Abstract
Currently available inhibitory optogenetic tools provide short and transient silencing of neurons, but they cannot provide long-lasting inhibition because of the requirement for high light intensities. Here we present an optimized blue-light-sensitive synthetic potassium channel, BLINK2, which showed good expression in neurons in three species. The channel is activated by illumination with low doses of blue light, and in our experiments it remained active over (tens of) minutes in the dark after the illumination was stopped. This activation caused long periods of inhibition of neuronal firing in ex vivo recordings of mouse neurons and impaired motor neuron response in zebrafish in vivo. As a proof-of-concept application, we demonstrated that in a freely moving rat model of neuropathic pain, the activation of a small number of BLINK2 channels caused a long-lasting (>30 min) reduction in pain sensation.
Remote manipulation of ion channels by light is a powerful method to control neuronal activities. Light-gated proton (archaerhodopsin)1 and chloride pumps (halorhodopsin)2–4 and anion-selective channelrhodopsins (ACRs)5–8 are established optogenetic tools for the inhibition of neuronal activity. Given their fast on and off kinetics, pumps are well suited for millisecond-precision applications2,9,10, but they have limitations when long-lasting inhibition (for seconds to minutes) is required. Under prolonged activity, pumps substantially affect ion-concentration gradients across the plasma membrane11,12 and eventually lead to paradoxical effects with activation instead of inactivation13. Longer inhibition can be achieved with ACRs that have a slow off kinetics8,14; however, their effectiveness depends directly on the chloride reversal potential (ECl−), which can vary among neurons. Immature neurons15 and axon initial segments16 of mature neurons can have a positively shifted ECl−, which will promote activation rather than inhibition after ACR channel opening.
Given the universal role of K+ conductance in the termination of action potentials and its major contribution to the resting potential, there is interest in engineering light-gated K+ channels. We previously engineered BLINK1, in which a LOV217,18 photoreceptor domain reversibly controls a K+ channel (KcvPBCV1)19 in response to blue light (455 nm)20. BLINK1 has favorable properties for optogenetics: a low light requirement that avoids tissue heating and damage, a large unitary conductance (>100 pS) to counteract excitatory currents, and a lack of inactivation during prolonged illumination. In vivo experiments with zebrafish embryos highlighted the use of BLINK1 as an inhibitory tool. However, BLINK1 has low surface expression, which hampers its wider use. We present here an improved version of the channel, BLINK2, which showed higher surface expression in neurons compared with that of BLINK1, as well as efficient inhibition of firing in three animal models: zebrafish, rat and mouse. Unique to BLINK2 is its post-illumination activity, which lasts tens of minutes. This property is advantageous for achieving long neuronal inhibition without toxic exposure to prolonged illumination, for instance, in the case of neuropathic pain or in behavioral animal experiments. As proof of principle, we show that BLINK2 activation by light reduced pain for more than 30 min in a rat model and inhibited the touch-evoked escape response in zebrafish. This durable light-off activity of BLINK2 paves the way for optogenetic interventions in chronic applications.
Results
Improving surface expression of BLINK1
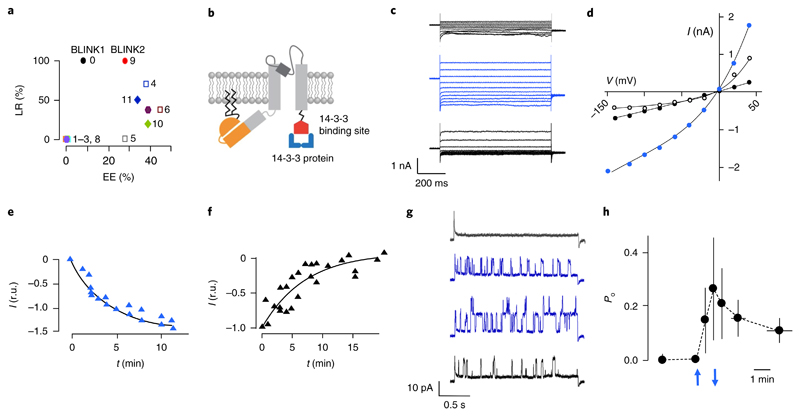
We improved BLINK1 trafficking to the plasma membrane by adding C-terminal signal sequences that promote forward trafficking in eukaryotic K+ channels21. We tested, alone and in combination, the endoplasmic reticulum export motif and trafficking signals of Kir2.121 and the 14-3-3 binding sites of TASK1-322 and KAT123–25. We coexpressed the constructs (Supplementary Table 1) with GFP in HEK293T cells and measured light-regulated currents by patch clamp. We evaluated two parameters: expression efficiency (the percentage of GFP+ cells with a measurable BLINK1-like K+ current) and light regulation (the percentage of cells with no K+ current in the dark). Most clones showed an increase in expression efficiency to >25%, compared with 8% for BLINK1 (Fig. 1a). High expression efficiency, however, resulted in a loss of light regulation. In clone 4, for instance, the addition of Kir2.1 trafficking signal increased the expression efficiency to about 40% but decreased light regulation from 100% to ~70%, thus making the construct unsuitable as an optogenetic tool. Only clone 9, which we renamed BLINK2, showed improved expression efficiency (~28%) and 100% light regulation. BLINK2 (Fig. 1b) has the same topology as BLINK1 and the C terminus of KAT1 (amino acids 506–677, KAT1 numbering) (Supplementary Table 1). This KAT1 sequence ends with the binding motif 673YFSDN677 for 14-3-3 proteins, a class of adaptors that promote KAT1 surface expression25. Figure 1c shows exemplary whole-cell recordings from a BLINK2-transfected COS7 cell in which dark/light transition activated a particularly high current, which is normally in the range of 200–500 pA. In the dark, we measured low currents in BLINK2-transfected cells that were similar to those in untransfected or GFP-transfected cells (Supplementary Fig. 1), indicating that the channel was closed. Inhibition by BaCl2 showed that the dark current was an endogenous potassium conductance of COS7 cells. 3 min of blue light illumination (455 nm, 90 μW/mm2) elicited a voltage-independent current increase, which reverted after 5 min of darkness (Fig. 1c,d). We estimated a ton of 2.7 min (n = 5) and a toff of 7.4 min (n = 6) (Fig. 1e,f).
Fig. 1. Engineering and characterization of BLINK2.
a, Surface expression and light regulation of BLINK1 derivatives. Expression efficiency (EE) was defined as the percentage of cells with measurable BLINK1-like current. Light regulation (LR) represents the percentage of cells that did not show dark current. Clones are numbered according to Supplementary Table 1. b, Cartoon representation of BLINK2 showing the KcvPBCV1channel (gray), LOV2 domain (orange), N-terminal myristoylation and palmitoylation sites (zigzagging black lines) and a fragment of Arabidopsis thaliana KAT1 protein (GenBank AED95356.1) (red) for binding of 14-3-3 proteins (blue). c, Whole-cell recordings from a COS7 cell transfected with BLINK2 in response to voltage steps from +60 to –140 mV in the dark (top black traces), 5 min after the start of blue light illumination (blue traces) and 5 min after returning to darkness (bottom black traces). Similar results were obtained in n=9 cells from 10 independent experiments. d, I/V relationship from measurements in c in the dark (black solid circles), in blue light (blue circles) and after a return to dark conditions (open black circles). e,f, Activation kinetics of BLINK2 current in blue light (e) and after deactivation in the dark (f). Currents were recorded at –100 mV and normalized to t = 5 and t = 0 min for activation and deactivation, respectively (r.u., relative units). Data were fitted with a single exponential (solid line). g, Single-channel recordings from cell-attached measurement of BLINK2 in COS7 cells. The traces show the current response to a voltage step from 0 mV to+40 mV in a dark-adapted cell (top black trace), after 1.5 and 2 min of blue light (blue traces) and 1 min after turning the light off (bottom black trace). Similar results were obtained in n=4 cells from 4 independent experiments. h, Open probability (Po) changes of BLINK2 single channels in response to dark/light transitions. Recordings were done at + 40mV in the cell-attached configuration. Blue arrows indicate the time of light on (upward-facing arrow) and light off (downward-facing arrow). Data shown are the mean ± s.d. of the time at which measurements were performed in 4 experiments. In all experiments reported in this figure, the blue light (455 nm) intensity was 90 μW/mm2.
BLINK2 is activated specifically by blue light (Supplementary Fig. 2). We measured BLINK2 single-channel currents in cell-attached recordings (Fig. 1g). Blue light gradually increased channel activity within 2 min. A return to darkness reduced channel activity after 1 min. The unitary conductance of the light-activated channel is about 130 pS (102 mM K+out). Comparison of the i/V relationship of BLINK2 with those of BLINK1 and KcvPBCV1 (Supplementary Fig. 3) showed that BLINK2 retained a large unitary conductance (>100 pS in 100 mM K+)20,26.
We determined the dynamics of BLINK2 open probability (Po) during light/dark transitions from cell-attached recordings (Fig. 1h; n = 4). An increase in Po was measurable after only 30 s of light exposure and increased further during 60 s of illumination. Deactivation in the dark was slower and highly variable. For example, in one case the current kept increasing over a short time window in the dark, and in another case we measured residual channel activity 20 min after the light was turned off. Single-channel data confirmed that the light-induced macroscopic current (Fig. 1c) was generated by the large conductance BLINK2 channel. The slow deactivation kinetics in the dark suggests that BLINK2 can be used as a tool for sustained inhibition after cessation of illumination.
BLINK2 localization in rat hippocampal primary neurons
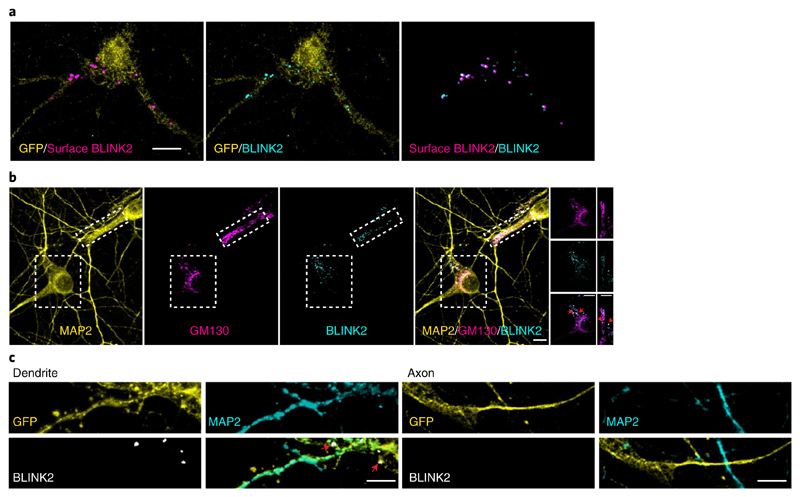
We infected rat primary hippocampal neuronal cultures with an adeno-associated virus (AAV) expressing BLINK2 (AAV-hSyn-BLINK2-IRES-eGFP). An immunofluorescence-based antibody assay27 showed that BLINK2 was expressed at the cell surface with a punctate staining pattern (Fig. 2a). We found no immunofluorescence for an intracellular protein (MAP2) in nonpermeabilized cells, which demonstrates the reliability of our assay and the specificity of BLINK2 membrane staining (Supplementary Fig. 4a). The percentage of cells surface-stained for BLINK2 versus the total number of GFP+ cells (n = 14) was 66.14% ± 3.23% (all values are ± s.e.m. unless stated otherwise).
Fig. 2. BLINK2 expression in rat hippocampal neurons.
a, BLINK2 expression. Left, BLINK2 at the cell surface (magenta). Center, total BLINK2 (turquoise). Right, merged image. GFP is shown in yellow. Scale bar, 10 μm. Similar results were obtained in n= 27cells from 3 independent experiments. b, From left to right, staining for MAP2, Golgi marker GM130 (magenta) and BLINK2 (turquoise), and merged images. The rightmost images are cropped views of the regions outlined by boxes in the other images in the row; red arrowheads indicate colocalization between BLINK2 and GM130. Scale bars, 10 μm. Similar results were obtained in n=15 cells from 3 independent experiments. c, GFP (yellow), MAP2 (turquoise) and BLINK2 (white) in dendrites (MAP2+) and axons (MAP2–). Scale bars, 5 μm. Similar results were observed in n=16 cells from 3 independent experiments. All images were acquired from cultured rat hippocampal neurons infected with AAV1/2-hSyn-BLINK2-IRES-eGFP. Neurons in c were also transfected with a GFP expression plasmid.
BLINK2 intracellularly colocalized with the Golgi marker GM130 in the soma and in dendritic Golgi outposts (Fig. 2b), which suggests that it is sorted along the secretory pathway. The average value for surface versus total staining was 32.73% ± 2.34% (n = 27). Thus, about one-third of BLINK2 protein expressed by a neuron reaches the plasma membrane. To determine the localization of BLINK2 in axonal and somatodendritic domains, we used the MAP2 marker. BLINK2 clusters were detectable in the dendritic compartment (MAP2+) but not in the axon (MAP2–) (Fig. 2c). We further assessed colocalization of BLINK2 clusters with the presynaptic marker Bassoon and the postsynaptic protein PSD-95. BLINK2 clusters partially colocalized with the synaptic markers Bassoon and PSD-95, indicating the presence of BLINK2 in some synapses (Supplementary Fig. 4b). The percentage of synapses in which BLINK2 localized was 26.35% ± 6.16% (n = 8 cells from two independent experiments) for Bassoon staining and 20.04% ± 2.98% (n = 7 cells from two independent experiments) for PSD-95 staining. In conclusion, our data show that BLINK2 is transported along the secretory pathway and expressed at the plasma membrane in hippocampal neurons, preferentially in the somatodendritic compartment.
Ex vivo recordings from mouse brain
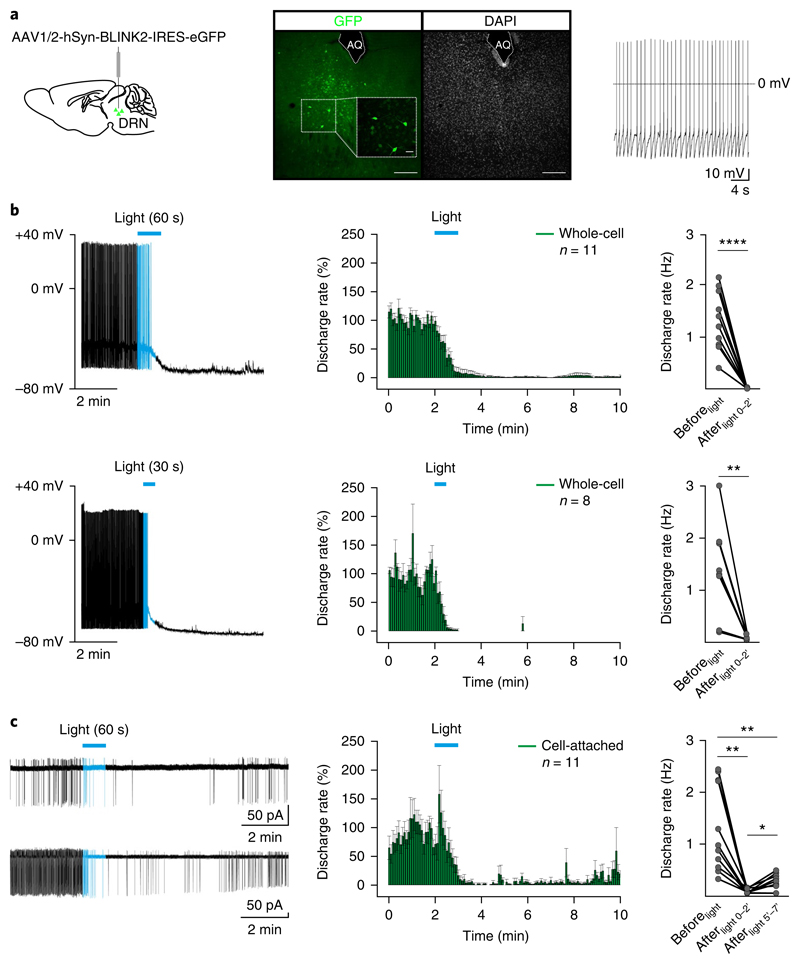
To test neuronal silencing by BLINK2 in brain slices, we injected the AAV-hSyn-BLINK2-IRES-eGFP virus in the dorsal raphe nucleus (DRN) of the midbrain (Fig. 3a). BLINK2-expressing neurons (GFP+) recorded at the soma in the dark showed basal tonic firing (0.2–7 Hz; Fig. 3a), similar to the activity in untransfected acute DRN slices28–30. After transfection with a control GFP-expressing virus (AAV1/2-hSyn-eGFP), none of the recorded cells (n = 7) showed inhibition of firing activity (Supplementary Fig. 5a,b). Passive and active properties of BLINK2-expressing cells were indistinguishable from those of controls in the dark (Supplementary Fig. 6a,b), which indicates that the channel is closed in the dark. Furthermore, the number of GFP+ cells did not vary 2, 4 and 8 weeks after infection, thus indicating that BLINK2 expression does not interfere with cell viability (Supplementary Fig. 7).
Fig. 3. BLINK2-mediated silencing of tonic firing activity in mouse DRN neurons.
a, Left, diagram indicating the virus injection site. Middle, sample confocal image showing expression of the AAV1/2-hSyn-BLINK2-IRES-eGFP virus in the mouse DRN (green, GFP; gray, DAPI). Scale bars, 200 μm or 40 μm (inset). AQ, aqueduct. n = 21 mice. Right, representative (n = 19 from 11 mice) current-clamp recording of tonic firing activity in DRN GFP+ neurons. b, Left, representative whole-cell current-clamp recordings of the firing response before and after 60 s (top) and 30 s (bottom) of blue light stimulation (duration indicated by horizontal blue bars) (top, n = 11 independent recordings in 5 mice; bottom, n = 8 independent recordings in 6 mice). Middle, time course of the effect of 60 s (top) and 30 s (bottom) of blue light stimulation (blue bar) on the firing discharge rate (5-s binning). Right, summary plots indicating the mean firing discharge rate 2 min before light (Beforelight; baseline) and 2 min after light-off (Afterlight 0–2′) (60 s, Beforelight versus Afterlight 0–2′, n = 11, P < 0.0001, t = 7.9, df = 10, two-sided paired t-test; 30 s, Beforelight versus Afterlight 0–2′, n = 8, P = 0.004, t = 4.2, df = 7, two-sided paired t-test). c, Left, representative cell-attached voltage-clamp recordings of firing responses before and after 60 s of blue light stimulation (blue bar) (n = 11 independent recordings; n = 10 mice). Middle, time course of the effect of 60 s of blue light stimulation (blue bar) on the firing discharge rate (5-s binning). Right, summary plot indicating the mean firing discharge rate 2 min before light (Beforelight) and at 2 (Afterlight 0–2′) and 5 (Afterlight 5′–7′) min after the end of light exposure (n = 11; repeated measures one-way ANOVA, F10,2 = 22, P = 0.0007; post hoc, Beforelight versus Afterlight 0–2′, P = 0.002; Beforelight versus Afterlight 5′–7′, P = 0.003; Afterlight 0–2′ versus Afterlight 5′–7′, P = 0.02; multiple comparison and Tukey’s P value correction) (*P < 0.05, **P < 0.01, ****P < 0.0001). Data in time course plots are presented as mean ± s.e.m. Blue light was delivered through the microscope objective (40× at 470 nm, 8.7 mW/mm2).
After exposure to 60 s or 30 s of blue light (470 nm, 8.7 mW/mm2), light-responsive neurons hyperpolarized and stopped firing within 2 min after the light was switched off (for 60-s exposure (n = 11), 1.32 ± 0.17 Hz Beforelight and 0.005 ± 0.003 Hz Afterlight 0-2′; for 30-s exposure (n = 8), 1.39 ± 0.2 Hz Beforelight and 0.016 ± 0.007 Hz Afterlight 0-2′) (Fig. 3b). Firing did not recover for at least 20–30 min after stimulation (data not shown). We reasoned that dialysis of intracellular constituents during whole-cell recordings might represent a caveat, and therefore we analyzed the discharge rate of tonically active GFP+ neurons in a cell-attached configuration28,31. Although light stimulation (60 s) substantially reduced the firing discharge rate (Beforelight, 1.3 ± 0.3 Hz; Afterlight 0-2′, 0.03 ± 0.01 Hz), we observed a slight recovery of activity (Afterlight 5′-7′, 0.18 ± 0.05 Hz) in 7 out of 11 cells in the dark (n = 11; repeated measures one-way ANOVA, F10,2 = 22, P = 0.0007; post hoc, Beforelight versus Afterlight 0-2′, P = 0.002; Beforelight versus Afterlight 5′-7′, P = 0.003; Afterlight 0-2′ versus Afterlight 5'-7′, P = 0.02; multiple comparison and Tukey’s P value correction) (Fig. 3c).
To compare BLINK2 to the opsin-based chloride pump eNphR3.0, we coinjected AAV1-hsyn-Cre and AAV5-EF1α-DIO-eNpHR3.0-eYFP viruses into the DRN (Supplementary Fig. 8a). In YFP+ cells, 60 s of yellow light (585 nm, 17 mW/mm2) induced rapid (< 5 s) silencing of firing activity (Supplementary Fig. 8b); this inhibitory effect faded within the illumination period. The firing rate returned to control levels within 2 min of dark onset (Beforelight, 2.7 ± 0.8 Hz; Light, 1.3 ± 0.7 Hz; Afterlight 0-2′, 2.3 ± 0.9 Hz; Supplementary Fig. 8c). To provide a quantitative comparison between the eNpHR3.0-mediated and BLINK2-mediated effects, we calculated the duration of firing inhibition as the ‘time below threshold’ (a detailed definition is presented in the Methods section), which was 71 ± 28 s for eNpHR3.0 (n = 9) and 420 ± 148 s for BLINK2 (n = 10) (Supplementary Fig. 8d). Thus eNpHR3.0-induced inhibition was faster than that of BLINK2 and did not persist in the dark.
In vivo validation of BLINK2
Next we validated BLINK2 for in vivo application in a zebrafish model. In a touch-evoked escape-response assay, embryos are gently touched on the tail to elicit an escape-type swimming episode. We reasoned that BLINK2 photoactivation would prevent or impair this behavior. In blue light, 2-d-old larvae injected at the one-cell stage with BLINK2 RNA showed an altered escape response to touch compared with that of controls; we did not observe a significant difference between experimental and control specimens when we repeated the experiment in the dark (GFP dark, 4.2% ± 4.2%; BLINK2 dark, 10.4% ± 5.3%; GFP light, 7.2% ± 3.7%; BLINK2 light, 46.4% ± 5.2%) (P = 0.028 and 0.0023 for BLINK2 dark versus light and GFP light versus BLINK2 light, respectively). The percentage of affected larvae (Supplementary Fig. 9a) was similar to that reported for BLINK120, with the exception that a subpopulation of BLINK2 embryos (13 of 91 embryos) required more touches than BLINK1 embryos in order for an observable response to be elicited (Supplementary Fig. 9b). The light-driven effect on escape developed with a half-time of 15–20 min and reverted in the dark with a similar kinetics (Supplementary Fig. 9c). The mutation Q513D, which accelerates dark recovery in the isolated LOV2 domain32, did not affect the kinetics of BLINK2 (Supplementary Fig. 9c).
Transgenic zebrafish line expressing BLINK2
Expression of BLINK2 under the control of UAS regulatory sequences in a transgenic zebrafish line allowed targeting of the channel in genetically defined populations of neurons through crossing with Gal4 reporter lines33.
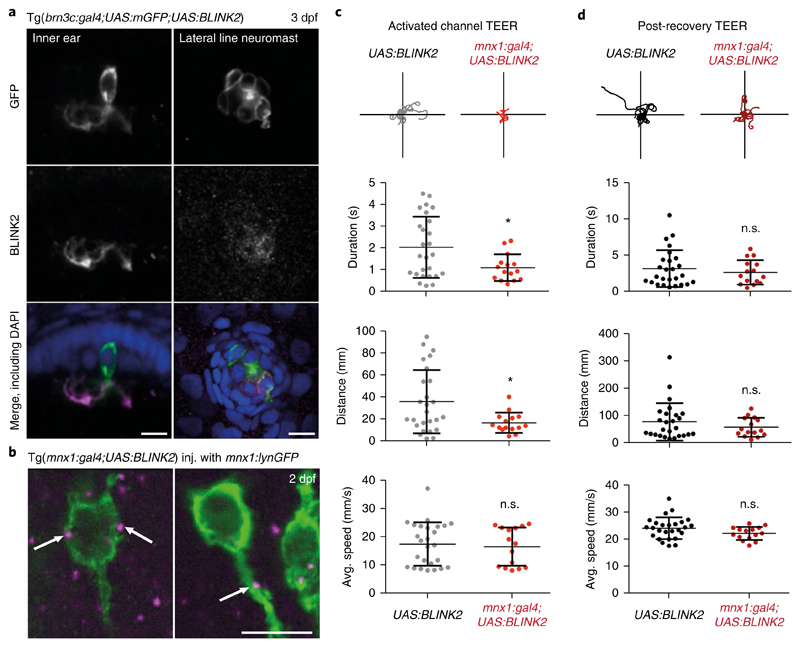
We expressed BLINK2, together with a membrane-bound fluorophore (mGFP), in hair cells of the zebrafish ear and lateral line neuromasts by crossing our specimens into the brn3c:gal4 background (Tg(brn3c:gal4;UAS:BLINK2;UAS:mGFP)). Whole-mount immunohistochemistry with anti-BLINK2 showed expression of the channel preferentially at the level of the apical cilia in hair cells (Fig. 4a). Its expression in neuromasts seemed most prominent in the cell body in GFP+ cells.
Fig. 4. BLINK2 expression and functional silencing in zebrafish.
a, Left, immunohistochemistry in 3-dpf (days post-fertilization) embryos, showing a hair cell of the inner ear labeled for membrane-targeted GFP (green in the merged image) and the BLINK2 channel (magenta in the merged image), both expressed under the control of brn3c:gal4. Right, neuromast cells from the same Tg(brn3c:gal4;UAS:mGFP) line labeled in the same way. Embryos were counterstained with DAPI (blue). Scale bars, 10 μm. Similar results were obtained in 3 independent experiments. b, Immunohistochemistry on whole 2-dpf embryos showing cell bodies and part of the axons of primary motor neurons stained by membrane-targeted GFP (green) and BLINK2 (magenta). White arrows indicate BLINK2 immunoreactivity at the plasma membrane and axonal tract. Genotypes are as indicated. GFP was expressed in subsets of motor neurons only. Scale bars, 10 μm. Similar results were obtained in 3 independent experiments. c, Touch-evoked escape response assay (TEER) in Tg(mnx1:gal4;UAS:BLINK2) and Tg(UAS:BLINK2) embryos. Embryos were assayed after a 20-min activation of the channel with blue light. Swim duration, distance and average speed were 2.03 ± 0.28 s, 35.80 ± 5.65 mm and 17.49 ± 1.50 mm/s, respectively, in control animals and 1.10 ± 0.16 s, 16.62 ± 2.38 mm and 16.60 ± 1.75 mm/s in BLINK2-expressing animals. Traces for 10 escape episodes are shown for each condition. n = 26 larvae for Tg(UAS:BLINK2) and n = 15 larvae for Tg(mnx1:gal4;UAS:BLINK2). Data are presented as the average (center line) ± s.d.; P values are, respectively, 0.019, 0.017 and 0.071. d, TEER assay in the same animals as in c after 1 h of rest in the dark. Swim duration, distance and average speed were 3.18 ± 0.50 s, 77.12 ± 13.42 mm and 24.03 ± 0.79 mm/s, respectively, in control animals and 2.67 ± 0.44 s, 57.03 ± 8.90 mm and 22.08 ± 0.63 mm/s in BLINK2-expressing animals. P values are, respectively, 0.48, 0.29 and 0.099. *P ≤ 0.05 (two-sided t-test). n.s., not significant.
To express BLINK2 in primary motor neurons of the spinal cord, we crossed the UAS:BLINK2 carrier zebrafish into the mnx1:gal4 background34 to obtain Tg(mnx1:gal4;UAS:BLINK2) embryos. We injected transgenic embryos from this cross with a construct containing the mnx1 promoter35 driving lynGFP, which encodes a membrane-bound fluorophore36, in order to visualize cell membranes and allow localization of the channel in this cell type. Whole-mount immunohistochemistry revealed channel puncta at the level of the membrane in motor neuron cell bodies and at the axon (Fig. 4b).
To functionally test silencing of neurons by BLINK2, we exposed Tg(mnx1:gal4;UAS:BLINK2) embryos to light from a blue LED (447 nm, 80 μW/mm2) for 20 min and then carried out the touch-evoked escape-response assay. The evoked behavior relies on spinal cord primary motor neurons and was affected by activation of the channel (Fig. 4c). We dissected the response into three parameters: duration, distance and average speed (Fig. 4c). Tg(mnx1:gal4;UAS:BLINK2) embryos exhibited a reduced escape duration and distance but a conserved instant maximum speed as compared with that of Tg(UAS:BLINK2) embryos, which indicates that BLINK2 did not inactivate muscles. We then left embryos for 1 h in the dark to allow BLINK2 channel-closing before we repeated the behavioral assay. The embryos with closed BLINK2 channels performed as the controls did, with no significant difference in their escape behavior (P = 0.48, 0.29 and 0.099 for duration, distance and average speed, respectively; two-sided t-tests) (Fig. 4d). This demonstrates that BLINK2 activation in this specific neuronal population was sufficient to reversibly impair function with a measurable behavioral defect.
BLINK2 stimulation relieves chemotherapy-induced neuropathic pain in a rat model
A property of BLINK2 is its prolonged activity after cessation of light exposure, which can last for several minutes (Figs. 1 and 3). Therefore BLINK2 is a candidate tool for optogenetic applications that require long-lasting inhibition, such as pain relief in peripheral neural circuits.
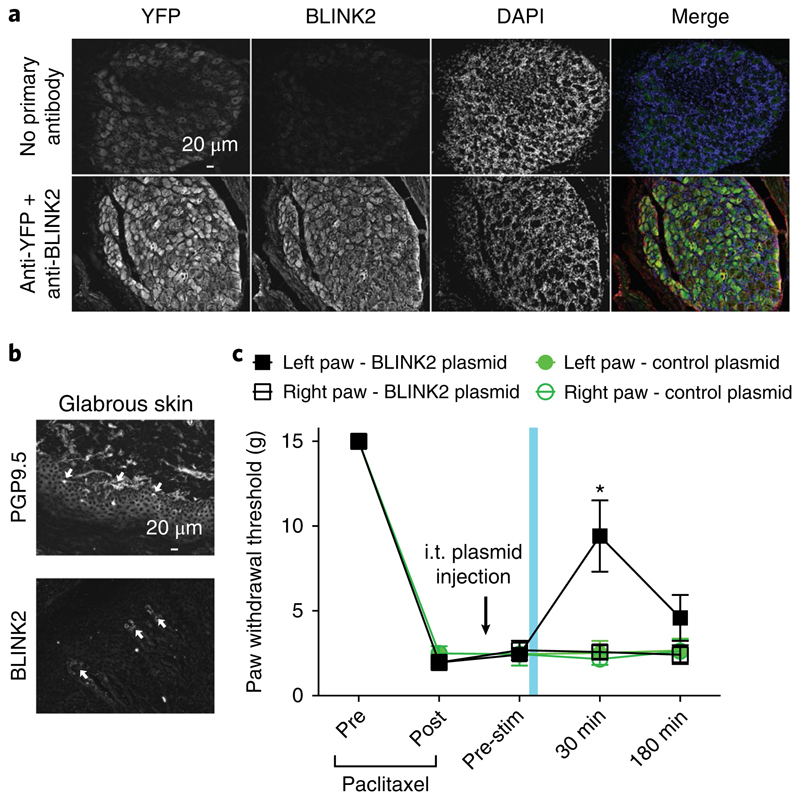
As proof of concept, we tested the effect of BLINK2 on neuropathic pain in rats. We expressed BLINK2 in dorsal root ganglia (containing the primary afferent neurons) by in vivo transfection via intrathecal injection. Twenty-four hours after injection, YFP and BLINK2 were expressed in sensory neurons in L4–L6 dorsal root ganglia (Fig. 5a) and nerve terminals of the glabrous skin (Fig. 5b). We then tested BLINK2-mediated silencing of nociceptive neurons in a preclinical model of chemotherapy-induced neuropathic pain. Rats injected with paclitaxel develop tactile allodynia (Fig. 5c), which is caused by ectopic firing of sensory neurons and increased nociceptive signal transmission37. Opening of a K+ channel should hyperpolarize the nociceptive neurons and prevent firing of action potentials.
Fig. 5. BLINK2-mediated reversal of chemotherapy-induced neuropathic pain in rats.
a, Fluorescent micrographs of 12-μm sections of adult dorsal root ganglia from animals that received intrathecal (i.t.) injection of BLINK2–YFP expression plasmid, immunostained 24 h after injection for YFP and BLINK2 (bottom row). In the images in the top row, no primary antibody control was used to visualize YFP fluorescence. Presented data are from three independent animals that yielded similar results. b, Fluorescent micrographs of 12-μm sections of glabrous skin from animals that received i.t. injection of BLINK2–YFP expression plasmid, immunostained 24 h after injection for PGP9.5 (nerve terminals) or BLINK2. White arrows indicate the nerve terminals in the glabrous skin stained with PGP9.5 or BLINK2. Presented data are from three independent animals that yielded similar results. c, Paw withdrawal thresholds for rats with chemotherapy-induced neuropathic pain (paclitaxel) and i.t. injection of BLINK2 plasmid (4.5 μg per rat; n = 6). Blue light illumination was applied for 1 min to the left paw only. *P < 0.05 for the left paw compared with the right paw. P = 0.0001 (two-way ANOVA with Student–Neuman–Kuels post hoc test). Data were analyzed by nonparametric two-way ANOVA, where time was the within-subject factor and treatment was the between-subjects factor. Data are presented as the average ± s.e.m.
After inducing tactile allodynia, we injected rats intrathecally with a BLINK2 plasmid for in vivo transfection. We expected protein expression and physiological consequence to peak about 24 h after injection38. On the day after plasmid injection, we illuminated BLINK2 by exposing the left paw to blue light for 1 min. The right paw was not illuminated and was used as an internal control. Illumination of the left paw reduced nociception for at least 30 min, as indicated by an increased threshold for paw withdrawal after touch, which resolved after 3 h (Fig. 5c). We observed this effect only in the left paw; the right paw did not show an increased paw withdrawal threshold. This measured the force needed to elicit a response in the rats (Fig. 5c). Rats injected with an empty plasmid were insensitive to blue light (Fig. 5c). These experiments show that a specific effect of blue light is to trigger BLINK2 activation and silencing of ectopic nociceptive inputs in chemotherapy-induced neuropathic pain.
Discussion
We have shown that the light-gated K+ channel BLINK2 is an inhibitory tool in long-lasting optogenetic experiments. Because BLINK2 is not activated by wavelengths greater than 500 nm, it can be combined with green-excitable labels and tools with minimal cross-talk.
Compared with BLINK120, BLINK2 shows slower activation and deactivation kinetics (on the order of minutes). The lasting inhibition is presumably due to the high channel conductance that prevents depolarizing inputs even if only a small number of channels remain active. We observed more severe inhibition in whole-cell experiments than in cell-attached experiments, which we presently do not understand and which may depend on the dilution of cytosolic factors during prolonged whole-cell measurements. However, the full recovery observed in our in vivo experiments indicates that the system is in principle reversible and does not cause severe stress to the cells. BLINK2 should have minimal effects on cells, as it exploits an inherent mechanism for hyperpolarization, namely, K+ efflux. We expect that BLINK2 will provide inhibition in all cell types and in many model organisms. Moreover, the combination of large unitary conductance and prolonged light-off activity allows cellular inhibition in a time range inaccessible to other inhibitory tools, such as the opsin-based chloride pump eNpHR3.0. In our experimental conditions, eNpHR3.0 inhibited firing transiently for no longer than tens of seconds.
BLINK2 is suitable for in vivo experiments that require very long inhibition times. BLINK2 may be used to dissect the role of genetically defined neuronal populations in behavioral experiments or for silencing of neurons during the development of neural circuits, where it is necessary to silence neurons for hours or days. This may be achievable with BLINK2 by light pulses of low frequency and intensity, which should prevent the unwanted tissue heating often associated with prolonged inhibition by other optogenetic tools39.
The slow post-illumination recovery of BLINK2 is a beneficial property for silencing peripheral neural circuits in the control of neuropathic pain. This is a high-priority issue in therapeutics because of inadequate responses to drug therapy40. In our hands, reduced pain sensation in a rodent model did not require constant light but was achieved with a brief transdermal light pulse and without the need for fiber-optic implantation. This avoids negative consequences of high-intensity illumination such as local tissue heating41 and facilitates potential clinical translation.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41592-018-0186-9.
Methods
Engineering of channel constructs
Constructs in Fig. 1a were prepared by overlapping PCR42. The IDs of the sequences used are as follow: AsPhototropin1 (Avena sativa), GeneBank AAC05083.1; mKir2.1 (Mus musculus), NCBI gene 16518; mTASK1 (M. musculus), NCBI gene 16527; mTASK3 (M. musculus), NCBI gene 223604; KAT1 (Arabidopsis thaliana), NCBI gene 834666.
QuikChange Lightning (Agilent Technologies) was used to introduce point mutations. BLINK2 used in all experiments except those in transgenic zebrafish contained the mutation Q513D in the LOV2 domain32 (AsPhot1 numbering).
Electrophysiology in cell lines
Cell culture and transfection protocol
HEK293T or COS7 cells were cultured in Dulbecco’s modified Eagle’s medium (Euroclone) supplemented with 10% FBS (Euroclone), 100 IU/ml penicillin, 100 μg/ml streptomycin and stored in a 37 °C humidified incubator with 5% CO2. Transfections were performed with TurboFect transfection reagent (Thermo Scientific) according to the supplier’s protocol: BLINK2 inserted in pcDNA3.1+ was cotransfected with a plasmid encoding GFP and incubated in the dark. For viral infection we added the virus directly to the cell culture medium. Currents were recorded after 2–3 d in GFP+ cells.
Patch-clamp recordings
One to two days after transfection, cells were dispersed by trypsin–EDTA treatment and seeded on 35-mm plastic petri dishes to allow single-cell measurements. GFP+ cells were selected for patch-clamp measurements. Membrane currents were recorded in the whole-cell configuration with a Dagan 3900A amplifier and digitized with a Digidata 1322A controlled by pCLAMP 9.2. The pipette resistance was about 2 MΩ. The pipette solution contained 10 mM NaCl, 130 mM KCl, 2 mM ATP–magnesium salt, 1 mM EGTA and 5 mM HEPES–KOH buffer, pH 7.2. The extracellular bath solution contained 100 mM KCl, 80 mM d-mannitol, 1.8 mM CaCl2, 1 mM MgCl2 and 5 mM HEPES–KOH buffer, pH 7.4. K+ concentrations were 101.7 mM for the extracellular solution and 133.7 mM for the pipette solution. The calculated Nernst reversal potential for K+ is –6.89 mV. The voltage protocol consisted of 20-mV steps from +60 to –140 mV. For cell-attached measurements, the pipette resistance was 2 MΩ and the pipette solution was the same as the extracellular solution. Transfected cells were kept in the dark before the assays, and all preliminary operations were performed under red light illumination (MRH2060–20T, LUXEON Rebel LEDs Red-Orange (617 nm)). Blue light illumination was provided by an LED (Royal Blue, 455 nm, High-Power LED; Thorlabs) or monochromatic light from a 75-W Xenon Arc lamp (PTI DeltaRem X, Photon Technology International) delivered through the 60× objective of a fluorescent Nikon Eclipse Ti-U microscope with an oil-immersion lens. In both cases, the light intensity measured with a power meter (Thorlabs) at the position of the sample was about 90 μW/mm2.
Statistical analysis
Significance was calculated by one-way ANOVA and Tukey post hoc test using GraphPad Prism for Windows (GraphPad Software, La Jolla, CA, USA; https://www.graphpad.com).
Viral expression of BLINK2
Cloning of AAV plasmids
BLINK2 cDNA was amplified from Q513D pGEMT-BLINK2 by PCR with primers containing BglII recognition sites (A^GATC). With the use of BglII restriction, pAAV1/2-hSyn-IRES-eGFP was linearized and BLINK2 cDNA was subsequently ligated into the linearized vector to produce pAAV1/2-hSyn-BLINK2-IRES-eGFP.
Virus production
HEK293T cells (ATCC, UK) were cultured in Iscove’s modified Dulbecco’s medium (Sigma-Aldrich, Germany) supplemented with 10% (v/v) FCS (Sigma-Aldrich) and penicillin–streptomycin–glutamine (Sigma-Aldrich) in 5 × 150 mm dishes. After 80% confluency was reached, cell were transfected in serum-free medium with the helper plasmids pRV1, pH21 and pDFΔ6 and pAAV1/2-hSyn-BLINK-IRES-eGFP or pAAV1/2-hSyn-IRES-eGFP at a molar ratio of 1:1 with CaCl2. On the next day, the medium was replaced with serum-containing medium, and 48 h after transfection cells were harvested, pelleted and resuspended in lysis solution (150 mM NaCl, 20 mM Tris, pH 8). Next, cells were subjected to a freeze–thaw cycle, and after the addition of NaDOC (0.5% v/v), the solution was incubated with Benzonase (Sigma-Aldrich; 50 units/ml) for 60 min at 37 °C. After centrifugation (3,000g at 4 °C for 10 min), the supernatant was frozen. The next day, we carried out ion-exchange chromatography with 1-ml HiTrapQ columns (GE Healthcare, UK). Viral particles were washed and eluted with solutions of 20 mM Tris, pH 8, with increasing NaCl concentrations (100–500 mM NaCl). Eluate was transferred to an Amicon Ultra-4 filter (Millipore, USA) to concentrate the viral particles and exchange the buffer for PBS. The purified virus was then aliquoted and stored at −80 °C. The titer was determined by real-time quantitative PCR on a 7900HT Fast Real-Time PCR system (Applied Biosystems Inc., USA) using primers against GFP (forward, AAGCTGACCCTGAAGTTCATCTGC; reverse, CTTGTAGTTGCCGTCGTCCTTGAA) and the GoTaq RT-qPCR kit (Promega, USA).
BLINK2 immunolocalization in rat primary neurons
Cell cultures and transfections
Hippocampal neuronal primary cultures were prepared from embryonic day 18–19 (E18– E19) rat hippocampi as previously described43. All the experiments were approved by the Institutional Animal Care and Use Committee of University of Milan and by the Italian Ministry of Health (#326/2015). Neurons were transfected at 7 days in vitro (DIV7) via the calcium-phosphate precipitation method with 4 μg of plasmid DNA for GFP for the experiments assessing the axonal and dendritic distribution of BLINK2 reported in Fig. 2c. Neurons were infected with AAV1/2-hSyn-BLINK2-IRES-eGFP at DIV10 and fixed at DIV12 for the immunocytochemistry assays.
Immunocytochemistry
For colocalization experiments, cells were fixed with 4% paraformaldehyde (PFA)–4% sucrose in PBS solution at 4 °C and washed several times with PBS. Cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature and then blocked with 5% BSA in PBS for 45 min at room temperature. Cells were then labeled with antibodies for intracellular epitopes overnight at 4 °C. Cells were washed and incubated with secondary antibodies for 1 h at room temperature. Cells were washed in PBS and mounted on glass slides with Fluoromount mounting medium (Sigma-Aldrich, USA).
To evaluate surface and total staining of BLINK2, neurons were fixed with 4% PFA–4% sucrose in PBS solution at 4 °C, and then incubated with anti-BLINK2 8D6 custom-made monoclonal antibody. This antibody, originally raised against the potassium channel Kcv, recognizes BLINK channels too20,44. To visualize surface expression, we blocked cells with 5% BSA in PBS and incubated them with an Alexa Fluor 555–conjugated secondary antibody. Afterward, cells were permeabilized with 0.1% Triton X-100 for 10 min, and intracellular expression was determined after incubation with 8D6 antibody and labeling of the total receptor fraction with an Alexa Fluor 405–conjugated secondary antibody.
Fluorescence images were acquired with the Zeiss Confocal LSM510 Meta system with a sequential acquisition setting at 1,024 × 1,024 pixel resolution; for each image two to four 0.5-μm sections were acquired and a z projection was obtained45. Images were acquired with signals in a linear range and without any saturated pixel, for reliable quantification and appropriate comparison of all experimental conditions.
For quantification of surface and total expression intensities, images were acquired with the same settings. The average intensity of surface fluorescence staining was determined after cell tracing and was normalized to the total intensity to correct for differences in expression. We obtained surface ratios by dividing the background-subtracted fluorescence intensities.
Antibodies
We used antibodies to MAP2 (Millipore; AB5222), GM130 (BD Bioscience; 610822) and GFP (Millipore; AB16901). Alexa Fluor fluorescently labeled antibodies were purchased from Thermo Fisher.
Ex vivo electrophysiology
Animals
All procedures involving animals were carried out in accordance with the Italian Ministry of Health’s directives (D.lgs 26/2014) regulating animal research. Animal experiments were designed in accordance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines, with a commitment to refinement, reduction and replacement, so as to minimize the number of mice used. C57BL/6J male mice were maintained in standard cages with food and water ad libitum at 22 ± 1 °C under an artificial 12/12-h light/dark cycle.
Stereotaxic injections
C57BL/6J male mice (4–6 weeks old) were anesthetized with a mixture of isoflurane (1–2%) and O2. Mice were positioned in a stereotaxic frame (Kopf Instruments) and their body temperature was maintained at 37 °C. We injected 0.5 μl (titer 1013) of AAV1/2-hSyn-Blink2-IRES-eGFP, AAV1/2-hSyn-eGFP or a 1:1 mixture of AAV1-hsyn-Cre (pENN.AAV.hSyn.Cre.WPRE.hGH, a gift from James M. Wilson (Perelman School of Medicine, University of Pennsylvania); Addgene viral prep # 105553-AAV1) and AAV5-EF1α-DIO-eNpHR3.0-eYFP (Stanford Virus Core) into the DRN (mediolateral, +1.15 mm, anteroposterior, –4.4 mm, dorsoventral, –3.6 mm under an angle of 20° from bregma; or mediolateral, +0.5 mm, anteroposterior, –4.36 mm, dorsoventral, –3 mm from bregma) at a speed of 0.1 μl/min. Ex vivo electrophysiology was performed at least 2 weeks after surgeries.
Immunofluorescence
Mice were killed 2, 4 or 8 weeks after the injection. Anesthetized mice were transcardially perfused with PBS followed by 4% PFA. Brains were dissected and post-fixed in 4% PFA overnight at 4 °C. 50-μm coronal sections were obtained with a vibratome (Leica Microsystems). Antigen retrieval was performed as follows: sections were incubated for 30 min at 80 °C in 50 mM sodium acetate solution. Then the slices were washed three times in a PBTriton 0.1% solution. Sections were incubated with chicken anti-GFP (Abcam; 1:500) primary antibody overnight at 4 °C and then rinsed in PBTriton 0.1%. Alexa Fluor 488–conjugated goat anti-chicken IgG (1:500; Life Technologies) was used overnight at 4 °C as the secondary antibody. The next day, sections were washed three times with PBTriton 0.1% solution and counterstained with DAPI. High-power confocal images in the injection site of the DRN region were obtained on a Nikon A1 confocal microscope with a 10× or 40× plan-apochromat.
Slice preparation
Mice were killed under isoflurane anesthesia, after which their brains were dissected out and transferred to ice-cold modified artificial cerebrospinal fluid (aCSF) containing 110 mM choline chloride, 2.5 mM KCl, 1.25 mM NaH2PO4, 7 mM MgCl2, 0.5 mM CaCl2, 25 mM NaHCO3, 25 mM d-glucose and 11.6 mM ascorbic acid, saturated with 95% O2 and 5% CO2. Coronal slices containing the DRN (250-μm thickness) were prepared with a Vibratome 1000S slicer (Leica) and transferred to aCSF containing 115 mM NaCl, 3.5 mM KCl, 1.2 mM NaH2PO4, 1.3 mM MgCl2, 2 mM CaCl2, 25 mM NaHCO3 and 25 mM d-glucose, aerated with 95% O2 and 5% CO2. After 20 min of incubation at 32 °C, slices were kept at 22–24 °C. During electrophysiological experiments, slices were continuously superfused with aCSF at a rate of 2 ml/min at 28 °C.
Electrophysiological recordings
Electrophysiology recordings were performed on coronal brain slices containing the DRN. The DRN was first visualized under infrared differential interference contrast to allow for subsequent identification of GFP+ or YFP+ neurons by epifluorescence microscopy. Patch pipettes (4–6 MΩ) were filled with a solution containing 135 mM NaCl, 10 mM HEPES, pH 7.2–7.3, for cell-attached recording or 130 mM KMeSO4, 5 mM KCl, 5 mM NaCl, 10 mM HEPES, 2 mM MgCl2, 0.1 mM EGTA, 0.05 mM CaCl2, 2 mM Na2ATP and 0.4 mM Na3GTP, pH 7.2–7.3 (280–290 mOsm/kg), for whole-cell recordings. Cell-attached experiments were performed in the voltage clamp configuration with GFP+ or YFP+ neurons held at the potential that gave a holding current of 0 pA (ref. 31.), whereas whole-cell experiments were performed in the current-clamp configuration, without current injection. Light (470 nm for BLINK2 activation, 8.7 mW/mm2; 585 nm for eNpHR3.0 activation, 17 mW/mm2) emitted by an LED (CoolLED) was delivered to the specimen through the microscope objective (IR-ACHROPLAN 40×/0.8-NA (numerical aperture); Zeiss). Data were acquired with a Multiclamp 700B amplifier controlled by pClamp 10 software (Molecular Devices) filtered at 10 kHz and sampled at 50 kHz (current clamp and voltage clamp) (Digidata 1322; Molecular Devices). We generated time-course plots by averaging the discharge firing rate every 5 s; values were normalized to 1 min of baseline recording before light illumination. All data are reported without corrections for liquid junction potentials. Data where the access resistance (Ra) changed by >20% were excluded from further analyses.
To identify light-responsive cells, we applied a threshold-based criterion: the threshold (Th) was set as the mean discharge rate minus 2 s.d., and the mean firing rate was calculated on values (5-s binning) computed over 1 min prior to light illumination. Cells were considered light responsive when their mean discharge rate fell below Th, or to zero, in at least two consecutive 5-s bins. ‘Time below threshold’ (Timeth) was measured as the interval between the time point at which the discharge rate fell below Th in at least two consecutive 5-s bins and the time point at which the discharge rate increased above Th in at least two consecutive 5-s bins.
Statistics
Appropriate parametric statistics were used to test our hypothesis, unless data did not meet the assumptions of the intended parametric test (normality test). In that case, appropriate nonparametric tests were used. Power analysis assumptions were as follows: power, 0.9; alpha, 0.5; two-tailed and expected difference 50% greater than the observed s.d. Data were analyzed by one-way repeated measures ANOVA for comparisons within a group, and by one-way ANOVA for between-group comparisons (GraphPad Prism 6 software). Post hoc analysis (Tukey or Dunnet, as indicated) was performed only when ANOVA yielded a significant main effect. Two groups were tested for statistical significance by two-population t-test and Mann–Whitney U nonparametric test (GraphPad Prism 6 software). Statistical details of experiments are shown in the results, figures and figure legends. Data are reported as mean ± s.e.m., unless stated otherwise.
Zebrafish experiments
Zebrafish husbandry
The zebrafish were housed and maintained at 28.5 °C according to standard procedures45. Experiments were done in compliance with European and French animal welfare guidelines.
Microinjections
Zygotes were injected with mnx1:lynGFP constructs to label single primary motor neuron membranes in the spinal cord in the Tg(mnx1:GAL4;UAS:BLINK2) background.
Transgenic BLINK2 zebrafish generation
To express BLINK2 under the control of the Gal4 trans-activator in stable transgenic zebrafish, we cloned the BLINK2 coding sequence in a p10UAS vector containing a cmcl2:eGFP cassette to visualize transgenic animals by heart GFP fluorescence using standard molecular biology techniques46. The plasmid also contained Tol2 flanking sites for efficient transgenesis in zebrafish and was named p10UAS:BLINK2-tol2;cmcl2:eGFP. This plasmid was injected at the one-cell stage with tol2 mRNA via standard transgenesis protocols47. Transgenic F1 larvae were identified by heart GFP expression, and BLINK2 gene insertion was verified by genomic PCR.
Whole-mount immunohistochemistry
Embryos at 48 h post-fertilization (hpf) were fixed overnight at 4 °C in 4% PFA diluted in PBS, then thoroughly rinsed in PBST (PBS with 0.1% Triton X-100). The fixed embryos were incubated with 1 mg/ml collagenase for 20 min, then rinsed in PBST before 1 h of incubation with block solution (PBS with 1% BSA, 2% normal goat serum, 1% DMSO, 0.1% Triton X-100). The embryos were then incubated sequentially with the primary antibodies (anti-GFP (1:300; Genetex), anti-RFP (1:200; AbCam), anti-BLINK2 (8D6) and DAPI (1:500; Life Technologies)) in fresh block solution, thoroughly rinsed in PBST and incubated with secondary antibodies (goat anti-rabbit Alexa Fluor 488 and goat anti-mouse Alexa Fluor 568 (both from Life Technologies)) also diluted in fresh blocking solution.
Microscopy
Embryos were embedded in 1% low-melting-point agarose in a glass-bottom tissue culture dish (Fluorodish; World Precision Instruments, USA).
Inner ear cells were imaged on an inverted laser scanning confocal microscope with spectral detection (LSM700; Zeiss) with a long-working-distance oil-immersion 25×/0.8-NA W GLY DIC LD LCI PL APO (UV) VIS-IR (420852-9870) lens. Acquisitions were done via the Zen software (Zeiss).
Spinal cord primary motor neurons were imaged on a Roper confocal spinning disk head mounted on a Zeiss upright microscope, using a long-working-distance water-immersion 40×/1-NA W DIC PL APO VIS-IR (421462-9900) lens. Acquisitions were done with a CoolSNAP HQ2 CCD (charge-coupled device) camera (Photometrics, USA) through the MetaMorph software (Molecular Devices, USA).
Touch-evoked escape response assay
Embryos at 48 hpf were staged, dechorionated and exposed to a blue LED light (Royal-Blue LED, λ 447 ± 10 nm, LUXEON Rebel LED) for 20 min (λ = 447, 80 μW/mm2) to activate the BLINK2 channel. The embryos were then placed in the center of an open petri dish filled with embryo medium. The escape response was elicited by a light touch on the tail with blunt forceps, and the resulting swimming episode was recorded with an Olympus FE-5000 camera at 30 Hz. The embryos were then left in the dark for 1 h to allow the inactivation of BLINK2, and the assay was subsequently performed again to test for recovery of locomotion. The videos were analyzed in ImageJ (NIH) using the Manual Tracking plugin (Fabrice Cordelières, Institut Curie-Orsay, France).
Statistics
Data were compiled in GraphPad Prism (Windows version 6.01) and t-tests were run to determine significance, set at P ≤ 0.05.
Rat pain model and intrathecal injection of BLINK2
Animals
Pathogen-free adult male and female Sprague Dawley rats (150–200 g; Envigo) were housed in temperature-controlled (23 ± 3 °C) and light-controlled (12-h light/12-h dark cycle; lights on 07:00–19:00) rooms with standard rodent chow and water available ad libitum. The Institutional Animal Care and Use Committee of the College of Medicine at the University of Arizona approved all experiments. All procedures were conducted in accordance with the Guide for Care and Use of Laboratory Animals published by the National Institutes of Health and the ethical guidelines of the International Association for the Study of Pain. Animals were randomly assigned to treatment or control groups for the behavioral experiments. Animals were initially housed three per cage but were individually housed after the intrathecal cannulation on a 12-h light-dark cycle with food and water ad libitum. All behavioral experiments were performed by experimenters who were blinded to the experimental groups and treatments
Paclitaxel-induced neuropathy model
Rats were given paclitaxel (P-925-1; Goldbio) based on the protocol described by Polomano et al.48. In brief, pharmaceutical-grade paclitaxel (Taxol) was resuspended at a concentration of 2 mg/ml in 30% 1:1 Cremophor EL:ethanol, 70% saline and given to the rats at 2 mg/kg intraperitoneally every other day for a total of four injections (days 0, 2, 4 and 6), resulting in a final cumulative dose of 8 mg/kg. No abnormal spontaneous behavioral changes in the rats were noted during or after the treatment. Animals developed mechanical hyperalgesia within 10 d after the first paclitaxel injection.
Implantation of intrathecal catheter
For intrathecal drug administration, rats were chronically implanted with catheters as described49. Rats were anesthetized with halothane and placed in a stereotactic head holder. The occipital muscles were separated from their occipital insertion and retracted caudally to expose the cisternal membrane at the base of the skull. Polyethylene tubing was passed caudally from the cisterna magna to the level of the lumbar enlargement. Animals were allowed to recover and were examined for evidence of neurologic injury. Animals with evidence of neuromuscular deficits were excluded.
In vivo transfection of BLINK2 plasmid
For in vivo transfection, the BLINK2 plasmid was diluted to 0.3 μg/μl in 5% sterile glucose solution as done previously38. Then, Turbofect in vivo transfection reagent (R0541; Thermo Fisher Scientific, Waltham, MA) was added according to the manufacturer’s instructions. Finally, 15 μl of the plasmid complexes were injected intrathecally in Sprague Dawley rats.
Testing of allodynia
The assessment of tactile allodynia (i.e., a decreased threshold for paw withdrawal after probing with normally innocuous mechanical stimuli) consisted of testing the withdrawal threshold of the paw in response to probing with a series of calibrated fine (von Frey) filaments. Each filament was applied perpendicularly to the plantar surface of the paw of rats held in suspended wire mesh cages. We determined the withdrawal threshold by sequentially increasing and decreasing the stimulus strength (the ‘up and down’ method), and we analyzed data using the nonparametric method of Dixon, as described by Chaplan et al.50, with results expressed as the mean withdrawal threshold.
Illumination of the paw was performed with blue LED light (Royal-Blue LED, λ 455 ± 9 nm, LUXEON Rebel LED) for 1 min (35.6 μW/mm2). The light was measured at a distance of 1–1.5 cm from the paw.
Immunohistofluorescence and epifluorescence imaging
L4–L6 dorsal root ganglia were dissected from adult rats and then fixed with 4% PFA overnight at 4 °C. Dorsal root ganglia were next transferred into a 30% sucrose solution and left at 4 °C until sinking of the tissues could be observed (~3 d). Tissues were cut at 12-μm thickness with a Bright OTF 5000 microtome cryostat (Hacker Instruments and Industries, Inc.), fixed onto charged glass slides and kept at –20 °C until use. Prior to antibody staining, slides were dried at room temperature for 30 min and rehydrated in PBS for 5 min. For glabrous skin staining, slides were incubated in ice-cold methanol for 5 min and left to dry at room temperature. The slices were permeabilized and saturated with PBS containing 3% BSA, 0.3% Triton X-100 solution for 30 min at room temperature, and then antibodies diluted in PBS, 3% BSA were added overnight at room temperature. Primary antibodies used were anti-GFP (AB3080; Millipore), anti-PGP9.5 (NB600-1160; Novus Biologicals) and anti-Blink2 8D6. The slices were then washed three times in PBS and incubated with PBS, 3% BSA containing secondary antibodies (Alexa Fluor 488 goat anti-rabbit or Alexa Fluor 594 goat anti-mouse secondary antibodies (Life Technologies)) for at least 3 h at room temperature. After three washes (PBS, 10 min, room temperature), DAPI was used to stain the nuclei of cells. Slides were mounted and stored at 4 °C until analysis. Immunofluorescent micrographs were acquired on a Nikon Eclipse Ti-U (Nikon Instruments Inc.) with a Plan Apo 10×/0.45-NA objective controlled by NIS Elements software (version 4.51; Nikon Instruments). The freeware image-analysis program ImageJ (https://imagej.nih. gov/ij/) was used to remove background and generate merged images. All images were obtained with identical acquisition parameters by individuals blinded to the staining conditions.
Statistical analyses
Behavioral threshold values were statistically analyzed for each foot separately, and the significance of differences was assessed between the averages of at least two pre-injection tests and the mean obtained for each post-injection test. In all tests, baseline data were obtained before and after paclitaxel treatment. Within each treatment group, post-administration means were compared with the contralateral values by nonparametric two-way ANOVA, where time was the within-subjects factor and treatment was the between-subjects factor, followed by post hoc pairwise comparisons (Student–Newman–Keuls method). A P value of <0.05 indicated statistical significance between treatment and nontreatment groups. Data were analyzed and plotted with Graphpad Prism 7.
Supplementary Material
Supplementary information is available for this paper at https://doi.org/10.1038/s41592-018-0186-9.
Acknowledgements
We thank S. Guazzi and M. Festa for technical help with cloning and zebrafish expression. We acknowledge M. Pesce and A. Gino for help with immunohistochemistry. pENN-AAV-hSyn-Cre-WPRE-hGH was a gift from J.M. Wilson (Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA). This work was supported by the 2016 Schaefer Research Scholars Program of Columbia University (to A. Moroni), MIUR PRIN (Programmi di Ricerca di Rilevante Interesse Nazionale; 494 2015, 2015795S5W to A. Moroni), the European Research Council (ERC; 2015 Advanced Grant 495 (AdG) n. 695078 noMAGIC to A. Moroni and G.T.), DFG priority program SPP1926 (to G.T.), the Fondazione Istituto Italiano di Tecnologia (to A.L., A.C., A.J.B. and R.T.), and AIRAlzh Onlus-COOP Italia (fellowship to S.P.).
Footnotes
Laura Alberio: 0000-0002-2887-3343
Andrea Locarno: 0000-0003-0640-1510
Shahad Albadri: 0000-0002-3243-7018
Silvia Moleri: 0000-0003-2277-1378
Aubin Moutal: 0000-0003-4268-1206
Monica Beltrame: 0000-0001-5094-3494
Filippo Del Bene: 0000-0001-8551-2846
Massimo Pasqualetti: 0000-0002-0844-8139
Raffaella Tonini: 0000-0003-1652-4709
Anna Moroni: 0000-0002-1860-406X
Reporting Summary. Further details on research design are available in the Nature Research Reporting Summary linked to this article.
Data availability
Raw data generated and analyzed during the current study are available from the corresponding author on reasonable request. Data have been deposited under the following accession codes: AddGene 117075; GenBank submission MH937726. Source data for Fig. 1 and Supplementary Fig. 9 are available online.
Author contributions
L.A. designed and prepared channel constructs, performed whole-cell patch-clamp experiments in vitro and analyzed the data; A.S. contributed to the design of the final BLINK2 clone; A.P. conducted and analyzed some electrophysiological recordings in vitro; G.R. produced the anti-BLINK2 antibody; G.T. and H.M.C. performed the single-channel in vitro patch experiments and analyzed the data; S.P., E.M. and M.D.L. designed, conducted and analyzed the immunolocalization experiments in rat primary neurons; A.L. designed, performed and analyzed the ex vivo mouse patch-clamp experiments; A.J.B. and A.C. designed and produced the BLINK2 viral constructs; A.L., N.B. and A.J.B. performed intracerebral viral injections; N.B. and M.P. carried out immunofluorescence analysis; E.R., V.B., S.A., F.S., S.M., M.B. and F.D.B. designed, performed and analyzed the zebrafish experiments; K.K. and G.T. designed, performed and analyzed the artificial bilayer measurements; S.L., A. Moutal, Y.J. and R.K. designed, performed and analyzed the pain experiments in rats; R.T. designed and supervised the electrophysiological ex vivo experiments and the production of BLINK2 and GFP-control viral constructs; A. Moroni conceived the study, coordinated research and wrote the manuscript; and G.T., F.D.B., E.M., M.B., R.K. and R.T. contributed to the writing.
Competing interests
The authors declare no competing interests
Reprints and permissions information is available at www.nature.com/reprints.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS One. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 4.Chuong AS, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat Neurosci. 2014;17:1123–1129. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wietek J, et al. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344:409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- 6.Berndt A, Lee SY, Ramakrishnan C, Deisseroth K. Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science. 2014;344:420–424. doi: 10.1126/science.1252367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science. 2015;349:647–650. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wietek J, et al. An improved chloride-conducting channelrhodopsin for light-induced inhibition of neuronal activity in vivo. Sci Rep. 2015;5 doi: 10.1038/srep14807. 14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue K, Kato Y, Kandori H. Light-driven ion-translocating rhodopsins in marine bacteria. Trends Microbiol. 2015;23:91–98. doi: 10.1016/j.tim.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2011;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alfonsa H, et al. The contribution of raised intraneuronal chloride to epileptic network activity. J Neurosci. 2015;35:7715–7726. doi: 10.1523/JNEUROSCI.4105-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raimondo JV, Kay L, Ellender TJ, Akerman CJ. Optogenetic silencing strategies differ in their effects on inhibitory synaptic transmission. Nat Neurosci. 2012;15:1102–1104. doi: 10.1038/nn.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahn M, Prigge M, Ron S, Levy R, Yizhar O. Biophysical constraints of optogenetic inhibition at presynaptic terminals. Nat Neurosci. 2016;19:554–556. doi: 10.1038/nn.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berndt A, et al. Structural foundations of optogenetics: determinants of channelrhodopsin ion selectivity. Proc Natl Acad Sci USA. 2016;113:822–829. doi: 10.1073/pnas.1523341113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaila K, Price TJ, Payne JA, Puskarjov M, Voipio J. Cation-chloride cotransporters in neuronal development, plasticity and disease. Nat Rev Neurosci. 2014;15:637–654. doi: 10.1038/nrn3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szabadics J, et al. Excitatory effect of GABAergic axo-axonic cells in cortical microcircuits. Science. 2006;311:233–235. doi: 10.1126/science.1121325. [DOI] [PubMed] [Google Scholar]
- 17.Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 18.Christie JM, Gawthorne J, Young G, Fraser NJ, Roe AJ. LOV to BLUF: flavoprotein contributions to the optogenetic toolkit. Mol Plant. 2012;5:533–544. doi: 10.1093/mp/sss020. [DOI] [PubMed] [Google Scholar]
- 19.Plugge B, et al. A potassium channel protein encoded by chlorella virus PBCV-1. Science. 2000;287:1641–1644. doi: 10.1126/science.287.5458.1641. [DOI] [PubMed] [Google Scholar]
- 20.Cosentino C, et al. Engineering of a light-gated potassium channel. Science. 2015;348:707–710. doi: 10.1126/science.aaa2787. [DOI] [PubMed] [Google Scholar]
- 21.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuzarte M, et al. Intracellular traffic of the K+ channels TASK-1 and TASK-3: role of N- and C-terminal sorting signals and interaction with 14-3-3 proteins. J Physiol (Lond) 2009;587:929–952. doi: 10.1113/jphysiol.2008.164756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sottocornola B, et al. The potassium channel KAT1 is activated by plant and animal 14-3-3 proteins. J Biol Chem. 2006;281:35735–35741. doi: 10.1074/jbc.M603361200. [DOI] [PubMed] [Google Scholar]
- 24.Sottocornola B, et al. 14-3-3 proteins regulate the potassium channel KAT1 by dual modes. Plant Biol (Stuttg) 2008;10:231–236. doi: 10.1111/j.1438-8677.2007.00028.x. [DOI] [PubMed] [Google Scholar]
- 25.Saponaro A, et al. Fusicoccin activates KAT1 channels by stabilizing their interaction with 14-3-3 proteins. Plant Cell. 2017;29:2570–2580. doi: 10.1105/tpc.17.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagliuca C, et al. Molecular properties of Kcv, a virus encoded K+ channel. Biochemistry. 2007;46:1079–1090. doi: 10.1021/bi061530w. [DOI] [PubMed] [Google Scholar]
- 27.Marcello E, Gardoni F, Di Luca M, Pérez-Otaño I. An arginine stretch limits ADAM10 exit from the endoplasmic reticulum. J Biol Chem. 2010;285:10376–10384. doi: 10.1074/jbc.M109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao S, et al. Cell type–specific channelrhodopsin-2 transgenic mice for optogenetic dissection of neural circuitry function. Nat Methods. 2011;8:745–752. doi: 10.1038/nmeth.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giorgi A, et al. Brain-wide mapping of endogenous serotonergic transmission via chemogenetic fMRI. Cell Rep. 2017;21:910–918. doi: 10.1016/j.celrep.2017.09.087. [DOI] [PubMed] [Google Scholar]
- 30.Mlinar B, Montalbano A, Piszczek L, Gross C, Corradetti R. Firing properties of genetically identified dorsal raphe serotonergic neurons in brain slices. Front Cell Neurosci. 2016;10:195. doi: 10.3389/fncel.2016.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkins KL. Cell-attached voltage-clamp and current-clamp recording and stimulation techniques in brain slices. J Neurosci Methods. 2006;154:1–18. doi: 10.1016/j.jneumeth.2006.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pudasaini A, El-Arab KK, Zoltowski BD. LOV-based optogenetic devices: light-driven modules to impart photoregulated control of cellular signaling. Front Mol Biosci. 2015;2:18. doi: 10.3389/fmolb.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baier H, Scott EK. Genetic and optical targeting of neural circuits and behavior—zebrafish in the spotlight. Curr Opin Neurobiol. 2009;19:553–560. doi: 10.1016/j.conb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Böhm UL, et al. CSF-contacting neurons regulate locomotion by relaying mechanical stimuli to spinal circuits. Nat Commun. 2016;7 doi: 10.1038/ncomms10866. 10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flanagan-Steet H, Fox MA, Meyer D, Sanes JR. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development. 2005;132:4471–4481. doi: 10.1242/dev.02044. [DOI] [PubMed] [Google Scholar]
- 36.Yoo SK, Starnes TW, Deng Q, Huttenlocher A. Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature. 2011;480:109–112. doi: 10.1038/nature10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, et al. Dorsal root ganglion neurons become hyperexcitable and increase expression of voltage-gated T-type calcium channels (Cav3.2) in paclitaxel-induced peripheral neuropathy. Pain. 2017;158:417–429. doi: 10.1097/j.pain.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moutal A, et al. Blocking CRMP2 SUMOylation reverses neuropathic pain. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiegert JS, Mahn M, Prigge M, Printz Y, Yizhar O. Silencing neurons: tools, applications, and experimental constraints. Neuron. 2017;95:504–529. doi: 10.1016/j.neuron.2017.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finnerup NB, et al. Pharmacotherapy for neuropathic pain in adults: a systematic review and meta-analysis. Lancet Neurol. 2015;14:162–173. doi: 10.1016/S1474-4422(14)70251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stachniak TJ, Ghosh A, Sternson SM. Chemogenetic synaptic silencing of neural circuits localizes a hypothalamus→midbrain pathway for feeding behavior. Neuron. 2014;82:797–808. doi: 10.1016/j.neuron.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryksin AV, Matsumura I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques. 2010;48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piccoli G, et al. Proteomic analysis of activity-dependent synaptic plasticity in hippocampal neurons. J Proteome Res. 2007;6:3203–3215. doi: 10.1021/pr0701308. [DOI] [PubMed] [Google Scholar]
- 44.Romani G, et al. A virus-encoded potassium ion channel is a structural protein in the chlorovirus Paramecium bursaria chlorella virus 1 virion. J Gen Virol. 2013;94:2549–2556. doi: 10.1099/vir.0.055251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) 4th ed. Univ. of Oregon Press; Eugene, OR: 2000. [Google Scholar]
- 46.Malinverno M, et al. Synaptic localization and activity of ADAM10 regulate excitatory synapses through N-cadherin cleavage. J Neurosci. 2010;30:16343–16355. doi: 10.1523/JNEUROSCI.1984-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suster ML, Sumiyama K, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics. 2009;10:477. doi: 10.1186/1471-2164-10-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polomano RC, Mannes AJ, Clark US, Bennett GJ. A painful peripheral neuropathy in the rat produced by the chemotherapeutic drug, paclitaxel. Pain. 2001;94:293–304. doi: 10.1016/S0304-3959(01)00363-3. [DOI] [PubMed] [Google Scholar]
- 49.Yaksh TL, Rudy TA. Chronic catheterization of the spinal subarachnoid space. Physiol Behav. 1976;17:1031–1036. doi: 10.1016/0031-9384(76)90029-9. [DOI] [PubMed] [Google Scholar]
- 50.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.