Abstract
The Sequence Distance Graph (SDG) framework works with genome assembly graphs and raw data from paired, linked and long reads. It includes a simple deBruijn graph module, and can import graphs using the graphical fragment assembly (GFA) format. It also maps raw reads onto graphs, and provides a Python application programming interface (API) to navigate the graph, access the mapped and raw data and perform interactive or scripted analyses. Its complete workspace can be dumped to and loaded from disk, decoupling mapping from analysis and supporting multi-stage pipelines. We present the design and
implementation of the framework, and example analyses scaffolding a short read graph with long reads, and navigating paths in a heterozygous graph for a simulated parent-offspring trio dataset.
SDG is freely available under the MIT license at https://github.com/bioinfologics/sdg
Keywords: Genome graph, genome assembly
Introduction
Sequence graphs are the core representation of genome assemblers 1– 3 Their use has increased lately thanks to the graphical fragment assembly (GFA) format for graph exchange 4, tools to work with genome variation graphs 5, and sequence to graph mappers 6– 10 But a lack of inter operation between graph-based tools, and limited tools for downstream graph-based analysis, contribute to a perceived complexity which maintains linear sequences as the typical unit of exchange. This flattening of graph representations within pipelines with multiple steps, that use different types of sequencing in an iterative fashion, produces ever-longer linear genome sequences through an information loss process. As a result, genome assembly projects are prone to error propagation and difficult to reproduce and control. These problems can be addressed developing graph-based frameworks to integrate the analysis of hybrid datasets.
The Sequence Distance Graph (SDG) framework implements a SequenceDistanceGraph representation that defines sequences in nodes and their adjacency in links, and an associated Workspace containing raw data and mappings. This provides an integrated working environment to use multiple sources of information to navigate and analyse genome graphs. Datastores allow random access to short, linked, and long read sequences on disk. A mapper on each datastore contains methods to map the reads to the graph and access the mapping data. KmerCounters provide functions to compute k-mer coverage over the graph from sequencing data, enabling coverage analyses. Additional DistanceGraphs, typically representing longer-range information and different linkage levels, define alternative topologies over the SequenceDistanceGraph nodes. Finally, a NodeView abstraction provides a proxy to a node, with methods to navigate the graph and access its mapped data. This comprehensive framework can be used to explore genome graphs interactively or to create processing methods for assembly or downstream analysis.
Here we describe the SDG implementation and basic tools, providing examples of use cases that highlight its analytic flexibility. First, we show how to create a hybrid assembly by integration of long reads linkage into a short-read graph. Then we analyse a simulated parent-child trio and show how the coverage of the parent datasets can be used to navigate the graph topology. These are only two of the multiple ways integrating data and genome graphs can be used to perform simple but powerful analyses.
Methods
Implementation
The C++ core library implements SDG’s data structures and methods for WorkSpaces, graphs, datastores and mappers. Its main goal is to provide a straightforward interface to project information from raw datasets onto graphs, and enable easy access and analysis of the graph-data combination. It uses OpenMP for parallel processing, and SWIG 4.0 to export a Python API to enable interactive data analysis.
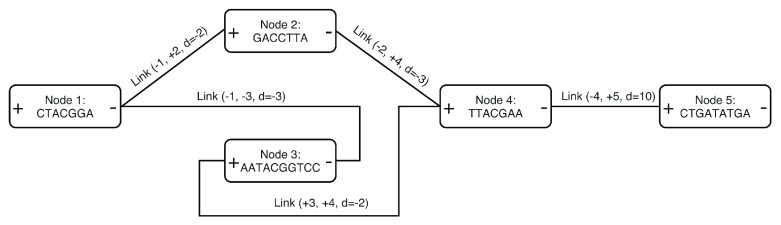
The SequenceDistanceGraph class contains a vector of nodes defining DNA sequences, and a vector of links. Every node has a positive and a negative end, and links are defined between these node ends. Links with positive distances represent gaps between linked sequences and negative distances represent overlaps. This representation, shown in Figure 1, is similar to those presented in 2, 11 but unifies the concept of overlap and gap. Paths can be defined as list of nodes, with the sign of the first end in the walk. Graphs can be read and written to GFA and GFA2 files.
Figure 1. A simple Sequence Distance Graph with 5 nodes, including links with d<0, representing overlaps, and a link representing a gap of 10bp.
Sequences appear in only one direction and their reverse complement can be obtained by traversing the node in opposite direction, from - to +. The two largest possible paths are [1, 2, 4, 5] and [1, -3, 4, 5], and their reverse complements [-5, -4, -2, -1] and [-5, -4, 3, -1] respectively.
The DistanceGraph class contains a set of links over the nodes of a SequenceDistanceGraph object. It is used to represent alternative sources of linkage information, such as longer range linkage produced by mapped reads for scaffolding.
The WorkSpace contains a single SequenceDistanceGraph, multiple DistanceGraphs, datastores and mappers, and its structure in memory represents the status of the SDG framework. It can be dumped and loaded from disk, providing persistence and checkpoints between different steps on SDG-based pipelines. Raw reads and k-mer counts are kept in separate files, pointed from the WorkSpace, to avoid duplication when using multiple WorkSpaces around the same dataset.
The DataStores and Mappers provide access and management to raw data and its mapping on the graph. Datastores do not load read data into memory, but rather provide random access to the on-disk data. The PairedRedMapper and LinkedReadMapper classes use a unique k-mer index to map reads to single nodes, with single reads mapping to multiple nodes not being mapped 3, 12 The LongReadMapper class generates multiple mappings from each read to nodes, using a short non-unique k-mer index (k=15 by default) 13, 14 Long read mapping filtering is left to later stages of the processing.
The KmerCounters creates an index with all the k-mers at a given k up to k=31 and counts occurrences of these k-mers on the graph, allowing then to count occurrences in datastores or fastq files. These counts, persisted in the KmerCounter with a name, can be then accessed to perform k-mer coverage analyses. Projections of raw k-mer coverage in the reads and the assembly over a particular sequence for a node or path, similar to those produce by the "sect" tool of K-mer Analysis Toolkit (KAT) 15 are valuable for content analysis. Spectra analysis of these frequencies can provide further insight into genome composition and representation on the assembly.
Two processing classes, LinkageUntangler and LinkageMaker, work with alternative linkage configurations. The LinkageMaker is used to condense information via one of its make_linkage* methods, from evidence in the WorkSpace into links in a DistanceGraph. The LinkageUntangler class works on a DistanceGraph to simplify, condense and/or linearise its linkage. In the second use case below it can be seen how a combination of LinkageMaker and LinkageUntangler can be used for scaffolding with long reads.
Finally, the NodeView class, and its associated LinkViews, provide a single-entry point for node-centric analyses. A NodeView from either a DistanceGraph or SequenceDistanceGraph is a wrapper containing a pointer to the graph and a node id, and will provide access to its nodes’ previous and next linked nodes, mapped reads, or k-mer coverage. A user with good understanding of the NodeView class should be able to access most information in the WorkSpace through it, making it the default choice for analysing the graph.
Operation
Requirements and installation. SDG can be run on Linux and MacOS, and requires enough RAM to hold the WorkSpace completely in memory, which will depend on the dataset. Space to hold the uncompressed sequences on the datastores on disk will also be required.
SDG can be installed via pre-compiled binaries from https://github.com/bioinfologics/sdg/releases. The binaries have been built using Python3 and GCC version 6 from the Ubuntu package manager for the Linux version. The MacOS version dependencies were obtained using Homebrew (Python3, GCC-6 and SWIG). SDG can be compiled using CMake, Python3, SWIG version 4 and GCC version 6 onwards. Detailed instructions can be found at https://bioinfologics.github.io/sdg/sdg/README.html#installation.
Typical workflow. Working with SDG typically involves two different stages: creating a WorkSpace with the data and mappings, and analysing this WorkSpace. SDG includes command line tools to create DataStores, KmerCounts, and WorkSpaces, and map reads within a WorkSpace.
sdg-datastore: creates a Datastore from raw reads and can process paired, 10x or long reads. An output prefix is specified as a parameter and a <prefix>.prseq, <prefix>.lrseq or <prefix>.loseq file is generated.
sdg-kmercounter: creates a KmerCounter indexing a graph from a WorkSpace or GFA, or works with an already generated one. A count can be added directly from raw reads or from a datastore. The KmerCounter is persisted on file with extension ’sdgkc’.
sdg-workspace: creates a WorkSpace from a base graph or works with an already generated one. Datastores and KmerCounters can be added. The WorkSpace is persisted on file with extension ’sdgws’.
sdg-dbg: creates a WorkSpace from a PairedReadDatastore by building a deBruijn graph and using this as the base graph. Counts for the k-mers from the graph and raw reads are added too.
sdg-mapper: maps reads within a WorkSpace. An updated WorkSpace is produced and dumped to the specified prefix.
WorkSpaces can also be instantiated with an empty graph, and the graph populated through the add_node and add_link methods. The following example on a python session shows how the simple graph from Figure 1 can be created from scratch, navigated through a NodeView instance and sequence from its paths extracted.
>>> import pysdg as SDG version 0.1 master b4d3f02 >>> ws=SDG.WorkSpace() >>> ws.sdg.add_node("CTACGGA") 1 >>> ws.sdg.add_node("GACCTTA") 2 >>> ws.sdg.add_node("AATACGGTCC") 3 >>> ws.sdg.add_node("TTACGAA") 4 >>> ws.sdg.add_node("CTGATATGA") 5 >>> ws.sdg.add_link(-1, 2, -2) >>> ws.sdg.add_link(-1, -3, -3) >>> ws.sdg.add_link(-2, 4, -3) >>> ws.sdg.add_link(3, 4, -2) >>> ws.sdg.add_link(-4, 5, 10) >>> nv=ws.sdg.get_nodeview(1) >>> nv <NodeView: Node 1 in SDG> >>> nv.next() <Vector: 2 LinkViews> >>> print(nv.next()) [ <LinkView: -3bp to Node -3>, <LinkView: -2bp to Node 2> ] >>> nv = nv.next()[0].node() >>> nv <NodeView: Node -3 in SDG> >>> print(nv.prev()) [ <LinkView: -3bp to Node 1> ] >>> nv.sequence() 'GGACCGTATT' >>> SDG.SequenceDistanceGraphPath(ws.sdg, [1, -3, 4, 5]).sequence() 'CTACGGACCGTATTACGAANNNNNNNNNNCTGATATGA'
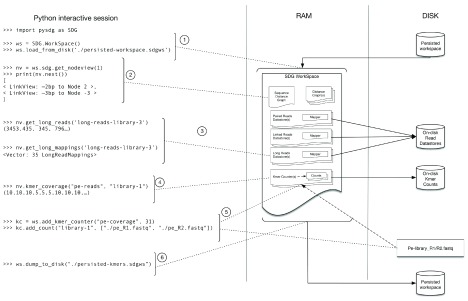
Typically, as shown in Figure 2, the API is used to explore a larger WorkSpace, with the methods accessing both in-memory and on-disk data, and modifying the status of the WorkSpace.
Figure 2. Structure of a WorkSpace and access via an Python interactive session.
The WorkSpace holds the information for a project and contains the graphs, the mappers and k-mer counts. From Python, a previously saved WorkSpace is loaded from disk (1). The NodeView object is centred on a specific node and can be used to access node characteristics (ie. size and sequence), graph topology from the perspective of the node you are on (i.e. neighbours in both directions (2)) and can also retrieve information projected onto the selected node (ie. mappings (3) and k-mer coverage (4)). Operations such as adding a KmerCounter to the WorkSpace and adding a count (5) can be performed, and the WorkSpace can be saved back to disk (6). Once loaded, the bulk of the WorkSpace is held in memory for fast access with the raw read data from the DataStores remaining on disk accessible through random access.
Example use cases
To illustrate the use of SDG, we have reproduced a short version of two examples from http://bioinfologics. github.io/sdg_examples.
All paired end datasets are available on https://zenodo.org/record/3363871#.XUwyVy2ZN24 16, and the PacBio reads are from NCBI accession PRJNA194437 17 For simplicity, we have also made the datasets available on https://opendata.earlham.ac.uk/opendata/data/sdg_datasets/ as ready-to-use ’fastq.gz’ files.
Hybrid assembly of short and long reads
This example is based on an E. coli dataset combining PacBio reads from 17 and Illumina Miseq 2x300bp reads subsampled from a test run. It uses the long reads to scaffold a short read based graph produced by sdg-dbg. Graphs are dumped to GFA files at different stages, and visualised using Bandage v0.8.1 18
First, we use the command line tools to create datastores for both long and short reads and an initial WorkSpace containing a DBG assembly:
sdg-datastore make -t paired -o ecoli_pe ../ecoli_pe_r1.fastq.gz -2 ../ecoli_pe_r2.fastq.gzsdg-datastore make -t long -o ecoli_pb -L ../ecoli_pb_all.fastq.gz sdg-dbg -p ecoli_pe.prseq -o ecoli_assm
From this point on, we use the python SDG library. First, we load the workspace, add a long read datastore and map its reads using a k=11 index.
importpysdgasSDG # Load sdg-dbg's workspace from disk, add the pacbio datastore ws=SDG.WorkSpace('ecoli_assm.sdgws') lords=ws.add_long_reads_datastore('ecoli_pb.loseq') # Map long reads lords.mapper.k= 11 lords.mapper.map_reads() ws.sdg.write_to_gfa1('initial_graph.gfa')
The graph, as shown in Figure 3A contains multiple unresolved repeats.
Figure 3.
Linkage at different stages of the long read scaffolding example, visualised using Bandage: A) SequenceDistanceGraph generated by sdg-dbg from short reads, B) DistanceGraph generated after using make_nextselected_linkage on the long read data, linking all nodes of 1100bp and more, C) DistanceGraph after eliminating all nodes with multiple connections (repeats).
We can use the LinkageMaker to create linkage using the long reads datastore. We do this by selecting the nodes between which to analyse possible linkage, in this case all nodes of 1100bp or more, and then calling the make_longreads_multilinkage method, with alignment filtering parameters of 1000bp and 10% id.
lm=SDG.LinkageMaker(ws.sdg) lm.select_by_size(1100) mldg=lm.make_longreads_multilinkage(ws.long_reads_datastores[0].mapper, 1000, 10)
This multi-linkage can be collapsed using the LinkageUntangler. The make_nextselected_linkage method links every selected node to its closest selected neighbours on each direction, aggregating the distances via a simple median calculation:
lu=SDG.LinkageUntangler(mldg) lu.select_by_size(1100) ns_dg=lu.make_nextselected_linkage() ns_dg.write_to_gfa1('ns_collapsed.gfa')
The new graph we dumped, as shown in Figure 3B, has disconnected the repeats and introduced long read linkage which skips over them, but it is still not fully solved. We can improve this further by getting rid of repetitive nodes that will be connected to multiple neighbours, as each of them belongs in more than one place. We do that by just turning these nodes’ selection off in the LinkageUntangler, which will then skip them in the solution.
fornvinns_dg.get_all_nodeviews(): if len(nv.prev())> 1orlen(nv.next())> 1: lu.selected_nodes[nv.node_id()]=False ns_nr_dg=lu.make_nextselected_linkage() ns_nr_dg.write_to_gfa1('ns_nr_final.gfa')
The last graph is now a circle, with all the repeats disconnected from any linkage.
Analysing a simulation of heterozygous parent-child trio with short reads
We created a simulation of a trio dataset for this example using the synthetic genome creation and sequencing package Pseudoseq.jl v0.1.0 19 Chromosomes 4 and 5 of the reference genome of the yeast strain S288C were used as templates to create a diploid, genome for each parent with 1% heterozygous sites. Each homologous pair of chromosomes was crossed over and recombined and the child inherited one chromosome from the first parent at random, and one chromosome from the second parent at random. Simulated paired end reads were generated for each genome, using an average fragment length of 700bp and a read length of 250bp, and an expected coverage of 70x with error rate was set to 0.1%.
First we used the command line tools to create a graph from the child reads using sdg-dbg, and add k-mer counts for both parents into the datastore.
sdg-datastore make -t paired -1 child/child-pe-reads_R1.fastq.gz -2 child/child-pe-reads_R2.fastq.gz -o child_pe
sdg-dbg sdg-dbg -p child_pe.prseq -o sdg_child
sdg-kmercounter add -c main.sdgkc -n p1 -f p1/p1-pe-reads_R1.fastq.gz -f p1/p1-pe-reads_R2.fastq.gz -o main
sdg-kmercounter add -c main.sdgkc -n p2 -f p2/p2-pe-reads_R1.fastq.gz -f p2/p2-pe-reads_R2.fastq.gz -o main
We now open the WorkSpace and use the NodeView::parallels method to look for the largest bubble structure in the graph, which should be formed by two parallel nodes with haplotypes coming from each parent.
import pysdgas SDG ws = SDG.WorkSpace('sdg_child.sdgws') #Largest node with one parallel node, and its parallel maxbubble = 0 for nv in ws.sdg.get_all_nodeviews(): if nv.size() > maxbubble andlen(nv.parallels()) == 1: maxbubble=nv.size() bubble_nvs=(nv,nv.parallels()[0])
Since each side should be a haplotype from a different parent, we should see a loss of k-mer coverage on the parent that didn’t contribute that haplotype. To check this, we create a plotting function to plot the output from the NodeView::kmer_coverage method.
defplot_kcov(nv): '''Plot kmer coverage across the three read sets. Requires pylab.''' figure();suptitle("Coverage for "+str(nv)); subplot(3,1,1);ylim((0,120)) plot(nv.kmer_coverage("main","PE"), label="child"); legend(loc=1); subplot(3,1,2);ylim((0,120)) plot(nv.kmer_coverage("main","p1"),"red", label="parent 1"); legend(loc=1); subplot(3,1,3);ylim((0,120)) plot(nv.kmer_coverage("main","p2"),"blue", label="parent 2"); legend(loc=1); plot_kcov(bubble_nvs[0]) plot_kcov(bubble_nvs[1])
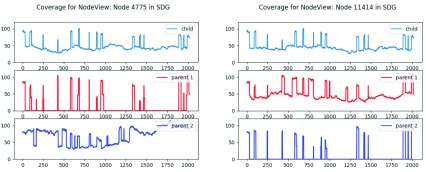
The plots, shown in Figure 4, reflect how Node 4775 contains content inherited from parent 2 and its parallel node 11414 contains content inherited from parent 1. We can create a function to extend these parent-specific regions by walking forward and backward as long as only one link takes us to a node that is fully covered by the content of the parent we are following.
Figure 4. Trio analysis: k-mer coverage for each side of the largest bubble structure in the child’s assembly by each of the three read sets.
Coverage drops to 0 on the opposite parent for k-mers that are unique to a parent.
defextend_parent_covered_path(starting_node,target_parent): ifws.sdg.get_nodeview(starting_node).kmer_coverage("main", target_parent).count(0)!= 0: returnSDG.SequenceDistanceGraphPath(ws.sdg,[]) p=SDG.SequenceDistanceGraphPath(ws.sdg,[starting_node]) forxin[0, 1]: nv=ws.sdg.get_nodeview(p.nodes[-1]) whilenv.next(): next_node= 0 fornlinnv.next(): ifnl.node().kmer_coverage("main", target_parent).count(0)== 0: ifnext_nodeornl.node().node_id()inp.nodes: next_node= 0 break else: next_node=nl.node().node_id() ifnext_node== 0:break p.nodes.append(next_node) nv=ws.sdg.get_nodeview(next_node) p.reverse() returnp path1=extend_parent_covered_path(11414, "p1") path2=extend_parent_covered_path(4775, "p2")
After using this function, path1 contains 49 nodes yielding 8672bp of sequence inherited from parent 1, and path2 contains 139 nodes yielding 26351bp of sequence inherited from parent 2. It is important to note that the difference in node count and sequence length arises because the extension function is haplotype-specific and its results depend in the topology of each haplotype graph.
Summary
The Sequence Distance Graph framework provides a unified workspace for different sequencing technologies using the genome graph as the basis of integration. It enables analyses across the graph topology, the raw data and its projections to the graph. We have shown how the NodeView class can be used through the Python API to produce interactive analyses that are both powerful and easy to follow. We expect this will be a useful codebase for all levels of users, not only for the construction of graph-based analysis but also for their teaching and dissemination.
Data availability
Source data
The PacBio, E. coli reads are deposited on NCBI accession PRJNA194437 from Koren et al. 17
E. coli K12 Re-sequencing with PacBio RS and 454: Accession number PRJNA194437, https://identifiers.org/ncbi/bioproject:PRJNA194437
Underlying data
The datasets used in the examples are available from: https://opendata.earlham.ac.uk/opendata/data/sdg_datasets/ and archived in Zenodo Zenodo: SDG Paper Datasets. http://doi.org/10.5281/zenodo.3363871 16
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Software availability
Software documentation: https://bioinfologics.github.io/sdg
Source code available from: http://github.com/bioinfologics/sdg
Archieved source code at time of publication: https://zenodo.org/record/3363165#.XUw1yy2ZN25 20
License: MIT License
Acknowledgements
We would like to thank Richard Harrison for helpful discussions about SDG’s results and continued support through the OctoSeq project. We thank James Cuff for input about design principles and continuous encouragement. We thank Kat Hodgkinson for her feedback and patience as an early user of the rough alpha version of SDG. We thank Camilla Ryan for enduring and joining never-ending discussions about graph representations and the design of the framework.
Funding Statement
This work was strategically funded by the BBSRC Core Strategic Programme Grant [BBS/E/T/000PR9818]. Work by GGA and BJC was also partially funded by the BBSRC grant "OctoSeq: Sequencing the octoploid strawberry"[BB/N009819/1].
[version 1; peer review: 2 approved
References
- 1. Pevzner PA, Tang H, Waterman MS: An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci U S A. 2001;98(17):9748–9753. 10.1073/pnas.171285098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Medvedev P, Brudno M: Maximum likelihood genome assembly. J Comput Biol. 2009;16(8):1101–1116. 10.1089/cmb.2009.0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Butler J, MacCallum I, Kleber M, et al. : ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18(5):810–820. 10.1101/gr.7337908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jackman SD, Myers EW, Gonella G: The GFA Specification. Reference Source [Google Scholar]
- 5. Garrison E, Sirén J, Novak AM, et al. : Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat Biotechnol. 2018;36(9):875–879. 10.1038/nbt.4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rautiainen M, Mäkinen V, Marschall T: Bit-parallel sequence-to-graph alignment. bioRxiv. 2018;323063 10.1101/323063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sirén J, Garrison JE, Novak AM, et al. : Haplotype-aware graph indexes. bioRxiv. 2019. 10.1101/559583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Novak AM, Garrison E, Paten B: A graph extension of the positional Burrows-Wheeler transform and its applications. Algorithms Mol Biol. 2017;12(1):18. 10.1186/s13015-017-0109-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jain C, Dilthey A, Misra S, et al. : Accelerating Sequence Alignment to Graphs. bioRxiv. 2019. 10.1101/651638 [DOI] [Google Scholar]
- 10. Limasset A, Flot JF, Peterlongo P: Toward perfect reads: self-correction of short reads via mapping on de Bruijn graphs. Bioinformatics. 2019; pii: btz102. 10.1093/bioinformatics/btz102 [DOI] [PubMed] [Google Scholar]
- 11. Paten B, Zerbino DR, Hickey G, et al. : A unifying model of genome evolution under parsimony. BMC Bioinformatics. 2014;15(1):206. 10.1186/1471-2105-15-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Batzoglou S, Jaffe DB, Stanley K, et al. : ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12(1):177–189. 10.1101/gr.208902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sović I, Šikić M, Wilm A, et al. : Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat Commun. 2016;7:11307. 10.1038/ncomms11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pavetić F, Katanić I, Matula G, et al. : Fast and simple algorithms for computing both LCS k and LCS k+ . arXiv: 1705.07279 [cs],2017. Reference Source [Google Scholar]
- 15. Mapleson D, Garcia Accinelli G, Kettleborough G, et al. : KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics. 2017;33(4):574–576. 10.1093/bioinformatics/btw663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yanes L, Garcia Accinelli G, Ward BJ, et al. : Sdg paper datasets.2019. https://zenodo.org/record/3363871 [Google Scholar]
- 17. Koren S, Harhay GP, Smith TP, et al. : Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14(9):R101. 10.1186/gb-2013-14-9-r101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wick RR, Schultz MB, Zobel J, et al. : Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31(20):3350–3352. 10.1093/bioinformatics/btv383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ward BJ: bioinfologics/pseudoseq.jl: First release.2019. 10.5281/zenodo.2656743 [DOI] [Google Scholar]
- 20. Yanes L, Garcia Accinelli G, Ward BJ, et al. : bioinfologics/sdg: Release candidate.2019;7 https://zenodo.org/record/3363165 [Google Scholar]