the appropriate molecular biomarkers are eligible for effective precision treatment [3], identification, stratification and evaluation of better prognostic/predictive markers are in great need [4]. Nowadays, breast cancer systemic treatment strategies are guided by molecular subtypes based on estrogen receptor (ER), progesterone receptor (PR) and epidermal growth factor receptor 2 (HER2) statuses [5], and clinically useful biomarkers are demanded in predicting a patient’s response and long-term outcomes. Some potential indicators have been found in the diagnosis and therapeutic monitoring of patients with breast cancer, such as SASH1, cystatin C and activin A [6–8].
Sperm-associated antigen 5 (SPAG5, also named DEEPEST, MAP126 or hMAP126), located on chromosome 17q11.2, was up-regulated in M-phase cells and played a vital role in cell mitosis and cell cycle checkpoint regulation [9]. By binding to microtubules, it regulated the timing of spindle organization as well as separation of sister chromatids [10]. In addition, SPAG5 protected cells from apoptosis via the mTOR signaling pathway [9, 11]. Knockdown of SPAG5 could significantly suppress proliferation and invasion of prostate cancer cells in vitro, along with inhibiting the growth and metastasis of tumor in vivo [10].
Previous studies indicated that the overexpression of SPAG5 gene might act as a potential biomarker which predicted poor prognosis in patients with lung cancer and cervical cancer [11, 12]. However, few studies focused on the prognostic value of SPAG5 in breast cancer patients. A recent study [13] reported that the transcript and protein products of SPAG5 might be independent prognostic and predictive biomarkers for chemotherapy sensitivity, particularly in ER negative (ER-) breast cancer. One stated the prognostic association of SPAG5 in ER+ breast cancer [14]. In addition, SPAG5 module was found to be involved in the mitotic checkpoint and associated with proliferation and progression of male breast cancer (MBC) [15].
To comprehensively assess the association of SPAG5 gene expression with clinical outcomes in patients with different breast cancer subtypes, including those undergoing systematic treatment (endocrine therapy and/or chemotherapy), we used a large public database containing pure transcriptomic data of more than 5000 breast cancer patients and validated SPAG5 as a prognostic candidate gene.
Methods
Breast cancer microarray database
Kaplan–Meier Plotter (http://www.kmplot.com) is an online public database evaluating the effect of 54,675 genes on patient clinical outcomes, using 10,293 samples of lung, breast, gastric or ovarian cancers. This online tool is handled by a PostgreSQL server that could simultaneously integrate gene expression and clinical data [16, 17]. Gene expression data and the survival information are derived from the Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA) and European Genome-phenome Atlas (EGA) (see Additional file 3: Table S1).
Data retrieval
We performed data retrieval from the online tool from July 2016 to October 2016. The database contained information of 5667 patients with breast cancer, with a median follow-up of 69 months. It allowed for filtering by ER, PR and HER2 statuses, lymph node statuses (positive or negative), grade (I, II or III) and TP53 statuses (mutated or wild type) [18]. In addition, analyses could be restricted to cohorts that only included patients with endocrine treatment or chemotherapy. Biased arrays were excluded. Although not all clinic-pathological data and survival outcomes were obtainable in each patient, we reported all available data.
Statistical analysis
We compared SPAG5 gene expression level using Kruskal-Wallis test (multi-group comparisons) or Mann-Whitney U test (two-cohort comparison). Mean expression value, 95% Confidence Interval (CI), standard error and standard deviation were analyzed. For the prognostic value of gene SPAG5, we plotted the Kaplan–Meier curves for SPAG5 (Affymetrix ID: 203145_at) in different breast cancer subtypes. The cutoff value of gene expression was chosen as median which split the patient samples into two groups and plots generated accordingly. The two patient cohorts were then compared, and we performed univariate Cox regression to calculate the hazard ratio (HR) with 95% confidence intervals (CIs) and log rank P-value. As not every patient’s data was included in the database that we needed to perform multiple Cox regression analyses, it was the best to do the multiple hypothesis testing [19, 20].
In addition, ROC analysis was performed by splitting the population into good and poor-outcome based on RFS, and we checked whether SPAG5 expression recognizes poor/good survival. We run the analysis for RFS of the entire dataset, ER-positive population and ER-positive population treated with endocrine therapy at 5 years and 10 years, respectively. Evaluation of gene SPAG5 with relapse free survival (RFS), overall survival (OS) and distant metastasis-free survival (DMFS) was performed. We also used this Kaplan-Meier Plotter to stratify breast cancer patient microarray data by ER, PR, HER2, lymph node status, histological grade and TP53 status, and explored the prognostic value of SPAG5 in those different breast cancer subtypes. We explored the survival of patients with different treatment strategies (hormonal therapy and/or chemotherapy). P-value < 0.05 was considered to be a statistically significant difference.
Results
SPAG5 gene expression in breast cancer patients
The Kaplan–Meier Plotter surveyed public microarray data repositories for survival among 5667 patients with breast cancer. Mean SPAG5 expression value was higher in ER- than ER+ breast cancer patients (mean value 434.48 vs. 602.64, p < 0.001), similar trend was also observed in PR- and HER2+ breast cancer patients. In addition, SPAG5 expression was progressively higher in more aggressive grades/subtypes of the disease (see Additional file 3: Tables S2 and S3 and Additional file 1: Figure S1).
SPAG5 gene expression was associated with breast cancer progression and poor prognosis
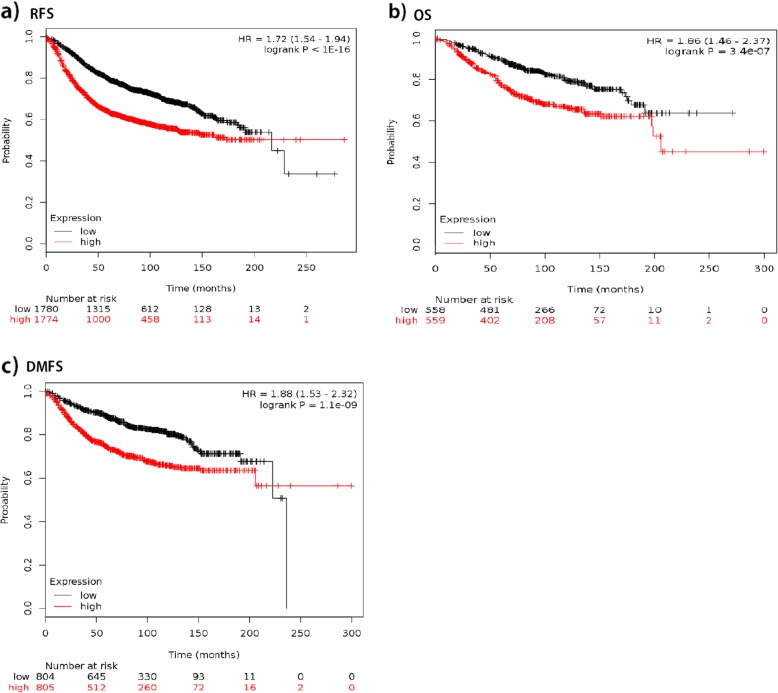
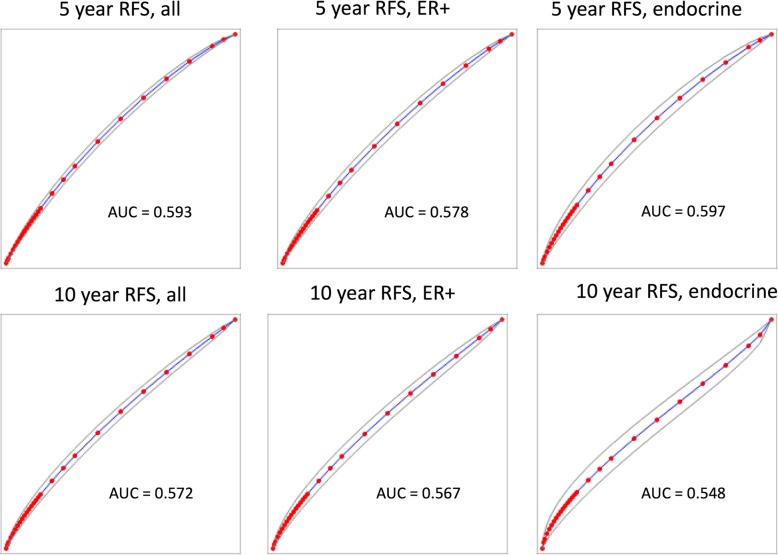
We plotted the Kaplan-Meier survival curves for SPAG5 using the web-based curator. The results showed that higher expression of SPAG5 was associated with worse RFS (n = 3557, HR = 1.72, 95% CI 1.54–1.94, p < 0.001), OS (n = 1117, HR = 1.86, 95% CI 1.46–2.37, p < 0.001), and DMFS (n = 1610, HR = 1.88, 95% CI 1.53–2.32, p < 0.001) in patients with breast cancer. Table 1 and Fig. 1 present the prognostic effect of the expression of SPAG5. In addition, we compared and correlated SPAG5 with other markers of progression, such as p53, AURKA, MKI67 and BIRC5, to assess independent value, and results showed that similar to AURKA, MKI67, BUB1, TOP2A which had statistically significant results for RFS, SPAG5 was associated with breast cancer progression. There was a significant association (coefficient over 0.25, p < 0.001) of SPAG5 with TOP2A, BIRC5, AURKA and BUB1. The association with PCNA and TP53 was significant, but the effect was too small to be meaningful (Additional file 3: Table S4). Based on ROC analysis, SPAG5 is a suitable prognostic marker of poor survival (see Fig. 2).
Table 1.
PH Cox regression univariate analyses for the association of gene SPAG5 with cancer progression and prognosis in different breast cancer subtypes
| Breast cancer subtypes | RFS | OS | DMFS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n | HR | P-value | n | HR | P-value | n | HR | P-value | |
| Total | 3557 | 1.72 (1.54–1.94) | < 0.001 | 1117 | 1.86 (1.46–2.37) | < 0.001 | 1610 | 1.88 (1.53–2.32) | < 0.001 |
| ER status | |||||||||
| ER+ | 2766 | 1.77 (1.55–2.03) | < 0.001 | 377 | 2.74 (1.74–4.33) | < 0.001 | 577 | 2.89 (1.95–4.29) | < 0.001 |
| ER- | 788 | 1.03 (0.82–1.28) | 0.81 | 142 | 0.91 (0.52–1.6) | 0.74 | 170 | 1.04 (0.62–1.73) | 0.89 |
| PR status | |||||||||
| PR+ | 525 | 2.02 (1.38–2.94) | < 0.001 | 0 | – | – | 122 | 1.35 (0.41–4.46) | 0.62 |
| PR- | 483 | 1.42 (1.04–1.93) | 0.027 | 2 | – | – | 95 | 2.24 (1.04–4.85) | 0.035 |
| HER2 status | |||||||||
| HER2+ | 168 | 0.78 (0.46–1.32) | 0.36 | 28 | 0.59 (0.19–1.83) | 0.36 | 66 | 1.54 (0.61–3.91) | 0.36 |
| HER2- | 756 | 1.78 (1.36–2.34) | < 0.001 | 62 | 0.92 (0.32–2.62) | 0.87 | 82 | 2.43 (0.63–9.39) | 0.18 |
| ER+/PR+/HER2+ | 76 | 1.53 (0.33–7.09) | 0.58 | 36 | 3.54 (0.41–30.58) | 0.22 | 45 | 1.83 (0.36–9.47) | 0.46 |
| ER+/PR−/HER2+ | 26 | 0.47 (0.09–2.35) | 0.35 | – | – | – | – | – | – |
| ER+/PR+/HER2- | 339 | 2.41 (1.48–3.93) | < 0.001 | 39 | 2.04 (0.18–22.51) | 0.55 | 79 | 1.66 (0.33–8.22) | 0.53 |
| ER+/PR−/HER2- | 77 | 1.47 (0.65–3.33) | 0.35 | – | – | – | – | – | – |
| ER−/PR−/HER2- | 255 | 1.45 (0.9–2.34) | 0.13 | – | – | – | 43 | 3.33 (0.67–16.58) | 0.12 |
| LN status | |||||||||
| LN+ | 945 | 1.63 (1.3–2.03) | < 0.001 | 197 | 1.38 (0.84–2.28) | 0.2 | 337 | 1.74 (1.14–2.65) | 0.009 |
| LN- | 1813 | 1.67 (1.4–1.99) | < 0.001 | 425 | 2.41 (1.56–3.74) | < 0.001 | 896 | 2.42 (1.79–3.27) | < 0.001 |
| Grade | |||||||||
| 1 | 308 | 2.52 (1.4–4.54) | 0.0014 | 135 | 2.45 (0.86–6.96) | 0.083 | 172 | 2.35 (0.94–5.84) | 0.059 |
| 2 | 724 | 1.9 (1.45–2.49) | < 0.001 | 287 | 2.92 (1.76–4.86) | < 0.001 | 495 | 1.93 (1.34–2.78) | < 0.001 |
| 3 | 723 | 1.17 (0.91–1.51) | 0.23 | 347 | 0.9 (0.6–1.34) | 0.6 | 391 | 1.12 (0.77–1.64) | 0.54 |
| TP53 status | |||||||||
| Mutated | 188 | 0.91 (0.57–1.47) | 0.71 | 111 | 0.95 (0.45–2.04) | 0.91 | 83 | 0.8 (0.33–1.93) | 0.62 |
| Wild type | 273 | 1.49 (0.97–2.28) | 0.064 | 187 | 2.16 (1.1–4.23) | 0.022 | 109 | 3.44 (1.44–8.22) | 0.0031 |
RFS Relapse free survival, OS Overall survival, DMFS Distant metastasis-free survival, HR Hazard ratio, − Ddata not available, LN Lymph node
Fig. 1.
The prognostic effect of the expression of SPAG5 in www.kmplot.com. a RFS b OS c DMFS
Fig. 2.
ROC analysis was performed by splitting the population into good and poor-outcome based on RFS, the analysis was run for RFS of the entire dataset, ER+ population and ER+ population treated with endocrine therapy at 5 years and 10 years, respectively
The expression of gene SPAG5 in patients receiving systematic therapy
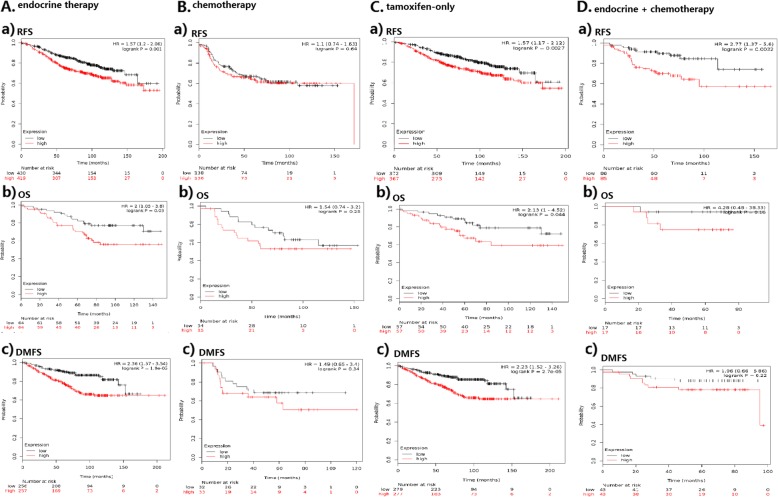
In patients with endocrine therapy, low SPAG5 transcript expression was significantly associated with longer RFS (HR = 1.57, 95% CI 1.2–2.06, p = 0.001) and OS (HR 2, 95% CI 1.05–3.8, p = 0.03) than was high SPAG5 expression. However, in patients who received chemotherapy, no significant difference existed between low and high SPAG5 transcript expressions in RFS (HR = 1.1, 95% CI 0.74–1.63, p = 0.64) and OS (HR = 1.54, 95% CI 0.74–3.2, p = 0.25) of breast cancer patients. Similar results were also seen for DMFS in patients with endocrine therapy (HR = 2.36, 95% CI 1. 57–3.54, p < 0.001) and chemotherapy (HR = 1.49, 95% CI 0.65–3.4, p = 0.34) (see Table 2 and Fig. 3). Tamoxifen was the most common drug used in hormonal therapy, and in patients receiving tamoxifen-only therapy, SPAG5 overexpression was associated with decreased RFS (HR = 1.57, 95% CI 1.17–2.12, p = 0.0027), OS (HR = 2.13, 95% CI 1.00–4.52, p = 0.044) and DMFS (HR = 2.23, 95% CI 1.52–3.26, p < 0.001). In 171 patients receiving both hormonal therapy and chemotherapy, SPAG5 overexpression was associated with decreased RFS (HR = 2.77, 95% CI 1.37–5.6, p = 0.0032) and data for OS and DMFS among those patients were not enough to draw a concrete conclusion (see Table 2 and Fig. 3).
Table 2.
PH Cox regression univariate analyses for the association of gene SPAG5 with endocrine therapy and chemotherapy
| RFS | OS | DMFS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| n | HR | P-value | n | HR | P-value | n | HR | P-value | |
| Systemic therapy subtypes | |||||||||
| Endo | 849 | 1.57 (1.2–2.06) | 0.001 | 128 | 2 (1.05–3.8) | 0.03 | 513 | 2.36 (1.57–3.54) | < 0.001 |
| Tamoxifen-only | 739 | 1.57 (1.17–2.12) | 0.0027 | 114 | 2.13 (1–4.52) | 0.044 | 556 | 2.23 (1.52–3.26) | < 0.001 |
| Chemo | 274 | 1.1 (0.74–1.63) | 0.64 | 69 | 1.54 (0.74–3.2) | 0.25 | 65 | 1.49 (0.65–3.4) | 0.34 |
| Endo + chemo | 171 | 2.77 (1.37–5.6) | 0.0032 | 34 | 4.28 (0.48–38.33) | 0.16 | 86 | 1.96 (0.66–5.86) | 0.22 |
Endo Endocrine therapy, chemo Chemotherapy
Fig. 3.
SPAG5 in patients with systematic therapy in univariate analysis. a. Association of SPAG5 with survival outcomes in patients with endocrine therapy. b. Association of SPAG5 with survival outcomes in patients with chemotherapy. c. SPAG5 expression was predictive of relapse in patients with tamoxifen-only therapy. d. SPAG5 expression and survival outcomes in patients with both endocrine therapy and chemotherapy
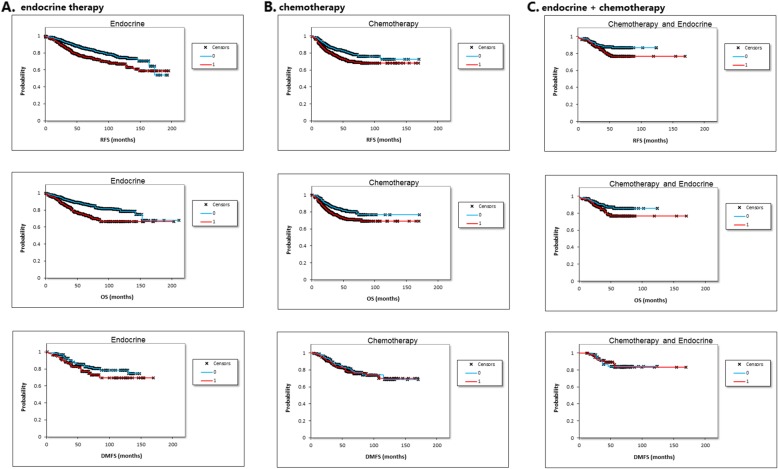
In multiple hypothesis testing, the association remained significant in patients with endocrine therapy with poor RFS (HR = 1.61, 95% CI 1.26–2.04, p < 0.001) and OS (HR = 1.95, 95% CI 1.47–2.60, p < 0.001). Data for DMFS and tamoxifen-only therapy was not enough for multivariate analysis (Table 3 and Fig. 4).
Table 3.
Multiple hypothesis testing of the association of gene SPAG5 with endocrine therapy and chemotherapy
| RFS | OS | DMFS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Systemic therapy subtypes | |||||||||
| Endocrine therapy | 1.61 | 1.26–2.04 | < 0.001 | 1.95 | 1.47–2.60 | < 0.001 | 1.45 | 0.85–2.48 | ns,0.17 |
| Chemotherapy | 1.54 | 1.21–1.98 | < 0.001 | 1.57 | 1.16–2.12 | 0.0033 | 1.10 | 0.70–1.71 | ns,0.68 |
| Endo + chemo | 1.69 | 1.02–2.82 | ns,0.0428 | 1.67 | 1.01–2.80 | ns,0.051 | 0.99 | 0.34–2.86 | ns,0.988 |
ns Not significant after correction for multiple hypothesis testing, endo Endocrine therapy, chemo Chemotherapy
Fig. 4.
SPAG5 in patients with systematic therapy in multiple hypothesis testing. a Association of SPAG5 with survival outcomes in patients with endocrine therapy. b Association of SPAG5 with survival outcomes in patients with chemotherapy. c SPAG5 expression and survival outcomes in patients with both endocrine therapy and chemotherapy
The prognostic value of SPAG5 expression in breast cancer with different molecular subtypes, histological grades and TP53 statuses
When patients were differentiated based on ER expression statuses, we plotted RFS, OS and DMFS curves for the ER+ and ER- subsets. We observed that high SPAG5 expression was associated with a significant increase in risk of relapse among ER+ (HR = 1.77, 95% CI 1.55–2.03, p < 0.001), but not ER- breast cancer patients (HR = 1.03, 95% CI 0.82–1.28, p = 0.81). Similarly, SPAG5 gain or amplification was associated with shorter OS (HR = 2.74, 95% CI 1.74–4.33, p < 0.001) and DMFS (HR = 2.89, 95% CI 1.95–4.29, p < 0.001) in the ER+ subgroup, but not ER- subgroup (p = 0.74 and p = 0.89, respectively) (see Table 1 and Additional file 2: Figure S2). Likewise, in multiple hypothesis testing, higher expression of SPAG5 was not associated with poorer survival in ER- subgroup, but the association was significant in ER+ patients with a reduction in RFS (HR = 1.85), OS (HR = 2.61) and DMFS (HR = 2.92) (see Table 4). In ER+/PR+/HER2- subgroup, SPAG5 expression was associated with shorter RFS (HR = 2.41, 95% CI 1.48–3.93, p < 0.001) (see Table 1). We further stratified ER+ patients according to PR, HER2, lymph node status, histological grade and TP53 statuses and the results were listed in Additional file 3: Table S5.
Table 4.
Multiple hypothesis testing of factors associated with survival
| Breast cancer subtypes | RFS | OS | DMFS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | |
| ER status | |||||||||
| ER+ | 1.85 | 1.57–2.18 | < 0.001 | 2.61 | 1.86–3.68 | < 0.001 | 2.92 | 2.09–4.07 | < 0.001 |
| ER- | 1.18 | 0.9–1.18 | 0.23 | 1.24 | 0.8–1.92 | 0.34 | 1.4 | 0.9–2.17 | 0.13 |
| PR status | |||||||||
| PR+ | 3.4 | 2.09–5.55 | < 0.001 | 6.34 | 1.26–31.88 | ns, 0.011 | 3.92 | 1.15–13.32 | ns, 0.018 |
| PR- | 1.51 | 1.11–2.05 | ns, 0.0081 | 2.79 | 0.99–7.82 | ns, 0.042 | 3.39 | 1.88–6.11 | < 0.001 |
| HER2 status | |||||||||
| HER2+ | 1.73 | 1.17–2.56 | ns, 0.0053 | 3.42 | 1.52–7.72 | 0.0017 | 1.78 | 0.94–3.39 | ns, 0.073 |
| HER2- | 2.12 | 1.79–2.51 | < 0.001 | 1.7 | 1.37–2.12 | < 0.001 | 1.93 | 1.58–2.36 | < 0.001 |
| Lymph node status | |||||||||
| Lymph node+ | 2.19 | 1.67–2.88 | < 0.001 | 1.72 | 1.18–2.5 | ns, 0.0044 | 2.03 | 1.4–2.94 | < 0.001 |
| Lymph node- | 1.82 | 1.52–2.17 | < 0.001 | 3 | 1.88–4.78 | < 0.001 | 2.39 | 1.8–3.19 | < 0.001 |
| Grade | |||||||||
| 1 | 3.12 | 1.8–5.42 | < 0.001 | 3.19 | 1.28–7.96 | ns, 0.0087 | 2.79 | 1.23–6.33 | ns, 0.011 |
| 2 | 1.99 | 1.56–2.55 | < 0.001 | 2.55 | 1.67–3.89 | < 0.001 | 2.61 | 1.85–3.68 | < 0.001 |
| 3 | 1.35 | 1.07–1.7 | ns, 0.011 | 1.25 | 0.89–1.77 | 0.19 | 1.75 | 1.13–2.73 | ns, 0.012 |
| TP53 status | |||||||||
| Mutated | 0.77 | 0.48–1.26 | 0.3 | 0.69 | 0.34–1.42 | 0.32 | 0.56 | 0.26–1.22 | 0.14 |
| Wild type | 1.82 | 1.16–2.85 | ns, 0.0077 | 2.56 | 1.38–4.76 | 0.0021 | 3.67 | 1.76–7.63 | < 0.001 |
ns Not significant after correction for multiple hypothesis testing; bold faced: remained significant
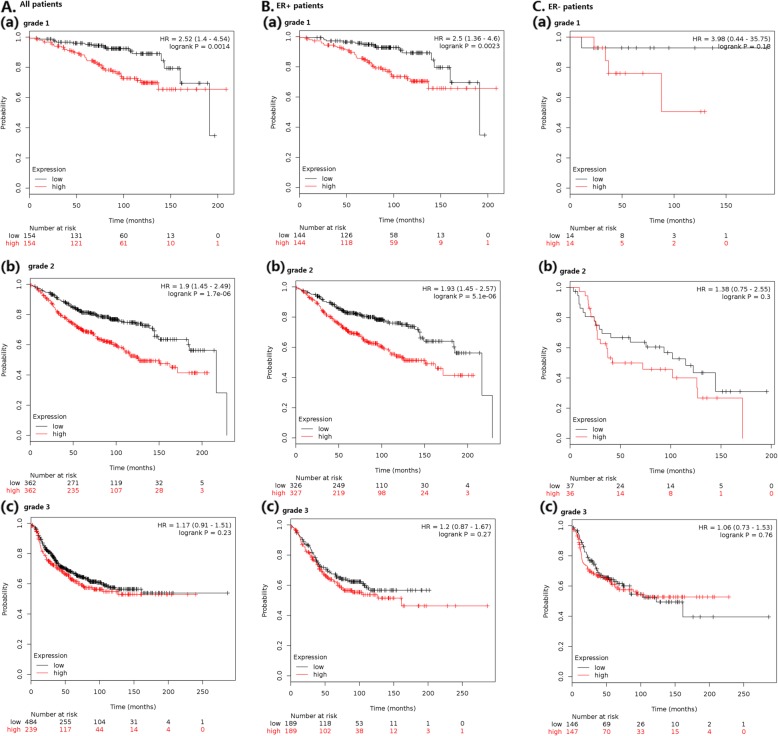
Among patients with grade 1 breast cancer, high SPAG5 expression was associated with a great increase in risk of recurrence (HR = 2.52, 95% CI 1.4–4.54, p = 0.0014). In more advanced cancers, high expression of SPAG5 indicated less of an association with RFS in grade 2 cancer (HR = 1.9, 95% CI 1.45–2.49, p < 0.001), and only a moderate tendency with no statistical difference toward shorter RFS was seen among patients with grade 3 cancer (HR = 1.17, 95% CI 0.91–1.51, p = 0.23) (see Table 1 and Fig. 5). In TP53 wild-type breast carcinomas, RFS (HR = 1.49, 95% CI 0.97–2.28, p = 0.064), OS (HR = 2.16, 95% CI 1.1–4.23, p = 0.022) and DMFS (HR = 3.44, 95% CI 1.44–8.22, p = 0.0031) were better in patients with low-expressed SPAG5, however, the survival curves did not show a significant difference in RFS of TP53-mutated breast cancer patients (p = 0.71). RFS was low in HER2- patients (HR = 1.78, 95% CI 1.36–2.34, p < 0.001), but this prognostic association was not obvious in HER2+ patients (HR = 0.78, 95% CI 0.46–1.32, p = 0.36). Results of SPAG5 expression in different PR statuses and lymph node statuses were also exhibited in Table 1. Multiple hypothesis testing supported the prognostic association of SPAG5 in these different subgroups of patients (see Table 4).
Fig. 5.
Expression of SPAG5 with RFS among A. all patients; B. ER+ breast cancer patients; C. ER- patients with grade 1–3 breast cancers
Discussion
In recent years, more and more attention has been attached on precision medicine, and there is a growing need for identification of prognostic biomarkers. SPAG5, originally identified as a microtubule-associated protein, with dual centrosome and kinetochores localization [21], has been reported to act as a promoter in tumorigenesis and progression [12]. In our study, data mining of 5667 publically available gene expression microarrays showed that elevated SPAG5 expression in breast cancer predicted a poor prognosis by the Kaplan-Meier method. We found that high SPAG5 expression was associated with lower RFS, OS, and DMFS, and SPAG5 might act as an important marker in systematic therapy, especially in ER+ breast cancer patients who received hormonal therapy.
SPAG5 was reported to be up-regulated in M-phase cells and play a vital role in cell mitosis and cell cycle checkpoint regulation [9]. In previous studies, SPAG5 was found increasing in many tumors and considered as a predominant oncogene in tumor promotion and metastasis [2]. Therefore, the identification of patients with higher SPAG5 expression before treatment would be important for personalized treatment. In our study, using a large combined cohort, we demonstrated that SPAG5 expression was significantly higher in patients with hormone negative (ER- and PR-) breast cancer. Meanwhile, we found that SPAG5 expression was increased in HER2 positive, poor differentiated, lymph node positive and TP53 mutated breast cancer subtypes all of which were strongly associated with tumor progression. Since the oncogenic potential of SPAG5 was also reported in prostate cancer [10], we hypothesized that SPAG5 could serve as a marker in predicting breast cancer proliferation and progression.
Systemic therapy for patients with early-stage breast cancer (ie, stages IA, IB, IIA, IIB, and IIIA) included chemotherapy, endocrine therapy, and targeted therapy [22]. It was important to choose certain biomarkers that could predict response to therapy and clinical outcomes. Recently a research team applied an artificial neural network performing data mining functions on SPAG5 and found that SPAG5 expression products were independent predictors for response to chemotherapy in breast cancer [13]. Similarly, we found that SPAG5 could predict prognosis of breast cancer patients with systemic treatment. However, our results suggested that in patients who received chemotherapy, SPAG5 had a moderate impact on survival outcomes including RFS (HR = 1.1), OS (HR = 1.54) and DMFS (HR = 1.49) in univariate analysis and the survival curves did not show a significant difference. As was referred in Hayes’s study [23], a HR of less than 2 meant that the clinical value was uncertain. A previous research suggested that SPAG5 could affect chemotherapy sensitivity of taxol in cell lines [2]. The causes of the inconsistency might be attributed to the different chemotherapy regimens and varying methodological qualities.
Endocrine therapy abrogating estrogen dependent cell proliferation has been shown to reduce recurrence and death [24] for most patients with ER+ breast cancer. Tamoxifen is a Selective Estrogen Receptor Modulator (SERM) widely used for adjuvant therapy [25] and could reduce 15-year risks of breast cancer recurrence and mortality rates after surgery [26] in ER+ breast cancer patients [27]. However, resistance to tamoxifen is frequent, and patients receiving adjuvant tamoxifen may eventually suffer recurrence or progression or even death from metastases [28]. We found that when patients received both hormonal therapy and chemotherapy, high SPAG5 expression could predict poor prognosis with HRs for RFS, OS and DMFS of 2.77, 4.28 and 1.96, respectively, although for OS and DMFS the difference was not statistically significant. Therefore, we assumed that high SPAG5 expression in breast cancer was potentially more relevant to malignant prognosis in hormonal therapy. Further, in hormonal therapy only, patients with high SPAG5 expression suffered decreased RFS, OS and DMFS in both univariate and multiple hypothesis testing. We considered that SPAG5 was correlated with mTOR signaling pathway activity during breast cancer treatment [2], and the cross-talk between the estrogen receptor and mTOR signaling pathway, the most well-known mechanism of endocrine resistance, led to poor prognosis of patients [29]. Therefore, SPAG5 contributed to the development of hormonal therapy resistance in ER+ breast cancer and the expression level was predictive on the survival outcomes of patients undergoing endocrine therapy. Further laboratory studies and clinical trials are needed to fully establish the association of SPAG5 in endocrine and tamoxifen-based therapy.
Choosing biomarkers based on different breast cancer subtypes to predict survival is vital for both doctors and patients. In clinical practice, ER, PR and HER2 statuses are biologic markers considered to be crucial factors for treatment [30]. In our study, the large cohort with 2766 samples proved apparent statistically significant difference between SPAG5-high and SPAG5-low expressions in ER+, but not ER- breast cancer, meaning that the expression level of SPAG5 could serve as a survival predictor in ER+ rather than ER- breast cancer patients. It might be because almost all ER+ patients received hormonal therapy and SPAG5 expression predicted survival of patients in hormonal therapy. In some breast cancer subtypes like PR+/ER+ breast cancer, positive SPAG5 expression presented a strong trend toward being associated with lower RFS (Additional file 3: Table S5). Moreover, SPAG5 was an important determinant of survival in HER2 negative rather than HER2 positive breast cancer patients.
Also in our study, RFS, OS and DMFS were better in TP53 wild-type breast carcinomas patients with low-expressed SPAG5, while the survival curves did not show a significant difference in the survival outcomes of TP53-mutated breast cancer patients. As mutations in TP53 might lead to overexpression of SPAG5, which was essential for promoting and regulating several aspects of mitosis, such as inactivating Separase which maintained the cohesion of sister chromatids, stabilizing mitotic spindle, enhancing the fidelity of chromosome segregation, and silencing spindle assembly checkpoint [31], G2/M phase transition and permanent cell cycling [32] could be triggered. Studies have reported that mutant TP53 was strongly associated with endocrine therapy resistance and agents dramatically increasing wild-type p53 levels could induce cell cycle arrest and apoptosis in cancer cells [33]. All these were in accordance with our hypothesis described previously that SPAG5 was related to the development of hormone resistance in breast cancer.
Histological grade is an important factor that affected the prognosis in breast cancer. In our study, we found that SPAG5 expression was not predictive in high grade (poorly differentiated) breast cancer patients, perhaps because poorly differentiated breast cancer cells proliferated fast and had a poor response to all kinds of therapies including hormonal therapy [34]. On the contrary, expression was strongly associated with survival outcomes in low histological grade/proliferative status. As is already known, SPAG5 is associated with cell cycle progression and formation of malignancies [10]. Considering the function of SPAG5 in progression of mitosis [35], these results might imply that early in the etiology of ER+ breast cancer subtypes, SPAG5 contributed to disease progression [36]. The gradual loss of this effect might be caused by the activation of parallel oncogenic pathways [37], and therefore weakened the influence of SPAG5 [38].
The potential of SPAG5 as a therapeutic target of breast cancer has been highlighted in some experiments. Down-regulation of SPAG5 exerted an antitumor effect. A study indicated that when silencing the expression of SPAG5 protein with RNA interference, multipolar and highly disorganized spindles were formed, inducing mitotic arrest [31] and apoptosis [39] through cell cycle deregulation and mitotic catastrophe. In cervical cancer cell lines, SPAG5 down-regulation resulted in inhibition of cell growth and proliferation by inducing G2/M phase cell cycle arrest [40]. What’s more, due to the loss of microtubule-binding ability of SPAG5, suppression of cell migration and invasion also occurred [41]. Thus, in addition to a potential prognostic biomarker, SPAG5 might act as a therapeutic target for breast cancer.
To our knowledge, this is the largest up-to-date research on the prognostic association of SPAG5 in different subtypes of breast cancer. We analyzed different subtypes of breast cancer comprehensively (including poor differentiated type, HER2 positive or TP53 mutated breast cancer), which was not reported previously. Our work presented that for chemotherapy, the survival of patients did not show a significant difference between low and high SPAG5 transcript expressions, but the prognostic association of SPAG5 in endocrine therapy and tamoxifen-only therapy was explored. We offered the potential to discriminate ER+ breast cancer patients at higher risks of relapse, as well as providing opportunities to customize therapies.
Our work has limitations. First, the molecular mechanism and association of SPAG5 in tumorigenesis and progression have not yet been fully identified. Second, the data of survival outcomes of new drugs for ER+ breast cancers including palbociclib [42] were lacking. Third, the optimal cutoff points of SPAG5 for survival prediction in breast cancer patients still merit further investigation. Therefore, further researches on the role of SPAG5 in breast cancer are mandatory in the future.
Conclusions
In conclusion, as a progression-driving oncogene, SPAG5 was closely related to disease progression and malignant prognosis of ER+ breast cancer patients undergoing endocrine therapy, and might act as a therapeutic target for breast cancer.
Supplementary information
Additional file 1: Figure S1. SPAG5 gene expression in all breast cancer patients with different subtypes.
Additional file 2: Figure S2. Survival curves for the ER+ and ER- breast cancer subset. A. ER+ breast cancer patients; B. ER- breast cancer patients.
Additional file 3: Table S1. Datasets used for the analysis. Table S2. SPAG5 expression in all breast cancer patients with different subtypes. Table S3. The comparison of gene expression level using Mann-Whitney U test or Kruskal-Wallis test. Table S4. The comparison and correlation of SPAG5 with other markers of progression in assessing independent value. Table S5. Subgroup analyses of SPAG5 gene in association with RFS in ER+/- breast cancer subtype.
Acknowledgements
We thank Dr. Liangqun Rong for his help with the collection of the data.
Abbreviations
- DMFS
Distant metastasis-free survival
- ER-
ER negative
- ER
Estrogen receptor
- ER+
ER positive
- HER2
Epidermal growth factor receptor 2
- HR
Hazard ratio
- OS
Overall survival
- PR
Progesterone receptor
- RFS
Relapse free survival
- SPAG5
Sperm-associated antigen 5
Authors’ contributions
CZ and XH provided the idea. CZ wrote the article. OM collected data, performed the statistical analysis and helped with the generation of figures in the manuscript. BG interpreted the data and substantively modified the article. XH interpreted data and helped with the final revision of the article. All authors reviewed the manuscript, agreed to be personally accountable for their own contributions and approved the final manuscript.
Funding
None.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Kaplan–Meier Plotter (http://www.kmplot.com). All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chenjing Zhu, Phone: +86 258 328 3560, Email: drchenjingzhu@gmail.com.
Otilia Menyhart, Email: vicabuba@gmail.com.
Balázs Győrffy, Email: zsalab2@yahoo.com.
Xia He, Phone: +86 258 328 3560, Email: hexiabm@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12885-019-6260-6.
References
- 1.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Yuan LJ, Li JD, Zhang L, Wang JH, Wan T, Zhou Y, Tu H, Yun JP, Luo RZ, Jia WH, Zheng M. SPAG5 upregulation predicts poor prognosis in cervical cancer patients and alters sensitivity to taxol treatment via the mTOR signaling pathway. Cell Death Dis. 2014;5:e1247. doi: 10.1038/cddis.2014.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diamandis M, White NM, Yousef GM. Personalized medicine: marking a new epoch in cancer patient management. Mol Cancer Res. 2010;8:1175–1187. doi: 10.1158/1541-7786.MCR-10-0264. [DOI] [PubMed] [Google Scholar]
- 4.Peer D. Precision medicine--delivering the goods. Cancer Lett. 2014;352:2–3. doi: 10.1016/j.canlet.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Abramovitz M, Krie A, Dey N, De P, Williams C, Leyland-Jones B. Identifying biomarkers to select patients with early breast cancer suitable for extended adjuvant endocrine therapy. Curr Opin Oncol. 2016;28(6):461–468. doi: 10.1097/CCO.0000000000000324. [DOI] [PubMed] [Google Scholar]
- 6.Burgess JT, Bolderson E, Saunus JM, Zhang SD, Reid LE, McNicol AM, Lakhani SR, Cuff K, Richard K, Richard DJ, O'Byrne KJ. SASH1 mediates sensitivity of breast cancer cells to chloropyramine and is associated with prognosis in breast cancer. Oncotarget. 2016;7(45):72807–72818. doi: 10.18632/oncotarget.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leto G, Incorvaia L, Flandina C, Ancona C, Fulfaro F, Crescimanno M, Sepporta MV, Badalamenti G. Clinical Impact of Cystatin C/Cathepsin L and Follistatin/Activin A Systems in Breast CancerProgression: a Preliminary Report. Cancer Investig. 2016:1–9. PMID: 27636861 [DOI] [PubMed]
- 8.Lei B, Zhang XY, Zhou JP, Mu GN, Li YW, Zhang YX, Pang D. Transcriptome sequencing of HER2-positive breast cancer stem cells identifies potential prognostic marker. Tumour Biol. 2016;37(11):14757–14764. doi: 10.1007/s13277-016-5351-0. [DOI] [PubMed] [Google Scholar]
- 9.Friese A, Faesen AC, Huis i’VPJ, Fischböck J, Prumbaum D, Petrovic A, Raunser S, Herzog F, Musacchio A. Molecular requirements for the inter-subunit interaction and kinetochore recruitment of SKAP and Astrin. Nat Commun. 2016;7:11407. doi: 10.1038/ncomms11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Li S, Yang X, Qiao B, Zhang Z, Xu Y. miR-539 inhibits prostate cancer progression by directly targeting SPAG5. J Exp Clin Cancer Res. 2016;35:60. doi: 10.1186/s13046-016-0337-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Välk K, Vooder T, Kolde R, Reintam MA, Petzold C, Vilo J, Metspalu A. Gene expression profiles of non-small cell lung cancer: survival prediction and new biomarkers. Oncology. 2010;79:283–292. doi: 10.1159/000322116. [DOI] [PubMed] [Google Scholar]
- 12.Thedieck K, Holzwarth B, Prentzell MT, Boehlke C, Kläsener K, Ruf S, Sonntag AG, Maerz L, Grellscheid SN, Kremmer E, Nitschke R, Kuehn EW, Jonker JW, Groen AK, Reth M, Hall MN, Baumeister R. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell. 2013;154:859–874. doi: 10.1016/j.cell.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 13.Abdel-Fatah TM, Agarwal D, Liu DX, Russell R, Rueda OM, Liu K, Xu B, Moseley PM, Green AR, Pockley AG, Rees RC, Caldas C, Ellis IO, Ball GR, Chan SY. SPAG5 as a prognostic biomarker and chemotherapy sensitivity predictor in breast cancer: a retrospective, integrated genomic, transcriptomic, and protein analysis. Lancet Oncol. 2016;17:1004–1018. doi: 10.1016/S1470-2045(16)00174-1. [DOI] [PubMed] [Google Scholar]
- 14.Buechler S. Low expression of a few genes indicates good prognosis in estrogen receptor positive breast cancer. BMC Cancer. 2009;9:243. doi: 10.1186/1471-2407-9-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The landscape of candidate driver genes differs between male and female breast cancer. PloS One. 2013:e78299. PMID:24194916. [DOI] [PMC free article] [PubMed]
- 16.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 17.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, Senn HJ. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almudevar A. Multiple hypothesis testing: a methodological overview. Methods Mol Biol. 2013;972:37–55. doi: 10.1007/978-1-60327-337-4_3. [DOI] [PubMed] [Google Scholar]
- 20.Bertolino F, Cabras S, Castellanos ME, Racugno W. Unscaled Bayes factors for multiple hypothesis testing in microarray experiments. Stat Methods Med Res. 2015;24:1030–1043. doi: 10.1177/0962280212437827. [DOI] [PubMed] [Google Scholar]
- 21.Abdel-Fatah TM, Perry C, Dickinson P, Ball G, Moseley P, Madhusudan S, Ellis IO, Chan SY. Bcl2 is an independent prognostic marker of triple negative breast cancer (TNBC) and predicts response to anthracycline combination (ATC) chemotherapy (CT) in adjuvant and neoadjuvant settings. Ann Oncol. 2013;24:2801–2807. doi: 10.1093/annonc/mdt277. [DOI] [PubMed] [Google Scholar]
- 22.Teven CM, Schmid DB, Sisco M, Ward J, Howard MA. Systemic therapy for early-stage breast cancer: what the plastic surgeon should know. Eplasty. 2017;17:e7. [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia. 2001;6:375–392. doi: 10.1023/A:1014778713034. [DOI] [PubMed] [Google Scholar]
- 24.Niwa T, Shinagawa Y, Asari Y, Suzuki K, Takanobu J, Gohno T, Yamaguchi Y, Hayashi SI. Estrogen receptor activation by tobacco smoke condensate in hormonal therapy-resistant breast cancer cells. J Steroid Biochem Mol Biol. 2016. PMID:27632897. [DOI] [PubMed]
- 25.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 26.Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, Wang YC, Dowsett M, Ingle J, Peto R. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei L, Wang X, Wu XD, Wang Z, Chen ZH, Zheng YB, Wang XJ. Association of CYP2D6*10 (c.100C>T) polymorphisms with clinical outcome of breast cancer after tamoxifen adjuvant endocrine therapy in Chinese population. Am J Transl Res. 2016;8:3585–3592. [PMC free article] [PubMed] [Google Scholar]
- 28.Salazar MD, Ratnam M, Patki M, Kisovic I, Trumbly R, Iman M, Ratnam M. During hormone depletion or tamoxifen treatment of breast cancer cells the estrogen receptor apoprotein supports cell cycling through the retinoic acid receptor α1 apoprotein. Breast Cancer Res. 2011;13:R18. doi: 10.1186/bcr2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Won HS, Lee KM, Oh JE, Nam EM, Lee KE. Inhibition of β-catenin to overcome endocrine resistance in tamoxifen-resistant breast cancer cell line. PLoS One. 2016;11:e0155983. doi: 10.1371/journal.pone.0155983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson E. The role of oestrogen and progesterone receptors in human mammary development and tumorigenesis. Breast Cancer Res. 2002;4:197–201. doi: 10.1186/bcr452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thein KH, Kleylein-Sohn J, Nigg EA, Gruneberg U. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J Cell Biol. 2007;178:345–354. doi: 10.1083/jcb.200701163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertucci F, Viens P, Birnbaum D. SPAG5: the ultimate marker of proliferation in early breast cancer. Lancet Oncol. 2016;17:863–865. doi: 10.1016/S1470-2045(16)30092-4. [DOI] [PubMed] [Google Scholar]
- 33.Ellis MJ, Perou CM. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov. 2013;3:27–34. doi: 10.1158/2159-8290.CD-12-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stone A, Cowley MJ, Valdes-Mora F, McCloy RA, Sergio CM, Gallego-Ortega D, Caldon CE, Ormandy CJ, Biankin AV, Gee JM, Nicholson RI, Print CG, Clark SJ, Musgrove EA. BCL-2 hypermethylation is a potential biomarker of sensitivity to antimitotic chemotherapy in endocrine-resistant breast cancer. Mol Cancer Ther. 2013;12:1874–1885. doi: 10.1158/1535-7163.MCT-13-0012. [DOI] [PubMed] [Google Scholar]
- 35.Chiu SC, Chen JM, Wei TY, Cheng TS, Wang YH, Ku CF, Lian CH, Liu CC, Kuo YC, Yu CT. The mitosis-regulating and protein-protein interaction activities of astrin are controlled by aurora-A-induced phosphorylation. Am J Physiol Cell Physiol. 2014;307:C466–C478. doi: 10.1152/ajpcell.00164.2014. [DOI] [PubMed] [Google Scholar]
- 36.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, Liu XS. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams MD, Nguyen T, Carriere PP, Tilghman SL, Williams C. Protein kinase CK2 expression predicts relapse survival in ERα dependent breast cancer, and modulates ERα expression in vitro. Int J Environ Res Public Health. 2016;13:ijerph13010036. doi: 10.3390/ijerph13050515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Li T, Pignon JC, Wang B, Wang J, Shukla SA, Dou R, Chen Q, Hodi FS, Choueiri TK, Wu C, Hacohen N, Signoretti S, Liu JS, Liu XS. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat Genet. 2016;48:725–732. doi: 10.1038/ng.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katayama K, Yasuda H, Tochigi Y, Suzuki H. The microtubule-associated protein astrin transgenesis rescues spermatogenesis and renal function in hypogonadic (hgn/hgn) rats. Andrology. 2013;1:301–307. doi: 10.1111/j.2047-2927.2012.00032.x. [DOI] [PubMed] [Google Scholar]
- 40.Kersten FF, van Wijk E, Hetterschijt L, Bauβ, Peters TA, Aslanyan MG, van der Zwaag B, Wolfrum U, Keunen JE, Roepman R, Kremer H. The mitotic spindle protein SPAG5/Astrin connects to the Usher protein network postmitotically. Cilia. 2012;1:2. doi: 10.1186/2046-2530-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fitzgerald CJ, Oko RJ, van der Hoorn FA. Rat Spag5 associates in somatic cells with endoplasmic reticulum and microtubules but in spermatozoa with outer dense fibers. Mol Reprod Dev. 2006;73:92–100. doi: 10.1002/mrd.20388. [DOI] [PubMed] [Google Scholar]
- 42.Owsley J, Jimeno A, Diamond JR. Palbociclib:CDK4/6 inhibition in the treatment of ER-positive breast cancer. Drugs Today (Barc) 2016;52:119–129. doi: 10.1358/dot.2016.52.2.2440528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. SPAG5 gene expression in all breast cancer patients with different subtypes.
Additional file 2: Figure S2. Survival curves for the ER+ and ER- breast cancer subset. A. ER+ breast cancer patients; B. ER- breast cancer patients.
Additional file 3: Table S1. Datasets used for the analysis. Table S2. SPAG5 expression in all breast cancer patients with different subtypes. Table S3. The comparison of gene expression level using Mann-Whitney U test or Kruskal-Wallis test. Table S4. The comparison and correlation of SPAG5 with other markers of progression in assessing independent value. Table S5. Subgroup analyses of SPAG5 gene in association with RFS in ER+/- breast cancer subtype.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Kaplan–Meier Plotter (http://www.kmplot.com). All data generated or analyzed during this study are included in this published article and its supplementary information files.