Abstract
Background
Soybean (Glycine max (L.)) is one the most important oil-yielding cash crops. However, the soybean production has been seriously restricted by salinization. It is therefore crucial to identify salt tolerance-related genes and reveal molecular mechanisms underlying salt tolerance in soybean crops. A better understanding of how plants resist salt stress provides insights in improving existing soybean varieties as well as cultivating novel salt tolerant varieties. In this study, the biological function of GmNHX1, a NHX-like gene, and the molecular basis underlying GmNHX1-mediated salt stress resistance have been revealed.
Results
We found that the transcription level of GmNHX1 was up-regulated under salt stress condition in soybean, reaching its peak at 24 h after salt treatment. By employing the virus-induced gene silencing technique (VIGS), we also found that soybean plants became more susceptible to salt stress after silencing GmNHX1 than wild-type and more silenced plants wilted than wild-type under salt treatment. Furthermore, Arabidopsis thaliana expressing GmNHX1 grew taller and generated more rosette leaves under salt stress condition compared to wild-type. Exogenous expression of GmNHX1 resulted in an increase of Na+ transportation to leaves along with a reduction of Na+ absorption in roots, and the consequent maintenance of a high K+/Na+ ratio under salt stress condition. GmNHX1-GFP-transformed onion bulb endothelium cells showed fluorescent pattern in which GFP fluorescence signals enriched in vacuolar membranes. Using the non-invasive micro-test technique (NMT), we found that the Na+ efflux rate of both wild-type and transformed plants after salt treatment were significantly higher than that of before salt treatment. Additionally, the Na+ efflux rate of transformed plants after salt treatment were significantly higher than that of wild-type. Meanwhile, the transcription levels of three osmotic stress-related genes, SKOR, SOS1 and AKT1 were all up-regulated in GmNHX1-expressing plants under salt stress condition.
Conclusion
Vacuolar membrane-localized GmNHX1 enhances plant salt tolerance through maintaining a high K+/Na+ ratio along with inducing the expression of SKOR, SOS1 and AKT1. Our findings provide molecular insights on the roles of GmNHX1 and similar sodium/hydrogen exchangers in regulating salt tolerance.
Keywords: Soybean, GmNHX1, Salt stress, VIGS, K+/Na+ ratio
Background
Plants are subjected to various biotic as well as abiotic stresses during their growth. Due to increasingly exacerbated salinization worldwide [1], compounded with the abiotic stresses such as cold and drought, the damage caused by salt stress has been significantly worse. Plants reserve many ways to tolerate salt stress, including efflux of salt and intracellular partitioning [2]. Salt tolerance, similar to many other metabolic processes, requires the proper control of cellular pH [3]. Na+/H+ exchangers (NHXs) are integral membrane transporters that catalyze the electroneutral exchange of K+ or Na+ for H+ and are implicated in cell expansion [4], development [5], ion homeostasis [6] and salt tolerance [7]. The Arabidopsis genome contains eight NHX homolog-encoding genes which are grouped based on their sequence similarity and localization into three distinct classes, those enriches in plasma membrane (NHX7/SOS1 and NHX8), endosomal/vesicular (NHX5, NHX6), and vacuolar membrane (NHX1, NHX2, NHX3, NHX4), respectively [2]. In Arabidopsis, NHXs that localize in vacuolar and plasma membrane are generally considered critical for maintaining Na+/K+ homeostasis [8]. NHX5 and NHX6, however, function as pH regulators of Golgi, trans-Golgi network, and pre-vacuolar compartments, regulating the sorting of newly synthesized peptides and the direction of Golgi-cargo movement [9].
Despite that Glycine soja (wild soybean) is often unsusceptible to salt stress [10], its close relative Glycine max (soybean) is typically osmotic sensitive [11]. Thus, the identification and characterization of endogenous genes that are involved in salt tolerance regulation would substantially benefit genetic breeding of soybeans. Overexpression of GmNAC15, a member of the NAC transcription factor family in soybean, enhances salt tolerance in soybean hairy roots [12]. Besides, overexpression of GmSK1, one of the multi-subunit E3 ligases, enhances tolerance to high salinity and drought stress when transformed into tobacco (Nicotiana tobacum) plants [13]. Overexpression of GmBIN2, a serine/threonine kinase related to brassinosteroid sensitivity, increases cellular Ca2+ content and reduces Na+ content, together enhances salt tolerance in transgenic Arabidopsis plants [14]. Previous studies in our lab have demonstrated that the overexpression of GmNHX1 (Gene ID: LOC100816746) is able to complement the defect of the Saccharomyces cerevisiae ena1–4, nhx1 and nha1 mutant and reduce the hindering effect of salt stress on cell growth [15]. In this study, we extend our understanding of the function of GmNHX1 gene and its roles in regulating salt tolerance. We uncover its subcellular localization as well as how its transcription responds to salt stress in soybeans. We further use Arabidopsis ecotype Col-0, a model plant allele that has been used in numerous physiology studies, as a host to evaluate the function and mechanism of GmNHX1 under salt stress condition. We uncover the role of GmNHX1 by utilizing this model organism, which is to maintain a higher Na+ efflux rate and K+/Na+ ratio under salt stress. Our findings provide important implications for understanding the molecular basis underlying salt tolerance in plants.
Results
GmNHX1 gene is related to salt stress resistance in plants
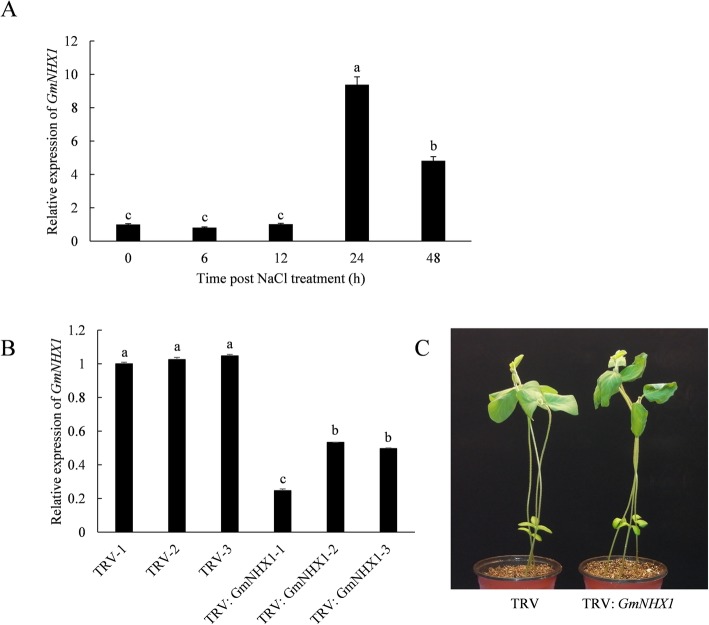
In order to examine the relation between GmNHX1 and the response to salt stress in soybeans, we examined the transcription level of GmNHX1 in soybean variety “Jidou-7”. We observed an increase of GmNHX1 gene expression after salt treatment and the peak of its expression was at 24 h post treatment (hpt), reaching approximately 10-fold of that of before treatment (0 hpt). The expression of GmNHX1 then began to decrease to approximately 5-fold of that of 0 hpt at 48 hpt (Fig. 1a). The upregulation of GmNHX1 expression under salt stress implies its relevance to salt stress response in soybeans and its potential role in regulating salt tolerance in soybeans.
Fig. 1.
GmNHX1 positively regulates plant salt tolerance. Transcription Level of GmNHX1 under salt stress in Soybean was detected by RT-qPCR, ACTIN gene was used as a reference gene. The data shows the mean ± S.E. of triplicate experiments. Columns with different letters indicate significant differences at P < 0.05 (a). The silencing efficiency of GmNHX1 in silenced plants using VIGS were detected by RT-qPCR, using ACTIN as reference gene (b). Phenotype of TRV- VIGS plants under 200 mM NaCl solution treatment for 24 h (c)
We then picked a specific fragment in the GmNHX1 coding region, to construct the TRV-VIGS vector, and the expression of GmNHX1 was examined using RT-qPCR after gene silencing. The result showed a satisfactory silencing efficiency after a routine period of TRV-VIGS in soybean plants. Compared to EV (unsilenced plants), TRV vector carrying GmNHX1 specific fragment reduced the expression level of GmNHX1 in infected soybeans by nearly 50% (Fig. 1b). As a result of the silencing of GmNHX1, the top of the plant drooped and the leaves wilted after 200 mM NaCl solution treatment compared to EV plant (Fig. 1c), suggesting that GmNHX1 is a critical gene that is involved in the process of plant adaptation to salt stress.
GmNHX1 enhances salt tolerance in Arabidopsis
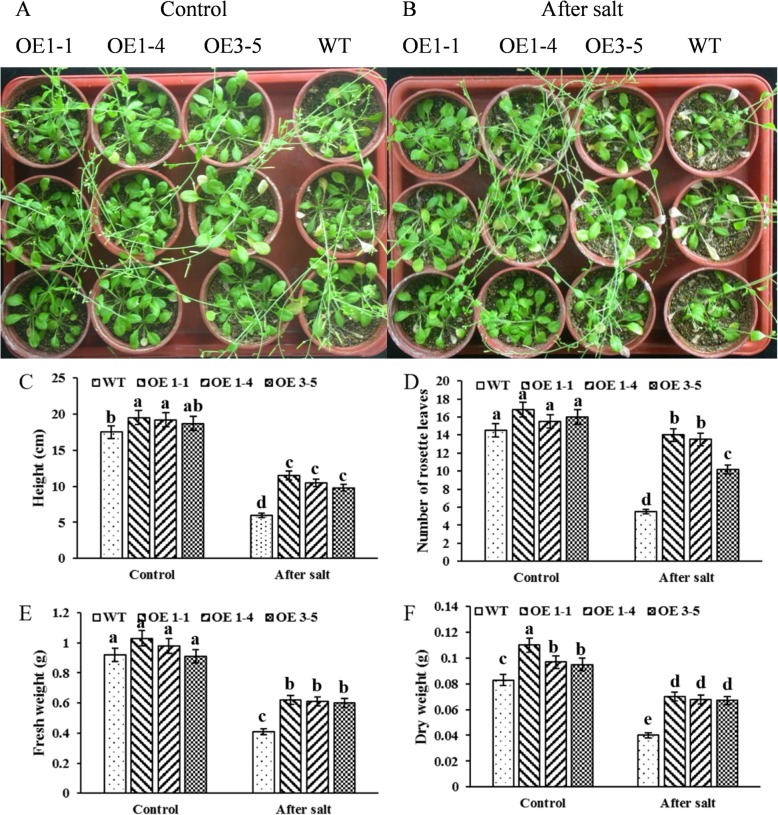
Since the silence of GmNHX1 reduces salt stress resistance in soybean, we wonder if exogenous expression of this gene could cause the opposite. We constructed the coding sequence of GmNHX1 gene into a T-DNA within which the GmNHX1 expression is driven by a CaMV 35S promoter, and the construct was then transformed into Arabidopsis Col-0. The expression of GmNHX1 in transformed plants was verified by RT-PCR. We observed significant expression of the gene in transformed plants but undetectable level of the expression in untransformed col-0 (Fig. 2). In order to investigate whether GmNHX1 overexpression is able to enhance salt tolerance, 21-day-old Arabidopsis plants expressing GmNHX1 were irrigated with 170 mM NaCl solution for 20 days, and plants that were irrigated with water were set as control. Wilting and chlorosis phenotypes were observed in wild-type plants after salt stress treatment, whereas only slight chlorosis was observed in all three lines that express GmNHX1 exogenously (Fig. 3a, b). Parameters such as stem length, number of rosette leaves, fresh weight, and dry weight were also measured. Plants expressing GmNHX1 showed significantly higher values in the measurements of stem length (Fig. 3c), rosette leaves (Fig. 3d), fresh weight (Fig. 3e) and dry weight (Fig. 3f), compared to wild-type under salt stress treatment. Taken together, we concluded that GmNHX1 enhanced plant resistance to salt stress condition.
Fig. 2.
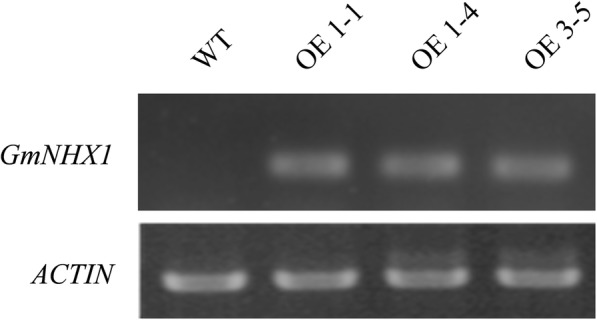
Expression analysis of GmNHX1 in transformed Arabidopsis. Using ACTIN as reference gene, RT-PCR was performed to detect the expression of GmNHX1
Fig. 3.
Effect of salt stress treatment on Arabidopsis expressing GmNHX1. a, b Arabidopsis phenotype before and after salt treatment; Statistical analysis was performed in measurements of stem length (c), number of rosette leaves (d), fresh weight (e) and dry weight (f) before and after salt treatment. The data shows the mean ± S.E. of triplicate experiments. Columns with different letters indicate significant differences at P < 0.05
GmNHX1 enhances salt tolerance through maintaining K+/Na+ ratio in root
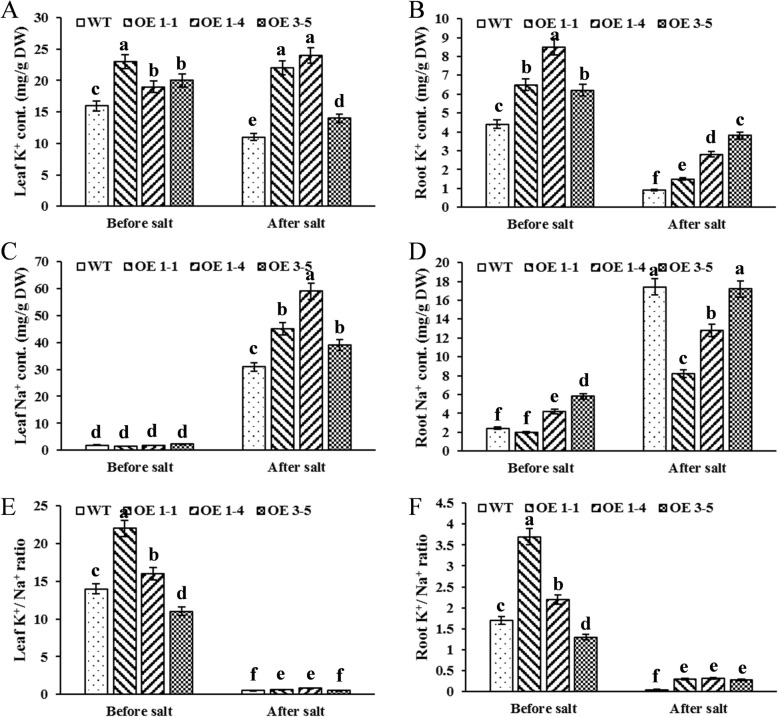
After revealing the positive role of GmNHX1 in salt resistance, we wondered the molecular mechanism underlying the GmNHX1-mediated salt resistance. GmNHX1 has a typical NHX domain, which encodes a functional unit that pumps Na+ against its concentration pressure in exchange for a proton. We therefore measured K+ and Na+ content in GmNHX1 expressing plants before and after salt stress treatment. K+ content in all three GmNHX1 expressing lines were significantly higher in roots and leaves both before and after salt stress treatment (Fig. 4a, b). As for Na+ content, all the three lines expressing GmNHX1 had no significant difference compared to wild-type plants in leaves before salt stress treatment (Fig. 4c), however all the three lines showed higher Na+ level in leaves after salt stress treatment than wild-type (Fig. 4c). In the meantime, Na+ content was significantly lower after salt stress treatment in roots of two of the three lines which express GmNHX1 (OE 1–1 and OE 1–4) compared to wild-type plants (Fig. 4d). These results suggested that exogenous expression of GmNHX1 might elevate Na+ transportation to leaves and reduce Na+ content in roots.
Fig. 4.
Concentration of Na+ and K+ in Arabidopsis before and after Salt treatment. Total concentration of K+ (a, b) and Na+ (c, d) was measured within the leaf (a, c) and root tissue (b, d). Relative K+/Na+ ratio in the leaf (e) and root tissue (f) was calculated respectively. The data shows the mean ± S.E. of triplicate experiments. Columns with different letters indicate significant differences at P < 0.05
The maintenance of a high K+/Na+ ratio within plant cells is one of the key factors that mediate salt tolerance in plants [16], especially for the root tissue. Compared to K+/Na+ ratio in leaves (Fig. 4e), K+/Na+ ratio in roots is significantly higher in all three GmNHX1 expressing lines compared to wild-type after salt stress treatment (Fig. 4f). GmNHX1 therefore contributes to the high K+/Na+ ratio in roots, in accordance with our results above.
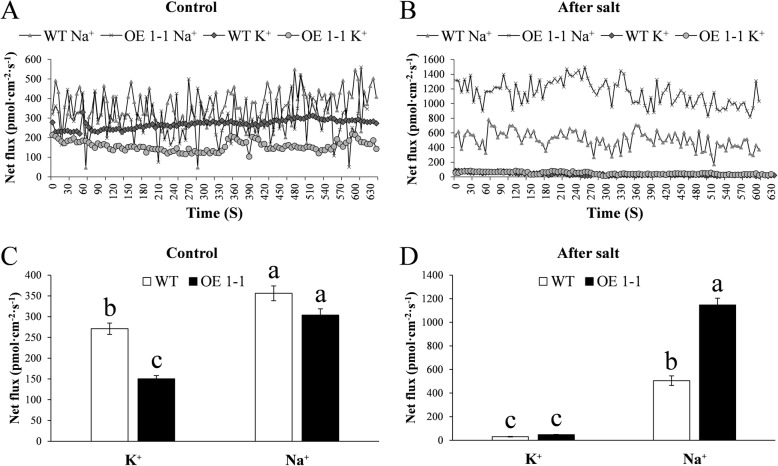
Vacuolar membrane-localized GmNHX1 regulates K+ and Na+ efflux
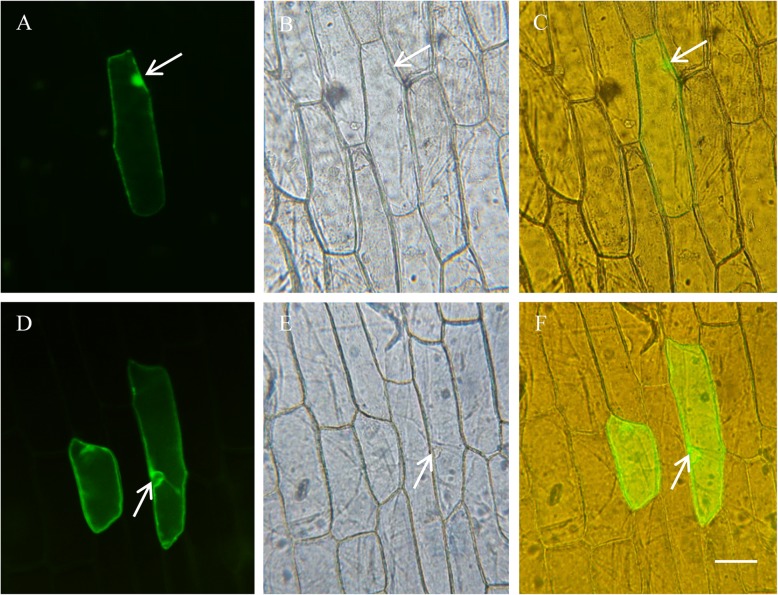
Plants overcome salt stress by means of regulating Na+ efflux and the partitioning of Na+ into vacuolar [2]. Given the observations above, we wondered whether GmNHX1 regulated salt tolerance via adjusting the efflux of these two ion molecules, or modifying the cellular ion storage. To address this question, subcellular localization of GmNHX1 was examined. An eGFP was fused to the C-terminal of GmNHX1 and the resulted fusion protein was constitutively expressed under the control of a CaMV 35S promoter. Transformed onion bulb endothelium cells showed a pattern of GFP fluorescence in which signals were enriched in the vacuolar membranes (Fig. 5). To explore the absorption law of Na+ and K+ under salt stress in Arabidopsis alleles expressing GmNHX1, we used NMT to detect the flow changes of Na+ and K+ after 25 h of 100 mM NaCl treatment. The result showed that the efflux of K+ in transformed plants were significantly fewer than that of wild-type, with no significant difference for Na+ efflux between those before and after salt treatment. The K+ efflux of both transformed and wild-type Arabidopsis after salt treatment were fewer than that of before salt treatment, but the rate of Na+ efflux of transformed plants was greater than that of wild-type. The rate of Na+ efflux of both transformed and wild-type Arabidopsis after salt treatment was significantly greater than that of before salt treatment, and the rate of Na+ efflux of transformed plants after salt treatment was significantly higher than that of wild-type (Fig. 6). This result suggested that Arabidopsis plants expressing GmNHX1, a vacuolar membrane-localized protein, maintain K+/Na+ ratio via elevating Na+ efflux rate in roots, along with reducing Na+ accumulation, which thereby avoiding the toxic effects of excessive salt in cells.
Fig. 5.
Subcellular localization of GmNHX1. Onion bulb endothelium cells expressing eGFP (a-c) and GmNHX1-eGFP (d-f) was analyzed under fluorescent microscopy, images were acquired using the 488 nM excitation (a, d) and light (b, e). Superimposed images were generated in c and f, respectively. Arrows indicate the nucleus regions. Bar = 100 μm
Fig. 6.
K+ and Na+ fluxes in root tissues of Arabidopsis before and after Salt treatment. Plant materials were subjected to 100 mM NaCl treatment for 24 h. K+ and Na+ fluxes in roots of wild-type and transformed plants without (a) or with (b) salt stress treatment are shown. The mean fluxes of K+ and Na+ without or with salt stress treatment were shown in c and d, respectively. The data shows the mean ± S.E. of triplicate experiments. Columns with different letters indicate significant differences at P < 0.05
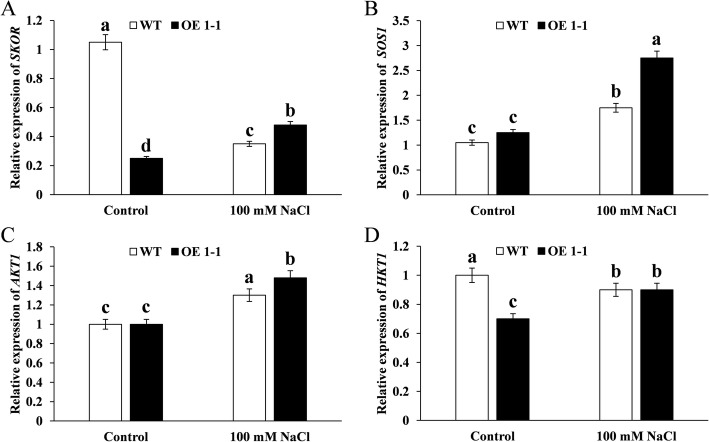
GmNHX1 regulates stress responsive genes
The seemingly contradictory observations above, in which GmNHX1 localized in vacuolar membranes whereas it was able to mediate Na+ efflux, prompted us to wonder if there were other mechanisms involved. The process of salt stress defense in plants relies on regulation of a series of stress responsive genes [17]. To reveal the relationship between GmNHX1 and stress responsive genes, we employed RT-qPCR to quantify the expression of AKT1, HKT1, SOS1 and SKOR, which are closely related to salt stress response [2]. Before salt stress treatment, the expression of SKOR and HKT1 in transformed plants were significantly lower than those of wild-type, whereas SOS1, AKT1 showed no significant difference between the two (Fig. 7). Salt stress treatment elevated the expression of SOS1 and AKT1, and reduced the expression of SKOR and HKT1. After salt treatment, GmNHX1 upregulated the expression of SKOR, SOS1 and AKT1, but not HKT1.
Fig. 7.
Expression of stress responsive genes in GmNHX1-expressing Arabidopsis. Arabidopsis expressing GmNHX1 and wild-type plants were treated with 100 mM NaCl for 24 h, relative expression of SKOR (a), SOS1 (b), AKT1 (c) and HKT1 (d) were detected using RT-qPCR. The data shows the mean ± S.E. of triplicate experiments. Columns with different letters indicate significant differences at P < 0.05
Discussion
Na+/H+ exchangers (NHX) function as regulators of intracellular ion homeostasis, mainly by increasing Na+ excretion of cells (such as SOS1) [18] or compartmenting Na+ into vacuolar, such as AtNHX1, to improve plant salt tolerance [19]. Apse et al. suggested that AtNHX1 is critical to the resistance to salt stress in plants [20]. Since GmNHX1 possesses classic sodium/hydrogen exchanger (NHX) features in its sequence, we therefore wonder if GmNHX1 is also related to salt stress resistance. In this work, we use VIGS to investigate the function of GmNHX1 in soybeans under salt stress condition. VIGS is a fast, simple and reliable approach that has been used in many functional biology studies, yet is still limited due to the difficulty in finding compatible plant virus stains [21]. TRV (Tobacco rattle virus)-mediated VIGS has been widely used in many functional biology studies [22], and is applicable in soybeans according to Liu et al. [23]. We found that the salt resistance has been reduced in GmNHX1-silenced plants, suggesting that this gene is closely related to plant salt stress resistance. Previous studies suggest that heterologous expression of chrysanthemum DgNHX1 is able to improve salt tolerance in tobacco, causing an increase of Na+ and K+ accumulation in tobacco leaves [24]. ZxNHX1 and ZxVP1–1 could increase the salt and drought resistance of roots, as well as increase the accumulation of Na+, K+ and Ca2+ in leaves [25]. Similarly, Heterologous expression of salt-tolerant plant hippocampus SbNHX1 can improve salt tolerance of Jatropha curcas, increase Na+ content and decrease K+ content in leaves when exposed to 200 mM NaCl [26]. Arachis hypogaea plants expressing AtNHX1 are resistant to drought, and the content of Na+ and K+ in leaves are increased [27]. Stress-inducible expression of TaNHX2 significantly improves growth performance as well as Na+ and K+ content from the leaf and root tissue of T2 transgenic eggplants (Solanum melongena L.) under salt stress, compared to non-transformed plants [28]. NHX in sweet sorghum is mainly involved in the transportation of Na+, facilitating Na+ homeostasis in response to the increase of salt concentration [29]. Our work showed that the contents of Na+ and K+ in roots have been significantly increased and the K+/Na+ ratio also increases significantly in plants expressing GmNHX1 under salt stress, in accordance with these reports.
Previous studies have shown that SOS1, membrane Na+/H+ exchanger protein, mediates the efflux of Na+ in roots [30], previous research shown that the high efficiency K+ channel protein HKT1 is located in the membrane, which plays an important role in maintaining the homeostasis of K+ and Na+ on the aboveground part of the plant [31–33]. SOS1 and HKT1 are located in the membrane, which play key role in regulating K+ absorbance from soil to the root cells, and the K+ and Na+ homeostasis [34]. AKT1 encodes an internal rectifier K+ channel protein, mainly regulates internal K+ flow into the root cells [35]. SKOR as an external rectifier K+ channel protein mainly involved in K+ loading from the column cell to the xylem [36]. Yuan et al. [37] prove that the ZxNHX1 regulates the whole plant K+/ Na+ homeostasis, and the expression of ion transport protein genes such as SKOR, SOS1, AKT1 and HKT1 were significantly down-regulated. In the NMT result, the net flux rate of K+ in transformed plants under normal conditions is significantly lower than that in the WT plants, consistent with the RT-qPCR results that GmNHX1 down-regulated the expression of SKOR and HKT1 genes in Arabidopsis under normal conditions, but had little effect on SOS1 and AKT1. Salt stress induced the expression of SOS1 and AKT1, and decreased the expression of SKOR and HKT1. Under salt stress, compared with WT, SKOR, SOS1 and AKT1 in GmNHX1 transformed plants increased significantly, but not HKT1, thich may explain why efflux of Na+ in the transformed plants after salt treatment is significantly greater than that in the WT plants.
We show that GmNHX1 overexpression enhances salt tolerance of Arabidopsis. We speculate that, on one hand, by increasing the efflux of Na+ in root cells, Na+ content is consequently reduced in roots and the K+/Na+ ratio increases; on the other hand, the Na+ is transported to leaves through the xylem, accumulating in the vacuolar in leaves, and K+/Na+ ratio is maintained stable. When under salt stress, the efflux rate of Na+ in roots is greatly increased, and the possible reason is that the expression of intermembrane Na+/H+ exchanger SOS1 is induced by GmNHX1. Our results further show that the salt tolerance of the transgenic Arabidopsis is mainly achieved by regulating the Na+ distribution in plants.
Conclusions
In summary, soybean Na+/H+ exchanger GmNHX1 responds to and regulates plant tolerance to salt stress. In transformed Arabidopsis which expresses GmNHX1, GmNHX1 changes the flow rate of K+ and Na+ in root cells by altering the expression of SKOR and SOS1, in order to regulate the accumulation of K+ and Na+ in roots and leaves, as well as the maintenance of a high K+ /Na+ ratio in roots, together improve the tolerance to salt stress in plants.
Methods
Cultivation and salt treatment of plant materials
Soybean cv. Jidou-7 was obtained from the Institute of Grain and Oil Crops, Hebei Academy of Agricultural and Forestry Sciences, and was cultivated in a greenhouse with a 14 h light/10 h dark cycle at a constant temperature of 25 °C and 700 μmol photons m− 2 s− 1. The 10-day-old soybean seedlings were transferred to Hoagland nutrient solution for 24 h, then transferred to Hoagland nutrient solution containing 170 mM NaCl, and sampled at 0 h, 6 h, 12 h, 24 h and 48 h, before RT-qPCR analysis. Arabidopsis ecotype Col-0 was obtained from the Arabidopsis Biological Resource Center (ABRC; http://abrc.osu.edu). The wild-type and transformed Arabidopsis seeds were surface sterilized and planted on MS medium, then transferred to vermiculite for 20 days and watered in Hoagland nutrient solution. For salt treatment, 21-day-old Arabidopsis plants were irrigated with 170 mM NaCl solution, biomass measurement including plant height, rosette number, fresh weight and dry weight, measurement of K+ and Na+ content and expression quantitation of salt stress related genes were performed 20 days after salt stress treatment initiation.
Virus induced gene silence
A specific fragment of GmNHX1 was amplified using primer pair GmNHX1-F and GmNHX1-R (Table 1), with PrimeSTAR HS DNA Polymerase (TaKaRa). The virus vector that is used to silence GmNHX1, pTRV2-GmNHX1, was constructed by inserting the amplified fragment of GmNHX1 into pTRV2 vector between BamH I and Kpn I recognition sites. TRV-VIGS was performed according to the previous report [22]. After infection, soybean seedlings were treated with 170 mM NaCl solution for 24 h, then GmNHX1 silencing efficiency was determined by RT-qPCR.
Table 1.
Synthetic DNA oligo used in this research
| Oligo name | Sequence (5`- 3`) | Application |
|---|---|---|
| GmNHX1-F | acgttgcacgggatcccccttcatgccatgggaca | Construction of TRV induced GmNHX1 silencing vector. |
| GmNHX1-R | ctagctagggggtacctccagaggaccaacatccaac | |
| RT GmNHX1 F | actgcgaagcaatgcaatca | Detection of transcriptional level of GmNHX1 and using RT-PCR. |
| RT GmNHX1 R | ggccattacgttcagttggtg | |
| RT ACTIN F | atggctgatggtgaagacattc | |
| RT ACTIN R | tccatgctcaatagggtacttg | |
| OE GmNHX1 F | ggtaccatggtttttgaaatcagttc | Construction of binary vector pCAMBIA1300-GmNHX1. |
| OE GmNHX1 R | tctagatcaacgccattgatggcca | |
| GFP GmNHX1 F | tgcccatgggacaaaatggtttttgaaatc | Construction of GFP fused vector pCAMBIA1300-GmNHX1-GFP. |
| GFP GmNHX1 R | cgccccgggacgccattgatgg | |
| qRT AtSKOR F | accgaaacaaactcggtaggaa | Detection of transcriptional level of salt stress related genes using RT-qPCR. |
| qRT AtSKOR R | ttagcacggatagagacaggaatg | |
| qRT AtSOS1 F | gtgaagcaatcaagcggaaa | |
| qRT AtSOS1 R | tgcgaagaaggcgtagaaca | |
| qRT AtHKT1 F | gatttgtccccacgaatgaga | |
| qRT AtHKT1 R | caaaaccaagaagcaagggaac | |
| qRT AtAKT1 F | aaaggtctcactcatcaacaacga | |
| qRT AtAKT1 R | tcggcaaaagaggcaaaataag | |
| qRT ACTIN F | gcaccgccagagagaaaatac | |
| qRT ACTIN R | caccaccacgaaccagataaga |
RT-PCR and RT-qPCR
Total RNA was isolated from plant material using UNlQ-10 Column Trizol Total RNA Isolation Kit (Sangon), and reverse transcript with PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa). RT-PCR and RT-qPCR was performed using Ex Taq DNA polymerase (TaKaRa) and SYBR Premix DimerEraser (TaKaRa) according to user manual, respectively, using ACTIN as reference gene. Primers used in RT-PCR and RT-qPCR experiments have been listed in Table 1.
Arabidopsis transformation
Full length CDS of GmNHX1 was PCR amplified using primer pair OE GmNHX1 F/ R (Table 1), and constructed into binary vector pCAMBIA1300 between the restriction enzyme recognize site of Kpn I and Xba I, under control of CaMV 35S promoter.
In vivo measurement of K+ and Na+
Dried plant materials were ground into fine powder. Concentrated sulfuric acid was added to the ground powder, and the mixture was boiled at 170 °C for 20 min. A few drops of 30% hydrogen peroxide were added till a large amount of white smoke appeared, followed by digestion at 220 °C for 40 min, then 330 °C for 2 h. The content of Na+ and K+ ions were measured using a flame spectrophotometer (Sherwood M410).
Subcellular localization of GmNHX1
Full-length CDS of GmNHX1 was PCR amplified using primer pair GFP GmNHX1 F/ R (Table 1), and constructed into pCAMBIA1300-GFP, between the restriction enzyme recognize sites Nco I and Sma I, and was fused to the N` terminal of GFP, resulting a fusion protein that is expressed under the control of CaMV 35S promoter. Purified pCAMBIA1300-GmNHX1-GFP plasmid was bombarded with a particle gun (BioRad PDS- 1000/He). Transformed onion bulb endothelium cells were cultivated in 1/2 MS medium for 24 h, before analyzed under the fluorescence microscopy (Olympus BX53).
Measurement of Na+ and K+ flow rate
Fluxes of Na+ and K+ ion was measured using NMT. 7-day-old Arabidopsis seedlings were transferred to MS medium containing 100 mM NaCl, NMT test was performed by Xuyue (Beijing) Sci.& tech. co., ltd., in accordance with previous report [38].
Acknowledgements
We thank Clara Elizabeth of the Pacific Lutheran University, Tacoma, and Zhou-Liang YU of the University of California, Berkeley, for their help in polishing the language of this article.
Abbreviations
- hpt
Hours post treatment
- NHX
Na+/H+ exchanger
- NMT
Non-invasive micro-test technique
- PCR
Polymerase chain reaction
- TRV
Tobacco rattle virus
- VIGS
Virus-induced gene silencing
Authors’ contributions
TJS wrote this manuscript, TJS, LF and JY performed the experiment, RZC and CYY provided technical support, JZ and DMW conceived and designed the experiments. All authors have read and approved the final manuscript.
Funding
The sources of funding for this study are provided by China Ministry of Agriculture and Hebei Provincial Department of Science and Technology through the following projects:
(1) Major scientific and technological projects for breeding new varieties of genetically modified organisms (2009ZX08004-001B), Dong-Mei Wang;
(2) Major scientific and technological projects for breeding new varieties of genetically modified organisms (2014ZX0800402B-001), Dong-Mei Wang;
(3) Introducing Foreign Student Funding Program (CN201706), Jie Zhang.
The funding bodies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tian-Jie Sun and Long Fan contributed equally to this work.
Contributor Information
Tian-Jie Sun, Email: suntianjie91@163.com.
Long Fan, Email: 360704708@qq.com.
Jun Yang, Email: yjun0504@126.com.
Ren-Zhi Cao, Email: caora@plu.edu.
Chun-Yan Yang, Email: chyyang66@163.com.
Jie Zhang, Email: zhangjiezhaolh@163.com.
Dong-Mei Wang, Email: dongmeiwang63@126.com.
References
- 1.Xu Y. Envirotyping for deciphering environmental impacts on crop plants. Theor Appl Genet. 2016;129:653–673. doi: 10.1007/s00122-016-2691-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19:371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Bao H, Guo J, Jia W, Tai F, Nie L, et al. Na+/H+ exchanger 1 participates in tobacco disease defence against Phytophthora parasitica var. nicotianae by affecting vacuolar pH and priming the antioxidative system. J Exp Bot. 2014;65:6107–6122. doi: 10.1093/jxb/eru351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker DJ, Leigh RA, Miller AJ. Potassium homeostasis in vacuolate plant cells (cytosolic K+/cytosolic pH/plant vacuole) Plant Biol. 1996;93:10510–10514. doi: 10.1073/pnas.93.19.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassil E, Ohto M, Esumi T, Tajima H, Zhu Z, Cagnac O, et al. The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell. 2011;23:224–239. doi: 10.1105/tpc.110.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida DM, Margarida Oliveira M, Saibo NJM. Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet Mol Biol. 2017;40:326–345. doi: 10.1590/1678-4685-gmb-2016-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J, Sun J, Cao P, Ren L, Liu C, Chen S, et al. Variation in tissue Na+ content and the activity of SOS1 genes among two species and two related genera of Chrysanthemum. BMC Plant Biol. 2016;16:1–15. doi: 10.1186/s12870-015-0700-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassil E, Tajima H, Liang Y-C, Ohto M-A, Ushijima K, Nakano R, et al. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell. 2011;23:3482–3497. doi: 10.1105/tpc.111.089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reguera M, Bassil E, Tajima H, Wimmer M, Chanoca A, Otegui MS, et al. pH regulation by NHX-type Antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell. 2015;27:1200–1217. doi: 10.1105/tpc.114.135699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji W, Li Y, Li J, Dai CH, Wang X, Bai X, et al. Generation and analysis of expressed sequence tags from NaCl treated Glycine soja. BMC Plant Biol. 2006;6:4. doi: 10.1186/1471-2229-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Guo R, Jiao Y, Jin X, Zhang H, Shi L. Comparison of salt tolerance in Soja based on metabolomics of seedling roots. Front Plant Sci. 2017;8:1101. doi: 10.3389/fpls.2017.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ming LI, Zheng HU, Jiang QY, et al. GmNAC15 overexpression in hairy roots enhances salt tolerance in soybean. J Integr Agric. 2018;17(3):530–538. doi: 10.1016/S2095-3119(17)61721-0. [DOI] [Google Scholar]
- 13.Chen Y, Chi Y, Meng Q, et al. GmSK1, an SKP1 homologue in soybean, is involved in the tolerance to salt and drought. Plant Physiol Biochem. 2018;127:25–31. doi: 10.1016/j.plaphy.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Ling-Shuang W, Qing-Shan C, Da-Wei X, et al. Overexpression of GmBIN2, a soybean glycogen synthase kinase 3 gene, enhances tolerance to salt and drought in transgenic Arabidopsis and soybean hairy roots. Sci Agric Sin. 2018;17(9):1959–1971. [Google Scholar]
- 15.Fan L, Sun T, Yang J, Zhang J, Wang D. Cloning and functional characterization of a vacular Na+/ H+ antiporter gene (GmNHX1) from soybean. J Agric Univ Hebei. 2015;38:7–12,24. [Google Scholar]
- 16.Li J, Bao S, Zhang Y, Ma X, Mishra-Knyrim M, Sun J, et al. Paxillus involutus strains MAJ and NAU mediate K (+)/Na (+) homeostasis in ectomycorrhizal Populus x canescens under sodium chloride stress. Plant Physiol. 2012;159:1771–1786. doi: 10.1104/pp.112.195370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villarino GH, Bombarely A, Giovannoni JJ, Scanlon MJ, Mattson NS. Transcriptomic analysis of Petunia hybrida in response to salt stress using high throughput RNA sequencing. PLoS One. 2014;9:1–13. doi: 10.1371/journal.pone.0094651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci U S A. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apse MP. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ Antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi T, Apse MP, Shi H, Blumwald E. Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc Natl Acad Sci U S A. 2003;100:12510–12515. doi: 10.1073/pnas.2034966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramegowda V, Mysore KS, Senthil-Kumar M. Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front Plant Sci. 2014;5:1–12. doi: 10.3389/fpls.2014.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senthil-Kumar M, Mysore KS. Tobacco rattle virus–based virus-induced gene silencing in Nicotiana benthamiana. Nat Protoc. 2014;9:1549–1562. doi: 10.1038/nprot.2014.092. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Liu N, Li F, Wu L, Zhang J, Wang D. Establishment of TRV-mediated transient gene-silencing system in soybean. Sci Agric Sin. 2015;48:2479–2486. [Google Scholar]
- 24.Liu QL, Xu KD, Zhong M, Pan YZ, Jiang BB, Liu GL, et al. Cloning and characterization of a novel vacuolar Na+/H + antiporter gene (DgNHX1) from chrysanthemum. PLoS One. 2013;8:1–7. doi: 10.1371/journal.pone.0083702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu G-Q, Feng R-J, Wang S-M, Wang C-M, Bao A-K, Wei L, et al. Co-expression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1–1 confers enhanced salinity tolerance in chimeric sugar beet (Beta vulgaris L.) Front Plant Sci. 2015;6:1–11. doi: 10.3389/fpls.2015.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jha B, Mishra A, Jha A, Joshi M. Developing transgenic Jatropha using the SbNHX1 gene from an extreme halophyte for cultivation in saline wasteland. PLoS One. 2013;8(8):e71136. doi: 10.1371/journal.pone.0071136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banjara Manoj, Zhu Longfu, Shen Guoxin, Payton Paxton, Zhang Hong. Expression of an Arabidopsis sodium/proton antiporter gene (AtNHX1) in peanut to improve salt tolerance. Plant Biotechnology Reports. 2011;6(1):59–67. doi: 10.1007/s11816-011-0200-5. [DOI] [Google Scholar]
- 28.Yarra R, Kirti PB. Expressing class I wheat NHX (TaNHX2) gene in eggplant (Solanum melongena L.) improves plant performance under saline condition. Funct Integr Genomics. 2019;19:541–554. doi: 10.1007/s10142-019-00656-5. [DOI] [PubMed] [Google Scholar]
- 29.Gu WT, Zhou LB, Liu RY, Jin WJ, Qu Y, et al. Synergistic responses of NHX, AKT1, and SOS1 in the control of Na+ homeostasis in sweet sorghum mutants induced by 12C6+-ion irradiation. Nucl Sci Tech. 2018;29:10. doi: 10.1007/s41365-017-0341-5. [DOI] [Google Scholar]
- 30.Yamaguchi T, Hamamoto S, Uozumi N. Sodium transport system in plant cells. Front Plant Sci. 2013;4:410. doi: 10.3389/fpls.2013.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus A, Tester M. The Na+ transporter AtHKT1;1 controls retrieval of Na + from the xylem in Arabidopsis. Plant Cell Environ. 2007;30:497–507. doi: 10.1111/j.1365-3040.2007.01637.x. [DOI] [PubMed] [Google Scholar]
- 32.Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003;22:2004–2014. doi: 10.1093/emboj/cdg207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002;531:157–161. doi: 10.1016/S0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- 34.Assaha DVM, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol. 2017;8:509. [DOI] [PMC free article] [PubMed]
- 35.Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, et al. A protein kinase, interacting with two Calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 36.Liu K, Li L, Luan S. Intracellular K+ sensing of SKOR, a shaker-type K+ channel from Arabidopsis. Plant J. 2006;46:260–268. doi: 10.1111/j.1365-313X.2006.02689.x. [DOI] [PubMed] [Google Scholar]
- 37.Yuan HJ, Ma Q, Wu GQ, Wang P, Hu J, Wang SM. ZxNHX controls Na+ and K+ homeostasis at the whole-plant level in Zygophyllum xanthoxylum through feedback regulation of the expression of genes involved in their transport. Ann Bot. 2015;115(3):495–507. doi: 10.1093/aob/mcu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J, Chen S, Dai S, Wang R, Li N, Shen X, et al. NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiol. 2008;149:1141–1153. doi: 10.1104/pp.108.129494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.