Abstract
Background
The current spread of carbapenemase-producing Enterobacterales (CPE) is a great concern.
Methods
We recovered 198 CPE from 162 patients admitted in our Hospital (March 2014-March 2016) during the R-GNOSIS European Project. Microbiological features and plasmid characteristics of CPE recovered from patients co-colonized with multiple CPE were studied.
Findings
Thirty patients (18.5%; CI 95%= 12.5%–24.5%) presented co-colonization with multiple CPE producing the same (CPE-SC) (15.4%) or a different carbapenemase (CPE-DC) (4.3%). OXA-48 (83.3%) was the most frequent carbapenemase, followed by VIM-1 (26.7%), NDM-1 (10%) and KPC-3 (3.3%). CPE-DC-patients had longer admissions [63 days (20–107)] than the other patients. Moreover, hospital stay until CPE detection was lower [9 days (5–14)] (p = 0.0052) in CPE-SC-patients than in those with a single colonization; 56% showed co-colonization in the first positive sample, although most of them had previous admissions and had received multiple antibiotic treatments. CPE were more frequently recovered in clinical samples from co-colonized [CPE-DC (28.6%), CPE-SC (24%)] patients than from patients with a single CPE (15.2%). Among CPE-SC—OXA-48 [80% (p = 0.11)], K. pneumoniae [88% (p = 0.006)] and E. coli [84% (p < 0.001)] were the most frequent species. In 60% of patients, K. pneumoniae and E. coli species were simultaneously recovered, frequently after a single OXA-48-K. pneumoniae colonization. High-risk clones (ST11, ST15, ST307) were detected in OXA-48-K. pneumoniae but a higher clonal diversity was found among E. coli. A frequent in-vivo cross-species plasmid transmission was shown, due to a dominant plasmid (IncL-pOXA-48), but also involving related or unrelated blaVIM-1-, blaNDM-1- and blaKPC-3-encoding plasmids.
Interpretation
CPE co-colonization status should be monitored during epidemiological surveillance cultures, as these patients might be at a higher risk for infection.
Funding
European Commission Framework Programme 7 and Instituto de Salud Carlos III, Spain
Keywords: CPE co-colonization, OXA-48-producing E. coli and K. pneumoniae, in vivo cross-species transmission, IncL-pOXA-48
Research in context
Evidence before this study
The increasing prevalence of carbapenemase-producing Enterobacterales (CPE) has become a major threat to healthcare facilities. A higher incidence of OXA-48-producing E. coli has been reported in our geographic area, including our Hospital. The role of epidemic high-risk clones of Klebsiella pneumoniae as ST11, ST307 or ST405 seems to be critical in the maintenance and spread of highly transferable plasmids such as the worldwide disseminated IncL/M-pOXA-48a.
Added value of this study
During the R-GNOSIS European Project (March 2014-April 2016), we detected 30 patients (18.5%) co-colonized with multiple CPE species (or clones of the same species) producing the same or a different carbapenemase. According to our results, patients co-colonized with isolates producing different carbapenemases had longer admissions and patients carrying multiple CPE producing the same carbapenemase enzyme had lower hospital stays until CPE detection. Moreover, CPE isolates were more frequently recovered in samples from infective sites in both subgroups of co-colonized patients than among patients with a single CPE. Major finding revealed that co-colonization with OXA-48 producers was the most frequent event and that K. pneumoniae and Escherichia coli were the predominant species. Our results highlight a successful horizontal inter-species dissemination of blaOXA-48 though a dominant plasmid (IncL-pOXA-48), likely from hospital-adapted K. pneumoniae high-risk clones to unrelated E. coli clones.
Implications of all the available evidence
Intestinal colonization is the key step in CPE epidemiology and most clinical cases are secondary to the selection of CPE in the gut. Our results suggest that multiple colonization with CPE might also increase the risk of infection with these multidrug-resistant microorganisms. Moreover, CPE co-colonization is more frequently due to cross-species transfer of an IncL-pOXA-48 plasmid than to the sequential acquisition of different CPE isolates. In addition, the increased prevalence of OXA-48-producing E. coli could contribute in the endemicity of OXA-48 carbapenemase in the hospital setting, but also in the community, as previously happened with CTX-M-15-producing E. coli. Implementation of active surveillance cultures are crucial for detect CPE co-colonization status and reduce the risk for infections.
1. Introduction
The occurrence and increased dissemination of carbapenemase-producing Enterobacterales (CPE) has been reported worldwide in the last decade, predominantly in hospital setting and more rarely in the community [1], [2]. Resistance to carbapenems can be due to loss or decreased expression of porins combined with plasmid-mediated extended-spectrum β-lactamases (ESBLs) or overexpression of chromosomal cephalosporinases (AmpC) and overproduction of efflux pumps, although the most frequent resistance mechanism is the carbapenemase production. Almost all carbapenemase-producers (except certain class D enzymes) show resistance to most β-lactam antibiotics and frequently exhibit co-resistance to other antimicrobial drugs. The fact that carbapenemase genes are usually contained on transmissible mobile genetic elements, such as plasmids, facilitate their spread among different bacterial species [2], [3].
In the last years, several nosocomial outbreaks of Enterobacterales harboring plasmid-acquired carbapenemases have been described in Spain, initially due to VIM and KPC groups, and more recently linked to OXA-48 [4], [5], [6], [7], [8]. NDM enzymes have been sporadically detected in our country, although are recently increasing [9], [10]. The epidemiology of OXA-48 enzyme has been largely associated with both global expansion of Klebsiella pneumoniae hospital-adapted high-risk clones and the dissemination of plasmids related to IncL/M-pOXA-48a [11], [12], [13]. Nevertheless, the current spread of blaOXA-48-encoding plasmids into other Gram-negative bacteria, especially E. coli, is contributing to the blaOXA-48 endemicity in health-care settings and depicts a great public health threat [8], [14], [15], [16].
The penetration and establishment of successful carbapenem-resistant clones into the human intestinal microbiota has become a matter of great concern. The improvement of surveillance screening programs for detecting multidrug-resistant microorganisms, the promotion of rapid microbiological diagnosis, as well as the reduction of excessive consumption of antimicrobials are necessary actions to prevent the emergence and global dissemination of CPE, including the community setting.
During an active surveillance-screening program for detecting ESBLs-carriers (R-GNOSIS European Project), we detected a relatively high number of patients co-colonized during the hospital stay with multiple CPE. The aim of our study was to define the microbiological features, the population structure and the plasmid content of CPE strains recovered from co-colonized patients and to associate these findings with their clinical characteristics.
2. Methods
2.1. Study design and patient's data
A total of 15,556 rectal swabs from 8209 patients admitted at Ramon y Cajal University Hospital (Madrid, Spain) were collected from March 4th, 2014 through March 31st, 2016, as part of an active surveillance-screening program for detecting ESBL-carriers (R-GNOSIS-FP7-HEALTH-F3-2011-282,512, www.r-gnosis.eu/). During this project, the CPE incidence was 2% (162/8209; CI 95%= 1.7%−2.3%) and 198 CPE isolates were recovered [16]. Most of the patients were colonized with carbapenemase-producing K. pneumoniae (102/162; 62.9%) and E. coli (37/162; 22.8%), and OXA-48 (104/162; 64.2%) was the most frequent carbapenemase followed by VIM-1 (50/162; 30.9%), NDM-1 (9/162; 5.6%) and KPC-3 (6/162; 3.7%).
In this work we have focused in the subset of patients colonized with multiple CPE. Accordingly, co-colonization was defined as the detection of ≥2 different species or clones within the same species of CPE in faecal cultures from the same host along their hospital stay [17]. Therefore, two subgroups were identified: patients co-colonized with multiple CPE producing the same carbapenemase (CPE-SC) and patients co-colonized with multiple CPE producing a different carbapenemase (CPE-DC).
According to the guidance of prevention and control against infection with CPE, infection control measures were implemented immediately after identification of each new case of colonization and/or infection [18].
Clinical and epidemiological characteristics were retrospectively reviewed and the following data were included: age, gender, patient location (hospital ward), length of stay (LOS), LOS until the first (LOS-1) and second (LOS-2) positive culture for carbapenemase production, infection and colonization sites, antibiotic treatment and underlying diseases. The study was approved by the Ramón y Cajal University Hospital Ethics Committee (Reference 251/13).
2.2. Screening protocol and bacterial identification
Rectal samples were obtained from 8209 patients within 72 h of ward admission and at discharge in those with a hospital stay ≥3 days. Additional samples were recovered weekly in patients hospitalized ≥7 days. Samples were directly plated on ChromoID-ESBL and –CARB/OXA-48 selective agar media (BioMérieux, Marcy l'Etoile, France) and growing colonies were identified by MALDI-TOF MS (Bruker Daltonics, Bremen, Germany). Carbapenemase production was confirmed with KPC/MBL/OXA-48 Confirm Kit test (Rosco Diagnostica, Taastrup, Denmark) and the Modified Hodge Test (MHT) [16].
2.3. Susceptibility testing
Antimicrobial susceptibility testing was determined with the MicroScan automated system (Beckman Coulter, CA, USA). Results were interpreted according to EUCAST guidelines (EUCAST breakpoint v7.1, www.eucast.org). Isolates categorized as intermediate were considered as resistant.
2.4. Resistance genes characterization
Presence of the blaVIM, blaKPC, blaNDM, blaOXA-48 and blaESBL genes (blaSHV, blaTEM and blaCTX−M) was demonstrated by multiplex PCR [16]. All PCR products were sequenced and compared with available sequences in the GenBank database.
2.5. Clonal relatedness and diversity analysis
Epidemiological relatedness among all CPE isolates was established by PFGE-XbaI digestion following standard procedures and clustering analysis of genotyping was carried out with the BioNumerics software (Applied Maths NV, Sint-Martens-Latem, Belgium). K. pneumoniae and E. coli PFGE clusters were also studied by MLST. The Simpson Diversity Index (SDI) based on clones identified by the PFGE method was used to evaluate the K. pneumoniae and E. coli populations diversity. The SDI is expressed between the values 0 (maximum clonality) and 1 (maximum diversity).
2.6. Transferability of carbapenemase genes and plasmid characterization
In order to characterize the carbapenemase-containing plasmids, conjugation and transformation assays were performed in non-duplicated bacterial isolates per patient. In vitro genetic transfer of carbapenemase genes was tested by filter mating assays using the azide-resistant E. coli J53 as recipient strain at a donor:recipient ratio of 1:2. Transconjugants were selected in MacConkey agar plates containing sodium azide (100 µg/ml) and ertapenem (0.5 µg/ml). Plasmid DNA purification with Qiagen Plasmid Midi Kit (Qiagen, Düsseldorf, Germany) and subsequent heat shock transformation using DH5-α E. coli as recipient were also performed in those strains in which conjugation was not effective. Transconjugants and transformants were confirmed by PCR and sequencing. The plasmid size was studied by S1 nuclease-digestion. Southern-blot DNA transfer and hybridization were also performed. Plasmid DNA in all transconjugants was also extracted using Qiagen Plasmid Midi Kit. Plasmid incompatibility groups were investigated by the PCR-based replicon typing (PBRT) scheme. The repA, traU and parA genes were detected by PCR to relate OXA-48-encoding plasmids to the IncL/M-pOXA-48a-plasmid backbone. Plasmid similarity was assessed comparing DraI- and HpaI-digested plasmid DNA profiles in all transconjugants/transformants.
2.7. Statistical analysis
Differences concerning patient characteristics were determined using the Kruskal-Wallis, ANOVA, Student's T and Fisher's Exact tests. Fisher's Exact test was also used to estimate differences in co-colonization rates of different carbapenemase types and the prevalence of different CPE species. Student's T and Z-tests were used to calculate 95% confidence intervals. All tests were performed using R software (RStudio, Boston, MA, http://www.rstudio.com/) and P-value <0.05 was considered statistically significant. Each patient contributed to the study once and the statistical analysis included the episode in which co-colonization with CPE was detected.
3. Results
3.1. Frequency of co-colonization with CPE and characteristics of patients
During the studied period, 30 patients (18.5%, CI 95% = 12.5% − 24.5%) showed co-colonization with multiple CPE. Twenty-five patients (25/162; 15.4%, CI 95%= 10%−21%) (patients 1–25) were co-colonized with two (21/25, 84.0%) or three (4/25, 16.0%) different species [or clones within the same species (2/25, 8.0%)] of CPE producing the same carbapenemase (CPE-SC). Additionally, co-colonization with multiple CPE each of which produced a different carbapenemase (CPE-DC) was also demonstrated in seven patients (7/162; 4.3%, CI 95%= 1.2%−7.4%) (patients 24–30). It should be noted that both co-colonization events were identified in two cases (patients 24 and 25). Among CPE isolates recovered from co-colonized patients, OXA-48 (25/30, 83.3%) was the most frequent carbapenemase, followed by VIM-1 (8/30, 26.7%), NDM-1 (3/30, 10%) and KPC-3 (1/30, 3.3%) producers (Table 1).
Table 1.
Data of patients co-colonized with carbapenemase-producing Enterobacterales.
| First positive culture |
Second positive culture |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Age | Gender | Ward | Underlying | Previous | ATB in the | AS | LOS | Previous | Bacterial | CP | LOS-1 | Previous | Bacterial | CP | LOS-2 | Clinical |
| diseases | admissions/CPE | previous year | (days) | ATB | ID | (days) | ATB | ID | (days) | sample | |||||||
| 1 | 81 | M | P | COPD | NO | – | NG | 9 | AMC, CLR | Ka + Ro | OXA-48 | 9 | – | – | – | – | – |
| 2 | 98 | M | U | Pneumonia, CRD | YES | LEV, AMC, CXM | PS | 15 | AMC | Kp + Ec | OXA-48 | 2 | AMC | Kp + Ec | OXA-48 | 8 | Kp (Urine and BS) |
| 3 | 40 | F | N | Spondylodiscitis | YES | VAN | NG | 35 | CTX, VAN | Kp | OXA-48 | 13 | CTX, VAN | Kp + Ec | OXA-48 | 19 | – |
| 4 | 88 | F | G | Diverticulitis | YES | LEV, CIP, FOS | PS | 28 | MTR, CIP | Ec | OXA-48 | 3 | MTR, VAN | Kp | OXA-48 | 14 | Kp (Urine) |
| 5 | 80 | M | P | Pneumonia | YES | AMC, TZP | PS | 29 | Ka | OXA-48 | 1 | TZP, VAN, MER, MTR | Kp + Ec | OXA-48 | 15 | – | |
| 6 | 65 | F | P | COPD | YES | LEV | NR | 161 | CRO, CD, CXM, AMC | Kp | OXA-48 | 15 | TZP | Kp + Ec | OXA-48 | 26 | Kp + Ec (Wound) |
| 7 | 83 | M | P | Pulmonary fibrosis | YES | GEN, MET, LEV, ERT, AZM, FEP | NG | 81 | GEN, MTR | Kp + Ko | OXA-48 | 17 | MER | Kp + Cf | OXA-48 | 81 | – |
| 8 | 38 | M | P | Alveolar proteinosis | NO* | MER, CIP | PS | 9 | – | Kp + Ec | OXA-48 | 2 | MER | Kp + Ec | OXA-48 | 9 | – |
| 9 | 78 | M | P | Pulmonary neoplasia | YES/Ec | LEV | NR | 30 | TZP, AMC | Kp + Cf | OXA-48 | 16 | AMC | Ec + Cf | OXA-48 | 23 | – |
| 10 | 52 | F | G | ERCP(SI) | YES/Ec | LEV | PS | 1 | – | Ec + Ko | OXA-48 | 1 | – | – | – | – | – |
| 11 | 85 | F | G | COPD | YES | CRO, AMC, AMP, LEV, FOS, CIP, MTR | PS | 20 | – | Ec | OXA-48 | 1 | LEV | Kp + Ec | OXA-48 | 11 | – |
| 12 | 86 | F | P | Pneumonia | NO* | – | PS | 3 | CRO y CD | Kp + Ec | OXA-48 | 2 | – | – | – | – | – |
| 13 | 94 | F | U | CRD | YES | MER, COL, CXM, LEV, AMC | PS | 6 | AMC | Kp + Ec | OXA-48 | 2 | – | Kp + Ec | OXA-48 | 6 | – |
| 14 | 84 | M | G | ARD | NO | – | NG | 23 | MET, CIP, AZT, TIG, DAP | Kp + Ec | OXA-48 | 21 | – | – | – | – | – |
| 15 | 87 | M | U | Bladder tumor infiltration | YES/Kp | CZ, AMC, CRO, CIP, TZP | PS | 19 | CRO | Kp + Ec | OXA-48 | 1 | AMC | Kp + Ec | OXA-48 | 4 | – |
| 16 | 68 | M | N | Brain aneurysm | NO | – | NG | 84 | AMC | Kp | OXA-48 | 45 | AMC | Kp + Ec | OXA-48 | 53 | Kp (Urine) |
| 17 | 49 | M | U | Cholangitis | YES / Kp + Ec | AMC, TZP, MER, AMK | PS | 5 | AMC, TZP | Kp + Ec | OXA-48 | 3 | – | – | – | – | Kp(prosthesis) |
| 18 | 87 | M | U | Cholangitis | YES | CIP, MTR | PS | 20 | AMC, GEN, CD | Kp + Ec | OXA-48 | 1 | – | Kp + Ec | OXA-48 | 6 | – |
| 19 | 71 | F | P | Pneumonia | YES | CZ, AMC | PS | 11 | LEVO | Kp + Ec | OXA-48 | 5 | LEV | Kp + Ec | OXA-48 | 11 | – |
| 20 | 90 | F | G | Cholangitis | YES/ Kp + Ec | MER, ERT, AMK, FEP, AMC, CIP | NR | 9 | TZP | Kp + Ec | OXA-48 | 9 | – | – | – | – | – |
| 21 | 72 | M | G | Septic shock of biliary origin | YES | TZP, MER, LIN, CIP | NR | 41 | LZD, MER | Kp | VIM-1 | 14 | LZD | Ec | VIM-1 | 14 | – |
| 22 | 64 | M | U | SI (Endoscopy) | YES | CZ, AMC, CX, TZP, CIP LZD, MER | NR | 79 | TZP | Cf | VIM-1 | 27 | Ko | VIM-1 | 79 | Ko (Urine) | |
| 23 | 36 | M | N | Cervical pain | NO* | – | NG | 58 | AMC, CZ | Kp | NDM-1 | 27 | AMC, CIP | Kp | NDM-1 | 58 | – |
| 24 | 66 | M | P | Pneumonia | YES | TZP | PS | 14 | – | Ec | KPC-3 | 1 | MER | Kp Ec | KPC-3 VIM-1 | 14 | – |
| 25 | 49 | M | N | Subarachnoid hemorrhage | NO | – | NG | 88 | VAN, MER, LZD | Kp | NDM-1 VIM-1 | 29 | – | Kp + Ec | VIM-1 | 79 | – |
| 26 | 92 | M | N | Cranioencephalic trauma | YES | TZP | NG | 92 | CIP | Kp | NDM-1 | 41 | TZP | Kp | NDM-1 OXA-48 | 52 | Kp NDM-1 (Urine) |
| 27 | 80 | F | G | Esophageal prosthesis (SI) | YES/ Kp + Ec | TZP, CZ, ERT, MER, VAN, LZD, SXT | PS | 6 | – | Ec | VIM-1 | 1 | – | Kp | OXA-48 | 2 | – |
| 28 | 75 | M | N | Cerebral cavernoma | NO | – | NG | 114 | VAN, CIP, AMK, CD, AZT | Ec | OXA-48 | 68 | CIP, AZT | Kp | VIM-1 | 71 | – |
| 29 | 76 | M | P | Pulmonary adenocarcinoma | YES | – | NG | 106 | CXM, AMC, MER, LZD, AMK, TZP, VAN | Ecl | VIM-1 | 80 | – | Kp | OXA-48 | 88 | Ecl (Urine) |
| 30 | 44 | M | P | Pulmonary adenocarcinoma | NO | – | NG | 24 | AMC, CX | Kp | VIM-1 | 10 | TZP | Ec | OXA-48 | 24 | – |
CPE = Carbapenemase producing Enterobacterales; ATB = Antibiotics; AS = Admission sample; CP = Carbapenemase; F = Female; M = Male; G = Gastroenterology; P = Pneumology; N = Neurosurgery; U = Urology; LOS = Length of stay; LOS-1 = Length of stay until first positive CPE culture; LOS-2 = Length of stay until second positive CPE culture; NG = Negative; PS = Positive; NR = Not registered; SI = Scheduled income; COPD = Chronic obstructive pulmonary disease; CRD = Chronic renal disease; ARD = acute renal disease; ERCP = Endoscopic retrograde cholangiopancreatography; Ka = Kluyvera spp.; Ro = R. ornithinolytica; Kp = K. pneumoniae; Ec = E. coli; Ecl = E. cloacae; Ko = K. oxytoca; Cf = C. freundii; AMC = Amoxicillin-clavulanic acid; AMK = Amikacin; AMP = Ampicillin; AZM = Azithromycin; AZT = Aztreonam; CD = Clindamycin; CIP = Ciprofloxacin; CLR = Clarithromycin; COL = Colistin; CRO = Ceftriaxone; CTX = Cefotaxime; CX = Cloxacillin; CXM = Cefuroxime; CZ = Cefazoline; DAP = Daptomycin; ERT = Ertapenem; FEP = Cefepime; FOS = Fosfomycin; GEN = Gentamicin; LEV = Levofloxacin; LZD = Linezolid; MER = Meropenem; MTR = Metronidazol; STX = Trimethoprim sulfamethoxazole;TIG = Tigecycline; TZP = Piperacillin-tazobactam; VAN = Vancomycin.
Previous admission in other health care center.
Even though the low number of cases precludes statistical significance (p = 0.11), overall, patients co-colonized with CPE-DC had longer LOS [63 days, CI 95% (20–107 days)] than CPE-SC-patients [35 days, CI 95% (20–51 days)] (p = 0.20) and patients colonized with a single CPE [32 days, CI 95% (25–38 days)] (p = 0.09). Conversely, LOS-1 was lower in CPE-SC-patients [9 days, CI 95% (5–14 days)] than among patients with CPE-DC [33 days, CI 95% (3–62 days)] (p = 0.15) or with a single CPE [22 days, CI 95% (17–26 days)] (p = 0.0052). Furthermore, LOS-2 was longer in CPE-DC-patients [47 days, CI 95% (16–79 days)] than in CPE-SC-patients [27 days, CI 95% (15–39 days)] (p = 0.19). It should be noted that CPE were more frequently recovered in samples from infective sites in both co-colonized subgroups [CPE-DC (28.6%) (p = 0.35; odds ratio = 0.51; CI 95%= 0.08–5.59) and CPE-SC (24%) (p = 0.39; odds ratio = 0.61; CI 95%= 0.20–2.08] than among patients with a single CPE (15.2%) (p = 0.11; odds ratio=0.46; CI 95%= 0.17–1.38) (Table 2). Overall, urine samples were the most frequent (6/9, 66.7%).
Table 2.
Patient characteristics during the R-GNOSIS Project.
| TOTAL | Co-colonization with CPE-SC |
Co-colonization with CPE-DC |
Colonization with a single CPE |
P- value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | CI 95% | Mean | CI 95% | Mean | CI 95% | Mean | CI 95% | ||
| Age (y)a | 71 | 69–74 | 73 | 66–80 | 69 | 53–85 | 71 | 69–73 | 0.744 |
| LOS (d)a | 33 | 28–39 | 35 | 20–51 | 63 | 20–107 | 32 | 25–38 | 0.113 |
| LOS-1 (d)b | 20 | 16–25 | 9* | 5–14 | 33 | 3–62 | 22* | 17–26 | 0.016 |
| LOS-2 (d)c | – | – | 27 | 15–39 | 47 | 16–79 | – | – | 0.194 |
| N | % | N | % | N | % | N | % | ||
| Malesd | 97 | 59.9 | 16 | 64.0 | 6 | 85.7 | 77 | 58.3 | >0.05 |
| Clinical sampled | 28 | 17.3 | 6 | 24.0 | 2 | 28.6 | 20 | 15.2 | >0.05 |
| TOTAL | 162 | 2.0 | 25 | 15.4 | 7 | 4.3 | 132 | 81.5 | |
CPE-SC = CPE producing the same carbapenemase.
CPE-DC = CPE producing a different carbapenemase.
ANOVA Test was used to determine the P-value.
Kruskal-Wallis Test was used to determine the P-value.
Student's T-Test was used to determine the P-value.
Fisher's Exact Test was used to determine the P-value.
P-value (Mann Whitney Test) ≤ 0.0052.
A sample at admission was recovered in most of the CPE-SC-patients (20/25) and 65% (13/20) of them had a positive CPE culture. Most of them had a previous admission in our hospital (18/25, 72%) or in other health care centers (3/25, 12%) and had been previously treated with multiple antibiotics. Quinolones (65.2%) and β-lactam-β-lactamase inhibitor combinations (65.2%) were the antimicrobials most frequently used, followed by carbapenems (43.5%) and cephalosporins (43.5%). Finally, most of the CPE-DC-patients (71.4%, 5/7) had a negative CPE result in the admission sample. All of them received multiple antibiotics during hospitalization until the CPE detection and the β-lactam-β-lactamase inhibitor combinations (60%), carbapenems (40%) and cephalosporins (40%) were the groups most frequently used (Table 1).
3.2. Events and dynamics of co-colonization
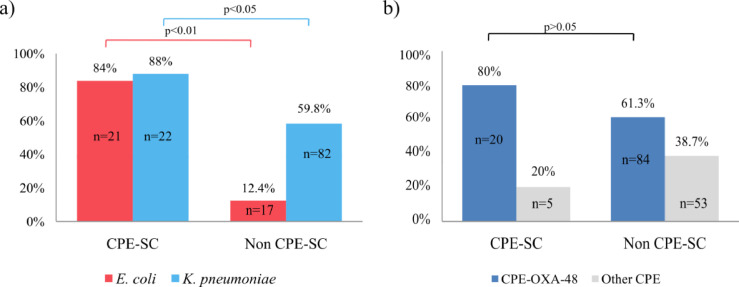
Among CPE-SC-patients, K. pneumoniae (22/25, 88%) and E. coli (21/25, 84%) were the predominant species. In 20 cases (20/25, 80%), both species were detected growing together (16/20) or in subsequent positive cultures (4/20) along the hospital stay (Table 1). K. pneumoniae (88%) or E. coli (84%) colonization was significantly more frequent in these CPE-SC patients than among the rest of the patients [K. pneumoniae: 82/137, 59.8%; (p = 0.006) (odds ratio = 0.20; CI 95% 0.40–0.73) E. coli: 17/137, 12.4%; (p < 0.001), (odds ratio = 0.03; CI 95% 0.01–0.09)] (Fig. 1a). OXA-48-co-colonization (20/25, 80%) was predominant with respect to other carbapenemases (p = 0.11) (odds ratio = 0.39; CI 95% 0.11–1.18) (Fig. 1b). Interestingly, OXA-48-co-colonization with K. pneumoniae and E. coli was the most frequent combination (15/20) and colonization with OXA-48-K. pneumoniae in a previous admission was demonstrated in four of these cases (4/15). It was remarkable that both species producing OXA-48 were persistently isolated in most patients in whom subsequent cultures were available [K. pneumoniae (10/11, 90.9%) and E. coli (7/8, 87.5%)]. K. pneumoniae and E. coli were also recovered coexisting along with other OXA-48 producing species but in a lower proportion (Table 1). The presence of ESBLs in CPE was demonstrated in 21 patients (84%). CTX-M-15 was the most frequent ESBL (18/21, 85.7%) and was mainly associated with OXA-48-K. pneumoniae (13/18, 72.2%) (Table S1).
Fig. 1.
(a) Frequency of intestinal recovery of E. coli or K. pneumoniae in patients co-colonized with carbapenemase-producing-Enterobacterales (CPE). CPE producing the same carbapenemase (CPE-SC), and non-CPE-SC patients (co-colonized with CPE producing different carbapenemases and with a single CPE); (b) Frequency of intestinal recovery of OXA-48 carbapenemase strains/clones compared with other carbapenemases, in CPE-SC and non-CPE-SC patients. P-value was calculated using the Fisher's Exact Test.
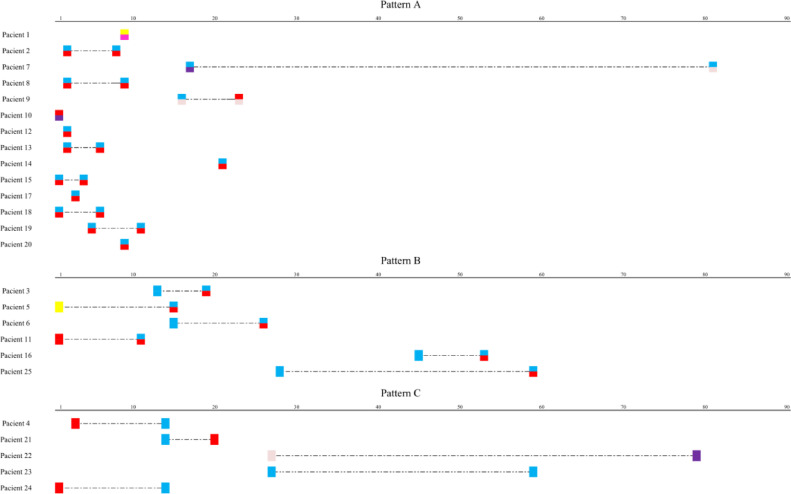
Three co-colonization patterns were identified among CPE-SC-patients: (i) co-colonization detected in the first positive culture (pattern A), (ii) co-colonization detected in the second positive culture, after the first CPE detection (pattern B) and (iii) colonization with different CPE in the first and subsequent cultures (pattern C). Pattern A was detected in 14/25 (56%) patients and all CPE isolates were OXA-producers. Most of them had previous admissions in our hospital (10/14, 71.4%) or in other health care centers (2/14, 14.3%). Pattern B was identified in six patients, five of whom were OXA-48 carriers. In three cases, OXA-48-K. pneumoniae strains were recovered in the first positive culture. Finally, pattern C was mostly identified among patients co-colonized with VIM-1, NDM-1 and KPC-3 producers (Fig. 2).
Fig. 2.
Dynamics of patients’ co-colonization with different carbapenemase producing Enterobacterales (CPE), producing the same carbapenemase during the R-GNOSIS Project. Numbers scale indicates the colonization or co-colonization day from the admission. Dashed line represents the LOS (length of hospital stay).
On the other hand, K. pneumoniae isolates were found in all CPE-DC-patients, while E. coli strains were identified in 71.4% (5/7) of them. Both species were also significantly more prevalent in this subgroup of co-colonized patients than among the rest of the patients [K. pneumoniae: 95/155, 61.3% (p = 0.047) (odds ratio = 0.0; CI 95% 0.0–1.15), E. coli: 32/155, 20.6% (p = 0.007) (odds ratio = 0.11; CI 95% 0.01–0.68)]. Each of these species was detected in the first positive culture in three cases, but persistence along subsequent cultures was not demonstrated (Table 1).
3.3. Molecular typing and clonal diversity
Among the CPE-SC patients, two cases of co-colonization with two different K. pneumoniae clones were identified (patients 23 and 25). In the remaining patients, identical PFGE profiles were observed among CPE isolates of the same bacterial species recovered in subsequent positive cultures. Among patients co-colonized with K. pneumoniae, ST11 (8/22, 36.4%), ST15 (3/22, 13.6%), ST101 (2/22, 9%) and ST307 (2/22, 9%) high-risk clones were identified. Conversely, high-genetic diversity was found among E. coli isolates (19 PFGE profiles) and clonal complex 10 (clone A) was the most frequent (4/21, 19%) (table S1). Differences were observed in terms of clonal diversity between K. pneumoniae (SDI = 0.89) and E. coli (SDI = 0.98). For the other species, PFGE-XbaI revealed a single Kluyvera spp. clone; two Citrobacter freundii clones [A (n = 2) and B (n = 1)] and three distinct clones of Klebsiella oxytoca (A-C) (Table S1).
Among CPE-DC-patients, K. pneumoniae ST11 (2/7, 28.6%) and ST101 (2/7, 28.6%) high-risk clones were also detected. Additionally, the K. pneumoniae ST54-VIM-1 clone was also identified (3/7, 42.8%). Conversely, all E. coli isolates were grouped into different PFGE patterns and STs (Table S1).
3.4. Transferability of carbapenemase genes and plasmid characterization
All blaOXA-48, blaVIM-1 and blaKPC-3 genes were successfully transferred to the azide-resistant E. coli J53 strain, while blaNDM-1 transconjugants were only obtained from a single clone (ST101). From Kluyvera spp., two transformants carrying blaOXA-48 were obtained (Table S1).
blaOXA-48, blaKPC-3 and most blaVIM-1 were located on a ca. 60 kb transferable plasmid in clinical isolates and transconjugant/transformant strains. All ca. 60 kb blaOXA-48- and blaVIM-1- encoding plasmids were assigned to the broad host-range IncL group. RFLP patterns showed comparable restriction profiles (profile A) and repA, parA and traU genes were amplified, indicating a close relation with the IncL/M-pOXA-48a plasmid previously described. [12] The blaCTX−M-15 gene was also located on these plasmids in both clinical and transconjugant isolates in only three K. pneumoniae isolates (cases 4, 13 and 26). In the K. pneumoniae ST54 clone a ca. 90 kb pVIM-1 + SHV-12 plasmid, nontypeable by PBRT, was identified. Concerning the blaKPC-3-harboring plasmid, the incompatibility group IncN was assigned following the PRBT scheme. Moreover, the restriction profiles (profile B) were different from those of blaOXA-48- and blaVIM-1-containing plasmids. Amplification for repA, traU and parA (genes related to the IncL/M-pOXA-48a backbone) was not demonstrated in blaVIM-1- (ca. 90 kb), blaKPC-3- (ca. 60 kb) and blaNDM-1- (ca. 120 kb) containing plasmids (Table S1).
Overall, OXA-48-harboring transconjugants/transformants exhibited different levels of non-susceptibility to carbapenems (58% ertapenem, 25% imipenem and 17% meropenem). In contrast to clinical isolates, these transconjugants/transformants showed high susceptibility to broad-spectrum cephalosporins and aztreonam. Moreover, all transconjugants carrying pNDM-1, pVIM-1 or pKPC-3 showed high resistance to carbapenems and broad-spectrum cephalosporins (Table 3).
Table 3.
Antibiotic non-susceptibility (resistant plus intermediate).
| OXA-48 |
VIM-1 |
KPC-3 |
NDM-1 |
|||||
|---|---|---|---|---|---|---|---|---|
| CI | TC | CI | TC | CI | TC | CI | TC | |
| No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | |
| Amoxicillin-Clavulanate | 48 (100) | 48 (100) | 12 (100) | 12 (100) | 2 (100) | 2 (100) | 4 (100) | 3 (100) |
| Piperacillin- tazobactam | 48 (100) | 48 (100) | 12 (100) | 12 (100) | 2 (100) | 2 (100) | 4 (100) | 3 (100) |
| Cefotaxime | 34 (70.8) | 10 (20.8) | 12 (100) | 12 (100) | 2 (100) | 2 (100) | 4 (100) | 3 (100) |
| Ceftazidime | 30 (62.5) | 10 (20.8) | 12 (100) | 12 (100) | 2 (100) | 2 (100) | 4 (100) | 3 (100) |
| Cefepime | 27 (56.2) | 6 (12.5) | 12 (100) | 12 (100) | 2 (100) | 2 (100) | 4 (100) | 3 (100) |
| Aztreonam | 28 (58.3) | 8 (16.7) | 5 (41.7) | 3 (25) | 2 (100) | 2 (100) | 4 (100) | 0 (0) |
| Imipenem | 9 (18.7) | 12 (25) | 6 (50) | 5 (41.7) | 2 (100) | 2 (100) | 4 (100) | 3 (100) |
| Meropenem | 31 (64.6) | 8 (16.7) | 7 (58.3) | 2 (16.7) | 2 (100) | 2 (100) | 4 (100) | 3 (100) |
| Ertapenem | 48 (100) | 28 (58.3) | 8 (66.7) | 3 (25) | 2 (100) | 2 (100) | 4 (100) | 3 (100) |
| Gentamicin | 25 (52.1) | 2 (4.2) | 4 (33.3) | 0 (0) | 0 (0) | 0 (0) | 4 (100) | 3 (100) |
| Tobramycin | 25 (52.1) | 1 (2.1) | 11 (91.6) | 0 (0) | 0 (0) | 0 (0) | 4 (100) | 3 (100) |
| Amikacin | 1 (2.1) | 0 (0) | 1 (8.3) | 0 (0) | 0 (0) | 0 (0) | 3 (75) | 3 (100) |
| Ciprofloxacin | 32 (66.7) | 1 (2.1) | 7 (58.3) | 1 (8.3) | 2 (100) | 0 (0) | 4 (100) | 0 (0) |
| Sulfamethoxazole trimethoprim | 31 (64.6) | 1 (2.1) | 8 (66.7) | 1 (8.3) | 2 (100) | 0 (0) | 1 (25) | 0 (0) |
| Tigecycline | 2 (4.2) | 1 (2.1) | 2 (16.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Fosfomycin | 36 (75) | 0 (0) | 7 (58.3) | 0 (0) | 2 (100) | 0 (0) | 3 (75) | 0 (0) |
| Colistin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
(CI = Clinical isolates; TC = Transconjugant).
4. Discussion
In recent years, emergence and occurrence of CPE has been increasing worldwide, including Spain [2], [19]. During the R-GNOSIS Project, the incidence of CPE intestinal carriage was 2% and was mainly due to OXA-48 producers [16]. According to previous publications, prolonged stays in healthcare institutions is recognized as a risk factor for rectal CPE colonization [20], [21]. In the present study, we retrospectively observed a high prevalence of co-colonization involving CPE producing different carbapenemases (4.3%), particularly in those patients with longer hospital stays. Nevertheless, our work indicates that intra-patient transmission of the same carbapenemase gene among different CPE species is probably the most frequent event resulting in co-colonization (15.4%). In subgroup CPE-SC-patients, the mean LOS until the detection of CPE (LOS-1) was significantly lower (9 days) than in patients carrying a single CPE. It should be noted that CPE-SC co-colonization was directly detected in the first positive sample in 56% of these patients, since most of them had previous admissions in our hospital (71.4%) or in other healthcare centers (14.3%) and had received multiple antibiotic treatments. Therefore, our results suggest that previous hospital admissions, prolonged LOS and antibiotic treatments could be a risk factor for co-colonization with CPE producing the same or a different carbapenemase. A recent study has evidenced the inter-species transmission of plasmid-encoding carbapenemases under antibiotic pressure [22]. Colonization with carbapenemase producing isolates is also considered a risk factor for infection [20], [21], [23]. In our study, CPE in infective sites were more frequently detected in co-colonized patients than in those colonized with a single CPE. Although further studies are required to support this finding, our results suggest that colonization with more than one CPE could increase the risk of infection with these multidrug-resistant microorganisms. A limitation of this retrospective study was the lack of a full set of swabs in each patient (admission, weekly and discharge samples). In fact, we cannot rule out that a higher number of cases involving patients co-colonized with multiple CPE could have occurred.
During our study, the incidence of K. pneumoniae and E. coli in patients co-colonized with CPE producing the same or a different carbapenemase was significantly higher than among the rest of the patients. Moreover, we observed that co-colonization with different OXA-48-producing Enterobacterales species was the most frequent event. Overall, K. pneumoniae was the main reservoir of blaOXA-48, although the number of studies reporting cases due to other OXA-48 producing Enterobacterales species is increasing worldwide, including those performed in our hospital [16], [24], [25], [26], [27]. In our study, OXA-48-producing K. pneumoniae and E. coli were recovered simultaneously in the same sample in most patients but, recurrently, after a prior colonization with a single OXA-48-K. pneumoniae. According to our results, we suggest that a diversity of E. coli clones act as the recipients of blaOXA-48 gene which is usually transferred by well-adapted hospital clones of K. pneumoniae. In Spain, the main candidates acting as blaOXA-48 donors are the high-risk K. pneumoniae clones ST11-OXA-48 and ST15-OXA-48 [8], [25]. Efficient in-vivo conjugative transfer of blaOXA-48 from K. pneumoniae to E. coli and the spread among different E. coli clones, might explain the early detection and high frequency of co-colonization with both Enterobacterales species during hospital stay. In our collection, the successful intra-patient horizontal transfer of blaOXA-48 was due to the spread of a dominant and highly transmissible ca. 60 kb IncL plasmid related to the previously described and worldwide disseminated IncL/M-pOXA-48a [12], [28]. The conjugation efficiency of IncL/M-pOXA-48a has been related to the insertion of the composite transposon Tn1999 containing blaOXA-48 into the tir gene [29], [30]. Note that most K. pneumoniae strains also carried blaESBLs genes (76.2%) but transfer to E. coli was non frequent if compared with carbapenemases genes, particularly blaOXA-48, suggesting that probably genes encoding-ESBL are chromosomally located (data not shown). Intra-patient lateral transmission of related pOXA-48 plasmids from high-risk clones of K. pneumoniae to unrelated E. coli isolates has also been occasionally described in other studies [4], [31], [32], [33]. It was remarkable that the cross-species transfer of related IncL VIM-1-containing plasmids was also detected over time during our study. Additionally, strains harboring these IncL pVIM-1 and pOXA-48 plasmids co-existed with other Enterobacterales containing unrelated IncN plasmids carrying blaKPC-3 gene and non-typeable plasmids carrying blaNDM-1 gene, which could contribute to exchange genetic material and facilitate the acquisition of other resistance genes. A recent study suggested that the rate of in vivo horizontal transfer of blaOXA-48 is underestimated and the development of in vivo models rather than in vitro experiments could help to highlight the real magnitude of acquisition and spread of carbapenemase-encoding plasmids [32].
Although, co-colonization with different CPE isolates in our study might be driven by our local epidemiological scenario dominated by OXA-48-producing Enterobacterales, our results suggest that CPE co-colonization is more frequently due to horizontal gene transfer between species than to the sequential acquisition of different CPE isolates. Under antibiotic exposure, CPE co-colonization probably increases in absolute numbers in the intestinal compartment. This fact might facilitate translocation or migration to the urinary tract and we found that co-colonized patients have CPE more frequently in infective sites [23]. Moreover, CPE co-colonization status should be monitored during epidemiological surveillance cultures, as these patients might be at a higher risk for systemic or local (mostly urinary tract) infections. Limitation of the use of antibiotics, shorter antibiotic treatments and reduction of hospital stay might also help to limit intestinal co-colonization with different CPE isolates.
Contributors
MH-G undertook the investigation, methodology, data curation and writing the original draft. PR-G and RC performed the funding acquisition, conceptualization, supervision and writing, reviewing and editing the manuscript. BP-V provided technical support to MH-G during the methodology. CN-S contributed to the data curation, writing, reviewing and editing the draft. FB and MM contributed to the supervision and writing, reviewing and editing the final version of the manuscript. All authors have read and approved the final version of this manuscript.
Declaration of Competing Interest
Authors declare no conflict of interest with the content of this article.
Acknowledgments
Acknowledgements
MH-G was supported with a contract from Instituto de Salud Carlos III, Spain (iP-FIS program, ref. IFI14/00022). We acknowledge financial support from European Commission (grants R-GNOSIS-FP7-HEALTH-F3-2011-282512) and Plan Nacional de I + D + i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0011) cofinanced by European Development Regional Fund “A way to achieve Europe” (ERDF), Operative program Intelligent Growth 2014–2020. The funders did not had any role in study design, data collection, data analysis, interpretation, or writing of the report. We thank the Preventive Medicine Service at Ramón y Cajal University Hospital for collecting the rectal swabs as a part of the R-GNOSIS European Project. We also thank Mrs. Mary Harper for English correction of the manuscript.
Ethics committee approval
This study was approved by the Ramón y Cajal University Hospital Ethics Committee (Reference 251/13).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.eclinm.2019.09.005.
Appendix. Supplementary materials
References
- 1.Nordmann P., Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect [Internet] 2014;20(9):821–830. doi: 10.1111/1469-0691.12719. Available from: [DOI] [PubMed] [Google Scholar]
- 2.Cantón R., Akóva M., Carmeli Y., Giske C.G., Glupczynski Y., Gniadkowski M. Rapid evolution and spread of carbapenemases among enterobacteriaceae in Europe. Clini Microbiol Infect. 2012;18(5):413–431. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 3.Nordmann P., Naas T., Poirel L. Global spread of carbapenemase-producing enterobacteriaceae. Emerg Infect. Dis. [Internet] 2011;17(10):1791–1798. doi: 10.3201/eid1710.110655. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3310682&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruiz-Garbajosa P., Hernández-García M., Beatobe L., Tato M., Méndez M.I., Grandal M. A single-day point-prevalence study of faecal carriers in long-term care hospitals in Madrid (Spain) depicts a complex clonal and polyclonal dissemination of carbapenemase-producing enterobacteriaceae. J Antimicrob Chemother [Internet] 2015;71(October 2015):348–352. doi: 10.1093/jac/dkv355. http://www.jac.oxfordjournals.org/lookup/doi/10.1093/jac/dkv355 Available from: [DOI] [PubMed] [Google Scholar]
- 5.Pitart C., Sole M., Roca I., Fa A., Vila J., Marco F. First outbreak of a plasmid-mediated carbapenem-hydrolyzing OXA-48 NL -Lactamase in Klebsiella pneumoniae in Spain. 2011;55(9):4398–401. [DOI] [PMC free article] [PubMed]
- 6.Ruiz-Garbajosa P., Curiao T., Tato M., Gijón D., Pintado V., Valverde A. Multiclonal dispersal of KPC genes following the emergence of non-ST258 KPC-producing Klebsiella pneumoniae clones in Madrid, Spain. J Antimicrob Chemother. 2013;68(11):2487–2492. doi: 10.1093/jac/dkt237. [DOI] [PubMed] [Google Scholar]
- 7.Gijón D., Curiao T., Baquero F., Coque T.M., Cantón R. Fecal carriage of carbapenemase-producing enterobacteriaceae : a hidden reservoir in hospitalized and nonhospitalized patients. J Clin Microbiol. 2012;50(5):1558–1563. doi: 10.1128/JCM.00020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oteo J., Ortega A., Bartolomé R., Bou G., Conejo C., Fernández-Martínez M. Prospective multicenter study of carbapenemase-producing enterobacteriaceae from 83 hospitals in Spain reveals high in vitro susceptibility to colistin and meropenem. Antimicrob Agents Chemother. 2015;59(6):3406–3412. doi: 10.1128/AAC.00086-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seara N., Oteo J., Carrillo R., Pérez-Blanco V., Mingorance J., Gómez-Gil R. Interhospital spread of NDM-7-producing Klebsiella pneumoniae belonging to ST437 in Spain. Int J Antimicrob Agents [Internet] 2015;46(2):169–173. doi: 10.1016/j.ijantimicag.2015.04.001. Available from: [DOI] [PubMed] [Google Scholar]
- 10.Barrado L., Pérez-Vázquez M., del Pozo J.L., Martín-Salas C., Leiva J., Mazón A. Clonal transmission of NDM-5-producing Escherichia coli belonging to high-risk sequence type ST405. Int J Antimicrob Agents [Internet] 2018;52(1):123–124. doi: 10.1016/j.ijantimicag.2018.05.018. Available from: [DOI] [PubMed] [Google Scholar]
- 11.Brañas P., Villa J., Viedma E., Mingorance J., Orellana M., Chaves F. Molecular epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital in Madrid: successful establishment of an OXA-48 ST11 clone. Int J Antimicrob Agents. 2016;46(February 2014):111–116. doi: 10.1016/j.ijantimicag.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Poirel L., Bonnin R.A., Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother. 2012;56(1):559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carattoli A., Seiffert S.N., Schwendener S., Perreten V., Endimiani A. Differentiation of INCL and INCM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS ONE. 2015;10(5):1–14. doi: 10.1371/journal.pone.0123063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernańdez J., Montero I., Fleites A., Rodicio M.R. Cluster of Escherichia coli isolates producing a plasmid-mediated OXA-48 beta-lactamase in a Spanish hospital in 2012. J Clin Microbiol. 2014;52(9):3414–3417. doi: 10.1128/JCM.01271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortega A., Sáez D., Bautista V., Fernández-Romero B., Lara N., Aracil B. Carbapenemase-producing Escherichia coli is becoming more prevalent in Spain mainly because of the polyclonal dissemination of OXA-48. J Antimicrob Chemother. 2016;71(8):2131–2138. doi: 10.1093/jac/dkw148. [DOI] [PubMed] [Google Scholar]
- 16.Hernández-García M., Pérez-Viso B., Carmen Turrientes M., Díaz-Agero C., López-Fresneña N., Bonten M. Characterization of carbapenemase-producing enterobacteriaceae from colonized patients in a university hospital in Madrid, Spain, during the R-gnosis project depicts increased clonal diversity over time with maintenance of high-risk clones. J Antimicrob Chemother. 2018;73(July):3039–3043. doi: 10.1093/jac/dky284. [DOI] [PubMed] [Google Scholar]
- 17.Snyder G.M., O'Fallon E.D.E. Co-colonization with multiple different species of multidrug-resistant gram-negative bacteria. Am J Infect Control. 2012;39(6):506–510. doi: 10.1016/j.ajic.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plan de Prevención y control frente a la infección por EPC en la Comunidad de Madrid. Versión 1. [Internet]. Available from:http://www.madrid.org/cs/Satellite?cid=1354274526068&language=%0Aes&pagename=PortalSalud%2FPage%2FPTSA_pintarContenidoFinal&%0Avest=1354274526068.
- 19.Albiger B., Glasner C., Struelens M.J., Grundmann H., Monnet D.L. Carbapenemase-producing enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Eurosurveillance. 2015;20(45):30062. doi: 10.2807/1560-7917.ES.2015.20.45.30062. [DOI] [PubMed] [Google Scholar]
- 20.Akova M., Daikos G.L., Tzouvelekis L., Carmeli Y. Interventional strategies and current clinical experience with carbapenemase-producing Gram-negative bacteria. Clin Microbiol Infect [Internet] 2012;18(5):439–448. doi: 10.1111/j.1469-0691.2012.03823.x. Available from: [DOI] [PubMed] [Google Scholar]
- 21.Paño Pardo J.R., Villar S.S., Ramos Ramos J.C., Pintado V. Infections caused by carbapenemase-producing enterobacteriaceae: risk factors, clinical features and prognosis. Enferm Infecc Microbiol Clin [Internet] 2014;32(S4):41–48. doi: 10.1016/S0213-005X(14)70173-9. Available from: [DOI] [PubMed] [Google Scholar]
- 22.Rooney C.M., Sheppard A.E., Clark E., Davies K., Hubbard A.T.M., Sebra R. Dissemination of multiple carbapenem resistance genes in an in vitro gut model simulating the human colon. J Antimicrob Chemother [Internet] 2019 Jul 1;74(7):1876–1883. doi: 10.1093/jac/dkz106. http://www.ncbi.nlm.nih.gov/pubmed/30989197 [cited 2019 Aug 13]Available from: [DOI] [PubMed] [Google Scholar]
- 23.Ruppé E., Andremont A. Causes, consequences, and perspectives in the variations of intestinal density of colonization of multidrug-resistant enterobacteria. Front Microbiol. 2013;4(MAY):1–10. doi: 10.3389/fmicb.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimou V., Dhanji H., Pike R., Livermore D.M., Woodford N.Characterization of enterobacteriaceae producing OXA-48-like carbapenemases in the UK. 2013;(April 2012):1660–5. [DOI] [PubMed]
- 25.Palacios-Baena Z., Oteo J., Conejo C., Larrosa M., Fernández-Martínez M., González-López J. Comprehensive clinical and epidemiological assessment of colonisation and infection due to carbapenemase-producing enterobacteriaceae in Spain. J Infect. 2016;72(2):152–160. doi: 10.1016/j.jinf.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Arana D.M., Saez D., García-Hierro P., Bautista V., Fernández-Romero S., De la Cal M. Concurrent interspecies and clonal dissemination of OXA-48 carbapenemase. Clin Microbiol Infect. 2015;21(2) doi: 10.1016/j.cmi.2014.07.008. 148.e1-148.e4. [DOI] [PubMed] [Google Scholar]
- 27.Hernández-García M., León-Sampedro R., Pérez-Viso B., Morosini M.I., López-Fresneña N., Díaz-Agero C. First report of an OXA-48- and CTX-M-213-producing Kluyvera species clone recovered from patients admitted in a university hospital in Madrid, Spain. Antimicrob Agents Chemother. 2018;62(11):1–10. doi: 10.1128/AAC.01238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira P.S., Felipe C., Araujo M.De, Seki L.M., Zahner V., Paula A., et al. Update of the molecular epidemiology of KPC-2-producing Klebsiella pneumoniae in Brazil: spread of clonal complex 11 (ST11, ST437 and ST340). 2013;11(October 2012):312–6. [DOI] [PubMed]
- 29.Carrër A., Poirel L., Yilmaz M., Akan Ö.A., Feriha C., Cuzon G. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother. 2010;54(3):1369–1373. doi: 10.1128/AAC.01312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potron A., Poirel L., Nordmann P. Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob Agents Chemother. 2014;58(1):467–471. doi: 10.1128/AAC.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skalova A., Chudejova K., Rotova V., Medvecky M., Studentova V., Chudackova E. Molecular characterization of OXA-48-like-producing enterobacteriaceae in the Czech Republic and evidence for horizontal transfer of pOXA-48-like plasmids. Antimicrob Agents Chemother [Internet] 2017;61(2) doi: 10.1128/AAC.01889-16. pii: e01889-16. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Göttig S., Gruber T.M., Stecher B., Wichelhaus T.A., Kempf V.A.J. In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin Infect Dis. 2015;60(12):1808–1815. doi: 10.1093/cid/civ191. [DOI] [PubMed] [Google Scholar]
- 33.Willemsen I., van Esser J., Kluytmans-van den Bergh M., Zhou K., Rossen J.W., Verhulst C. Retrospective identification of a previously undetected clinical case of OXA-48-producing K. pneumoniae and E. coli: the importance of adequate detection guidelines. Infection. 2016;44(1):107–110. doi: 10.1007/s15010-015-0805-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.