Abstract
The amyloid aggregation of peptides and proteins is a hallmark of neurological disorders and type 2 diabetes. Human islet amyloid polypeptide (IAPP), co-secreted with insulin by pancreatic β-cells, plays dual roles in both glycemic control and the pathology of type 2 diabetes. While IAPP can activate the NLRP3 inflammasome and modulate cellular autophagy, apoptosis and extracellular matrix metabolism, no data is available concerning intracellular protein expression upon exposure to the polypeptide. More surprisingly, how intracellular protein expression is modulated by nanoparticle inhibitors of protein aggregation remains entirely unknown. In this study, we first examined the changing proteomes of βTC6, a pancreatic β-cell line, upon exposure to monomeric, oligomeric and fibrillar IAPP, and detailed cellular protein expression rescued by graphene quantum dots (GQDs), an IAPP inhibitor. We found that 29 proteins were significantly dysregulated by the IAPP species, while majority of these proteins were nucleotide-binding proteins. Collectively, our liquid chromatography tandem–mass spectrometry, fluorescence quenching, helium ion microscopy, cytotoxicity and discreet molecular dynamics simulations data revealed a remarkable capacity of GQDs in regulating aberrant protein expression through H-bonding and hydrophobic interactions, pointing to nanomedicine as a new frontier against human amyloid diseases.
Keywords: IAPP, oligomer, amyloid, protein expression, graphene quantum dot
Graphical Abstract
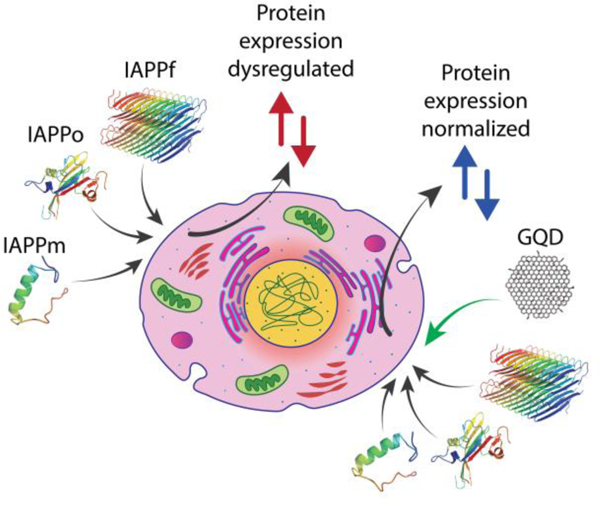
Exposure to monomeric and aggregating human islet amyloid polypeptide induced differential dysregulation of protein expression in pancreatic β-cells, which was effectively mitigated by two-dimensional graphene quantum dots.
Summary of the work:
Exposure to human islet amyloid polypeptide in different aggregation states induced differential dysregulation of protein expression in pancreatic β-cells, which was rescued by graphene quantum dots.
1. Introduction
Human islet amyloid polypeptide (IAPP) is a 37-residue hormone co-secreted and co-stored with insulin in pancreatic β-cells, and is co-released with the latter for satiety signaling [1]. Environmental factors of pH (4.5–5 within β-cells and 7.2 extracellularly), physiological metals (such as Ca2+ and Zn2+), glucose homeostasis, insulin, chaperones and chaperone-like proteins (e.g., serum albumin) can trigger the aberrant aggregation of IAPP from monomers to oligomers, protofibrils and amyloid fibrils, thereby eliciting toxicity to β-cells. The implications of IAPP toxicity range from cell degeneration to insulin deficiency and type 2 diabetes (T2D), a disease debilitating 370 million people worldwide [2–5].
Mounting evidence has implicated the oligomers and protofibrils, among all forms of amyloid protein aggregates, as the most toxic [6, 7]. One working mechanism is that the toxicity of amyloid proteins is rendered through their transformations from random to alpha helical- and then β-sheet-rich conformations, upon their N-termini initiated association with cell membranes [1, 8]. As a result, oligomeric proteins can increase porosity and fluidity of the membranes, impact concentration gradients and transport of biometals and nutrients, and compromise integrity of cell molecular machines and organelles to trigger a host of cellular responses ranging from reactive oxygen species (ROS) production, autophagy and apoptosis, to extracellular matrix metabolism and, ultimately, cell death [1].
To date, only a handful of studies have detailed the adverse effects of amyloid proteins on cellular protein and gene expressions [9, 10]. Among the data available, it has been shown that amyloid beta (Aβ) impacted pathways important for neuronal physiology and dysregulation in Alzheimer’s disease (AD), including cell adhesion, vesicle trafficking, actin cytoskeleton dynamics and insulin signaling. Compared with Aβ, little known about the effects of IAPP aggregation species on the proteome of pancreatic β-cells, and no data is available concerning the effects of IAPP aggregation inhibitors on intracellular protein expression, two glaring knowledge deficiencies which hinder the development of new therapeutics against T2D.
Among current mitigation strategies, aggregation inhibition with small molecules (epigallocatechin-3-gallate or resveratrol) [11, 12] and nanomaterials (OH-terminated PAMAM dendrimers, chiral silica nanohelices, beta-lactoglobulin-coated gold nanoparticles, silver nanoparticles, iron oxide nanoparticles, and carbon nanotubes, etc.) has been shown effective for the prevention of IAPP toxicity in vitro (buffer or beta cells), ex vivo (beta-cell islets) or in vivo (zebrafish embryos) [13–16]. Graphene quantum dots (GQDs), specifically, are a miniaturized derivative of graphene sheets possessing no toxicity up to 500 µg/mL [17] and a superior translocation efficacy across the blood-brain barrier [18]. Carboxylated GQDs, for example, have been recently demonstrated as potent inhibitors against IAPP amyloidogenesis in zebrafish embryos [19] as well as synucleinopathies in the brain of a Parkinson’s disease (PD) mouse model [18]. The unique amphiphilic 2D structure and large surface area of GQDs are especially suited for interfacing amphiphilic amyloid proteins than serum proteins, as amyloid proteins evolve from disordered monomers to increasingly hydrophobic and toxic oligomers and protofibrils [1]. Such increasing hydrophobicity entails high affinity of amyloid proteins for the hydrophobic GQD moieties, while the OH groups of the GQDs further mediate H-bonding with amyloid proteins to enhance their association [16]. The autofluorescence of GQDs, resulting from their quantum confinement effect, may be utilized for tracking the biodistribution of the nanomaterial in vivo, among other applications.
In recent years, high-throughput proteomic techniques have been used to evaluate the effect of amyloidogenesis on the initiation and progression of amyloid diseases such as AD [20, 21]. To facilitate the development of nanomedicines as a new frontier against amyloid diseases, in this study we used liquid chromatography tandem–mass spectrometry (LC-MS/MS) to characterize the protein expression profiles of the βTC6 pancreatic β-cell line exposed to three structurally representative IAPP species, namely, IAPP monomers (IAPPm), oligomers (IAPPo) and amyloid fibrils (IAPPf). Driven by the findings we further studied protein expression regulated by hydroxylated GQDs (20 μg/mL) subsequent to β-cell exposure to the IAPP species to explore the potential of the nanomaterial against cell degeneration in T2D. We found proteins which regulate gene expression, especially nucleotide-binding proteins, were most prone to the toxic forms of IAPP, while the hydroxylated GQDs partially reversed the dysregulation effects of IAPPm and IAPPo through H-bonding and hydrophobic interactions for the majority of these proteins, implicating the potential of the nanomaterial against cell degeneration in T2D.
2. Results and discussion
2.1. Characterization of GQDs and three IAPP aggregation states
Transmission electron microscopy (TEM) imaging indicated that GQDs were relatively uniform and monodisperse disks of ~3.8 ± 0.5 nm in lateral dimensions (Fig. S1a, Electronic Supplementary Material or ESM). The zeta potential of the hydroxylated GQDs was charge neutral at 0.78 ± 0.1 mV in aqueous solutions, which differed from the negative charge of carboxylated GQDs used in the study by Wang et al. against IAPP amyloidogenesis in vivo [19]. The use of hydroxylated GQDs in the present study intended to utilize their capacity in forming H-bonding with IAPP for aggregation and toxicity inhibition. The hydrophobic moieties of GQDs also favored binding with the hydrophobic and toxic IAPP species to drive their aggregation off pathway. The Fourier transform infrared (FTIR) spectrum of the GQDs indicated the presence of oxygen-functional groups, confirming the enrichment of hydroxyl groups (-OH) with a peak at wavenumber of ~3,400 cm−1, and the main components of C=C, C-O and CH bonds at wavenumbers of ~1,560, 1,040, and 2,900 cm−1, respectively (Fig. S2a). Furthermore, thermogravimetric analysis (TGA) of GQDs (Fig. S2b) indicated decreases in mass as a function of rising temperature. The TGA curve exhibited two steps of weight losses between 50 and 150 °C under N2, with a ~50% weight loss occurring at 100 °C. These weight losses corresponded to the detachment of abundant oxygen-containing groups such as hydroxyls and C-O within the GQDs, in addition to evaporation of solvent at the early stage of the weight loss. The maximum fluorescence excitation and emission of GQDs occurred at 350 and 440 nm, respectively (Fig. S3a).
Freshly dissolved hexafluoro-2-propanol (HFIP)-treated IAPP in Milli-Q water assumed the monomeric form (IAPPm). Oligomeric IAPP (IAPPo) was prepared by incubating the HFIP-treated IAPP in Milli-Q water at room temperature for 1 h, while full-length IAPP mature fibrils (IAPPf) were obtained by incubating the peptide at room temperature for more than 24 h. All three fibrillization states were confirmed by TEM imaging (Fig. S1c–e) and by atomic force microscopy in our previous experimental studies [11, 22].
IAPPf remodelling by the GQDs was imaged by TEM (Fig. S1f). FiberApp [23] was used to quantify the key mesoscopic parameters, including the contour length and persistence length of the fibrils w/o the GQDs. Specifically, IAPPf were micrometers in length and ~10–15 nm in diameter (Fig. S1e inset), consistent with the literature[1]. Upon interaction with the GQDs, the preformed fibrils assumed heterogeneous length distribution with major fraction (40%) being 400 nm in length (Fig. S1f inset) as determined by FiberApp. Such analysis was not feasible for IAPPm and IAPPo, due to the ineffectiveness of FiberApp for analyzing non-tubular morphologies.
2.2. Fluorescence quenching revealed IAPP-GQD association
To probe the interactions between GQDs and IAPP, the quenching of GQD autofluorescence by the three IAPP aggregation states of different concentrations (10–30 µM) was recorded. At fixed excitation wavelength of 350 nm and GQD concentration of 300 μg/mL, the peak emission fluorescence intensity of GQDs at 440 nm decreased linearly for IAPPm, IAPPo and IAPPf (R2 = 0.985, R2 = 0.932, R2 = 0.999) with increasing IAPP concentrations (Fig. S3b–g). These reductions in GQD fluorescence intensity, at 7% for IAPPm, 11% for IAPPo and 9% for IAPPf each of 20 µM (the concentration used for viability, ROS and proteomics assays), suggest associations of the monomeric, protofibrils and fibrils of IAPP during aggregation. The binding of the GQDs and the IAPP species was further studied by computer simulations (Figs. 1&S4).
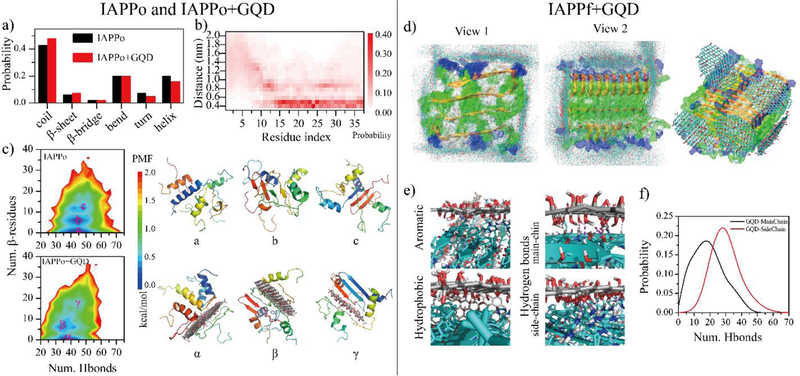
Figure 1.
DMD simulations of GQD interacting with IAPPo and IAPPf. (a) Secondary structure of IAPPo in the absence and presence of the GQD. (b) Distance probability distribution of each IAPPo residue to the GQD. (c) Two-dimensional potential mean force (2D-PMF) of the IAPPo w/o the GQD. Three different types of IAPPo structures – including helical (a/α), partial β-sheet (b/β) and β-sheet rich (c/γ) - of the IAPPo (top right) and the IAPPo with the GQD (bottom right) were also presented with the coordinates labelled in the PMF on the left. DMD simulation of GQDs interacting with IAPPf. (d) Structures of GQD sheets bound on the surface of an IAPPf, obtained from different independent DMD trajectories. (d) Distribution of GQD nanosheet atoms (carbons in cyan and oxygens in red) on the surface of the IAPPf by overlaying snapshots obtained from ten independent DMD trajectories. A representative GQD-IAPPf complex structure is shown to the right. (e) The GQD-IAPPf binding interfaces highlight the dominant inter-molecular interactions. (f) Probability distribution of the number of hydrogen bonds formed by GQD with the main-chains or side-chains of the IAPPf.
2.3. Discrete molecular dynamics (DMD) simulations of IAPP-GQD association
To complement the quenching assay, molecular details of the interactions between IAPP and GQDs were further studied by discrete molecular dynamics (DMD) simulations (Figs. 1&S4). The IAPPm mainly adopted unstructured conformation (i.e., random coils and bends) with some partial helical and transient β-sheet structures (Fig. S4a). In the presence of a GQD nanosheet, the structured conformation (helix and β-sheet) of IAPPm was completely destructed. The distribution of hydrogen bonds formed by main-chain atoms of IAPPm in both systems revealed the hydrogen bonds in IAPPm were significantly cleaved off by the GQD (Fig. S4b). Moreover, IAPPm had a strong propensity of forming hydrogen bonds with the GQD. The minimum distance probability distribution of each IAPPm residue relative to the GQD revealed that all of the residues in IAPPm adsorbed onto the GQD surface with a most populated distance occurring at 0.4~0.5 nm (Fig. S4c). The binding site distribution of IAPPm on the GQD surface indicated that IAPPm mainly bound to the nanosheet surface rather than the edges (Fig. S4d). Overall, the structured conformation (helix or partial β-sheet, Fig. S4e) of IAPPm was nearly completely converted into unstructured conformation (Fig. S4f) in the presence of the GQD.
To investigate the effects of GQD on the conformations of IAPPo, we simulated four IAPP peptides with and without a GQD nanosheet. The random coil and β-sheet structures were slightly enhanced while the helical structures were weakly reduced in the presence of the GQD (Fig. 1a). Different from IAPPm, only residues 14–37 of IAPPo displayed high binding probabilities with the GQD. In addition to the first residue layer at ~0.5 nm from the nanosheet surface, a second layer of residues at ~0.9 nm from the nanosheet was also observed, though with a lower population. Examination of the simulation snapshots revealed that residues in this region could form some β-sheets perpendicular to the GQD (Fig. 1b). The N-terminal residues 1–10 were mainly unbound. The conformational distribution of IAPPo was also analyzed by computing 2D-PMF (i.e., two-dimensional potential of mean force) as a function of the total number of hydrogen bonds formed by main-chain atoms (Num. Hbonds) and the number of residues adopting the β-sheet structure (Num. β-sheet residues). For both systems, IAPPo predominantly adopted a helical-rich structure with a large number of hydrogen bonds and a low β-sheet content (Fig. 1c–(a), the α state). Besides, β-sheet containing conformations were also observed (Fig. 1c–(b&c), the β and γ states). Similarly to IAPPm, the total number hydrogen bonds in IAPPo was also suppressed with the GQD. Overall, these results indicate that GQDs displayed distinct effects on the secondary structure of IAPPm and IAPPo. In addition, the N-terminal residues in IAPPo (Fig. 1b) display much weaker binding strength with than those residues in IAPPm (Fig. S4c).
To understand the conformation of IAPPf interacting with a GQD, we simulated five GQD nanosheets binding with a 20-peptide IAPP fibril (Fig. 1d–f). Ten independent DMD simulations which started with different initial structures and velocities were performed. The GQD nanosheet could bind both the elongation and secondary nucleation surfaces of the IAPPf to hinder the fibril growth (Fig. 1d). The driving forces of the binding were mainly aromatic, hydrophobic and hydrogen bonds (Fig. 1e). The GQD displayed a high propensity to form hydrogen bonds with both the main-chains and side-chains of the IAPPf (Fig. 1f). The strong binding affinity was indicated by a high potential energy gain of ~320 kcal/mol upon binding five GQD nanosheets averaged over ten different independent simulations (Fig. S5). In addition, the coating of GQDs on the surface of IAPPf also reduced direct interaction of the fibril with cells.
2.4. Graphene quantum dots mitigated IAPP toxicity in vitro
As with Aβ and alpha synuclein, oligomeric/protofibrillar forms are also known as the most toxic IAPP species associated with β-cell degeneration [14, 24, 25]. To quantify the toxicities of IAPP structures with and without GQDs to βTC6 cells, viability (Fig. 2a, for 15 h of incubation; Fig. S6, for 24 h of incubation in complete Dulbecco’s modified Eagle’s medium with 15% fetal bovine serum) and ROS assays were performed. As expected, the most potent IAPP species in inducing cell death, membrane damage as well as intracellular ROS generation were IAPPo (Fig. 2; Figs. S6&S7). Neither IAPPm nor IAPPf caused significant cell death or membrane damage, however, IAPPm induced comparable levels of intracellular ROS with IAPPo (Fig. 2j), suggesting a higher potential for toxicity of IAPPm compared to IAPPf. Helium ion microscopy (HIM) further demonstrated, consistently with the viability and ROS assays, that IAPPo treated cells acquired a severely deformed morphology and membrane blebbing compared to untreated control cells and IAPPm and IAPPf treated cells (Fig. 2b, d, f, h; Fig. S7). However, the morphology of IAPPm treated cells was more similar to IAPPo-incubated cells, likely caused by IAPPm induced oxidative stress as indicated by relatively high intracellular ROS levels (Fig. 2j). Co-incubation of pancreatic β-cells with IAPPm or IAPPo and GQDs significantly mitigated IAPP-induced toxicity. In the case of IAPPm, GQDs were potent in alleviating IAPP toxicity by ~34%. With IAPPo, the GQDs reduced IAPP cytotoxicity by 76%, confirming a stronger binding affinity between the two species. (Fig. 2a). Consistent with the viability assay, significant reduction in intracellular ROS (24.5% for IAPPm and 45.4% for IAPPo) was observed for IAPP with the GQDs, indicating the mitigation power of the nanoparticles against ROS production by the IAPP species (Fig. 2j), especially by IAPPo. This may be attributed to the relatively high hydrophobicity of IAPPo and protofibrils [24, 25], which should bind preferably with the hydrophobic moieties of the GQDs than with other cellular proteins to sequester the peptide from aggregation and eliciting toxicity. In comparison, no cell damage was noticeable in the HIM images when the cells were exposed to IAPPm or IAPPo in the presence of the GQDs, and, as expected, no cell deformation was recorded after exposure to IAPPf with the GQDs (Fig. 2b–i). Consistently, confocal fluorescence microscopy indicated more cell damage (red fluorescence spots) by IAPPm and IAPPo than IAPPf, and the presence and cellular distributions of the IAPP aggregates (green fluorescence spots) (Fig. S8).
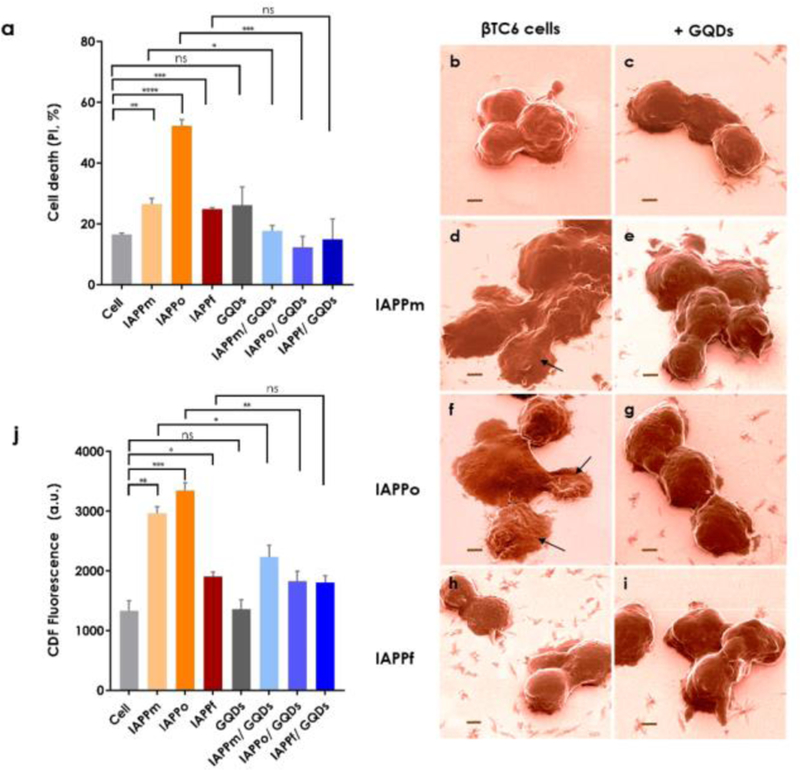
Figure 2.
Viability, morphology and ROS generation of βTC6 pancreatic β-cells exposed to the IAPP species with or without GQDs. (a) βTC6 cell toxicities upon exposure to IAPPm, IAPPo and IAPPf in the presence and absence of GQDs. The GQDs were effective in reducing IAPP toxicity of all structures, especially IAPPo as the most toxic species, indicating a strong hydrogen and hydrophobic binding affinity between GQDs and IAPP. PI: propidium iodide. (b-i) Helium ion microscopy images as a visual evidence on toxicity of IAPP species: (b) βTC6 cells as control, treated with (c) GQDs and (d, f, h) IAPPm, IAPPo and IAPPf and (e, g, i) IAPPm, IAPPo and IAPPf withGQDs, respectively. Arrows in d and f show the deformation of cell membranes induced by IAPPm and IAPPo, respectively. In the cases of IAPPf and mixture of IAPPf with GQDs no damage was observed. Scale bars: 2 µm. Cells were incubated with IAPP species, with or without GQDs, in the viability assay for 15 h and before HIM for 1 h. (j) Intracellular ROS levels upon exposure to IAPPm, IAPPo and IAPPf in the presence and absence of GQDs. Incubation: 1 h. The experiments were carried out in triplicate, error bars show standard deviations and q-value performed by 1% FDR rate approach by using the two-stage step-up method of benjamini krieger and yekutieli (ns: q ≥ 0.01, *: q < 0.01, **: q ≤ 0.001, ***: q ≤ 0.0001 and ****: q ≤ 0.00001). IAPP concentration: 20 µM.
2.5. Effects of IAPP on the proteome of pancreatic β-cells
Following the experimental design of proteomics analysis (Fig. 3a), we identified 1,608 proteins across all the samples (Fig. 3b). Label-free quantification analysis showed that IAPPm induced an over-expression of 11 proteins and under-expression of another 11 proteins. The addition of GQDs inhibited the IAPPm-induced over/under expression of 10 proteins, respectively. In comparison, IAPPo induced the over-expression of 10 proteins and under-expression of another 10 proteins, out of which the aberrant expressions of 19 proteins were mitigated by GQDs. The comparable capacities of IAPPm and IAPPo in protein dysregulation and ROS production (Figs. 2j&3c) may be attributed to the rapid aggregation of the peptide. One of the most amyloidogenic proteins known [26, 27], IAPPm could be converted to IAPPo in hours or less at micromolar concentrations [3, 11, 15, 28]. As expected, IAPPf, the least toxic form of IAPP, caused an over-expression of only one protein and under-expression of two other proteins, with no detectable changes in protein expression when cells were co-incubated with IAPPf and GQDs (Fig. 3c).
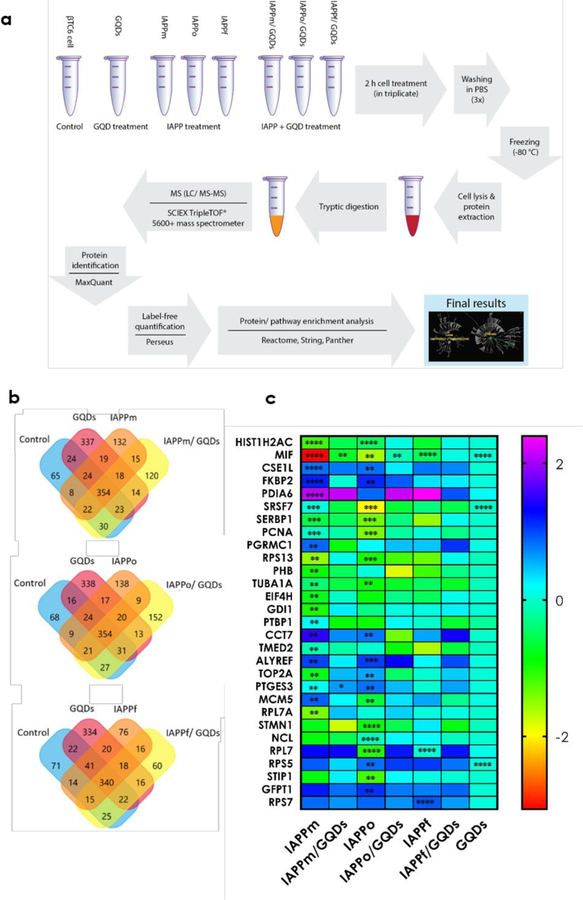
Figure 3.
Proteomics analysis of IAPP- and GQD-treated pancreatic β-cells. (a) Scheme of experimental procedures for quantifying up- and down-regulation of intracellular protein expression in β-cells exposed to IAPP species in the absence and presence of GQDs. (b) More than 1,600 proteins were identified across all replicates with more than 50% overlap within each condition. (c) In total, expressions of 29 proteins were altered by the different forms of IAPP treatments. Statistical analysis was done based on the multiple t-test for the average abundance of proteins in each condition compared to the untreated control sample. **, ***, and **** indicate adjusted p-values of p <0.01, p <0.001, and p <0.0001 respectively. The color gradient is the indicator of fold changes compared to the control sample. Based on the adjusted p-value analysis, GQDs alone induced no significant up or down protein expression. In the case of RPL7, both IAPPo and IAPPf showed significant dysregulation (p <0.0001). However, IAPPo and IAPPf with GQDs recovered from significant changes.
2.6. Pathway classification of differentially expressed proteins
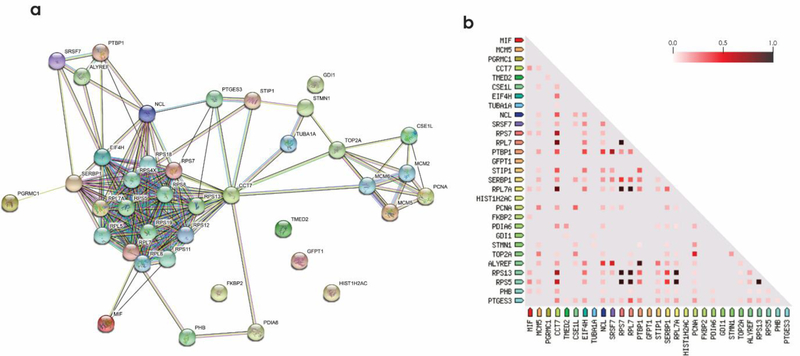
Biochemical pathway analysis was used to classify differentially expressed proteins from the β-cells treated with the three IAPP species. To achieve this, pathway classification was performed on the differentially expressed proteins identified in each comparison test by Reactome [29]. The canonical pathway analysis (Figs. 4&S9) of three treatments revealed that DNA-repair and replication, cell cycle, gene expression and metabolism of proteins were the pathways mainly affected by IAPPm and IAPPo. Unsurprisingly, the fewest number of pathways were affected by IAPPf. IAPPo has been reported to cause highest membrane disorders and cytotoxicity [24, 30]. Consistently, and for the first time, IAPPo were the most destructive structure in dysregulating the proteins pathways. We also found proteins in the same pathway could be regulated in different ways (up/down). Based on the differential expression of proteins following exposure to IAPP aggregates we visualized changes to protein networks using STRING [31, 32], as presented in Fig. 5 and Tables S1&S2.
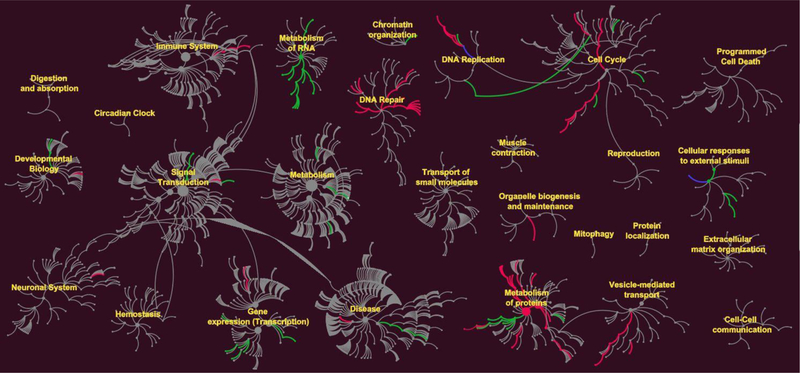
Figure 4.
Effects of IAPPo on up and down regulation of intracellular pathways. Reactome pathways nodes are represented as filled circles. Connections between pathways and sub-pathways (edges) are represented by lines. The green edges represent up regulation and red edges down regulation. In some pathways both up and down regulations of proteins were recorded, shown as blue lines. DNA repair, metabolism of proteins, gene expression and cell cycle pathways were dysregulated by IAPPo.
Figure 5.
Protein-protein interaction analysis and co-expression of dysregulated proteins by all three IAPP species. (a) STRING database was used for protein-protein interaction analysis with a minimum interaction score of 0.400. (b) Co-expression analysis of IAPP dysregulated proteins. The color code indicates the highest correlation of proteins as 1.0 (black) and the least as 0.0 (white). Enrichment analysis, molecular action legends and co-expression scores are shown in Table S1.
2.7. Nucleic acid binding proteins as most affected by IAPP
We used Panther Gene Ontology software [33] to determine the proteins mostly altered by IAPP in each condition. A total of 15 out of 29 altered proteins were “binding proteins”. Interestingly, 13 proteins were “nucleic acid-binding proteins”, including 3 DNA binding and 10 RNA binding (Table S1).
We found six ribosomal proteins, including RPL7, RPS5, RPS13, RPL7A, RPS7 and CCT7, differentially regulated by the IAPP species. A ribosomal protein is any of the proteins that, in conjunction with rRNA, makes up the ribosomal subunits involved in the cellular process of translation. Although ribosomal proteins are known for playing an essential role in ribosome assembly and protein translation, ribosome-independent functions have also been documented [34]. The roles of ribosomal proteins in amyloid diseases such as AD have been studied. For example, a new study shows that Aβ induced changes in multiple proteins involved in ribosomal machinery [35]. In addition, investigation on the chaperone activity of CCT7 for oxidative stress in neuronal apoptosis has been performed [36, 37]. In our study, specifically, the expressions of RPS13 and CCT7 increased in the β-cells treated with IAPPm and IAPPo. Although the regulation of RPS5 by IAPPm was not statistically significant, its expression was significantly decreased by IAPPo. Previously, the overexpression of RPS13 and RPS5 have been reported in AD [38]. We noted significant decreases in the expression of RPL7 by IAPPm and IAPPf and of RPL7A by IAPPm. However, to our knowledge, there are no reports on the dysregulation of these 60s ribosomal proteins in amyloidogenesis.
Alternative splicing has recently become a new mechanism for deciphering different diseases such as diabetes [39, 40] and AD, with RNA-splicing as a major genetic footprint to the diseases [41]. Here our results indicated IAPP could interfere with the regulation of mRNA splicing factor proteins, including nucleolin and SRSF7. SRSF7 was upregulated after treatment with IAPPm but downregulated by IAPPo.
There is increasing evidence that many RNA-binding proteins (RBPs) and RBP-regulated RNA networks are disrupted under diabetic conditions [39]. Here we found two altered RBPs in the β-cells exposed to the toxic forms of IAPP: both IAPPm and IAPPo increased the expression of SERBP1 but decreased the expression of Tho4.
All three IAPP species induced the expression of macrophage migration inhibitory factor (MIF) protein. MIF is a key proinflammatory cytokine involved in many inflammatory reactions and disorders. In addition to its inflammatory roles, MIF can form amyloid fibrils associated with amyloidogenic proteins such as Aβ and alpha synuclein during acid denaturing conditions [42]. Previous studies have suggested that binding of Aβ to microtubules may explain, in part, the mechanism of amyloid induced toxicity [43, 44]. In addition, the cross-interactions between Aβ with IAPP and insulin have been reported [45–47], reinforcing the importance of the findings in this study. Here we found that TUBA1A (a major constituent of microtubules) and STMN1 (involved in the regulation of microtubules) were regulated by IAPPm and IAPPo. Both IAPPm and IAPPo increased the expression of TUBA1A, while IAPPo decreased the expression of STMN1.
2.8. GQDs prevented IAPP-induced toxicity
The effects of GQDs on the inhibition of the fibrillization and toxicity of Aβ and IAPP have been shown in previous studies [19, 48]. In this study, we observed the remarkable effect of the hydroxylated GQDs on the inhibition of protein dysregulation, especially for the most toxic form of IAPPo. The GQD concentration used in this study, at 20 µg/mL, was one order of magnitude lower than its toxic concentration [17, 49, 50], but was adequate for mitigating the toxicity of IAPP at µM concentrations, a condition which triggers β-cell degeneration in vivo [1]. GQDs prevented or significantly decreased the level of protein dysregulation (compared with the untreated control) induced by all three forms of IAPP. As shown in Fig. 3c, 20 out of the 22 proteins that affected by IAPPm, 19 out of the 20 proteins affected by IAPPo and all dysregulation caused by IAPPf were mitigated by GQDs. The nanostructures themselves did not cause significant changes (or with low fold changes) on the protein expression of β cells. Interestingly, GQDs could prevent the adverse effects on the MIF proteins induced by all three IAPP species. PDIA6 protein catalyzes the formation, reduction, and isomerization of disulfide bonds in proteins and also acts as a chaperone in the inhibition of protein misfolding [51]. This protein was remarkably upregulated by IAPPm, but with GQDs the level of dysregulation induced by IAPPm was decreased to with no significant difference from untreated cells (the control). The dysregulation of the IAPP structures in contrast to the constructive effects of GQDs are consistent with the in vitro toxicity assay of IAPP on pancreatic β-cells (Fig. 2a).
3. Conclusions
Protein expression and cellular pathways regulated by IAPPm and the two major IAPP aggregating species IAPPo and IAPPf were studied along with the impact of hydroxylated GQDs on amyloidosis. Using a comprehensive LC-MS/MS approach, a total of 1,608 proteins were identified in all samples, where 29 proteins showed significant over/under expression, and among them over 30% were RNA and DNA binding proteins. Such aberrant protein expression was mitigated by the GQDs to various extents, among which the most significant effect was shown for IAPPo, the most toxic species of the three aggregation states. These protein expression profiles showed a good correlation with biophysical (fluorescence quenching and HIM imaging) and toxicity (viability and ROS assays) characterizations, where IAPPo elicited the most damage to β-cells and which was ameliorated by GQDs. The binding between the IAPP species and GQDs was a result of H-bonding and hydrophobic interaction, as revealed by atomistic DMD simulations. Together, this study has demonstrated the potency of nanoparticles in restoring protein expression against the toxicity elicited by amyloid protein aggregation, thereby facilitating the development and application of novel therapeutics against a range of amyloid diseases.
Supplementary Material
Acknowledgements
This work was conceived by PCK, and was by supported by ARC Project No. CE140100036 (Davis), NSF CAREER CBET-1553945 (Ding), NIH MIRA R35GM119691 (Ding), AFTAM Research Collaboration Award (Davis and Ke), the National Natural Science Foundation of China (No. 11904189) (Sun), and the Juvenile Diabetes Research Foundation (Purcell and Faridi). A.W.P. is supported by a Principal Research Fellowship from the Australian NHMRC. TEM imaging was performed at Bio21 Advanced Microscopy Facility, University of Melbourne. HIM imaging was performed at the MCFP platform, University of Melbourne by Dr. Anders Barlow.
Footnotes
Electronic Supplementary Material: Supplementary material (Materials and Methods, Figures S1-S9 in ESM and Tables S1 and S2 in ESM) is available in the online version of this article at https://doi.org/10.1007/s12274–019-2520–7.
References
- [1].Ke PC; Sani MA; Ding F; Kakinen A; Javed I; Separovic F; Davis TP; Mezzenga R Implications of peptide assemblies in amyloid diseases. Chem. Soc. Rev 2017, 46, 6492–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhang X; Clair JR; London E; Raleigh DP Islet amyloid polypeptide membrane interactions: effects of membrane composition. Biochemistry 2017, 56, 376–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cao P; Abedini A; Wang H; Tu LH; Zhang X; Schmidt AM; Raleigh DP Islet amyloid polypeptide toxicity and membrane interactions. Proc Natl Acad Sci USA 2013, 110, 19279–19284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Knowles TP; Vendruscolo M; Dobson CM The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol 2014, 15, 384–396. [DOI] [PubMed] [Google Scholar]
- [5].Sudhakar S; Kalipillai P; Santhosh PB; Mani E Role of surface charge of inhibitors on amyloid beta fibrillation. J. Phys. Chem. C 2017, 121, 6339–6348. [Google Scholar]
- [6].Haataja L; Gurlo T; Huang CJ; Butler PC Islet amyloid in type 2 diabetes, and the toxic oligomer hypothesis. Endoc. Rev 2008, 29, 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen X; Mao SS Titanium dioxide nanomaterials: synthesis, properties, modifications, and applications. Chem. Rev 2007, 107, 2891–2959. [DOI] [PubMed] [Google Scholar]
- [8].Fusco G; Chen SW; Williamson PTF; Cascella R;Perni M; Jarvis JA; Cecchi C; Vendruscolo M; Chiti F; Cremades N; Ying L; Dobson CM; De Simone A Structural basis of membrane disruption and cellular toxicity by alpha-synuclein oligomers. Science 2017, 358, 1440–1443. [DOI] [PubMed] [Google Scholar]
- [9].Sebollela A; Freitas-Correa L; Oliveira FF; Paula-Lima AC; Saraiva LM; Martins SM; Mota LD; Torres C; Alves-Leon S; de Souza JM; Carraro DM; Brentani H; De Felice FG; Ferreira ST Amyloid-beta oligomers induce differential gene expression in adult human brain slices. J. Biol. Chem 2012, 287, 7436–7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kong L.-n.; Zuo P.-p.; Mu L; Liu Y.-y.; Yang N Gene expression profile of amyloid beta protein-injected mouse model for Alzheimer disease. Acta Pharmacologica Sinica 2005, 26, 666. [DOI] [PubMed] [Google Scholar]
- [11].Kakinen A; Adamcik J; Wang B;Ge X; Mezzenga R; Davis TP; Ding F; Ke PC Nanoscale inhibition of polymorphic and ambidextrous IAPP amyloid aggregation with small molecules. Nano Res 2018, 11, 3636–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nedumpully-Govindan P; Kakinen A; Pilkington EH; Davis TP; Ke PC; Ding F Stabilizing off-pathway oligomers by polyphenol nanoassemblies for IAPP aggregation inhibition. Sci. Rep 2016, 6, 19463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gurzov EN; Wang B; Pilkington EH; Chen P; Kakinen A; Stanley WJ; Litwak SA; Hanssen EG; Davis TP; Ding F; Ke PC Inhibition of hIAPP amyloid aggregation and pancreatic beta-cell toxicity by OH-terminated PAMAM dendrimer. Small 2016, 12, 1615–1626. [DOI] [PubMed] [Google Scholar]
- [14].Pilkington EH; Lai M;Ge X; Stanley WJ; Wang B;Wang M; Kakinen A; Sani MA; Whittaker MR; Gurzov EN; Ding F; Quinn JF; Davis TP; Ke PC Star polymers reduce islet amyloid polypeptide toxicity via accelerated amyloid aggregation. Biomacromolecules 2017, 18, 4249–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Javed I; Yu T;Peng G; Sánchez-Ferrer A; Faridi A; Kakinen A;Zhao M; Mezzenga R; Davis TP; Lin S; Ke PC In vivo mitigation of amyloidogenesis through functional–pathogenic double-protein coronae. Nano Lett 2018, 18, 5797–5804. [DOI] [PubMed] [Google Scholar]
- [16].Wang M; Sun Y; Cao X; Peng G; Javed I; Kakinen A; Davis TP; Lin S; Liu J; Ding F; Ke PC Graphene quantum dots against human IAPP aggregation and toxicity in vivo. Nanoscale 2018, 10, 19995–20006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nurunnabi M; Khatun Z; Huh KM; Park SY; Lee DY; Cho KJ; Lee Y-K In vivo biodistribution and toxicology of carboxylated graphene quantum dots. ACS Nano 2013, 7, 6858–6867. [DOI] [PubMed] [Google Scholar]
- [18].Kim D; Yoo JM; Hwang H; Lee J; Lee SH; Yun SP; Park MJ; Lee M; Choi S; Kwon SH; Lee S; Kwon S-H; Kim S; Park YJ; Kinoshita M; Lee Y-H; Shin S; Paik SR; Lee SJ; Lee S; Hong BH; Ko HS Graphene quantum dots prevent α-synucleinopathy in Parkinson’s disease. Nat. Nanotechnol 2018, 13, 812–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang M; Kakinen A; Pilkington EH; Davis TP; Ke PC Differential effects of silver and iron oxide nanoparticles on IAPP amyloid aggregation. Biomaterials Sci 2017, 5, 485–493. [DOI] [PubMed] [Google Scholar]
- [20].Portelius E; Zetterberg H; Gobom J; Andreasson U; Blennow K Targeted proteomics in Alzheimer’s disease: focus on amyloid-beta. Expert Rev. Proteomics 2008, 5, 225–237. [DOI] [PubMed] [Google Scholar]
- [21].Savas JN; Wang YZ; DeNardo LA; Martinez-Bartolome S; McClatchy DB; Hark TJ; Shanks NF; Cozzolino KA; Lavallee-Adam M; Smukowski SN; Park SK; Kelly JW; Koo EH; Nakagawa T; Masliah E; Ghosh A; Yates JR Amyloid accumulation drives proteome-wide alterations in mouse models of Alzheimer’s disease-like pathology. Cell Rep 2017, 21, 2614–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sun Y; Kakinen A; Xing Y; Pilkington EH; Davis TP; Ke PC; Ding F Nucleation of beta-rich oligomers and beta-barrels in the early aggregation of human islet amyloid polypeptide. Biochim. Biophys. Acta, Mol. Basis Dis 2019, 1865, 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Usov I; Mezzenga R FiberApp: An Open-Source Software for tracking and analyzing polymers, filaments, biomacromolecules, and fibrous objects. Macromolecules 2015, 48, 1269–1280. [Google Scholar]
- [24].Lin CY; Gurlo T; Kayed R; Butler AE; Haataja L; Glabe CG; Butler PC Toxic human islet amyloid polypeptide (h-IAPP) oligomers are intracellular, and vaccination to induce anti-toxic oligomer antibodies does not prevent h-IAPP-induced beta-cell apoptosis in h-IAPP transgenic mice. Diabetes 2007, 56, 1324–1332. [DOI] [PubMed] [Google Scholar]
- [25].Faridi A; Sun Y; Okazaki Y; Peng G; Gao J; Kakinen A; Faridi P; Zhao M; Javed I; Purcell AW; Davis TP; Lin S; Oda R; Ding F; Ke PC Mitigating human IAPP amyloidogenesis in vivo with chiral silica nanoribbons. Small 2018, 14, e1802825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kayed R; Head E; Thompson JL; McIntire TM; Milton SC; Cotman CW; Glabe CG Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science, 2003, 300, 486–489. [DOI] [PubMed] [Google Scholar]
- [27].Lopes DH; Colin C; Degaki TL; de Sousa AC; Vieira MN; Sebollela A; Martinez AM; Bloch C Jr.; Ferreira ST; Sogayar MC Amyloidogenicity and cytotoxicity of recombinant mature human islet amyloid polypeptide (rhIAPP). J. Biol. Chem 2004, 279, 42803–42810. [DOI] [PubMed] [Google Scholar]
- [28].Krotee P; Rodriguez JA; Sawaya MR; Cascio D; Reyes FE; Shi D; Hattne J; Nannenga BL; Oskarsson ME; Philipp S; Griner S; Jiang L; Glabe CG; Westermark GT; Gonen T; Eisenberg DS Atomic structures of fibrillar segments of hIAPP suggest tightly mated β-sheets are important for cytotoxicity. eLife 2017, 6, e19273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Joshi-Tope G; Gillespie M; Vastrik I; D’Eustachio P; Schmidt E; de Bono B; Jassal B; Gopinath GR; Wu GR; Matthews L; Lewis S; Birney E; Stein L Reactome: a knowledgebase of biological pathways. Nucleic Acids Res 2005, 33, D428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cremades N; Cohen SIA; Deas E; Abramov AY; Chen AY; Orte A; Sandal M; Clarke RW; Dunne P; Aprile FA; Bertoncini CW; Wood NW; Knowles TPJ; Dobson CM; Klenerman D Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell 2012, 149, 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Szklarczyk D; Franceschini A; Wyder S; Forslund K; Heller D; Huerta-Cepas J; Simonovic M; Roth A; Santos A; Tsafou KP; Kuhn M; Bork P; Jensen LJ; von Mering C STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015, 43, D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Szklarczyk D; Morris JH; Cook H; Kuhn M; Wyder S; Simonovic M; Santos A; Doncheva NT; Roth A; Bork P; Jensen LJ; von Mering C The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017, 45, D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mi H; Huang X; Muruganujan A; Tang H; Mills C; Kang D; Thomas PD PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 2017, 45, D183–d189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhou X; Liao W-J; Liao J-M; Liao P; Lu H Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol 2015, 7, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Deng L; Pushpitha K; Joseph C; Gupta V; Rajput R; Chitranshi N; Dheer Y; Amirkhani A; Kamath K; Pascovici D; Wu JX; Salekdeh GH; Haynes PA; Graham SL; Gupta VK; Mirzaei M Amyloid β induces early changes in the ribosomal machinery, cytoskeletal organization and oxidative phosphorylation in retinal photoreceptor cells. Front. Mol. Neurosci 2019, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Anantharam V; Lehrmann E; Kanthasamy A; Yang Y; Banerjee P; Becker KG; Freed WJ; Kanthasamy AG Microarray analysis of oxidative stress regulated genes in mesencephalic dopaminergic neuronal cells: relevance to oxidative damage in Parkinson’s disease. Neurochem. Int 2007, 50, 834–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pavel M; Imarisio S; Menzies FM; Jimenez-Sanchez M; Siddiqi FH;Wu X;Renna M;O’Kane CJ;Crowther DC; Rubinsztein DC CCT complex restricts neuropathogenic protein aggregation via autophagy. Nat. Comm 2016, 7, 13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Garcia-Esparcia P; Sideris-Lampretsas G; Hernandez-Ortega K; Grau-Rivera O; Sklaviadis T; Gelpi E; Ferrer I Altered mechanisms of protein synthesis in frontal cortex in Alzheimer disease and a mouse model. Am. J. Neurodegener. Disease 2017, 6, 15–25. [PMC free article] [PubMed] [Google Scholar]
- [39].Nutter CA; Kuyumcu-Martinez MN Emerging roles of RNA-binding proteins in diabetes and their therapeutic potential in diabetic complications. Wiley Interdiscip. Rev. RNA 2018, 9, e1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Juan-Mateu J; Villate O; Eizirik DL MECHANISMS IN ENDOCRINOLOGY: Alternative splicing: the new frontier in diabetes research. European J. Endocrinology 2016, 174, R225–R238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Love JE; Hayden EJ; Rohn TT Alternative splicing in Alzheimer’s disease. J. Parkinson Dis. Alzheimer Dis 2015, 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lashuel HA; Aljabari B; Sigurdsson EM; Metz CN; Leng L; Callaway DJ; Bucala R Amyloid fibril formation by macrophage migration inhibitory factor. Biochem. Biophys. Res. Comm 2005, 338, 973–980. [DOI] [PubMed] [Google Scholar]
- [43].Gevorkian G; Gonzalez-Noriega A; Acero G; Ordoñez J; Michalak C; Munguia ME; Govezensky T; Cribbs DH; Manoutcharian K Amyloid-beta peptide binds to microtubule-associated protein 1B (MAP1B). Neurochem. Int 2008, 52, 1030–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pianu B; Lefort R; Thuiliere L; Tabourier E; Bartolini F The Abeta1–42 peptide regulates microtubule stability independently of tau. J. Cell Sci 2014, 127, 1117–1127. [DOI] [PubMed] [Google Scholar]
- [45].Luo J; Warmlander SK; Graslund A; Abrahams JP Cross-interactions between the Alzheimer disease amyloid-beta peptide and other amyloid proteins: a further aspect of the Amyloid cascade hypothesis. J. Biol. Chem 2016, 291, 16485–16493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lim YA; Rhein V; Baysang G; Meier F; Poljak A; Raftery MJ; Guilhaus M; Ittner LM; Eckert A; Gotz J Abeta and human amylin share a common toxicity pathway via mitochondrial dysfunction. Proteomics 2010, 10, 1621–1633. [DOI] [PubMed] [Google Scholar]
- [47].Götz J; Lim Y-A; Eckert A Lessons from two prevalent amyloidoses-what amylin and Aβ have in common. Front. Aging Neurosci 2013, 5, 38–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Liu Y; Xu LP; Dai W; Dong H; Wen Y; Zhang X Graphene quantum dots for the inhibition of beta amyloid aggregation. Nanoscale 2015, 7, 19060–19065. [DOI] [PubMed] [Google Scholar]
- [49].Wu C; Wang C; Han T; Zhou X;Guo S; Zhang J Insight into the cellular internalization and cytotoxicity of graphene quantum dots. Adv. Healthc. Mater 2013, 2, 1613–1619. [DOI] [PubMed] [Google Scholar]
- [50].Chong Y; Ma Y; Shen H; Tu X; Zhou X; Xu J; Dai J; Fan S; Zhang Z The in vitro and in vivo toxicity of graphene quantum dots. Biomaterials 2014, 35, 5041–5048. [DOI] [PubMed] [Google Scholar]
- [51].Matsusaki M; Kanemura S; Kinoshita M; Lee Y-H; Inaba K; Okumura M The protein disulfide isomerase family: From proteostasis to pathogenesis. Biochim. Biophys. Acta, Gen. Subj 2019, DOI: 10.1016/j.bbagen.2019.04.003. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.