Abstract
Few invertebrates can survive cryopreservation in liquid nitrogen, and the mechanisms by which some species do survive are underexplored, despite high application potential. Here, we turn to the drosophilid Chymomyza costata to strengthen our fundamental understanding of extreme freeze tolerance and gain insights about potential avenues for cryopreservation of biological materials. We first use RNAseq to generate transcriptomes of three C. costata larval phenotypic variants: those warm-acclimated in early or late diapause (weak capacity to survive cryopreservation), and those undergoing cold acclimation after diapause entry (extremely freeze tolerant, surviving cryopreservation). We identify mRNA transcripts representing genes and processes that accompany the physiological transition to extreme freeze tolerance and relate cryopreservation survival to the transcriptional profiles of select candidate genes using extended sampling of phenotypic variants. Enhanced capacity for protein folding, refolding and processing appears to be a central theme of extreme freeze tolerance and may allow cold-acclimated larvae to repair or eliminate proteins damaged by freezing (thus mitigating the toxicity of denatured proteins, endoplasmic reticulum stress and subsequent apoptosis). We also find a number of candidate genes (including both known and potentially novel, unannotated sequences) whose expression profiles tightly mirror the change in extreme freeze tolerance status among phenotypic variants.
Keywords: insect, cryopreservation, transcriptome, cryoprotectant, cold acclimation
1. Introduction
The mechanisms by which overwintering insects survive at sub-zero body temperatures have been investigated for more than a century (see, for instance, reviews in [1–7]). Specific molecular, biochemical and physiological adjustments underlying this survival are typically assigned to one of three cold hardiness ‘strategies’ based on the phase behaviour of body water during cold exposure: (i) supercooling (maintenance of body water in the liquid phase), (ii) cryoprotective dehydration (loss of a substantial proportion of the body water by evaporation) and (iii) freeze tolerance (conversion of body water to the solid phase—ice crystals). Freeze tolerance evolved multiple times in a diverse array of insects [7,8]. Although most freeze-tolerant insects perish at temperatures below a species-specific threshold (the lower lethal temperature; LLT) typically between −5°C and −40°C [4,9], some insects and other invertebrates seem to have no LLT and withstand freezing to the temperature of liquid nitrogen (−196°C), even in a fully hydrated state; examples include the nematode Anguillula silusiae [10], rotifer Philodina acuticornis [11], poplar sawfly Trichiocampus populi [12], malt fly Chymomyza costata [13] and leech Ozobranchus jantseanus [14]. These extremely freeze-tolerant animals may hold the key to advancing techniques in cryopreservation of tissues, organs or even complex organisms [15]. More specifically for insects, such techniques would improve the management of stock populations for research and application [16–21].
In order to build up extreme (or high) freeze tolerance, malt fly larvae (and other adapted insects, respectively) typically enter diapause and then undergo cold acclimation [7,22,23]. Diapause is a seasonal phenotype associated with arrested development and deep metabolic suppression [24,25], while cold acclimation further increases resistance to environmental stress [24,26]. Both adaptive complexes—diapause and cold acclimation—are based on the massive alteration of -omic profiles from the transcriptome [27–30], through proteome [31,32], to metabolome [33,34]. One of the key mechanisms underlying extreme freeze tolerance in insects is the accumulation of high concentrations of low molecular mass cryoprotective compounds (CPs). For instance, diapausing cold-acclimated T. populi prepupae accumulate trehalose (up to 202 mmol kg−1 of total body water) [12] while diapausing cold-acclimated malt fly larvae accumulate proline (up to 339 mmol kg−1 total body water) [23,35,36]. The CPs act through various mechanisms including a colligative reduction in the relative amount of ice [37,38], stabilization of lipid bilayers [39], stabilization of proteins' native structures [40,41] and/or promoting vitrification [23,42]. It is well known in practical cryogenics that both natural and engineered CPs facilitate the storage of cells and tissues at ultra-low temperatures [42]. However, the application of CPs to non-adapted (e.g. tropical) and/or non-acclimated (e.g. non-diapause, warm-acclimated) insects is only partially successful in improving their freeze tolerance [23,35,43].
In insects with extreme freeze tolerance, we speculate that CPs work in synergy with macromolecular protective systems that are seasonally stimulated. Indeed, a number of proteins with different cryoprotective roles are specifically enhanced in abundance during diapause and cold acclimation, including ice-binding proteins, heat shock proteins (HSPs) and late embryogenesis abundant (LEA) proteins. IBP adsorption to ice planes slows or limits their growth and prevents ice recrystallization damage [44,45], while HSPs assist in protein folding, localization and degradation [46]. Small HSPs (sHSPs) and LEA proteins act as ‘molecular shields’ which prevent protein aggregation during desiccation and freezing [47–50]. These macromolecules can also cooperate with CPs to protect cellular and protein structure under thermal or desiccation stress. For example, the interaction of sHSP p26 with trehalose significantly improves both mammalian cell survival during drying [51] and protection/reactivation of thermally inactivated citrate synthase in brine shrimp [52]. Cryophilic diatom-derived IBP reduces freezing damage to human blood cells only in the presence of glycerol [53]. LEA proteins act synergistically with trehalose to prevent protein aggregation during dehydration stress [54]. Still, the role of such macromolecules in insect extreme freeze tolerance remains unknown as they have yet to be assessed in the few species capable of surviving deep cryopreservation. Extremely freeze-tolerant insects may further possess unique (novel) proteins that behave similarly to CPs by preventing unwanted interactions between other macromolecules, complexes and organelles in frozen (i.e. tightly packed, dehydrated) cells.
Here, we aimed to identify candidate macromolecules and processes that potentially contribute to the survival of malt fly larvae in liquid nitrogen. Specifically, we employed a non-targeted transcriptomics approach (RNAseq) to broadly compare gene expression among three larval phenotypes differing in freeze tolerance: larvae in early and late diapause (under warm conditions) are moderately freeze tolerant, while larvae that are cold-acclimated after diapause entry develop extreme freeze tolerance (including storability in liquid nitrogen). To validate RNAseq results and further resolve the candidate proteins whose transcriptional patterns most closely associate with extreme freeze tolerance, we performed qPCR for select sequences over an extended sampling plan (additional time points throughout diapause maintenance and cold acclimation). The transcriptomics approach is a rapid, sensitive and technically feasible means of comparing among multiple phenotypes and time points and will be complemented by forthcoming metabolomic and proteomic studies (in preparation). Cryoprotective proteins must be present in high abundance to act as molecular shields, and although mRNA transcript expression does not necessarily reflect protein abundance [55], this rough estimate of relative protein abundance allows us to focus on transcripts with large fold changes across larval variants.
2. Material and methods
(a). Malt fly rearing and acclimation
We reared malt fly colonies (‘Sapporo’ strain, originating from Japan [56]) on artificial diet in MIR 154 incubators (Sanyo Electric, Osaka, Japan) as described previously [57]. For experiments, we generated three different malt fly larvae phenotypic variants (details in electronic supplementary material, figure S1) according to our earlier protocols [22,27,58]: (1) non-diapause ‘LD’ larvae—which pupate after approximately three weeks and emerge as adults after approximately one month; (2) diapausing, warm-acclimated ‘SD’ larvae—which do not pupate but maintain diapause until death; and (3) cold-acclimated ‘SDA’ larvae—which develop extreme freeze tolerance, gradually terminate their diapause within two months and remain in quiescence [23,35].
(b). RNAseq: library preparation, sequencing, alignment and annotation
We used three biological replicates (each comprising 10 pooled larvae) for transcriptome sequencing of three malt fly variants: early diapause (SD6); late diapause (SD11) and cold acclimated (SDA11) (see electronic supplementary material, figure S1 for explanation). The pools of larvae were homogenized in 400 µl RNA Blue (Top-Bio, Vestec, Czech Republic) for 30 s on ice. Total RNA was extracted according to manufacturer's instructions for RNA blue, treated with DNase I (Invitrogen, ThermoFisher Scientific, Prague, Czech Republic) and diluted to a final concentration of 0.1 µg RNA per µl in PCR Ultra H2O. RNA quality checking, cDNA library preparation and 80 bp, single-end read sequencing on the Illumina NextSeq 500 (Illumina, San Diego, CA) were performed by the EMBL Genomics Core Facility (Heidelberg, Germany). Sequencing of the nine cDNA libraries across the three malt fly variants yielded a total of 357 million reads with an average GC content of 47.5%. We assessed the quality of sequenced libraries by FastQC software (v. 0.11.5) [59] and used Trimmomatic [60] via the Galaxy Web service [61] to remove adapter sequences and discard sequences shorter than 36 nucleotides or containing unknown bases. Approximately 304 million reads remained after quality control. Through the Galaxy Web service, we used Bowtie2 [62] to align reads to a previously assembled reference malt fly transcriptome derived from both LD and SD larvae (a full assembly of 113 447 contigs, which was further refined to 21 326 non-redundant contigs [63]). Just over 99% of our reads aligned to the full reference assembly. We then used Cufflinks [64] to assemble transcripts (contigs) and estimate read counts. These normalized libraries contained a total of 22 872 contigs with non-zero read counts (representing 86.1% of the refined reference assembly; [63]). Libraries were further filtered to remove contigs with low abundance (those with fewer than five read counts per million in three of the six libraries per comparison) [65], yielding approximately 6800 contigs per library.
(c). Differential expression analyses and annotation
We compared relative transcript abundance among malt fly variants using the edgeR Bioconductor package [65] for R statistical software (v. 3.2.2) [66,67]. Criteria for differential expression analyses were conservative; genes were considered to be differentially expressed (DE) if the p-value (adjusted for false discovery rate; FDR) was less than 0.05 and the absolute fold change between treatments was greater than or equal to 4 (log2 fold change greater than or equal to 2). Biological coefficients of variation did not exceed 0.3 (i.e. gene expression among biological replicates varied by less than 30%). DE transcripts were annotated with Blast2GO software (v. 4.1.9, Oracle Corp.) [68]. Putative identities were assigned by BLASTx against the non-redundant (nr) protein database (NCBI, November 2017) (E-value threshold = 1 × 10−3, accepting five hits for each transcript), mapped for Gene Ontology (GO) terms and annotated (E-value threshold = 1 × 10−6). Approximately 65% of the contigs had putative identities, 5% were predicted or uncharacterized proteins and 30% had no putative identity. We attempted to identify the uncharacterized or predicted proteins via a less-stringent BLASTx search against the nr database (E-value threshold = 1, accepting 100 hits for each transcript) using Geneious software (v. 10.1.3, Biomatters Ltd.). In this way, we gained putative identities for an additional 50 transcripts. Finally, we assigned all annotated DE genes to one of 10 custom categories: ‘Development’, ‘Cuticle’, ‘Metabolism’, ‘Detoxification’, ‘Membranes’, ‘Cytoskeleton’, ‘Transcription’, ‘Protein processing’, ‘Other’ and ‘Uncharacterized’ (electronic supplementary material, spreadsheet S1) based on functional information from the literature and online databases (UniProt and FlyBase).
We characterized the coordinated differential expression of pathways (functionally related gene sets) according to KEGG (Kyoto Encylopedia of Genes and Genomes; [69]). KEGG identities for contigs were retrieved from the KEGG Automatic Annotation Server [70] based on non-species-specific reference pathways. Approximately 21% of the contigs with non-zero read counts were assigned to KEGG pathways. Differential pathway expression among malt fly variants was analysed based on read counts for transcripts with KEGG identities using the Generally Applicable Gene-set Enrichment (GAGE) and Pathview Bioconductor packages [71,72] in R. We accepted pathways as DE if the FDR-adjusted p-value was less than 0.05. Because GAGE assesses coordinated expression change, these pathway analyses identified more DE genes relative to our more strict criteria (greater than or equal to fourfold change) for individual genes assessed via edgeR (above).
(d). Direct validation
Direct validation of edgeR analysis was performed (a) from the same aliquots of total RNA samples used to generate transcriptomes (technical validation) and (b) in an independent set of new biological replicates (a new generation of flies) conditioned exactly as in electronic supplementary material, figure S1 (biological validation). We used quantitative real-time PCR (qPCR) on a CFX96 PCR cycler (BioRad, Philadelphia, PA, USA) to amplify 10 select DE sequences against four reference genes (sequences coding for Ribosomal proteins RpL32 (Rp49), RpL19, RpS11 and RpS27A [27]) (see electronic supplementary material, figure S2 for methods details). All PCR primer sequences can be found in electronic supplementary material, spreadsheet S1.
(e). Extended validation and extreme freeze tolerance assays
An extended validation of the RNAseq results was performed with the aim to retrieve the best candidate sequences with transcriptional patterns most closely associated with extreme freeze tolerance. For this purpose, we prepared a new, extended set of total RNA samples from larvae representing different phenotypes (electronic supplementary material, spreadsheet S1) in four biological replicates, each comprised of 10 pooled larvae. Total RNA samples were processed as described in RNASeq (above). Next, we performed qPCR analysis (as in Direct validation, above) for an arbitrary selection of 15 candidate genes based on results of RNAseq DE analyses (all target and PCR primer sequences can be found in electronic supplementary material, spreadsheet S1). (a) We took six genes coding for HSPs (Hsp22, Lethal(2)efl, Hsp27, Hsp40, Hsp70 and Hsp83) in order to have representatives of the most clearly upregulated functional category 8 (Protein processing). We then took DE sequences scoring relatively high in log fold change (logFC), log counts per million (logCPM) and sequence length among those upregulated in cold-acclimated larvae (SDA11) relative to those in diapause maintenance (SD6 and SD11). This way we added (b) four sequences, coding for Yellow d (or Major royal jelly 1, Seq3773), Glutamic acid-rich protein (GARP, Seq93436), Larval serum protein 2 (Lsp2, Seq55855) and Companion of reaper (Corp, Seq5725) (functional category 9 ‘Other’), (c) four uncharacterized protein sequences: Seq102667, Seq80983, Seq3519 and Seq4228 and finally (d) one non-annotated (N/A) sequence: Seq93437.
In parallel, we characterized the association between the transcriptomic profiles of the 15 candidate genes (above) and larval extreme freeze tolerance (capacity to survive after exposure to liquid nitrogen) for select LD, SD and SDA variants (electronic supplementary material, figure S2) using the optimum cryopreservation protocol described earlier [23]. Briefly, groups of 20 larvae were slowly cooled to −30°C, plunged into liquid nitrogen for 60 min, then returned to −30°C before rewarming to 5°C. Thawed larvae were transferred to fresh larval diet and maintained thereafter at 18°C and a long-day photoperiod. Dead larvae were removed 12 h later while all living larvae were maintained for a subsequent six weeks and the emergence of adult flies was scored as the ultimate criterion of survival. For each variant, extreme freeze tolerance was measured in 100–300 larvae and expression of each candidate gene was measured in four larvae.
3. Results and discussion
To our knowledge, this is the first transcriptomic characterization of an insect capable of surviving prolonged cryopreservation in liquid nitrogen. We used the comparative transcriptomic approach as an hypothesis-generating first step in seeking new candidate cryoprotectants and to further understand the physiological mechanisms of extreme freeze tolerance; however, we acknowledge that other forms of gene and protein regulation which may be important (e.g. miRNAs or post-translational modifications) [73] are not captured by our methods and therefore warrant investigation in future.
(a). Differentially regulated genes and pathways
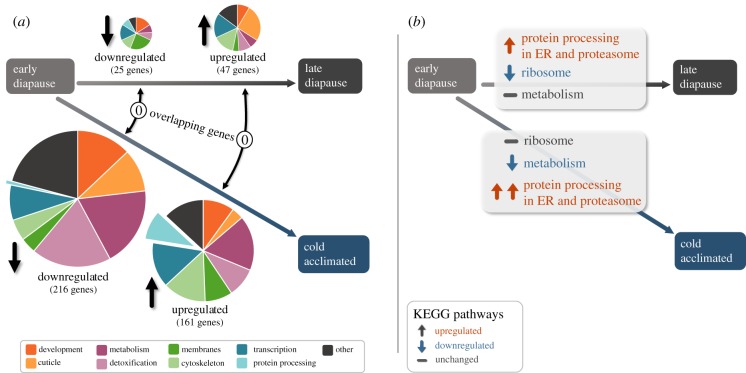
We observed fewer DE genes between early and late diapause phenotypes (190 transcripts) than between early diapause and cold-acclimated phenotypes (776 transcripts). A full list of DE genes is available in electronic supplementary material, spreadsheet S1. Of those genes with putative identities, 25 and 47 were down- and upregulated in late diapause (respectively), while 216 and 161 were down- and upregulated with cold acclimation, respectively). Only a single DE gene (Seq109825; downregulated but with no putative identity) overlapped between late diapause and cold acclimation (relative to early diapause), suggesting that cold acclimation and development of extreme freeze tolerance may involve a unique set of physiological processes distinct from those linked to diapause progression.
In a recent review of insect freeze tolerance, Toxopeus & Sinclair [7] hypothesized that five broad mechanisms are involved: (1) control of ice formation; (2) reduction of ice content; (3) stabilization of macromolecules; (4) management of biochemical processes/reduction of harmful metabolite damage and (5) post-thaw repair and recovery. We did not expect to see a reflection of mechanism 1 in this study, as no gene annotations in the C. costata transcriptome [63] had identifiers such as ice binding, nucleation or thermal hysteresis. Moreover, our knowledge suggests that C. costata has limited capacity to control ice formation: neither supercooling capacity nor vulnerability to ice inoculation changed much with acclimation [23,35], and there is no sign of thermal hysteresis activity in larval haemolymph (V. Koštál & J. Rozsypal 2017, unpublished observations). Ice content (related to mechanism 2) differs only slightly among C. costata larval variants with decreasing temperatures (described previously by [23]). Mechanism 3 probably involves cryoprotectant accumulation as a non-colligative means of stabilizing macromolecular structures, and despite that cryoprotectant accumulation is a hallmark of C. costata freeze tolerance [23,35], we found little direct reflection of it in the transcriptome. Similar lack of evidence for direct transcriptional control of cryoprotectant synthesis (myo-inositol, proline and trehalose) was reported for the freeze-tolerant cricket, Gryllus veletis [43]. The reasons for such results may relate to the importance of post-transcriptional control mechanisms [43], general problems with interpretation of metabolism using -omics approaches [74] and/or technical limitations such as insufficient resolution at time and tissue levels. Mechanism 3 also includes the upregulation of molecular chaperones (a common phenomenon of insect thermal tolerance, e.g. [75,76]), which was clearly supported in the C. costata transcriptome (discussed further in the next section). Mechanism 4 was reflected in the form of global downregulation of processes in C. costata larvae linked to active metabolism, including oxidative phosphorylation (figure 1b; a full list of the pathways provided in electronic supplementary material, figure S3). Still, ribosomal transcription was generally maintained during cold acclimation (figure 1; electronic supplementary material, figure S3–S5). The aspect of mechanism 4 relating to the reduction of damage from harmful metabolites showed rather a trend of downregulation (electronic supplementary material, spreadsheet S1) and no systematic reflection in KEGG pathways. Mechanism 5 includes some elements overlapping with mechanism 3 (e.g. protein processing machinery), which appears to be a central theme of the transcriptomic transformation in extremely freeze-tolerant C. costata larvae.
Figure 1.
Summary of DE genes with putative identities (a) and select DE KEGG pathways (b) for third instar malt fly larvae in the states of late diapause or cold acclimation relative to early diapause. Genes were sorted according to custom functional categories and the numbers of DE genes in different categories are reflected in pie diagram size (the total number of DE genes given below). A full list of DE genes (including the description of functional categories) and KEGG pathways are given in electronic supplementary material, spreadsheet S1 and figure S3, respectively. (Online version in colour.)
Expression of genes related to the category ‘Protein processing’ was the most contrasting between larval variants; compared to early diapause, two genes were downregulated in late diapause, while 15 genes were upregulated with cold acclimation (figure 1b; electronic supplementary material, spreadsheet S1). Some pathways appeared to be statistically rather than biologically upregulated with progression from early to late diapause. For example, upregulation of KEGG pathway ko04141 (Protein processing in the endoplasmic reticulum) in late relative to early diapause was driven primarily by only a few genes; secretory 61 (Sec61), translocon-associated protein (TRAP) and Hsp70 (electronic supplementary material, figure S6). Similarly, the upregulation of pathway ko03050 (Proteasome) during late diapause was extremely weak (based on two genes, each in only one of the three biological replicates; see electronic supplementary material, figure S7). By contrast, the upregulation of both these pathways with cold acclimation was supported by a large number of genes (electronic supplementary material, figures S8 and S9, spreadsheet S1). The relevance of enhanced protein processing capabilities for extreme freeze tolerance is discussed in detail below.
(b). Roles of protein processing in extreme cold tolerance
As both ice formation and low temperatures threaten the stability and function of proteins [77], extreme freeze tolerance should largely rely on enhanced protein protection (stabilization and chaperoning), repair (refolding) and/or degradation of denatured or misfolded proteins. Indeed, a growing body of literature highlights the importance of protein management for insect freeze tolerance [7]. Based on transcriptomic analysis, however, we cannot conclusively distinguish between upregulated responses representing (i) the direct response to proteins that failed to fold properly (or were partially denatured) during cold acclimation, and (ii) the removal of excess proteins that are no longer needed when the rates of all biological processes were drastically reduced by low temperature. We hypothesize that at least part of the upregulation represents (iii) an ‘anticipatory’ response (i.e. adaptive preparation for cold and freeze-dehydration stresses most likely to be endured during the overwintering period, which is supported by previous studies in flesh flies [78,79]).
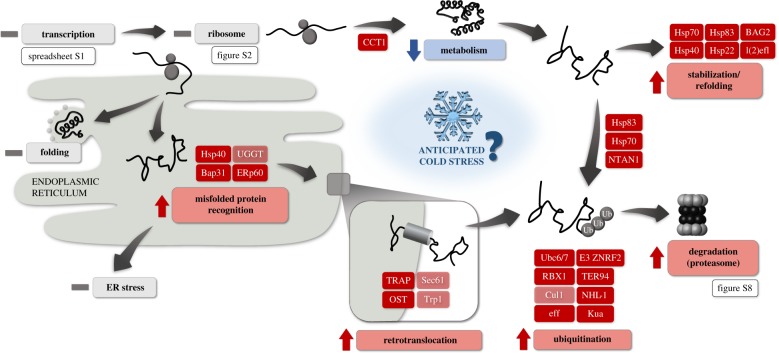
Cold and freeze-dehydration stresses may impact the process of nascent proteins' folding or mature proteins’ higher-order structures, causing their misfolding or unfolding, respectively. In both cases, the aberrant proteins elicit a complex network of responses including recognition, targeting, transport, refolding or elimination by degradation. A visual synthesis (based on results of DE analysis and KEGG analysis) of this enhanced machinery accompanying cold acclimation (i.e. acquisition of extreme freeze tolerance) is provided in figure 2 (see also electronic supplementary material, figure S8 for the relevant KEGG pathway). We comment only briefly on the major elements of this machinery below and provide more specific descriptions of the roles of the individual genes in electronic supplementary material, figure S10).
Figure 2.
Upregulation of gene transcripts involved in protein processing machinery associated with the acquisition of extreme freeze tolerance in malt fly larvae. Genes/pathways in red are upregulated (with weaker red indicating relatively weaker upregulation), blue are downregulated and grey are unchanged for cold-acclimated larvae relative to those in early diapause. Differential expression was based on both KEGG pathways (electronic supplementary material, figures S3 and S8) and individual gene expression analyses (electronic supplementary material, spreadsheet S1) (see text for more details). See electronic supplementary material, figure S10 for full a description of the upregulated genes and processes.
In the endoplasmic reticulum (ER), misfolded proteins are eliminated by ER-associated degradation (ERAD). In this process (as summarized by [80]), misfolded proteins are exported (retrotranslocated) to the cytosol and ubiquitinated, which targets them for degradation by the 26S proteasome. Accumulation of misfolded proteins above the ER folding capacity (ER stress) initiates the unfolded protein response (UPR), which increases chaperone production and either allows recovery from ER stress or may result in apoptotic cell death. A single gene (coding for ATF4) was downregulated among three pathways that sense ER stress (i.e. those initiated by PERK, ATF6 and IRE1; electronic supplementary material, figure S8). This lack of differential expression related to actual ER stress indirectly supports the hypothesis that anticipatory enhancement of chaperones and other effectors of UPR occurs to deal with misfolded protein load that likely comes after future cold shock. Four genes upregulated in extremely freeze-tolerant larvae encoded proteins involved in misfolded protein recognition and targeting for ERAD: Hsp40, ERp60, Bap31 and UGGT. Another four genes encoded proteins involved in retrotranslocation of ERAD-targeted proteins to the cytosol: TRAP, Sec61, OST and Trp1 (figure 2). Cold acclimation also resulted in increased expression of multiple genes involved in protein ubiquitination (electronic supplementary material, figure S8), as well as genes encoding proteasome subunits (electronic supplementary material, figure S9).
In extremely freeze-tolerant larvae, six genes encoding chaperones involved in mature protein stabilization and refolding were upregulated: Hsp83, Hsp70, Hsp40, BAG2 and two sHSPs (Hsp22 and Lethal(2)efl) (figure 2). Hsp70 (with co-chaperones BAG2 and Hsp40) and Hsp83 may additionally cooperate with co-chaperone CHIP to direct substrate proteins for degradation [80–86]. Similarly, the gene coding for Chaperonin-containing T-complex 1 (CCT1), which facilitates the folding of cytoskeletal (and other) proteins [87], was also upregulated. The importance of cytoskeletal protection and repair for maintaining cell structure, tissue integrity and transport function at low temperatures is supported by a growing number of studies in insects, including the malt fly [88–92]. Lethal(2)efl and mitochondrial Hsp22 stabilize and facilitate denatured protein refolding, preventing protein aggregation [93,94]. Through this enhanced protein processing, cold-acclimated malt fly larvae likely have an improved ability to remove damaged proteins before they reach toxic levels in the cytosol and/or before they accumulate to levels that induce ER stress, thereby avoiding induction of apoptotic pathways and subsequent cell death during and after cold exposure [80]. Taken together, this machinery is likely protective against protein crowding and denaturation at extremely low temperatures and/or upon cellular freeze dehydration.
(c). Candidate cryoprotective genes
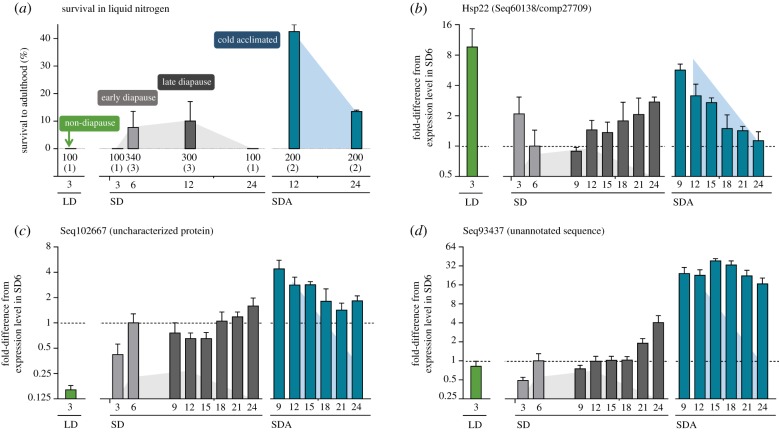
Cryopreservation survival analyses (figure 3a) confirmed that non-diapause larvae (LD3) and relatively young larvae destined to diapause (SD3) have no ability to survive in liquid nitrogen. A moderate capacity for such survival occurred in diapause maintenance phenotypes (SD6, SD12) but was lost with long-term maintenance (SD24). Truly extreme freeze tolerance (the highest capacity for survival in liquid nitrogen) was found in cold-acclimated larvae at 12 weeks (SDA12), which perfectly confirmed our earlier results [23,35], but this was again partially lost with increasing time of storage at low temperature (SDA24). Examples of transcriptional profiles for three selected sequences are depicted in figure 3b–d, while all profiles are shown in electronic supplementary material, figures S11–S13. The transcriptional profiles of lethal(2)efl (electronic supplementary material, figure S11B) and hsp70 (electronic supplementary material, figure S11E) matched relatively well with extreme freeze tolerance. Among the heat shock genes, the mitochondrial hsp22 (figure 3b) stood out as a particularly interesting candidate considering its 7.5-fold increase in expression with cold acclimation (SDA9) relative to early diapause (SD6). Although expression of hsp22 in non-diapause larvae (LD3) was also very high, the gene need not be necessarily removed from the list of candidate cryoprotective macromolecules. Hsp22 localizes to mitochondria [92] and may play very different roles in the two distinct physiological contexts: LD (direct development, rapid growth, high activity) versus SDA (diapause, deep metabolic suppression, no activity). The rapid metabolism of LD larvae must be supported by high activity of their mitochondria, and (in D. melanogaster) hsp22 expression is upregulated with ageing and oxidative stress [95] as well as in response to rising ecdysone titres just prior to pupation [96,97]. These stimuli for hsp22 upregulation are absent in the C. costata SDA phenotype. We also know that C. costata mitochondria are particularly susceptible to cryoinjury in LD larvae but are robust to freezing challenges in SDA larvae (T. Štětina & V. Koštál 2018, unpublished results). Potentially, Hsp22 could act (synergistically with other cryoprotectants and DE chaperones) in SDA larvae to preserve mitochondrial structure during freeze-dehydration insult.
Figure 3.
Freeze tolerance (a) and expression patterns of select sequences (b–d) in third instar malt fly larvae. In freeze tolerance assays (a), larvae were plunged into liquid nitrogen for 1 h using the optimum protocol described earlier [23]. After thawing, larvae were moved to long days at 18°C for six weeks and the emergence of adult flies was scored as the ultimate criterion of survival. Bars show means ± s.d. and the numbers below each bar show the total number of larvae subjected to the assay (with the number of different fly generations assayed given in parentheses). Examples of transcriptional profiles are shown for three select sequences (b–d). Each bar (representing a particular phenotypic variant, figure S1) is the mean ± s.d. (n = 4) relative expression of the target sequence compared to four reference genes (via RTqPCR). The relative expression in phenotypic variant SD6 (early diapause) is arbitrarily set to 1 and the expressions in all other variants are normalized to it. Shaded areas behind the bars indicate survival in liquid nitrogen for SD larvae (grey shading) and SDA larvae (blue shading), as derived from (a) (note that survival was zero for non-diapause LD larvae and also SD3 larvae).
Transcriptional patterns of four other annotated but functionally poorly characterized genes are shown in electronic supplementary material, figure S12. Lsp2 was characterized by extremely high expression in non-diapause LD3 larvae (more than 1000-fold higher than in early diapause), corresponding well to the proposed function of larval serum proteins as amino acid stores that rapidly build up in the haemolymph of larvae just prior to pupation [98]. Interestingly, lsp2 transcripts also gradually accumulated and reached considerably high levels (up to 100-fold) in cold-acclimated larvae relative to those in early diapause (electronic supplementary material, figure S12A). As ice forms in the extracellular space of malt fly larvae [35], it is possible that haemolymph-abundant Lsp2 protein may somehow interfere with ice crystal growth. The expression of Corp (which is induced by DNA damage in D. melanogaster [99]) increased more than 10-fold in malt fly larvae upon cold acclimation (electronic supplementary material, figure S12B). Corp protein inhibits the pro-apoptotic activity of p53 and may thus mitigate apoptotic cell death for cold-acclimated larvae following a freezing challenge [99]. The transcripts of yellow d (electronic supplementary material, figure S12C) and GARP (electronic supplementary material, figure S12D) showed strong and persistent upregulation response to cold acclimation. The yellow family contains functionally poorly characterized genes that are most likely involved in melanin formation [100]. As development at lower temperatures increases melanin deposition in drosophilids (e.g. [101]), this might partially explain our results. GARP is a structural homologue of the functionally uncharacterized D. melanogaster gene CG43106 (putative GARP A0A0B4K825), but the relevance of this protein for extreme freeze tolerance is currently unclear.
Over half of the DE genes significantly up- or downregulated with cold acclimation coded for uncharacterized proteins or had no putative identities at all. Transcriptional profiles of five selected unidentified candidates are shown in electronic supplementary material, figure S13. Ideally, this set of unidentified genes contains novel and potentially important cryoprotectants. For instance, the uncharacterized Seq102667 (figure 3c) showed almost a perfect match to the profile of extreme freeze tolerance, while the unannotated Seq93437 (figure 3d) showed extremely strong and persistent response to cold acclimation. For select unannotated candidates, we are now conducting more comprehensive searches for their putative identities, raising antibodies for localization and quantification of gene products, and performing functional validation assays (e.g. for enzyme activity in vitro and survival of transfected cells post-freezing, results in a forthcoming study).
4. Conclusion
The transition from moderate to extreme freeze tolerance in malt fly larvae is accompanied by an enhanced protein processing capacity, probably in anticipation of oncoming cellular freeze-dehydration stress. With this ‘prophylactic’ response, cold-acclimated larvae may become better able to both protect and stabilize proteins, and repair or eliminate those which become damaged, thereby avoiding excessive misfolded protein load, ER stress and associated apoptosis. In addition to classical chaperones, we identified a number of functionally uncharacterized sequences in which the transcriptional profile perfectly matches the profile of larval extreme freeze tolerance (i.e. strong upregulation in response to cold acclimation). Identification of the protein products of these sequences, and their functional analysis, may be crucial for achieving not only a fundamental understanding of freeze tolerance plasticity in insects but also for obtaining new means of freeze tolerance manipulation for cryopreservation of cells, tissues and whole organisms.
Supplementary Material
Supplementary Material
Acknowledgements
We would like to thank Jaroslava Korbelová for preparing larvae and total RNA samples for RNAseq, Rodolphe Poupardin for providing the de novo assembled malt fly transcriptome and guidance for bioinformatics, and Jan Provazník for guidance with sequence library assessment and alignment.
Data accessibility
The sequencing datasets supporting the results of this article are available via the NCBI Sequence Read Archive (accession number: PRJNA499074).
Authors' contributions
V.K. conceptualized the study, P.H. prepared larvae and total RNA samples and performed qPCR validations and survival analysis, L.E.D.M. processed the transcriptomic data, performed the statistical analyses, prepared the figures and wrote the original manuscript draft, V.K. edited the final manuscript.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by a Czech Science Foundation (grants 16-06374S and 19-13381S to V.K.). The funding body had no role in the study design, collection, analysis or interpretation of the data, nor the writing of the report or decision to submit the report for publication.
References
- 1.Bachmetjew P. 1901. Experimentelle entomologische studien vom physikalisch-chemischen standpunkt aus. Leipzig, Germany: Verlag von Wilhelm Engelmann. [Google Scholar]
- 2.Payne NM. 1927. Measures of insect cold hardiness. Biol. Bull. 52, 449–457. ( 10.2307/1536906) [DOI] [Google Scholar]
- 3.Salt R. 1961. Principles of insect cold-hardiness. Annu. Rev. Entomol. 6, 55–74. ( 10.1146/annurev.en.06.010161.000415) [DOI] [Google Scholar]
- 4.Asahina E. 1970. Frost resistance in insects. Adv. Insect Physiol. 6, 1–49. [Google Scholar]
- 5.Lee RE. 1991. Principles of insect low temperature tolerance. In Insects at low temperature (eds Lee RE, Denlinger DL), pp. 17–46. Berlin, Germany: Springer. [Google Scholar]
- 6.Teets NM, Denlinger DL. 2013. Physiological mechanisms of seasonal and rapid cold-hardening in insects. Physiol. Entomol. 38, 105–116. ( 10.1111/phen.12019) [DOI] [Google Scholar]
- 7.Toxopeus J, Sinclair BJ. 2018. Mechanisms underlying insect freeze tolerance. Biol. Rev. 93, 1891–1914. ( 10.1111/brv.12425) [DOI] [PubMed] [Google Scholar]
- 8.Chown S, Sinclair B. 2010. The macrophysiology of insect cold hardiness. In Low temperature biology of insects (eds Denlinger DL, Lee RE Jr), pp. 191–222. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 9.Sinclair BJ. 1999. Insect cold tolerance: how many kinds of frozen? Eur. J. Entomol. 96, 157–164. [Google Scholar]
- 10.De Coninck L. 1951. On the resistance of the free-living nematode Anguillula silusiae to low temperatures. Biodynamica 7, 77–84. [PubMed] [Google Scholar]
- 11.Koehler JK. 1967. Studies on the survival of the rotifer Philodina after freezing and thawing. Cryobiology 3, 392–399. ( 10.1016/S0011-2240(67)80134-2) [DOI] [Google Scholar]
- 12.Tanno K. 1971. Frost injury and resistance in the poplar sawfly, Trichiocampus populi Okamoto. Contrib. Inst. Low Temp. Sci. 16, 1–41. ( 10.1016/s0011-2240(69)80014-3) [DOI] [Google Scholar]
- 13.Moon I, Fujikawa S, Shimada K. 1996. Cryopreservation of Chymomyza larvae (Diptera: Drosophilidae) at −196°C with extracellular freezing. Cryo-Lett. 17, 105–110. [Google Scholar]
- 14.Suzuki D, Miyamoto T, Kikawada T, Watanabe M, Suzuki T. 2014. A leech capable of surviving exposure to extremely low temperatures. PLoS ONE 9, e86807 ( 10.1371/journal.pone.0086807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahy GM, Wowk B. 2015. Principles of cryopreservation by vitrification. In Cryopreservation and freeze-drying protocols, 3rd edn (eds Wolkers WF, Oldenhof H), pp. 21–82. Berlin, Germany: Springer. [DOI] [PubMed] [Google Scholar]
- 16.Leopold R. 2007. Colony maintenance and mass-rearing: using cold storage technology for extending the shelf-life of insects. In Area-Wide control of insect pests (eds Vreysen MJB, Robinson AS, Hendrichs J), pp. 149–162. Berlin, Germany: Springer. [Google Scholar]
- 17.Mazur P, Schneider U, Mahowald AP. 1992. Characteristics and kinetics of subzero chilling injury in Drosophila embryos. Cryobiology 29, 39–68. ( 10.1016/0011-2240(92)90005-M) [DOI] [PubMed] [Google Scholar]
- 18.Colinet H, Boivin G. 2011. Insect parasitoids cold storage: a comprehensive review of factors of variability and consequences. Biol. Control 58, 83–95. ( 10.1016/j.biocontrol.2011.04.014) [DOI] [Google Scholar]
- 19.Wasylyk JM, Tice AR, Baust JG. 1988. Partial glass formation: a novel mechanism of insect cryoprotection. Cryobiology 25, 451–458. ( 10.1016/0011-2240(88)90053-3) [DOI] [Google Scholar]
- 20.Steponkus P, et al. 1990. Cryopreservation of Drosophila melanogaster embryos. Nature 345, 170 ( 10.1038/345170a0) [DOI] [PubMed] [Google Scholar]
- 21.Leopold RA, Rinehart JP. 2010. A template for insect cryopreservation. In Low temperature biology of insects (eds Denlinger DL, Lee RE Jr), pp. 325–341. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 22.Koštál V, Mollaei M, Schöttner K. 2016. Diapause induction as an interplay between seasonal token stimuli, and modifying and directly limiting factors: hibernation in Chymomyza costata. Physiol. Entomol. 41, 344–357. ( 10.1111/phen.12159) [DOI] [Google Scholar]
- 23.Rozsypal J, Moos M, Šimek P, Koštál V. 2018. Thermal analysis of ice and glass transitions in insects that do and do not survive freezing. J. Exp. Biol. 221, 170464 ( 10.1242/jeb.170464) [DOI] [PubMed] [Google Scholar]
- 24.Denlinger DL. 1991. Relationship between cold hardiness and diapause. In Insects at low temperature (eds Lee RE, Denlinger DL), pp. 174–198. Berlin, Germany: Springer. [Google Scholar]
- 25.Hahn DA, Denlinger DL. 2011. Energetics of insect diapause. Annu. Rev. Entomol. 56, 103–121. ( 10.1146/annurev-ento-112408-085436) [DOI] [PubMed] [Google Scholar]
- 26.Pullin AS. 1996. Physiological relationships between insect diapause and cold tolerance: coevolution or coincidence. Eur. J. Entomol. 93, 121–130. [Google Scholar]
- 27.Koštál V, Štětina T, Poupardin R, Korbelová J, Bruce AW. 2017. Conceptual framework of the eco-physiological phases of insect diapause development justified by transcriptomic profiling. Proc. Natl Acad. Sci. USA 114, 8532–8537. ( 10.1073/pnas.1707281114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang B, Liu X-J, Shi Z-K, Shen Q-D, Xu Y-X, Wang S, Zhang F, Wang S-G. 2017. Transcriptome analysis and identification of induced genes in the response of Harmonia axyridis to cold hardiness. Comp. Biochem. Physiol. D 22, 78–89. ( 10.1016/j.cbd.2017.01.004) [DOI] [PubMed] [Google Scholar]
- 29.Des Marteaux LE, McKinnon AH, Udaka H, Toxopeus J, Sinclair B. 2017. Effects of cold-acclimation on gene expression in Fall field cricket (Gryllus pennsylvanicus) ionoregulatory tissues. BMC Genomics 18, 357 ( 10.1186/s12864-017-3711-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragland GJ, Keep E. 2017. Comparative transcriptomics support evolutionary convergence of diapause responses across Insecta. Physiol. Entomol. 42, 246–256. ( 10.1111/phen.12193) [DOI] [Google Scholar]
- 31.Colinet H, Renault D, Charoy-Guével B, Com E. 2012. Metabolic and proteomic profiling of diapause in the aphid parasitoid Praon volucre. PLoS ONE 7, e32606 ( 10.1371/journal.pone.0032606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kristensen TN, Kjeldal H, Schou MF, Nielsen JL. 2016. Proteomic data reveal a physiological basis for costs and benefits associated with thermal acclimation. J. Exp. Biol. 219, 969–976. ( 10.1242/jeb.132696) [DOI] [PubMed] [Google Scholar]
- 33.MacMillan HA, Knee JM, Dennis AB, Udaka H, Marshall KE, Merritt TJ, Sinclair BJ. 2016. Cold acclimation wholly reorganizes the Drosophila melanogaster transcriptome and metabolome. Sci. Rep. 6, 28999 ( 10.1038/srep28999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michaud MR, Denlinger DL. 2007. Shifts in the carbohydrate, polyol, and amino acid pools during rapid cold-hardening and diapause-associated cold-hardening in flesh flies (Sarcophaga crassipalpis): a metabolomic comparison. J. Comp. Physiol. B 177, 753–763. ( 10.1007/s00360-007-0172-5) [DOI] [PubMed] [Google Scholar]
- 35.Koštál V, Zahradnickova H, Simek P. 2011. Hyperprolinemic larvae of the drosophilid fly, Chymomyza costata, survive cryopreservation in liquid nitrogen. Proc. Natl Acad. Sci. USA 108, 13 041–13 046. ( 10.1073/pnas.1107060108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimada K, Riihimaa A. 1988. Cold acclimation, inoculative freezing and slow cooling: essential factors contributing to the freezing-tolerance in diapausing larvae of Chymomyza costata (Diptera: Drosophilidae). Cryo. Lett. 9, 5–10. [Google Scholar]
- 37.Zachariassen KE. 1985. Physiology of cold tolerance in insects. Physiol. Rev. 65, 799–832. ( 10.1152/physrev.1985.65.4.799) [DOI] [PubMed] [Google Scholar]
- 38.Muldrew K, Acker JP, Elliott JA, McGann LE. 2004. The water to ice transition: implications for living cells. In Life in the frozen state (eds Fuller BJ, Lane N, Benson EE), pp. 93–134. Boca Raton, FL: CRC Press. [Google Scholar]
- 39.Crowe JH, Clegg JS, Crowe LM. 1998. Anhydrobiosis: the water replacement hypothesis. In The properties of water in foods ISOPOW 6 (ed. Reid DS.), pp. 440–455. Berlin, Germany: Springer. [Google Scholar]
- 40.Arakawa T, Timasheff SN. 1985. Theory of protein solubility. Methods Enzymol. 114, 49–77. ( 10.1016/0076-6879(85)14005-X) [DOI] [PubMed] [Google Scholar]
- 41.Rudolph AS, Crowe JH. 1986. A calorimetric and infrared spectroscopic study of the stabilizing solute proline. Biophys. J. 50, 423–430. ( 10.1016/S0006-3495(86)83478-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elliott GD, Wang S, Fuller BJ. 2017. Cryoprotectants: a review of the actions and applications of cryoprotective solutes that modulate cell recovery from ultra-low temperatures. Cryobiology 76, 74–91. ( 10.1016/j.cryobiol.2017.04.004) [DOI] [PubMed] [Google Scholar]
- 43.Toxopeus J, Koštál V, Sinclair Brent J. 2019. Evidence for non-colligative function of small cryoprotectants in a freeze-tolerant insect. Proc. R. Soc. B 286, 20190050 ( 10.1098/rspb.2019.0050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duman JG. 2015. Animal ice-binding (antifreeze) proteins and glycolipids: an overview with emphasis on physiological function. J. Exp. Biol. 218, 1846–1855. ( 10.1242/jeb.116905) [DOI] [PubMed] [Google Scholar]
- 45.Bar Dolev M, Braslavsky I, Davies PL. 2016. Ice-binding proteins and their function. Annu. Rev. Biochem. 85, 515–542. ( 10.1146/annurev-biochem-060815-014546) [DOI] [PubMed] [Google Scholar]
- 46.King AM, MacRae TH. 2015. Insect heat shock proteins during stress and diapause. Annu. Rev. Entomol. 60, 59–75. ( 10.1146/annurev-ento-011613-162107) [DOI] [PubMed] [Google Scholar]
- 47.Tunnacliffe A, Wise MJ. 2007. The continuing conundrum of the LEA proteins. Naturwissenschaften 94, 791–812. ( 10.1007/s00114-007-0254-y) [DOI] [PubMed] [Google Scholar]
- 48.Hand SC, Menze MA, Toner M, Boswell L, Moore D. 2011. LEA proteins during water stress: not just for plants anymore. Annu. Rev. Physiol. 73, 115–134. ( 10.1146/annurev-physiol-012110-142203) [DOI] [PubMed] [Google Scholar]
- 49.Bahrndorff S, Tunnacliffe A, Wise MJ, McGee B, Holmstrup M, Loeschcke V. 2009. Bioinformatics and protein expression analyses implicate LEA proteins in the drought response of Collembola. J. Insect Physiol. 55, 210–217. ( 10.1016/j.jinsphys.2008.11.010) [DOI] [PubMed] [Google Scholar]
- 50.Li S, Chakraborty N, Borcar A, Menze MA, Toner M, Hand SC. 2012. Late embryogenesis abundant proteins protect human hepatoma cells during acute desiccation. Proc. Natl Acad. Sci. USA 109, 20 859–20 864. ( 10.1073/pnas.1214893109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma X, et al. 2005. A small stress protein acts synergistically with trehalose to confer desiccation tolerance on mammalian cells. Cryobiology 51, 15–28. ( 10.1016/j.cryobiol.2005.04.007) [DOI] [PubMed] [Google Scholar]
- 52.Viner RI, Clegg JS. 2001. Influence of trehalose on the molecular chaperone activity of p26, a small heat shock/α-crystallin protein. Cell Stress Chaperon 6, 126 () [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang J-S, Raymond JA. 2004. Reduction of freeze-thaw-induced hemolysis of red blood cells by an algal ice-binding protein. Cryoletters 25, 307–310. [PubMed] [Google Scholar]
- 54.Goyal K, Walton LJ, Tunnacliffe A. 2005. LEA proteins prevent protein aggregation due to water stress. Biochem. J. 388, 151–157. ( 10.1042/BJ20041931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gygi SP, Rochon Y, Franza BR, Aebersold R. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730. ( 10.1128/MCB.19.3.1720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riihimaa AJ, Kimura MT. 1988. A mutant strain of Chymomyza costata (Diptera: Drosophilidae) insensitive to diapause-inducing action of photoperiod. Physiol. Entomol. 13, 441–445. ( 10.1111/j.1365-3032.1988.tb01128.x) [DOI] [Google Scholar]
- 57.Kostal V, Noguchi H, Shimada K, Hayakawa Y. 1998. Developmental changes in dopamine levels in larvae of the fly Chymomyza costata: comparison between wild-type and mutant-nondiapause strains. J. Insect Physiol. 44, 605–614. ( 10.1016/S0022-1910(98)00043-2) [DOI] [PubMed] [Google Scholar]
- 58.Koštál V, Shimada K, Hayakawa Y. 2000. Induction and development of winter larval diapause in a drosophilid fly, Chymomyza costata. J. Insect Physiol. 46, 417–428. ( 10.1016/S0022-1910(99)00124-9) [DOI] [PubMed] [Google Scholar]
- 59.Andrews S. 2018. FastQC: a quality control tool for high throughput sequence data. See https://github.com/s-andrews/FastQC.
- 60.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 ( 10.1186/gb-2010-11-8-r86) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 9, 357–359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poupardin R, Schöttner K, Korbelová J, Provazník J, Doležel D, Pavlinic D, Beneš V, Koštál V. 2015. Early transcriptional events linked to induction of diapause revealed by RNAseq in larvae of drosophilid fly, Chymomyza costata. BMC Genomics 16, 720 ( 10.1186/s12864-015-1907-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. ( 10.1038/nprot.2012.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. ( 10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R Core Team. 2018. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 67.Risso D, Ngai J, Speed TP, Dudoit S. 2014. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902. ( 10.1038/nbt.2931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. ( 10.1093/bioinformatics/bti610) [DOI] [PubMed] [Google Scholar]
- 69.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. ( 10.1093/nar/28.1.27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. ( 10.1093/nar/gkm321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo W, Brouwer C. 2013. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831. ( 10.1093/bioinformatics/btt285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. 2009. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinf. 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dykes IM, Emanueli C. 2017. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genom. Proteom. Bioinf. 15, 177–186. ( 10.1016/j.gpb.2016.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suarez RK, Moyes CD. 2012. Metabolism in the age of ‘omes’. J. Exp. Biol. 215, 2351–2357. ( 10.1242/jeb.059725) [DOI] [PubMed] [Google Scholar]
- 75.Colinet H, Overgaard J, Com E, Sørensen JG. 2013. Proteomic profiling of thermal acclimation in Drosophila melanogaster. Insect Biochem. Mol. Biol. 43, 352–365. ( 10.1016/j.ibmb.2013.01.006) [DOI] [PubMed] [Google Scholar]
- 76.Zhang G, Storey JM, Storey KB. 2011. Chaperone proteins and winter survival by a freeze tolerant insect. J. Insect Physiol. 57, 1115–1122. ( 10.1016/j.jinsphys.2011.02.016) [DOI] [PubMed] [Google Scholar]
- 77.Bhatnagar BS, Bogner RH, Pikal MJ. 2007. Protein stability during freezing: separation of stresses and mechanisms of protein stabilization. Pharm. Dev. Technol. 12, 505–523. ( 10.1080/10837450701481157) [DOI] [PubMed] [Google Scholar]
- 78.Ragland GJ, Denlinger DL, Hahn DA. 2010. Mechanisms of suspended animation are revealed by transcript profiling of diapause in the flesh fly. Proc. Natl Acad. Sci. USA 107, 14 909–14 914. ( 10.1073/pnas.1007075107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rinehart JP, Yocum GD, Denlinger DL. 2000. Developmental upregulation of inducible hsp70 transcripts, but not the cognate form, during pupal diapause in the flesh fly, Sarcophaga crassipalpis. Insect Biochem. Mol. Biol. 30, 515–521. ( 10.1016/S0965-1748(00)00021-7) [DOI] [PubMed] [Google Scholar]
- 80.Strudwick N, Schröder M. 2007. The unfolded protein response. In Systems biology (eds Al-Rubeai M, Fussenegger M), pp. 69–155. Berlin, Germany: Springer. [Google Scholar]
- 81.Gong WJ, Golic KG. 2006. Loss of Hsp70 in Drosophila is pleiotropic, with effects on thermotolerance, recovery from heat shock and neurodegeneration. Genetics 172, 275–286. ( 10.1534/genetics.105.048793) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Höhfeld J, Cyr DM, Patterson C. 2001. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2, 885–890. ( 10.1093/embo-reports/kve206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McDonough H, Patterson C. 2003. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperon. 8, 303 () [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alderson TR, Kim JH, Markley JL. 2016. Dynamical structures of Hsp70 and Hsp70-Hsp40 complexes. Structure 24, 1014–1030. ( 10.1016/j.str.2016.05.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qiu X-B, Shao Y-M, Miao S, Wang L. 2006. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570. ( 10.1007/s00018-006-6192-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dai Q, et al. 2005. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J. Biol. Chem. 280, 38 673–38 681. ( 10.1074/jbc.M507986200) [DOI] [PubMed] [Google Scholar]
- 87.Pavel M, et al. 2016. CCT complex restricts neuropathogenic protein aggregation via autophagy. Nat. Commun. 7, 13821 ( 10.1038/ncomms13821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Des Marteaux LE, Stinziano JR, Sinclair BJ. 2018. Effects of cold acclimation on rectal macromorphology, ultrastructure, and cytoskeletal stability in Gryllus pennsylvanicus crickets. J. Insect Physiol. 104, 15–24. ( 10.1016/j.jinsphys.2017.11.004) [DOI] [PubMed] [Google Scholar]
- 89.Des Marteaux LE, Štětina T, Koštál V. 2018. Insect fat body cell morphology and response to cold stress is modulated by acclimation. J. Exp. Biol. 221, jeb189647 ( 10.1242/jeb.189647) [DOI] [PubMed] [Google Scholar]
- 90.Kim M, Robich RM, Rinehart JP, Denlinger DL. 2006. Upregulation of two actin genes and redistribution of actin during diapause and cold stress in the northern house mosquito, Culex pipiens. J. Insect Physiol. 52, 1226–1233. ( 10.1016/j.jinsphys.2006.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kayukawa T, Ishikawa Y. 2009. Chaperonin contributes to cold hardiness of the onion maggot Delia antiqua through repression of depolymerization of actin at low temperatures. PLoS ONE 4, e8277 ( 10.1371/journal.pone.0008277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.MacMillan HA, Yerushalmi GY, Jonusaite S, Kelly SP, Donini A. 2017. Thermal acclimation mitigates cold-induced paracellular leak from the Drosophila gut. Sci. Rep. 7, 8807 ( 10.1038/s41598-017-08926-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vos MJ, Carra S, Kanon B, Bosveld F, Klauke K, Sibon OC, Kampinga HH. 2016. Specific protein homeostatic functions of small heat-shock proteins increase lifespan. Aging Cell 15, 217–226. ( 10.1111/acel.12422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morrow G, Heikkila JJ, Tanguay RM. 2006. Differences in the chaperone-like activities of the four main small heat shock proteins of Drosophila melanogaster. Cell Stress Chaperon 11, 51 ( 10.1379/CSC-166.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morrow G, Le Pécheur M, Tanguay RM. 2016. Drosophila melanogaster mitochondrial Hsp22: a role in resistance to oxidative stress, aging and the mitochondrial unfolding protein response. Biogerontology 17, 61–70. ( 10.1007/s10522-015-9591-y) [DOI] [PubMed] [Google Scholar]
- 96.Amin J, Mestril R, Voellmy R. 1991. Genes for Drosophila small heat shock proteins are regulated differently by ecdysterone. Mol. Cell. Biol. 11, 5937–5944. ( 10.1128/MCB.11.12.5937) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mason P, Hall L, Gausz J. 1984. The expression of heat shock genes during normal development in Drosophila melanogaster (heat shock/abundant transcripts/developmental regulation). Mol. Gen. Genet. 194, 73–78. ( 10.1007/BF00383500) [DOI] [Google Scholar]
- 98.Lepesant J, Levine M, Garen A, Lepesant-Kejzlarvoa J, Rat L, Somme-Martin G. 1982. Developmentally regulated gene expression in Drosophila larval fat bodies. J. Mol. Appl. Genet. 1, 371–383. ( 10.1016/b978-0-12-068350-5.50020-x) [DOI] [PubMed] [Google Scholar]
- 99.Chakraborty R, Li Y, Zhou L, Golic KG. 2015. Corp regulates P53 in Drosophila melanogaster via a negative feedback loop. PLoS Genet. 11, e1005400 ( 10.1371/journal.pgen.1005400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Drapeau MD. 2001. The family of yellow-related Drosophila melanogaster proteins. Biochem. Biophys. Res. Commun. 281, 611–613. ( 10.1006/bbrc.2001.4391) [DOI] [PubMed] [Google Scholar]
- 101.Shearer PW, West JD, Walton VM, Brown PH, Svetec N, Chiu JC. 2016. Seasonal cues induce phenotypic plasticity of Drosophila suzukii to enhance winter survival. BMC Ecol. 16, 11 ( 10.1186/s12898-016-0070-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing datasets supporting the results of this article are available via the NCBI Sequence Read Archive (accession number: PRJNA499074).