Abstract
Computational fluid dynamics (CFD) models are emerging tools for assisting in diagnostic assessment of cardiovascular disease. Recent advances in image segmentation have made subject-specific modelling of the cardiovascular system a feasible task, which is particularly important in the case of pulmonary hypertension, requiring a combination of invasive and non-invasive procedures for diagnosis. Uncertainty in image segmentation propagates to CFD model predictions, making the quantification of segmentation-induced uncertainty crucial for subject-specific models. This study quantifies the variability of one-dimensional CFD predictions by propagating the uncertainty of network geometry and connectivity to blood pressure and flow predictions. We analyse multiple segmentations of a single, excised mouse lung using different pre-segmentation parameters. A custom algorithm extracts vessel length, vessel radii and network connectivity for each segmented pulmonary network. Probability density functions are computed for vessel radius and length and then sampled to propagate uncertainties to haemodynamic predictions in a fixed network. In addition, we compute the uncertainty of model predictions to changes in network size and connectivity. Results show that variation in network connectivity is a larger contributor to haemodynamic uncertainty than vessel radius and length.
Keywords: haemodynamics, fluid dynamics, pulmonary circulation, uncertainty quantification, image segmentation
1. Introduction
Definitive diagnosis of pulmonary hypertension (PH), defined as a mean pulmonary arterial blood pressure greater than or equal to 25 mmHg, requires a series of medical tests including invasive right heart catheterization and non-invasive computed topography (CT) imaging of the heart and lungs [1]. Diagnostic protocols interpret each data source independently to make an ultimate decision about disease classification and severity [2], but recent studies [3,4] have proposed assimilation of haemodynamics and imaging data with computational fluid dynamics (CFD) modelling, providing insight into the structure and function of the pulmonary system.
Medical imaging and image segmentation have emerged as powerful non-invasive tools for disease diagnostics [5–7], providing an abundance of data for analysing the cardiovascular system under physiological and pathological conditions [1]. Advances in image segmentation include semi- and fully automated algorithms for geometric reconstruction of complex vascular regions [8,9]. However, inherent uncertainty is present as most image segmentation software require manual specification of the image intensity thresholds (pre-segmentation parameters) between background and foreground. For example, van Horssen et al. [10] showed that variation in image resolution affected the cumulative volume of a cast of the coronary arterial tree after segmentation. Rempfler et al. [11] compared segmentation algorithms on retinal images, showing that posterior probability estimates for foreground pixels varied with different segmentation techniques when compared with the true segmentation or so-called ground truth. In contrast with the aforementioned studies, in vivo images are only captured up to a finite resolution, which makes ground truth rendering impossible. These two studies quantified variability in segmented networks but did not investigate how this uncertainty affected pulsatile haemodynamics.
Haemodynamic predictions (e.g. cross-sectional averaged flow and pressure) in the pulmonary vasculature are often computed using either three-dimensional (3D) [12] or one-dimensional (1D) [3] CFD models. Three-dimensional models predict local flow patterns with more precision [4] but are computationally expensive, making it difficult to perform multiple forward model evaluations for uncertainty quantification, i.e. UQ [13]. For instance, Sankaran et al. [14] computed 3D CFD model sensitivity to coronary stenosis diameters, using surrogate model approximations to combat high computational cost. However, they did not account for possible changes in network connectivity nor for the uncertainty from the initial segmentations of the vasculature. By contrast, 1D models are more computationally efficient, reducing the need for surrogates and allowing for investigations into variability of network connectivity. Moreover, a recent study [15] of the coronary vasculature showed that 1D models attain similar haemodynamic predictions as 3D when using appropriate boundary conditions. Recent studies analysed 1D systemic arterial models [10,16] to understand how uncertainty in network structure impacts haemodynamics. Fossan et al. [17] devised an optimization strategy to determine the number of vessels needed to match haemodynamic predictions in the coronary arteries, and Huberts et al. [13] used polynomial chaos expansion to quantify the sensitivity of flow predictions to variations in vessel radius. In contrast with the systemic circulation, the pulmonary vasculature is more compliant, branches more rapidly and operates at a much lower mean pressure, indicating that results from the systemic circulation may not be valid for comparison.
The total uncertainty in the haemodynamic prediction is a combination of uncertainty in the model parameters and uncertainty from the modelling framework. As noted above, several previous studies have studied uncertainty with respect to prescribed haemodynamic parameters and the 1D approximation, but to our knowledge, this is the first known investigation of the impact of uncertainties in network reconstruction on CFD simulations in the pulmonary vasculature. Specifically, our study examines how pre-segmentation parameters impact estimated vessel radius, vessel length and network connectivity, and propagate this uncertainty to haemodynamic predictions in the pulmonary circulation. To do so, we analyse multiple segmentations of a microcomputed tomography (micro-CT) image from a mouse pulmonary arterial tree. We propagate this uncertainty using a 1D CFD model by constructing the model domain from each segmentation. Inverse UQ is performed by estimating probability density functions (PDFs) for vessel radii and length, and then propagating uncertainties (forward UQ) using Monte Carlo sampling. Uncertainty in haemodynamic predictions is quantified by analysing three sets of predictions (depicted in figure 1): (i) predictions using 25 segmented networks (total variation); (ii) predictions from a representative network with fixed connectivity when drawing realizations of length and radius perturbations (parameter variation); and (iii) predictions from the same representative network when geometric parameters are fixed, but connectivity and network size are varied (network variation). We argue that UQ is an essential component of the model analysis when computational models are integrated into clinical protocols. The animal dataset used here [18,19] serves as a preliminary step in understanding disease progression and has potential for extrapolation to human PH.
Figure 1.
Workflow for UQ of haemodynamics. Multiple segmentations are performed to construct the segmented networks (SNs), of which one network is selected as the represented network (RN). Inverse UQ is performed on the 25 SNs by estimating PDFs for vessel radius and length. The 25 SNs are used in model simulations to understand the total variation, while the PDFs for the vessel dimensions are used to propagate uncertainty in the parameter variation study. Lastly, we change the structure of the RN to understand the variation induced by network connectivity. Pressure and flow predictions are then compared from the three sources of variation.
2. Material and methods
2.1. Experimental data
This study uses existing micro-CT and haemodynamic data from two male C57BL6/J control mice aged 10–12 weeks. A detailed description of experimental protocols for the imaging and haemodynamic data can be found in Vanderpool et al. [18] and Tabima et al. [19], respectively. Briefly, haemodynamic data include a flow waveform ensembled over 20 cardiac cycles measured using an inline flow meter (Transonic Systems, Ithaca, NY, USA) in the main pulmonary artery (MPA). The imaging data are obtained after euthanization and inflation of the mouse lung at 17.2 mmHg. A cannula with outer diameter of 0.127 cm and inner diameter of 0.086 cm is attached to the MPA before 360° imaging and reconstruction to DICOM 3.0 files. Both procedures were approved by the University of Wisconsin-Madison Institutional Animal Care and Use Committee.
2.2. Image analysis
2.2.1. Image segmentation
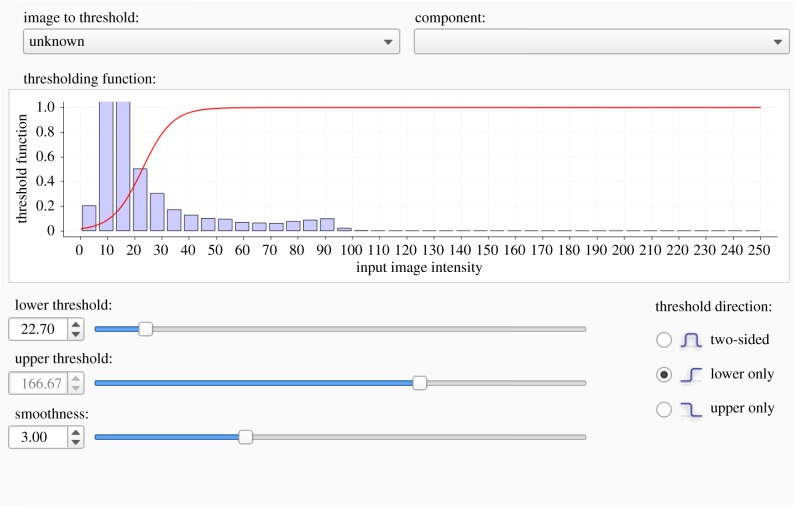
The micro-CT image is stored as a DICOM 3.0 file with voxel dimensions 497 × 497 × 497. The grey-scale image (shown in figure 4a) is transformed to a binary map identifying the vascular (foreground) and non-vascular (background) regions using global thresholding and image segmentation in ITK–SNAP [20]. Global thresholding is a pre-segmentation technique requiring a priori selection of thresholds to specify the image intensity bounds of the foreground. Threshold bounds are traditionally selected in an ad hoc manner to ensure that the foreground is captured [3,21,22]. In addition, ITK–SNAP requires specification of a smoothing parameter to determine the boundary between the foreground and background (figure 2). Due to the experimental protocol and use of perfused contrast, the image segmented in this study does not contain high-intensity voxels from other anatomical features (e.g. the veins or the heart) within the region of interest. Therefore, only the lower threshold (θ1) and smoothing (θ2) pre-segmentation parameters require specification.
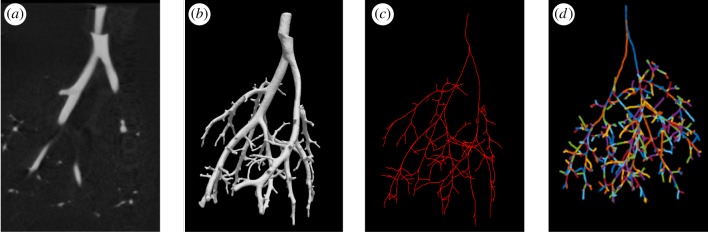
Figure 4.
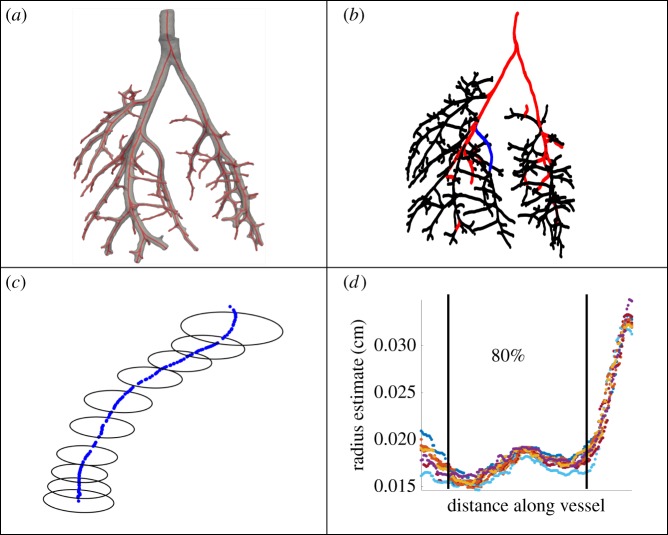
Image to network workflow. (a) The foreground visible in the image file; (b) the 3D rendering of the vascular foreground; (c) centrelines obtained using VMTK; (d) a graphical representation of the network used in the 1D model with vessels (edges) and bifurcations (nodes) identified using custom MATLAB algorithms. (Online version in colour.)
Figure 2.
ITK–SNAP interface for prescribing (θ1, θ2). Voxel intensities in the histogram are mapped to foreground and background based on thresholding function (red curve) and pre-segmentation parameters. Here, we only assume a lower threshold on image intensities, as shown by the constant value of 1 in the threshold function for all values greater than the lower threshold. (Online version in colour.)
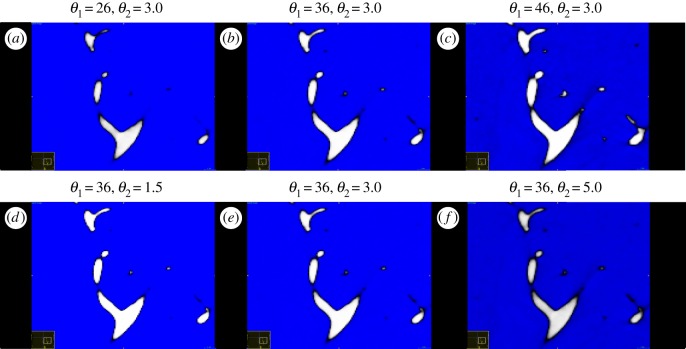
Acceptable intervals for (θ1, θ2) are determined to preserve the foreground for the large vessels across segmentations. To study segmentation-induced uncertainty, we assume a uniform distribution for the two parameters, with and , and draw 25 realizations of pre-segmentation parameter sets (θ1, θ2) (given in table 1) using the rand function in MATLAB (Mathworks, Natick, MA, USA). As shown in figure 3, the foreground for distal vascular segments changes significantly when (θ1, θ2) are varied, but maintains features for the large, proximal vessels.
Table 1.
Summary of pre-segmentation parameters and network features.
| pre-segmentation parameters (θ1, θ2) | number of vessels | number of generations | number of terminal vessels | total volume (cm3) |
|---|---|---|---|---|
| (22, 5.0) | 276 | 15 | 149 | 21.0871 |
| (25, 6.0) | 422 | 17 | 226 | 21.3407 |
| (26, 4.7) | 415 | 17 | 219 | 22.3524 |
| (26, 4.8) | 425 | 18 | 227 | 22.8591 |
| (26, 5.1) | 441 | 17 | 234 | 22.7031 |
| (27, 5.8) | 450 | 17 | 240 | 22.9599 |
| (28, 6.0) | 333 | 15 | 178 | 20.6542 |
| (30, 4.6) | 428 | 16 | 230 | 21.7283 |
| (30, 5.7) | 461 | 17 | 245 | 23.0039 |
| (30, 6.5) | 476 | 18 | 252 | 23.1922 |
| (30, 8.0) | 409 | 16 | 220 | 21.7642 |
| (31, 5.6) | 462 | 18 | 246 | 23.3346 |
| (31, 6.1) | 310 | 15 | 164 | 18.2311 |
| (32, 4.1) | 419 | 16 | 220 | 22.2851 |
| (33, 4.2) | 446 | 18 | 239 | 23.0664 |
| (33, 5.1) | 505 | 18 | 269 | 24.6089 |
| (34, 3.3) | 495 | 18 | 265 | 24.1804 |
| (34, 3.4) | 474 | 17 | 257 | 24.2923 |
| (35, 3.6) | 459 | 17 | 242 | 23.2488 |
| (35, 4.8) | 470 | 17 | 250 | 23.0868 |
| (35, 6.8) | 404 | 17 | 214 | 22.7536 |
| (36, 4.0) | 419 | 17 | 226 | 22.0391 |
| (36, 4.1) | 376 | 16 | 197 | 22.5833 |
| (37, 3.9) | 409 | 17 | 221 | 21.6596 |
| (44, 7.6) | 185 | 12 | 98 | 20.4368 |
Figure 3.
Qualitative differences in foreground (white) of distal vascular segments when changing the lower threshold (θ1) and the smoothing parameter (θ2). (a–c) Changes in foreground with θ1; (d–f) changes in foreground with θ2. (Online version in colour.)
We use active contour evolution, a semi-automated segmentation algorithm available in ITK–SNAP, to segment the micro-CT image (see the electronic supplementary material, §S.1). We consistently use 2000 iterations of the contour evolution, ensuring that the largest arteries carrying the majority of the blood volume are captured. The imaging protocol described in Vanderpool et al. [18] has a spatial resolution between 30 and 40 µm, providing a lower bound of 40 µm for the measurement uncertainty diameter (20 µm for radius).
2.2.2. Network reconstruction
Segmented geometries are exported as surface meshes and converted to VTK polygonal files using Paraview [23] (Kitware, Clifton Park, NY, USA). Surface mesh VTK files were imported into VMTK (www.vmtk.org) [6] to extract vessel segment centrelines, lengths and radii using native scripts. We developed a custom MATLAB algorithm to extract the network connectivity from the extracted quantities and identify all the vessels in each network. Subsequently, we use a recursive algorithm to construct a connectivity matrix identifying the geometry of the tree used in the 1D model. Further details of the reconstruction are given in the electronic supplementary material, §S.2. Figure 4 illustrates the workflow starting from the micro-CT image segmentation and ending with the connected network representation.
Voxels are converted to centimetres using a scaling factor based on the known diameter of the cannula (0.086 cm). The MPA radius is estimated using measurements distal to the cannula before the left pulmonary arterial (LPA) and right pulmonary arterial (RPA) bifurcation. Figure 5 shows an example network with radii estimates at each point along the network and within a single vessel. Measured values for radii vary within each vessel segment, limiting our inference of tapering. To proceed with calculations, we fix the vessel radius as the mean over the centre 80% of the individual estimates, which mitigates the impact of extreme diameters in the ostium regions at either end of each segment.
Figure 5.
Components of an arterial tree. (a) Three-dimensional segmentation of network; (b) centreline representation of a tree with the 32 vessel subset (red and blue); (c) magnification of the representative vessel in blue from (b) depicting radius estimates; (d) radius estimates along the representative vessel in (c), where the centre 80% of points are used to calculate the mean radius. Actual radius values obtained in (d) are calculated at orthogonal centreline slices in VMTK, while (c) shows non-orthogonal radii estimates for illustrative purposes. (Online version in colour.)
We construct connected graphs using the centreline data and create a connectivity matrix linking vessels, represented by their length and radius, and bifurcations. In addition, we capture global network features including the number of vessels, the number of bifurcations (i.e. generations) and the total vascular volume. The CFD model used for haemodynamics modelling assumes a binary structure, with each generation of the tree being formed by a new set of vessels. The resulting networks were found to have no loops within the region of interest studied here. To characterize branching properties, we compute morphometric indices for each network including structured tree scaling parameters, asymmetry and area ratios, and Murray's exponent (see the electronic supplementary material, §S.7, for details).
2.3. Haemodynamics modelling
2.3.1. Blood flow model
Similar to previous studies [3,24,25], we use a 1D CFD model to predict time-varying flow, pressure and area in each vessel. The model is derived under the assumptions that vessels are cylindrical, blood is incompressible, and flow is laminar, axisymmetric and Newtonian. The model equations are obtained by integrating over the cross-sectional area of the vessel, described in detail in [3]. Mass conservation and momentum balance are then given by
| 2.1 |
and
| 2.2 |
where x (cm) and t (s) denote the axial and temporal coordinates, A(x,t) (cm2) denotes the cross-sectional area, Q(x,t) (cm3 s−1) the volumetric flow rate, P(x,t) (mmHg) is the transmural blood pressure and R(x,t) (cm) the vessel radius. The blood density (g cm−3) and the kinematic viscosity (cm2 s−1) are assumed constant [26,27]. The right-hand side of equation (2.2) accounts for the frictional losses by assuming a flat velocity profile
| 2.3 |
with a linearly decreasing boundary layer with thickness (cm), where T (s) is the length of the cardiac cycle extracted from data [3,28]. To close the system of equations, we employ a constitutive law relating blood pressure and vessel cross-sectional area. We model vessels as thin walled, incompressible, homogeneous and orthotropic cylinders tethered in the longitudinal direction. Under these conditions, the linear stress–strain relation [3,29,30] is given by
| 2.4 |
where β = Eh/r0 = 37.5 mmHg denotes the arterial stiffness, E (mmHg) is Young's modulus in the circumferential direction, h (cm) is the wall thickness and (cm2) is the reference area obtained at the reference pressure P0 (mmHg).
The system (2.1)–(2.4) is solved using the two-step Lax–Wendroff finite difference scheme in C++ [24] with a temporal resolution of 1.3 × 10−5 (s) and a spatial resolution of 0.025 (cm), ensuring that the Courant–Friedrichs–Lewy (CFL) condition is not violated. To ensure the stability and convergence of the numerical scheme, the lengths of any vessels shorter than the spatial resolution of the solver are artificially augmented to be the size of one grid point.
2.3.2. Wave intensity analysis
Wave propagation in the pulmonary system can be characterized using wave intensity (WI) analysis [31]. Detailed derivations of WI quantities can be found in the electronic supplementary material, §S.3. Briefly, considering Q = AU, where U is the blood flow velocity (m s−1) in the vessel under the assumption of negligible frictional losses, we define the forward and backward components of WI as
| 2.5 |
where ‘+’ and ‘−’ indicate the direction of the local waves, and and are the associated pressure and velocity ‘wavefronts’ [3,32]. The wave separation depends on the local pulse wave velocity (PWV)
| 2.6 |
Local waves can either be compressive or decompressive [28,31]. Wave reflections are current clinical indicators for pulmonary vascular disease and PH progression and can be attributed to impedance mismatch at both proximal and distal vessel junctions [31,32].
2.3.3. Inflow, outflow and junction conditions
The system governed by equations (2.1)–(2.4) is hyperbolic with characteristics pointing in opposite directions, thus two boundary conditions are needed at each vessel inlet and outlet. At the network inlet (the MPA), we prescribe a measured flow waveform from a single cardiac cycle. At network bifurcations, we impose two conditions ensuring conservation of flow and a continuity of pressure
| 2.7 |
where the subscripts indicate the parent and daughter vessels and lp denotes the length of the parent vessel. Lastly, we impose a three-element Windkessel model at the outlet of terminal vessels [4] to characterize the downstream vasculature, which relates pressure and flow via an RCR circuit model
| 2.8 |
where R1 is the proximal resistance, R2 is the distal resistance and CT is the total compliance [29,33].
2.3.4. Parametrization
The haemodynamics modelling parameters include those describing the vascular structure (radius, length and stiffness), the fluid dynamics (including viscosity, density and the boundary layer thickness) and the inflow and outflow boundary conditions. We assume that inflow, viscosity, density and wall stiffness (β) are fixed and independent of the network geometry [3,28,34], while parameters specifying the vessel radius, length and Windkessel outflow boundary conditions depend on the network structure [3,17].
For each network, vessel radii and length are determined from the segmentation, while estimates are needed for Windkessel parameters. Similar to our previous study [3], we assume that the total compliance CT can be determined from the diastolic decay time constant , where is the total vascular resistance [3]. RT is computed as the ratio of mean pressure to mean flow, i.e. , and as discussed in our previous studies [3,30], a priori resistance values for each terminal vessel can be calculated using Poiseuille's equation, relating mean pressure and flow via the vessel dimensions. Both junction conditions in equation (2.7) are used together with Poiseuille's law to give the mean flow distribution relationship
| 2.9 |
where , consistent with Poiseuille's equation (see electronic supplementary material, §S.4, for details). Finally, we set and [3,30].
2.4. Inverse uncertainty quantification
We employ inverse UQ to estimate vessel length and radius PDFs over the 25 segmented networks. To compare measurements across segmentations, PDFs are computed for radius and length from a 32-vessel subset after data standardization. Two estimation techniques, kernel density estimation (KDE) and Gaussian process (GP) density estimation, are used to compare estimated PDFs. Weighted least-squares regression and GP regression are used to remedy the issues of non-constant variance, i.e. heteroscedasticity, in vessel dimensions.
2.4.1. Data standardization
A subset of 32 pulmonary vessels of various calibre (figure 5b) is selected from the 25 segmented networks. The 32 vessels are visible in all 25 networks and contain radius and length measurements that encompass the full range of measurements in the networks. Length and radius measurements are standardized using
| 2.10 |
where are the measured quantities from the ith vessel and jth segmentation, and and are the mean and the standard deviations of these quantities across the 25 networks.
2.4.2. Density estimation
KDE, a non-parametric technique [35], estimates the PDFs for radius and length. This technique requires specification of a bandwidth parameter, determining how influential each data point is in the density estimation. We consider both Silverman's rule of thumb [35] and maximum-likelihood leave-one-out cross-validation [36] for bandwidth estimation. These methods are compared to logistic GP density estimation [37] using the GP Stuff toolkit in MATLAB [38]. Due to space restrictions, the methodological details have been relegated to the electronic supplementary material, §S.5.
2.4.3. Statistical models for computing the length and radius variance
The PDFs constructed from the 32-vessel subset are representative of the overall variation in the length and radius across all the segmented networks. However, the magnitudes of and vary from vessel to vessel and need to be modelled explicitly before forward UQ. We use the coefficient of variation, , to compare these measurements' variability.
The statistical model relates the average measurements of radius and length across segmentations to their coefficient of variation. The variance of the measurements exhibits heteroscedasticity, as smaller vessel segments are more sensitive to pre-segmentation parameters leading to non-constant variance. This violates the assumptions of ordinary linear regression; hence, we consider weighted least-squares regression and GP regression with input-dependent noise [39]. Deterministic weighted least-squares regression iteratively fits regression models by updating weights for each data point. The optimal weights (optimal in a maximum-likelihood sense) are given by the inverse of the variance of the response [40]. Since this variance is unknown, we approximate the weights as , where is the residual from the unweighted regression model, reducing the impact of highly variable observations on the regression prediction. We consider exponential, logarithmic, square root and linear weighted least-squares regression models. For GP regression, we employ a GP for the response, , and for the latent variance of . We use the Matérn covariance function [41] with a smoothness parameter in the GP formulation (see electronic supplementary material, §S.6, for more details).
2.5. Forward uncertainty quantification
Forward UQ propagates model and parameter uncertainties to simulated quantities of interest. To analyse the posterior variation in model predictions, we pursue three sets of simulations determining (i) the total variation of haemodynamic predictions associated with segmentation, (ii) the variation to changes in vessel radius and length, and (iii) the variation to changes in network size and connectivity. The first set of simulations (i) use the 25 segmented networks, whereas the last two (ii–iii) are conducted in a representative network.
2.5.1. Total variation
We predict haemodynamics using each of the 25 segmented networks to quantify the total variation of flow and pressure predictions in the MPA, LPA and RPA. The observed variation is attributed to several sources of uncertainty, including the parameters of the model and the size and connectivity of the network. Once the total variation is calculated, we quantify the relative contributions from the parameter and network variation.
2.5.2. Representative network
A representative network is used to examine the variation in vessel radius and length as well as changes in network size and connectivity. We first compute the MPA pressure waveform for each of the 25 segmented networks, then we ensemble average these to determine a mean MPA pressure. The network with the smallest least-squares cost between its MPA pressure waveform and the ensemble-averaged waveform is designated as the representative network.
2.5.3. Parameter variation
As mentioned in §2.3, we assume that density, viscosity and vessel stiffness are constant while parameters impacted by image segmentation, including vessel length and radius, vary. The outflow boundary conditions are dependent on vessel length and radius; thereby, we analyse the variation in model predictions associated with changes in vessel dimensions.
We compute inverse cumulative distribution functions (CDFs) for the length and radius PDFs. The inverse CDF, , is a non-decreasing function defined on the interval [0,1] that provides values from the original PDF, allowing for inverse transform sampling for forward UQ [13]. Briefly, let u be a realization from a uniform distribution, , and define the realization from the inverse CDF as . There exists a mapping from the realization to the inverse CDF for the radius and the length via and ; hence, we draw samples from the inverse CDF to provide standardized measurements l* and r* for length and radius.
We define a mapping from the inverse CDF of in vessel i to the perturbed values (in units of cm). Writing and , where is the statistical model found from §2.4.3, we get
| 2.11 |
for each average measurement in vessel i. The values are used as the dimensions for each vessel in the 1D model when doing the forward UQ. We set , where are the original measurements from the representative network. To ensure the convergence of the posterior predictions [29], we draw M = 104 realizations using Monte Carlo sampling. The pseudo-algorithm for UQ propagation is as follows:
1. Draw a random sample .
2. Map the sample to and .
3. Perturb the nominal radius and length by using equation (2.11).
4. Run 1D CFD model with new radius and length values.
5. Repeat steps 1–4 M times.
2.5.4. Network variation
We simulate the effect of network truncation by iteratively eliminating terminal vessel pairs from the representative network. To do so, we calculate the total volume of each terminal vessel (i.e. ) and remove vessels with the smallest volume. Total resistance, total compliance and total mean flow in the network are conserved during this process, the details of which are in electronic supplementary material, §S.3.
3. Results
We analyse the total variation of flow and pressure predictions and identify the relative contributions from variations in vessel parameters versus variations in network size and connectivity. Variation in model predictions is quantified by comparing simulations in the MPA, LPA and RPA.
3.1. Network statistics
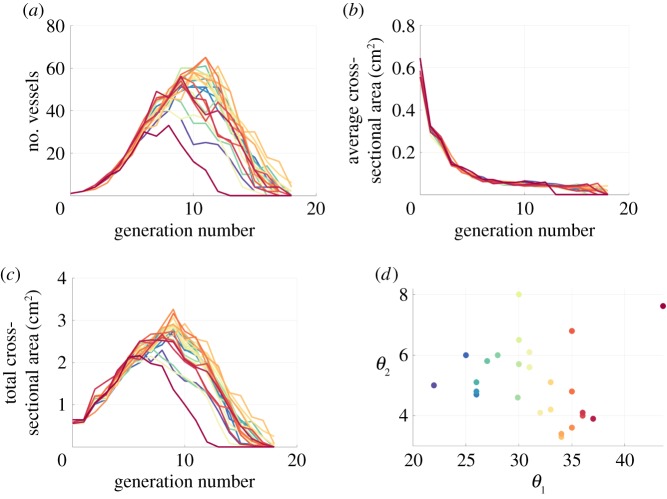
Figure 6 summarizes network characteristics obtained from the 25 segmented networks, including number of vessels (figure 6a), average cross-sectional area (figure 6b) and total cross-sectional area (figure 6c) in each generation. The average total number of vessels in each segmented network is 437 with a standard deviation of 76 and the mean number of generations across segmentations is approximately 17. The number of vessels and total cross-sectional area of the networks are consistent across segmentations up until the sixth generation, after which the results deviate. Most segmentations achieve a maximum number of vessels and cross-sectional area between generations 8 and 14, while the average cross-sectional area rapidly decreases until the fifth generation, and then remains fairly constant afterward. Analysis across all networks in figure 6d shows that one network (corresponding to ) is an outlier, having significantly fewer vessels and a lower total cross-sectional area. Table 1 includes all pre-segmentation parameter sets used in the repeated segmentations as well as network-level features. Results of calculating morphometric indices show that the parent to daughter area ratio is greater than 1 and that Murray's exponent is approximately 3 (see electronic supplementary material, §S.7), consistent with literature findings [30,42].
Figure 6.
Morphometric features from the 25 segmentations marked by different coloured lines. The number of vessels (a) is consistent between segmentations until the fifth generation. The average cross-sectional area (b) decreases rapidly after the first generation, while the total cross-sectional area (c) varies significantly between segmentations. The segmentation parameters are plotted against each other in (d), with a clear outlier present at (44, 7.6) (in pink) indicating a set of pre-segmentation parameters that have marked effects on the network structure. The outlier, located in the top right-hand corner of (d), has lower number of vessels and total cross-sectional area as depicted in the pink curve in (a,c). (Online version in colour.)
3.2. Inverse uncertainty quantification
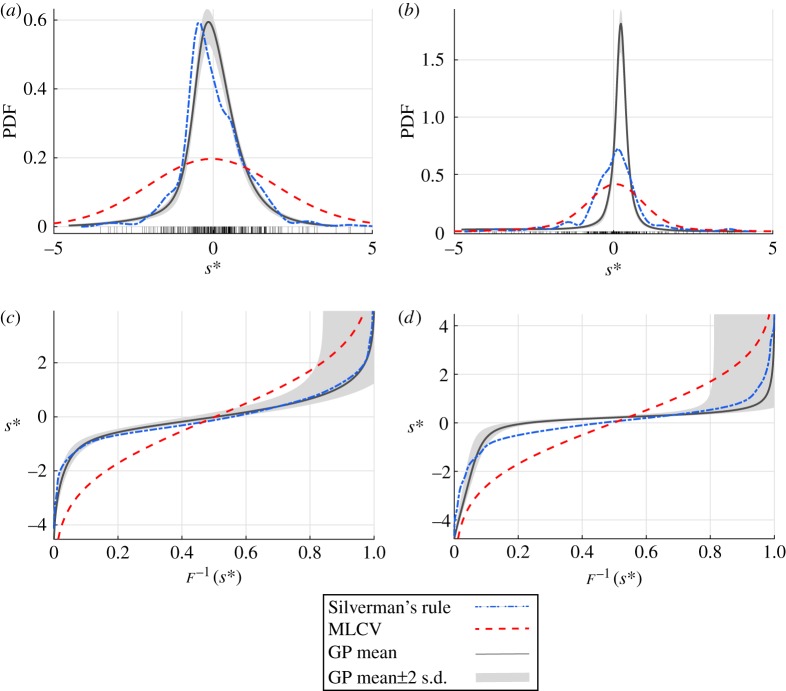
Figure 7 shows the length and radii PDF for the 32 representative vessels computed using Silverman's rule, maximum-likelihood cross-validation and GPs. Before density estimation, the standard deviation for each of the 32 vessels is used to normalize the data (see equation (2.7)). The maximum coefficient of variation is 21% for the radius and 49% for the length estimate. The bandwidth estimates for Silverman's rule are and while for the maximum-likelihood cross-validation, the estimated bandwidths are and for the length and radius densities, respectively. Computations using Silverman's rule exhibit overfitting, while the maximum-likelihood cross-validation over-smooths the density relative to the GP. In summary, the GP density estimation provides the most robust approximation for the PDF, which is therefore chosen in the forward uncertainty propagation in §3.3.
Figure 7.
Density estimates (a,b) and inverse CDFs (c,d) for the standardized radius and length values, respectively, measured in the 32-vessel subset. The bandwidth parameters used for the length and radius KDEs were determined using Silverman's rule (blue, dash-dot) and maximum-likelihood cross-validation (MLCV, red, dashed). The GP mean and 95% confidence interval are shown as a solid curve with grey bands. Standardized values are denoted by the black tick marks in (a) and (b). (Online version in colour.)
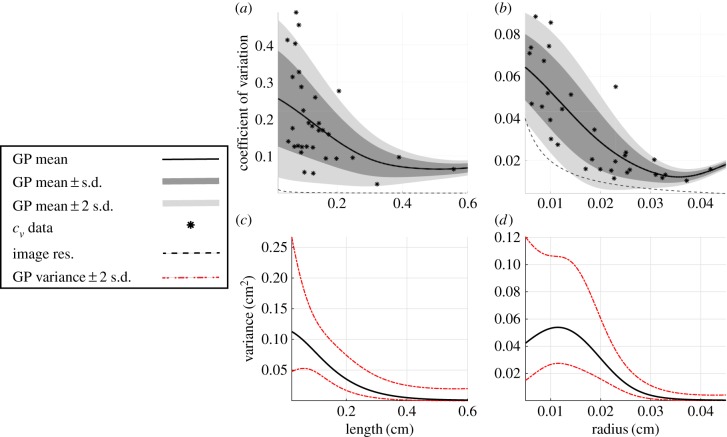
Weighted least squares with exponential, logarithmic, square root and linear regression functions are unable to resolve the heteroscedastic nature of the data (plots not shown). Instead, we use the GP regression model with input-dependent noise to estimate . Figure 8a,b shows the GP regression for and , respectively, while figure 8c,d depicts the latent variance. The coefficient of variation for vessel measurements across segmentations increases as vessels get smaller. The mean variance for increases as the length decreases, yet the mean variance of has a sharp decrease for the smallest vessels. Both GP models stay above the minimum variability of 20 µm.
Figure 8.
GP regression using non-constant variance for the relationship between length and radius and their coefficient of variation (cv). The GP means and standard deviations are computed from the cv data obtained from the 32-vessel subset (asterisks) and plotted against the analytical bound of the image resolution (black, dash-dot curve). The mean of the GPs and ±1 and 2 s.d. from the mean are shown in (a,b) in black, dark grey and light grey, respectively. The variance of the GPs in (c,d) are predicted using an additional GP and provide a mean (black) and variance (dashed curve) for the variance estimate. Both mean curves in (a,b) are above the uncertainty bound of the imaging protocol. (Online version in colour.)
3.3. Forward uncertainty quantification
The MPA flow data are used as an inflow boundary condition; hence, they do not change in any of the simulations. The ensemble-averaged pressure predictions in the MPA, LPA and RPA along with ±2 s.d. are shown in the first column of figure 9. The mean, systolic, diastolic and pulse pressure and max flow, min flow and total volume are given in table 2. The flow distribution to the LPA is much larger than the RPA, a consequence of the larger radius of the LPA. This deviation in flow is apparent in the WI plots in figure 10, showing a more complex series of wave reflections in the RPA than the LPA and MPA. The ensemble-averaged pressure waveform calculated from the 25 networks identifies the network generated by as the representative network.
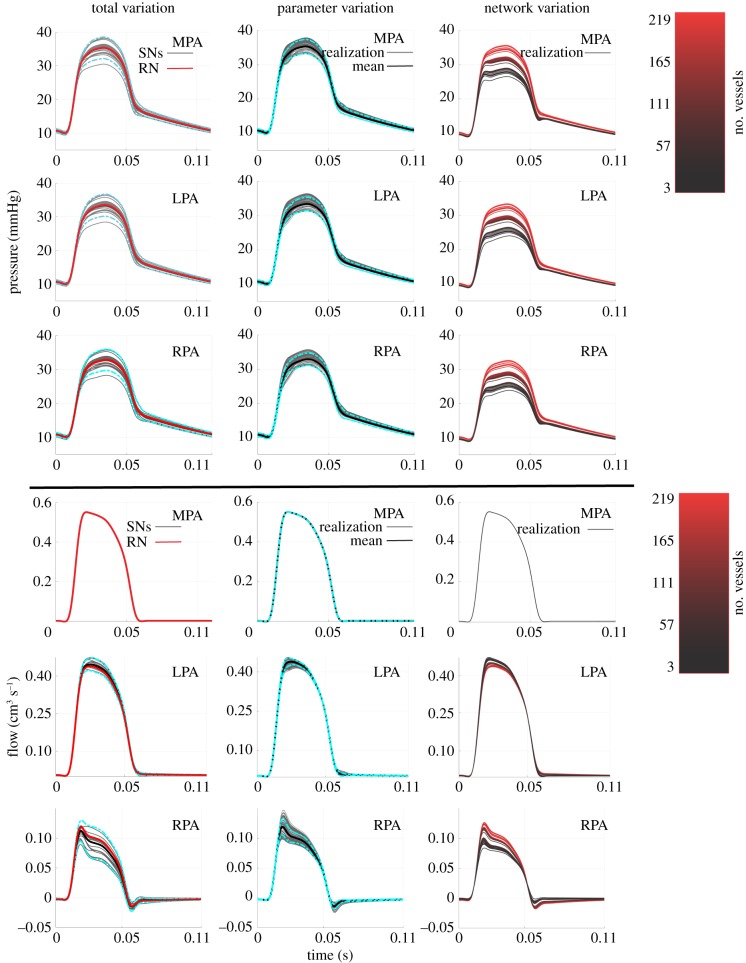
Figure 9.
Pressure and flow predictions in the first pulmonary bifurcation when studying total variation, parameter variation and network variation. Predictions from the total variation include simulations in the 25 segmented networks, the representative network (in red) and ±2 s.d. from the mean (cyan, dash-dot). The parameter variation plots (second column) show the 10 000 Monte Carlo realizations (grey) along with the mean (black) ±2 s.d. from the mean (cyan, dash-dot). Lastly, the network variation predictions (third column) show the predictions when using 219 vessels in the network (bright red) up until the network is reduced to the MPA, LPA and RPA (black). (Online version in colour.)
Table 2.
Results from simulations. Values are expressed as means ± s.d. Pressure values are in units of mmHg, flow values are in units of cm3 s−1 and volume values are in units of cm3.
| pressure |
||||
|---|---|---|---|---|
| mean pressure | systolic pressure | diastolic pressure | pulse pressure | |
| total variation | ||||
| MPA | 20.36 ± 0.78 | 35.35 ± 1.63 | 10.02 ± 0.27 | 25.33 ± 1.39 |
| LPA | 19.66 ± 0.79 | 33.46 ± 1.67 | 10.00 ± 0.27 | 23.45 ± 1.43 |
| RPA | 19.52 ± 0.78 | 32.83 ± 1.60 | 10.10 ± 0.28 | 22.74 ± 1.34 |
| parameter variation | ||||
| MPA | 20.38 ± 0.54 | 35.37 ± 1.03 | 10.04 ± 0.23 | 25.33 ± 0.82 |
| LPA | 19.68 ± 0.53 | 33.46 ± 0.99 | 10.02 ± 0.23 | 23.43 ± 0.78 |
| RPA | 19.56 ± 0.50 | 33.46 ± 0.90 | 10.11 ± 0.24 | 22.80 ± 0.69 |
| network variation | ||||
| MPA | 18.29 ± 0.84 | 31.70 ± 2.07 | 9.08 ± 0.18 | 22.63 ± 1.91 |
| LPA | 17.44 ± 0.86 | 29.34 ± 2.13 | 9.08 ± 0.17 | 20.27 ± 1.97 |
| RPA | 17.31 ± 0.83 | 28.71 ± 1.96 | 9.15 ± 0.20 | 19.56 ± 1.77 |
| flow |
||||
|---|---|---|---|---|
| mean flow | max flow | min flow | volume | |
| total variation | ||||
| LPA | 0.142 ± 0.004 | 0.447 ± 0.013 | −0.000 ± 0.000 | 0.016 ± 0.000 |
| RPA | 0.027 ± 0.004 | 0.113 ± 0.009 | −0.015 ± 0.004 | 0.003 ± 0.000 |
| parameter variation | ||||
| LPA | 0.140 ± 0.001 | 0.439 ± 0.006 | 0.000 ± 0.015 | 0.015 ± 0.000 |
| RPA | 0.029 ± 0.001 | 0.119 ± 0.007 | −0.014 ± 0.002 | 0.003 ± 0.000 |
| network variation | ||||
| LPA | 0.141 ± 0.001 | 0.447 ± 0.009 | −0.001 ± 0.001 | 0.016 ± 0.000 |
| RPA | 0.027 ± 0.001 | 0.009 ± 0.010 | −0.014 ± 0.004 | 0.003 ± 0.000 |
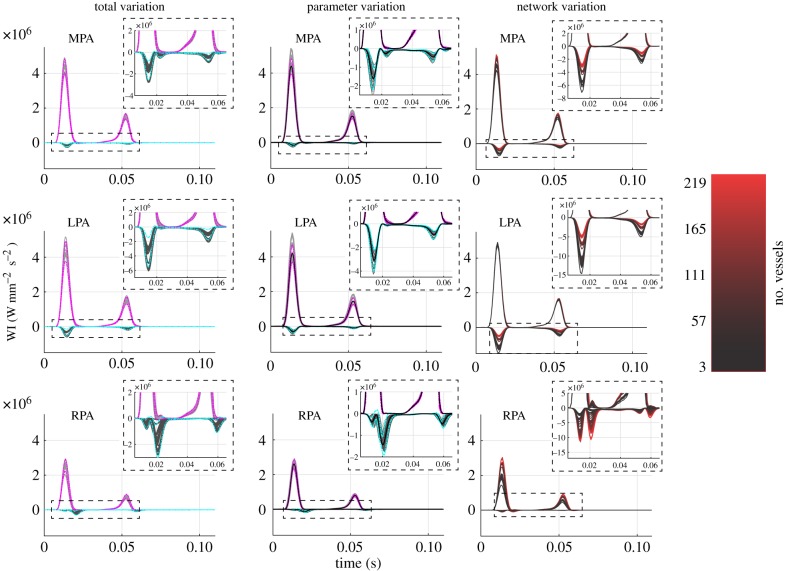
Figure 10.
WI analysis in the first pulmonary bifurcation. Forward (positive values) and backward (negative values) waves are calculated for the total variation and parameter variation (grey lines) along with ±2 s.d. from the mean (forward waves, magenta; backward waves, cyan). Predictions from the network variation study are colour coded according to network size, as described in figure 9. (Online version in colour.)
For the parameter variation component of the study, we use the inverse sampling methodology defined in §2.5.3 to propagate 104 realizations of perturbed radius and length values in the representative network. The second column of figure 9 shows the model predictions along with the mean and ±2 s.d. from the mean. The variation in the MPA, LPA and RPA systolic and pulse pressure is significantly larger than the mean and diastolic pressures (table 2). The flow predictions in the LPA and RPA have larger variability in the mean and max flow in comparison to the minimum flow. The variation attributed to network size and connectivity is calculated by fixing each vessel's radius and length in the representative network before reducing the full network iteratively. As described in §2.5.4, we reduce the network by starting at the smallest branches and moving towards the proximal vasculature while ensuring that Windkessel boundary conditions are adjusted for each simulation (figure 9, third column). Overall, reducing the number of vessels from 219 in the largest network to 3 in the smallest network introduces a discrepancy of approximately 10 mmHg in the pressure predictions of all three pulmonary arteries.
WI analysis results in figure 10 show that all vessels have two forward waves at the beginning and end of systole. The MPA and LPA have a pronounced backward wave during peak flow and the MPA and RPA have a secondary backward wave later in systole. The RPA also has an additional backward wave that occurs towards the end of diastole. The connectivity study in the last column of figure 10 shows that the MPA and LPA achieve their largest forward and backward WI value for the smallest network, while the opposite is true for the RPA. Time-averaged PWV in the MPA, as calculated using equation (2.6), was for the total variation, for the parameter variation and for the network variation. The mean PWV was 0.2% larger in the network variation study versus the total and parameter variation studies, a minimal relative difference.
4. Discussion
Recent advances in image segmentation have made subject-specific modelling of PH feasible, yet the modelling process still comprises segmentation-induced uncertainty that propagates through to simulation results. This is the first known study to explicitly quantify the variability of 1D CFD blood flow and pressure predictions arising from uncertainty in pre-segmentation parametrization. We investigate three types of segmentation-induced variations: the total variation arising from changes in pre-segmentation parameters, variation due to changes in vessel length and radius and variation with respect to network connectivity and size. Results suggest that variation in network structure is the greater contributor to uncertainty in haemodynamic predictions, consistent with what is known of the pulmonary vascular physiology. Moreover, the methodology developed herein can be used to generate and analyse a 1D model network for any vascular system.
4.1. Segmentation and construction of network graphs
Results show that pre-segmentation parameters drastically influence the number of vessels in the network, while the number of generations attainable remains relatively consistent. It is apparent that the network obtained from image segmentation is strongly linked to the range of image intensities considered in the foreground via choice of . Most notably, the segmentation parameter set (31, 6.1) gives a volume that is significantly smaller than networks with similar pre-segmentation parameters, suggesting that even slight changes in pre-segmentation parameters can reduce the number and size of small vessels captured. The largest vascular tree used in this study contains 500 vessels, a small fraction of the thousands of blood vessels that comprise the full pulmonary arterial system [4,9]. We expect the trends seen in figure 5 to continue if more vessels are obtained from the segmentation. Our techniques study uncertainty induced by global thresholding, but could be applied when other pre-segmentation techniques are used, as global thresholding is commonly used [3,21,22] but is only one of many segmentation methods.
The variability in the total number of vessels for a given network highlights the variation attributed to segmentation. This is expected in other networks that exhibit dispersive branching patterns, such as the coronary arteries [10] or cerebral vasculature [33]. We employed a generation-based ordering scheme to describe the branching structure, where each bifurcation is considered a new generation of blood vessels. By contrast, other authors [43] have used other ordering systems, e.g. Strahler [42] schemes, to identify structural properties of the pulmonary system, though these methods are not as compatible with CFD network models.
4.2. Inverse uncertainty quantification
KDEs and GPs are commonly used techniques [37,44], but this study is the first to use GPs in density estimation for vascular measurements. Forward UQ is typically carried out by assuming a parametric parameter distribution a priori, forcing prior assumptions on the unknown parameter distributions. By estimating the density directly from repeated measurements, we construct a non-parametric, representative density describing the uncertainty of the measurements across segmentations without prior assumptions.
The standardized measurements allow us to generalize the uncertainty of the 32-vessel subset to the entire vascular network, increasing the robustness of the density estimate. As shown in figure 8, the three density estimates are similar in the mode of the distribution (approx. 0); however, the GP density estimation allows for additional UQ in both the density and CDF estimates [37]. We construct marginal density estimates for the PDFs of radius and length, which assumes independence among the two quantities. PDF estimation methods that account for correlation between radius and length measurements should be investigated further.
GP regression is necessary for the data considered, as weighted least squares cannot correct the heteroscedastic variance. The coefficient of variation for the measurements increased as the measured dimensions decreased in value, suggesting that smaller vessels are subject to larger fluctuations in estimated dimensions when varying pre-segmentation parameters. Similar conclusions have been made in simulations predicting the fractional flow reserve in coronary crowns [10], as the smaller regions of the vasculature were susceptible to higher segmentation error. However, our work is the first to consider estimated, non-parametric densities for UQ propagation, and does not require a priori distribution assumptions.
4.3. Total variation of model simulations
The total network size obtained from the segmentation procedure has several effects on the model output. As shown in table 2, changes in network topology due to segmentation induced a variation in systolic pressure that was nearly six times larger than the variation of diastolic pressure. Moreover, we observe that the total variation standard deviation for the systolic and pulse pressure is larger in comparison to the mean and diastolic pressure. All four of these pressure metrics are typically used in diagnostic tools of diseases such as PH [2]. Though systolic pressure and pulse pressure have a small standard deviation (approx. 5% relative to the mean), studies investigating coronary-related mortality found that these pressure quantities were important for risk assessment in patients with congenital heart disease [45]. This further indicates a need for UQ when using these models for cardiovascular disease diagnostics and risk assessment.
4.4. Parameter variation
The standard deviation of diastolic pressure resulting from parameter (radius and length) variation is greater than that resulting from network (size and connectivity) variation. This suggests that changes in vessel dimensions and nominal boundary conditions can ultimately raise the diastolic pressure of the system, which is expected in the case of chronic vascular remodelling [2]. The standard deviation of the pulse pressure for the parameter variation only accounted for approximately 30% of the total variation value and had less of an effect on all other pressure and flow quantities when compared with the network variation. Larger networks encompassing the entire pulmonary tree will increase the parameter uncertainty, as they correspond to more vessels and more uncertain estimates of radius and length. This would in turn bias haemodynamic parameter estimates, since network predictions would be based on the initial segmentation results [14].
4.5. Network variation
The largest effects on pressure and flow waveform predictions in the network are attributed to changes in network connectivity and size, as seen in figures 9 and 10. Quantitative metrics provided in table 2 also show that network variation produces larger standard deviations in systolic and pulse pressures versus parameter variation, suggesting that the configuration of vessels in the pulmonary system may play an important role in haemodynamic predictions. It is known that the highly compliant pulmonary system uses its rapidly branching structure to perfuse the lung under a low-pressure gradient and varying cardiac outputs, and that network remodelling is common with pulmonary vascular disease [2,4]. For example, Rol et al. [43] concluded that changes in vessel diameter with PH cannot solely explain the increase in vascular resistance, and further hypothesized that network rarefaction may be a larger contributor to increased pulmonary arterial pressure. Olufsen et al. [25] investigated this computationally by altering structured tree boundary conditions, effectively reducing the size of the microvasculature and increasing pulmonary artery pressure. Our model analysis agrees with clinical hypotheses that vascular rarefaction and associated changes in network size and connectivity contribute more to changes in pulmonary arterial pressure than vessel narrowing.
The Poiseuille relation used to distribute network resistance introduces an impedance mismatch at each terminal vessel. Reflected pressure waves due to this mismatch become more prevalent as successive vessels are added to the system, leading to an increased pressure [16,46]. While other studies have considered non-reflective boundary conditions [15,47], it is hypothesized that wave reflections occur in the pulmonary system when PH is present [31], illustrating the appropriateness of reflective boundary conditions in the model.
Our results show three instances where reducing part of the network causes a larger change in pressure, which agree with a previous investigation by Epstein et al. [47] that showed a critical threshold in the number of vessels that lead to larger discrepancy in haemodynamic predictions. It is often the case that haemodynamic data are only available in select locations of the vascular system [3,29,30], a drawback when performing parameter estimation. Changes in network size will lead to changes in optimal parameter values during parameter inference, making the problem ill-posed as estimated parameters describing stiffness, compliance and vascular resistance will depend on the size of the network used in CFD simulations. This further indicates that uncertainty in the network structure must be taken into account when using 1D CFD models for clinical decision-making [30].
4.6. Limitations and future directions
Several potential limitations of this study can be addressed in future investigations. First, we assume negligible tapering in each vessel, which could play a role in proximal artery dynamics. Second, our model assumptions ignore radius-dependent stiffness and the effects of wall viscoelasticity, which may be important in pulmonary arteries [46] and could change the model sensitivity to network size and vessel dimensions. However, the focus of this study was to quantify how changes in the model domain attributed to pre-segmentation parameters impact haemodynamic predictions for fixed material parameters. Similar to previous studies [29,30,33], nominal parameters are calculated under the assumption of steady flow and that pressure does not drop in the large vessels. However, in the 1D domain, we solve nonlinear equations, which account for inertial and viscous losses resulting in a pressure drop along the vessels. Yet, no experimental studies have measured pressure beyond the first few generations; hence, validation of this assumption in the pulmonary circulation is difficult.
Additional limitations involved model construction and quantified results. The length of the smallest vessels was augmented during simulations to ensure the CFL condition is not violated. The CFL condition could also be met by either increasing the number of time-steps, which increases computational cost, or devising a numerical scheme using adaptive time-stepping, extending the scope of the study. We also provided conventional mean and standard deviation calculations as familiar metrics for comparison. An alternative approach is to perform formal global sensitivity analysis. State-of-the-art methods are based on Sobol indices defined via conditional variances of different order [13,14,30,43]; however, their computation via Monte Carlo or quasi-Monte Carlo simulations is computationally expensive. This computational complexity is aggravated by the fact that the image segmentation includes manual inspection and the parameter space can therefore only be sampled at a coarse level. A potential way to alleviate this problem is to use statistical emulation, e.g. using GPs, to compute first order and total effects indices. This can, in principle, follow the method described in [43], by adapting and extending existing approaches and software tools; see https://github.com/samcoveney/maGPy. However, this exploration is beyond the scope of the present study and provides an interesting direction for future research.
We consider the frequently used three-element Windkessel model as the boundary condition for the 1D model, yet this model greatly simplifies the physiological resistance beyond the segmented vessels. By contrast, structured tree boundary conditions [24,25,28] can provide an additional level of complexity for approximating downstream resistance and attempt to capture network structure beyond the limits of image segmentation. In addition, the experimental protocol inhibited the same mouse from being used for both the haemodynamic and imaging data. While this is a limitation for possible parameter inference, our methodology still captures variability in model predictions due to uncertainty in the vessel dimensions and network structure. Future human-based studies could incorporate non-invasive flow and imaging data from the same patient in the model. Finally, future subject-specific models of the pulmonary vasculature would be enhanced by allowing for trifurcations and considering branching angles in the vascular tree, thus accounting for more of the physiological traits of the network.
5. Conclusion
The uncertainty of model predictions must be accounted for in the absence of ground truth geometries and haemodynamic data. We have presented the first known investigation of the impact of uncertainties in imaging-based network reconstruction on CFD simulations in the pulmonary vasculature. This work identifies the uncertainties pertaining to image pre-segmentation parameters by explicitly measuring the variation in radius and length measurements of a subset of vascular segments. Another novelty of this work is in estimating densities of radius and length from data obtained using state-of-the-art non-parametric techniques, rather than assuming a fixed and potentially biased functional form of the distribution a priori. Moreover, our study is the first to perform UQ on the dimensions and network topology of a 1D CFD model in an expansive pulmonary vascular network. Our results show that the network variation has the most influence on predictions of blood pressure and flow, while changes in vessel length and radius have less impact on haemodynamic predictions.
Supplementary Material
Acknowledgements
We thank Dr Diana Tabima for her review and input on the experimental protocol.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material. Software/code implementing the developed methodology can be found at https://github.com/mjcolebank.
Authors' contributions
M.J.C. designed the study, developed algorithms, performed all haemodynamic simulations, pre- and post-processing of data, and drafted the manuscript. L.M.P. and D.H. carried out statistical analyses with GPs and helped revise the manuscript. N.C. designed the experimental protocol and gave clinical insight. M.U.Q., M.S.O and L.E.F conceived and coordinated the study and helped with revisions of the manuscript. All authors gave final approval for publication.
Competing interests
We have no competing interests.
Funding
This work was supported by the NSF-DMS (grant nos 1246991 and 1615820), the EPSRC (grant no. EP/N014642/1), the Royal Society of Edinburgh (grant no. 62335), the NIH (grant no. R01 HL-086939) and the American Heart Foundation (grant no. 19PRE34380459).
References
- 1.Lang IM, Plank C, Sadushi-Kolici R, Jakowitsch J, Klepetko W, Maurer G. 2010. Imaging in pulmonary hypertension. JACC Cardiovasc. Imaging 3, 1287–1295. ( 10.1016/j.jcmg.2010.09.013) [DOI] [PubMed] [Google Scholar]
- 2.Fukumoto Y. 2015. Diagnosis and treatment of pulmonary hypertension, vol. 101, pp. 311–319. Singapore: Springer. [Google Scholar]
- 3.Qureshi MU, Colebank MJ, Paun M, Ellwein L, Chesler N, Haider MA, Hill NA, Husmeier D, Olufsen MS. 2019. Hemodynamic assessment of pulmonary hypertension in mice: a model based analysis of the disease mechanism. Biomech. Model. Mechanobiol. 18, 219–243. ( 10.1007/s10237-018-1078-8) [DOI] [PubMed] [Google Scholar]
- 4.Tawhai MH, Clark AR, Burrowes KS. 2011. Computational models of the pulmonary circulation. Pulm. Circ. 1, 224–238. ( 10.4103/2045-8932.83452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidoiu V, Hadjilucas L, Teh I, Smith NP, Schneider JE, Lee J. 2016. Evaluation of noise removal algorithms for imaging and reconstruction of vascular networks using micro-CT. Biomed. Phys. Eng. Express 2, 045015 ( 10.1088/2057-1976/2/4/045015) [DOI] [Google Scholar]
- 6.Antiga L, Piccinelli M, Botti L, Ene-Iordache B, Remuzzi A, Steinman DA. 2008. An image-based modeling framework for patient-specific computational haemodynamics. Med. Biol. Eng. Comput. 46, 1097–1112. ( 10.1007/s11517-008-0420-1) [DOI] [PubMed] [Google Scholar]
- 7.Payer C, Pienn M, Bálint Z, Shekhovtsov A, Talakic E, Nagy E, Olschewski A, Olschewski H, Uschler M. 2016. Automated integer programming based separation of arteries and veins from thoracic CT images. Med. Image Anal. 34, 109–122. ( 10.1016/j.media.2016.05.002) [DOI] [PubMed] [Google Scholar]
- 8.Van Rikxoort EM, Van Ginneken B.. 2013. Automated segmentation of pulmonary structures in thoracic computed tomography scans. Phys. Med. Biol. 58, R187–R220. ( 10.1088/0031-9155/58/17/R187) [DOI] [PubMed] [Google Scholar]
- 9.Helmberger M, Pienn M, Urschler M, Kullnig P, Stollberger R, Kovacs G, Olschewski A, Olschewski H, Bálint Z. 2014. Quantification of tortuosity and fractal dimension of the lung vessels in pulmonary hypertension patients. PLoS ONE 9, e87515 ( 10.1371/journal.pone.0087515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Horssen P, van Lier MG, van den Wijngaard JP, VanBavel E, Hoefer IE, Spaan JA, Siebes M. 2016. Influence of segmented vessel size due to limited imaging resolution on coronary hyperemic flow prediction from arterial crown volume. Am. J. Physiol. 310, H839–H846. ( 10.1152/ajpheart.00728.2015) [DOI] [PubMed] [Google Scholar]
- 11.Rempfler M, Andres B, Menze BH. 2017. Uncertainty estimation in vascular networks. In Graphs in biomedical image analysis, computational anatomy and imaging genetics (eds M Cardoso et al.). Lecture Notes in Computer Science, vol. 10551, pp. 42–52. Cham, Switzerland: Springer. ( 10.1007/978-3-319-67675-3_5) [DOI]
- 12.Kheyfets VO, et al. 2015. Patient-specific computational modeling of blood flow in the pulmonary arterial circulation. Comput. Methods Programs Biomed. 120, 88–101. ( 10.1016/j.cmpb.2015.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huberts W, Donders WP, Delhaas T, van de Vosse FN.. 2014. Applicability of the polynomial chaos expansion method for personalization of a cardiovascular pulse wave propagation model. Int. J. Numer. Meth. Biomed. Eng. 30, 1679–1704. ( 10.1002/cnm.2695) [DOI] [PubMed] [Google Scholar]
- 14.Sankaran S, Grady L, Taylor CA. 2015. Fast computation of haemodynamic sensitivity to lumen segmentation uncertainty. IEEE Trans. Med. Imaging 34, 2562–2571. ( 10.1109/TMI.2015.2445777) [DOI] [PubMed] [Google Scholar]
- 15.Alastruey J, Xiao N, Fok H, Schaeffter T, Figueroa CA. 2016. On the impact of modelling assumptions in multi-scale, subject-specific models of aortic haemodynamics. J. R. Soc. Interface 13, 20160073 ( 10.1098/rsif.2016.0073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivolo S, et al. 2016. Impact of coronary bifurcation morphology on wave propagation. Am. J. Physiol. 311, H855–H870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fossan FE, Mariscal-Harana J, Alastruey J, Hellevik LR. 2018. Optimization of topological complexity for one-dimensional arterial blood flow models. J. R. Soc. Interface 15, 20180546 ( 10.1098/rsif.2018.0546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanderpool RR, Kim AR, Molthen RC, Chesler NC. 2010. Effects of acute Rho kinase inhibition on chronic hypoxia-induced changes in proximal and distal pulmonary arterial structure and function. J. Appl. Physiol. 1609, 188–198. ( 10.1152/japplphysiol.00533.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabima DM, Roldan-Alzate A, Wang Z, Hacker TA, Molthen RC, Chesler NC. 2012. Persistent vascular collagen accumulation alters haemodynamic recovery from chronic hypoxia. J. Biomech. 45, 799–804. ( 10.1016/j.jbiomech.2011.11.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. 2006. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–1128. ( 10.1016/j.neuroimage.2006.01.015) [DOI] [PubMed] [Google Scholar]
- 21.Newberry MG, Ennis DB, Savage VM. 2015. Testing foundations of biological scaling theory using automated measurements of vascular networks. PLoS Comput. Biol. 11, e1004455 ( 10.1371/journal.pcbi.1004455) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellwein LM, Marks DS, Migrino RQ, Foley WD, Sherman S, LaDisa JF. 2016. Image-based quantification of 3D morphology for bifurcations in the left coronary artery. Catheter Cardiovasc. Interv. 87, 1244–1255. ( 10.1002/ccd.26247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Utkarsh A. 2015. The Paraview guide: a parallel visualization application. Clifton Park, NY: Kitware, Inc. [Google Scholar]
- 24.Olufsen MS, Peskin CS, Kim WY, Pedersen EM, Nadim A, Larsen J. 2000. Numerical simulation and experimental validation of blood flow in arteries with structured-tree outflow conditions. Ann. Biomed. Eng. 28, 1281–1299. ( 10.1114/1.1326031) [DOI] [PubMed] [Google Scholar]
- 25.Olufsen MS, Hill NA, Vaughan GDA, Sainsbury C, Johnson M. 2012. Rarefaction and blood pressure in systemic and pulmonary arteries. J. Fluid Mech. 705, 280–305. ( 10.1017/jfm.2012.220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riches AC, Sharp JG, Thomas DB, Smith SV. 1973. Blood volume determination in the mouse. J. Physiol. 228, 279–284. ( 10.1113/jphysiol.1973.sp010086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Windberger U, Bartholovitsch A, Plasenzetti R, Korak KJ, Heinze G. 2003. Whole blood viscosity, plasma viscosity and erythrocyte aggregation in nine mammalian species. Exp. Physiol. 88, 431–440. ( 10.1113/eph8802496) [DOI] [PubMed] [Google Scholar]
- 28.Qureshi MU, Vaughan GDA, Sainsbury C, Johnson M, Peskin CS, Olufsen MS, Hill NA. 2014. Numerical simulation of blood flow and pressure drop in the pulmonary arterial and venous circulation. Biomech. Model. Mechanobiol. 13, 1137–1154. ( 10.1007/s10237-014-0563-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paun LM, Qureshi MU, Colebank MJ, Hill NA, Olufsen MS, Haider MA, Husmeier D. 2018. MCMC methods for inference in a mathematical model of pulmonary circulation. Stat. Neerl. 72, 306–338. ( 10.1111/stan.12132) [DOI] [Google Scholar]
- 30.Colebank MJ, Qureshi MU, Olufsen MS. In press. Sensitivity analysis and uncertainty quantification of 1D models of the pulmonary hemodynamics in mice under control and hypertensive conditions. Int. J. Numer. Meth. Biomed. Eng. ( 10.1002/cnm.3242) [DOI] [PubMed] [Google Scholar]
- 31.Su J, Hilberg O, Howard L, Simonsen U, Hughes AD. 2016. A review of wave mechanics in the pulmonary artery with an emphasis on wave intensity analysis. Acta Physiol. 218, 239–249. ( 10.1111/apha.12803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes AD, Parker KH. 2009. Forward and backward waves in the arterial system. Med. Biol. Eng. Comput. 47, 207–210. ( 10.1007/s11517-009-0444-1) [DOI] [PubMed] [Google Scholar]
- 33.Reymond P, Merenda F, Perren F, Ru D. 2009. Validation of a one-dimensional model of the systemic arterial tree. Am. J. Physiol. 297, 208–222. [DOI] [PubMed] [Google Scholar]
- 34.Krenz GS, Dawson CA. 2003. Flow and pressure distributions in vascular networks consisting of distensible vessels. Am. J. Physiol. 284, H2192–H2203. ( 10.1152/ajpheart.00762.2002) [DOI] [PubMed] [Google Scholar]
- 35.Silverman BW. 1998. Density estimation for statistics and data analysis. Boca Raton, FL: Chapman & Hall/CRC Press. [Google Scholar]
- 36.Guidoum AC. 2015. kedd: Kernel estimator and bandwidth selection for density and its derivatives. R package version 1.0.3. See http://CRAN.R-project.org/package=kedd (accessed August 2018).
- 37.Riihimäki J, Vehtari A. 2014. Laplace approximation for logistic Gaussian process density estimation and regression. Bayesian Anal. 9, 425–448. ( 10.1214/14-BA872) [DOI] [Google Scholar]
- 38.Vanhatalo J, Riihimäki J, Hartikainen J, Jylänki P, Tolvanen V, Vehtari A. 2013. GPstuff: Bayesian modeling with Gaussian processes. J. Mach. Learn. Res. 14, 1175–1179. [Google Scholar]
- 39.Goldberg P, Williams C, Bishop C. 1997. Regression with input-dependent noise. Adv. Neural Inf. Process Syst. 10, 493–499. [Google Scholar]
- 40.Hastie T, Tibshirani R, Friedman J. 2009. The elements of statistical learning: data mining, inference, and prediction. New York, NY: Springer. [Google Scholar]
- 41.Rasmussen CE, Williams CKI. 2006. Gaussian processes for machine learning. Cambridge, MA: The MIT Press. [Google Scholar]
- 42.Rol N, Timmer EM, Faes TJC, Noordegraaf AV, Grünberg K, Bogaard HJ, Westerhof N. 2017. Vascular narrowing in pulmonary arterial hypertension is heterogeneous: rethinking resistance. Physiol. Rep. 5, e13159 ( 10.14814/phy2.13159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang W, Yen RT, McLaurine M, Bledsoe G. 1996. Morphometry of the human pulmonary vasculature. J. Appl. Physiol. 81, 2123–2133. ( 10.1152/jappl.1996.81.5.2123) [DOI] [PubMed] [Google Scholar]
- 44.Melis A, Clayton RH, Marzo A. 2017. Bayesian sensitivity analysis of a 1D vascular model with Gaussian process emulators. Int. J. Numer. Meth. Biomed. Eng. 33, e2882 ( 10.1002/cnm.2882) [DOI] [PubMed] [Google Scholar]
- 45.Kind T, Faes TJC, Vonk-Noordegraaf A, Westerhof N. 2011. Proportional relations between systolic, diastolic and mean pulmonary artery pressure are explained by vascular properties. Cardiovasc. Eng. Technol. 2, 15–23. ( 10.1007/s13239-010-0027-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Lakes RS, Golob M, Eickhoff JC, Chesler NC. 2013. Changes in large pulmonary arterial viscoelasticity in chronic pulmonary hypertension. PLoS ONE 8, e78569 ( 10.1371/journal.pone.0078569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Epstein S, Willemet M, Chowienczyk PJ, Alastruey J. 2015. Reducing the number of parameters in 1D arterial blood flow modeling. Am. J. Physiol. 309, H222–H234. ( 10.1152/ajpheart.00857.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gounley J, Vardhan M, Randles A. 2017. A computational framework to assess the influence of changes in vascular geometry on blood flow. In Proc. Platform for Advanced Scientific Computing Conf., Lugano, Switzerland, 26–28 June 2017, article 2. New York, NY: ACM ( 10.1145/3093172.3093227) [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material. Software/code implementing the developed methodology can be found at https://github.com/mjcolebank.