Abstract
Background
Acinetobacter baumannii is one of the most important opportunistic pathogens responsible for hospital acquired infections. It displays multi-drug resistance profile and has the ability to colonize surfaces and persist under harsh conditions. A. baumannii two-component signal transduction system BfmRS, consisting of response regulator BfmR and sensor kinase BfmS, has been implicated in the control of various virulence-related traits and has been suggested to act as a global modulator of A. baumannii physiology.
Results
Here, we assessed the role of BfmR regulator in pellicle formation and bacterial competition, features important for the establishment of A. baumannii in clinical environment. We show that BfmR is required for the pellicle formation of A. baumannii, as ΔbfmRS mutant lacked this phenotype. The loss of bfmRS also greatly reduced the secretion of A. baumannii Hcp protein, which is a component of T6SS secretion system. However, T6SS-mediated killing phenotype was not impaired in ΔbfmRS mutant. On the contrary, the same mutation resulted in the transcriptional activation of contact-dependent inhibition (CDI) system, which A. baumannii used to inhibit the growth of another clinical A. baumannii strain and a closely related species Acinetobacter baylyi.
Conclusions
The obtained results indicate that BfmR is not only required for the pellicle phenotype induction in A. baumannii, but also, due to the down-regulation of a CDI system, could allow the incorporation of other A. baumannii strains or related species, possibly increasing the likelihood of the pathogens’ survival.
Keywords: Acinetobacter, BfmR, Pellicle, T6SS, CDI
Background
Acinetobacter baumannii is clinically important Gram-negative opportunistic pathogen responsible for the broad range of severe nosocomial infections in critically ill patients [1]. Due to its multi-drug resistance, the ability to form biofilms and various mechanisms allowing persistence under harsh environmental settings such as the presence of disinfectants, prolonged periods of desiccation, or oxidative stress, A. baumannii has become a threat to human health [2].
A. baumannii BfmR regulator along with histidine kinase BfmS, comprises two-component signal transduction system (TCS) BfmRS [3]. TCSs are widely distributed among prokaryotes allowing them to effectively adapt to ever-changing environment conditions. The membrane anchored sensor kinase, via phosphate transfer, controls its cognate response regulator. The latter, either regulates gene transcription directly or binds to target proteins eliciting a specific response of its host [4].
It has been shown that one of the clinically important A. baumannii traits, biofilm formation, is controlled by the BfmR [3]. Further studies provided some insights into the role of BfmRS in A. baumannii pathogenesis by showing that the loss of BfmS results in a significant reduction of motility [5]. The sensor BfmS has been shown to be required for the biofilm modulation, adhesion to epithelial cells, and increased sensitivity to serum killing [6]. The bfmS mutant showed increased secretion of membrane proteins, including OmpA, which is considered as one of the virulence factors of A. baumannii [6]. Further studies showed that BfmR is required for A. baumannii persistence in a murine lung infection model [7], for growth in human ascites fluid and for serum resistance [8, 9], while BfmS rather than BfmR was required for the successful growth in Galleria mellonella larvae [10].
BfmR has been also shown to control tolerance to desiccation and responses to oxidative stress [11]. In addition, via the up-regulation of β-lactamase production and cell envelope synthesis, resistance to β-lactams is also modulated by the BfmR [9]. Moreover, the whole bfmRS operon has been shown to be required for the regulation of the K locus, which is responsible for the capsular exopolysaccharide expression [12]. All observations discussed above indicate that BfmR is involved in the control of the genes responsible for a variety of phenotypes in A. baumannii, although its precise role is far from being fully elucidated.
In this study, we describe novel phenotypes that are reciprocally regulated by the BfmR. We demonstrate that the regulator is required for the pellicle formation, where bacteria form tightly packed biomass on the surface of culture media. At the same time, BfmR represses the contact-dependent inhibition (CDI) system, leading to the inability to suppress the growth of competing related strains. These observations suggest that A. baumannii via BfmR may modulate cooperative behavior against closely related strains during the pellicle formation, which may be advantageous for the establishment of this opportunistic pathogen in clinical environment.
Results
BfmR regulates A. baumannii pellicle formation
The pellicle is characterized as a form of biofilm that is floating on the surface of culture media and allows bacteria to acquire favorable ecological niche and directly access high concentrations of oxygen and nutrients from the air and liquid, respectively [13]. A. baumannii BfmR is responsible for the up-regulation of the csuA/BABCDE operon leading to the biofilm formation on solid surfaces [3]. It has also been determined that the CsuA/B pilin is the most abundant component of A. baumannii pellicle [14]. Therefore, we were interested whether the BfmR is involved in the manifestation of this phenotype.
From the collection of clinical A. baumannii isolates, characterized previously [15], the isolate V15, which showed a pellicle forming phenotype, has been selected. The deletion of ΔbfmRS operon was generated as described in the Methods. The decision to obtain a mutant with the deletion of the whole bfmRS operon was based on the previously published results indicating that the sensor kinase BfmS acts as a negative regulator of BfmR, and that only bfmR can fully complement whole ΔbfmRS mutant to WT levels [9]. The deletion was confirmed by sequencing and by performing qPCR analysis of bfmR and bfmS genes using total RNA. For complementation experiments, the plasmids carrying the genes encoding BfmR (pbfmR), BfmS (pbfmS), or both proteins (pbfmRS) were constructed and introduced into A. baumannii V15 as described in the Methods.
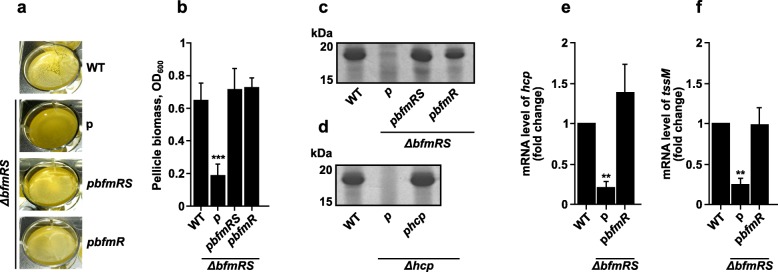
We then performed the pellicle formation assay by growing the strains in TSB media under the stationary conditions at 30 °C for 30 h as described in the Methods. These growth conditions were previously suggested to generally promote pellicle formation [13, 16]. Compared to the WT strain, which formed a thick and uniform pellicle morphology (Fig. 1a), the ability of ΔbfmRS mutant to develop a pellicle was impaired as only some biomass (white structures) was located near the walls of the wells (Fig. 1a and b). The inability of the mutant to form a pellicle was fully complemented with the bfmR allele or the whole bfmRS operon, when supplemented in trans (Fig. 1a and b). It must be noted that due to the toxicity of bfmRS and bfmR constructs with the native upstream sequences, we used plasmids with a leaky Ptac promoter (transcripts were observed without additional supplementation of IPTG), which allowed the basal expression of bfmR at the level comparable to that of WT (approximately 2.86 ± 0.89 fold up-regulation compared to WT) and did not interfere with the growth of the strains. Finally, we observed no effect of the wild-type bfmS allele on the restoration of the pellicle phenotype in ΔbfmRS mutant, even using induction with IPTG up to 0.1 mM (Additional file 3: Figure S1a). These results show that BfmR is responsible for the pellicle phenotype manifestation, also they are consistent with the previously published data indicating that ΔbfmRS mutants may be complemented solely by the bfmR construct [9].
Fig. 1.
BfmR is required for A. baumannii pellicle formation and secretion of Hcp into culture media. a and b Pellicles formed by A. baumannii V15 (WT), ΔbfmRS mutant, and ΔbfmRS strain complemented with the plasmids pbfmRS or pbfmR: Macroscopic images of formed pellicles (a); Quantitative evaluation of pellicles with values normalized by the total growth volume (b). c and d 12.5% SDS-PAGE gel view of TCA-precipitated total protein fraction from culture media of WT, ΔbfmRS, and ΔbfmRS complemented with the plasmids pbfmRS or pbfmR (c); and WT, Δhcp mutant, and Δhcp complemented with a plasmid phcp (d). Proteins were precipitated with TCA, separated by 12.5% SDS-PAGE, and visualized by staining with Coomassie blue. Numbers on the left denote molecular mass in kDa. e and f Expression levels of A. baumannii V15 transcripts hcp (e) and tssM (f) in WT, ΔbfmRS, and ΔbfmRS mutant complemented with pbfmR. Expression data in the graphs are displayed as a fold change compared to the WT which is set at 1. hcp gene in plasmid phcp was induced using IPTG concentration of 0.1 mM. Error bars represent standard deviation. **, p < 0.01; ***, p < 0.001, relative to WT.
Hcp secretion into the culture media is regulated by the BfmR
Pellicle formation requires various secreted proteins, polysacharides, and/or DNA to stabilize the structure [13]. Currently, only a few A. baumannii proteins have been linked to a pellicle formation [14]. However, there was an observation that during this process, the expression of multiple virulence factors changes [17]. To investigate, whether BfmR is involved in the regulation of pellicle phenotype-associated components, we first compared the electrophoretic profiles of precipitated total protein fractions from culture media of the WT and ΔbfmRS strains.
As can be seen in Fig. 1c, SDS-PAGE analysis of proteins precipitated from the culture media, showed a single band, migrating as a ~ 18 kDa entity, which was significantly reduced in the ΔbfmRS mutant. The secretion of the protein was restored after complementation with either bfmRS or bfmR alleles (Fig. 1c), correlating with the restoration of the pellicle phenotype. The band was identified by mass spectrometry as A. baumannii Hcp protein. The protein is a structural component of the bacterial Type VI secretion system (T6SS), which together with the additional proteins assembles into a needle-like apparatus that is used to puncture adjacent cells and to deliver effectors (toxins) into target cell [18, 19].
To confirm that the secreted protein was indeed Hcp, we generated Δhcp deletion in A. baumannii V15 strain, which resulted in the loss of Hcp secretion into culture media (Fig. 1d). The secretion was readily complemented with a copy of hcp gene cloned under the inducible promoter in the plasmid phcp, when induced with 0.1 mM IPTG (Fig. 1d). The complementation under inducing conditions only, could be explained by the fact that before being secreted, the Hcp must assemble into tubular structure made from multiple copies of Hcp monomers [19].
The examination of the hcp-specific mRNA levels in the ΔbfmRS mutant showed an approximately five-fold reduction, when compared to the WT strain (Fig. 1e). The introduction of the plasmid pbfmR resulted in the restoration of transcription level. In parallel to the examination of hcp gene transcription, we assessed the transcription of tssM gene. The latter codes for the subunit of the membrane-anchoring complex, which is also essential for the assembly of T6SS apparatus [20]. As can be seen from the results presented in Fig. 1f, tssM mRNA levels in the ΔbfmRS were decreased four-fold, when compared to the WT. This indicates that the loss of bfmR might lead to the down-regulation of the whole T6SS system, resulting in the reduced secretion of Hcp into culture media.
The abundance of Hcp in culture media and the correlation between the Hcp secretion and the formation of pellicle exhibited by the WT strain, prompted us to test whether Hcp is required for the pellicle formation. The Hcp, secreted into media could be embedded into pellicle matrix, potentially reinforcing the structure. However, our results showed the deletion of hcp did not interfere with the manifestation of the phenotype (Additional file 3: Figure S1b). This shows that Hcp is not required for the pellicle formation of A. baumannii.
Loss of bfmRS does not affect T6SS-mediated inter-genus killing
The findings above suggest that the down-regulation of T6SS might impact the killing phenotype of A. baumannii as it is known that Hcp secretion is the indication of a functional T6SS [18]. Previously, it has been demonstrated that A. baumannii is able to eliminate competing bacteria in a T6SS-dependent manner [21–24]. Therefore, we performed competition assays using E. coli MC4100 strain as a prey. Remarkably, while A. baumannii V15 was able to significantly reduce E. coli MC4100 numbers by 50–250-fold, the ΔbfmRS strain did not display any impairment in the killing phenotype (Fig. 2a). The phenotype remained mainly unchanged in the ΔbfmRS strain complemented with either bfmRS, or bfmR alleles (Fig. 2a). To confirm that the observed killing phenotype against E. coli was due to the function of T6SS, we investigated killing capacity of V15 Δhcp and the double mutant ΔbfmRSΔhcp. The results showed that both mutants exhibited reduction in killing of E. coli MC4100 (Fig. 2b and Additional file 4: Figure S2a). The phenotype of Δhcp and ΔbfmRSΔhcp mutants was readily complemented with the wild-type hcp allele (phcp) under inducing (0.1 mM IPTG) conditions (Fig. 2b). We also evaluated whether there is a difference in killing efficiency against clinical strains of Pseudomonas aeruginosa (P16) and Klebsiella pneumoniae (K39). As can be seen in Additional file 5: Figure S3a-d, both strains can be killed via T6SS of A. baumannii V15. However, the results also show that the loss of bfmRS, apparently, does not influence the reduction of the killing efficiency of A. baumannii. These data indicate that despite clearly affecting the expression of the T6SS apparatus and significantly impairing the secretion of Hcp into the supernatant, the BfmRS system does not affect A. baumannii T6SS-mediated killing of E. coli, P. aeruginosa or K. pneumoniae.
Fig. 2.
BfmR modulates A. baumannii V15 competitiveness against E. coli MC4100 and A. baylyi ADP1. Quantitative evaluation of inter-bacterial competition assay displaying a recovered number of prey: E. coli MC4100 (a and b) and A. baylyi ADP1 (c and d). Competition was performed with the following strains used as the aggressors: A. baumannii V15 (WT), ΔbfmRS, and ΔbfmRS complemented with the plasmids pbfmRS or pbfmR (a and d); WT, Δhcp, Δhcp strain complemented with the plasmid phcp, ΔbfmRSΔhcp mutant, and ΔbfmRSΔhcp mutant complemented with the plasmid phcp (b and c). E. coli DH5α was used as a negative non-competitive control to enumerate bacteria numbers if there were no competition. Error bars represent standard deviation. The horizontal lines represent mean value. Values were calculated from at least three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant. Hcp gene in plasmid phcp was induced using IPTG concentration of 0.1 mM
BfmR regulates T6SS-independent killing mechanism against closely related species
Next, we evaluated A. baumannii aggressiveness against more closely related species. For this purpose, we used Acinetobacter baylyi ADP1 strain as a prey. As can be seen in Fig. 2c, ADP1 strain was significantly (approximately 70–200-fold) out-competed by A. baumannii V15 and the killing was T6SS-dependent as the Δhcp mutant displayed significantly reduced killing efficiency (Fig. 2c). The killing phenotype of the mutant was readily restored to the WT level with the hcp allele under inducing conditions (Fig. 2c). Remarkably, in contrast to the results obtained with E. coli MC4100 strain, the double mutant ΔbfmRSΔhcp, lacking an active T6SS, still significantly reduced ADP1 numbers at the efficiency comparable to that of WT strain (Fig. 2c). Interestingly, we observed that the ΔbfmRS mutant was able to significantly reduce ADP1 numbers as well, and displayed even more aggressive killing phenotype than the WT (approximately 10-fold) (Fig. 2d). The observed killing phenotype of ΔbfmRS and ΔbfmRSΔhcp mutants could not be complemented with either pbfmR or pbfmRS (Fig. 2d and Additional file 4: Figure S2b). These results suggest, that bfmRS deletion leads to the activation of T6SS-independent killing mechanism that is effective against A. baylyi ADP1 but not E. coli MC4100.
BfmR negatively regulates contact-dependent inhibition system of A. baumannii
We hypothesized that the observed killing of A. baylyi ADP1 but not E. coli MC4100 could be explained by the activation of currently poorly defined A. baumannii contact-dependent inhibition mechanism, which requires its receptor on the target cell and is classified as a type of Type V Secretion System (T5SS) and was shown to be functional in A. baumannii [25, 26].
CDI systems are composed of three components belonging to the cdiBAI gene cluster. The first two genes (cdiB and cdiA) encode a two-partner secretion system, which allows a large CdiA hemagglutinin-repeat protein to be displayed on the surface of bacterial cell. The last gene (cdiI) encodes an immunity protein, which binds and neutralizes the cognate toxin [25]. Previous work indicated that Acinetobacter sp. might contain up to two functional CDI systems [26, 27].
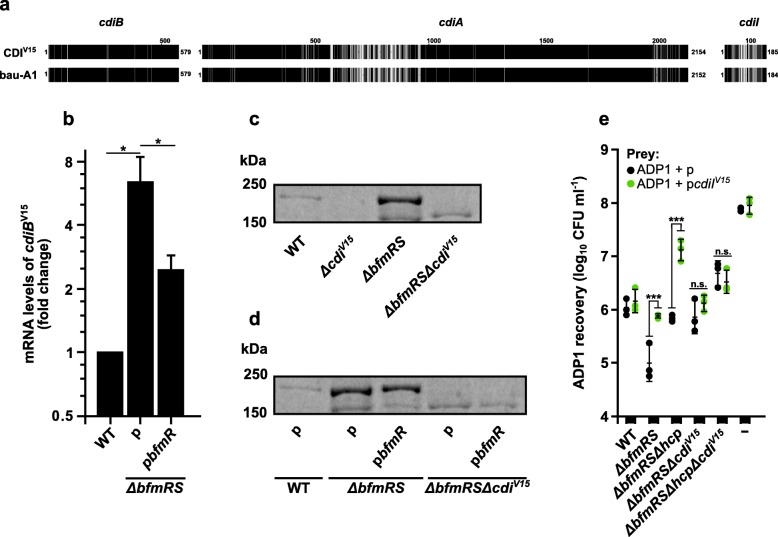
Therefore, based on the previous classification of A. baumannii CDI systems [28], we created a set of 3 primer pairs targeting a rather conserved cdiB genes and managed to identify and sequence the CDI locus of A. baumannii V15 strain (CDIV15). The results indicated that CDIV15 is a type-I CDI system with a CdiA protein, most identical to bau-A1/pit-A3 type CdiA proteins (90%) (Fig. 3a). Other CDIV15 proteins, namely CdiB and CdiI, were 98% and 66% identical to their counterparts in bau-A1 system, respectively (Fig. 3a). It is worth to note that some A. baumannii strains, namely AR_0037 (GenBank accession: MPBX01000005.1/bau-D9 CdiA type [28]), 1295549 (JFXB01000002.1/bau-B2), 426863 (JFYF01000002.1/bau-B2), ATCC19606 (JMRY01000015.1/bau-B2) contained nearly identical immunity proteins in the genome regions, which did not have a CDI system nearby, indicating that these strains are potentially immune to CDIV15.
Fig. 3.
A. baumannii V15 contains a functional CDI system. a Multiple amino acid sequence alignment of each of the CDIV15 loci encoded proteins with the most similar type-I CDI system (bau-A1/Genbank accession number: AMFH01000034.1). The most similar CDI was determined by performing multiple sequence comparisons with the recently characterized Acinetobacter CDI systems [28]. Alignments are displayed as sequence fingerprints using alignment shading software Texshade (version 1.25) [29]. Identical amino acids are shaded black. Unique residues are shaded gray. Annotations above alignments indicate the aligned CDI proteins. b Expression of A. baumannii cdiB in WT, ΔbfmRS, and ΔbfmRS mutant complemented with pbfmR. The data are displayed as a fold change compared to the WT which is set at 1. c and d Part of 10% SDS-PAGE gel view showing the presence of the CdiA protein found in the total protein fraction from culture media of WT, ΔcdiV15, ΔbfmRS, ΔbfmRSΔcdiV15, and ΔbfmRS and ΔbfmRSΔcdiV15 complemented with pbfmR. Proteins were precipitated with TCA, separated by 10% SDS-PAGE, and visualized by staining with Coomassie blue. Numbers on the left denote molecular mass in kDa. e Quantitative evaluation of inter-bacterial competition assay displaying a recovered number of A. baylyi ADP1 with or without plasmid pcdiIV15 containing A. baumannii V15 immunity gene cdiI. Competition was performed with A. baumannii V15 mutants ΔbfmRS, ΔbfmRSΔhcp, ΔbfmRSΔcdiV15, ΔbfmRSΔhcpΔcdiV15 as the aggressors. E. coli DH5α was used as a negative non-competitive control to enumerate bacteria numbers if there were no competition. Error bars represent standard deviation. The horizontal lines represent mean value. Values were calculated from at least three independent experiments. *, p < 0.05; ***, p < 0.001; n.s., not significant. cdiIV15 gene in plasmid pcdiIV15 was induced using IPTG concentration of 5 mM
Having identified that A. baumannii V15 contains an intact cdi locus, we further investigated, whether the observed T6SS-independent killing mechanism is indeed caused by the CDIV15 system. By comparing the mRNA levels of the cdiB gene between A. baumannii V15 ΔbfmRS mutant and WT we observed that the mutant displayed increased transcript levels of cdiB by ~ 6.4-fold. Additionally, complementation of ΔbfmRS mutant with the bfmR allele displayed a ~ 2.5-fold increase in the transcript levels of cdiB, when compared to the WT (Fig. 3b). This indicates that the complemented strain displayed an intermediate transcription level of the CDIV15. This result was consistent with the observed killing phenotype displayed against A. baylyi ADP1 strain, where the aggressiveness level of ΔbfmRS mutant could not be complemented with the bfmR allele to that of WT.
A. baumannii uses CDI to out-compete A. baylyi ADP1
To confirm our observation that the CDIV15 system is activated in the bfmRS mutant, a partial deletion of A. baumannii cdiV15 locus was generated in the WT and ΔbfmRS strains. In the resulting mutants, the cdiB gene was left intact but approximately 90% of the cdiAV15 along with cdiIV15 were deleted. This was due to the size of the genomic region that prevented us from obtaining the whole CDIV15 operon deletion. It is interesting to note, that during the characterization of A. baumannii ΔbfmRSΔcdiV15 and ΔcdiV15 mutants, we have observed the loss of ~ 200 kDa band in the SDS-PAGE gel of precipitated total protein fraction from culture media (Fig. 3c). The band corresponds to the predicted molecular weight of CdiA protein (~ 231 kDa), and its identity was subsequently confirmed by mass spectrometry. We have also noticed that, consistently with the mRNA data (Fig. 3b), the predicted CdiA band in WT was of lower intensity when compared to the ΔbfmRS strain (Fig. 3c). Additionally, ΔbfmRS strain complemented with the pbfmR displayed the predicted CdiA band of intermediate intensity, compared to the WT and ΔbfmRS (Fig. 3d).
We then investigated the aggressiveness of the mutants against A. baylyi ADP1. In addition, a plasmid containing the cdiIV15 allele under the inducible promoter was created and introduced into ADP1 strain. The cdiIV15 codes for the putative immunity gene, which should protect the host from the aggressor, if the latter uses the CDIV15 system for the killing. By performing the killing assays, we found that the deletion of cdiV15 in the ΔbfmRS and ΔbfmRSΔhcp mutants resulted in a low (~ 10-fold) but significant reduction of aggressiveness, when compared to the parent mutant (Fig. 3e). Interestingly, ΔbfmRSΔhcpΔcdiV15 mutant still displayed a killing phenotype (Fig. 3e). The reasons that caused this are currently unknown. However, it could be explained by the activation of a secondary CDI system that we were unable to detect with our PCR screen, as it is known that some Acinetobacter sp. strains contain two active CDI systems [26, 27].
The introduction of the cdiIV15 allele into A. baylyi ADP1 strain reduced the susceptibility to the killing by A. baumannii ΔbfmRSΔhcp and ΔbfmRS, but not by the ΔbfmRSΔcdiV15 and ΔbfmRSΔhcpΔcdiV15 strains (Fig. 3e), further confirming our findings that the ΔbfmRS and ΔbfmRSΔhcp mutants activate T6SS-independent killing mechanism, and show that this phenotype could be attributed to the activation of CDIV15 locus.
CDI-mediated A. baumannii intra-species competition
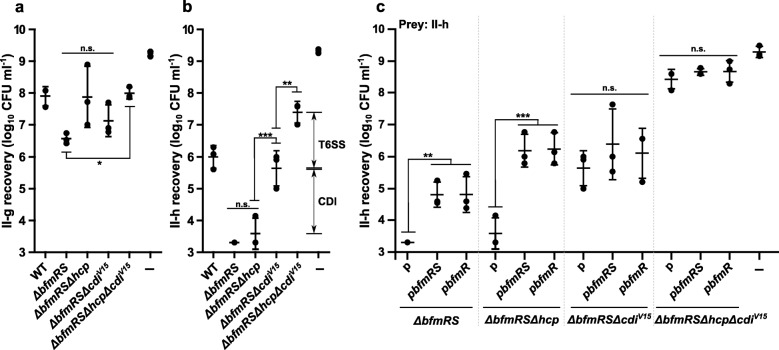
Bacterial inter-species killing via CDI system was observed only in a few cases and showed a very low efficiency, therefore it was suggested that the primary role of CDI is to differentiate sibling cells from other closely related bacteria from the same species [30, 31]. This could explain a rather modest changes in the killing efficiency that we observed towards A. baylyi ADP1. This prompted us to investigate the CDI-mediated killing phenomenon within the A. baumannii species. For this purpose, using primer pairs targeting cdiB genes of all known A. baumannii CDI systems, we have screened clinical A. baumannii isolates, representing different genotypically related groups (pulsotypes) of strains (n = 15) belonging to international clonal lineage II (IC II) by PCR [32]. The results showed that two clinical A. baumannii strains II-g and II-h did not contain a known cdiB gene. Therefore, these strains were selected for further competition experiments.
As can be seen in Fig. 4a and b, II-h strain was highly susceptible to the killing by A. baumannii V15, while II-g strain displayed a low susceptibility to this phenotype (Fig. 4a and Additional file 6: Figure S4a). The total numbers of the susceptible strain II-h were reduced ~ 107-fold by the ΔbfmRS and ΔbfmRSΔhcp mutants (Fig. 4b). When the competition was performed with the triple mutant ΔbfmRSΔhcpΔcdiV15, the recovery of II-h increased by a factor of 104, compared to ΔbfmRS. The strain containing only functional T6SS (ΔbfmRSΔcdiV15), displayed an intermediate phenotype (Fig. 4b). It is worth to mention that the WT displayed mostly T6SS-dependent killing phenotype (Additional file 6: Figure S4b). These results indicate, that the ΔbfmRS mutant kills II-h strain via both mechanisms – T6SS and CDI, while WT strain uses only T6SS-dependent killing.
Fig. 4.
CDI mediated A. baumannii intra-species competition. a-c Quantitative evaluation of inter-bacterial competition assay displaying a recovered number of A. baumannii clinical strains II-g (a) and II-h (b and c). Competition was performed with the following A. baumannii V15 mutants used as the aggressors: ΔbfmRS, ΔbfmRSΔhcp, ΔbfmRSΔcdiV15, ΔbfmRSΔhcpΔcdiV15 (a and b). The II-h strain was also challenged with the ΔbfmRS, ΔbfmRSΔhcp, ΔbfmRSΔcdiV15, ΔbfmRSΔhcpΔcdiV15 mutants, which were complemented with either pbfmRS or pbfmR (c). E. coli DH5α was used as a negative non-competitive control to enumerate bacteria numbers if there were no competition. Error bars represent standard deviation. The horizontal lines represent mean value. Values were calculated from at least three independent experiments. ***, p < 0.001; **, p < 0.01; n.s., not significant
Additionally, we investigated, whether the complementation with the bfmR allele resulted in an inhibition of the CDI-mediated killing against II-h. Notably, the results showed that when ΔbfmRS mutant was complemented with either pbfmR or pbfmRS, the recovery numbers of II-h increased by ~ 30-fold (Fig. 4c). Interestingly, when the ΔbfmRSΔhcp mutant was complemented with the same alleles, the reduction of killing phenotype was ~ 500-fold (Fig. 4c). Lastly, as expected, the complementation of either ΔbfmRSΔcdiV15 or ΔbfmRSΔhcpΔcdiV15 with the bfmR or bfmRS alleles did not influence any change in the killing phenotype of the mutants (Fig. 4c). These results were consistent with the observed intermediate transcriptional up-regulation of CDIV15 system in the ΔbfmRS strain complemented with the bfmR, when compared to the WT and ΔbfmRS strains (Fig. 3b). Additionally, bfmR or bfmRS complemented strains ΔbfmRSΔhcp, ΔbfmRSΔcdiV15, containing either functional CDI or T6SS, respectively, displayed an intermediate killing phenotype, compared to the ΔbfmRS and ΔbfmRSΔhcpΔcdiV15 (Fig. 4c). These results confirm observation that BfmR acts as a negative regulator of the CDIV15 system of A. baumannii V15.
Discussion
In this study, we aimed to further characterize the response regulator BfmR and its role in A. baumannii physiology. We were able to show that the presence of BfmR induces pellicle formation, while at the same time the regulator acts negatively on the contact-dependent inhibition system. The ability to form pellicle varies among A. baumannii strains considerably [16, 32] and seems to be affected by the production of secondary signaling molecule cAMP [16], indicating that regulatory mechanisms play a great role in the manifestation of this phenotype. Given the fact that pellicle formation is considered as an important factor for the persistence and transmission of the pathogenic species [13, 14], the understanding of how it is regulated in A. baumannii is important. Our results, showing that BfmR is responsible for the formation of this structure, suggests that there is a specific currently unknown signal or signals that the BfmRS system responds to, which leads to the induction of the pellicle. Currently, the best known inducer of the pellicle phenotype is oxygen gradient that emerges when bacteria grow to a high cell density [13]. Also, the fact that BfmR is responsible for the surface biofilm formation [3] and the observation that pellicles were slightly attached to the walls of the tubes [14] suggests that surface biofilms may be the initial stage of pellicle formation. This speculation is also based on the previous findings showing that the four-day mature pellicle, compared to the one-day pellicle, contains ~ 4.5-fold increased BfmR levels [17], indicating that during pellicle formation the levels of the protein gradually increase. Since we were unable to reliably dissect pellicle and biofilm formation phenotypes apart, we could not reject the possibility that pellicle formation is a secondary effect due to the inability to form surface biofilms.
Biofilm formation is characterized by some level of bacterial organization, that includes morphologically and genetically diverse individuals, which generate and respond to signaling by the nearby cells and/or the surroundings [33, 34]. Since biofilm is a densely packed structure, competition for limited resources and space occurs. In bacteria, the antagonism is exerted mainly via two different contact-dependent inter-bacterial competition systems: non-specific type VI secretion system (T6SS) and receptor-specific contact-dependent growth inhibition system (CDI) [35].
Our results show that A. baumannii BfmR is involved in the regulation of both of them. Firstly, we determined that BfmR is required for the extensive secretion of Hcp protein into media and up-regulates transcription of hcp and tssM genes, which are essential for T6SS activity [20]. However, despite the fact that Hcp secretion is the hallmark of a functional Type VI secretion system [18], we did not observe the reduction in T6SS-dependent killing phenotype. This suggests that either the inhibition of killing effect was too small, or the Hcp secretion plays a role in other, killing independent activities such as metal ion acquisition or in contribution to pathogenesis [36–38]. Our results are consistent with the previous observations indicating that a mature four-day A. baumannii pellicle displays up-regulation of some T6SS locus genes, compared to the planktonic cultures [17]. This suggests that during the unfavourable conditions A. baumannii may choose to activate T6SS. The presence of this phenotype in the pellicle could be crucial for bacteria to defend from other competing species and prevent from the rise of mutants in a population, which do not contribute to the secretion of stabilizing materials. However, there seems to be a great heterogeneity among different A. baumannii strains in terms of ability to secrete Hcp and in T6SS regulation mechanisms in general [20, 22, 23, 39].
We also determined that the BfmR negatively regulates yet poorly characterized contact-dependent inhibition system. Such systems, allow some species of Gram-negative bacteria to deliver toxic effectors to neighboring bacteria and inhibit their growth [25, 26, 40]. It is known that CDI requires a receptor to deliver a toxin to its target, therefore the inhibitory activity is restricted to closely related bacteria, and, according to some suggestions, could be used to distinguish self from non-self [35, 41]. Our results are consistent with these observations as we detected CDI activity against A. baumannii and A. baylyi ADP1 but not against E. coli MC4100, P. aeruginosa, and K. pneumoniae.
The negative regulation of the CDIV15 system by the BfmR suggests that CDI is not required for the pellicle/biofilm formation of A. baumannii V15. Indeed, our data show that the deletion of CDIV15 did not impact the ability of the strain to form a pellicle or biofilm (unpublished observation). Our results support previous findings indicating that A. baylyi ADP1 also does not require CDI system to form biofilms [27]. Recently, evolution experiments with P. aeruginosa indicated that biofilm cultures, compared to the planktonic bacteria, experienced higher mutability rates, which led to the diversification of biofilm population with clone variants that contributed to the population’s ability to colonize the surface [34]. The turned off CDI system in these populations could allow their preservation while at the same time the active T6SS could prevent the invading species.
It must be noted that out of two clinical A. baumannii isolates, which were selected for the competition experiments, only one was susceptible to the CDIV15-mediated killing, despite both of them lacking CDI cluster and belonging to the same international clonal lineage II [42, 43]. The resistant strain may possess similar orphan immunity gene, which could protect it from the CDIV15 system, as we observed the presence of these orphan genes dispersed throughout at least some of the A. baumannii strains. Additionally, we noticed, that these regions may contain more than one immunity module (unpublished observation), indicating that some strains may posses innate immunity protecting them from the attack executed by a strain containing a different CDI system. Moreover, our recent work indicates that the susceptible strain II-h is capsule-deficient [32]. This observation could also explain the susceptibility phenotype as the required receptor for the CDI-mediated killing may be hidden in the capsule-positive strain II-g, thereby effectively restricting the A. baumanii V15 CDIV15 attack.
Conclusions
It is well known that BfmRS system promotes biofilm formation which is an important feature of microbial persistence under unfavourable conditions. However, the regulatory pathways of A. baumannii global regulator BfmR are still poorly understood. Here we provide evidence that A. baumannii via the BfmR, is able to promote pellicle formation, and Hcp secretion into culture media. However, at the same time, BfmR acts as a negative regulator of a CDI system of A. baumannii, which is used to inhibit the growth of related bacteria. Therefore, our results suggest that during these conditions A. baumannii via the BfmRS system may allow a cooperative behavior towards related bacteria, which could improve the survival chances of A. baumannii.
Methods
Bacterial strains and growth conditions
E. coli JM107 was used for all cloning experiments and DNA manipulations. Bacillus spp. was used as the source for sacB gene. Acinetobacter sp., Pseudomonas aeruginosa (P16), and Klebsiella pneumoniae (K39) strains were grown in tryptic soy broth (TSB) (Oxoid). All E. coli strains were grown in Luria-Bertani (LB) media. All A. baumannii strains used in the work were previously characterized [15, 32]. Bacteria were grown at 37 °C, unless indicated otherwise. Growth media was supplemented with antibiotics, where appropriate: ampicillin 100 μg mL− 1, gentamicin 10 μg mL− 1, streptomycin 100 μg mL− 1, ceftazidime 10 μg mL− 1. Bacterial strains and plasmids used in this study are listed in the Additional file 1: Table S1.
Plasmid construction
All molecular biology procedures were performed using reagents obtained from Thermo Fisher Scientific and according the manufacturer’s recommendations. Primers were obtained from Metabion or Thermo Fisher Scientific and are listed in the Additional file 2: Table S2. All final constructs were verified by sequencing. The suicide plasmid pUC19_sacB was constructed by amplifying the levansucrase gene sacB using primers sacB_F/sacB_R and cloning the PCR product into pUC19 via XbaI and PaeI. The A. baumannii/E. coli IPTG inducible shuttle expression plasmid pUC_gm_AcORI_Ptac_gfp_TER was obtained by cloning of Ptac promoter, terminator sites (TER) (both from pKK223–3), and the gfp gene (from pAcGFP1-C3) into the pUC19 plasmid containing Acinetobacter sp. ORI from pWH1266 [44]. The plasmid was then inverse amplified with primers M13_rwd/Aac3I_seqR to remove the gentamicin cassette and blunt ligated to the lacIq gene amplified from BL21(DE3) strain genomic DNA to generate A. baumannii/E. coli shuttle expression plasmid pUC_AcORI_Ptac_gfp_TER_lacIq2.
Complementation plasmids were constructed by replacing the gfp gene in the IPTG inducible shuttle expression plasmid with the required gene amplified from A. baumannii V15 genomic DNA. Control plasmid pUC_AcORI_Ptac_TER_lacIq2 was obtained by removing the gfp gene and is denoted in figures as “p”, where relevant. The complementation plasmid containing the cdiIV15 gene was further modified by replacing the bla gene with the aac3I cassette. The created plasmids were used to complement the relevant strains of A. baumannii (Additional file 1: Table S1).
Generation of A. baumannii mutant strains
A modified marker-less gene deletion technique from Oh et al. [45] was used to obtain mutant strains of A. baumannii. Approximately 1 kb long upstream and downstream regions of the genes to be deleted were amplified separately from the genomic DNA of A. baumannii V15 and joined via the overlap PCR with gentamicin resistance cassette aac3I using primer pairs described in the Additional file 2: Table S2. The resulting DNA fragments were cloned into pUC19_sacB plasmid. The relevant A. baumannii strains were electroporated with the obtained plasmids and selected on LB agar plates with 10 μg mL− 1 of gentamicin. Then, a single colony was inoculated into LB media without antibiotics and grown overnight at 37 °C with shaking. Serial dilutions of the overnight culture were plated onto LB agar plates containing 10% sucrose and grown overnight at 37 °C. Mutants were identified by PCR with specific primers (Additional file 2: Table S2) and confirmed by sequencing. All obtained mutants and their variants complemented with various constructs were tested for growth impairments by inoculating overnight cultures of A. baumannii into the wells of 96-well polystyrene plates at a density of 106 CFU mL− 1 in 0.25x TSB and then measuring growth at 37 °C until stationary phase. IPTG were added at the early logarithmic phase (OD600 = 0.25–0.3) where necessary to determine the maximum concentration that did not induce growth impairments due to the presence of constructs.
Pellicle formation assays
The pellicle formation was evaluated by inoculating overnight cultures of A. baumannii grown in 1x TSB media at 37 °C into the wells of a flat-bottom 12 well polystyrene microplate at a density of 106 CFU mL− 1 in a total volume of 3 mL. The cultures were incubated stationary at 30 °C for 30 h. To collect the pellicles from the surface of the media, 200 μL of isopropanol were added to the each well, which allowed to easily remove almost all pellicle material. The removed pellicles were resuspended in 500 μL of 10 mM NaOH, followed by a quick neutralization with HCl and the OD600 of suspension was measured and normalized to the total volume of culture so as to be comparable to the planktonic OD600 readings.
Protein secretion assay
Total protein content from culture media used in the pellicle formation assay was precipitated using 100% (w/v) trichloroacetic acid (TCA) as follows. Firstly, the collected culture media was centrifuged at 13000 g for 10 min at 4 °C and subsequently filtered through 0.22 μm filter to remove remaining biomass. Then, in the resulting supernatant TCA was added to a final concentration of 10% (w/v), and centrifuged at 13000 g for 45 min at 4 °C. The resulting pellet was washed twice with ice-cold acetone and dried by incubating tube at 95 °C for a few minutes before being re-suspended with Laemmli-SDS-PAGE sample buffer. The samples were analyzed using the Laemmli-SDS-PAGE system. After electrophoresis, gels were stained with Coomassie brilliant blue. Approximately 3.5 μg of the total protein were loaded into each well. The PageRuler™ unstained broad range protein ladder (7.5 μl) was used as marker. The whole gels are shown in Additional file 7: Figure S5a-S5d. Protein identification by MALDI-TOF mass spectrometry was undertaken at Proteomics Department of Vilnius University Life Sciences Center.
RNA isolation and analysis of gene expression by qPCR
Overnight cultures of A. baumannii grown in 1x TSB media at 37 °C were diluted with 0.25x TSB and inoculated into the wells of a flat-bottom 96 well polystyrene microplate at a density of 106 CFU mL− 1 in 0.25x TSB. The cultures were grown until the logarithmic phase (OD600 = 0.35–0.4). Total RNA was isolated, DNA removed and cDNA synthesized as recommended by the supplier (Thermo Fisher Scientific). RNA integrity and contamination with DNA was checked by agarose gel electrophoresis. qPCR was performed using primer pairs listed in the Additional file 2: Table S2 (all primers exhibited 95–107% amplification efficiency (with > 0.99 coefficient of determination) at used concentrations). Product specificity was investigated by melting curve analysis. The changes in gene expression were calculated as ΔΔCt, using rpoB as a house-keeping gene. At least three biological replicates, each with two technical replicates, were performed.
Inter-bacterial competition assay
The assay was performed as described previously [26] with some modifications. Briefly, the strains grown overnight in TSB media at 37 °C were washed twice with the fresh TSB to remove residual antibiotics. Then, bacteria were diluted with the fresh TSB to a final concentration of ~ 108 CFU mL− 1 and mixed at aggressor (A. baumannii V15):prey ratio of 10:1, 10:1, 20:1, when the competition was performed with E. coli strain MC4100, A. baylyi strain ADP1, or all other strains, respectively. Five microlitre of resulting suspension was placed onto TSB media containing 1.5% agar and allowed to dry. The competitions were performed at 37 °C for 6 h. To quantitatively evaluate the number of surviving bacteria, spots were excised from the plate, vigorously re-suspended in TSB broth, serially diluted, and plated on TSB agar plates containing selective antibiotics: streptomycin (100 μg mL− 1) for E. coli strain MC4100, ampicillin (100 μg mL− 1) for all A. baumannii V15 mutants, ceftazidime (10 μg mL− 1) for all other clinical A. baumannii strains, gentamicin (10 μg mL− 1) for A. baylyi ADP1, P. aeruginosa, and K. pneumoniae. All strains had a natural resistance or were transformed with a plasmid containing appropriate marker allowing for a selective isolation. All experiments contained control reactions, which consisted of non-aggressive E. coli strain DH5α mixed with each of the strain to obtain a total number bacteria if there were no competition between strains. The obtained number of colonies was calculated as CFU per mL of culture.
Statistical analyses
All statistical comparisons were performed using one-way ANOVA (p = 0.05) with a Tukey HSD post-hoc test. Inter-bacterial competition assay was calculated as follows: CFU per mL was normalized by taking first the decadic logarithm and using these values for statistical analysis. Changes in gene expression experiments were considered significant if the differences were more than 2-fold. Asterisks in the figures denote the statistically significant difference between the groups (n.s., not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). The analyses were performed using R package (version 3.2.3). Graphs were drawn using QtiPlot.
GenBank accession number
The sequence of A. baumannii V15 cdiBAI locus has been deposited in GenBank under the accession number MK405474.
Supplementary information
Additional file 1: Table S1. Bacterial strains and plasmids used in the study.
Additional file 2: Table S2. Primers used in the study.
Additional file 3: Figure S1. BfmS and functional T6SS are not required for pellicle production. Quantitative evaluation of pellicles formed by: (a) A. baumannii V15 (WT), ΔbfmRS mutant, and ΔbfmRS mutant, complemented with the plasmid pbfmS; (b) WT, Δhcp mutant, and Δhcp mutant, complemented with the plasmid phcp. Pellicle values were normalized by the total growth volume. Error bars represent standard deviation. IPTG denotes induction conditions using 0 or 0.1 mM IPTG concentration.
Additional file 4: Figure S2. Loss of BfmRS system activates T6SS-independent killing phenotype of A. baumannii V15 against A. baylyi ADP1 only. Quantitative evaluation of inter-bacterial competition assay displaying a recovered number of prey: (a) E. coli MC4100 and (b) A. baylyi ADP1. Competition was performed with the following strains used as the aggressors: WT, ΔbfmRSΔhcp mutant, ΔbfmRSΔhcp strain complemented with the plasmids pbfmRS or pbfmR. E. coli DH5α was used as a negative non-competitive control to enumerate bacteria numbers if there were no competition. Error bars represent standard deviation. The horizontal lines represent mean value. Values were calculated from at least three independent experiments. ***, p < 0.001; n.s., not significant.
Additional file 5: Figure S3. Quantitative evaluation of inter-bacterial competition assays displaying the recovered numbers of clinical strains that were used as a prey: (a and b) Pseudomonas aeruginosa P16 and (c and d) Klebsiella pneumoniae K39. Competition was performed with the following strains used as the aggressors: WT, ΔbfmRS, Δhcp, ΔbfmRSΔhcp. The hcp mutants were also complemented with the wild-type hcp allele (plasmid phcp). E. coli DH5α was used as a negative non-competitive control to enumerate bacteria numbers if there were no competition. Error bars represent standard deviation. The horizontal lines represent mean value. Values were calculated from at least three independent experiments.*, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant. The hcp gene in plasmid phcp was induced using IPTG concentration of 0.1 mM.
Additional file 6: Figure S4. T6SS is used as the main mechanism for species antagonism by A. baumannii V15 (WT) strain. Quantitative evaluation of inter-bacterial competition assay displaying a recovered number of CDI lacking A. baumannii strains used as a prey: (a) II-g; (b) II-h. Competition was performed with the following strains used as the aggressors: WT, Δhcp mutant, Δhcp mutant complemented with the wild-type hcp allele (plasmid phcp). E. coli DH5α was used as a negative non-competitive control to enumerate bacteria numbers if there were no competition. Error bars represent standard deviation. The horizontal lines represent mean value. Values were calculated from at least three independent experiments.*, p < 0.05; n.s., not significant. Hcp gene in plasmid phcp was induced using IPTG concentration of 0.1 mM.
Additional file 7: Figure S5. The whole gels showing TCA-precipitated total protein fraction from culture media. Related to Fig. 1c-d and Fig. 3c-d. Proteins separated by 12.5% (a and b) or 10% (c and d) SDS-PAGE and visualized by staining with Coomassie blue. PageRuler™ Unstained Broad Range Protein Ladder (Thermo Fisher) was used as marker. Numbers on the left of the gels denote molecular mass in kDa.
Acknowledgements
We thank Algirdas Mikšys for helpful discussions and comments during the writing of the manuscript and Audrius Gegeckas for providing a Bacillus spp. strain.
Abbreviations
- cAMP
Cyclic adenosine monophosphate
- CDI
Contact-dependent inhibition system
- cDNA
Complementary DNA
- CFU
Colony Formation Unit
- HCl
Hydrochloric acid
- IC
International clonal lineage
- IPTG
Isopropyl β-D-thiogalactopyranoside
- kb
Kilobase
- kDa
Kilodalton
- LB
Luria-Bertani media
- MALDI-TOF
Matrix-assisted laser desorption/ionization time of flight mass spectrometry
- NaOH
Sodium hydroxide
- PCR
Polymerase Chain Reaction
- qPCR
Quantitative polymerase chain reaction
- SDS-PAGE
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- T5SS
Type V Secretion System
- T6SS
Type VI Secretion System
- TCA
Trichloroacetic acid
- TCS
Two-component signal transduction system
- TSB
Tryptic soy broth
- w/v
Weight by volume
- WT
Wild-type
Authors’ contributions
Conceived and designed the experiments: RK, ES; Performed the experiments: RK, JS, JA; Analyzed the data: RK, JS, JA, ES; Wrote the paper: RK, ES. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The sequence of A. baumannii V15 cdiBAI locus has been deposited in GenBank under the accession number MK405474.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Renatas Krasauskas, Email: renatas.krasauskas@gf.vu.lt.
Jūratė Skerniškytė, Email: jurate.skerniskyte@gf.vu.lt.
Julija Armalytė, Email: julija.armalyte@gf.vu.lt.
Edita Sužiedėlienė, Email: edita.suziedeliene@gf.vu.lt.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12866-019-1621-5.
References
- 1.Antunes LCS, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71(3):292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 2.Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology. 2008;154(Pt 11):3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 4.Zschiedrich Christopher P., Keidel Victoria, Szurmant Hendrik. Molecular Mechanisms of Two-Component Signal Transduction. Journal of Molecular Biology. 2016;428(19):3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemmer KM, Bonomo RA, Rather PN. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology. 2011;157(Pt 9):2534–2544. doi: 10.1099/mic.0.049791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liou M-L, Soo P-C, Ling S-R, Kuo H-Y, Tang CY, Chang K-C. The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. J Microbiol Immunol Infect. 2014;47(4):275–281. doi: 10.1016/j.jmii.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Wang N, Ozer EA, Mandel MJ, Hauser AR. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio. 2014;5(3):e01163–14. [DOI] [PMC free article] [PubMed]
- 8.Russo TA, Manohar A, Beanan JM, Olson R, MacDonald U, Graham J, et al. The response regulator BfmR is a potential drug target for Acinetobacter baumannii. mSphere. 2016;1(3):e00082-16. [DOI] [PMC free article] [PubMed]
- 9.Geisinger E, Mortman NJ, Vargas-Cuebas G, Tai AK, Isberg RR. A global regulatory system links virulence and antibiotic resistance to envelope homeostasis in Acinetobacter baumannii. PLoS Pathog. 2018;14(5):e1007030. doi: 10.1371/journal.ppat.1007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebhardt MJ, Gallagher LA, Jacobson RK, Usacheva EA, Peterson LR, Zurawski DV, et al. Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in Acinetobacter baumannii. MBio. 2015;6(6):e01660–e01615. doi: 10.1128/mBio.01660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrow JM, Wells G, Pesci EC. Desiccation tolerance in Acinetobacter baumannii is mediated by the two-component response regulator BfmR. PLoS One. 2018;13(10):e0205638. doi: 10.1371/journal.pone.0205638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geisinger E, Isberg RR. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015;11(2):e1004691. doi: 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armitano J, Méjean V, Jourlin-Castelli C. Gram-negative bacteria can also form pellicles. Environ Microbiol Rep. 2014;6(6):534–544. doi: 10.1111/1758-2229.12171. [DOI] [PubMed] [Google Scholar]
- 14.Nait Chabane Y, Marti S, Rihouey C, Alexandre S, Hardouin J, Lesouhaitier O, et al. Characterisation of pellicles formed by Acinetobacter baumannii at the air-liquid interface. PLoS One. 2014;9(10):e111660. doi: 10.1371/journal.pone.0111660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Povilonis J, Seputiene V, Krasauskas R, Juskaite R, Miskinyte M, Suziedelis K, et al. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J Antimicrob Chemother. 2013;68(5):1000–1006. doi: 10.1093/jac/dks499. [DOI] [PubMed] [Google Scholar]
- 16.Giles SK, Stroeher UH, Eijkelkamp BA, Brown MH. Identification of genes essential for pellicle formation in Acinetobacter baumannii. BMC Microbiol. 2015;15:116. doi: 10.1186/s12866-015-0440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kentache T, Ben Abdelkrim A, Jouenne T, Dé E, Hardouin J. Global dynamic proteome study of a pellicle-forming Acinetobacter baumannii strain. Mol Cell Proteomics. 2017;16(1):100–112. doi: 10.1074/mcp.M116.061044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pukatzki S, McAuley SB, Miyata ST. The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol. 2009;12(1):11–17. doi: 10.1016/j.mib.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Brunet YR, Hénin J, Celia H, Cascales E. Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep. 2014;15(3):315–321. doi: 10.1002/embr.201337936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber BS, Miyata ST, Iwashkiw JA, Mortensen BL, Skaar EP, Pukatzki S, et al. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One. 2013;8(1):e55142. doi: 10.1371/journal.pone.0055142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carruthers MD, Nicholson PA, Tracy EN, Munson RS. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One. 2013;8(3):e59388. doi: 10.1371/journal.pone.0059388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Repizo GD, Gagné S, Foucault-Grunenwald M-L, Borges V, Charpentier X, Limansky AS, et al. Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS One. 2015;10(9):e0138265. doi: 10.1371/journal.pone.0138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A. 2015;112(30):9442–9447. doi: 10.1073/pnas.1502966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber BS, Hennon SW, Wright MS, Scott NE, de Berardinis V, Foster LJ, et al. Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. MBio. 2016;7(5):e01253-16. [DOI] [PMC free article] [PubMed]
- 25.Willett JLE, Ruhe ZC, Goulding CW, Low DA, Hayes CS. Contact-dependent growth inhibition (CDI) and CdiB/CdiA two-partner secretion proteins. J Mol Biol. 2015;427(23):3754–3765. doi: 10.1016/j.jmb.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harding CM, Pulido MR, Venanzio GD, Kinsella RL, Webb AI, Scott NE, et al. Pathogenic Acinetobacter species have a functional type I secretion system and contact-dependent inhibition systems. J Biol Chem. 2017;292(22):9075–9087. doi: 10.1074/jbc.M117.781575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Gregorio E, Esposito EP, Zarrilli R, Di Nocera PP. Contact-dependent growth inhibition proteins in Acinetobacter baylyi ADP1. Curr Microbiol. 2018;75(11):1434–1440. doi: 10.1007/s00284-018-1540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Gregorio E, Zarrilli R, Di Nocera PP. Contact-dependent growth inhibition systems in Acinetobacter. Sci Rep. 2019;9(1):154. doi: 10.1038/s41598-018-36427-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beitz E. TeXshade: shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics. 2000;16(2):135–139. doi: 10.1093/bioinformatics/16.2.135. [DOI] [PubMed] [Google Scholar]
- 30.Beck CM, Morse RP, Cunningham DA, Iniguez A, Low DA, Goulding CW, et al. CdiA from Enterobacter cloacae delivers a toxic ribosomal RNase into target bacteria. Structure. 2014;22(5):707–718. doi: 10.1016/j.str.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koskiniemi S, Garza-Sánchez F, Edman N, Chaudhuri S, Poole SJ, Manoil C, et al. Genetic analysis of the CDI pathway from Burkholderia pseudomallei 1026b. PLoS One. 2015;10(3):e0120265. doi: 10.1371/journal.pone.0120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skerniškytė J, Krasauskas R, Péchoux C, Kulakauskas S, Armalytė J, Sužiedėlienė E. Surface-related features and virulence among Acinetobacter baumannii clinical isolates belonging to international clones I and II. Front Microbiol. 2019;9:3116. doi: 10.3389/fmicb.2018.03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poltak SR, Cooper VS. Ecological succession in long-term experimentally evolved biofilms produces synergistic communities. ISME J. 2011;5(3):369–378. doi: 10.1038/ismej.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flynn KM, Dowell G, Johnson TM, Koestler BJ, Waters CM, Cooper VS. Evolution of ecological diversity in biofilms of Pseudomonas aeruginosa by altered cyclic Diguanylate signaling. J Bacteriol. 2016;198(19):2608–2618. doi: 10.1128/JB.00048-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia EC. Contact-dependent interbacterial toxins deliver a message. Curr Opin Microbiol. 2018;42:40–46. doi: 10.1016/j.mib.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapitein N, Mogk A. Deadly syringes: type VI secretion system activities in pathogenicity and interbacterial competition. Curr Opin Microbiol. 2013;16(1):52–58. doi: 10.1016/j.mib.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, et al. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun. 2017;8:14888. doi: 10.1038/ncomms14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Zhou Z, He F, Ruan Z, Jiang Y, Hua X, et al. The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS One. 2018;13(2):e0192288. doi: 10.1371/journal.pone.0192288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Venanzio G, Moon KH, Weber BS, Lopez J, Ly PM, Potter RF, et al. Multidrug-resistant plasmids repress chromosomally encoded T6SS to enable their dissemination. Proc Natl Acad Sci U S A. 2019;116(4):1378–1383. doi: 10.1073/pnas.1812557116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruhe ZC, Low DA, Hayes CS. Bacterial contact-dependent growth inhibition. Trends Microbiol. 2013;21(5):230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruhe ZC, Wallace AB, Low DA, Hayes CS. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. MBio. 2013;4(4):e00480-13. [DOI] [PMC free article] [PubMed]
- 42.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect. 2007;13(8):807–815. doi: 10.1111/j.1469-0691.2007.01759.x. [DOI] [PubMed] [Google Scholar]
- 43.Karah N, Sundsfjord A, Towner K, Samuelsen Ø. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist Updat. 2012;15(4):237–247. doi: 10.1016/j.drup.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Armalytė J, Jurėnas D, Krasauskas R, Čepauskas A, Sužiedėlienė E. The higBA toxin-antitoxin module from the opportunistic pathogen Acinetobacter baumannii – regulation, activity, and evolution. Front Microbiol. 2018;9:732. doi: 10.3389/fmicb.2018.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh MH, Lee JC, Kim J, Choi CH, Han K. Simple method for Markerless gene deletion in multidrug-resistant Acinetobacter baumannii. Appl Environ Microbiol. 2015;81(10):3357–3368. doi: 10.1128/AEM.03975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Bacterial strains and plasmids used in the study.
Additional file 2: Table S2. Primers used in the study.
Additional file 3: Figure S1. BfmS and functional T6SS are not required for pellicle production. Quantitative evaluation of pellicles formed by: (a) A. baumannii V15 (WT), ΔbfmRS mutant, and ΔbfmRS mutant, complemented with the plasmid pbfmS; (b) WT, Δhcp mutant, and Δhcp mutant, complemented with the plasmid phcp. Pellicle values were normalized by the total growth volume. Error bars represent standard deviation. IPTG denotes induction conditions using 0 or 0.1 mM IPTG concentration.
Additional file 4: Figure S2. Loss of BfmRS system activates T6SS-independent killing phenotype of A. baumannii V15 against A. baylyi ADP1 only. Quantitative evaluation of inter-bacterial competition assay displaying a recovered number of prey: (a) E. coli MC4100 and (b) A. baylyi ADP1. Competition was performed with the following strains used as the aggressors: WT, ΔbfmRSΔhcp mutant, ΔbfmRSΔhcp strain complemented with the plasmids pbfmRS or pbfmR. E. coli DH5α was used as a negative non-competitive control to enumerate bacteria numbers if there were no competition. Error bars represent standard deviation. The horizontal lines represent mean value. Values were calculated from at least three independent experiments. ***, p < 0.001; n.s., not significant.
Additional file 5: Figure S3. Quantitative evaluation of inter-bacterial competition assays displaying the recovered numbers of clinical strains that were used as a prey: (a and b) Pseudomonas aeruginosa P16 and (c and d) Klebsiella pneumoniae K39. Competition was performed with the following strains used as the aggressors: WT, ΔbfmRS, Δhcp, ΔbfmRSΔhcp. The hcp mutants were also complemented with the wild-type hcp allele (plasmid phcp). E. coli DH5α was used as a negative non-competitive control to enumerate bacteria numbers if there were no competition. Error bars represent standard deviation. The horizontal lines represent mean value. Values were calculated from at least three independent experiments.*, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant. The hcp gene in plasmid phcp was induced using IPTG concentration of 0.1 mM.
Additional file 6: Figure S4. T6SS is used as the main mechanism for species antagonism by A. baumannii V15 (WT) strain. Quantitative evaluation of inter-bacterial competition assay displaying a recovered number of CDI lacking A. baumannii strains used as a prey: (a) II-g; (b) II-h. Competition was performed with the following strains used as the aggressors: WT, Δhcp mutant, Δhcp mutant complemented with the wild-type hcp allele (plasmid phcp). E. coli DH5α was used as a negative non-competitive control to enumerate bacteria numbers if there were no competition. Error bars represent standard deviation. The horizontal lines represent mean value. Values were calculated from at least three independent experiments.*, p < 0.05; n.s., not significant. Hcp gene in plasmid phcp was induced using IPTG concentration of 0.1 mM.
Additional file 7: Figure S5. The whole gels showing TCA-precipitated total protein fraction from culture media. Related to Fig. 1c-d and Fig. 3c-d. Proteins separated by 12.5% (a and b) or 10% (c and d) SDS-PAGE and visualized by staining with Coomassie blue. PageRuler™ Unstained Broad Range Protein Ladder (Thermo Fisher) was used as marker. Numbers on the left of the gels denote molecular mass in kDa.
Data Availability Statement
The sequence of A. baumannii V15 cdiBAI locus has been deposited in GenBank under the accession number MK405474.