Abstract
Meiotic drivers are selfish genetic elements that bias their transmission into gametes, often to the detriment of the rest of the genome. The resulting intragenomic conflicts triggered by meiotic drive create evolutionary arms races and shape genome evolution. The phenomenon of meiotic drive is widespread across taxa but is particularly prominent in the Drosophila genus. Recent studies in Drosophila have provided insights into the genetic origins of drivers and their molecular mechanisms. Here, we review the current literature on mechanisms of drive with an emphasis on sperm killers in Drosophila species. In these systems, meiotic drivers often evolve from gene duplications and targets are generally linked to heterochromatin. While dense in repetitive elements and difficult to study using traditional genetic and genomic approaches, recent work in Drosophila has made progress on the heterochromatic compartment of the genome. Although we still understand little about precise drive mechanisms, studies of male drive systems are converging on common themes such as heterochromatin regulation, small RNA pathways, and nuclear transport pathways. Meiotic drive systems are therefore promising models for discovering fundamental features of gametogenesis.
Keywords: meiotic drive, Drosophila, genetic conflict, heterochromatin
1. Introduction
Nearly a century has passed since Gershenson [1] discovered the first documented case of meiotic drive in Drosophila obscura. While conducting genetic crosses from flies he had caught near Moscow, he noticed that some males sired only female or primarily female progeny. A careful analysis then allowed Gershenson to conclude that the responsible gene (hereafter ‘the driver’) was located on the X chromosome and caused ‘spermatozoa with the Y chromosome to participate in the fertilization process in but an insignificant number’. This phenomenon, called ‘sex-ratio distortion’ (hereafter SR), is widespread, particularly in Drosophila. In its broader definition, a meiotic driver is a selfish genetic element that biases its own transmission into the functional gametes of heterozygous individuals at the expense of the alternative allele. There are two ways to be over-represented: gamete killing in males (disruption of alternative sperm) and gonotaxis in females (preferential transmission into the ovule). Meiotic drive systems are sex-specific and most known meiotic drive systems in Drosophila act in males through sperm killing. During spermatogenesis, the driver prevents the formation of functional sperm that do not have a copy of itself. This efficient strategy is referred to as ‘interference’ because it is basically a game for (at least) two players: a driver and its target (reviewed in [2]).
The emergence of a gamete killer is by its very nature detrimental for the ‘fair’ genes that ensure their evolutionary success by being advantageous for their carriers. It sets the stage for an intragenomic conflict between, on one side, the driver and linked genomic regions and, on the other side, the rest of the genome. Drive enhancers promote transmission of the driver and drive suppressors restore fair segregation. This arms race triggers rapid evolution towards a new genomic balance.
Since Gershenson's seminal paper, meiotic drivers have been reported in a range of animals, plants and fungi [3]. However, with few exceptions, we lack even basic information about their identity and how they operate. There are multiple reasons for this. First, genetic mapping is often difficult, because for many known systems, the drive locus (or loci) lies within a chromosomal inversion or other non-recombining regions [4–6]. Second, as a consequence of intragenomic conflict, drive systems typically involve multiple genetic elements with epistatic interactions, making it difficult to identify individual components [7–9]. Finally, drive systems involve processes and genomic regions that are still poorly understood, such as heterochromatin.
Here, we review recent advances in our understanding of meiotic drive systems, with a focus on sperm killers in Drosophila. We discuss the origins of meiotic drivers and the role of gene duplication. We review recent developments on drive mechanisms and note emerging themes involving small RNA pathways, heterochromatin formation and nuclear transport. Finally, we discuss possible evolutionary consequences of meiotic drive.
2. Genetic origins of drivers
(a). Where do drivers emerge?
In males, a meiotic drive system consists of at least two loci: a driver and a target. In heterozygotes, the driver skews transmission rates in its favour by destroying target-bearing gametes in trans. This requires that the two loci are genetically linked, but on opposite homologous chromosomes to avoid creating a ‘suicide’ chromosome where the driver causes its own destruction. Therefore, drivers are typically linked to a resistant (insensitive) allele of the target.
Since tight linkage between these loci is critical for the success of drivers, these systems tend to arise in low- or non-recombining regions of the genome (e.g. sex chromosomes and heterochromatin), or acquire modifiers of recombination (e.g. chromosomal inversions) to create or reinforce linkage. Most known meiotic drive systems are X-linked (electronic supplementary material, table S1), where the lack of recombination between X and Y chromosomes provides fertile grounds for driver systems to emerge. Autosomal drivers, however, can occur in heterochromatic regions around the centromere and telomere, where there is little recombination. For example, the autosomal Segregation Distorter system of Drosophila melanogaster (SD) involves at least two loci: the driver (Segregation distorter, or Sd) is in the proximal region of chromosome 2L, and its target (Responder or Rsp) corresponds to a large block of tandem satellite repeats in the pericentric heterochromatin of chromosome 2R [8].
Most known meiotic drive systems are more complicated and involve additional modifiers of drive. Suppressors counteract the driver to restore Mendelian transmission ratios, and enhancers increase the strength of the driver. While suppressors can be unlinked from the driver and the target, enhancers must be linked to the driver (figure 1b). Chromosomal inversions are often associated with meiotic drive systems [4–6], as they may either suppress recombination between the drivers and their target [8] or reinforce the linkage between the drivers and enhancers [9].
Figure 1.
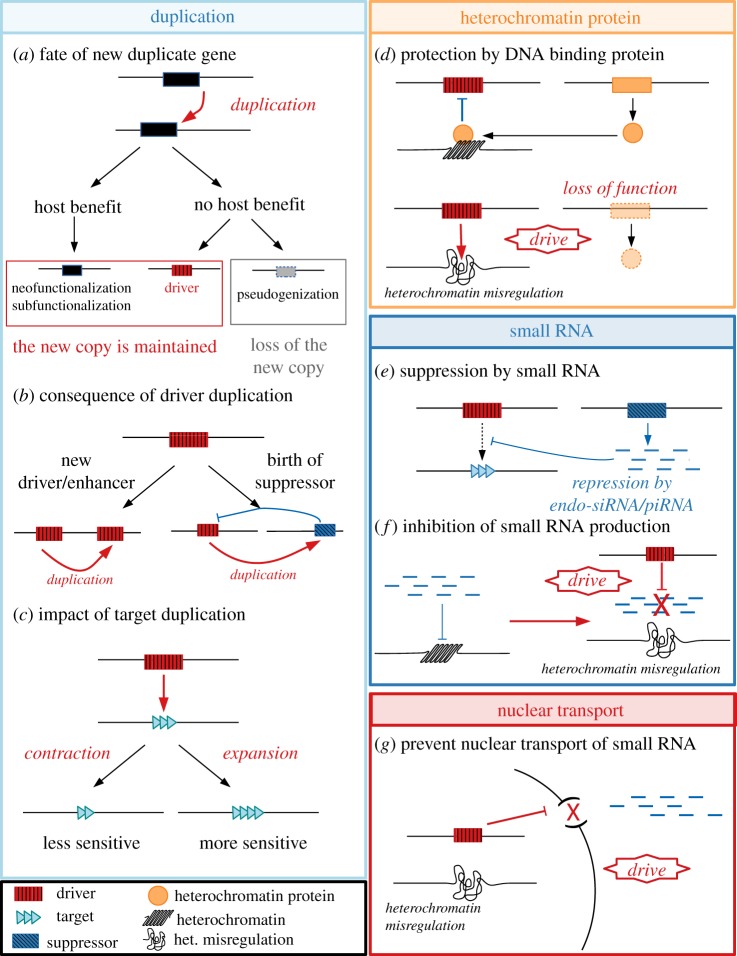
The schematics in the left panel show how duplication events can contribute to meiotic drive system evolution: (a) a duplicated gene can give rise to the emergence of a new driver; (b) driver duplication could give rise to a new driver/enhancer or to a suppressor element; (c) repetitive targets can expand or contract, which could affect their sensitivity to the driver. The schematics in the right panel show the different mechanisms involved in drive: (d) heterochromatin-binding proteins can protect the target from the driver. Small RNAs can be involved drive in multiple ways: (e) they can be suppressors of drive; (f) the misregulation of small RNAs can induce cause drive directly; or (g) disrupted nuclear transport can prevent the proper processing or targeting of small RNAs. (Online version in colour.)
The disparity in the number of known sex-linked and autosomal drivers (electronic supplementary material, table S1) is in part due to ascertainment bias: it is easier to detect distorted sex chromosome transmission because it manifests as a deviation from the Fisherian sex ratio (1 : 1) [10]. Distorted autosomal transmission can go unnoticed in the absence of a visible or scored molecular marker linked to the distorter. For example, the autosomal SD system in D. melanogaster was discovered because of the unexpected lack of recovery of visibly marked chromosomes in a genetic screen unrelated to meiotic drive [11]. It is easier to detect drivers that are currently segregating, as the effects of a driver are only apparent in heterozygotes. Fixed drivers will be cryptic unless they are introduced into a population or species naive to the driver.
(b). Molecular origins of drivers
The molecular identities of loci that cause meiotic drive are unknown in many systems. However, we find a pattern among the known systems in Drosophila (six partially known systems among the 19 identified, see electronic supplementary material, table S1): most of these drivers originate in gene duplication events. Meiotic drivers correspond to duplicated genes in the SD system of D. melanogaster [8], Winters SR system of Drosophila simulans [12] and a similar origin is suspected in an SR system in Drosophila neotestacea [13]. Newly duplicated genes are often deleterious due to a dosage imbalance, while their redundancy also allows them to evolve a new beneficial function (neofunctionalization). In the case of drive systems, some duplicated genes can become selfish and survive despite having a fitness cost (figure 1a). Many known suppressors of drive also originated from gene duplications [14,15] (figure 1b). Nearly all known meiotic drive systems in Drosophila involve gene duplication, with one exception in Drosophila pseudoobscura, where a meiotic drive locus (Overdrive) arose from a rapidly evolving single copy gene [16]. While gene duplication is a common theme among drive systems, the genetic basis of drive is often more complex, and involves epistatic interactions among multiple loci [7,17].
3. Cytological defects during spermatogenesis
There are two categories of drivers: those that induce meiotic failure and those whose defects are observed at post-meiotic stages (electronic supplementary material, table S1). The cytological phenotype of the Paris SR system in D. simulans is one of the best described. In SR males, the first cytological defects occur during meiosis as aberrant segregation of the Y chromosome during anaphase II. In some cases, the two Y chromosome sister chromatids migrate together to the same meiotic spindle pole. In other cases, they appear stretched between the poles, the two daughter nuclei remain connected by a chromosome bridge and the resulting spermatids fail to elongate [18]. Meiotic defects also occur in D. pseudoobscura and Drosophila athabasca SR males, where the Y chromosome ‘degenerates’ during anaphase II, appearing as a chromatin mass with no centromeric activity [19]. Finally, meiotic defects are inferred when SR males give rise to XO sons (lacking a Y chromosome and usually sterile) in Drosophila subobscura [20] and D. paramelanica [4].
In other Drosophila drive systems, the observed cellular defects are post-meiotic and occur as spermatids differentiate into mature sperm. In the D. simulans Winters SR system, half of the early spermatids (inferred to be Y-bearing sperm) display a nuclear condensation defect [12]. Defects during spermatid differentiation are also observed in D. affinis [21] and D. neotestacea [13]. In D. melanogaster males heterozygous for SD, about half of the post-elongation spermatids show defects in chromatin condensation [22] and the histone-to-protamine transition [23].
It is important to note that the stage at which cytological defects are observed can differ from when the driver acts. For example, while defects occur during meiosis II in the Paris SR system, the driver is only expressed in spermatogonia [17]. In D. melanogaster, although the first visible defects of SD occur post-meiosis, temperature sensitivity assays suggest that the driver acts early in meiosis [24]. However, the timing of SD may vary between SD chromosomes [25], and suppressors and drivers may differ in their timing [26,27]. Moreover, cellular phenotypes are often highly variable [18,21]. Many drive systems are influenced by factors like temperature [24–26,28,29] or age [30–33].
4. Molecular mechanisms of drive
(a). Heterochromatin-binding proteins
The first implication of a heterochromatin protein in meiotic drive was in the D. simulans Paris SR system where the distortion results from an interaction between two X-linked drive loci. The first drive locus maps to a 37 kb tandem duplication containing six genes; however, the molecular identity of the causal element within this duplication is still unknown [7,34]. The second driver corresponds to HP1D2 [17], a young and rapidly evolving member of the HP1 (heterochromatin protein 1) gene family that is involved in heterochromatin organization. HP1D2 originated from a duplication of HP1D/rhino 15–22 Mya ago in the Sophophora subgenus [35]. HP1D2 is expressed in premeiotic male germ cells and specifically binds the Y chromosome, which is entirely heterochromatic in D. simulans. The Paris SR system requires both the tandem duplication and a dysfunctional allele of HP1D2 to drive. This suggests that HP1D2 prepares the Y chromosome for meiosis (figure 1d). Heterochromatin-binding proteins, including rhino, are essential for the regulation of some piRNA clusters in D. melanogaster ovaries [36]. Rhino has strongly female-biased expression, primarily in the germline [37]. PiRNA regulation is not well understood in the male germline; however, it is possible that HP1D2 plays a role. The other drive locus in the Paris SR system (the duplication) contains part of Trf2, a core transcription initiation factor that interacts with rhino to facilitate piRNA cluster expression in D. melanogaster [7,38]. Both the deletion of HP1D2 and duplication of Trf2 support the hypothesis that the piRNA pathway is involved in the Paris SR system (figure 1f).
(b). Small RNA pathways
The role of small RNAs in drive is well studied in the D. simulans Winters SR system, where the duplication of the X-linked driver called Dox gave rise to an autosomal suppressor called Nmy [12]. Nmy makes a hairpin RNA (hpRNA) that is processed into endogenous short-interfering RNAs (endo-siRNAs) that target the driver [14]. Similarly, another autosomal suppressor of the D. simulans Durham SR system, Tmy, encodes an hpRNA targeting the Winters X-linked driver [14], which suggests that these two D. simulans SR systems may not be independent. The Stellate system is suspected to be a cryptic drive system in D. melanogaster [39] (but see [40]). Stellate (Ste) is an X-linked multicopy gene whose product, if left unsuppressed, leads to the accumulation of crystal-like structures in testes premeiotic germ cells [41]. Suppressor of Stellate (Su(Ste)) is a multicopy Y-linked gene family paralogous to Ste. Su(Ste) loci generate small Piwi-interacting RNAs (piRNAs) that suppress Ste [42]. Ste and Su(Ste) independently amplified to high copy number on the X and Y chromosomes, respectively [43]. Deletions of the Y-linked Su(Ste) region cause low fertility or sterility in males, depending on the X-linked Ste copy number, and also have skewed sex ratios towards female offspring [44]. However, it is unclear whether the deletion of the Su(Ste) region itself or the overexpression of Ste causes the meiotic drive [40]. In both the Winters and Stellate systems, small RNAs generated from duplications of the driver suppress drive, but through different silencing pathways (piRNA and endo-siRNA, respectively, figure 1e). Interestingly, mutations in piRNA pathway genes enhance drive in the SD system of D. melanogaster [45], suggesting that piRNAs may also be involved in this drive system.
(c). Nuclear transport
One of the best-studied autosomal drive systems, SD in D. melanogaster, may involve a nuclear transport defect. The driver is a truncated duplication of Ran GTPase Activating Protein or RanGAP (the driver is referred to as Sd-RanGAP). Sd-RanGAP is missing a nuclear export signal (NES) site and a SUMO domain [46] and thus is mislocalized to the nucleus [47]. Normally, the cytoplasmic localization of RanGAP and the nuclear localization of its corresponding guanine nucleotide exchange factor RanGEF (or RCC1 in Drosophila) are critical for establishing the Ran gradient. The Ran gradient controls many important cellular processes from nuclear transport of proteins and some RNAs, to nuclear envelope assembly, and heterochromatin formation [48]. Interestingly, both the overexpression of wild-type RanGAP [49] and multiple copies of an unknown enhancer can cause RanGAP mislocalization and drive even in the absence of the Sd-RanGAP duplication [49,50]. The catalytic activity of Sd-RanGAP is required for drive, and the drive phenotype is rescued by overexpressing RanGEF or Ran [47], suggesting that drive is a result of disrupting the Ran gradient, perhaps due to defects in nuclear transport. Nuclear transport is also implicated in D. neotestacea, where an X-linked duplicated copy of importin-α is overexpressed in SR males and is a promising candidate for the driver [13].
As nuclear transport is important for both small RNA pathways and the histone-to-protamine transition in spermatogenesis, perturbations could lead to the chromatin condensation defects found in several drive systems (figure 4a). In the case of SD, the target is a pericentromeric satellite repeat, which produces small RNAs [45]; however, it is still unclear how Sd-RanGAP interacts with the satellite repeat. It is possible that the production of small RNAs coming from the target is disrupted (figure 1f). Alternatively, dysfunctional nuclear transport may interfere with heterochromatin regulation by preventing some critical RNAs or proteins from interacting with their target (figure 1g). These possibilities are not mutually exclusive.
5. Consequences of drive
(a). Deleterious effects of drivers
While meiotic drivers gain an evolutionary advantage by biasing their own transmission rate, they are inherently deleterious to the host genome. Because of the loss of Y-bearing sperm, driver males produce about half the functional sperm of non-driver males [51–54]. The impact on male fertility can be proportionally greater than sperm loss, especially for species with multiple mating and sperm competition [53,55–57]. Another consequence of SR drive is that abnormal segregation of the Y chromosome can lead to the production of XO males, which are sterile. This occurs in D. simulans [18], D. pseudoobscura [58], D. paramelanica [4], D. testacea [59] and D. neotestacea [60]. There is one known exception in D. affinis, where XO males are fertile and produce only sons [21,61] in the presence of SR, suggesting that the production of XO males in this species is associated with a resistance mechanism. Besides fertility costs, the fixation of SR meiotic drivers may result in extreme scarcity or complete loss of one sex and therefore the extinction of the population [10].
(b). Arms races via rapid evolution or gene duplication
In most cases, suppressors that counteract drivers should be favoured by natural selection, as they restore host fitness and/or re-balance skewed segregation [10]. However, drivers that overcome suppression will quickly spread in the population and the reduction in host fitness will subsequently lead to selection for mutations that re-establishes suppression, and so on.
Arms races are expected to occur between loci with antagonistic relationships. The clearest case is in the co-amplification of a pair of antagonistic mouse genes [62,63]: the X-linked multicopy genes (Slx and Slxl1) and the Y-linked multicopy gene (Sly). SLX/SLXL1 are activators of sex chromosome-encoded genes while SLY is a repressor [64]. Interestingly, knockdown of SLX/SLXL1 in the testes causes a male-biased sex ratio, and reciprocally, knockdown of SLY causes a female-biased sex ratio [64]. These genes may have co-amplified as a result of a conflict over sex chromosome transmission. Increases in Slx/Slxl1 copy number lead to increases in expression that could overcome Sly-induced repression, thus favouring X chromosome transmission. Increases in Sly copy number would either restore sex ratio or distort transmission in favour of the Y chromosome. The sex chromosomes of primates, cats and pigs also have multicopy genes [65–68], which could have resulted from similar conflicts. Similar systems exist in Drosophila species such as the Ste and Su(Ste) loci in D. melanogaster [39,69] (figure 1b). On the neo-sex chromosomes of D. miranda [70], some neo-Y genes are more highly amplified, resulting in a neo-Y chromosome nearly twice the size of their homologous neo-X chromosome. Neo-sex chromosomes are generally more gene-rich than ancient sex chromosomes, and therefore may be hot spots for drivers and their suppressors.
Recurrent positive selection on drivers and suppressors are likely consequences of arms races. In D. melanogaster, both longitudinal and population genetic data suggest that SD chromosomes turnover rapidly, with new variants displacing old ones over short time periods [71–73]. Different SD haplotypes in European [73] and African populations [72,73] experienced independent selective sweeps. The target satellite repeat (Rsp) also evolves rapidly—the organization of the repeat suggests that they originated before the divergence between D. melanogaster and the D. simulans clade species; however, the target of drive is missing in the D. simulans clade [74]. In the D. simulans Winters SR system, both the driver Dox and its suppressor Nmy show signatures of recurrent selection consistent with the model of repeated rounds of drive and suppression of drive [75].
Unlike the SD system, the targets of SR drive systems remain uncharacterized because a high repeat content makes the Y chromosome genetically intractable. Nevertheless, Y-linked repeats such as satellite DNA, turnover rapidly [76]. Being a target of drive may facilitate the rapid evolution of satellite DNA. Rsp copy number in D. melanogaster's SD system correlates with drive sensitivity, where large copy number alleles are more sensitive to drive [77]. In addition, resistant Y chromosomes are known in multiple SR drive systems [4,21,78,79]. These observations are consistent with a model where repeats are in a continuous struggle to escape or counteract drive (figure 1c).
Intragenomic conflicts can also cause the rapid evolution of some gene families. The HP1 gene family is rapidly evolving, with recurrent gene gains and losses, mostly with testis-restricted expression [35,80]. One of the clearest cases is HP1D2, part of the Paris SR system [17]. We suspect that the initial function of HP1D2 was to regulate Y-linked chromatin (figure 1d). Outside of the Paris SR system, conflict is implicated in the duplication of other HP1 proteins. Oxpecker also arose from a duplication of HP1D/rhino, and is involved in transposable element (TE) silencing [80]. Another duplicated gene, Umbrea, is an HP1B paralogue [35] that became essential through neofunctionalization [81,82]. Umbrea localizes to the centromere and is involved in chromosome segregation, but in a species-specific manner, suggesting that there is coevolution between centromeric DNA satellites and duplicated genes [83]. These heterochromatin proteins share a common structure: a chromo domain, which mediates interactions with histones and a chromoshadow domain involved in protein–protein interactions. Heterochromatin proteins are central players in the centromere drive hypothesis proposed by Malik & Henikoff [84]. This model proposes that centromeres have the capacity to be selfish and bias their transmission through the female germline. Heterochromatin-binding proteins may evolve rapidly to mitigate this bias and restore parity in female meiosis [85,86] (see box 1).
Box 1. Female drive systems.
The asymmetry in female meiosis presents an opportunity for conflicts, as only one of the four meiotic products becomes the egg. Centromeres can be selfish and gain a transmission advantage during female meiosis, biasing their segregation to the egg side of the spindle pole. There are parallels between male and female drive systems—they both seem to involve gene duplications, heterochromatin and heterochromatin-binding proteins. Female drive in plants and animals fit into two categories:
-
(1)
True centromere drive where centromeres compete based on ‘strength’ [85,87]: differences in satellite repeats in and near the centromeres can take advantage of a spindle asymmetry in female meiosis [88] to achieve biased segregation [86,87,89].
-
(2)
Driving mutations outside the native centromeres: one of the best-studied cases of female drive is of large heterochromatic ‘knobs’ consisting of tandem repeats on maize chromosomes. On abnormal chromosome 10, a non-recombining region at the distal tip of the chromosome contains a locus that converts its knobs into meiotic drivers that cheat by moving towards the spindle pole faster than their homologues [90] resulting in their biased segregation [91,92]. The causal locus is a multicopy gene called Kinesin driver (Kindr) that localizes to the knobs and facilitates their accelerated movement [93].
Genetic conflicts induced by meiotic drivers can have strong impacts on genome evolution. These conflicts can drive the evolution of gene families [94], and leave signatures of selection that range from a genic to a chromosome scale. Examining these patterns as potential consequences of genetic conflict may lead to the identification of new cryptic drive systems.
(c). Hybrid incompatibility
The meiotic drive theory of hybrid incompatibility [95] posits that the observed bias of hybrid disruption towards the heterogametic sex, known as Haldane's rule, may result from the expression of divergent cryptic meiotic drivers that become unsuppressed in the hybrids. The arms race between drivers and suppressors may fuel the divergence between species and contribute to hybrid male sterility. This idea is the subject of active debate and has long suffered from a lack of empirical data (summarized in [96]).
The first direct link between incompatibilities and drive came from D. pseudoobscura pseudoobscura and D. pseudoobscura bogotana, two subspecies that diverged 155–230 Kya [97]. Hybrid males between D. p. pseudoobscura males and D. p. bogotana females are nearly sterile and produce progeny that are almost all daughters [98]. Both phenotypes are caused by a single X-linked testis expressed gene, Overdrive, which encodes a polypeptide with an MADF DNA-binding domain [16]. Interestingly, another MADF domain-containing protein (HMR, for hybrid male rescue) is involved in a different hybrid incompatibility between D. melanogaster and sibling species [99]. In D. p. bogotana, Overdrive has autosomal suppressors not present in D. p. pseudoobscura, leading to de-repression of the driver in hybrids [98]. The QTL mapping of loci involved in sterility and segregation distortion revealed that both phenotypes share similar genetic basis [100]. Similarly, overlapping QTLs for hybrid incompatibility and meiotic drive were found between D. albomicans and D. nasuta [15] and D. persimilis and D. pseudoobscura [101].
In D. subobscura, the frequency of the SR chromosome is around 15–20% in a North African populations, but they are almost absent in a South European population [102,103]. The variation in SR frequency could be due to hybrid incompatibility [103]. This hypothesis was recently supported by Verspoor et al. [104] in an independent genetic background, suggesting that hybrid male incompatibility prevents the spread of SR between the North African population and the European population.
Cryptic drive systems are well studied in the D. simulans clade in the context of hybrid incompatibilities. Introgression of the D. mauritiana Tmy locus, an autosomal suppressor of the D. simulans Durham SR system, into a D. simulans background causes both meiotic drive and male sterility when homozygous [105]. Meiklejohn et al. [106] identified an unknown cryptic X-linked driver in D. mauritiana that induces SR distortion in a D. simulans genetic background. However, they also detected a recent introgression of D. simulans cryptic drivers, Dox and MDox into D. mauritiana, causing a local reduction in interspecific divergence, which impedes the evolution of hybrid sterility [106]. This case presents an alternative outcome of meiotic drive in the presence of gene flow for complex speciation events.
6. Conclusion: emerging themes in meiotic drive mechanisms
When we compare the key players and phenotypes involved in male meiotic drive systems across Drosophila, we see common themes emerge, suggesting that drivers may target similar processes in spermatogenesis. Known drive phenotypes appear to be associated with heterochromatin formation, either by targeting heterochromatic sequences directly (e.g. SD in D. melanogaster) or by involving heterochromatin-binding proteins (e.g. Paris SR of D. simulans). The three drive mechanisms that we reviewed here—heterochromatin regulation, small RNA pathways and nuclear transport—are inter-dependent [38,107]. These complicated interactions make it difficult to disentangle the causes and consequences of meiotic drive. Considering how these pathways are connected may provide more insight into how meiotic drivers may have shaped the evolution of conserved germline features and pathways.
Germlines have defence mechanisms against selfish genetic elements. Most widely known are the RNA interference pathways that use small RNAs (e.g. endo-siRNAs and piRNAs) to target TEs. These small RNA pathways are well studied in the D. melanogaster female germline [108] but poorly understood in males, where many small RNAs are not TE-derived. Cryptic drive systems in Drosophila species involve suppressors that generate small RNAs, for example, the hpRNAs that generate endo-siRNAs in D. simulans [14,29] and the piRNAs that suppress a cryptic X-linked driver in D. melanogaster [108]. It is possible that one common function of small RNA pathways in the male germline is to suppress meiotic drivers.
Small RNAs need to reach their target genetic elements for silencing, and proteins that make up or interact with the nuclear pore are important for piRNA production [109–111]. Nuclear pore complexes are the gatekeepers controlling what enters and exits the nucleus [112]. Processes that mediate the transport across the nuclear pore can end up at the centre of conflicts between, for example, pathogens (reviewed in [113]), TEs (e.g. [114]) and meiotic drivers [115,116]. These conflicts may trigger the rapid evolution of nuclear pore complex proteins [116] that can cause hybrid incompatibilities between closely related species [117]. In this way, meiotic drive may have consequences for species divergence and speciation.
The evidence from across Drosophila drive systems connecting heterochromatin regulation, small RNAs and nuclear transport pathways suggests that selfish genetic elements may expose common vulnerabilities in spermatogenesis. While their mechanisms differ, male and female drive systems have interesting parallels. For example, gene duplication and heterochromatin-binding proteins also feature in plant female drive systems (see box 1). Studying molecular mechanisms of drive in both males and females may lend insights into some basic features of gametogenesis.
This review is focused on the Drosophila genus, as most known male drive systems occur in fruit flies. However, this enrichment is likely to be, at least in part, due to ascertainment bias because of the rich history of Drosophila genetics research in the past century. Meiotic drivers have been discovered in a wide range of organisms, suggesting that they may be pervasive, but cryptic, or go undetected without any obvious phenotypic effect. Drosophila are powerful models for studying drive systems and their mechanisms. Advances in long-read sequencing and genome editing technologies provide opportunities to extend these studies to new systems [94,118].
Supplementary Material
Acknowledgements
We thank Anna Lindholm, Nina Wedell and Tom Price for the invitation to contribute to this special issue and the two anonymous reviewers for helpful comments on the manuscript.
Data accessibility
This article has no additional data.
Competing interests
We declare that we have no competing interests.
Funding
This work was supported by the National Institutes of Health (grant no. NIH R35-GM119515) and National Science Foundation (grant no. NSF MCB 1844693) to A.M.L. A.M.L. is supported by a Stephen Biggar and Elisabeth Asaro fellowship in Data Science. C.M.-M. was supported by the Centre Nationale de la Recherche Scientifique (UMR 9191). C.C. is funded by a French ministerial scholarship.
References
- 1.Gershenson S. 1928. A new sex-ratio abnormality in Drosophila obscura. Genetics 13, 488–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burt A, Trivers R. 2009. Genes in conflict: the biology of selfish genetic elements. Cambridge, MA: Harvard University Press. [Google Scholar]
- 3.Lindholm AK, et al. 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evol. 31, 315–326. ( 10.1016/j.tree.2016.02.001) [DOI] [PubMed] [Google Scholar]
- 4.Stalker HD. 1961. The genetic systems modifying meiotic drive in Drosophila paramelanica . Genetics 46, 177–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobzhansky T, Sturtevant AH. 1938. Inversions in the chromosomes of Drosophila pseudoobscura . Genetics 23, 28–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pieper KE, Dyer KA. 2016. Occasional recombination of a selfish X-chromosome may permit its persistence at high frequencies in the wild. J. Evol. Biol. 29, 2229–2241. ( 10.1111/jeb.12948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montchamp-Moreau C, Ogereau D, Chaminade N, Colard A, Aulard S. 2006. Organization of the sex-ratio meiotic drive region in Drosophila simulans. Genetics 174, 1365–1371. ( 10.1534/genetics.105.051755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larracuente AM, Presgraves DC. 2012. The selfish Segregation Distorter gene complex of Drosophila melanogaster . Genetics 192, 33–53. ( 10.1534/genetics.112.141390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu CI, Beckenbach AT. 1983. Evidence for extensive genetic differentiation between the sex-ratio and the standard arrangement of D. pseudoobscura and D. persimilis and identification of hybrid sterility factors. Genetics 105, 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton WD. 1967. Extraordinary sex ratios. Science 156, 477–488. ( 10.1126/science.156.3774.477) [DOI] [PubMed] [Google Scholar]
- 11.Sandler L, Hiraizumi Y, Sandler I. 1959. Meiotic drive in natural populations of Drosophila melanogaster. I. The cytogenetic basis of segregation-distortion. Genetics 44, 233–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao Y, Araripe L, Kingan SB, Ke Y, Xiao H, Hartl DL. 2007. A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. PLoS Biol. 5, e293 ( 10.1371/journal.pbio.0050293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pieper KE, Unckless RL, Dyer KA. 2018. A fast-evolving X-linked duplicate of importin-α2 is overexpressed in sex-ratio drive in Drosophila neotestacea. Mol. Ecol. 27, 5165–5179. ( 10.1111/mec.14928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin CJ, Hu F, Dubruille R, Vedanayagam J, Wen J, Smibert P, Loppin B, Lai EC. 2018. The hpRNA/RNAi pathway is essential to resolve intragenomic conflict in the Drosophila male germline. Dev. Cell 46, 316–326. ( 10.1016/j.devcel.2018.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Sun T, Woldesellassie F, Xiao H, Tao Y. 2015. Sex ratio meiotic drive as a plausible evolutionary mechanism for hybrid male sterility. PLoS Genet. 11, e1005073 ( 10.1371/journal.pgen.1005073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phadnis N, Orr HA. 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323, 376–379. ( 10.1126/science.1163934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helleu Q, Gerard PR, Dubruille R, Ogereau D, Prud'homme B, Loppin B, Montchamp-Moreau C. 2016. Rapid evolution of a Y-chromosome heterochromatin protein underlies sex chromosome meiotic drive. Proc. Natl Acad. Sci. USA 113, 4110–4115. ( 10.1073/pnas.1519332113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cazemajor M, Joly D, Montchamp-Moreau C. 2000. Sex-ratio meiotic drive in Drosophila simulans is related to equational nondisjunction of the Y chromosome. Genetics 154, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novitski E, Peacock WJ, Engel J. 1965. Cytological basis of ‘Sex Ratio’ in Drosophila pseudoobscura. Science 148, 516–517. ( 10.1126/science.148.3669.516) [DOI] [PubMed] [Google Scholar]
- 20.Hauschteck-Jungen E, Jungen H, Muller M. 1972. Karyotype and meiosis in wild and sex-ratio Drosophila melanogaster males. Rev. Suisse Zool. 79, 297–305. [PubMed] [Google Scholar]
- 21.Unckless RL, Larracuente AM, Clark AG. 2015. Sex-ratio meiotic drive and Y-linked resistance in Drosophila affinis . Genetics 199, 831–840. ( 10.1534/genetics.114.173948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokuyasu KT, Peacock WJ, Hardy RW. 1977. Dynamics of spermiogenesis in Drosophila melanogaster: VII. Effects of Segregation Distorter (SD) chromosome. J. Ultrastruct. Res. 58, 96–107. ( 10.1016/S0022-5320(77)80011-7) [DOI] [PubMed] [Google Scholar]
- 23.Hauschteck-Jungen E, Hartl DL. 1982. Defective histone transition during spermiogenesis in heterozygous segregation distorter males of Drosophila melanogaster. Genetics 101, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mange EJ. 1968. Temperature sensitivity of segregation-distortion in Drosophila melanogaster . Genetics 58, 399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews K, Mortin MA. 1983. SD-72 has a temperature-sensitive period during spermiogenesis. Can. J. Genet. Cytol. 25, 662–667. ( 10.1139/g83-097) [DOI] [Google Scholar]
- 26.Hihara YK. 1971. Genetic analysis of modifying system of segregation distortion in Drosophila melanogaster II. Two modifiers for SD system on the system on the second chromosome of D. melanogaster. Jpn. J. Genet. 49, 209–222. ( 10.1266/jjg.49.209) [DOI] [Google Scholar]
- 27.Hiraizumi Y. 1993. Temperature sensitivity of negative segregation distortion in Drosophila melanogaster . Genetics 135, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denell RE, Judd BH, Richardson RH. 1969. Distorted sex ratios due to segregation distorter in Drosophila melanogaster . Genetics 61, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao Y, Masly JP, Araripe L, Ke Y, Hartl DL. 2007. A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. PLoS Biol. 5, e292 ( 10.1371/journal.pbio.0050292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiraizumi Y, Watanabe SS. 1969. Aging effect on the phenomenon of segregation distortion in Drosophila melanogaster . Genetics 63, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandler L, Hiraizumi Y. 1961. Meiotic drive in natural populations of Drosophila melanogaster. VIII. A heritable aging effect on the phenomenon of segregation-distortion. Can. J. Genet. Cytol. 3, 34–36. ( 10.1139/g61-009) [DOI] [Google Scholar]
- 32.Montchamp-Moreau C, Cazemajor M. 2002. Sex-ratio drive in Drosophila simulans: variation in segregation ratio of X chromosomes from a natural population. Genetics 162, 1221–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Carvalho AB, Klaczko LB.. 1992. Age and sex-ratio expression in Drosophila mediopunctata. Genetica 87, 107–111. ( 10.1007/BF00121000) [DOI] [PubMed] [Google Scholar]
- 34.Fouvry L, Ogereau D, Berger A, Gavory F, Montchamp-Moreau C. 2011. Sequence analysis of the segmental duplication responsible for Paris sex-ratio drive in Drosophila simulans . G3 (Bethesda) 1, 401–410. ( 10.1534/g3.111.000315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine MT, McCoy C, Vermaak D, Lee Y.CG, Hiatt MA, Matsen FA, Malik HS. 2012. Phylogenomic analysis reveals dynamic evolutionary history of the Drosophila heterochromatin protein 1 (HP1) gene family. PLoS Genet. 8, e1002729 ( 10.1371/journal.pgen.1002729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klattenhoff C, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138, 1137–1149. ( 10.1016/j.cell.2009.07.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volpe AM, Horowitz H, Grafer CM, Jackson SM, Berg CA. 2001. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics 159, 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersen PR, Tirian L, Vunjak M, Brennecke J. 2017. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 549, 54–59. ( 10.1038/nature23482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurst LD. 1996. Further evidence consistent with Stellate's involvement in meiotic drive. Genetics 142, 641–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belloni M, Tritto P, Bozzetti MP, Palumbo G, Robbins LG. 2002. Does Stellate cause meiotic drive in Drosophila melanogaster? Genetics 161, 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bozzetti MP, et al. 1995. The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the beta subunit of casein kinase 2. Proc. Natl Acad. Sci. USA 92, 6067–6071. ( 10.1073/pnas.92.13.6067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. 2004. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 24, 6742–6750. ( 10.1128/MCB.24.15.6742-6750.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kogan GL, Epstein VN, Aravin AA, Gvozdev VA. 2000. Molecular evolution of two paralogous tandemly repeated heterochromatic gene clusters linked to the X and Y chromosomes of Drosophila melanogaster . Mol. Biol. Evol. 17, 697–702. ( 10.1093/oxfordjournals.molbev.a026348) [DOI] [PubMed] [Google Scholar]
- 44.Palumbo G, Bonaccorsi S, Robbins LG, Pimpinelli S. 1994. Genetic analysis of Stellate elements of Drosophila melanogaster . Genetics 138, 1181–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gell SL, Reenan RA. 2013. Mutations to the piRNA pathway component aubergine enhance meiotic drive of segregation distorter in Drosophila melanogaster . Genetics 193, 771–784. ( 10.1534/genetics.112.147561) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merrill C, Bayraktaroglu L, Kusano A, Ganetzky B. 1999. Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science 283, 1742–1745. ( 10.1126/science.283.5408.1742) [DOI] [PubMed] [Google Scholar]
- 47.Kusano A, Staber C, Ganetzky B. 2001. Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Dev. Cell 1, 351–361. ( 10.1016/S1534-5807(01)00042-9) [DOI] [PubMed] [Google Scholar]
- 48.Quimby BB, Dasso M. 2003. The small GTPase Ran: interpreting the signs. Curr. Opin Cell Biol. 15, 338–344. ( 10.1016/S0955-0674(03)00046-2) [DOI] [PubMed] [Google Scholar]
- 49.Kusano A, Staber C, Ganetzky B. 2002. Segregation distortion induced by wild-type RanGAP in Drosophila. Proc. Natl Acad. Sci. USA 99, 6866–6870. ( 10.1073/pnas.102165099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Temin RG. 1991. The independent distorting ability of the Enhancer of Segregation Distortion, E(SD), in Drosophila melanogaster . Genetics 128, 339–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Policansky D, Ellison J. 1970. ‘Sex ratio’ in Drosophila pseudoobscura: spermiogenic failure. Science 169, 888–889. ( 10.1126/science.169.3948.888) [DOI] [PubMed] [Google Scholar]
- 52.Hauschteck-Jungen E, Maurer B. 1976. Sperm dysfunction in sex ratio males of Drosophila subobscura. Genetica 46, 459 ( 10.1007/BF00128092) [DOI] [Google Scholar]
- 53.Angelard C, Montchamp-Moreau C, Joly D. 2008. Female-driven mechanisms, ejaculate size and quality contribute to the lower fertility of sex-ratio distorter males in Drosophila simulans. BMC Evol. Biol. 8, 326 ( 10.1186/1471-2148-8-326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hartl DL, Hiraizumi Y, Crow JF. 1967. Evidence for sperm dysfunction as the mechanism of segregation distortion in Drosophila melanogaster . Proc. Natl Acad. Sci. USA 58, 2240–2245. ( 10.1073/pnas.58.6.2240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price TA, Bretman AJ, Avent TD, Snook RR, Hurst GD, Wedell N. 2008. Sex ratio distorter reduces sperm competitive ability in an insect. Evolution 62, 1644–1652. ( 10.1111/j.1558-5646.2008.00386.x) [DOI] [PubMed] [Google Scholar]
- 56.Atlan A, Joly D, Capillon C, Montchamp-Moreau C. 2004. Sex-ratio distorter of Drosophila simulans reduces male productivity and sperm competition ability. J. Evol. Biol. 17, 744–751. ( 10.1111/j.1420-9101.2004.00737.x) [DOI] [PubMed] [Google Scholar]
- 57.Sutter A, Lindholm AK. 2016. Meiotic drive changes sperm precedence patterns in house mice: potential for male alternative mating tactics? BMC Evol. Biol. 16, 133 ( 10.1186/s12862-016-0710-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henahan J, Cobbs G. 1983. Origin of X/O progeny from crosses of sex-ratio trait males of Drosophila pseudoobscura . J. Hered. 74, 145–148. ( 10.1093/oxfordjournals.jhered.a109752) [DOI] [PubMed] [Google Scholar]
- 59.Keais GL, Hanson MA, Gowen BE, Perlman SJ. 2017. X chromosome drive in a widespread Palearctic woodland fly, Drosophila testacea. J. Evol. Biol. 30, 1185–1194. ( 10.1111/jeb.13089) [DOI] [PubMed] [Google Scholar]
- 60.James AC, Jaenike J. 1990. ‘Sex ratio’ meiotic drive in Drosophila testacea. Genetics 126, 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voelker RA, Kojima KI. 1972. Fertility and fitness of XO males in Drosophila. II. Quantitative analysis. Evolution 26, 560–573. ( 10.1111/j.1558-5646.1972.tb01964.x) [DOI] [PubMed] [Google Scholar]
- 62.Cocquet J, Ellis PJ, Yamauchi Y, Mahadevaiah SK, Affara NA, Ward MA, Burgoyne PS. 2009. The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 7, e1000244 ( 10.1371/journal.pbio.1000244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cocquet J, et al. 2010. Deficiency in the multicopy Sycp3-like X-linked genes Slx and Slxl1 causes major defects in spermatid differentiation. Mol. Biol. Cell 21, 3497–3505. ( 10.1091/mbc.E10-07-0601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cocquet J, Ellis PJ, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS. 2012. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 8, e1002900 ( 10.1371/journal.pgen.1002900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lucotte EA, Skov L, Jensen JM, Macia MC, Munch K, Schierup MH. 2018. Dynamic copy number evolution of X- and Y-linked ampliconic genes in human populations. Genetics 209, 907–920. ( 10.1534/genetics.118.300826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brashear WA, Raudsepp T, Murphy WJ. 2018. Evolutionary conservation of Y chromosome ampliconic gene families despite extensive structural variation. Genome Res. 28, 1841–1851. ( 10.1101/gr.237586.118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skinner BM, et al. 2016. The pig X and Y chromosomes: structure, sequence, and evolution. Genome Res. 26, 130–139. ( 10.1101/gr.188839.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nam K, Munch K, Hobolth A, Dutheil JY, Veeramah KR, Woerner AE, Hammer MF, Mailund T, Schierup MH. 2015. Extreme selective sweeps independently targeted the X chromosomes of the great apes. Proc. Natl Acad. Sci. USA 112, 6413–6418. ( 10.1073/pnas.1419306112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellison C, Bachtrog D. 2019. Recurrent gene co-amplification on Drosophila X and Y chromosomes. PLoS Genet. 15, e1008251 ( 10.1371/journal.pgen.1008251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mahajan S, Wei KH, Nalley MJ, Gibilisco L, Bachtrog D. 2018. De novo assembly of a young Drosophila Y chromosome using single-molecule sequencing and chromatin conformation capture. PLoS Biol. 16, e2006348 ( 10.1371/journal.pbio.2006348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Temin RG, Marthas M. 1984. Factors influencing the effect of segregation distortion in natural populations of Drosophila melanogaster . Genetics 107, 375–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Presgraves DC, Gerard PR, Cherukuri A, Lyttle TW. 2009. Large-scale selective sweep among segregation distorter chromosomes in African populations of Drosophila melanogaster . PLoS Genet. 5, e1000463 ( 10.1371/journal.pgen.1000463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brand CL, Larracuente AM, Presgraves DC. 2015. Origin, evolution, and population genetics of the selfish Segregation Distorter gene duplication in European and African populations of Drosophila melanogaster. Evolution 69, 1271–1283. ( 10.1111/evo.12658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larracuente AM. 2014. The organization and evolution of the Responder satellite in species of the Drosophila melanogaster group: dynamic evolution of a target of meiotic drive. BMC Evol. Biol. 14, 233 ( 10.1186/s12862-014-0233-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kingan SB, Garrigan D, Hartl DL. 2010. Recurrent selection on the Winters sex-ratio genes in Drosophila simulans . Genetics 184, 253–265. ( 10.1534/genetics.109.109587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei KH, Lower SE, Caldas IV, Sless TJS, Barbash DA, Clark AG. 2018. Variable rates of simple satellite gains across the Drosophila phylogeny. Mol. Biol. Evol. 35, 925–941. ( 10.1093/molbev/msy005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu C-I, Lyttle TW, Wu M-L, Lin G-F. 1988. Association between a satellite DNA sequence and the responder of segregation distorter in D. melanogaster . Cell 54, 179–189. ( 10.1016/0092-8674(88)90550-8) [DOI] [PubMed] [Google Scholar]
- 78.Montchamp-Moreau C, Ginhoux V, Atlan A. 2001. The Y chromosomes of Drosophila simulans are highly polymorphic for their ability to suppress sex-ratio drive. Evolution 55, 728–737. ( 10.1554/0014-3820(2001)055[0728:TYCODS]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 79.Branco AT, Tao Y, Hartl DL, Lemos B. 2013. Natural variation of the Y chromosome suppresses sex ratio distortion and modulates testis-specific gene expression in Drosophila simulans . Heredity (Edinb) 111, 8–15. ( 10.1038/hdy.2013.5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levine MT, Vander Wende HM, Hsieh E, Baker EP, Malik HS. 2016. Recurrent gene duplication diversifies genome defense repertoire in Drosophila. Mol. Biol. Evol. 33, 1641–1653. ( 10.1093/molbev/msw053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen S, Zhang YE, Long M. 2010. New genes in Drosophila quickly become essential. Science 330, 1682–1685. ( 10.1126/science.1196380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Joppich C, Scholz S, Korge G, Schwendemann A. 2009. Umbrea, a chromo shadow domain protein in Drosophila melanogaster heterochromatin, interacts with Hip, HP1 and HOAP. Chromosome Res. 17, 19–36. ( 10.1007/s10577-008-9002-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ross RJ, Weiner MM, Lin H. 2014. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 505, 353–359. ( 10.1038/nature12987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malik HS, Henikoff S. 2009. Major evolutionary transitions in centromere complexity. Cell 138, 1067–1082. ( 10.1016/j.cell.2009.08.036) [DOI] [PubMed] [Google Scholar]
- 85.Fishman L, Willis JH. 2005. A novel meiotic drive locus almost completely distorts segregation in Mimulus (monkeyflower) hybrids. Genetics 169, 347–353. ( 10.1534/genetics.104.032789) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fishman L, Saunders A. 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322, 1559–1562. ( 10.1126/science.1161406) [DOI] [PubMed] [Google Scholar]
- 87.Iwata-Otsubo A, et al. 2017. Expanded satellite repeats amplify a discrete CENP—a nucleosome assembly site on chromosomes that drive in female meiosis. Curr. Biol. 27, 2365–2373. ( 10.1016/j.cub.2017.06.069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Akera T, Chmatal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, Lampson MA. 2017. Spindle asymmetry drives non-Mendelian chromosome segregation. Science 358, 668–672. ( 10.1126/science.aan0092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pardo-Manuel de Villena F, Sapienza C. 2001. Female meiosis drives karyotypic evolution in mammals. Genetics 159, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rhoades MM. 1942. Preferential segregation in maize. Genetics 27, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buckler ES, Phelps-Durr TL, Buckler CS, Dawe RK, Doebley JF, Holtsford TP. 1999. Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153, 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hiatt EN, Dawe RK. 2003. Four loci on abnormal chromosome 10 contribute to meiotic drive in maize. Genetics 164, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dawe RK, et al. 2018. A kinesin-14 motor activates neocentromeres to promote meiotic drive in maize. Cell 173, 839–850. ( 10.1016/j.cell.2018.03.009) [DOI] [PubMed] [Google Scholar]
- 94.Lewis SH, Webster CL, Salmela H, Obbard DJ. 2016. Repeated duplication of Argonaute2 is associated with strong selection and testis specialization in Drosophila. Genetics 204, 757–769. ( 10.1534/genetics.116.192336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Frank SA. 1991. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution 45, 262–267. ( 10.1111/j.1558-5646.1991.tb04401.x) [DOI] [PubMed] [Google Scholar]
- 96.McDermott SR, Noor MA. 2010. The role of meiotic drive in hybrid male sterility. Phil. Trans. R Soc. B 365, 1265–1272. ( 10.1098/rstb.2009.0264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang RL, Wakeley J, Hey J. 1997. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics 147, 1091–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Orr HA, Irving S. 2005. Segregation distortion in hybrids between the Bogota and USA subspecies of Drosophila pseudoobscura. Genetics 169, 671–682. ( 10.1534/genetics.104.033274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barbash DA, Siino DF, Tarone AM, Roote J. 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl Acad. Sci. USA 100, 5302–5307. ( 10.1073/pnas.0836927100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Phadnis N. 2011. Genetic architecture of male sterility and segregation distortion in Drosophila pseudoobscura Bogota–USA hybrids. Genetics 189, 1001–1009. ( 10.1534/genetics.111.132324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McDermott SR, Noor MAF. 2012. Mapping of within-species segregation distortion in Drosophila persimilis and hybrid sterility between D. persimilis and D. pseudoobscura . J. Evol. Biol. 25, 2023–2032. ( 10.1111/j.1420-9101.2012.02581.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jungen H. 1968. Sex ratio in natural populations of Drosophila subobscura. Archiv der Julius Klaus-Stiftung fur Vererbungsforschung, Sozialanthropologie und Rassenhygiene 43, 52–57. [PubMed] [Google Scholar]
- 103.Hauschteck-Jungen E. 1990. Postmating reproductive isolation and modification of the ‘sex ratio’ trait in Drosophila subobscura induced by the sex chromosome gene arrangement A2 + 3+5 + 7. Genetica 83, 31–44. ( 10.1007/bf00774686) [DOI] [PubMed] [Google Scholar]
- 104.Verspoor RL, Smith JML, Mannion NLM, Hurst GDD, Price TAR. 2018. Strong hybrid male incompatibilities impede the spread of a selfish chromosome between populations of a fly. Evol. Lett. 2, 169–179. ( 10.1002/evl3.55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tao Y, Hartl DL, Laurie CC. 2001. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc. Natl Acad. Sci. USA 98, 13 183–13 188. ( 10.1073/pnas.231478798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meiklejohn CD, et al. 2018. Gene flow mediates the role of sex chromosome meiotic drive during complex speciation. eLife 7, e35468 ( 10.7554/eLife.35468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang F, et al. 2012. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151, 871–884. ( 10.1016/j.cell.2012.09.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027. ( 10.1016/S0960-9822(01)00299-8) [DOI] [PubMed] [Google Scholar]
- 109.Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. 2013. The genetic makeup of the Drosophila piRNA pathway. Mol. Cell 50, 762–777. ( 10.1016/j.molcel.2013.04.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon GJ. 2013. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol. Cell 50, 736–748. ( 10.1016/j.molcel.2013.04.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parikh RY, Lin H, Gangaraju VK. 2018. A critical role for nucleoporin 358 (Nup358) in transposon silencing and piRNA biogenesis in Drosophila. J. Biol. Chem. 293, 9140–9147. ( 10.1074/jbc.AC118.003264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wente SR. 2000. Gatekeepers of the nucleus. Science 288, 1374–1377. ( 10.1126/science.288.5470.1374) [DOI] [PubMed] [Google Scholar]
- 113.Le Sage V, Mouland AJ. 2013. Viral subversion of the nuclear pore complex. Viruses 5, 2019–2042. ( 10.3390/v5082019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rowley PA, Patterson K, Sandmeyer SB, Sawyer SL. 2018. Control of yeast retrotransposons mediated through nucleoporin evolution. PLoS Genet. 14, e1007325 ( 10.1371/journal.pgen.1007325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Presgraves DC. 2007. Does genetic conflict drive rapid molecular evolution of nuclear transport genes in Drosophila? Bioessays 29, 386–391. ( 10.1002/bies.20555) [DOI] [PubMed] [Google Scholar]
- 116.Presgraves DC, Stephan W. 2007. Pervasive adaptive evolution among interactors of the Drosophila hybrid inviability gene, Nup96. Mol. Biol. Evol. 24, 306–314. ( 10.1093/molbev/msl157) [DOI] [PubMed] [Google Scholar]
- 117.Tang S, Presgraves DC. 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323, 779–782. ( 10.1126/science.1169123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wei KH, Reddy HM, Rathnam C, Lee J, Lin D, Ji S, Mason JM, Clark AG, Barbash DA. 2017. A pooled sequencing approach identifies a candidate meiotic driver in Drosophila. Genetics 206, 451–465. ( 10.1534/genetics.116.197335) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.