Abstract
Carotenoids are primarily responsible for the characteristic red flesh coloration of salmon. Flesh coloration is an economically and evolutionarily significant trait that varies inter- and intra-specifically, yet the underlying genetic mechanism is unknown. Chinook salmon (Oncorhynchus tshawytscha) represents an ideal system to study carotenoid variation as, unlike other salmonids, they exhibit extreme differences in carotenoid utilization due to genetic polymorphisms. Here, we crossed populations of Chinook salmon with fixed differences in flesh coloration (red versus white) for a genome-wide association study to identify loci associated with pigmentation. Here, the beta-carotene oxygenase 2-like (BCO2-l) gene was significantly associated with flesh colour, with the most significant single nucleotide polymorphism explaining 66% of the variation in colour. BCO2 gene disruption is linked to carotenoid accumulation in other taxa, therefore we hypothesize that an ancestral mutation partially disrupting BCO2-l activity (i.e. hypomorphic mutation) allowed the deposition and accumulation of carotenoids within Salmonidae. Indeed, we found elevated transcript levels of BCO2-l in white Chinook salmon relative to red. The long-standing mystery of why salmon are red, while no other fishes are, is thus probably explained by a hypomorphic mutation in the proto-salmonid at the time of divergence of red-fleshed salmonid genera (approx. 30 Ma).
Keywords: genetic polymorphisms, flesh coloration, single nucleotide polymorphisms, genome-wide association study, carotenoid, BCO2-l
1. Introduction
Many animals acquire carotenoid pigments from their environment and use them for a variety of biological processes, ranging from enhancing immune function and antioxidant capacity to integument pigmentation for sexual selection [1]. Salmonids are one taxonomic group that are well-known for their allocation of carotenoids, resulting in the bright red coloration of many tissues [2]. Although carotenoid pigmentation of skin and eggs in fishes is not unique to salmon, flesh pigmentation has evolved almost exclusively in Salmonidae, specifically in only four genera including Oncorhynchus, Salvelinus, Salmo and Parahucho [2]. Those members of Salmonidae diverged before 30 Ma [3] and exhibit an anadromous life history along with active nest-building activity [2]. Thus, carotenoids stored in the flesh during their ocean phase may be used as a source of antioxidants during the spawning phase, when carotenoids are mobilized to enhance protection of somatic tissues during this demanding life stage and to serve as sexually dimorphic reproductive signals [2].
Flesh pigmentation is also a key commercial trait of wild and farmed salmon alike. Consumers often associate the degree of flesh redness with product quality [4], and in the aquaculture sector, carotenoid additives in food can account for up to 25% of feed costs [4–6]. Characterizing the underlying genetic architecture of red flesh coloration is thus a major goal in salmonid aquaculture [7], but is also important for understanding the evolution of pigmentation more generally. In teleosts, genome sequence analyses have revealed five carotenoid cleaving genes: two beta-carotene monooxygenase genes (BCMO1 and BCMO1-like) and three beta-carotene oxygenase 2 genes (BCO2a, BCO2b and BCO2-like) [8]. Previous studies in salmonids have found that flesh colour variation in red salmon is likely controlled by just a few major loci [7,9–13], and in Atlantic salmon (Salmo salar), inhibition of BCMO1 and BCMO1-like can result in increased carotenoid deposition [14]. Whether the role of pigmentation genes is conserved across all salmonids, and whether any of these genes are responsible for the unique ability of salmonids to deposit carotenoids at any level, remains unknown.
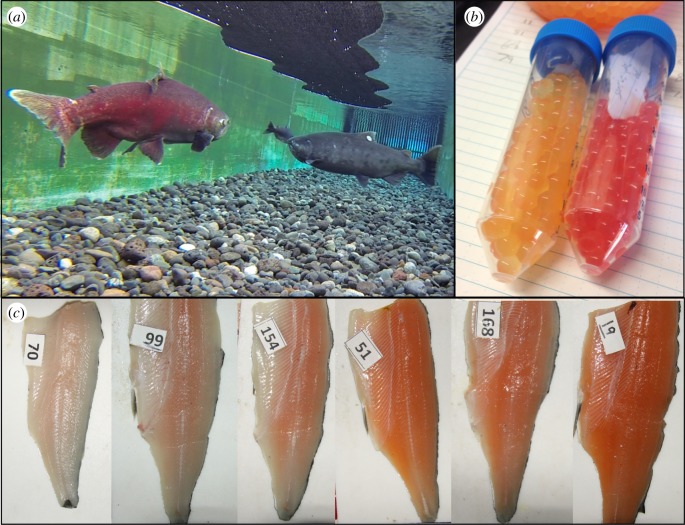
Chinook salmon (Oncorhynchus tshawytscha) is an ideal species to identify candidate genes associated with flesh colour because, unlike other salmonids, this species exhibits extreme intraspecific variation in carotenoid pigmentation of eggs, skin and flesh (figure 1). This carotenoid variation results in two flesh colour morphs (red and white) in Chinook salmon that vary in frequency among populations [15]. Phenotypic trade-offs associated with carotenoids exist between morphs, where, for example, carotenoids can improve immune function, mating success and embryo survival in salmon [2,16–18], but also lead to greater predation risk [19] and potentially toxic effects at high levels [20,21]. Despite the large fitness effect of this genetic colour polymorphism [10], no studies have yet identified gene loci involved in the variation of this trait. Nonetheless, breeding studies suggest pigmentation in Chinook salmon is controlled by a small number of loci with large effects [10,13], and it is hypothesized that these loci do not influence carotenoid absorption but rather the rate at which absorbed carotenoids are metabolized into colourless derivatives [22]. Here, we take advantage of this unique colour polymorphism in Chinook salmon by crossing populations with fixed differences in flesh coloration and genetically characterizing their offspring using traditional genome-wide associations (GWAs) to identify the mechanism(s) responsible for flesh pigmentation in salmon. In addition, the role of candidate gene(s) in driving flesh colour differences is further examined in our study through association analysis in a separate wild population and by measuring gene expression differences. Overall, this work will answer the long-standing question ‘why are salmon red?’.
Figure 1.
Colour variation in Chinook salmon due to genetic polymorphisms influencing carotenoid deposition. (a) Differences in spawning colour of red and white males underwater and (b) differences in egg colour between female Chinook salmon from the Quesnel River, British Columbia, Canada, where red- and white-flesh morphs occur in sympatry. (c) Photographs of the variation in flesh pigmentation generated using Chinook salmon derived from the backcrossing of the two flesh colour populations from the Harrison River and Big Qualicum River, BC. (Online version in colour.)
2. Material and methods
(a). Breeding design and sampling for flesh colour genome-wide association study
(i). Inbred strains for flesh colour
Individuals from two populations of Chinook salmon that exhibit fixed differences in flesh colour, including a pure red-fleshed strain from Big Qualicum River, British Columbia (BC), Canada and a pure white-fleshed strain from Harrison River, BC, were used to create backcrosses in our study. In both populations, the other flesh colour phenotype is absent; thus, individuals are expected to be homozygous at flesh colour loci. These two strains were crossed together in 2005 to generate F1 hybrids that were reared at the Fisheries and Oceans Canada (DFO) laboratory in West Vancouver, BC, Canada.
(ii). F2 backcrosses
In the autumn of 2008, a family of F2 backcrosses of Chinook salmon were bred at DFO by crossing an F1 hybrid male (Big Qualicum red-fleshed × Harrison white-fleshed) to a Harrison white-fleshed female. This backcross design was chosen for the association study to allow the segregation of red-fleshed alleles into a white-fleshed genomic background. Offspring were reared until weight ranged from 200 to 400 g (approx. 1.7 years post-fertilization; see electronic supplementary material for rearing details), and at this time, a fillet was taken to quantify the level of pigmentation in the muscle through visual observation. The fish were given a score between 1 and 5 (±0.5) as indicated by the Roche Colour Card for Salmonids (F. Hoffmann-La Roche, Switzerland). Liver samples were collected for genetic analysis. Individuals were also assessed for precocious maturation based on external coloration and gonad development, and all other individuals were sexed using PCR [23]. A total of 400 individuals were sampled for phenotype, and a subset of 183 individuals was selected for genotyping. The distribution of flesh colour phenotypes was the same for the original sample and the subset of genotyped individuals (electronic supplementary material, figure S1; Kolmogorov–Smirnov test p = 0.99).
(iii). Wild red and white Chinook salmon: Quesnel River population
A subsample of red and white adult Chinook salmon from a sympatric population in the Quesnel River, BC was also included in our design (n = 32; 17 white and 15 red). Chinook salmon captured by netting during the spawning season in 2013 and 2014 were characterized as red or white based on external spawning coloration (for details, see [18]), and colours were scored as binary values. Fin clips were collected for genetic analysis. Quesnel River individuals were included in the analysis to establish whether any significant loci detected in the backcrossed family (involving Big Qualicum and Harrison River strains) would also be present in a different wild population that exhibit both red and white phenotypes.
(b). Genotyping for flesh colour genome-wide association study
(i). DNA extraction, genotyping and single nucleotide polymorphism discovery
DNA was extracted from fin or liver tissue using a standard phenol/chloroform extraction method [24]. All genomic DNA samples were quantified using Quant-iT PicoGreen dsDNA assay kit (ThermoFisher Scientific Inc.) and screened for quality by visual assessment on agarose gels. All samples ranged between 50 and 100 ng µl−1 in concentration. Library preparation and genotype-by-sequencing (GBS) were carried out following the protocol described previously [25] at Cornell University's Institute of Biotechnology. Briefly, the library preparation and GBS protocol included the digestion of DNA with a restriction enzyme, the ligation of adaptors to resulting fragments, the pooling and cleaning of samples and PCR amplification of fragments followed by sequencing on an Illumina platform [25]. For our study, the restriction enzyme EcoT221 was used to digest genomic DNA during library preparation. Methods for single nucleotide polymorphism (SNP) discovery and alignment of SNPs to the Chinook genome [26] are provided in the electronic supplementary material.
(c). Statistical analyses for flesh colour association
(i). Genome-wide association study analyses in F2 generation
TASSEL v. 5.2.19 was used to conduct all GWA analyses using a mixed linear model (MLM) approach [27]. Prior to analyses, SNPs were filtered for minor allele frequency (MAF) of 0.05 and genotyped in at least 80% of individuals. No individuals were lost due to low numbers of reads during sequencing or due to low genotyping rate. The initial number of SNPs identified prior to filtering was 32 636 SNPs, although additional individuals not used in this study were included in the SNP discovery pipeline (see electronic supplementary material for details on SNP discovery). For the F2 family, a total of 18 753 SNPs were genotyped in greater than 80% of individuals, and after MAF filtering, a total of 15 974 SNPs were retained. However, 5114 SNPs were heterozygous in all F2 individuals and thus not informative for GWA analyses. Thus, a total of 10 860 SNPs were used for GWA analyses in the F2 family. We identified loci showing a significant association with flesh colour variation using MLM, where kinship was accounted for in the model. We did not include sex or weight as covariates in the model because, using a t-test, we found no significant difference in colour between sexes (p = 0.32) nor the distribution (Kolmogorov–Smirnov test) of colours between sexes (p = 0.73). We also found no significant relationship between weight and colour (p = 0.18). Genome-wide significance was determined by Bonferroni correction based on the total number of SNPs (α = 0.05/total number of SNPs tested). Given that Bonferroni correction is highly conservative, we also considered loci to be ‘suggestive’ if the model for the marker had an R2 greater than 0.05. Manhattan and Q–Q plots were created to visualize the results using qqman package [28] in R software [29]. Gene annotations for regions with significant SNPs were examined for potential candidate genes associated with pigmentation with the expectation that long-distance linkage disequilibrium (LD) would be present in the F2 generation. Gene annotations were downloaded for the Chinook salmon genome [26] from NCBI (GCA_002872995.1), and a custom R script was used to extract all genes near SNPs (i.e. within 10 000 bp).
(ii). Wild Quesnel population genome-wide association study analyses and outlier tests
Association analyses were also performed within a wild population of sympatric red and white Chinook salmon from the Quesnel River, BC. First, if the F2 analysis identified specific genomic regions of interest, we used association analysis within that specific genomic region (a 9.69 Mbp region on Chr 30 that included 48 SNPs after filtering; see Results) using the MLM approach with a kinship matrix calculated from the full SNP dataset with no covariates included. A less conservative α level of 0.05 was used for our targeted analysis given that long-distance LD present in the F2 generation may be absent within the wild population. Therefore, depending on genome coverage in the region of interest, significant loci may be more difficult to detect in the wild population, and there were fewer (n = 32) individuals analysed. Second, given that red and white individuals may differ at additional loci not directly related to carotenoid pigmentation but linked to the evolutionary differences between the phenotypes due to the diverse physiological role of carotenoids, we also examined outlier loci between the two groups across the genome. We used Arlequin 3.5.2.2 [30] to detect loci under selection using 20 000 simulations with 100 demes per group, with p-values adjusted for false discovery rate with the p.adjust function in R. We only considered loci under divergent selection between red and white to be outliers (upper tail of the FST distribution).
(d). Gene expression at BCO2-l gene
The above analyses identified a probable candidate gene responsible for flesh coloration variation in Chinook salmon. To evaluate differences in the activity of this gene (beta-carotene oxygenase 2-like, BCO2-l), we designed primers to measure transcription of BCO2-l in muscle tissue of red and white Chinook salmon groups reared as part of separate study examining physiological effects of carotenoid pigments (unpublished). Post-smolt Chinook salmon from the pure red-fleshed Big Qualicum River strain and from the pure white Harrison River strain were fed either a commercial pigmented or unpigmented diet over a three-month period. Fish were sampled pre- and post-diet manipulation. At the time of sampling, the Big Qualicum strain had deposited significantly more pigment in the flesh than did the Harrison River white strain. At the end of the trial, muscle tissue was preserved in RNAlater and RNA was isolated using standard Trizol method as described by the manufacturer (Thermo Fisher Scientific). First-strand cDNA was synthesized from total RNA (0.5 µg) using the High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Applied Biosystems, California, USA). Quantitative PCR was performed using primers specific to the BCO2-l gene: Forward, CACAGAGGTGTGTGGGACCAT; Reverse, GACTCACTGTTGGTTTCCGAACT. qPCR reactions were performed on a 7500 Fast Real-Time PCR System (Applied Biosystems) under Fast conditions with Fast SYBR™ Green Master Mix (Thermo Fisher Scientific). A reference gene, ubiquitin, was used to normalize mRNA levels (Forward primer, ACAGCTGGCCCAGAAGTACAA; Reverse primer, GGCGAGCGTAGCATTTGC). Relative mRNA expression levels were calculated using the 2−ΔΔCt method [31]. Data were log transformed to meet assumptions of normality for statistical analyses.
3. Results
(a). Genome-wide association study for carotenoid pigmentation
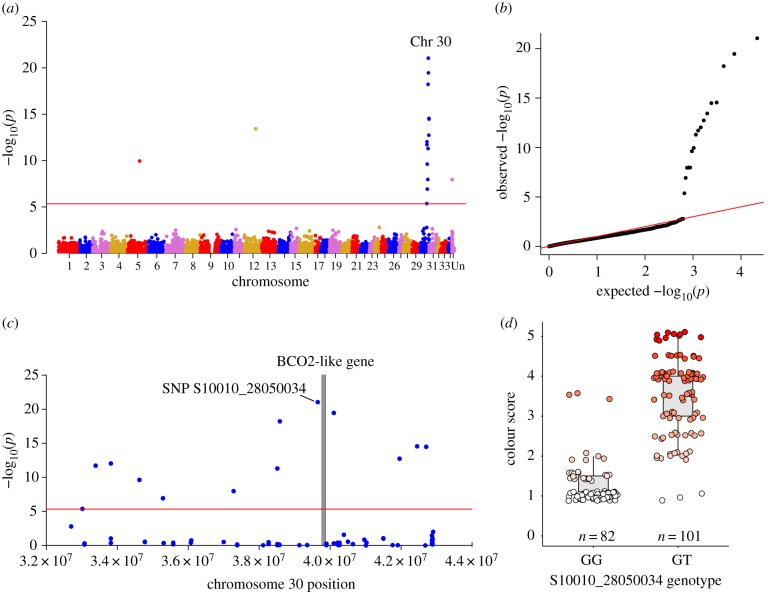
Using GWA analyses of offspring from one F2 backcross family (n = 183 offspring) with a range in flesh colour phenotypes (electronic supplementary material, figure S1), we found 17 SNPs (out of 10 860 SNPs) were significantly associated with flesh coloration at the genome-wide level (all p-values < 4.60 × 10−6; figure 2a,b) and were primarily localized to a region on chromosome 30 (figure 2a,c). The most significant SNP (S10010_28050034; p = 9.19 × 10−22) explained 66% of the variation in colour and was adjacent to the beta-carotene oxygenase 2-like locus (BCO2-l). BCO2 genes have been linked to carotenoid pigmentation in mammals and birds, where its disruption increases the accumulation of carotenoids in tissues [21,32–36]. In our study, the most significant SNP showed a clear association with flesh colour (figure 2c,d), although three fish for each of the GG and GT genotypes (figure 2d) were distinct from the main genotype associations for colour, suggesting that this SNP (S10010_28050034) locus is not causal, but is in LD with the causal BCO2-l gene. Additional significant SNPs on Chr 30 probably reflect an association with the same functional locus due to long-distance LD (figure 2c) due to the single generation of recombination between the red and white parents and low recombination rates in salmonids [37]. Indeed, the significance of these additional associated SNPs decreased with distance from BCO2-l gene (p = 0.007; R2 = 0.46; electronic supplementary material, figure S2).
Figure 2.
Genome-wide association study (GWAS) results for flesh colour in Chinook salmon. (a) Manhattan plot of all loci (SNPs) analysed for association with flesh pigmentation in an F2 family of Chinook salmon derived from the backcrossing of two flesh colour populations. SNPs located above the red line indicate genome-wide significance based on Bonferroni correction (p < 4.60 × 10−6). (b) The quantile–quantile plot obtained from the analysis. (c) The significant peak found on Chromosome 30 in more detail with the grey bar indicating the position of the carotenoid-associated gene (BCO2-l) that aligned next to the most significant SNP. (d) The phenotype association for genotypes GG and GT at the most significant SNP (S10010_28050034; p = 9.18 × 10−22, R2 = 0.66). Points are jittered horizontally and vertically for visualization. (Online version in colour.)
Only three of the 17 significant SNPs did not map to Chr 30 (figure 2a) and may represent spurious associations or misplaced SNPs due to off-target alignments or genome misassemblies, as no other SNPs were associated with flesh colour in those regions (expected with high LD in F2 generation). Additionally, the closest genes located near these significant SNPs were not associated with obvious functions related to carotenoid pigmentation, and these included Krueppel-like factor 2 on Chr 5 and WD repeat-containing protein 43-like on Chr 12, and the third SNP did not align to a physical position in the genome. Although our analysis did not identify other candidate loci, association analysis in the F3 backcross generation (not presented here) did not yield significant association with BCO2-l (or any other loci), suggesting either no other loci influence pigmentation in these families or that their effects are minor. It is possible that the F2 fish selected as parents to generate the F3 progeny groups possessed a pigmented phenotype that arose from multiple minor loci affecting carotenoid pigmentation that segregated among F3 progeny to generate weak pigmentation phenotypes that were individually not strongly associated with a specific chromosomal region. Further investigation of the effects of minor loci and their interactions acting on pigmentation in Chinook salmon is warranted.
(b). Genetic variation associated with pigmentation in a wild population of Chinook salmon
The above evidence suggests that BCO2-l is the probable candidate responsible for flesh coloration variation in Chinook salmon. Thus, we further explored the nature of pigmentation variation using a wild population of sympatric red and white salmon from the Quesnel River (n = 32), where we targeted the candidate region of Chr 30 and performed a MLM GWA analysis. Out of the 48 SNPs used in the analysis, five SNPs (from three sequence tags) within the region were significant at α level 0.05 (electronic supplementary material, figure S3). In addition, FST-based outlier analysis revealed low genome-wide divergence between red and white individuals (mean FST = −0.0008; n = 8940 SNPs) with only 35 SNPs considered outliers under divergent selection (electronic supplementary material, table S2 and figure S4A). No outliers were found on Chr 30, although notably, mean FST was highest for Chr 30 (mean FST = 0.007) relative to all other chromosomes (electronic supplementary material, figure S4B). Outliers identified here may highlight other loci indirectly related to differences between carotenoid phenotypes (electronic supplementary material, table S2), and more genome coverage may be needed in the BCO2-l region to detect loci of significant effect in the wild population because long-distance LD is not expected to be as strong due to extensive prior opportunity for genetic exchange.
(c). Gene expression differences at BCO2-l
In other taxa, the disruption or downregulation of the BCO2 gene has been linked to the accumulation of carotenoids in tissues [21,32–36,38,39], and consequently, we hypothesized that increased expression of BCO2-l should be exhibited in white Chinook salmon resulting in greater cleavage and reduced accumulation of coloured carotenoids. To assess this directly, we analysed gene transcript levels of BCO2-l in the muscle of red (Big Qualicum) and white (Harrison) salmon fed a pigmented and unpigmented diet under common garden conditions (Material and methods). Consistent with our prediction, relative expression of BCO2-l was found to be significantly greater in white relative to red Chinook salmon prior to diet manipulation (t-test: t = −2.43, d.f. = 7.95, p = 0.041) as well as post-diet manipulation (two-way ANOVA: F = 31.902, p = 2.05 × 10−6), with no effect of diet type (F = 0.045, p = 0.833) or interaction of diet and colour strain (F = 0.198, p = 0.659) on BCO2-l gene transcription (figure 3).
Figure 3.
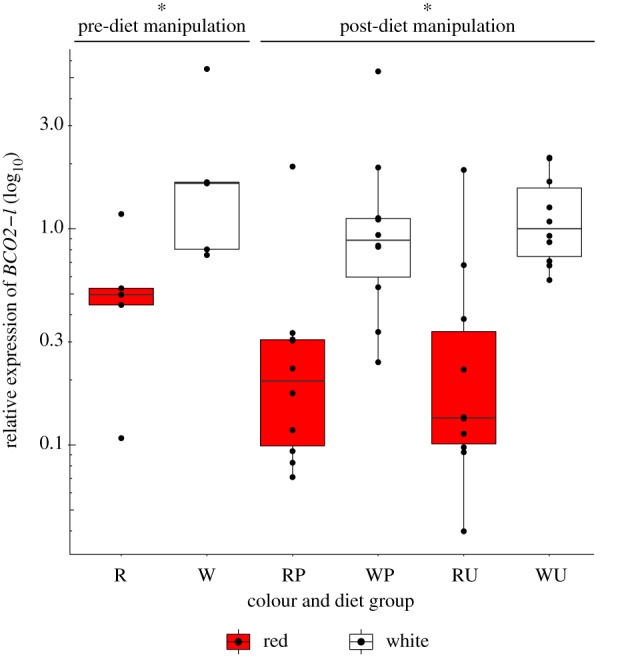
Boxplot of relative expression (log10 transformed) of BCO2-l gene in red (Big Qualicum) and white (Harrison) Chinook salmon strains reared under common garden conditions. Gene transcription was measured in muscle tissue prior to diet manipulation (red = R and white = W), and 3 months after a diet manipulation where each strain was fed both a pigmented (P) and unpigmented (U) diet. Asterisk (*) indicates significance between W and R phenotypes. (Online version in colour.)
4. Discussion
Carotenoid deposition is critical for the red coloration displayed by salmonids during courtship and additionally influences multiple fitness processes (e.g. antioxidation, crypsis and immunity) [2,19,40]. While carotenoids provide many benefits, carotenoid pigmentation of flesh has evolved almost exclusively in salmonid fishes [2], yet the underlying gene responsible for the ability to deposit carotenoids has not been identified until now. In our study, multiple lines of evidence indicate that BCO2-l is the gene responsible for variation in carotenoid accumulation in Chinook salmon, and by extension, other salmonids as well. We propose that an ancestral mutation partially disrupting BCO2-l (i.e. hypomorphic mutation [41]) in proto-salmonids provided an adaptation that allowed the group to exploit carotenoids as an antioxidant resource in their muscle tissue during stressful spawning migrations and nest building, thereby facilitating the evolution of anadromy [2]. Within Chinook salmon, a more recent mutation that elevated BCO2-l activity (i.e. hypermorphic mutation [41]) and increased the rate at which carotenoids were metabolized into colourless derivatives [22] probably drove the evolution of the white flesh phenotype prior to the end of the last glaciation [15]. The white flesh morph exists within many populations of Chinook salmon, and while different mutations can lead to convergent phenotypes [42], given the expected handicap associated with a loss of flesh carotenoids in salmon, it seems more likely that a mutation increasing the activity of the major candidate locus (BCO2-l) in muscle tissue evolved rarely but provided a sufficiently large advantage during post-glacial recolonization of particular river environments [15]. Indeed, the rarity of the mutation is supported by the lack of a derived white morph in other salmonid species. Furthermore, this is consistent with our finding of significant but weaker associations near BCO2-l in a different population. The weaker associations found in the Quesnel population may be a result of a smaller sample size and the need for higher density genomic markers in the BCO2-l region in wild populations with extensive opportunity for recombination to break down long-distance patterns of LD.
Hypermorphic mutations are often co-dominant and this mode of action, involving two loci [10], has previously been proposed for pigmentation of Chinook salmon. Our data support that variation at the BCO2-l locus arises from regulatory changes, and these results are comparable with a recent study in reptiles that found cis-acting regulatory changes in BCO2 could explain differences in carotenoid coloration [38]. Nonetheless, while regulatory changes are likely, changes in amino acid sequence that influence BCO2-l activity may also be possible. In addition, it is likely that there are other minor loci that, together with environmental influences, could explain the range of carotenoid variation found among and within pigmented salmonid species. Future genomic studies that examine the gene–phenotype associations across multiple Chinook salmon populations as well as among salmonid species with varying rates of carotenoid deposition would provide further information on the specific genetic changes that have occurred at the BCO2-l locus.
Consistent with our findings, studies in reptiles and birds have found that colour polymorphisms can often be explained by a single major effect locus [38,43]. The role of a single gene in driving differences between colour morphs is surprising given that morphs often show correlated differences in other key traits [38,43]. Indeed, differences in reproduction, predation, and physiology have been associated with carotenoid pigmentation in salmonids [16–19,44–47]. Therefore, fitness-related differences between morphs suggest that BCO2-l may have pleiotropic effects on other biological processes in Chinook salmon. The maintenance of this colour polymorphism in nature may thus be explained by multitrait divergence that can lead to balanced fitness trade-offs.
In Chinook salmon, the occurrence of the white phenotype has puzzled scientists given that higher carotenoid content in salmonids has demonstrated benefits on egg survival, immune function, mating success and antioxidant capacity [2,16,18,40,45,46]. However, the white phenotype may have evolved primarily due to advantages of reduced predation in fresh water [19], and potentially through reduced toxicity associated with high levels of carotenoids [20,21]. Under experimental conditions, red eggs experience greater predation relative to white eggs, and in a high predation environment, the advantage of reduced predation risk in white eggs is predicted to outweigh the expected cost of their lower incubation survival [19]. Furthermore, despite the expected immunity benefit of carotenoids, differences between red and white Chinook salmon in immune function (viral susceptibility) have not been found [40]; however, morphs exhibit significant functional genetic differences in immune genes, potentially highlighting the use of alternative mechanisms by morphs to enhance immunity [18]. In addition, morphs display differences in sexual selection, as evidence suggests that red females employ pre-spawning mate choice and white females use cryptic processes (post-spawning) to bias paternity of their eggs in favour of males of their own colour morph [17,18].
Overall, in Chinook salmon, the colour polymorphism associated with variation in BCO2-l appears to be maintained through the interplay of both natural and sexual selection processes, and the identification of the genomic mechanism underlying this fitness trait provides further insight into these evolutionary mechanisms. By investigating this polymorphism in salmon, our study reveals an interesting evolutionary history in Salmonidae and ultimately offers an answer to the long-standing question of ‘why are salmon red?’.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Sharon Mitchell and Katie Hyma at the Cornell University Institute of Biotechnology in the Genomic Diversity Facility for performing the GBS and TASSEL pipeline analysis for SNP identification. We thank Yellow Island Aquaculture Ltd and staff for providing care and facilities for experimental fish. We are grateful to the members of the Devlin lab, N. Antoniolli, S. Toews, J. Drown and E. Olague for assistance with fish sampling. We also thank the Quesnel River Research Center and staff as well as J. Laycock and M. Dender for their help with wild fish collection. Assistance from DFO hatchery staff Grant Ladouceur, Les Clint, Reid Schrul and Larry Kahl is also much appreciated. We thank the McGill University and Génome Québec Innovation Centre for their services in preparing the various sequencing libraries and performing the sequencing.
Ethics
All experiments conducted in the study involving salmon followed animal care guidelines and were approved by Fisheries and Oceans Canada Animal Care Committee.
Data accessibility
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.mn91560 [48].
Authors' contributions
R.H.D. conceived the project and designed the experiment. W.E.V., R.H.D., S.J.L., J.W.H., T.E.P. and D.D.H. contributed to experimental design, fish care and sample collection. R.H.D. and W.E.V. designed and executed the common garden diet experiment. S.J.L. performed DNA extractions and sample preparation for genotyping, performed statistical analyses and drafted the manuscript. D.S. performed qRT-PCR laboratory work and analyses. K.A.C. and B.F.K. contributed data (Chinook genome assembly), and K.A.C. aligned genomic data to the Chinook assembly. All authors contributed to the writing of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This project was supported by NSERC and Fisheries and Oceans Canada.
References
- 1.Svensson PA, Wong B. 2011. Carotenoid-based signals in behavioural ecology: a review. Behaviour 148, 131–189. ( 10.1163/000579510X548673) [DOI] [Google Scholar]
- 2.Rajasingh H, Våge DI, Pavey SA, Omholt SW. 2007. Why are salmonids pink? Can. J. Fish. Aquat. Sci. 64, 1614–1627. ( 10.1139/f07-119) [DOI] [Google Scholar]
- 3.Lecaudey LA, Schliewen UK, Osinov AG, Taylor EB, Bernatchez L, Weiss SJ. 2018. Inferring phylogenetic structure, hybridization and divergence times within Salmoninae (Teleostei: Salmonidae) using RAD-sequencing. Mol. Phylogenet. Evol. 124, 82–99. ( 10.1016/j.ympev.2018.02.022) [DOI] [PubMed] [Google Scholar]
- 4.Alfnes F, Guttormsen AG, Steine G, Kolstad K. 2006. Consumers' willingness to pay for the color of salmon: a choice experiment with real economic incentives. Am. J. Agric. Econ. 88, 1050–1061. ( 10.1111/j.1467-8276.2006.00915.x) [DOI] [Google Scholar]
- 5.Buttle L, Crampton V, Williams P. 2001. The effect of feed pigment type on flesh pigment deposition and colour in farmed Atlantic salmon, Salmo salar L. Aquac. Res. 32, 103–111. ( 10.1046/j.1365-2109.2001.00536.x) [DOI] [Google Scholar]
- 6.Torrissen O, Christiansen R. 1995. Requirements for carotenoids in fish diets. J. Appl. Ichthyol. 11, 225–230. ( 10.1111/j.1439-0426.1995.tb00022.x) [DOI] [Google Scholar]
- 7.Baranski M, Moen T, Våge DI. 2010. Mapping of quantitative trait loci for flesh colour and growth traits in Atlantic salmon (Salmo salar). Genet. Sel. Evol. 42, 17 ( 10.1186/1297-9686-42-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helgeland H, Sandve SR, Torgersen JS, Halle MK, Sundvold H, Omholt S, Våge DI. 2014. The evolution and functional divergence of the beta-carotene oxygenase gene family in teleost fish—exemplified by Atlantic salmon. Gene 543, 268–274. ( 10.1016/j.gene.2014.02.042) [DOI] [PubMed] [Google Scholar]
- 9.Araneda C, Neira R, Iturra P. 2005. Identification of a dominant SCAR marker associated with colour traits in Coho salmon (Oncorhynchus kisutch). Aquaculture 247, 67–73. ( 10.1016/j.aquaculture.2005.02.028) [DOI] [Google Scholar]
- 10.Withler R. 1986. Genetic variation in carotenoid pigment deposition in the red-fleshed and white-fleshed Chinook salmon (Oncorhynchus tshawytscha) of Quesnel River, British Columbia. Can. J. Genet. Cytol. 28, 587–594. ( 10.1139/g86-086) [DOI] [Google Scholar]
- 11.Houston R, Bishop S, Hamilton A, Guy D, Tinch A, Taggart J, Derayat A, McAndrew B, Haley C. 2009. Detection of QTL affecting harvest traits in a commercial Atlantic salmon population. Anim. Genet. 40, 753–755. ( 10.1111/j.1365-2052.2009.01883.x) [DOI] [PubMed] [Google Scholar]
- 12.Tsai H-Y, Hamilton A, Tinch AE, Guy DR, Gharbi K, Stear MJ, Matika O, Bishop SC, Houston RD. 2015. Genome wide association and genomic prediction for growth traits in juvenile farmed Atlantic salmon using a high density SNP array. BMC Genom. 16, 969 ( 10.1186/s12864-015-2117-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCallum IM, Cheng KM, March B. 1987. Carotenoid pigmentation in two strains of Chinook salmon (Oncorhynchus tshawytscha) and their crosses. Aquaculture 67, 291–300. ( 10.1016/0044-8486(87)90214-6) [DOI] [Google Scholar]
- 14.Lien S, Sodeland M, Moen T. 2015. Predicting the ability of Atlantic salmon to utilize dietary pigment based on the determination of polymorphisms. See www.freepatentsonline.com/y2017/0114399.html.
- 15.Hard JJ, Wertheimer AC, Johnson WF. 1989. Geographic variation in the occurrence of red-and white-fleshed Chinook salmon (Oncorhynchus tshawytscha) in western North America. Can. J. Fish. Aquat. Sci. 46, 1107–1113. ( 10.1139/f89-143) [DOI] [Google Scholar]
- 16.Tyndale ST, Letcher RJ, Heath JW, Heath DD. 2008. Why are salmon eggs red? Egg carotenoids and early life survival of Chinook salmon (Oncorhynchus tshawytscha). Evol. Ecol. Res. 10, 1187–1199. [Google Scholar]
- 17.Lehnert SJ, Heath DD, Devlin RH, Pitcher TE. 2017. Post-spawning sexual selection in red and white Chinook salmon (Oncorhynchus tshawytscha). Behav. Ecol. 28, 1–10. ( 10.1093/beheco/arw142) [DOI] [Google Scholar]
- 18.Lehnert SJ, Pitcher TE, Devlin RH, Heath DD. 2016. Red and white Chinook salmon: genetic divergence and mate choice. Mol. Ecol. 25, 1259–1274. ( 10.1111/mec.13560) [DOI] [PubMed] [Google Scholar]
- 19.Lehnert SJ, Devlin RH, Pitcher TE, Semeniuk CA, Heath DD. 2017. Redder isn't always better: cost of carotenoids in Chinook salmon eggs. Behav. Ecol. 28, 549–555. ( 10.1093/beheco/arw182) [DOI] [Google Scholar]
- 20.Wu L, et al. 2017. Lack of β, β-carotene-9′,10′-oxygenase 2 leads to hepatic mitochondrial dysfunction and cellular oxidative stress in mice. Mol. Nutr. Food Res. 61, 1600576 ( 10.1002/mnfr.201600576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amengual J, Lobo GP, Golczak M, Li HNM, Klimova T, Hoppel CL, Wyss A, Palczewski K, Von Lintig J.. 2011. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 25, 948–959. ( 10.1096/fj.10-173906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.March B, Hajen W, Deacon G, MacMillan C, Walsh M. 1990. Intestinal absorption of astaxanthin, plasma astaxanthin concentration, body weight, and metabolic rate as determinants of flesh pigmentation in salmonid fish. Aquaculture 90, 313–322. ( 10.1016/0044-8486(90)90255-L) [DOI] [Google Scholar]
- 23.Devlin R, Biagi C, Smailus D. 2001. Genetic mapping of Y-chromosomal DNA markers in Pacific salmon. Genetica 111, 43–58. ( 10.1023/A:1013759802604) [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 25.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6, e19379 ( 10.1371/journal.pone.0019379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen KA, Leong JS, Sakhrani D, Biagi CA, Minkley DR, Withler RE, Rondeau EB, Koop BF, Devlin RH. 2018. Chinook salmon (Oncorhynchus tshawytscha) genome and transcriptome. PLoS ONE 13, e0195461 ( 10.1371/journal.pone.0195461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. ( 10.1093/bioinformatics/btm308) [DOI] [PubMed] [Google Scholar]
- 28.Turner SD. 2014. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. BioRxiv.
- 29.R Core Development Team. 2016. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 30.Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10, 564–567. ( 10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- 32.dela Seña C, Sun J, Narayanasamy S, Riedl KM, Yuan Y, Curley RW, Schwartz SJ, Harrison EH. 2016. Substrate specificity of purified recombinant chicken β-carotene 9′,10′-oxygenase (BCO2). J. Biol. Chem. 291, 14 609–14 619. ( 10.1074/jbc.M116.723684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian R, Pitchford W, Morris C, Cullen N, Bottema C. 2010. Genetic variation in the β, β-carotene-9′,10′-dioxygenase gene and association with fat colour in bovine adipose tissue and milk. Anim. Genet. 41, 253–259. ( 10.1111/j.1365-2052.2009.01990.x) [DOI] [PubMed] [Google Scholar]
- 34.Våge DI, Boman IA. 2010. A nonsense mutation in the beta-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries). BMC Genet. 11, 10 ( 10.1186/1471-2156-11-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niu Y, et al. 2017. Biallelic β-carotene oxygenase 2 knockout results in yellow fat in sheep via CRISPR/Cas9. Anim. Genet. 48, 242–244. ( 10.1111/age.12515) [DOI] [PubMed] [Google Scholar]
- 36.Li B, et al. 2014. Inactivity of human β, β-carotene-9′,10′-dioxygenase (BCO2) underlies retinal accumulation of the human macular carotenoid pigment. Proc. Natl Acad. Sci. USA 111, 10 173–10 178. ( 10.1073/pnas.1402526111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allendorf F, Thorgaard GH. 1984. Tetraploidy and evolution of salmonid fishes. In Evolutionary genetics of fishes (ed. Turner B.), pp. 1–53. New York, NY: Plenum Press. [Google Scholar]
- 38.Andrade P, et al. 2019. Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc. Natl Acad. Sci. USA 116, 5633–5642. ( 10.1073/pnas.1820320116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, et al. 2019. A carotenoid oxygenase is responsible for muscle coloration in scallop. Biochim. Biophys. Acta 1864, 966–975. [DOI] [PubMed] [Google Scholar]
- 40.Lehnert SJ, Garver KA, Richard J, Devlin RH, Lajoie C, Pitcher TE, Heath DD. 2018. Significant differences in maternal carotenoid provisioning and effects on offspring fitness in Chinook salmon colour morphs. J. Evol. Biol. 31, 1876–1893. ( 10.1111/jeb.13383) [DOI] [PubMed] [Google Scholar]
- 41.Muller HJ. 1932. Further studies on the nature and causes of gene mutations. In Proceedings of the 6th International Congress of Genetics (ed. Jones DF.), vol. 1, pp. 213–255. Menasha, WI: Brooklyn Botanic Garden. [Google Scholar]
- 42.Steiner CC, Römpler H, Boettger LM, Schöneberg T, Hoekstra HE. 2008. The genetic basis of phenotypic convergence in beach mice: similar pigment patterns but different genes. Mol. Biol. Evol. 26, 35–45. ( 10.1093/molbev/msn218) [DOI] [PubMed] [Google Scholar]
- 43.Toomey MB, et al. 2018. A non-coding region near Follistatin controls head colour polymorphism in the Gouldian finch. Proc. R. Soc. B 285, 20181788 ( 10.1098/rspb.2018.1788) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyndale ST. 2005. Why are salmon eggs red? An investigation of the benefits of red carotenoid-based pigmentation in the eggs and offspring of Chinook salmon (Oncorhynchus tshawytscha). MSc thesis, University of Windsor, Windsor, ON. [Google Scholar]
- 45.Parolini M, Iacobuzio R, Possenti CD, Bassano B, Pennati R, Saino N. 2018. Carotenoid-based skin coloration signals antioxidant defenses in the brown trout (Salmo trutta). Hydrobiologia 815, 267–280. ( 10.1007/s10750-018-3571-6) [DOI] [Google Scholar]
- 46.Wilkins LG, Da Cunha LM, Menin L, Ortiz D, Vocat-Mottier V, Hobil M, Nusbaumer D, Wedekind C.. 2017. Maternal allocation of carotenoids increases tolerance to bacterial infection in brown trout. Oecologia 185, 1–13. ( 10.1007/s00442-017-3952-y) [DOI] [PubMed] [Google Scholar]
- 47.Craig JK, Foote CJ. 2001. Countergradient variation and secondary sexual color: phenotypic convergence promotes genetic divergence in carotenoid use between sympatric anadromous and nonanadromous morphs of sockeye salmon (Oncorhynchus nerka). Evolution 55, 380–391. ( 10.1111/j.0014-3820.2001.tb01301.x) [DOI] [PubMed] [Google Scholar]
- 48.Lehnert SJ, Christensen KA, Vandersteen WE, Sakhrani D, Pitcher TE, Heath JW, Koop BF, Heath DD, Devlin RH. 2019. Data from: Carotenoid pigmentation in salmon: variation in expression at BC02-l locus controls a key fitness trait affecting red coloration Dryad Digital Repository. ( 10.5061/dryad.mn91560) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Lehnert SJ, Christensen KA, Vandersteen WE, Sakhrani D, Pitcher TE, Heath JW, Koop BF, Heath DD, Devlin RH. 2019. Data from: Carotenoid pigmentation in salmon: variation in expression at BC02-l locus controls a key fitness trait affecting red coloration Dryad Digital Repository. ( 10.5061/dryad.mn91560) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.mn91560 [48].