Abstract
Introduction
Inherited susceptibility to lung cancer risk in never smokers is poorly understood. The major reason for this gap in knowledge is that this disease is relatively uncommon (except in Asians), making it difficult to assemble an adequate study sample. In this study we conducted a genome-wide association study (GWAS) on the largest, to date, set of European-descent never smokers with lung cancer.
Methods
We conducted a two-phase (discovery and replication) GWAS in never smokers of European descent. We further augmented the sample by performing a meta-analysis with never smokers from the recent OncoArray study, which resulted in a total of 3,636 cases and 6,295 controls. We also compare our findings with those in smokers with lung cancer.
Results
We detected three genome-wide statistically significant SNPs rs31490 (OR 0.769, 95% confidence interval (CI) [0.722–0.820], p-value 5.31×10−16), rs380286 (OR 0.770, 95% CI [0.723-0.820], p-value 4.32×10−16), and rs4975616 (OR 0.778, 95% CI [0.730–0.829], p-value 1.04×10” 14). All three mapped to Chromosome 5 CLPTM1L-TERT region, previously shown to be associated with lung cancer risk in smokers and in never smoker Asian women, and risk of other cancers including breast, ovarian, colorectal and prostate.
Conclusions
We found that genetic susceptibility to lung cancer in never smokers is associated to genetic variants with pan-cancer risk effects. The comparison with smokers shows that top variants previously shown to be associated with lung cancer risk only confer risk in the presence of tobacco exposure, underscoring the importance of gene-environment interactions in the etiology of this disease.
Introduction
Lung cancer is the leading cause of cancer mortality worldwide, accounting for over 1 million deaths each year 1. Although most lung cancer is preventable, since the majority of cases occur in tobacco smokers 2, around 10% of cases are seen in lifetime never-smokers. Even though lung cancer is diagnosed in a minority of never smokers it still ranks as the seventh to ninth most common cause of cancer death worldwide 2.
In never smokers, lung cancer has characteristics distinct from those associated with smoking, including different histology and mutation spectrum 3. The only well-established risk factors for lung cancer in never smokers are exposure to radon 4, secondhand smoke and dust 5, asbestos 6, and, notably, family history of cancer 5, 7,, which has provided evidence for inherited susceptibility.
To date, genome-wide association studies (GWAS) on lung cancer has largely been focused on ever smokers, and have identified 18 independent loci influencing risk 7, 8. While several GWAS studies in never smokers have been conducted, these have primarily been based on Asian women 9–12. Several environmental risk factors for lung cancer, including cooking fumes and air pollution, are highly prevalent in Asian populations 13, raising the possibility of effect modification. Identifying lung cancer susceptibility alleles among never smoking European populations has been limited to candidate gene analyses 14, 15 and small GWA studies 16–18. Reported here are the results of a large GWAS of lung cancer in never smokers of European descent, based on 3,636 cases and 6,295 controls.
Materials and Methods
Study design and samples
Never smokers were defined as individuals who had smoked less than 100 cigarettes over their lifetime. The study had a discovery and a replication series, both from studies participating in the International Lung Cancer Consortium (ILCCO; http://ilcco.iarc.fr). The discovery series, after quality control (Appendix), comprised 1,287 cases and 1,655 controls with European ancestry from seven centers (Table A.1). The replication series comprised 960 cases and 940 controls from 16 study centers, of which some centers (but not study subjects) participated also in the discovery phase (Table A.2). Comprehensive details of each series have been previously reported 17, 19–23. To increase statistical power, data on never smokers recently generated by the OncoArray lung cancer study from ILCCO 20 were also leveraged. After excluding samples overlapping between the OncoArray and the discovery set and between the OncoArray and the replication set, 1,149 cases and 1,144 controls from the discovery, 1,527 cases and 4,211 controls from the OncoArray, and 960 cases and 940 controls from the replication sets were included in the final analyses. Most of the lung cancer cases (76.7% in the discovery, 69.2% in the replication, and 63.1% in the OncoArray sets) had histologically confirmed adenocarcinoma, followed by squamous and small cell carcinoma (Tables A.1–A.3). Given that subtype-specific associations are likely to exist, adenocarcinomas were also analyzed separately. Table 1 presents the demographic characteristics of the final dataset.
Table 1.
Characteristics of never smoking lung cancer cases and controls included in the final dataset.
| Characteristic | Cases (n=3,636) | Controls (n=6,296) | |||
|---|---|---|---|---|---|
| Age, mean, SD | 63.6 | 12.4 | 61.9 | 11.9 | |
| Sex, n, % | Male | 1,156 | 31.8 | 2,595 | 41.2 |
| Female | 2,480 | 68.2 | 3,701 | 58.8 | |
| Histology, n, % | Adenocarcinoma | 2,509 | 69.0 | 6,296 | |
| Squamous cell carcinoma | 310 | 8.5 | 6,296 | ||
Genotyping and quality control
Both cases and controls from the discovery set were genotyped using Illumina Infinium OmniExpress-24 v1.2 BeadChips, with the exception of cases and controls from Harvard School of Public Health (HSPH), genotyped on Illumina Human660W-Quad BeadChip. Genotyping of the replication series for 384 selected SNPs was performed using Illumina GoldenGate technology. Genotyping quality control and SNP selection procedures are detailed in the Appendix. The OncoArray genotyping platform, the never smoker samples to which it was applied, and genotyping and quality control procedures are described in the Appendix and have been previously characterized in detail 20, 24
Data analysis
To harmonize data and address population stratification in the discovery set, the studies were grouped as follows. Provided they used the same genotyping array and study participants were from the similar geographic origin they were combined. This resulted in two groups: UK studies and North American studies. Since the HSPH samples were genotyped on a different platform, these were analyzed separately. Thus the following clusters were used: (i) HSPH, (ii) UK, and (iii) North America (see Table A.4 for more detail). Three separate GWAS analyses were ran based on the three groups. We applied logistic regression analyses with case-control status as the outcome and the SNP genotype as a predictor to identify risk-associated SNPs in these three groups. Additive models, with 0 for the reference allele homozygotes, 1 for heterozygotes, and 2 for variant allele homozygotes were used. Reference alleles were defined as in the hg19 reference genome. Age (continuous variable), sex, secondhand smoke exposure (SHS; from any venue at any period in a lifetime), education level, and study site within the group (if more than one site) were used as covariates. The definition of the education variables and more information on the SHS assessment are given in the Appendix. Missing values for SHS and education status were treated as a separate category. To offset potential effects of population stratification within clusters, SNP based principal components analyses (PCA) were performed 25 and the corresponding first five principal components were included as covariates, even though the PCA of these three GWAS clusters do not suggest population stratification (Figure A.1). An inverse variance fixed effects meta-analysis was used to combine the results for the three group-based GWASs 26.
A brief description of the OncoArray never smoker dataset is provided in the Appendix. To perform the joint analysis of the discovery and the OncoArray sets, inverse variance meta-analysis was used, whereby studies were grouped into five clusters (Discovery-North America,Discovery-UK, OncoArray-North America, OncoArray-UK, and OncoArray-Continental Europe), as detailed in Table A.5. This joint analysis was adjusted for age, sex, study site within the group, and the first five principal components, but not SHS or education level, as they were not available in the OncoArray set.
Criteria for SNP selection and the quality control procedures in the replication phase are described in the Appendix.
Results
We focus on the joint analysis of the discovery and OncoArray sets as having the largest sample size (the results for the discovery set separately are presented in the Appendix, Figure A.2 showing the Q-Q plot that demonstrates no indication of an inflation of type I error (λ=1.005), and Table A.6 presenting the list of the top SNPs derived from the discovery set (p<1×10−4)).
Figure 1 presents the scatter plot of the -log10p-values against the chromosome position (the so-called Manhattan plot) for the meta-analysis of the discovery and the OncoArray samples. The analysis identified 71 genome-wide statistically significant SNPs (P<5×10−8, the accepted genome-wide level of statistical significance 27), all of them mapping to the 5p15.33 CLPTM1L-TERT region. Table A.7 presents the 229 top SNPs at P<10−5. There is also a peak on Chromosome 9 in the CDKN2A region, but none of the SNPs in this regions attained statistical significance at the GWAS level.
Figure 1.
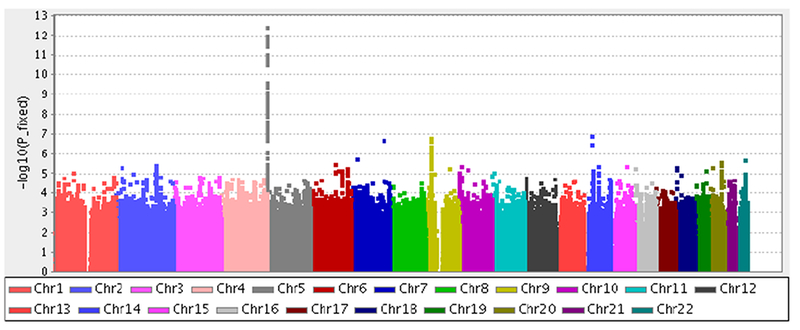
Manhattan plot of the association analysis of lung cancer in European ancestry never smokers performed jointly in the discovery set and the OncoArray samples. The x-axis is chromosomal position, and the y-axis is the statistical significance on a –log10 scale.
The principal component analysis of the replication samples showed no differences by the case-control status for the first five principal components (Figure A.3).
Table A.8 presents the list of nominally statistically significant (p<0.05) SNPs from the replication analysis. The most significant SNPs, rs380286 (p=3.88×10−7), rs31490 (p=4.68×10−7), and rs4975616 (p=2.50×10−6) were located in the 5p15.33 (CLPTM1L-TERT) region (Table 2). These three SNPs were significant after the Bonferroni correction for 370 tests resulting in the p-value of 1.35×10−4 to declare significance (the FDR approach identified the same three SNPs as statistically significant; Table A.8).
Table 2.
The three GWAS-significant (P<5×10−8) variants for lung cancer in European ancestry never smokers, found in the joint analysis of the original discovery set, the never smoker subset of the OncoArray set, and the replication set (6 clusters, 3636 cases, 6295 controls), adjusted for age, sex, and the first five principal components.
| SNP ID | CHR* | Position | Odds Ratio* | 95% CI | P-value* | Reference allele | Effect allele | EAF* | Gene symbol | |
|---|---|---|---|---|---|---|---|---|---|---|
| Lower boundary | Upper boundary | |||||||||
| rs380286** | 5 | 1320247 | 0.770 | 0.723 | 0.820 | 4.32×10−16 | A | G | 0.4169 | CLPTM1L |
| rs31490† | 5 | 1344458 | 0.769 | 0.722 | 0.820 | 5.31×10−16 | G | A | 0.4142 | CLPTM1L |
| rs4975616‡ | 5 | 1315660 | 0.778 | 0.730 | 0.829 | 1.04×10−14 | G | A | 0.4005 | CLPTM1L |
Adjusted for age, gender, and the first 5 principal components; CHR, chromosome; EAF, effect allele frequency
intronic variant
splice variant
downstream gene variant
The 370 candidate SNPs selected for the replication (see Appendix for the selection criteria) were analyzed using all three study population sets: the discovery, the replication, and the OncoArray (total 3,636 cases and 6,295 controls). The analysis identified three SNPs statistically significant at the genome wide level: rs380286 (P=1.6×10−14), rs31490 (P=5.1×10−14), and rs4975616 (P=5.8×10−14; Table 2). These three SNPs are from the CLPTM1L-TERT region and the association with the variant alleles was consistently negative (OR < 1). These SNPs belong to a wide LD block corresponding to the LD Region 2 marked by rs451360 as described in 28. The very high LD between the pairs of SNPs (0.925 for rs380286 and rs31490; 0.915 for rs380286 and rs4975616; 0.955 for rs31490 and rs4975616) did not allow identifying the leading SNP among the three, as there was very little variation in a SNP when the genotypes of the other two were fixed.
The results of the joint analysis of the discovery and replication sets without the OncoArray samples are shown in the Table A.9. In brief, the same 3 SNPs from the CLPTM1L-TERT region were identified as genome-wide statistically significant.
Analysis of only adenocarcinoma cases produced nearly identical results, with only CLPTM1L-TERT region SNPs showing statistical significance (Tables A.10, A.11).
Table 3 summarizes the comparisons between our study results and previous published findings reported in never smokers from genome-wide and candidate gene/SNP association studies in both individuals of European descent and Asians. Our study confirmed SNPs located in 5p15.33 (CLPTM1L-TERT) region. Notably, the direction of the association is highly concordant among the studies for the SNPs in this region. The results for 3q28 (TP63) and 6q22.2 (ROS1-DCBLD1) regions are suggestive in our analysis (P-values of ~10−4 for both these regions). The results from our study for the loci identified in the recently published largest-to-date lung cancer study that involved mostly smokers20 are shown in Table A.12.
Table 3.
Previous findings from the association analyses of lung cancer in never smokers, with a comparison to this study
| Previously Published Studies | This Study* | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Gene | RefSeq* | Study type | Pubmed ID | Histology | Ethnicity | Discovery cases | controls | Replication cases | controls | OR* | P-value | OR | P-value |
| 13q31.3 | GPC5 | rs2352028 | GWAS* | Li et al 17 | NSCLC | Mostly Eur. descent | 377 | 377 | 328 | 407 | 1.46 | 5.90E-06 | 0.99 | 0.95 |
| 5p15.33 | CLPTM1L | rs4975616 | Candidate | Wang et al 15 | NSCLC | Eur. descent | 239 | 553 | - | 0.69 | 7.90E-04 | 0.78 | 1.04E-14 |
| 5p15.33 | CLPTM1L-TERT | rs2736100 | GWAS | Hsiung et al 9 | Adeno | Asian women | 584 | 585 | 2184 | 12515 | 1.5 | 5.40E-11 | 1.3 | 2.66E-09 |
| 10q25.2 | VTI1A | rs7086803 | GWAS | Lan et al 10 | NSCLC | Asian women | 5547 | 4492 | 1085 | 2877 | 1.3 | 5.10E-17 | 1.3 | 0.011 |
| 6q22.2 | ROS1-DCBLD1 | rs9387478 | 0.85 | 7.80E-08 | 0.86 | 1.50E-04 | ||||||
| 6p21.32 | HLA II | rs2395185 | 1.16 | 2.60E-06 | 1.04 | 0.34 | ||||||
| 5p15.33 | CLPTM1L-TERT | rs2736100 | 1.38 | 4.20E-27 | 1.27 | 2.66E-09 | ||||||
| 5p15.33 | CLPTM1L-TERT | rs2853677 | GWAS | Shiraishi et al 12 | Adeno | Asians (Japanese) | 1695 | 5333 | 3328 | 8168 | 1.44 | 3.90E-23 | 1.28 | 1.12E-09 |
| 5p15.33 | CLPTM1L-TERT | rs2736100 | 1.37 | 9.90E-19 | 1.27 | 2.66E-09 | ||||||
| 3q28 | TP63 | rs10937405 | 1.28 | 2.00E-10 | 1.16 | 1.50E-04 | ||||||
| 17q24.3 | BPTF | rs7216064 | 1.21 | 1.50E-06 | 1.1 | 0.054 | ||||||
| 6p21.3 | BTNL2 | rs3817963 | 1.21 | 1.50E-07 | 1.06 | 0.2 | ||||||
| 1q25.1 | ACVR1B | rs10127728 | Candidate | Spitz et al 14 | NSCLC | Mostly Eur. descent | 451 | 508 | - | 1.68 | 3.00E-04 | 1.06 | 0.34 |
| 3q28 | TP63 | rs4488809 | Replication of GWAS findings | Seow et al 11 | Adeno | Asian women | 7448 | 7007 | 0.8 | 4.30E-17 | 0.82 | 8.52E-07 | |
| 5p15.33 | TERT | rs2736100 | 7505 | 7070 | 1.43 | 6.12E-43 | 0.79 | 2.66E-09 | |||||
| 6p21.1 | FOXP4 | rs7741164 | 10531 | 10648 | 1.17 | 3.96E-13 | 0.97 | 8.28E-01 | |||||
| 6p21.3 | BTNL2 | rs3817963 | 7255 | 6745 | 1.16 | 1.63E-07 | 1.06 | 1.97E-01 | |||||
| 6p21.32 | HLA-DPB1 | rs2179920 | 7457 | 7020 | 1.17 | 1.69E-05 | 1.08 | 9.42E-02 | |||||
| 6p21.32 | HLA class II | rs2395185 | 7757 | 9637 | 1.16 | 2.04E-09 | 1.04 | 3.91E-01 | |||||
| 6q22.2 | ROS1/DCBLD1 | rs9387478 | 8022 | 9970 | 0.86 | 5.25E-11 | 0.86 | 1.53E-04 | |||||
| 9p21.3 | rs72658409 | 10780 | 10938 | 0.76 | 2.37E-10 | 0.89 | 1.43E-01 | ||||||
| 10q25.2 | VTI1A | rs7086803 | 7964 | 9914 | 1.25 | 9.22E-17 | 1.31 | 1.12E-02 | |||||
| 12q13.13 | rs11610143 | 10267 | 10634 | 0.85 | 3.55E-13 | 0.97 | 4.88E-01 | ||||||
| 17q24.3 | BPTF | rs7216064 | 7720 | 8630 | 0.86 | 6.19E-09 | 1.10 | 5.43E-02 | |||||
”This study” pertains to the results of the meta-analysis of the discovery and OncoArray sets, except for rs4975616, for which the result from the meta-analysis of the discovery, OncoArray, and replication sets is shown; RefSeq, Reference sequence or SNP ID; GWAS, genome wide association study; OR, odds ratio; nominally significant p-values are shown in bold
A comparison of the regional association plots for the CLPTM1L-TERT region and 15q25 (CHRNA3) region in never smokers and smokers was also performed (whereby the smokers’ data were obtained from the lung OncoArray project) (Figure 2 a,b). We found that the risk association profile plotted as the -log10P for the SNPs in the CLPTM1L-TERT region in never smokers tightly followed that in smokers (Figure 2a). By contrast, the association profiles in the CHRNA3 region (implicated in nicotine dependence) are strikingly different in never and ever smokers, with very high -log10P values in smokers and a flat profile in never smokers (Figure 2b). Analogous comparisons for two other regions, TP63 and CDKN2A, are presented in the Figure A.4.
Figure 2.
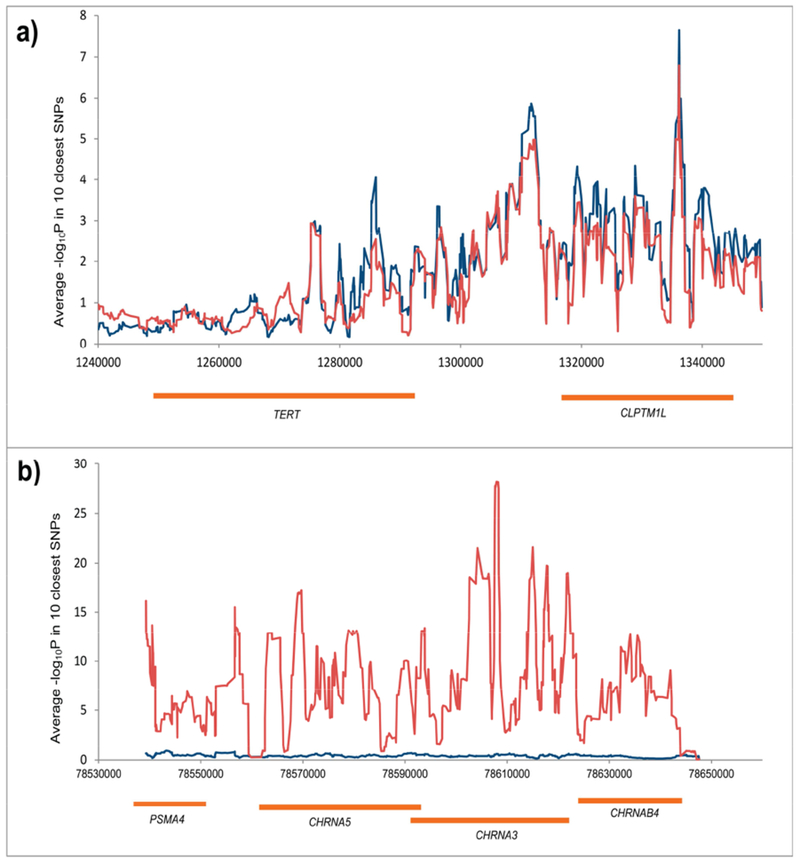
Regional association plots for smokers (red line) and never smokers (blue line) in CLPTM1L-TERT region (a) and CHRNA3-5 region (b). The y axis corresponds to –log10P for 650 SNPs in the CLPTM1L-TERT region and –log10P for 535 SNPs in CHRNA3-5 region. To aid visual representation we selected the 10 closest SNP and computed average –log10P- values.
The analyses of associations for the 3 most statistically significant SNPs from the CLPTM1L-TERT region stratified by the SHS exposure status are shown in the Appendix (Table A.13). There was no indication of SNP-SHS interaction effects or a SNP effect modification by the SHS exposure, as the interaction term was not significant for any of the SNPs.
Discussion
This is the largest lung cancer GWAS so far conducted in never smokers of European descent. However, only one region (CLPTM1L-TERT) strongly associated with lung cancer risk in this patient population was found. Our results for this region corroborate findings by earlier studies of lung cancer in never smokers (Table 3), showing consistent direction of effect. The 5p15.33 CLPTM1L-TERT region SNPs have also been reported to be associated with multiple cancers including lung cancer in smokers 16, breast cancer 29, glioma 30, nasopharyngeal cancer 31 and prostate cancer 32. TERT encodes the catalytic subunit of the telomerase reverse transcriptase, which takes part in adding nucleotide repeats to chromosome ends 33. While active in early development and germ cells, this gene is not expressed in most adult tissues, resulting in a shortening of telomeres with each cell division. When telomeres become critically short, the cell can no longer divide. However, cancer cells can upregulate telomerase, which enables them to continue dividing 34 The CLPTM1L gene is reported to be overexpressed in lung and pancreatic cancer where it promotes growth and survival 35, 36. Also there is a locus within the CLPTM1L gene that serves as a binding site for ZNF148, which promotes expression of TERT 37.
Functional annotation of the top identified SNPs using Encyclopedia of DNA Elements (ENCODE) 38 found that rs4975616 coincides with the binding site for three transcription factors: ELF1, ZEB1 and BCLAF1. Both TERT and CLPTM1L are among the many target genes for ELF1 and ZEB1; CLPTM1L (but not TERT) is among the target genes for BCLAF1. According to Ensemble regulatory database 39, SNP rs31490 is located in the region that acts as a promotor for CLPTM1L in the developing lung. In the Genotype-Tissue Expression (GTEx) 40 all three SNPs: rs31490, rs380286, and rs4975616 are reported as eQTLs for TERT in esophagus and CLPTM1L in skin tissue.
Previously, a fine-mapping study has been conducted on this locus to deeply investigate its association with lung cancer risk 41. The study included a limited number of never smokers and the novel loci identified did not show a significant effect specifically in never smokers. However, the direction of the effect was largely consistent with that in smokers, in line with what our study reports (Figure 2a).
For other SNPs, e.g. those reported by Li et al 17, no association in our study was detected. However, Li et al.’s study 17 used additional covariates (e.g. COPD, lung cancer family history) to adjust for in their analyses. This may have made a comparison of their results with our study less straightforward, because the data on these covariates were not available from the majority of the sites participating in our study. The SNPs rs10937405 for 3q28 and rs9387478 for 6q22.2, previously reported to be significant in Asian never smoking women (Table 3), showed at best a suggestive association (P-values of ~10−4 in both cases). These two regions have been shown also to be implicated in other cancer sites. SNPs in the TP63 region have been shown to be associated with lung adenocarcinoma in the UK population 8, acute lymphoblastic leukemia 42, bladder cancer 43 and pancreatic cancer 44 SNPs in the ROS1-DCBLD1 region have been shown to be associated with colorectal cancer 45. This further suggests that SNPs/regions associated with lung cancer risk in never smokers are not specific for this type of cancer but rather have pleiotropic effects.
Our analysis was designed to control for demographic variables (age and sex, as controls were slightly but statistically significantly younger (p<0.001) and had a higher proportion of men than cases (p<0.001)) as well as for known and potential risk factors, specifically, where possible, for education status and self-reported secondhand smoke exposure 46. To account for possible population stratification, the first five principal components and the study site were also adjusted. However, the information on radon exposure, asbestos, prior respiratory conditions, and diet was not available from most studies. As such, these established and putative risk factors were not accounted for in the analyses. A further limitation is the self-reported nature of the never smoker status. Differential misreporting of the smoking status, e.g., if a modest proportion of former or current smoker controls reported that they have never smoked, might lead to SNPs associated with smoking appear as protective. Unfortunately, the great majority of the participating studies did not verify it by cotinine measurements. However, SNPs in CHRNA3–5 or CYP2A6 regions, known to be associated with smoking 20, did not show any effect in this study (Figure 2b; Table A.11).
Latest GWASs of lung cancer in smokers have generated many more findings than did this study, which is not surprising given that the former are much larger. Most SNPs reported as statistically significant in smokers showed the same direction of effect in never smokers (Table A.12). Gene-smoking interaction may be another factor contributing to the higher number of positive findings among smokers than never smokers: some of the sequence variations that are neutral in the absence of tobacco smoking confer risk when smoking and the associated tissue and DNA damage are present.
High BMI 47 and alcohol exposure 48 are common and may also explain a proportion of the lung cancer risk in never smokers. It is possible that there are rare variants influencing risk that could not be detected by a GWAS that focuses on common variants. Additionally, gene-gene interactions that are beyond the scope of this study may in part explain variability in the incidence of lung cancer in never smokers. Very rarely, individuals can carry inherited mutations in TP53 increasing lung cancer risk 49. The availability of results from our GWAS will allow additional exposures to be studied using Mendelian Randomization approaches (as exemplified in 50), and developing models that can identify never smokers at highest risk for lung cancer development could improve early detection.
Supplementary Material
Acknowledgments
Disclosure of funding
This work was supported in part by National Institutes of Health (NIH) grants CA149462, CA209414, CA092824, ES00002, U01CA209414, U19CA203654, 1K07CA172294, P50CA119997, R01CA060691, R01CA87895, P30CA22453, P30CA008748, P30CA076292, U01CA164973, and Department of Health and Human Services grant HHSN261201300011; James & Esther King Biomedical Research Program Grant 09KN-15; Helmholtz-DAAD fellowship A/07/97379; the Society of Memorial Sloan Kettering Cancer Center through their annual appeal and Steps for Breath; Italian Ministry of Health grant for Institutional Research 2017–2018 and Associazione Italiana per la Ricerca sul Cancro grant IG2015/17564IO; Instituto de Salud Carlos III. PI15/01211 grant and Xunta de Galicia grant 10CSA208057PR; The Toronto study was supported by The Canadian Cancer Society Research Institute (020214), and the Alan Brown Chair and Lusi Wong Programs at the Princess Margaret Hospital Foundation; the LUCY study was funded in part by the Germany National Genome Research Network (NGFN), the DFG (BI576/2–1; BI 576/2–2, Bi 576/4–1; Bi 576/4–2; Wi 621/10–1; Wi 621/10–2), the Helmholtzgemeinschaft (HGF) and the Federal office for Radiation Protection (BfS:STSch4454); KORA Surveys were funded by the Helmholtz-Zentrum München (HMGU), which is funded by the German Federal Ministry of Education, Science, Research and Technology and the State of Bavaria. The Liverpool Lung Project is funded by the Roy Castle Lung Cancer Foundation. The Resource for the Study of Lung Cancer Epidemiology in North Trent (ReSoLuCENT) study was funded by the Sheffield Hospitals Charity, Sheffield Experimental Cancer Medicine Centre and Weston Park Hospital Cancer Charity. ILCCO data harmonization was supported by Canada Research Chair to R. J. H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that none of them has a conflict of interest
References
- 1.Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2018. [DOI] [PubMed] [Google Scholar]
- 2.Thun MJ, Hannan LM, Adams-Campbell LL, et al. Lung cancer occurrence in never-smokers: an analysis of 13 cohorts and 22 cancer registry studies. PLoS Med 2008;5:e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subramanian J, Govindan R. Lung cancer in ‘Never-smokers’: a unique entity. Oncology (Williston Park) 2010;24:29–35. [PubMed] [Google Scholar]
- 4.Torres-Duran M, Ruano-Ravina A, Parente-Lamelas I, et al. Residential radon and lung cancer characteristics in never smokers. Int J Radiat Biol 2015;91:605–610. [DOI] [PubMed] [Google Scholar]
- 5.Gorlova OY, Zhang Y, Schabath MB, et al. Never smokers and lung cancer risk: a case-control study of epidemiological factors. Int J Cancer 2006;118:1798–1804. [DOI] [PubMed] [Google Scholar]
- 6.Markowitz SB, Levin SM, Miller A, et al. Asbestos, asbestosis, smoking, and lung cancer. New findings from the North American insulator cohort. Am J Respir Crit Care Med 2013;188:90–96. [DOI] [PubMed] [Google Scholar]
- 7.Gorlova OY, Weng SF, Zhang Y, et al. Aggregation of cancer among relatives of never-smoking lung cancer patients. Int J Cancer 2007;121:111–118. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Broderick P, Matakidou A, et al. Variation in TP63 is associated with lung adenocarcinoma in the UK population. Cancer Epidemiol Biomarkers Prev 2011;20:1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiung CA, Lan Q, Hong YC, et al. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet 2010;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan Q, Hsiung CA, Matsuo K, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet 2012;44:1330–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seow WJ, Matsuo K, Hsiung CA, et al. Association between GWAS-identified lung adenocarcinoma susceptibility loci and EGFR mutations in never-smoking Asian women, and comparison with findings from Western populations. Hum Mol Genet 2017;26:454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiraishi K, Kunitoh H, Daigo Y, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet 2012;44:900–903. [DOI] [PubMed] [Google Scholar]
- 13.Liao Y, Xu L, Lin X, et al. Temporal Trend in Lung Cancer Burden Attributed to Ambient Fine Particulate Matter in Guangzhou, China. Biomed Environ Sci 2017;30:708–717. [DOI] [PubMed] [Google Scholar]
- 14.Spitz MR, Gorlov IP, Amos CI, et al. Variants in inflammation genes are implicated in risk of lung cancer in never smokers exposed to second-hand smoke. Cancer Discov 2011;1:420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Broderick P, Matakidou A, et al. Role of 5p15.33 (TERT-CLPTM1L), 6p21.33 and 15q25.1 (CHRNA5-CHRNA3) variation and lung cancer risk in never-smokers. Carcinogenesis 2010;31:234–238. [DOI] [PubMed] [Google Scholar]
- 16.Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet 2009;85:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, Sheu CC, Ye Y, et al. Genetic variants and risk of lung cancer in never smokers: a genomewide association study. Lancet Oncol 2010;11:321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landi MT, Chatterjee N, Caporaso NE, et al. GPC5 rs2352028 variant and risk of lung cancer in never smokers. The Lancet Oncology 2010;11:714–716; author reply 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisen T, Matakidou A, Houlston R, et al. Identification of low penetrance alleles for lung cancer: the GEnetic Lung CAncer Predisposition Study (GELCAPS). BMC Cancer 2008;8:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKay JD, Hung RJ, Han Y, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet 2017;49:1126–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz AG, Yang P, Swanson GM. Familial risk of lung cancer among nonsmokers and their relatives. Am J Epidemiol 1996;144:554–562. [DOI] [PubMed] [Google Scholar]
- 22.Wenzlaff AS, Cote ML, Bock CH, et al. GSTM1, GSTT1 and GSTP1 polymorphisms, environmental tobacco smoke exposure and risk of lung cancer among never smokers: a population-based study. Carcinogenesis 2005;26:395–401. [DOI] [PubMed] [Google Scholar]
- 23.Ugolini D, Neri M, Canessa PA, et al. The CREST biorepository: a tool for molecular epidemiology and translational studies on malignant mesothelioma, lung cancer, and other respiratory tract diseases. Cancer Epidemiol Biomarkers Prev 2008;17:3013–3019. [DOI] [PubMed] [Google Scholar]
- 24.Amos CI, Dennis J, Wang Z, et al. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol Biomarkers Prev 2017;26:126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 26.Viechtbauer. Conducting Meta-Analyses in R with the metafor Package. Journal of Statistical Software 2010;36:1–48. [Google Scholar]
- 27.Amos CI. Successful design and conduct of genome-wide association studies. Hum Mol Genet 2007;16 Spec No. 2:R220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Zhu B, Zhang M, et al. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the TERT-CLPTM1L region on chromosome 5p15.33. Hum Mol Genet 2014;23:6616–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haiman CA, Chen GK, Vachon CM, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet 2011;43:1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rajaraman P, Melin BS, Wang Z, et al. Genome-wide association study of glioma and metaanalysis. Hum Genet 2012;131:1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bei JX, Su WH, Ng CC, et al. A GWAS Meta-analysis and Replication Study Identifies a Novel Locus within CLPTM1L/TERT Associated with Nasopharyngeal Carcinoma in Individuals of Chinese Ancestry. Cancer Epidemiol Biomarkers Prev 2016;25:188–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kote-Jarai Z, Olama AA, Giles GG, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet 2011;43:785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung AL, Deng W. Telomere dysfunction, genome instability and cancer. Front Biosci 2008;13:2075–2090. [DOI] [PubMed] [Google Scholar]
- 34.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer 1997;33:787–791. [DOI] [PubMed] [Google Scholar]
- 35.James MA, Wen W, Wang Y, et al. Functional characterization of CLPTM1L as a lung cancer risk candidate gene in the 5p15.33 locus. PLoS One 2012;7:e36116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia J, Bosley AD, Thompson A, et al. CLPTM1L promotes growth and enhances aneuploidy in pancreatic cancer cells. Cancer Res 2014;74:2785–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang J, Jia J, Makowski M, et al. Functional characterization of a multi-cancer risk locus on chr5p15.33 reveals regulation of TERT by ZNF148. Nat Commun 2017;8:15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis CA, Hitz BC, Sloan CA, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 2018;46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zerbino DR, Achuthan P, Akanni W, et al. Ensembl 2018. Nucleic Acids Res 2018;46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kachuri L, Amos CI, McKay JD, et al. Fine mapping of chromosome 5p15.33 based on a targeted deep sequencing and high density genotyping identifies novel lung cancer susceptibility loci. Carcinogenesis 2016;37:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellinghaus E, Stanulla M, Richter G, et al. Identification of germline susceptibility loci in ETV6- RUNX1-rearranged childhood acute lymphoblastic leukemia. Leukemia 2012;26:902–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Figueroa JD, Ye Y, Siddiq A, et al. Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum Mol Genet 2014;23:1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Childs EJ, Mocci E, Campa D, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet 2015;47:911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters U, Jiao S, Schumacher FR, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology 2013;144:799–807 e724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Couraud S, Zalcman G, Milleron B, et al. Lung cancer in never smokers--a review. Eur J Cancer 2012;48:1299–1311. [DOI] [PubMed] [Google Scholar]
- 47.Gao C, Patel CJ, Michailidou K, et al. Mendelian randomization study of adiposity-related traits and risk of breast, ovarian, prostate, lung and colorectal cancer. International journal of epidemiology 2016;45:896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fehringer G, Brenner DR, Zhang ZF, et al. Alcohol and lung cancer risk among never smokers: A pooled analysis from the international lung cancer consortium and the SYNERGY study. Int J Cancer 2017;140:1976–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwang SJ, Cheng LS, Lozano G, et al. Lung cancer risk in germline p53 mutation carriers: association between an inherited cancer predisposition, cigarette smoking, and cancer risk. Hum Genet 2003;113:238–243. [DOI] [PubMed] [Google Scholar]
- 50.Wang C, Qin N, Zhu M, et al. Metabolome-wide association study identified the association between a circulating polyunsaturated fatty acids variant rs174548 and lung cancer. Carcinogenesis 2017;38:1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


