Abstract
Clustered regularly interspaced short palindromic repeats and associated proteins (CRISPR-Cas) of bacterial adaptive immunity have been adopted as a powerful and versatile tool for manipulation of the genome. This paradigm has been widely applied in biological research and treatments of animal or cellular disease models. A critical feature of CRISPR-Cas is the protospacer adjacent motif (PAM), which dictates the DNA target recognition mechanism of Cas proteins. While, direct identifying functional PAM sequences in human cells remains a challenge. Here, we developed a positive screen system termed PAM-DOSE (PAM Definition by Observable Sequence Excision) to delineate the functional PAMs in human cells. Specifically, the PAM libraries for CRISPR-Cas (SpCas9, SpCas9-NG, FnCas12a, AsCas12a, LbCas12a and MbCas12a) were generated and the corresponding CRISPR-Cas mediated cleaved fragments with functional PAM in human cells were harvested for DNA sequencing, which could be tracked and visualized with either florescence microscopy or flow cytometry analysis. With this system, we identified the functional PAMs of CRISPR-Cas members. We also found that spacer sequence affects the PAM preference of Cas proteins. This method will facilitate identification of functional PAMs for Cas-mediated human genome editing applications.
Keywords: CRISPR-Cas, PAM, Human, Next-generation Sequencing
1. Introduction
CRISPR-Cas triggered targeted genomic sequence changes into living cells and organisms provides a powerful tool for biological research as well as a potential avenue for improved therapy of genetic diseases [[1], [2], [3]]. This tool comprises Cas protein and CRISPR RNA. CRISPR provides nucleic acid-based immunity to viruses and plasmids containing sequences (protospacers) matching CRISPR spacers [4]. The PAM sequences are absent from the repeats which provides a mechanism for it being recognized as a self-genome (i.e., bacteria) or invading nucleic acids (i.e., bacteriophage) by Cas [5]. Thus, for efficient cleavage, the presence of the PAM sequence is essential for CRISPR-Cas mediated genome editing. The most commonly used Cas protein, Streptococcus pyogenes Cas9 (SpCas9) recognizes 5′-NGG-3′ as functional PAM [2]. Additional Cas proteins and their variants require different PAMs for the efficient cleavage as a genome editing tool to manipulate the human genome [2].
As the PAM sequence is the key parameter for the design of CRISPR-Cas mediated genome editing, different strategies have been adopted to identify the PAM of CRISPR-Cas. These include bioinformatics analysis of the spacer flanking sequence by aligning the spacer sequence of the CRISPR with the genome of the host and invading nucleic acids [6], incubating the purified recombinant Cas protein with RNA and a target sequence harboring PAM library [[7], [8], [9]], bacterial based selection system includes antibiotic-dependent depletion [[10], [11], [12], [13]], potent phage to infect cells containing the crRNA library [14] and PAM-SCANR [15]. CRISPR-Cas requires target gene in human cells for treatment of human diseases, thus, direct identification of functional PAM in human cells will facilitate its use in clinical treatment. So far, no method is available to directly elucidate functional PAMs in human cells.
In our previous studies, we designed CRISPR-Cas mediated genome editing to target loxP sequence [16], which is further utilized for comparing the activity of SpCas9, SaCas9 (Staphylococcus aureus Cas9) and FnCas12a (FnCpf1, Francisellanovicida U112. Cpf1, also designated as FnCas12a) [17]. We also demonstrated that the T base of 5′-NNGRRT-3′ PAM is dispensable for SaCas9 mediated efficient cleavage in human cells via this system [17]. As DNA cleavage events triggered by CRISPR-Cas9 could be captured with this system, theoretically, it may be harnessed for the identification of functional PAM in human cells. Through this concept, here we describe and demonstrate a PAM identification system termed PAM-DOSE (PAM Definition by Observable Sequence Excision) in human cells.
2. Results
2.1. Design and development of a dual fluorescence reporter library for identifying functional PAM preferences
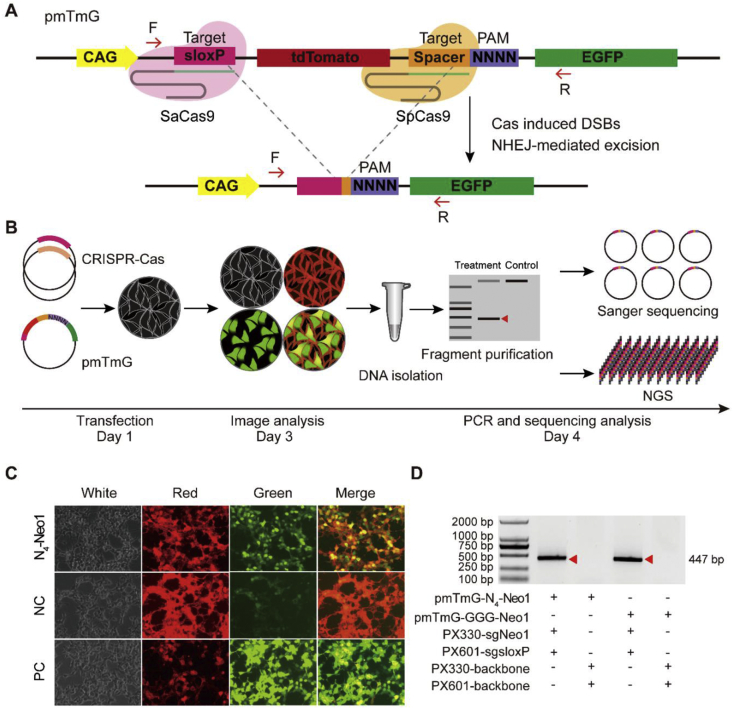
In our previous studies, we had demonstrated that the Cas proteins instead of Cre enzyme could be utilized for site-specific genetic manipulation at the loxP site [16], and also showed the loxP flanking the tdTomato cassette could be replaced with additional sequences [17]. In these studies, two identical sequences flanking the tdTomato gene were placed; it can't be directly harnessed due to the excessive combinations of two different sites with different PAM sequence (i.e., SaCas9, 46∗46 = 16777216). Then we selected two different sequences flanking the tdTomato gene, one for fixed enzyme (i.e., SaCas9 with its canonical PAM), the other for the tested Cas protein with PAM library. Initially, SpCas9 was selected for testing the system as its canonical PAM is well characterized.
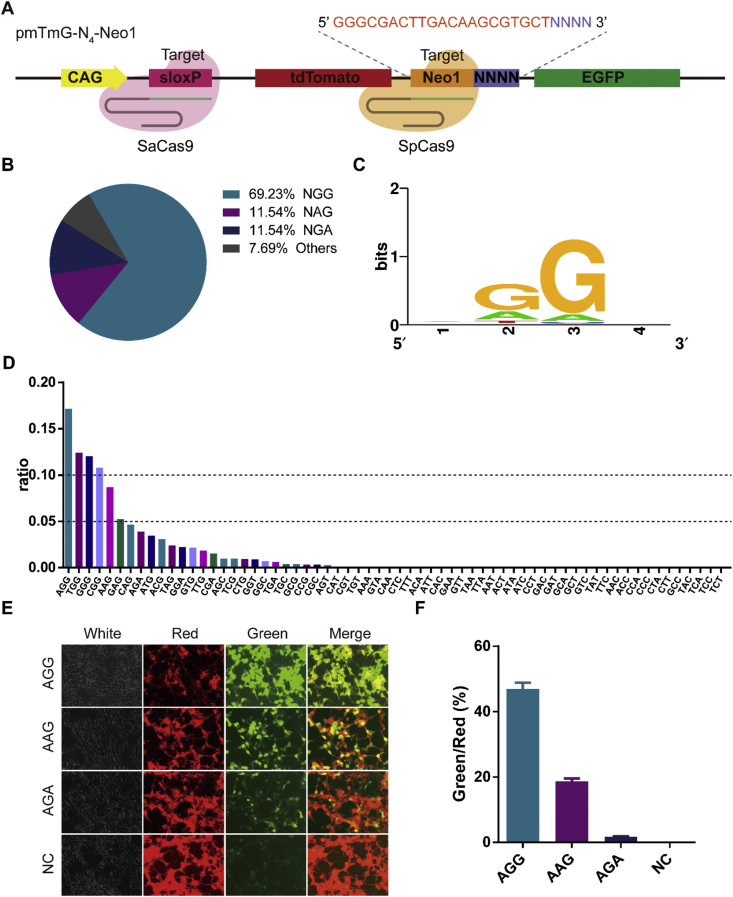
A plasmid (pmTmG) was generated that harbors two fluorescent protein genes (tdTomoto and EGFP) and two target sequences (sloxP and Spacer N4) (Fig. 1A). The sloxP sequence for the guide RNA spacer for SaCas9 with 5′-NNGRRT-3′ PAM was inserted upstream of the tdTomato gene. The Spacer N4 harbors spacer (Neo1) and 4 randomized nucleotides located downstream of the tdTomato cassette, was used to empirically determine the PAM recognition of Cas proteins in human cells (Fig. 1A). After the cleavage, the tdTomato cassette will be excised and the CAG promoter will drive the expression of the EGFP gene directly, thereby allowing for live visualization under the fluorescent microscope or detection via FCA (flow cytometry analysis, Fig. 1A–C). The randomized nucleotides may be retained after the excision and then their sequences could be obtained with sequencing via Sanger sequencing or NGS (next-generation sequencing) (Fig. 1D). Not surprisingly, we found NGG is the canonical PAM for SpCas9 via Sanger sequencing (Fig. 2A, B; Fig. S1). Due to low through-put of Sanger sequencing, we performed NGS (Fig. 2C, D), which revealed that SpCas9 recognizes 5′-NGG-3′ for PAMs with Neo1 spacer sequence. The proportion of 5′-AGG-3′ PAM is 17% in total, which is higher than another PAMs, 5′-AAG-3' (9%) and 5′-AGA-3' (4%). Besides, we selected three identified PAMs (5′-AGG-3′, 5′-AAG-3′ and 5′-AGA-3′) for quantitative comparison. The results showed it is indeed the case (Fig. 2E, F). Here we named this system as PAM-DOSE (PAM Definition by Observable Sequence Excision) as it could be tracked by the fluorescence microscope or FCA (Fig. 1, Fig. 2E). Collectively, we conclude that, using SpCas9 as an example, PAM-DOSE may prove to be useful for ultimately identifying functional PAM of CRISPR-Cas in human cells.
Fig. 1.
PAM Definition by Observable Sequence Excision (PAM-DOSE). (A) Illustration of PAM-DOSE system. A library of PAM sequences was introduced into the constructs. In the presence of a functional PAM, cleavage mediated tdTomato cassette excision was performed, which leads to the expression of EGFP and it could be tracked via fluorescent microscope. (B) Illustration of each step of PAM-DOSE. (C) Representative images showing visualization of tdTomato and EGFP with PAM-DOSE. (D) DNA fragments harboring functional PAM sequence. PX330-backbone and PX601-backbone represent the backbone of PX330(SpCas9) and PX601(SaCas9), respectively. PC and NC represent the positive control and negative control, respectively. The PCR products from the excision events were pointed out with a red triangle. PmTmG-GGG-Neo1 is a plasmid containing only GGG PAM as a positive control for SpCas9 or SpCas9-NG at Neo1 site.
Fig. 2.
Screening the PAM of SpCas9 in human cell. (A) Target sequence harboring a library of potential PAM sequences at N4-Neo1. (B) Pie chart for comprehensive screening of the PAM for SpCas9 at Neo1 site (via Sanger sequencing). (C–D) Sequence logo and ratio of recognized PAMs calculated based on NGS results at Neo1 site. (E) HEK-293 cells were transfected pmTmG-N4-Neo1 with PAM library and two different plasmids for the cleavage and images were obtained 48 h after transfection. (F) Relative activity of different PAM at Neo1 site was analyzed with FCA (flow cytometry analysis, red represents tdTomato signal; green represents EGFP signal).
2.2. Functional PAM preferences of SpCas9 enzyme with additional target sequences
The above results showed PAM-DOSE may be utilized to clarify the identity of functional PAM for CRISPR-SpCas9 in human cells. We sought to test whether additional target sequence would result in the same pattern. To answer this question, we tested two additional target sequences, N4-Zeo and N6-Neo2 (Figs. S2, S3). We observed that 5′-NGG-3′ PAM remains canonical PAM for SpCas9 with any of these three tested target sequences, including N4-Neo1, N4-Zeo and N6-Neo2. At these three target sites, the ratio of 5′-NAG-3′ PAM and 5′-NGA-3′ PAM to the total cleavage of PAMs was 0.21–0.23 and 0.08–0.14, respectively, and other types of PAM for a small portion at three target sequences. However, there are subtle differences in the PAM preferences of the Cas protein with the three spacer sequences. (Fig. 2B; Figs. S1, S2, S3). Specifically, different spacer sequences have different preferences for the third base G (Fig. 2C; Fig. S3). The cleavage efficiency of SpCas9 on 5′-NAG-3′ PAM is not as high as that of 5′-NGG-3′, which is consistent with the literature [18]. We also observed CRISPR/SpCas9 with 5′-GAG-3′ PAM at the Zeo site could trigger robust cleavage (Fig. S3; Fig. S4). Notably, the preference of 5′-NGG-3′ PAM for the first base is different at these sites (Fig. 2D; Figs. S3C, S3F). From these results of three different target sequences, we observed there are subtle differences in PAM preferences of SpCas9 in human cells via PAM-DOSE.
2.3. Identifying functional PAM preferences of Cas12a
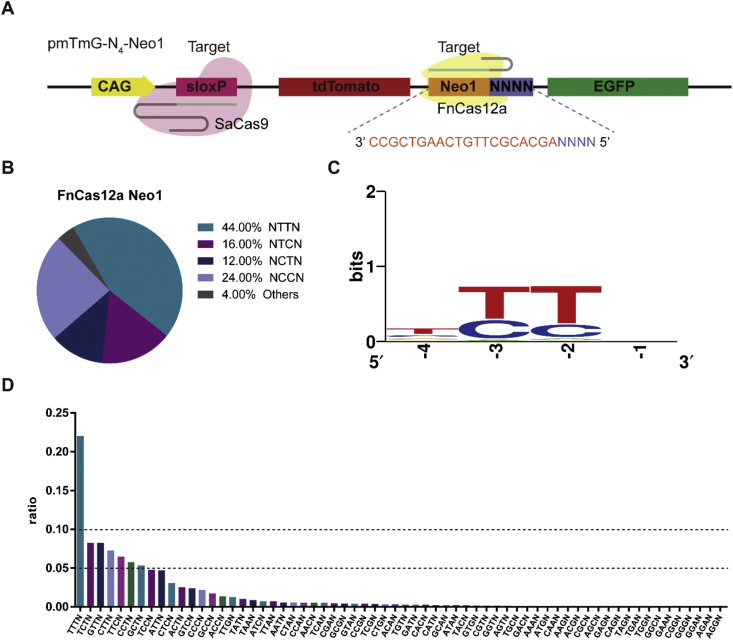
To determine whether PAM-DOSE could be utilized for identifying additional Cas proteins, we tested it with Cas12a (Cpf1). FnCas12a is a single RNA-guided endonuclease of class 2 CRISPR-Cas, which has robust DNA interference with features distinct from Cas9 and has been harnessed as a tool for human genome manipulation [7,19]. Studies showed that FnCas12a possesses high efficiency targeting with 5′-KYTV-3′ PAM in the human genome [20] or 5′-TTN-3′ PAM [7]. We co-transfected SaCas9-sloxP, FnCas12a-Neo1 or FnCas12a-Zeo and N4-Neo1 or N4-Zeo PAM library into HEK-293 cells (Fig. 3A). Because Cas12a is less active than SpCas9 as we previous reported [17], we observed relative fewer green cells after the transfection (Fig. S5). Sanger sequencing showed that including the 5′-NTTN-3′ PAM, a notable amount was present of 5′-NYYN-3’ (Fig. 3B; Figs. S5, S6). The functional PAM of FnCas12a as determined by PAM-DOSE slightly differs from previous studies [7,15]. Specifically, compared with results based on the bacterial reverse screening or PAM-SCANR [7,15], we observed a relatively high preference for C at the −2 and −3 positions, compared with other bases except for T (Fig. 3C, D; Figs. S6B, S6C). The results of fluorescence observation and FCA were consistent with the sequencing results (Fig. S7). Further, the ratios of 5′-NCTN-3′, 5′-NTCN-3′ and 5′–NCCN–3′ to the total cleavage of PAMs was 0.23, 0.13 and 0.10, respectively, which is consistent with the non-canonical PAM previously revealed through structural based analysis [7,21]. The results demonstrated the applicability of PAM-DOSE in PAM assay.
Fig. 3.
The PAM of FnCas12a in human cell. (A)Target harboring a library of PAM sequences at N4-Neo1. (B) Pie chart for comprehensive screening of the PAM for FnCas12a at Neo1 site (via Sanger sequencing). (C–D) Sequence logo and ratio of recognized PAMs calculated based on NGS results at Neo1 site.
In addition, we performed a study on AsCas12a, LbCas12a and MbCas12a with PAM-DOSE (Fig. S8, S9). In general, the results were consistent with previous literature reports [7]. The PAM-preferred logos of AsCas12a, LbCas12 and MbCas12a obtained in PAM-DOSE showed a slight decreased preference for the T-base at −2, and −3 sites, compared with the in vitro results [7]. The functional PAM and non-canonical PAM of AsCas12a and LbCas12a obtained from PAM-DOSE are in line with other studies [7,21]. As for MbCas12a, 5′-NYYN-3′, except for 5′-NTTN-3’ [22], may be harnessed as a non-canonical PAM for genome editing in human cells.
2.4. Identification of the functional PAM of the SpCas9 mutant (SpCas9-NG)
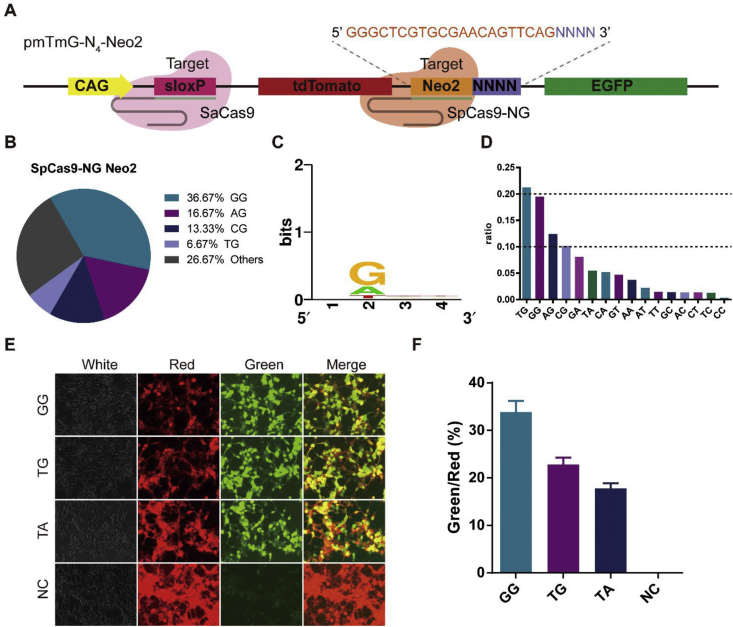
SpCas9 is one of the most commonly used Cas proteins for genome editing. It only recognizes the 5′-NGG-3′ sequence as PAM enabling efficient cleavage. This requirement is a key limitation to SpCas9 usage for genome editing and related applications (i.e., dSpCas9 for gene regulation). Recently, SpCas9-NG was identified as a variant of SpCas9 which recognizes 5′-NG-3′ PAM with high cleavage activity and fidelity [23]. We sought to know the PAM preference of SpCas9-NG in human cells. We selected Neo2 and Zeo as target sequences to investigate the PAM of SpCas9-NG (Fig. 4; Figs S10, S11). From the results of Sanger sequencing and NGS, we identified the functional PAM of SpCas9-NG is 5′-NG-3’ (Fig. 4B–D; Figs S10, S11), which is consistent with the PAM preference of SpCas9-NG obtained from in vitro studies [23]. We observed that the ratio of 5′-NA-3′ to all cleavage PAMs was 0.23, compared to 5′-NG-3' (0.63), which reveals 5′-NA-3′ could serve as non-canonical PAM in human cells (Fig. 4C, D; Figs S10, S11). We generated the target sequence harboring 5′-NA-3′ PAM. The fluorescence signal and FCA results demonstrate that SpCas9-NG has considerable cleavage activity with 5′-NA-3′PAM (Fig. 4E, F).
Fig. 4.
The PAM of SpCas9-NG in human cell. (A) Target harboring a library of PAM sequences at N6-Neo2. (B) Pie chart for comprehensive screening of the PAM for SpCas9-NG at Neo2 site (via Sanger sequencing). (C–D) Sequence logo and ratio of recognized PAMs calculated based on NGS data at Neo2 site. (E) HEK-293 cells were co-transfected pmTmG with single PAM and CRISPR-Cas systems. Images were obtained 48 h after transfection. (F) Relative activity of different PAM at Neo2 site was analyzed with FCA (red represents tdTomato; green represents EGFP).
3. Discussion
There are various in vitro and in vivo methods for identifying functional PAM, including in vitro recombinant Cas protein-based assays, plasmid clearance, and PAM-SCANR. The in vitro conditions may not fully simulate the cellular environment or affected by component changes, resulting in erroneous PAM assignments [9]. Most plasmid depletion methods rely on negative antibiotic resistance in vivo, which requires extensive library coverage to recognize deleted sequences, and the Cas protein must be expressed in a non-functional PAM host. PAM-SCANR is another method for defining PAM in E. coli strains, which uses forward screening. Catalytically dead Cas9 (usually labeled as dCas9) protein that lacks endonuclease activity was used in the PAM-SCANR method, which relies on gene repression to distinguish between functional and non-functional PAMs. As stated in the article, the drawback of gene repression is the lack of nuclease activity associated with fully functioning systems, potentially missing PAM-dependent allosteric changes that drive DNA cleavage or Cas3 recruitment. For human therapeutic purposes, there is no doubt that PAM identification studies should be performed using human target cells. In addition, optimizing the Cas catalytic activity to determine PAM in humans is dependent on using Cas rather than the dCas protein. The method described in this paper, PAM-DOSE, is performed in human cells. Furthermore, catalytically active Cas has been utilized, which may accurately reflect the Cas mediated DNA cleavage in human cells.
In our previous study, we took advantage of the GFP reporter cell line to study the PAM preference based on disrupting the GFP coding frame triggered by CRISPR-Cas [24]. However, the target selection spectrum of the GFP gene is very limited. For example, there are only three sites harboring a 5′-TTTN-3′ PAM sequence in the GFP gene that could be used for AsCpf1 and LbCpf1 mediated cleavage, including one near the stop codon [20]. Also, the target sequence itself affects the DNA cleavage efficiency, which is another factor that affects defining a functional PAM sequence. The PAM library harboring random nucleotides was rapidly generated using the gap filling method and at least two different target sequences via PAM-DOSE will minimize the locus bias effects. We acknowledge the PAM identification may be resolved with PAM library integrated into the genome via lentivirus, which may provide better interpretation of DNA cleavage and repair in chromosomes. While, each colony harboring one PAM of lentivirus PAM libraries would be integrated into the different location in the genome, which may possess locus bias for the PAM preference.
Here we developed PAM-DOSE for identifying functional PAM of CRISPR-Cas in human cells, including SpCas9, Cas12a (including FnCas12a, AsCas12a, LbCas12a and MbCas12a) and SpCas9-NG. We demonstrated the universality of the identification method using six different CRISPR-Cas configurations from two main types (Type II-A and V-A), including a variant of SpCas9. In addition, to avoid the target sequence bias in determining a preference for Cas protein, multiple sites were selected and evaluated. The results revealed that there are some differences in PAM preference at different target sequences for Cas protein. This method is meritorious because the PAM sequence was directly obtained from the functional cleavage fragment. After Sanger sequencing of a certain number (i.e., 20 colonies) of individual colonies harboring the cleavage fragment, the dominant functional (canonical) PAM sequence could be easily identified. Comprehensive PAM can be revealed by NGS. Furthermore, excision events can also be tracked (or visualized) with fluorescence microscopy or FCA (Fig. 1, Fig. 2E).
We observed additional PAMs (5′-GTG-3′ and 5′-GGT-3′) for SpCas9, which possesses certain cleavage activity. Similar results have been observed for FnCas12a, i.e., 5′-NYYN-3′ PAM (except 5′-NTTN-3′), which may extend the target selection of FnCas12a in human cells. In addition, we found that Cas protein has less preference for a certain base of PAM in human cells than that from in vitro or E. coli studies [7,23,25], which may be utilized for modest gene regulation with an inactive Cas form. The structural analyses will shed light on the molecular basis of PAM recognition with different protospacer sequences.
In summary, here we developed PAM-DOSE method for identifying functional PAMs for SpCas9, Cas12a (FnCas12a, AsCas12a, LbCas12a and MbCas12a) and SpCas9-NG, which are expected to facilitate optimizing nuclease mediated genome editing for human gene therapy.
4. Materials and methods
4.1. Plasmid construction
Plasmids PX330 and PX601 were gifts from Feng Zhang (Addgene plasmid # 42230 and # 61591). Plasmids PX330-NG for the expression of SpCas9-NG was a gift from Osamu Nureki (Addgene plasmid # 117919). SgRNA oligos were annealed and inserted into the PX330, PX601 and PX330-NG, respectively. Plasmids for the expression of FnCas12a, LbCas12a and MbCas12a were obtained from Addgene (Addgene plasmids # 69976, # 69982 and # 69986, respectively). CrRNA oligos annealed and inserted into PY094-FnCas12a. The crRNA expression cassette of AsCas12a, LbCas12a and MbCas12a were generated by insertion of PCR products into vector pJET1.2 (Thermo Fisher Scientific). Plasmid DNA was isolated by standard techniques. Detailed sequences of the primers used in the constructions are listed in Table S1. DNA sequencing confirmed the specific sequences required in the construct. Detailed sequences of the primers for vector sequencing are listed in Table S2.
4.2. Library generation
The vector plasmid pmTmG contains the following parts: tdTomato, EGFP, two loxP sites (Fig. 1A) has been described in our previous study [16]. To generate a PAM library of pmTmG at different target sites, we developed a method by random nucleotide gap filling (Fig. S12). We fist synthesized three single-strand oligonucleotides. After annealing, single-strand oligonucleotides were ligated into pmTmG backbone cut by EcoRI and MluI. Then the ligation products are transformed into DH10B (E. coli). The gap can be fill by E. coli DNA polymerase II [26]. More than 1400 N4-Neo1 colonies, 1100 N4-Zeo colonies and 15000 N6-Neo2 colonies were obtained (Fig. S13). The libraries have more than three times the number of possible combinations of PAM at three sites, which coverage was determined by Sanger sequencing and NGS sequencing (Fig. S14). The coverage of the libraries N4-Neo1, N4-Zeo, N6-Neo2 determined by NGS was 100%, 99.22%, and 96.12%, respectively. Detailed sequences of the sites are listed in Table S3.
4.3. Cell culture, transfection, images and FCA
On day 1, 1.8∗105 HEK-293 cells were seeded per well of a 12-well plate, and cultured in DMEM media supplemented with 10% FBS, penicillin–streptomycin, at 37 °C with 5% CO2. On day 2, HEK-293 cells were co-transfected with 400 ng pmTmG PAM library and its 0.75 times the molar number of plasmids expressing Cas and sgRNA (crRNA) using TurboFect (Thermo) (Fig. S15). The fresh medium was added in the wells of transfected HEK-293 cells on day 3. Images were obtained under an inverted fluorescence microscope on day 4. The transfected cells were analyzed using a Cytomics FC 500 MPL flow cytometer at day 4. Quantification was based on relative fluorescent frequencies. Green percentage/red percentage was determined by the formula (O2+O4)/(O1+O2), where O2 is the cells in both green and red, O1 is the cells only in red, and O4 is the cells only in green.
4.4. Sequence analysis
Purification of plasmids from cells was performed using standard protocols. PCR is used to amplify the sequences flanking the CRISPR target locus, and products were inserted into the vector pJET1.2. Detailed sequences of the primers used are listed in Table S4. The fragment flanking the target sequence was amplified for NGS via a two-round PCR harboring barcode for the amplicons. Detailed sequences of the primers used are listed in Table S4. Amplicons were then subjected to HiSeq 2500 sequencing (Illumina). All raw data are available through NCBI SRA (PRJNA533734: PAM-DOSE). PAM regions without indel within three bases near PAM were extracted. PAMs were counted and used to generate sequence logos [27].
Conflicts of interest
The authors declare that there is no conflicts of interest.
Acknowledgements
This study was supported by the grants from the Natural Science Foundation of China (81201181 to FG; 81473295 & 81670882, ZMS), Natural Science Foundation of Zhejiang Province, China (LQ16H040002,HT.X), Science Technology project of Zhejiang Province (2017C37176 to FG), Zhejiang provincial & Ministry of Health Research Fund for Medical Sciences (WKJ-ZJ-1828 to JZZ), Wenzhou City Grant (Y20160410 to CB.L.; Y20160008 & C20170007 to JZ.Z) and Lin He’s Academician Workstation of New Medicine and Clinical Translation (17331209 to CB.L.; 18331105 to JZ.Z).
Footnotes
Peer review under responsibility of Chinese Society for Cell Biology (CSCB).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cr.2019.08.002.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Jiang F., Doudna J.A. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 2.Komor A.C., Badran A.H., Liu D.R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J. CRISPR-Cas: adapting to change. Science. 2017;356 doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 5.Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 6.Biswas A., Gagnon J.N., Brouns S.J., Fineran P.C., Brown C.M. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol. 2013;10:817–827. doi: 10.4161/rna.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karvelis T., Gasiunas G., Young J., Bigelyte G., Silanskas A., Cigan M. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015;16:253. doi: 10.1186/s13059-015-0818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esvelt K.M., Mali P., Braff J.L., Moosburner M., Yaung S.J., Church G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue C., Seetharam A.S., Musharova O., Severinov K., Brouns S.J., Severin A.J. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res. 2015;43:10831–10847. doi: 10.1093/nar/gkv1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leenay R.T., Maksimchuk K.R., Slotkowski R.A., Agrawal R.N., Gomaa A.A., Briner A.E. Identifying and visualizing functional PAM diversity across CRISPR-cas systems. Mol Cell. 2016;62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang F., Liu C., Chen D., Tu M., Xie H., Sun H. CRISPR/Cas9-loxP-Mediated gene editing as a novel site-specific genetic manipulation tool. Mol Ther Nucleic Acids. 2017;7:378–386. doi: 10.1016/j.omtn.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie H., Tang L., He X., Liu X., Zhou C., Liu J. SaCas9 requires 5'-NNGRRT-3' PAM for sufficient cleavage and possesses higher cleavage activity than SpCas9 or FnCpf1 in human cells. Biotechnol J. 2018;13 doi: 10.1002/biot.201800080. [DOI] [PubMed] [Google Scholar]
- 18.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun H., Li F., Liu J., Yang F., Zeng Z., Lv X. A single multiplex crRNA array for FnCpf1-mediated human genome editing. Mol Ther: J Am Soc Gene Ther. 2018;26:2070–2076. doi: 10.1016/j.ymthe.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu M., Lin L., Cheng Y., He X., Sun H., Xie H. A 'new lease of life': FnCpf1 possesses DNA cleavage activity for genome editing in human cells. Nucleic Acids Res. 2017;45:11295–11304. doi: 10.1093/nar/gkx783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamano T., Zetsche B., Ishitani R., Zhang F., Nishimasu H., Nureki O. Structural basis for the canonical and non-canonical PAM recognition by CRISPR-Cpf1. Mol Cell. 2017;67:633–645. doi: 10.1016/j.molcel.2017.06.035. e633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toth E., Czene B.C., Kulcsar P.I., Krausz S.L., Talas A., Nyeste A. Mb- and FnCpf1 nucleases are active in mammalian cells: activities and PAM preferences of four wild-type Cpf1 nucleases and of their altered PAM specificity variants. Nucleic Acids Res. 2018;46:10272–10285. doi: 10.1093/nar/gky815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Ge X., Yang F., Zhang L., Zheng J., Tan X. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci Rep. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 26.Rangarajan S., Woodgate R., Goodman M.F. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:9224–9229. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.