Abstract
Recombinant adeno-associated viruses (rAAVs) are excellent vectors for gene delivery. However, current Sf9/Cap-Rep packaging cell line-dependent OneBac systems still lack versatility and flexibility for large-scale production of rAAVs. In this study, we developed an improved OneBac system that includes a novel dual-function baculovirus expression vector (BEV) termed BEV/Cap-(ITR-GOI) that carries both the AAV Cap gene and rAAV genome inverted terminal repeat (ITR) sequences flanking the gene of interest (GOI), a versatile Sf9-GFP/Rep packaging cell line that harbors silent copies of the AAV2 Rep gene that can be expressed after BEV infection, and constitutively expressed green fluorescent protein (GFP) reporter genes to facilitate cell line screening. The BEV/Cap-(ITR-GOI) construct allows flexibility to switch among different Cap gene serotypes using simple BEV reconstruction, and is stable for at least five serial passages. Furthermore, the Sf9-GFP/Rep stable cell line is versatile for production of different rAAV serotypes. The yield levels for rAAV2, rAAV8, and rAAV9 exceeded 105 vector genomes (VG) per cell, which is similar to other currently available large-scale rAAV production systems. The new Bac system-derived rAAVs have biophysical properties similar to HEK293 cell-derived rAAVs, as well as high quality and activity. In summary, the novel Sf9-GFP/Rep packaging cell line-dependent OneBac system can facilitate large-scale rAAV production and rAAV-based gene therapy.
Keywords: rAAV production, OneBac, recombinant baculovirus, Sf9 packaging cell line, gene therapy
Introduction
Recombinant adeno-associated viruses (rAAVs) have a unique genome that contains a gene of interest (GOI) flanked by the inverted terminal repeats (ITRs) of the AAV that can be packaged in viral capsids.1,2 rAAVs have many advantages including the ability to efficiently infect different cell types, diverse serotypes related to cell-type tropism, long and persistent expression periods, and low pathogenic risk, which together make rAAV a promising viral vector for gene therapy.3,4 The traditional method to produce rAAVs involves HEK293 cell-based triple-plasmid cotransfection.5 The three plasmids contain the ITR-GOI, Rep gene, Cap gene, and adenovirus helper genes. The AAV ITRs provide cis elements required for rAAV replication and packaging, whereas the AAV Rep gene, Cap gene, and helper virus genes are supplied in trans. After the approval of the rAAV-based gene therapy drugs Glybera to treat lipoprotein lipase deficiency, Luxturna to treat hereditary retinal dystrophy, and Zolgensma to treat lethal spinal muscular atrophy, in 2012, 2017, and 2019, respectively, the need for large amounts of rAAVs having different serotypes for use in clinical applications has been increasing.6–8
However, traditional methods are not suitable for large-scale production of rAAV9 and thus alternative baculovirus expression vector (BEV)-mediated rAAV production systems (Bac systems) have recently been developed. Highly efficient BEV infection provides advantages over traditional plasmid transfection.10 Moreover, insect Sf9 cells are better suited for large-scale and high-density serum-free suspension culture than HEK293 cells.11
The previous two decades have seen significant advances in large-scale rAAV production methods based on Bac systems. In 2002, Urabe et al. reported the first successful generation of rAAV upon coinfection of insect cells with three BEVs: BEV/Cap, BEV/Rep, and BEV/(ITR-GOI).12 However, this approach is not widely used due to low yields and genetic instability, particularly for BEV/Rep.13 To improve the genetic stability of the BEVs, the Rep and Cap expression cassettes were optimized. In 2008, Chen constructed a modified BEV/Cap-Rep to express AAV Rep and Cap proteins using artificial introns.14 In 2009, Smith et al. modified the AAV Rep and Cap expression cassettes using leaky ribosome scanning to construct another kind of BEV/Cap-Rep. The Rep or Cap proteins can both be translated from single mRNA transcripts.15 These dual-functional BEV/Cap-Rep constructs were stable and showed no apparent decrease in Cap and Rep protein expression after five or seven continuous passages.14,15 These two-Bac systems have gradually replaced the previously used three-Bac system. However, low coinfection ratios of multi-BEVs often result in defective rAAV and low yields.16
In 2009, Aslanidi et al. established an Sf9/Cap-Rep packaging cell line integrated with silent copies of both AAV Rep and Cap genes, which enable rAAV production by single BEV/(ITR-GOI) infection. The rAAV yield with this cell line exceeded 105 vector genomes (VG) per cell, which is ∼10 times higher than previous multi BEV-based Bac systems.16 In 2014, Mietzsch et al. further expanded this Sf9/Cap-Rep packaging cell line-dependent OneBac system to produce the full range of rAAV1-12 serotypes.17 In 2018, Wu et al. created a single BEV/Cap-(ITR-GOI)-Rep that integrated all of the elements for rAAV production in both cultured Sf9 cells and armyworm larvae.18 The rAAV yield level in this system was similar to the current highest state upon infection of Sf9 cells. Moreover, this research was the first to exploit a potential low-cost rAAV production method in insect larvae. However, the main concern with this system is that BEV stability could decrease significantly after four serial passages.
For the Sf9 packaging cell line-dependent OneBac system, recent progress mainly involved modifications to the AAV Rep and Cap expression cassettes used to construct Sf9/Cap-Rep packaging cell lines. In 2015 and 2017, Mietzsch et al. developed an improved OneBac 2.0 system19,20 that had 100-fold enhancement in rAAV5 infectivity through the use of a splicing-based strategy to raise the relative amounts of VP1 in AAV5 capsids. Meanwhile, collateral packaging of nonvector DNA was significantly suppressed in the absence of Rep-binding element (RBE) during the production of AAV1, AAV2, AAV5, and AAV8. In 2019, Joshi et al. developed a scalable and robust AAV5 production process through fed-batch bioreactor using the OneBac 2.0 system.21 However, the Sf9/Cap-Rep packaging cell line lacked versatility and flexibility for production of rAAVs having different serotypes. These disadvantages severely impeded the application of this Bac system.
In this study, we developed an improved Sf9 packaging cell line-dependent Bac system by optimizing the distribution of three rAAV packaging elements (i.e., the Rep gene, Cap gene, and ITR-GOI) between the BEV and Sf9 cell genomes. First, we constructed a novel dual-functional BEV/Cap-(ITR-GOI) that carries the Cap gene and the ITR-GOI. This construct provides flexibility to switch among different Cap gene serotypes using a simple BEV construct. Second, we constructed a modified Sf9-GFP/Rep packaging cell line that is more versatile and allows simple screening for green fluorescent protein (GFP) reporter gene expression using fluorescence-activated cell sorting (FACS). The novel Sf9-GFP/Rep packaging cell line-dependent OneBac system is versatile and flexible yet maintains high virus yields and thus can facilitate large-scale rAAV production and rAAV-based gene therapy.
Materials and Methods
Plasmid construction
The plasmids pAAV-MCS, pAAV-RC2, pAAV-RC8, pAAV-RC9, pAAV-GFP, pAAV-helper (Clontech), and pFast.Bac.dual as well as Escherichia coli. DH10Bac (Invitrogen) were maintained in our laboratory. The plasmid pIR-hr2-RBE-rep78 was a kind gift from Prof. Sergei Zolotukhin.16 The pFD/Rep, pFD/Cap2, pFD/Cap8, and pFD/Cap9 and pFD/Cap2-(ITR-GFP) constructs were described previously.18 In brief, the Cap gene was inserted downstream from the P10 promoter in the pFast.Bac.dual (pFD) plasmid, and translated using atypical initiation codons (CTG for VP1, ACG for VP2, and ATG for VP3) to generate VP1, VP2, and VP3 in a stoichiometry of around 1:1:10 from one transcript without alternate splicing.9,12 Meanwhile, we digested the pFD plasmid with MroI and SpeI to insert the ITR-GOI adjacent to the Cap gene. For simple detection of rAAV, we used a GFP reporter gene expression cassette driven by the cytomegalovirus (CMV) promoter. To generate this cassette, we first constructed a pFD/Cap2-(ITR-GFP) shuttle plasmid. A codon-modified AAV2 Rep gene expressing a dual-functional Rep78- and Rep52-encoding mRNA transcript termed pFD/Rep was artificially synthesized and inserted under control of the polyhedrin (PH) promoter in the pFD plasmid.15 The pFD/Cap8-(ITR-GFP) and pFD/Cap9-(ITR-GFP) constructs were created by substituting the Cap8 or Cap9 genes from pFD/Cap8 and pFD/Cap9 for the Cap2 gene in pFD/Cap2-(ITR-GFP) digested with PacI and NheI.
To insert the GFP gene fused to the C-terminus of the blasticidin (BSD) resistance gene and separated by a foot-and-mouth disease virus 2A proteinase (FMDV.2A) self-cleaving peptide into pIR-hr2-RBE-rep78, we synthesized a 498 bp fragment termed PT2AN (showed in Supplementary Data) from the psimpleT vector. Then, the GFP reporter gene was amplified by Nhe-GFP-F (5′-ACTGGCTAGCATGGTGAGCAAGGGCGAGGAG-3′) and GFP-Bsiw-R (5′-CAGTCGTACGTTACTTGTACAGCTCGTCCAT GCC-3′) primers and inserted into the psimpleT-PT2AN vector digested with NheI and BsiwI. Finally, the PT2AN-GFP fragment was inserted into pIR-hr2-RBE-rep78 digested with PvuII and NdeI to create pIR-hr2-RBE-Rep78-GFP. The BEVs were generated according to the Bac-to-Bac Baculovirus Expression System protocol (Invitrogen).
Cell culture
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (FBS) in 100 mm or six-well plates at 37°C, 5% CO2. Polyethylenimine (PEI) Max (Polysciences) was used for the transfection of HEK293 cells. Adherent cultured Sf9 cells were maintained in Grace's Insect Medium (Gibco) supplemented with 10% FBS in 100 mm or six-well plates at 28°C. Sf9 cells cultured in suspension were maintained in Sf-900II SFM (Gibco) in shake flasks at 28°C, 130 rpm. Cellfectin II reagent (Invitrogen) was used for Sf9 cells transfection.
Construction of Sf9-GFP/Rep stable packaging cell line and FACS
Sf9 cells were cultured in six-well plates to 70% confluence before transfection with the pIR-hr2-RBE-Rep78-GFP plasmid and culturing in medium with Blasticidin S antibiotic (25 μg/mL, BSD; Invitrogen). After 3 weeks of antibiotic selection, most of the surviving cells were positive for green fluorescence. The Sf9-GFP/Rep cells were then screened using green FACS with flow cytometry (BD FACSAria III). Cells were resuspended in phosphate-buffered saline (PBS, pH 7.0) supplemented with 0.1% pluronic acid F-68 (PF68; Gibco). The cell sorting rate was ∼2 × 106 cells per hour. Cells having green fluorescence intensity that was within the top 10% of all measured values were collected into a tube at 4°C using PBS supplemented with 0.1% PF68 as a running buffer. After sorting, cells were pelleted by centrifugation at 300 g for 10 min and maintained in culture medium with BSD. The sorted Sf9-GFP/Rep cells were propagated and expanded to establish the Sf9-GFP/Rep stable packaging cell line.
rAAV production and purification
For HEK293 cells, rAAV production was carried out using a traditional triple-plasmid transfection method as described previously.22 HEK293 cells at 80% confluence were cotransfected with pAAV-RC2, pAAV-GFP, and pAAV-helper using PEI reagent. For Sf9 and Sf9-GFP/Rep cells, adherent cultured insect cells at 80% confluence or a suspension of cultured insect cells at 3 × 106 cells/mL density was infected with BEV at a multiplicity of infection (MOI) of 3. Cells were harvested 3 days postinfection and used for downstream purification involving an iodixanol step density gradient centrifugation as described previously.23,24 In brief, cells were suspended in lysis buffer (50 mM Tris, 2 mM MgCl2, pH 7.5) at a density of 2 × 107 cells/mL, then lysed by three freeze–thaw cycles in liquid nitrogen and a 37°C bath. The crude lysates were digested with 50 U/mL benzonase (Sigma) with addition of 150 mM NaCl at 37°C for 1 h, then clarified by centrifugation at 2,500 g for 15 min and filtered. The iodixanol step density gradients (15%, 25%, 40%, and 58%) were diluted in PBS-MK buffer (1 × PBS, 1 mM MgCl2, 2.5 mM KCl) from 60% iodixanol (OptiPrep; Sigma) and then were successively underlaid in 39 mL Quick-Seal tubes (Beckman Coulter). Supernatants from the crude lysates were then gently overlaid onto the gradient. After centrifugation for 2 h in a 70 Ti rotor at 340,000 g and 37°C, the fraction obtained from the 40% phase was dialyzed against PBS and then ultrafiltered and concentrated with Amicon Ultra-15 centrifugal filter units (MWCO, 100 kDa; Millipore). The purified rAAVs were stored at −80°C.
Virus titration
Virus titers were analyzed by quantitative PCR assay using the iQ SYBR Green Supermix kit (Bio-Rad). Standard curves were generated with 10-fold serial dilutions of standard plasmids. The BEV sample treatment was an adaptation of a previously described alkaline polyethylene glycol-based method.16 The BEV titering primers were specific for the AcMNPV gene Ac-IE-01, Bac-F (5′-CCGTAACGGACCT CGTACTT-3′), and Bac-R (5′-CCGTTGGGATTTGTGGTAAC-3′). The purified rAAVs were used to evaluate the rAAV yield. Samples of purified rAAVs were treated as described previously.17 The rAAV titering primers were specific for the CMV promoter, CMV-F (5′-TCCGCGTTACATAACTTACGG-3′), and CMV-R (5′-GGGCGTACTTGGCATATGAT-3′).
In vitro transduction assay
HEK293 cells and Sf9 cells were seeded in 96-well plates at 80% confluence. Samples of rAAVs, BEVs, and supernatants from different cell lysates were heat treated in a water bath at 60°C for 30 min. The heat-treated and untreated control samples were then added to 96-well plates to infect either HEK293 cells or Sf9 cells. Two days postinfection, the GFP expression was observed by fluorescence microscopy.
Western blotting, silver staining, and electron microscopy
Samples were denatured and separated using 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The anti-Rep monoclonal antibody (clone 303.9; Progen) and anti-AAV VP1/VP2/VP3 monoclonal antibody (clone B1; Progen) were used to detect the AAV Rep and Cap proteins, respectively. GFPs were detected with a rabbit anti-GFP antibody (ab290; Abcam). β-Tubulin proteins were detected with a monoclonal antibody (66240-1-lg; Proteintech). For silver staining, fixed gels were stained with a Fast Silver Stain Kit (Beyotime). For electron microscopy, ∼5 μL purified rAAVs was loaded onto a 400-mesh carbon-coated copper grid for 2 min, and then removed by adsorption to filter paper. The grid was negatively stained with 2% phosphotungstic acid (pH 7.4) for 2 min, and then washed with a drop of water. The grids were air-dired and examined with a transmission electron microscope (TEM; Hitachi H-7000A).
rAAV in vivo transduction assay
All procedures were approved by the Animal Care and Use Committees at the Wuhan Institute of Physics and Mathematics, the Chinese Academy of Sciences. Virus transduction analysis was performed using 8-week-old male C57BL/6 mice. In brief, ∼200 nL purified rAAV (2.0 × 108 VG) was stereotaxically microinjected into the dentate gyrus (DG) of mouse brains. Three weeks after injection, the mice were sacrificed. The brains were removed, fixed in 4% PFA overnight, dehydrated in 30% sucrose, and then sectioned into 40-μm slices. The fixed slices were stained with DAPI and imaged using a virtual slide microscope (Olympus VS120).
Results
Generation of the novel BEV/Cap-(ITR-GFP)
Currently available inducible Sf9 packaging cell line-based rAAV production systems use a strategy in which expression of stably integrated Rep and Cap helper genes is silenced in the Sf9 cell line until infection with BEV/(ITR-GOI).16 This strategy avoids cell toxicity caused by constitutive expression of the Rep and Cap genes.25 However, integration of the Cap gene into the Sf9/Cap-Rep cell genome decreases the flexibility for production of rAAV having diverse Cap gene serotypes. Therefore, we aimed to improve the Sf9/Cap-Rep packaging cell line-dependent OneBac system by optimizing the distribution of rAAV packaging elements (namely, the Rep gene, Cap gene, and ITR-GOI between the BEV and Sf9 cell genomes. To optimize the BEV, we integrated the Cap gene and ITR-GOI into a single BEV, which avoids complex screening procedures needed to confirm integration of the Cap gene into the Sf9 stable packaging cell line.
To construct the novel dual-functional BEV/Cap-(ITR-GFP), we optimized the organization of the Cap gene expression cassette and ITR-GOI. Meanwhile, to facilitate the organization of the Cap and Rep gene open reading frames, we used the ribosome leaky scanning mechanism as described previously.15 The BEV/Rep was also constructed for rAAV production and functional validation with BEV/Cap-(ITR-GFP). Schematic diagrams of the two constructs are shown in Fig. 1A. Representative experiments used traditional rAAV2 for functional verification.
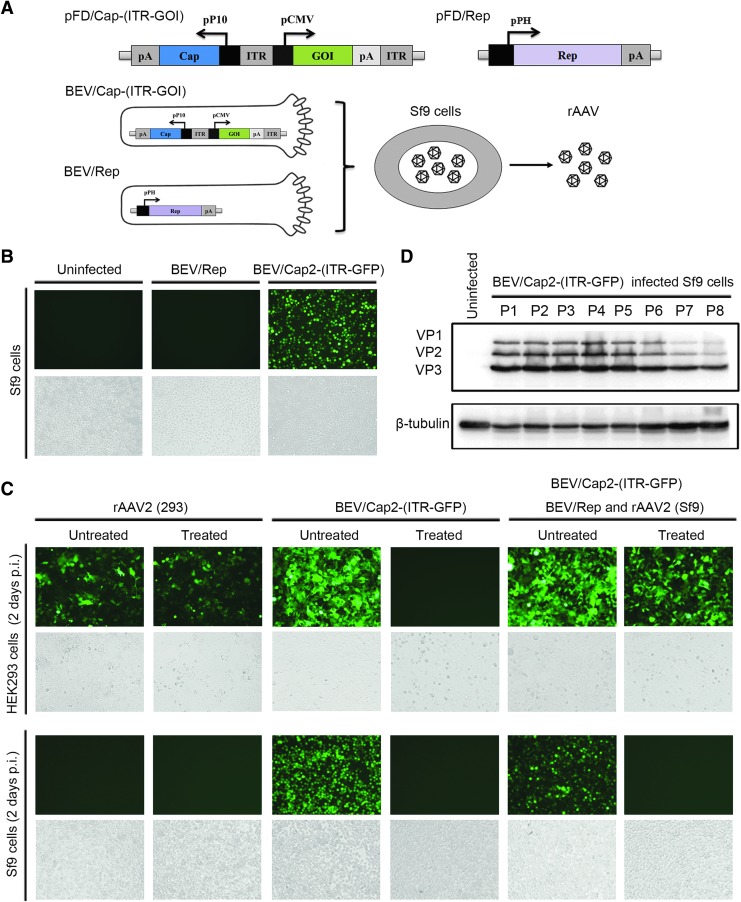
Figure 1.
Schematic of the BEV/Cap-(ITR-GFP) and BEV/Rep constructions and verification for the rAAV production upon the two BEVs coinfection of Sf9 cells. (A) Schematic of pFD/Cap-(ITR-GFP) and pFD/Rep constructs, and the production of rAAV upon BEV/Cap-(ITR-GFP)- and BEV/Rep-coinfected Sf9 cells. Functional elements are depicted as open boxes. (B) The images of Sf9 cells infected with BEV/Rep or BEV/Cap2-(ITR-GFP) were observed under fluorescence microscope. (C) The images of 96-well plate-cultured HEK293 and Sf9 cells infected with AAV2 (293 cell derived) at an MOI of 5,000, BEV/Cap2-(ITR-GFP) (10 μL culture supernatant per well), and supernatants of BEV/Cap2-(ITR-GFP)- and BEV/Rep-coinfected Sf9 cell lysate samples (10 μL crude lysate supernatant of 1 × 104 Sf9 cells per well) were observed under fluorescence microscope, respectively. p.i., postinfection; treated, 60°C treated for 30 min. Representative fluorescence and bright fields are shown. (D) Western blot analysis of Cap protein expressions upon serial passages of the BEV/Cap2-(ITR-GFP) from passage 1 (P1) to passage 8 (P8). The AAV Cap proteins and endogenous β-tubulin are indicated on the left. BEV, baculovirus expression vector; CMV, cytomegalovirus; GFP, green fluorescent protein; ITR, inverted terminal repeat; MOI, multiplicity of infection; pP10, baculovirus P10 promoter; pCMV, cytomegalovirus promoter; pPH, baculovirus polyhedron promoter; rAAV, recombinant adeno-associated virus. Color images available online.
To obtain BEV/Cap2-(ITR-GFP) and BEV/Rep, the DH10Bac strain was transformed with pFD/Cap2-(ITR-GFP) and pFD/Rep plasmids, respectively. Then, the positive Bacmid DNA was extracted and transfected into adherent cultured Sf9 cells. At 72 h post-transfection, the transfected Sf9 cells showed obvious cytopathic effects (CPE). Moreover, green fluorescence was visible in pFD/Cap2-(ITR-GFP)-transfected Sf9 cells (Fig. 1B). This result indicates that BEV/Cap2-(ITR-GFP) and BEV/Rep were successfully obtained.
The BEV/Cap-(ITR-GFP) is stable and capable of producing rAAV upon coinfection of Sf9 cells with BEV/Rep
We next used the two BEVs to verify their capacity for rAAV production (Fig. 1B, C). Sf9 cells were coinfected with BEV/Cap2-(ITR-GFP) and BEV/Rep at a 1:1 ratio (MOI = 3). At 72 h postinfection, the culture supernatants were collected and the cells were harvested. Differences in thermostability of nonenveloped AAV and enveloped BEV permitted the selective inactivation of BEV by heat treatment at 60°C for 30 min.26 We designed a simple HEK293 and Sf9 cell-based in vitro transduction assay by detecting GFP reporter gene expression to test the rAAV activity in crude cell lysates (Fig. 1C). For rAAV2 (293 cell derived) samples in 293 cell-based assays, both the heat-treated and untreated cells expressed GFP. This result indicates that rAAV2 is heat resistant. In Sf9 cell-based assays, neither untreated nor heat-treated cells expressed GFP, indicating that rAAV2 does not infect Sf9 cells. For BEV/Cap2-(ITR-GFP) samples in both 293 cell- and Sf9 cell-based assays, only the untreated cells expressed GFP, whereas the heat-treated cells did not. For crude lysate supernatants from BEV/Cap2-(ITR-GFP)- and BEV/Rep-coinfected cells that contain only a few nonsecreted BEVs and most of the rAAV, in 293 cell-based assays both untreated and treated cells expressed GFP, with slightly lower levels seen for the treated cells. These data indicate that GFP-expressing rAAV2 was present in large quantities. In Sf9 cell-based assays, the untreated cells expressed GFP, but the heat-treated cells did not (Fig. 1C). These results demonstrate that rAAV2 was produced in BEV/Cap2-(ITR-GFP)- and BEV/Rep-coinfected Sf9 cells. Taken together, these findings showed that the novel BEV/Cap2-(ITR-GFP) was sufficient for rAAV production.
Because the novel BEV/Cap2-(ITR-GFP) contained both the Cap gene and ITR-GOI, its stability is paramount for large-scale production of rAAVs. We investigated the stability of BEV/Cap2-(ITR-GFP) by analyzing Cap gene expression level upon amplification across serial passages from passage 1 (P1) to passage 8 (P8). For each passage, the Sf9 cells were infected with the BEVs at a lower MOI of 0.1, and the culture supernatants were then collected as BEV stocks at 72 h postinfection. Then, BEVs from P1 to P8 were used to infect Sf9 cells cultured in six-well plates at an MOI of 3. Western blotting of infected Sf9 cells at 72 h postinfection to detect Cap protein expression clearly showed three bands representing VP1/VP2/VP3 proteins (Fig. 1D). The Cap protein expression levels were stable from P1 to P5 before slightly decreasing after P5. Thus, BEV/Cap2-(ITR-GFP) was stable for at least five passages.
Generation of the Sf9-GFP/Rep stable packaging cell line
AAV2 Rep is typically used to produce rAAVs having different serotypes.27 Thus, a single packaging cell line that also provides Rep2 helper function would be more versatile for the production of different rAAVs. In addition, constitutive expression of the GFP reporter gene would facilitate screening of the Sf9 stable packaging cell line. Based on the previously described pIR-hr2-RBE-rep78 plasmid, which contains the Rep gene infection-inducible expression cassette,16 we constructed a modified plasmid pIR-hr2-RBE-rep78-GFP carrying an EGFP reporter gene fused with the FMDV.2A peptide downstream of the BSD resistance coding gene (Fig. 2A). A similar strategy to combine GFP and BSD resistance was used to select insect cell lines for recombinant protein expression.28 The GFP-BSD resistance fusion gene was also successfully used for selection of engineered recombinant poxviruses.29
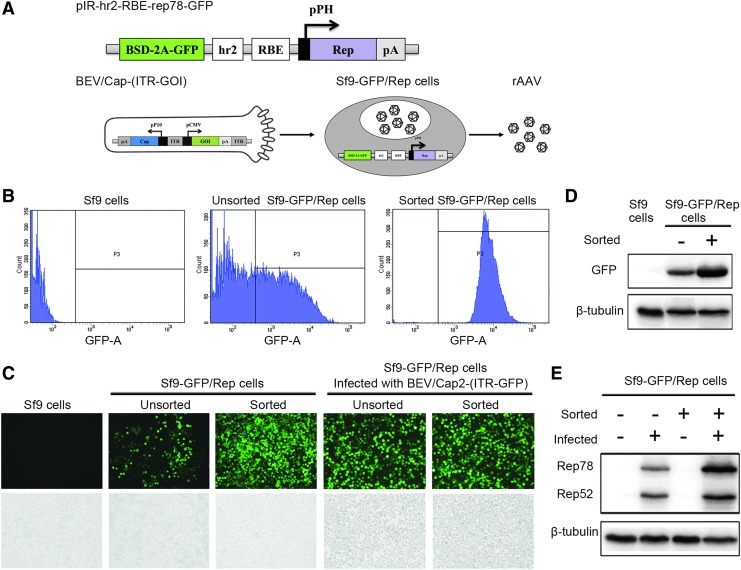
Figure 2.
Generation of the Sf9-GFP/Rep stable packaging cell line. (A) Schematic of the pIR-hr2-RBE-rep78-GFP construct and the production of rAAV upon BEV/Cap-(ITR-GFP) infection of Sf9-GFP/Rep cells. Functional elements are depicted as open boxes. BSD-2A-GFP, blasticidin and green fluorescent protein connected by an FMDV.2A self-cleavage peptide. (B) FACS analysis of Sf9 cells, unsorted Sf9-GFP/Rep cells, and sorted Sf9-GFP/Rep cells. (C) The images of Sf9 cells, unsorted Sf9-GFP/Rep cells, sorted Sf9-GFP/Rep cells, and BEV/Cap2-(ITR-GFP)-infected Sf9-GFP/Rep cells were observed under fluorescence microscope. Representative fluorescence and bright fields are shown. (D) Western blot analysis of GFP expression in unsorted and sorted Sf9-GFP/Rep cells. The GFP and endogenous β-tubulin are indicated on the left. (E) Western blot analysis of Rep protein-induced expression in unsorted and sorted Sf9-GFP/Rep cells upon BEV/Cap2-(ITR-GFP) infection. The Rep protein and endogenous β-tubulin are indicated on the left. FACS, fluorescence-activated cell sorting; FMDV.2A, foot-and-mouth disease virus 2A proteinase; RBE, Rep-binding element. Color images available online.
Sf9 cells were transfected with the plasmid pIR-hr2-RBE-rep78-GFP and cultured in medium with BSD selection. During the first 2 weeks with antibiotic screening, many cells died. After 3 weeks of BSD selection, most surviving cells had antibiotic resistance, and the proportion of GFP-positive cells increased before reaching stable levels (Fig. 2C).
In theory, the number of integrated Rep gene copies was consistent with both that of the BSD antibiotic resistance gene and GFP gene because these genes were integrated in the Sf9-GFP/Rep cell line genome upon transfection with the same pIR-hr2-RBE-rep78-GFP plasmid. To sort Sf9 cells having high number of copies of the integrated Rep gene and to establish a high-yield Sf9-GFP/Rep stable cell line, BSD-resistant Sf9 cells were sorted by FACS. In consideration of feasibility and operability, cells having green fluorescence intensity that fell within the top 10% of all fluorescence-positive cells were sorted once after a 3-week selection in BSD, and then expanded to establish the Sf9-GFP/Rep stable cell line. The FACS and microscope assays showed similar results wherein sorted cells were almost all GFP positive and had brighter green fluorescence relative to the unsorted cells (Fig. 2B, C). A Western blot assay also showed that the GFP expression level was substantially higher in the sorted cells than in the unsorted cells, whereas control Sf9 cells had no detectable GFP expression (Fig. 2D).
To verify the inducible expression of the integrated Rep gene, we analyzed the Rep protein levels by Western blot assay. Unsorted and sorted Sf9-GFP/Rep cells were first infected with BEV/Cap2-(ITR-GFP). At 72 h postinfection, the infected cells showed significant CPE (Fig. 2C). The Rep proteins were detectable in the infected Sf9-GFP/Rep cells (Fig. 2E, lanes 2 and 4), whereas the Rep protein was absent in the uninfected Sf9-GFP/Rep cells (Fig. 2E, lanes 1 and 3). The Rep protein expression was markedly higher in the sorted Sf9-GFP/Rep cells than in the unsorted Sf9-GFP/Rep cells (Fig. 2E, lanes 2 and 4). Together these results demonstrate the successful establishment of the Sf9-GFP/Rep stable packaging cell line.
rAAV yield from BEV/Cap-(ITR-GFP)-infected Sf9-GFP/Rep cells
To investigate whether rAAVs could be produced in BEV/Cap2-(ITR-GFP)-infected Sf9-GFP/Rep cells, we used a simple cell-based in vitro transduction assay with HEK293 and Sf9 cells to test the rAAV activity in crude cell lysates as described previously for Fig. 1C. At 3 days postinfection with BEV/Cap2-(ITR-GFP), Sf9-GFP/Rep cells were collected and an in vitro transduction assay was performed (Fig. 3A). Supernatants from BEV/Cap2-(ITR-GFP)-infected Sf9-GFP/Rep crude cell lysates contained a few nonsecreted BEVs and most of the rAAV. Meanwhile, in 293 cell-based assays, both heat-treated and untreated cells expressed GFP, with the treated cells showing slightly lower GFP fluorescence intensity than the untreated cells. This result indicated that a high yield of rAAV2 expressing GFP was generated. As expected, in Sf9 cell-based assays, the untreated cells expressed GFP but the heat-treated cells did not (Fig. 3A), which also indicates that rAAV2 could be successfully produced from BEV/Cap2-(ITR-GFP)-infected Sf9-GFP/Rep cells.
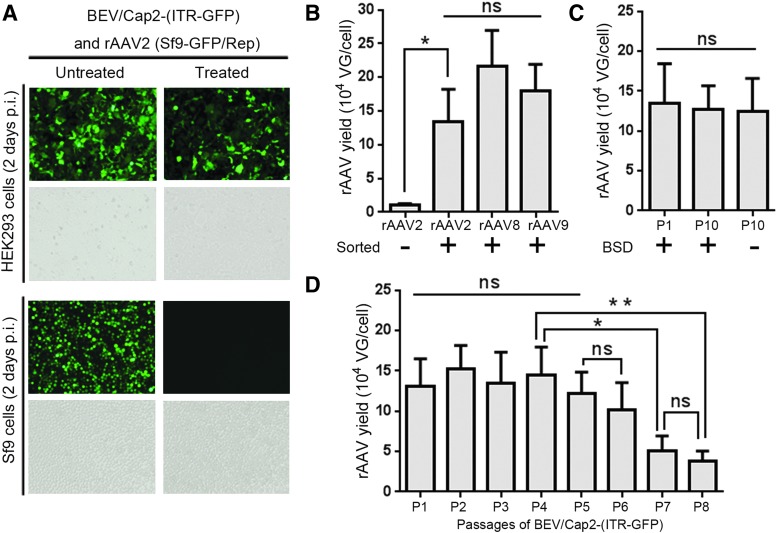
Figure 3.
Verification of rAAV production in BEV/Cap-(ITR-GFP)-infected Sf9-GFP/Rep cells and analysis of the rAAVs yields of the new OneBac system. (A) The images of 96-well plate-cultured HEK293 and Sf9 cells infected with supernatants of BEV/Cap2-(ITR-GFP)-infected Sf9-GFP/Rep cell lysate samples (10 μL crude lysate supernatants of 1 × 104 Sf9-GFP/Rep cells per well) were observed under fluorescence microscope, respectively. p.i., postinfection; treated, 60°C treated for 30 min. Representative fluorescence and bright fields are shown. (B) Yields analysis of rAAV2 derived from the BEV/Cap2-(ITR-GFP)-infected unsorted Sf9-GFP/Rep cells, and rAAVs derived from the BEV/Cap2-(ITR-GFP)-, BEV/Cap8-(ITR-GFP)-, and BEV/Cap9-(ITR-GFP)-infected sorted Sf9-GFP/Rep cells. (C) Yields analysis of rAAV2 derived from the BEV/Cap2-(ITR-GFP)-infected sorted Sf9-GFP/Rep cells (passage 1, P1) and Sf9-GFP/Rep cells (passage 10, P10). (D) Yields analysis of rAAV2 derived from the BEV/Cap2-(ITR-GFP)-infected sorted Sf9-GFP/Rep cells from passage 1 (P1) to passage 8 (P8) of the BEV. The suspension cultured Sf9-GFP/Rep cells were infected with BEVs (MOI = 3). After 72 h postinfection, the infected cells were harvested. The rAAV yields were determined as VG per cell. Experiments were performed in triplicate. All values are presented as mean ± standard deviation. Asterisks depict Tukey's multiple comparison test significance between groups after an ANOVA test, *p < 0.05 and **p < 0.01; BSD, blasticidin; ns, no significant differences; VG, vector genomes. Color images available online.
We speculated that the GFP expression level of the Sf9-GFP/Rep cell line might correlate well not only with integrated Rep gene copies but also with the rAAV yield. To examine this possibility, we investigated the yield of rAAV by infecting suspension-cultured Sf9-GFP/Rep cells with BEV/Cap2-(ITR-GFP). The rAAV2 yield from the sorted Sf9-GFP/Rep cells was 13.2-fold higher than that for unsorted Sf9-GFP/Rep cells (1.35 ± 0.46 × 105 VG/cell and 1.02 ± 0.24 × 104 VG/cell, respectively; Fig. 3B). Thus, the GFP expression level of Sf9-GFP/Rep cells could feasibly reflect the rAAV yield.
To evaluate the flexibility and rAAV yields of this new Sf9-GFP/Rep packaging cell line-dependent OneBac system, we made side-by-side comparisons of yields for rAAV2, rAAV8, and rAAV9 from 30 mL suspension-cultured Sf9-GFP/Rep cells infected with BEV/Cap2-(ITR-GFP), BEV/Cap8-(ITR-GFP), or BEV/Cap9-(ITR-GFP). The yields for rAAV2, rAAV8, and rAAV9 were 1.35 ± 0.46 × 105 VG/cell, 2.16 ± 0.53 × 105 VG/cell, and 1.80 ± 0.39 × 105 VG/cell, respectively (Fig. 3B). These results demonstrate that the new OneBac system is highly flexible and can accommodate a variety of serotypes for rAAV production with relatively high yields.
The rAAV yield stability of the new OneBac system is key for large-scale production, and is closely related to both the BEV/Cap2-(ITR-GFP) and Sf9-GFP/Rep stable cell line. We first verified the rAAV yield stability of the modified Sf9-GFP/Rep cell line. The original sorted Sf9-GFP/Rep cells (termed as passage 1) were cultured in serial suspensions using 1:8 dilutions for 10 continuous passages with or without BSD. After 10 passages, rAAV yields were tested and compared with the original rAAV yields (passage 1) upon infection with BEV/Cap2-(ITR-GFP). The rAAV yield level of the Sf9-GFP/Rep packaging cells remained stable after 10 passages (all >105 VG/cell), even in the absence of BSD selection (Fig. 3C). We next investigated the rAAV yield stability from BEV/Cap2-(ITR-GFP). Suspension-cultured Sf9-GFP/Rep cells (passage 3) were infected with BEV/Cap2-(ITR-GFP) at P1 to P8. The rAAV yields were maintained at high levels and showed no obvious decrease from P1 to P5 (all >105 VG/cell). After P5, the rAAV yields gradually decreased to 3.73 ± 1.32 × 104 VG/cell at P8 (Fig. 3D). These results demonstrated that the new OneBac system is highly stable for large-scale production of rAAV.
Characterization of rAAVs derived from the new OneBac system
To investigate the properties of rAAVs produced using the new OneBac system, we purified the newly generated rAAVs using iodixanol step density gradient centrifugation and analyses of biophysical properties and transduction activity. The purity of the rAAV was analyzed by SDS-PAGE with silver staining. The rAAVs exhibited high purity and Sf9 cell-derived rAAV2, rAAV8 and rAAV9 were similar to those derived from 293 cells. Three bands corresponding to the AAV VP1 (87 kDa), VP2 (73 kDa), and VP3 (62 kDa) capsid proteins were apparent on the gels, and had the expected ratio of ∼1:1:10 (Fig. 4A).
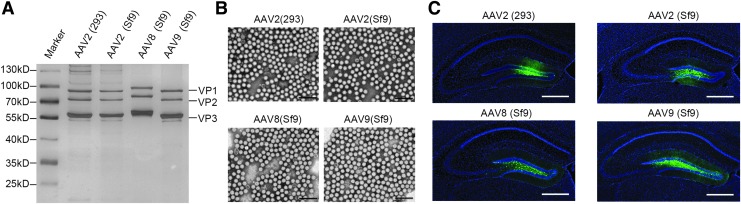
Figure 4.
Characterization of purified rAAVs derived from BEV/Cap-(ITR-GFP)-infected Sf9-GFP/Rep cells. (A) Silver stain of purified rAAVs. Samples of HEK293 cell-derived AAV2 (293), Sf9-GFP/Rep cell-derived AAV2 (Sf9), AAV8 (Sf9), and AAV9 (Sf9) were separated by 10% SDS-PAGE, and then visualized by silver staining. About 1.0 × 1010 VG rAAVs were loaded per lane. (B) The images of purified rAAVs were observed under electron microscope. Original magnification: × 100,000; scale bars: 100 nm. (C) Transduction activity analysis of the rAAVs produced by the new OneBac system. Fluorescence microscope observation of GFP expression in the mice brain DG regions. Green: GFP, Blue: DAPI, Scale bars: 500 μm. Representative fields are shown. DG, dentate gyrus; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Color images available online.
Negative-stained viral particles were also visualized by TEM. The 293 cell-derived rAAV2 and rAAV2, rAAV8 and rAAV9 derived from the new OneBac system had similar uniform spherical structures. The virions appeared as hexagonally shaped particles having a 20–25 nm in diameter. Empty particles were distinguished by an electron-dense central region of the capsid that was absent in full particles. The TEM images indicated that the population of particles that had full content after iodixanol gradient purification of rAAV2, rAAV8, and rAAV9 derived from the new OneBac system was very high at ∼92.3%, 93.9%, and 95.8%, respectively (Fig. 4B).
The rAAV transduction activities were next analyzed in brains of C57 mice. The rAAV genomes carried a CMV promoter-driven GFP reporter gene expression cassette. About 2.0 × 108 VG of each rAAV were injected into the mouse brain DG. Three weeks after infection, the mice were sacrificed for analysis of transduction activity. High levels of GFP expression and distinct structures of the GFP-labeled nerve cells were visible near the injection sites in mice treated with rAAV2, rAAV8, and rAAV9 derived from the new OneBac system, and the patterns were similar to those seen for mice injected with 293 cells (Fig. 4C). These results demonstrate that rAAVs produced using the new OneBac system had high quality and activity.
Discussion
The BEV-mediated rAAV production system (Bac system) has been intensively developed and promoted for preclinical and clinical gene therapy applications.9,30 In 2012, the first approved rAAV-based gene therapy drug Glybera was produced using the Bac system.6 In recent years, earlier multi-BEV infection-based Bac systems have been gradually replaced by single BEV infection-based Bac systems. The important advantage is that the single BEV infection overcomes the inherently low ratio of multi-BEV coinfection.12 According to the distributions of three rAAV packaging elements between the BEV and Sf9 cell genomes, current single BEV infection-derived Bac systems can be classified into two types: a packaging cell line-independent Bac system such as that described by Wu et al. that integrates all three rAAV packaging elements in a single BEV18 and the Sf9/Cap-Rep packaging cell line-dependent Bac system like that developed by Aslanidi et al. that involves a BEV infection-inducible Sf9/Cap-Rep packaging cell line.16 Both of these single BEV infection-derived Bac systems can produce high rAAV yields, exceeding 105 VG/cell. The major difference between the two types is the regulation mechanisms for Rep and Cap helper gene expression. Compared with the packaging cell line-independent Bac system, the current Sf9/Cap-Rep packaging cell line-dependent Bac system lacks versatility and flexibility. Moreover, for production of different rAAV serotypes, different cell lines that integrate both Rep and Cap genes are needed and screening of the packaging cell lines that allow efficient production of rAAV is challenging.17
To overcome the disadvantages of current Sf9/Cap-Rep packaging cell line-dependent Bac systems, we optimized transfer of the Cap gene from the Sf9 cells to the BEV. Here we successfully constructed a novel dual-functional BEV/Cap-(ITR-GFP) that carries both the Cap gene and the ITR-GOI, and verified its capacity to yield rAAV upon coinfection of Sf9 cells with BEV/Rep. The stability of the BEV is critical for large-scale production of rAAV. We demonstrated that Cap protein expression from BEV/Cap2-(ITR-GFP) was stable for at least five passages (Fig. 1D), which is similar to that previously reported for BEV/Cap-Rep that showed stable expression of Rep and Cap proteins over five or seven continuous passages, respectively.14,15 Furthermore, the rAAV yield from BEV/Cap2-(ITR-GFP) was stable and exhibited no apparent decrease for at least five passages (Fig. 3D). Compared with the previously reported BEV/Cap2-(ITR-GFP)-Rep, which was stable for only four passages,18 the BEV/Cap2-(ITR-GFP) has a better performance in terms of stability. The improved BEV stability might be due to the separation of ITR-GOI and Rep in that the ITR sequences are susceptible to Rep-mediated cleavage.31 Taken together, the higher stability of BEV/Cap-(ITR-GFP) will likely be beneficial for larger scale production of rAAV upon infection of Sf9-GFP/Rep packaging cells.
For the Sf9-GFP/Rep packaging cell line, the constitutively expressed GFP reporter gene allows the use of FACS that markedly reduces the difficulty of packaging cell line screening. FACS is widely used to select high-producing subpopulations of insect cells.32,33 Instead of time-consuming cell line screening methods involving limiting dilution and colony picking, our screening method requires only sorting of the population of cells having the highest (top 10%) green fluorescence intensity using FACS (Fig. 2B). The sorted cells were then expanded to establish the Sf9-GFP/Rep stable packaging cell line. Compared with the those of unsorted cells, expression levels of both GFP and infection-induced Rep expression showed apparent increases in the sorted Sf9-GFP/Rep cells (Fig. 2D, E). This result indicates that the GFP expression level correlates well with that of Rep gene expression induction. Furthermore, the rAAV2 yield was ∼13.2-fold higher from the sorted Sf9-GFP/Rep cells than from the unsorted cells (Fig. 3B). This result indicates that the GFP expression level can reflect the efficiency of the rAAV yield for Sf9-GFP/Rep cells. Thus, optimization of FACS parameters for Sf9-GFP/Rep cells could likely be used to further improve rAAV yield. The rAAV yield from Sf9-GFP/Rep stable cell lines was stable even in the absence of BSD for at least 10 passages (Fig. 3C). This result is consistent with findings for previously reported Sf9/Cap-Rep packaging cell lines that are genetically stable even in the absence of BSD, and can be cultured and passaged similarly to naive Sf9 cells.17 Furthermore, this finding suggests that sorted Sf9-GFP/Rep stable cell lines are suitable for successive scale-up.
Notably, the high-yield Sf9/Cap-Rep stable cell lines (F3 clone, B8 clone) often had a three- to nine-fold increase in the number of Cap gene copies relative to that for the Rep gene, as was previously reported.16,21 These data may indicate that the demand for Cap gene expression in Sf9 cells is higher during rAAV production. The different demands for Rep and Cap genes expression could introduce some complexity into screening of the high-yield Sf9/Cap-Rep cell line. Even if two different fluorescent reporter genes are used to monitor Cap and Rep expression levels, sorting high-yield Sf9/Cap-Rep packaging cell lines using FACS with undefined parameters for two genes can be difficult. However, moderate levels of Rep expression in Sf9 packaging cells could be sufficient for rAAV production. We, therefore, posit that relocating the Cap gene from the Sf9 cell genome to BEV is a better option, since, for the Sf9-GFP/Rep packaging cell line, the GFP reporter gene can easily reflect the Rep gene expression. Our results verified that the GFP expression level of the Sf9-GFP/Rep cell line might indeed correlate well with rAAV yield (Fig. 3B).
Modification of infection-inducible constructs for the Sf9/Cap-Rep cell line showed enhanced infectivity with rAAV5 production and minimal encapsidation of foreign DNA for production of rAAV1, rAAV2, rAAV5, and rAAV8 vectors,19,20 which demonstrates the unique advantages of Sf9 insect cells for good manufacturing practice-grade vector production relative to that for manufacture using HEK293 cells.34 In addition, a recently developed optogenetic gene expression system based on VP-EL222/C120 elements achieved light-gated transcription with spatial and temporal resolution in several mammalian cell lines and intact zebrafish embryos.35 We have verified that the VP-EL222/C120 elements can also regulate the expression of fluorescence reporter genes in Sf9 cells (Wu et al. unpublished data). The optogenetic gene expression strategy might thus be a potential approach for construction of an Sf9 packaging cell line having inducible Rep gene expression. The flexible optogenetic regulation of Rep helper gene expression both spatially and temporally could be helpful for rAAV production. Also, deletion of the baculoviral cathepsin (v-CATH) gene could avoid proteolytic degradation of AAV VP1/VP2 capsid proteins.36 Further efforts would be helpful for improving the Sf9-GFP/Rep packaging cell line-dependent OneBac system.
The new OneBac system described here produces rAAV2, rAAV8, and rAAV9 at relatively high yield levels, similar to the current state of large-scale rAAV production systems (Fig. 3B). The new OneBac system-derived rAAV2, rAAV8, and rAAV9 were of high quality, had similar biophysical properties to those described for rAAV2 produced from 293 cells, and showed high in vivo transduction activities (Fig. 4). In conclusion, by optimizing the distributions of rAAV packaging elements between the BEV and Sf9 cell genomes, we developed a novel modified Sf9-GFP/Rep packaging cell line and a novel BEV/Cap-(ITR-GOI)-based OneBac system that has improved versatility and flexibility. This study contributes to the new generation of Sf9 packaging cell line-dependent Bac systems for large-scale rAAV production.
Supplementary Material
Acknowledgments
We are grateful to Prof. Sergei Zolotukhin for providing the plasmid pIR-hr2-RBE-rep78, to Liting Luo and Lingling Xu (Core Facility Center, Wuhan Institute of Physics and Mathematics) for technical assistance with centrifugation and microscope imaging, and to Ding Gao and Juan Min (Core Facility Center, Wuhan Institute of Virology) for technical assistance with electron microscopy and FACS.
Authors' Contributions
Y.W. designed and performed the experiments, analyzed the data, and wrote and edited the article. T.M. and L.J. performed the experiments and acquired the data. Z.H. and R.D. carried out viral vector production. T.Y. contributed to animal experiments. F.X. supervised the project and edited the article.
Author Disclosure
No competing financial interests exist.
Funding Information
This study was supported financially by the National Natural Science Foundation of China (Grant No. 31500868 to Y.W. and Grant No. 31771156 to F.X.), the National Basic Research Program (973 Program) of China (Grant No. 2015CB755600 to F.X.), Strategic Priority Research Program (B) (Grant No. XDB0205005 to F.X.), and the State Key Program of National Natural Science Foundation of China (Grant No. 31830035 to F.X.).
Supplemenary Material
References
- 1. Hastie E, Samulski RJ. Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success-a personal perspective. Hum Gene Ther 2015;26:257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ayuso E, Mingozzi F, Bosch F. Production, purification and characterization of adeno-associated vectors. Curr Gene Ther 2010;10:423–436 [DOI] [PubMed] [Google Scholar]
- 3. Daya S, Berns KI. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 2008;21:583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet 2011;12:341–355 [DOI] [PubMed] [Google Scholar]
- 5. Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 1998;72:2224–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bryant LM, Christopher DM, Giles AR, et al. Lessons learned from the clinical development and market authorization of Glybera. Hum Gene Ther Clin Dev 2013;24:55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dias MF, Joo K, Kemp JA, et al. Molecular genetics and emerging therapies for retinitis pigmentosa: basic research and clinical perspectives. Prog Retin Eye Res 2018;63:107–131 [DOI] [PubMed] [Google Scholar]
- 8. Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet 2014;15:445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galibert L, Merten OW. Latest developments in the large-scale production of adeno-associated virus vectors in insect cells toward the treatment of neuromuscular diseases. J Invertebr Pathol 2011;107 Suppl:S80–S93 [DOI] [PubMed] [Google Scholar]
- 10. Kotin RM. Large-scale recombinant adeno-associated virus production. Hum Mol Genet 2011;20:R2–R6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cecchini S, Virag T, Kotin RM. Reproducible high yields of recombinant adeno-associated virus produced using invertebrate cells in 0.02- to 200-liter cultures. Hum Gene Ther 2011;22:1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Urabe M, Ding C, Kotin RM. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther 2002;13:1935–1943 [DOI] [PubMed] [Google Scholar]
- 13. Kohlbrenner E, Aslanidi G, Nash K, et al. Successful production of pseudotyped rAAV vectors using a modified baculovirus expression system. Mol Ther 2005;12:1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen H. Intron splicing-mediated expression of AAV Rep and Cap genes and production of AAV vectors in insect cells. Mol Ther 2008;16:924–930 [DOI] [PubMed] [Google Scholar]
- 15. Smith RH, Levy JR, Kotin RM. A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol Ther 2009;17:1888–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aslanidi G, Lamb K, Zolotukhin S. An inducible system for highly efficient production of recombinant adeno-associated virus (rAAV) vectors in insect Sf9 cells. Proc Natl Acad Sci U S A 2009;106:5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mietzsch M, Grasse S, Zurawski C, et al. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1–12 vectors for gene therapy. Hum Gene Ther 2014;25:212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu Y, Jiang L, Geng H, et al. A recombinant baculovirus efficiently generates recombinant adeno-associated virus vectors in cultured insect cells and larvae. Mol Ther Methods Clin Dev 2018;10:38–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mietzsch M, Casteleyn V, Weger S, et al. OneBac 2.0: Sf9 cell lines for production of AAV5 vectors with enhanced infectivity and minimal encapsidation of foreign DNA. Hum Gene Ther 2015;26:688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mietzsch M, Hering H, Hammer EM, et al. OneBac 2.0: Sf9 cell lines for production of AAV1, AAV2, and AAV8 vectors with minimal encapsidation of foreign DNA. Hum Gene Ther Methods 2017;28:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Joshi PRH, Cervera L, Ahmed I, et al. Achieving high-yield production of functional AAV5 gene delivery vectors via fedbatch in an insect cell-one baculovirus system. Mol Ther Methods Clin Dev 2019;13:279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nat Protoc 2006;1:1412–1428 [DOI] [PubMed] [Google Scholar]
- 23. Zolotukhin S, Byrne BJ, Mason E, et al. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther 1999;6:973–985 [DOI] [PubMed] [Google Scholar]
- 24. Strobel B, Miller FD, Rist W, et al. Comparative analysis of cesium chloride- and iodixanol-based purification of recombinant adeno-associated viral vectors for preclinical applications. Hum Gene Ther Methods 2015;26:147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Q, Chen F, Trempe JP. Characterization of cell lines that inducibly express the adeno-associated virus Rep proteins. J Virol 1994;68:4847–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu YK, Yang CJ, Liu CL, et al. Using a fed-batch culture strategy to enhance rAAV production in the baculovirus/insect cell system. J Biosci Bioeng 2010;110:187–193 [DOI] [PubMed] [Google Scholar]
- 27. Rabinowitz JE, Rolling F, Li C, et al. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J Virol 2002;76:791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zitzmann J, Schreiber C, Eichmann J, et al. Single-cell cloning enables the selection of more productive Drosophila melanogaster S2 cells for recombinant protein expression. Biotechnol Rep (Amst) 2018;19:e00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wong YC, Lin LC, Melo-Silva CR, et al. Engineering recombinant poxviruses using a compact GFP-blasticidin resistance fusion gene for selection. J Virol Methods 2011;171:295–298 [DOI] [PubMed] [Google Scholar]
- 30. Cecchini S, Negrete A, Kotin RM. Toward exascale production of recombinant adeno-associated virus for gene transfer applications. Gene Ther 2008;15:823–830 [DOI] [PubMed] [Google Scholar]
- 31. Li Z, Brister JR, Im DS, et al. Characterization of the adenoassociated virus Rep protein complex formed on the viral origin of DNA replication. Virology 2003;313:364–376 [DOI] [PubMed] [Google Scholar]
- 32. Kato T, Yoshizuka K, Park EY. New strategy for rapid isolation of stable cell lines from DNA-transformed insect cells using fluorescence activated cell-sorting. J Biotechnol 2010;147:102–107 [DOI] [PubMed] [Google Scholar]
- 33. Vidigal J, Dias MM, Fernandes F, et al. A cell sorting protocol for selecting high-producing sub-populations of Sf9 and High Five cells. J Biotechnol 2013;168:436–439 [DOI] [PubMed] [Google Scholar]
- 34. Kondratov O, Marsic D, Crosson SM, et al. Direct head-to-head evaluation of recombinant adeno-associated viral vectors manufactured in human versus insect cells. Mol Ther 2017;25:2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Motta-Mena LB, Reade A, Mallory MJ, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol 2014;10:196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galibert L, Savy A, Dickx Y, et al. Origins of truncated supplementary capsid proteins in rAAV8 vectors produced with the baculovirus system. PLoS One 2018;13:e0207414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.