Abstract
Studies have demonstrated that kallikrein-associated peptidase 11 (KLK11) is dysregulated in various cancers. However, the potential roles of KLK11 in esophageal squamous cell carcinoma (ESCC) are still unknown. In our study, we found that the expression of KLK11 in advanced ESCC was significantly down regulated than that in the adjacent tissues, and patients with higher KLK11 expression had markedly increased overall survival rates compared with those with lower KLK11 expression. In addition, up regulation of KLK11 decreased the proliferation capacity of TE-1 and EC18 cells, and down regulation of KLK11 increased the proliferation capacity. To explore the possible mechanism of KLK11 in regulating the proliferation of ESCC, the expression of the related factors in Wnt/β-catenin pathway and cell cycle-mediated factors, such as GSK-3β/p-GSK-3β, β-catenin, Ki67, p-Rb/Rb, CDK6, CDK4 and Cyclin D1, were determined. Furthermore, KLK11 was found to be negatively correlated with the expression of β-catenin in the nucleus, as showed by decreased expression of cyclin D1 and Ki67 through deactivation of the Wnt/β-catenin signaling pathway. XAV-939, a Wnt/β-catenin inhibitor, partially decreased the effects of KLK11 deficiency on ESCC cell proliferation. Finally, we validated that KLK11 inhibited ESCC proliferation in vivo. Our results showed that the inhibitory effects of KLK11 on the proliferation of TE-1 and EC18 cells might be associated with inhibition of Wnt/β-catenin signaling pathway. KLK11 played a key role in inhibiting ESCC carcinogenesis and progression and became a potential biomarker for poor prognosis in patients with ESCC.
Keywords: Esophageal squamous cell carcinoma, kallikrein-associated peptidase 11, Wnt/β-catenin signaling pathway, proliferation
Introduction
Esophageal cancer is one of the most predominant malignant tumors. According to data from the Global Burden of Disease Cancer Collaboration in 2015, more than 483,000 new cases of esophageal cancer and more than 439,000 deaths from esophageal cancer are documented, making it as the 11th most prevalent and the 6th deadliest cancer [1]. ESCC and esophageal adenocarcinoma (EAC) are the two main pathological subtypes of esophageal cancer. Unlike EAC which is prevalent in western countries, ESCC are more found in China. Specifically, Northern and central China, the parts of the ‘Asian belt’, have a very high incidence of ESCC, as showed by more than 100 cases out of the population of 100,000 [2]. With the development of treatment strategy, the 5-year survival rate of early esophageal cancer has improved dramatically, but the risk of death from advanced esophageal cancer remains high. Therefore, a novel biomarker for diagnosing early esophageal cancer needs to be identified.
Kallikrein-associated peptidase constitute a subgroup of 15 secreted trypsin and chymotrypsin-like serine proteases, clustering on chromosome 19q13.4 [3]. KLK proteins show the similarity in biological characteristics, such as the expression patterns and regulatory mechanisms, implicating in various processes in diseases [4]. Several KLK members have been reported to be up regulated in ovarian cancer [5]. KLK6 plays an important role in promoting the growth, migration, and invasion of gastric cancer cells [6]. KLK11 is initially isolated from the human hippocampus and later found to be expressed in a variety of tissues [7]. In addition, KLK11 is highly expressed in chemoresistant cell lines, especially oxaliplatin-resistant cells. Moreover, Knockdown of KLK11 can reverse oxaliplatin resistance by blocking PI3K/AKT signaling pathway [8]. KLK11 expression in laryngeal cancer with the primary or recurrent nature is significantly decreased compared with that in the non-malignant [9]. Serum specimens from 138 patients with non-small cell lung cancer (NSCLC) are collected and found that the expression of KLK11 in NSCLC is significantly higher compared to that in the control [10]. However, the mechanism of KLK11 in the carcinogenesis and progression of esophageal cancer remains unknown. These prompted us to explore the roles of KLK11 in human esophageal cancer.
Materials and methods
Clinical specimen collection
Tissue microarray and immunohistochemistry. A piece of ESCC tissue microarray was used to examine the expression of KLK11. The tissue microarray (Microarray Number: HEso-Squ180Sur-04, Chip lot Number: XT15-012) was purchased from Shanghai Outdo Biotech Co., Ltd. (Shanghai, China) and contained 100 pairs of ESCC samples and 80 pairs of their para-carcinoma tissues. Patients were selected based on a clear pathological diagnosis of early stage (Stages IA-IIIA) ESCC. All patients’ follow-up records were collected from January 2006 to July 2015.
A total of 8 cancer tissue samples of ESCC, along with matched samples of adjacent tissues, were collected from The First Affiliated Hospital of Gannan Medical College, Jiangxi province, China, from January to December in 2017. After the surgical operation, tissue specimens were frozen in liquid nitrogen immediately. None of patients had been treated with radiotherapy or chemotherapy. All patients were well informed, the processes were approved by Ethics Committee of The First Affiliated Hospital of Gannan Medical University, and written informed consent was obtained from each patient.
Immunohistochemical staining and scoring
Immunohistochemistry was performed following a previously published protocol [11]. Simply, after being dewaxed and hydrated, the tissue chip was antigenretrieved by microwaving in citrate buffer (10 mM citric acid, pH 6.0), blocked in 5% animal serum, and incubated with anti-KLK11 primary antibody (dilution, 1:1500) overnight at 4°C. Specimens were developed by DAB and the nuclei were counterstained by hematoxylin. The sections were photographed under a microscope (Zeis, Oberkochen, Germany).
Two senior pathologists were blinded to patients’ outcome and assigned to evaluate the immunoreactivity independently. Intensity of immunostaining was scored as 0 (no immunostaining), 1 (weak immunostaining), 2 (moderate immunostaining), and 3 (strong immunostaining). The percentage of immunoreactive cells scoring was documented as 0 (none), 1 (< 20%), 2 (20-50%), 3 (51-75%), and 4 (> 75%). And a final score was created to determine the cut-off value for low and high expression group by using grades of the extent × grades of intensity staining. The low expression was defined as a final score < 6 and the high expression with a final score ≥ 6.
Cells lines and cell culture
The primary cultures of human normal esophageal epithelial cells (HEEC) and human ESCC cell lines (KYSE-510, EC18, TE-1, KYSE-150, KYSE-410, KYSE-30, and ECA-109) were obtained from the Research Center of Clinical Medicine at Nanfang Hospital in Guangzhou, China. HEEC was cultured in keratinocyte serum free medium (Gibco, Invitrogen, Carlsbad, California, USA) supplemented with 10% (volume/volume) fetal bovine serum (FBS), 40 μg/mL bovine pituitary extract, 1.0 ng/mL epidermal growth factor EGF, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in a humidified incubator under 5% CO2 condition. All of the human ESCC cell lines were grown in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco), supplemented with 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin in the humid atmosphere containing 5% CO2 at 37°C.
Lentivirus vectors and transfection
Three pairs of short hairpin RNA (shRNA) targeting KLK11 lentivirus vectors (shKLK11#1) and negative controls (scramble, NC) were specifically synthesized by Synbio Technologies (Suzhou, China). EC18 cells (EC18-shKLK11#1, EC18-scramble) and TE-1 cells (TE-1-shKLK11#1, TE-1-scramble) were transfected with the lentivirus vectors carrying shKLK11#1 or scramble (Table S1A), respectively. Knockdown efficiency was measured at 48 h post-transfection by western blotting and qPCR. The shRNA with the highest knockdown efficiency was selected for subsequent experiments. KLK11 lentivirus vector (EC18-KLK11 and TE-1-KLK11) and control vector (EC18-vector and TE-1-vector) were purchased from Synbio Technologies (Suzhou, China). Instructions from the manufacturer were followed.
Total RNA extraction and quantitative real-time PCR (qPCR)
Total RNA was obtained from the cell lysates using the Trizol reagent (Invitrogen, Carlsbad, California, USA), according to the manufacturer’s instructions. KLK11 mRNA level was measured by qPCR with the SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Hercules, CA), and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference. The primers of KLK11, GAPDH, Ki67, cyclin D1, CDK4, CDK6, and c-myc were recorded in Table S1B. The qPCR procedures consisted of 50 cycles with an annealing temperature of 60°C. The data were calculated by the 2-ΔΔCt method.
Western blotting
The proteins in ESCC cells and tissues were extracted from the cell lysate using RIPA (Biyotime Biotechnology), and the concentration of protein was tested with a BCA Kit (Biyotime Biotechnology). Equal mass of protein was separated by SDS-PAGE (10% or 8%) and then transferred onto 0.45 µm PVDF membranes (Roche, Indianapolis, IN). The PVDF membranes were incubated in TBST blocking solution containing 5% skimmed milk at room temperature for 1 h. After incubation with the primary antibodies at 4°C overnight, the membrane was washed with TBST for 3 × 10 minutes, followed by incubation with the secondary antibodies at room temperature for 1 h, and with TBST again. Western blotting was performed according to the standard protocol with the following antibodies: anti-KLK11 antibody (Abcam, Cambridge, MA), anti-Ki67 (Abcam, Cambridge, MA), anti-GSK-3β (Abcam, Cambridge, MA), anti-Cyclin D1 antibody (Cell Signaling, Danvers, MA), anti-Rb antibody (Thermo, Waltham, MA), anti-p-Rb antibody (Bioworld Technology, Louis Park, MN, USA), anti-β-Catenin antibody (Cell Signaling, Danvers, MA), anti-CDK6 antibody (Bioworld Technology, Louis Park, MN, USA), anti-CDK4 antibody (Bioworld Technology, Louis Park, MN, USA), anti-GAPDH antibody (Bioworld Technology, Louis Park, MN, USA), anti-p84 antibody (Abcam, Cambridge, MA).
Cell proliferation assay
TE-1 and EC18 cells transfected with KLK11, vector, shKLK11#1, and scramble, respectively, were cultured in 96-well plates (2000 cells/well) for 24 h. According to the manufacturer’s protocol, 20 µl of 5 mg/mL MTT solution (MTT Cell Proliferation and Cytotoxicity Assay Kit, BOSTER, China) was added into the culture medium, and then incubated at 37°C for 4 h. The optical density at 570 nm (OD450) was measured. Each experiment was performed in triplicate.
Colony formation assay
The cells in the logarithmic growth phase were transfected with KLK11, vector, shKLK11#1, and scramble, respectively, and divided into 6-well plates (1000 cells/well). After 14 days culture, cells were fixed with 75% ethanol for 30 min and then stained with 0.2% crystal violet. Count the number of clones per well and take a photo. Each experiment was repeated 3 times.
Flow cytometry
To measure the dividing cells by flow cytometry, TE-1 and EC-18 cells transfected with KLK11, vector, shKLK11#1, and scramble, respectively, were fixed with 70% ethanol overnight. Then, propidium iodide and RNaseA were added, dyeing at 37°C for 30 min. Finally, the sample was detected by LSR FORTESSA (BD Biosciences, Franklin Lakes, NJ, USA). The results were analyzed using ModFit LT2.0 software (Verity Software House, Topsham, ME, USA).
Luciferase assays
TE-1 and EC-18 cells (3.5 × 104) transfected with KLK11, vector, shKLK11#1, and scramble, respectively, were seeded in 24-well plates for 24 h. About 100 ng of TOP Flash-luciferase plasmid (Clontech, Mountain View, CA) and FOP Flash-luciferase plasmid (Clontech, Mountain View, CA) plus 1 ng of pRL-TK renilla plasmid (Promega) were transfected into cells using the lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instructions. Luciferase and renilla signals were measured after transfection for 48 h using the Dual Luciferase Reporter Assay Kit (Promega). Three independent experiments were performed, and data are presented as mean ± SD.
In vivo experiments
Suspensions of 1 × 106 cells (TE-1-shKLK11#1, TE-1-scramble, TE-1-KLK11, and TE-1-vector) in 0.1 ml of PBS were injected subcutaneously into the backs of BALB/C nude mice (4-6 weeks, 18-22 g), which were obtained from Medical Laboratory Animal Center of Guangdong Province. Five mice were used in each experimental group. Tumor growth was assessed by measuring the xenografts in two dimensions once a week. Tumor volumes were calculated according to the following formula: (volume) = 1/2 × (long axis) × (short axis)2. After post-injection for 28 days, mice were sacrificed, and the tumors were carefully dissected. All animal experiments were performed in accordance with the principles and procedures outlined in the Nanfang Medical University Guide for the Care and Use of Animals under the assurance number SCXK (Guangdong) 2008-0002. Approval was obtained from the Nanfang Hospital Animal Ethics Committee.
Statistical analysis
Statistical analyses were performed using SPSS 22.0. Student’s t test or chi-squared test was used to analyze the data. Survival curves were constructed by using the Kaplan-Meier method and analyzed by the log-rank test. P < 0.05 was considered statistically significant.
Results
KLK11 expression was significantly down-regulated in esophageal cancer patients
We first analyzed microarray datasets from TCGA databases. The results showed that KLK11 expression was significantly reduced in tumor tissues compared with that in normal esophageal tissues (Figure 1A, 1B). Western blotting data showed that KLK11 protein expression was obviously decreased in ESCC tissues (T = 8) compared with that in the matched cancer-adjacent tissues (N = 8) (Figure 1C, 1D). Finally, immunohistochemistry analysis demonstrated that KLK11 expression was significantly lower in tumor tissues compared with that in the matched adjacent non-tumor tissues (Figure 1E, 1F). We found that the immunoreactivity of KLK11 was dysregulated in 67 (67%) or 33 (33%) of the 100 ESCC samples (Figure 1G).
Figure 1.
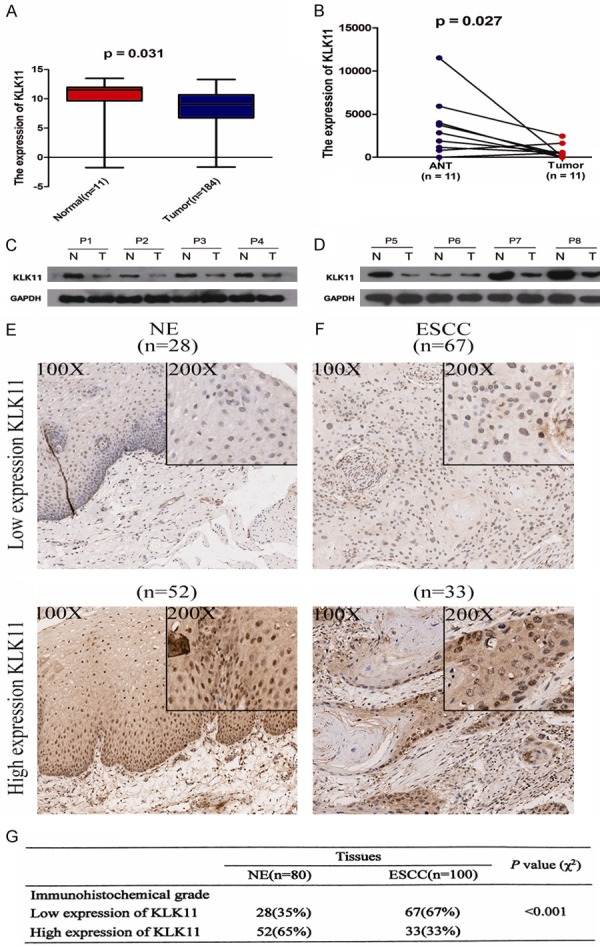
The expression of KLK11 is significantly lower in ESCC tissues. A and B. Indicated detection of KLK11 expression in dataset TCGA. C and D. Indicated KLK11 protein expression in 8 pairs of ESCC tissues and their para-carcinoma tissues by western blotting. KLK11 expression in ESCC tissues were significantly lower than those in the matched adjacent non-tumor tissues. E. KLK11 protein level was measured by immunohistochemical analysis in normal tissues. F. KLK11 protein level was measured by immunohistochemical analysis in ESCC tissues. Representative images are showed at 100 × and 200 × magnifications, respectively. G. KLK11 protein level in 100 ESCC tissues and matched normal tissues. Data were representative of three independent experiments (mean and SEM). *P < 0.05.
KLK11 was correlated with clinicopathological features and prognosis in ESCC patients
The relationship between clinicopathological characteristics and KLK11 expression in patients with ESCC was summarized in Table 1. KLK11 expression was prominently correlated with tumor classification (P = 0.011), lymphatic metastasis (P = 0.019), and TNM stage (P = 0.005). There was no significant relevance in ESCC patients with age, gender, and pathology. However, TNM stage (P = 0.002), lymph node metastasis (P = 0.012), tumor classification (P = 0.001), and KLK11 expression (P = 0.034) were statistically significant in ESCC patients, compared with those in the control group (Table 2). KLK11 expression was decreased in the advanced ESCC and negatively associated with TNM stage and metastasis. These indicated that KLK11 might inhibit the progression of ESCC.
Table 1.
KLK11 expression was significantly associated with clinical characteristics
| Characteristics | No. (n = 100) | KLK11 expression | P | |
|---|---|---|---|---|
|
| ||||
| High (n = 33) | Low (n = 67) | |||
| Gender | 1.000 | |||
| Male | 74 | 24 (72.7%) | 50 (74.6%) | |
| Female | 26 | 9 (27.3%) | 17 (25.4%) | |
| Age (years) | 0.398 | |||
| < 65 | 45 | 17 (51.5%) | 28 (41.8%) | |
| ≥ 65 | 55 | 16 (48.5%) | 39 (58.2%) | |
| Pathology | 0.479 | |||
| G1-G2 | 72 | 22 (66.7%) | 50 (74.6%) | |
| G3 | 28 | 11 (33.3%) | 17 (25.4%) | |
| Tumor classification | 0.011 | |||
| T1-2 | 18 | 13 (39.4%) | 5 (7.5%) | |
| T3-4 | 82 | 20 (60.6%) | 62 (92.5%) | |
| Lymph node metastasis | 0.019 | |||
| Absent | 46 | 21 (63.6%) | 25 (37.3%) | |
| Present | 54 | 12 (36.4%) | 42 (62.7%) | |
| TNM stage (AJCC) | 0.005 | |||
| I-II | 49 | 23 (69.7%) | 26 (38.8%) | |
| III | 51 | 10 (30.3%) | 41 (61.2%) | |
χ2 test was used. Statistical significance (P < 0.05) was showed.
Table 2.
Summary of univariate and multivariate Cox regression analyses of overall survival
| Parameters | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| P-value | HR | 95% CI | P-value | HR | 95% CI | |
| Gender (male vs. female) | 0.800 | 0.938 | 0.570-1.542 | |||
| Age (< 65 vs. ≥ 65) | 0.511 | 0.863 | 0.577-1.338 | |||
| Pathology (G1-2 vs. G3) | 0.057 | 1.582 | 0.986-2.536 | |||
| Lymph node metastasis (Absent vs. Present) | < 0.001 | 3.285 | 2.018-5.347 | 0.012 | 2.020 | 1.169-3.491 |
| Tumor classification (T1-2 vs. T3-4) | < 0.001 | 4.363 | 2.128-8.944 | 0.001 | 3.826 | 1.784-8.205 |
| TNM stage (I-II vs. III) | < 0.001 | 3.918 | 2.380-6.448 | 0.002 | 2.553 | 1.420-4.590 |
| KLK11 expression (Low vs. High expression) | 0.001 | 2.335 | 1.400-3.894 | 0.034 | 1.839 | 1.047-3.228 |
HR: Hazard ratio; CI: Confidence interval. The bold number represents the P-values with significant differences.
To investigate the prognostic value of KLK11 expression in ESCC, the association between KLK11 expression and patients’ survival rate was assessed using Kaplan-Meier analysis with the log-rank test (Figure 2). The results showed that patients with low expression of KLK11 had a higher survival rate than patients with high expression of KLK11. These findings revealed that KLK11 served as a potential prognosis predictor in ESCC.
Figure 2.
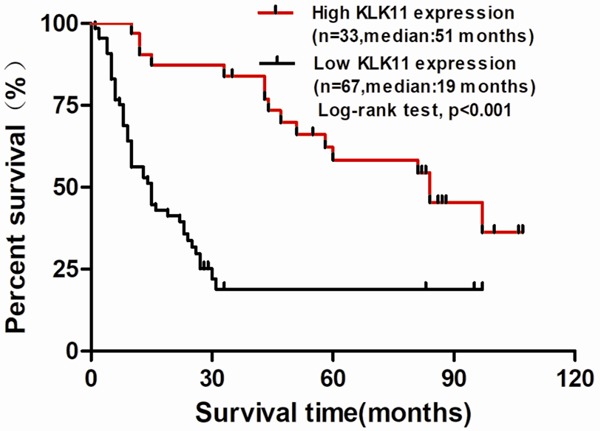
Low KLK11 expression was associated with poor prognosis in ESCC patients. ESCC samples from the tissue microarray were divided as two different groups, high KLK11 expression and low KLK11 expression group. Patients’ survival information was provided by the manufacture. Kaplan-Meier analysis of overall survival was done for the two group.
KLK11 inhibited ESCC cell proliferation and reduced colony formation in vitro
To explore the biological functions of KLK11 in regulation of ESCC proliferation, we measured the mRNA and protein expression of KLK11s in ESCC cell lines and HEEC. The expression of KLK11 is significantly lower in ESCC cell lines (Figure 3A, 3B). KLK11 shRNA was used to examine the functional roles of KLK11 in ESCC. The knockdown efficiency was confirmed by qPCR and Western blotting. The results demonstrated that shRNA1 exhibited the highest knockdown efficiency (Figure 3C, 3D). Therefore, shRNA1 was selected for subsequent experiments. Furthermore, qPCR and western blotting also confirmed successful restoration of KLK11 expression in four groups of TE-1 and EC18 cells (Figure 3E-H).
Figure 3.
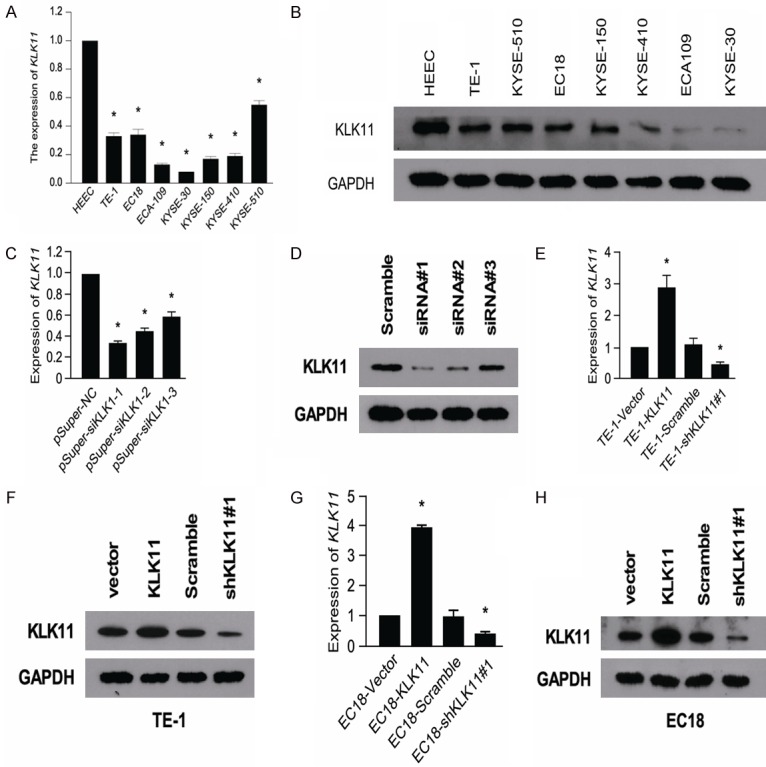
The expression of KLK11 is significantly lower in ESCC cell lines. QPCR (A) and western blotting (B) showed the expression of KLK11 in seven ESCC cell lines and HEEC. KLK11 knockdown efficiency was confirmed by qPCR (C) and western blotting (D) in ESCC cells. qPCR (E) and western blotting (F) confirmed the restoration of KLK11 expression in four groups of TE-1 cells. qPCR (G) and western blotting (H) confirmed the restoration of KLK11 expression in four groups of EC18 cells. Results were showed as mean ± SD (*P < 0.05) of triplicate determination from three independent experiments.
To verify the knockdown efficiency of shRNAs, shRNA#1 and shRNA#2 were selected for MTT assays and Western blotting. Results of MTT assays indicated that transfection of KLK11 shRNA#1 and shRNA#2 into TE-1 and EC18 cells, respectively, led to increased cell proliferation. Compared with KLK11 shRNA#1 group, KLK11 shRNA#2 group had a similar trend in cells proliferation in TE-1 and EC18 (Figure S1A, S1B). The protein expression of Ki67, cyclin D1, β-catenin (nucleus) and β-catenin (cytosol) in TE-1cells (shKLK11#1, shKLK11#2, and scramble) and EC18 cells (shKLK11#1, shKLK11#2, and scramble) were detected by Western blotting. We found that transfection of KLK11 shRNA#1 and shRNA#2 into TE-1 and EC18 cells led to increased expression of Ki67, cyclin D1, β-catenin (nucleus). KLK11 shRNA#1 group and KLK11 shRNA#2 group also had a similar trend (Figure S1C, S1D). Then, we chose shRNA#1 for the subsequent experiments.
MTT assays showed that KLK11 overexpression in TE-1 and EC18 cells resulted in significant inhibition of cell proliferation compared with the negative control group. In contrast, transfection of KLK11 shRNA into TE-1 and EC18 cells led to increased cell proliferation (Figure 4A, 4B). Similarly, increased expression of KLK11 significantly reduced the cell colony numbers in TE-1 and EC18 cells, and KLK11 knockdown had the opposite effects (Figure 4C). These results indicated that KLK11 inhibited the growth of ESCC cells in vitro. Analysis of cell cycle phase by flow cytometry revealed a G0/G1 arrest induced by KLK11. The percentage of KLK11 overexpression groups EC18-KLK11 and TE-1-KLK11 in G0/G1 were 71.05% and 70.26%, respectively. KLK11 knockdown group had the most percentage of cells in the S phase of cell cycle (Figure 4D). These indicated that KLK11 reduced the entry of ESCC cells into the S phase and the proliferation of ESCC cells.
Figure 4.
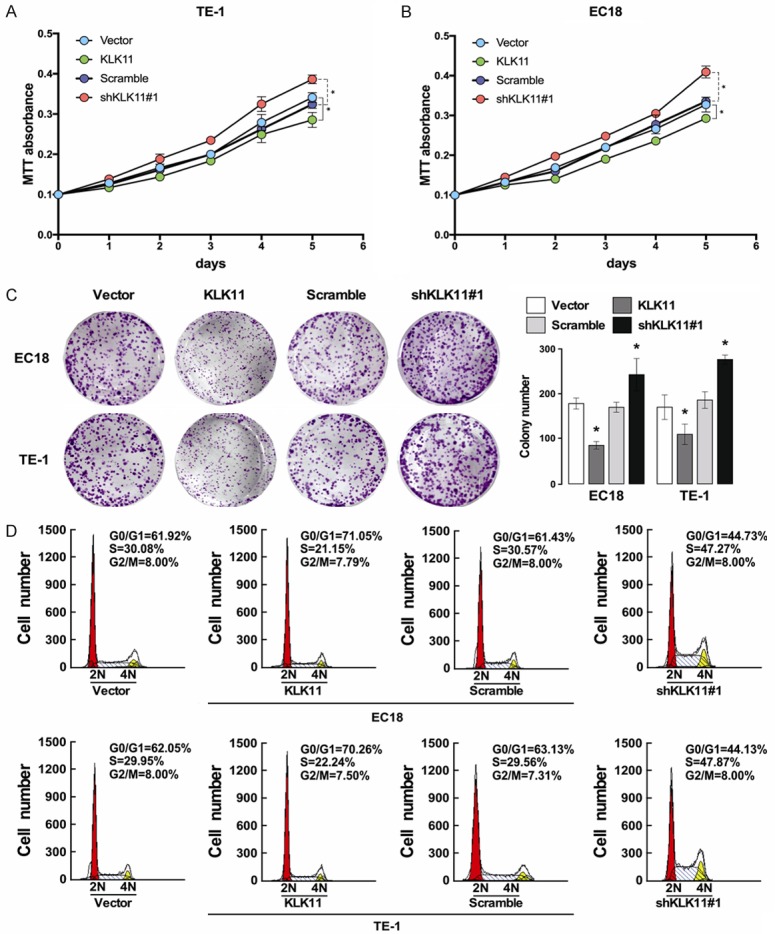
KLK11 inhibited the proliferation of ESCC cells. A. KLK11 inhibited the proliferation of TE-1 cells by MTT assays. B. KLK11 inhibited the proliferation of EC18 cells by MTT assays. C. Colony formation assays showed that KLK11 overexpression inhibited TE-1 and EC18 cells proliferation. These effects were abolished by treatment with shKLK11. D. Flow cytometry analysis of cell cycle phase revealed a G0/G1 arrest induced by KLK11. Data were representative of three independent experiments (mean and SEM). *P < 0.05.
KLK11 affected the expression of the related factors in Wnt/β-catenin pathway and cell cycle-mediated factors
To explore the possible mechanism of KLK11 in regulating the proliferation of ESCC, we investigated the expression of the related proteins in Wnt/β-catenin pathway. Compared with the scramble group, the TE-1-shKLK11#1 (EC18-shKLK11#1) cells group showed an upregulation in the phosphorylation levels of GSK-3β. Meanwhile, TE-1-KLK11 (EC18-KLK1) cells group showed lower expression of p-GSK-3β (Figure 5A). The dual luciferase assay showed that the TE-1-shKLK11#1 (EC18-shKLK11#1) cells group exhibited higher luciferase activity than the scramble cell group. In contrast, TE-1-KLK11 (EC18-KLK1) cells group showed lower luciferase activity (Figure 5B). These results suggested that KLK11 inhibited Wnt/β-catenin signaling pathway. The TE-1 and EC18 cells transfected with KLK11 lentivirus showed decreased expression of β-catenin in the nucleus, compared with that in the cytosol. These indicated that KLK11 could inhibit the translocation of β-catenin into the nucleus. Western blot assay showed that KLK11 was negatively correlated with the expression of cyclin D1, CDK4, CDK6, and p-Rb (Figure 5C). The mRNA expression of Ki67, cyclin D1, CDK4, CDK6, and c-myc were detected by qPCR in TE-1 cells (TE-1-shKLK11#1, TE-1-scramble, TE-1-KLK11, and TE-1-vector) and EC18 cells (EC18-shKLK11#1, EC18-scramble, EC18-KLK11, and EC18-vector). We found that KLK11 was negatively correlated with the mRNA expression of Ki67, cyclin D1, CDK4, CDK6, and c-myc (Figure 5D). These suggested that the inhibitory effects of KLK11 on the proliferation of ESCC cells might be associated with down regulation of Ki67, cyclin D1, CDK4, CDK6, and c-myc expression.
Figure 5.
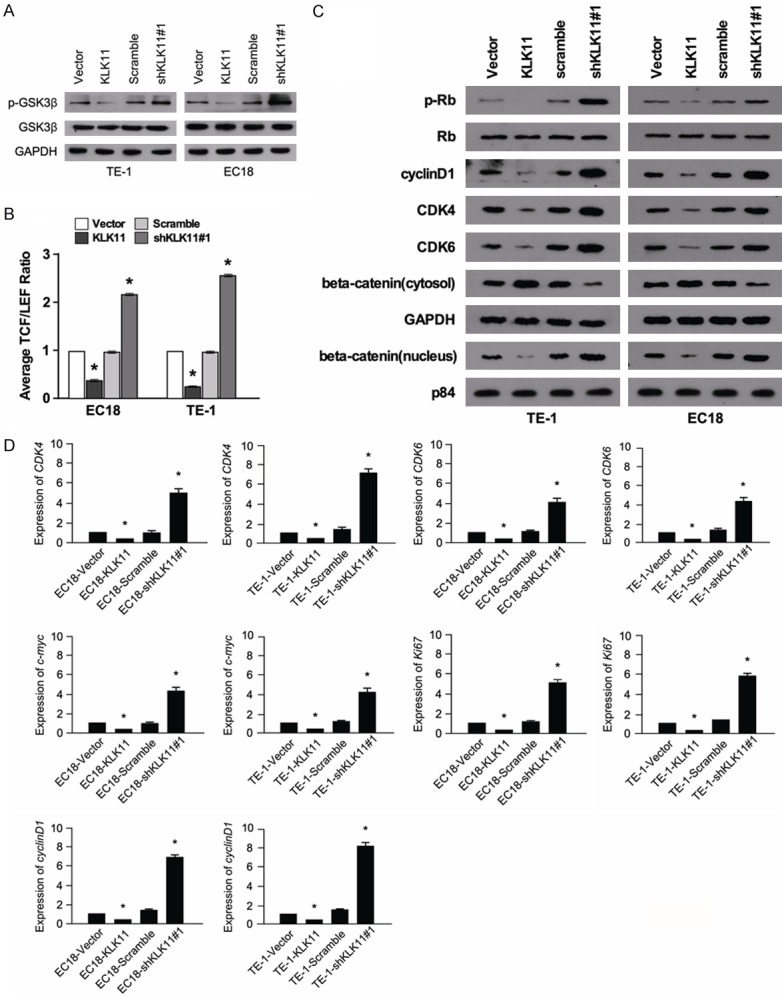
KLK11 affected the expression of the related factors in Wnt/β-catenin pathway and cell cycle-mediated factors. A. The expression of p-GSK-3β/GSK-3β in EC18 and TE-1 cells after lentivirus vectors delivery in each group as measured by western blot. B. The TCF/LEF luciferase ratio reported the activity of Wnt/β-catenin pathway in the indicated cells. C. Expression of CDK4, cyclin D1, CDK6, p-Rb, Rb, β-catenin (cytosol), and β-catenin (nucleus) were determined in TE-1 and EC18 cells transfected with KLK11. KLK11 inhibited the proliferation of ESCC by regulating the expression of the above proteins. D. Analysis of Ki67, cyclin D1, CDK4, CDK6, and c-myc expression in KLK11-overexpression or KLK11-knockdown ESCC cells by qPCR. Data were representative of three independent experiments (mean and SEM). *P < 0.05.
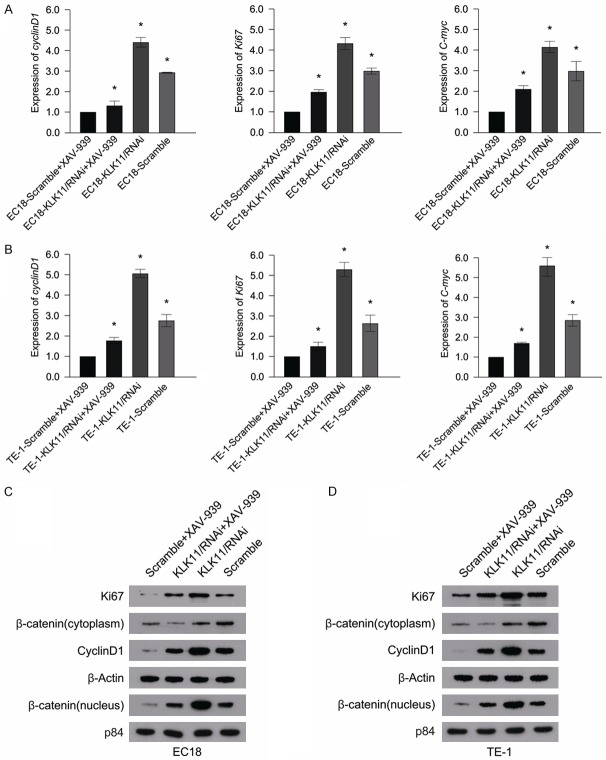
KLK11 down regulated the expression of cyclin D1 and Ki67 through deactivation of the Wnt/β-catenin signaling pathway
To explore the mechanism of KLK11 in mediating Wnt/β-catenin pathway, cells (TE-1-shKLK11, TE-1-scramble, EC18-shKLK11, and EC18-scramble) were treated with Wnt/β-catenin signaling inhibitor (XAV-939). The mRNA and protein expression of both ki67 and Cyclin D1 were suppressed by XAV-939 (Figure 6A-C). Therefore, we suggested that KLK11 down regulated the expression of ki67 and Cyclin D1 by inhibiting Wnt/β-catenin pathway. In addition, the proliferative capacity of ESCC cells treated with XAV-939 was lower than those in the control groups (Figure 7A-C). These suggested that KLK11 suppresses cellular proliferation via inhibition of Wnt/β-catenin signaling pathway in ESCC.
Figure 6.
KLK11 down regulated the expression of Ki67 and cyclin D1 through deactivation of the Wnt/β-catenin signaling pathway. A, B. ESCC cells were pre-treated with XAV-939 for 24 h before harvesting, and the detection of Ki67, cyclin D1, c-myc, and expression levels were performed by qPCR. C, D. Treatment of EC18 and TE-1 cells with XAV-939, the expression of Ki67, cyclin D1, β-catenin (cytosol), and β-catenin (nucleus) were significantly inhibited. Data were representative of three independent experiments (mean and SEM). *P < 0.05.
Figure 7.
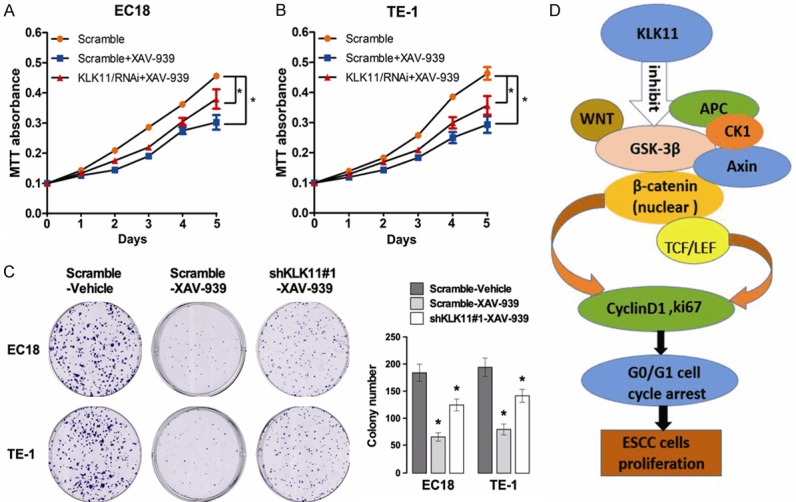
KLK11 suppressed ESCC cells proliferation through inhibiting the activity of the Wnt/β-catenin pathway. A, B. XAV-939 was added to TE-1 and EC18 cells transfected with shRNA, and the proliferation of TE-1 and EC18 cells was detected by MTT assays. C. XAV-939 was added to TE-1 and EC18 cells transfected with shRNA, and the proliferation of TE-1 and EC18 cells was detected by colony formation assays. D. The relationship between KLK11 and the Wnt/β-catenin pathway. Data were representative of three independent experiments (mean and SEM). *P < 0.05.
KLK11 inhibited ESCC proliferation in vivo
To determine whether KLK11 could inhibit tumor growth in vivo, four group cells (TE-1-shKLK11#1, TE-1-scramble, TE-1-KLK11, and TE-1-vector) were implanted subcutaneously into the flanks of nude mice. The growth of tumor cells in mice injected with TE-1-KLK11 cells was slower than those injected with TE-1-vector group. In contrast, the group injected with TE-1-shKLK11#1 cells formed significantly larger tumors in volume, compared with the group injected with TE-1-scramble cells (Figure 8A, 8B). Then, tumor tissues from these mice were obtained and prepared for qPCR and immunohistochemical analysis. We found that KLK11 was negatively correlated with Cyclin D1 expression (Figure 8C, 8D). Together, these results demonstrated that KLK11 might inhibit the activity of Wnt/β-catenin signaling pathway and suppress tumor proliferation in vivo.
Figure 8.
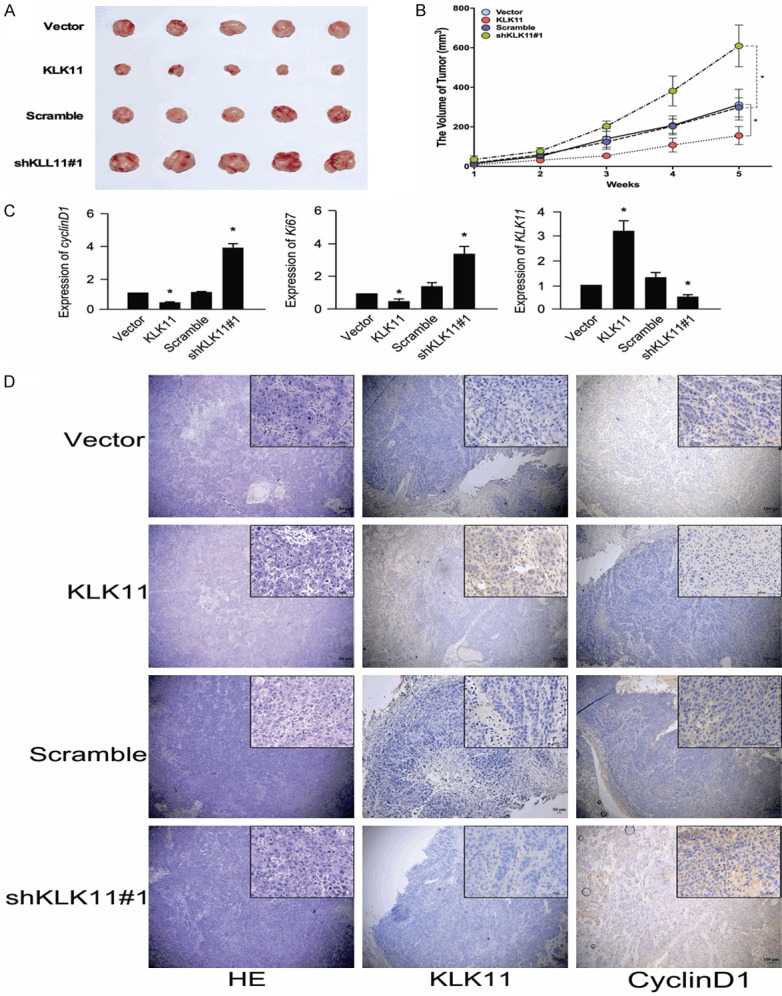
KLK11 inhibited the proliferation of ESCC in vivo. (A, B) Tumor xenograft volume (A) and tumor size (B) in shRNA-KLK11-treated nude mice were bigger than those in the scramble group. Tumor sizes in KLK11 overexpression nude mice were smaller than those in the vector group. (C) Analysis of Ki67, cyclin D1, and KLK11 expression in mice tumor tissues with KLK11 overexpression or KLK11 knockdown by qPCR. (D) Analysis of KLK11 and Cyclin D1 in xenografts by Immunohistochemistry. Data were representative of three independent experiments (mean and SEM). *P < 0.05.
Discussion
The mechanism of tumorigenesis and development of ESCC has not been fully understood. With the development of diagnostic and therapeutic techniques, the prognosis and survival of ESCC have been significantly improved, particularly for the early ESCC. Therefore, it is necessary to find a new tumor biomarker for the diagnosis and treatment of ESCC.
The relationship between KLK11 and ESCC has been rarely reported. This article is the first report to explore the mechanism of KLK11 in regulating ESCC proliferation. KLK11 was down regulated in ESCC and correlated with worse overall survival rates (OS) of the patients. The effects of KLK11 on ESCC proliferation were detected. KLK11 inhibited the activity of Wnt/β-catenin signaling pathway and suppressed ESCC proliferation.
Accumulating evidence has reported that KLK11 plays critical roles in suppressing tumor progression of many human malignant cancers [12]. Geng X found that elevated mRNA levels of KLK11 were significantly linked with prolonged overall survival (OS; P = 0.021) and progression-free survival (PFS; P = 0.008) in ovarian cancer [13]. Jamaspishvili T showed statistically significant differences for all studied proteins between benign prostatic hyperplasia and prostate cancer. Both KLK7 (P = 0.026) and KLK11 (P < 0.001) expressions were decreased in prostate cancer cells compared to those in normal/benign cells [14]. Thus, KLK11 exerts a tumor suppressing effect on these tumors. Controversially, Xu Z, et al found that knockdown of KLK11 increased the oxaliplatin chemosensitivity of colorectal cancer cells. KLK11 inhibition increased oxaliplatin-induced apoptosis, which might be associated with activation of caspase-3 cleavage and the apoptosis signaling pathway [15]. Higher levels of KLK11 in gastric carcinoma were related to worse overall survival (P = 0.008) [16]. Ehrenfeld P, et al demonstrated that up-regulation of KLK11 and KLK6 expression increased cell proliferation and invasion in breast cancer cells [17]. KLK11 presents different profiles in different tumors. These suggest that the mechanism by which KLK11 is involved in tumorigenesis and development may be complex.
We confirmed that KLK11 could attenuate the activity of Wnt/β-catenin signaling pathway and suppress tumor proliferation in ESCC. Zhang Y, et al proved that knockdown of KLK11 activated apoptosis via suppressing PI3K/AKT pathway in colorectal cancer cells [8]. In our studies, we found that KLK11 suppressed the translocation of β-catenin into the nucleus, leading to inhibition of Wnt/β-catenin signaling pathway. Wnt/β-catenin signaling plays key roles in tissue homeostasis and cell fate decisions in embryonic and post-embryonic development. Aberrant regulation of such pathway is associated with developmental disorders and cancers [18]. The signaling function of β-catenin/Armadillo in the nucleus is mediated via TCF/LEF transcription factors, which are in association with Wnt-mediated transcription [19]. Valenta T, et al suggested that imbalance in the structural and signaling properties of β-catenin resulted in development of diseases, including cancers [20]. Increased levels of β-catenin in the nucleus initiate the transcription of TCF/LEF, which leads to the proliferation of ESCC cells. Kang S, et al also found that ubiquitin-specifc protease 6 N-terminal-like protein exerted carcinogenesis via activation of Wnt/β-catenin pathway in colorectal cancer cell [21]. The nuclear Ki67 protein is exclusively expressed in the proliferating cells [22]. It has been reported that increased expression of Ki67 is related to a poor prognosis in cancers [23]. High proliferative activity is a hallmark of malignant tumors, and Ki67 has been regarded as a cancer biomarker. As expected, Ki67 expression is particularly high in the poorly differentiated cancer tissues [24]. Cyclin D1 has attracted widespread attention, due to the prevalence of its dysregulation in human cancers [25]. Reduced degradation and decreased cytoplasmic accumulation of β-catenin results in increased translocation into the nuclear, where it associates with LEF/TCF and drives the expression of downstream target genes. The CCND1 gene, which encodes cyclin D1, represents a key target. β-catenin/LEF-1 complexes bind to the motifs at -75 and -15 within the CCND1 promoter [26,27]. Treatment with Wnt/β-catenin signaling pathway inhibitor XAV-939, the expression of cyclin D1 and ki67 were decreased.
In conclusion, our results validated that KLK11 functioned as a tumor suppressor to attenuate the tumorigenesis and progression of ESCC by inhibition of the activity of Wnt/β-catenin signaling pathway (Figure 7D). We suggested that KLK11 could be a potential biomarker for poor prognosis in ESCC patients and become a therapeutic target in the strategy of managing ESCC.
Acknowledgements
This study was funded by the Natural Science Foundation of Jiangxi province, China (No. 20181BAB215008), the Science and Technology Plan Project of Ganzhou, Jiangxi, China (No. (2017) 179).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bollschweiler E, Plum P, Mönig SP, Hölscher AH. Current and future treatment options for esophageal cancer in the elderly. Expert Opin Pharmacother. 2017;18:1001–1010. doi: 10.1080/14656566.2017.1334764. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 3.Borgono CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer. 2004;4:876–90. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- 4.Yousef GM, Diamandis EP. The new human tissue kallikrein gene family: structure, function, and association to disease. Endocr Rev. 2001;22:184–204. doi: 10.1210/edrv.22.2.0424. [DOI] [PubMed] [Google Scholar]
- 5.Gong W, Liu Y, Seidl C, Dreyer T, Drecoll E, Kotzsch M, Bronger H, Dorn J, Magdolen V. Characterization of kallikrein-related peptidase 4 (KLK4) mRNA expression in tumor tissue of advanced high-grade serous ovarian cancer patients. PLoS One. 2019;14:e0212968. doi: 10.1371/journal.pone.0212968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu S, Shi J, Zhang S, Li Z. KLK6 promotes growth, migration, and invasion of gastric cancer cells. J Gastric Cancer. 2018;18:356–367. doi: 10.5230/jgc.2018.18.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida S, Taniguchi M, Suemoto T, Oka T, He X, Shiosaka S. cDNA cloning and expression of a novel serine protease, TLSP. Biochim Biophys Acta. 1998;1399:225–228. doi: 10.1016/s0167-4781(98)00116-x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Xu Z, Sun Y, Chi P, Lu X. Knockdown of KLK11 reverses oxaliplatin resistance by inhibiting proliferation and activating apoptosis via suppressing the PI3K/AKT signal pathway in colorectal cancer cell. Onco Targets Ther. 2018;11:809–821. doi: 10.2147/OTT.S151867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patsis C, Yiotakis I, Scorilas A. Diagnostic and prognostic significance of human kallikrein 11 (KLK11) mRNA expression levels in patients with laryngeal cancer. Clin Biochem. 2012;45:623–30. doi: 10.1016/j.clinbiochem.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Xu CH, Zhang Y, Yu LK. The diagnostic and prognostic value of serum human kallikrein-related peptidases 11 in non-small cell lung cancer. Tumour Biol. 2014;35:5199–203. doi: 10.1007/s13277-014-1674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rong Z, Yi L, Zhao JJ, Jun PH, Yu W, Xu FL, Wei BX, Xiao CW, Ji RZ, Qi EW, Yan FZ. EPB41L3 is a potential tumor suppressor gene and prognostic indicator in esophageal squamous cell carcinoma. Int J Oncol. 2018;52:1443–1454. doi: 10.3892/ijo.2018.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filippou PS, Karagiannis GS, Musrap N, Diamandis EP. Kallikrein-related peptidases (KLKs) and the hallmarks of cancer. Crit Rev Clin Lab Sci. 2016;53:277–91. doi: 10.3109/10408363.2016.1154643. [DOI] [PubMed] [Google Scholar]
- 13.Geng X, Liu Y, Diersch S, Kotzsch M, Grill S, Weichert W, Kiechle M, Magdolen V, Dorn J. Clinical relevance of kallikrein-related peptidase 9, 10, 11, and 15 mRNA expression in advanced high-grade serous ovarian cancer. PLoS One. 2017;12:e0186847. doi: 10.1371/journal.pone.0186847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamaspishvili T, Scorilas A, Kral M, Khomeriki I, Kurfurstova D, Kolar Z, Bouchal J. Immunohistochemical localization and analysis of kallikrein-related peptidase 7 and 11 expression in paired cancer and benign foci in prostate cancer patients. Neoplasma. 2011;58:298–303. [PubMed] [Google Scholar]
- 15.Xu Z, Chi P, Pan J, Shen S, Sun Y, Wang X, Lu X. Knockdown of KLK11 inhibits cell proliferation and increases oxaliplatin sensitivity in human colorectal cancer. Exp Ther Med. 2016;12:2855–2860. doi: 10.3892/etm.2016.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolin DL, Sy K, Rotondo F, Bassily MN, Kovacs K, Brezden-Masley C, Streutker CJ, Yousef GM. Prognostic significance of human tissue kallikrein-related peptidases 11 and 15 in gastric cancer. Tumour Biol. 2016;37:437–46. doi: 10.1007/s13277-015-3802-7. [DOI] [PubMed] [Google Scholar]
- 17.Ehrenfeld P, Manso L, Pavicic MF, Matus CE, Borquez C, Lizama A, Sarmiento J, Poblete MT, Bhoola KD, Naran A, Figueroa CD. Bioregulation of kallikrein-related peptidases 6, 10 and 11 by the kinin B1 receptor in breast cancer cells. Anticancer Res. 2014;34:6925–38. [PubMed] [Google Scholar]
- 18.Schaefer KN, Peifer M. Wnt/Beta-catenin signaling regulation and a role for biomolecular condensates. Dev Cell. 2019;48:429–444. doi: 10.1016/j.devcel.2019.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 20.Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31:2714–36. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun K, He SB, Yao YZ, Qu JG, Xie R, Ma YQ, Zong MH, Chen JX. Tre2 (USP6NL) promotes colorectal cancer cell proliferation via Wnt/β-catenin pathway. Cancer Cell Int. 2019;19:102. doi: 10.1186/s12935-019-0823-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–22. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 23.Karamitopoulou E, Perentes E, Tolnay M, Probst A. Prognostic significance of MIB-1, p53, and bcl-2 immunoreactivity in meningiomas. Hum Pathol. 1998;29:140–5. doi: 10.1016/s0046-8177(98)90224-6. [DOI] [PubMed] [Google Scholar]
- 24.Yang C, Zhang J, Ding M, Xu K, Li L, Mao L, Zheng J. Ki67 targeted strategies for cancer therapy. Clin Transl Oncol. 2018;20:570–575. doi: 10.1007/s12094-017-1774-3. [DOI] [PubMed] [Google Scholar]
- 25.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 26.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben-Ze’ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med (Berl) 2016;94:1313–1326. doi: 10.1007/s00109-016-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.