Abstract
This study assessed the biological functions of LIM-domain-only 4 (LMO4) in gastric cancer (GC) and investigated the underlying molecular mechanisms. It was found that the expression of LMO4 was significantly upregulated in GC tissues and closely associated with clinicopathological factors, overall survival and disease-free survival of patients. After knockdown of LMO4 in MGC-803 and SGC-7901 cells, invasion and proliferation were obviously suppressed. Furthermore, LMO4 knockdown suppressed the phosphorylation of phosphatidylinositol 3-kinase (PI3K), Akt and mammalian target of rapamycin (mTOR). Miltefosine, the inhibitor of PI3K/Akt, and dactolisib, the inhibitor of mTOR, abrogated recombinant LMO4-induced GC cell invasion and proliferation. These results suggest that LMO4 promotes GC cell invasion and proliferation mainly through PI3K-Akt-mTOR signaling. LMO4 may serve as a potential therapeutic target for GC in the future.
Keywords: LIM domain only 4, gastric cancer, invasion, proliferation, PI3K-Akt, mTOR
Introduction
Gastric cancer (GC) is the second most common type of malignancy worldwide. In developed Western countries, the five-year survival rate for GC is as low as 10-19% [1]. In China, GC is the most common type of malignancy according to the number of cases diagnosed annually [2]. It is currently elusive what factors contribute to the development, progression and metastasis of GC in geographic areas with high prevalence, including certain cities in Northwest China [3,4]. GC includes two distinct morphological subtypes: gastric intestinal-type adenocarcinoma and diffuse gastric adenocarcinoma [5-7]. In addition to tumor protein 53 mutations, studies have reported frequent inactivating mutations in genes associated with cell adhesion and chromatin remodeling [8,9]. Although certain essential factors that may serve as therapeutic targets have been identified in recent years, effective methods to treat GC or monitor disease progression in the clinic are still lacking [10-13].
LMO4 is a member of the LIM-only family of proteins (LMO). The main members include LMO1, LMO2, LMO3 and LMO4 [14]. It has been reported that overexpression of LMO1 or LMO2 was associated with T cell leukemia and was considered an oncogene in T cell leukemia [15-17]. LMO3 has been reported to be oncogenic in human neuroblastoma [18,19]. LMO4 was demonstrated to be an oncogene in T cell leukemia [20]. Additionally, LMO4 was associated with the malignant phenotype of pancreatic cancer, non-small-cell lung cancer, head and neck cancer, and other tumors [21-23]. Moreover, overexpression of LMO4 suppressed the differentiation of mammary epithelial cells and promoted the initiation and development of breast cancer [24-26]. However, the role of LMO4 and its related mechanism in GC remain unclear.
The present study reports that LMO4 expression was significantly upregulated in GC tissues and was closely associated with clinicopathological characteristics and patient prognosis. Knockdown of LMO4 significantly suppressed the invasion and proliferation of GC cells. Furthermore, the effects of LMO4 on the invasion and proliferation of GC cells were dependent on PI3K-Akt-mTOR signaling.
Materials and methods
Cell culture
Human GC cell lines, including AGS, BGC-823, HGC-27, MGC-803, MKN-45 and SGC-7901, were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). BGC-823, MGC-803, MKN-45 and SGC-7901 cells were cultured in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, MA, USA) supplemented with 10% (v/v) fetal calf serum (FCS) (Gibco, Thermo Fisher Scientific) and 1% antibiotics. AGS was cultured in DMEM F-12 (Gibco, Thermo Fisher Scientific) supplemented with 10% FCS and 1% antibiotics. HGC-27 was cultured in RPMI-1640 medium supplemented with 20% FCS and 1% antibiotics. All of the GC cells were cultured at 37°C in a humidified atmosphere containing 5% CO2.
Clinical samples
Human gastric tumor (n = 15) and normal tissues (n = 10), in which 10 gastric tumor and normal tissues were paired, were obtained from patients who were admitted at the Department of Gastroenterology, Shijiazhuang First Hospital. All of the patients provided written informed consent prior to enrollment, and the study was approved by the Research Ethics Committee of Shijiazhuang First Hospital. In addition, a human tissue microarray, which was also obtained from the Department of Gastroenterology, Shijiazhuang First Hospital, was used, which contained GC samples and paired normal tissues from 164 cases.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol (Takara Bio, Otsu, Japan) and reverse-transcribed with a PrimeScript RT-PCR kit (Takara Bio). Real-time qPCR analyses were performed with SYBR Premix Ex Taq (Takara Bio) on a 7500 real-time PCR system (Thermo Fisher Scientific, MA, USA) at the recommended thermal cycling settings: one initial cycle at 95°C for 30 sec, followed by 40 cycles of 5 sec at 95°C and 31 sec at 60°C. The results were quantified using the 2-ΔΔCq method [27]. The primer sequences used were as follows: LMO4, forward 5’-GGACAGTCGATTCCTGCGAG-3’ and reverse 5’-TGTAGTGAAACCGATCTCCCG-3’; and β-actin, forward 5’-CTCCATCCTGGCCTCGCTGT-3’ and reverse 5’-GCTGTCACCTTCACCGTTCC-3’.
Western blot analysis
Cells were lysed in lysis buffer (50 mM Tris-HCl; 150 mM NaCl; 1% Triton-X 100; 1 mM each of MgCl2; MnCl2, CaCl2, and 1 mM phenylmethylsulfonyl fluoride; and 10 mM sodium fluoride; Sangon, Shanghai, China). The protein concentration was determined using the BCA method. A total of 20 µg protein was loaded per lane, and proteins were separated by 6-12% SDS-PAGE under reducing conditions and transferred onto a nitrocellulose membrane (Thermo Fisher Scientific). The membrane was blocked in PBS/Tween-20 containing 5% bovine serum albumin (Sangon, Shanghai, China), followed by incubation with the antibodies for LMO4 (1:1,000, Abcam, MA, USA), phospho-PI3K p85 (1:2,000), total PI3K (1:2,000), phospho-Akt (1:2,000), total Akt (1:2,000), phospho-mTOR (1:2,000), total mTOR (1:2,000; all from Cell Signaling Technology, MA, USA) and GAPDH (1:1,000, Sigma-Aldrich, Darmstadt, Germany) at 4°C overnight. Subsequently, the membrane was washed and incubated with IRDye® 680 LT Goat anti-Rabbit IgG (H+L, 1:10,000) or IRDye® 800CW goat anti-mouse IgG (H+L, 1:10,000, LI-COR Biosciences, NE, USA) at room temperature for 1 h. The fluorescently labeled secondary antibodies were directly detected with the Odyssey imaging system (LI-COR Biosciences).
Small interfering (si) RNA transfection
siRNA duplexes for LMO4 were produced by Genepharma (Abcam, MA, USA). The LMO4 siRNA and scrambled siRNA were purchased from Abnova (Abnova, Taiwan, China). Transfection steps were performed according to the manufacturer’s protocol for the X-tremeGENE siRNA transfection reagent (Sigma-Aldrich, Darmstadt, Germany).
Recombinant (r) LMO4 protein and inhibitors
rLMO4 protein was purchased from Abcam. The inhibitor of PI3K/Akt (miltefosine) and the inhibitor of mTOR (dactolisib) were purchased from Selleck Chemicals (TX, USA). rLMO4 protein was added to BGC-823 and HGC-27 cells, and miltefosine or dactolisib was added to the above cells 2 h later. All cells were incubated at 37°C.
Invasion assay
MGC-803, SGC-7901, BGC-823 or HGC-27 cells were detached, resuspended in serum-free RPMI-1640 medium, and seeded at 2 × 104 cells per well in 100 µl in 8-µm Transwell inserts (EMD Millipore, MA, USA) with membranes coated in Matrigel (BD Biosciences, NJ, USA) on top of the wells of a 24-well plate. RPMI-1640 medium containing 5% FBS was added to the bottom chamber. Cells were incubated at 37°C for 48 h. Subsequently, filters were fixed and stained with 0.1% (w/v) crystal violet at room temperature for 20 min. Noninvading cells were removed from the upper side of the membrane, and invaded cells on the lower side were counted under a microscope at a magnification of × 400. At least three grids per field were counted, and the experiments were repeated twice.
Cell viability assay
Cell viability was determined using a standard Cell Counting Kit-8 (CCK-8) assay. MGC-803, SGC-7901, BGC-823 or HGC-27 cells were seeded into 96-well plates (100 μl per well) at a density of 2 × 104 cells per ml, and the total number of cells seeded in each well was 2 × 103. MGC-803 and SGC-7901 cells were transfected with LMO4 siRNA or scrambled siRNA before these cells were seeded. However, BGC-823 and HGC-27 cells were treated with rLMO4 protein, miltefosine or dactolisib after the cells had adhered to the bottom of dishes. After incubation for 12, 24, 48 or 72 h, 10 μl CCK-8 reagent (Dojindo, Kumamoto, Japan) was added to each well. After 2 h of incubation at 37°C, the optical density was measured using a microplate reader at a wavelength of 450 nm.
Animal experiments
Mice were housed and manipulated according to protocols approved by the Shijiazhuang First Hospital Animal Care Commission. All animals received humane care according to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals” prepared by the National Academy of Sciences and published by the National Institutes of Health (USA). Male NU/NU mice (5 weeks, n = 5 every group) were kept on a 12-hour day/night cycle with free access to food and water. MGC-803 cells were trypsinized, washed in PBS, and resuspended in serum-free RPMI-1640 medium. A total of 1 × 106 MGC-803 cells in 100 μl DMEM were injected subcutaneously in the lower back. Tumor growth was monitored for 2 weeks.
Statistical analysis
Values are expressed as the mean ± standard error of the mean. Survival time was analyzed with the Kaplan-Meier method. The association between LMO4 expression and the clinicopathological features of patients with gastric cancer was evaluated using Pearson’s Chi-square test. One-way analysis of variance was used for comparison between groups. Statistical analyses were performed using SPSS 16.0 for windows (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
LMO4 expression is closely associated with clinicopathological characteristics and prognosis of GC patients
To investigate the expression of LMO4 in GC tissues, 15 gastric tumors and 10 normal tissues were collected and analyzed by RT-qPCR. The expression of LMO4 was significantly upregulated in GC tissues (Figure 1A). In 10 paired gastric tumor and normal tissues within this cohort, LMO4 expression was also upregulated in GC tissues (Figure 1B). By western blotting, we found that LMO4 expression was upregulated in GC tissues in 5 paired gastric tumor and normal tissues (Figure 1C).
Figure 1.
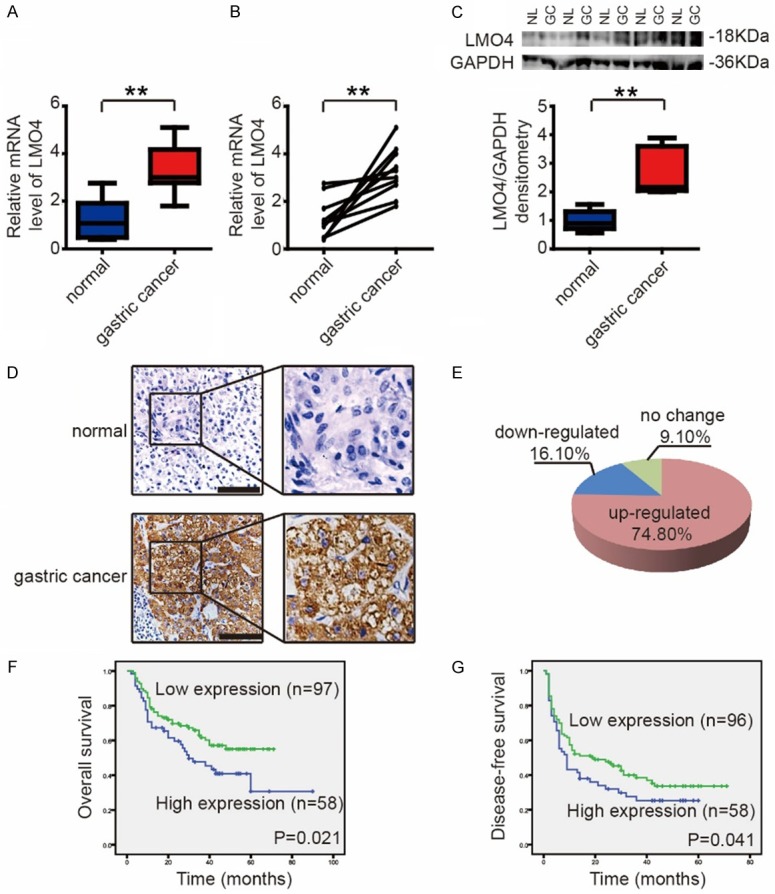
LMO4 expression is upregulated in GC and closely associated with patient prognosis. (A) The mRNA expression levels of LMO4 in 15 gastric tumors and 10 normal gastric tissues. **P<0.01. (B) The mRNA expression levels of LMO4 in 10 paired gastric tumor and normal tissues within this cohort. **P<0.01. (C) The protein expression levels of LMO4 in 5 paired gastric tumor and normal tissues. **P<0.01. (D) The immunohistochemical staining of LMO4 in gastric tumor and normal tissues (scale bar, 10 μm). (E) A GC tissue microarray (n = 164) indicated that the expression of LMO4 was upregulated in 74.80% of GC tissues. (F and G) Kaplan-Meier analysis of (F) OS (P = 0.021) and (G) DFS (P = 0.041) in GC patients from the microarray stratified by the expression of LMO4. Immunohistochemical staining was scored according to the ratio and intensity of positive-staining cells: 0-35% was designated as low expression; >36% was designated as high expression.
A GC tissue microarray (n = 164) was then used to investigate the correlations between LMO4 expression and clinicopathological characteristics and patient prognoses. Immunohistochemical staining was scored according to the ratio and intensity of positive-staining cells: 0-35% was designated as the low-expression group; >36% was designated as the high-expression group. The expression of LMO4 was upregulated in 74.80% of GC tissues (Figure 1D and 1E). Furthermore, the expression of LMO4 was closely associated with tumor size, differentiation, vascular embolism and TNM stage (Table 1), and high expression of LMO4 was positively correlated with poor overall survival (OS; P = 0.021) and disease-free survival (DFS; P = 0.041) (Figure 1F and 1G).
Table 1.
Correlation of clinicopathological factors with LMO4 expression
| Variable | LMO4 (n) | |||
|---|---|---|---|---|
|
|
||||
| High | Low | P | ||
| Age | ≤62 years | 29 | 48 | 0.713 |
| >62 years | 30 | 57 | ||
| Gender | Female | 11 | 12 | 0.635 |
| Male | 48 | 93 | ||
| Smoking history | Yes | 42 | 72 | 0.216 |
| No | 17 | 33 | ||
| Lauren subtype | Diffuse | 28 | 56 | 0.698 |
| Intestine | 30 | 49 | ||
| Location | Upper | 13 | 18 | 0.397 |
| Middle | 24 | 63 | ||
| Lower | 16 | 21 | ||
| Remnant stomach | 6 | 3 | ||
| Tumor size | ≤5 cm | 22 | 73 | <0.001 |
| >5 cm | 37 | 32 | ||
| Differentiation | Well | 5 | 2 | 0.003 |
| Moderate | 15 | 54 | ||
| Poor | 39 | 49 | ||
| Vascular embolism | Yes | 40 | 100 | <0.001 |
| No | 19 | 5 | ||
| TNM stage | Stage I | 33 | 84 | 0.003 |
| Stage II | 8 | 15 | ||
| Stage III/IV | 18 | 6 | ||
Pearson’s χ2 test was used for statistical analysis. TNM, tumor-nodes-metastasis; LMO4, LIM domain only 4.
LMO4 knockdown suppresses the invasion and proliferation of GC cells
To further investigate the biological functions of LMO4 in GC, the expression level of LMO4 was first detected in 6 GC cell lines. As shown in Figure 2A, LMO4 was highly expressed in MGC-803 and SGC-7901 cells compared with the other cell lines. Then, MGC-803 and SGC-7901 cell lines were used to knock down LMO4 by using siRNA (labeled si-LMO4-1 and si-LMO4-2). RT-qPCR and western blotting analysis demonstrated that LMO4 was successfully silenced in MGC-803 and SGC-7901 cells (Figure 2B and 2C).
Figure 2.
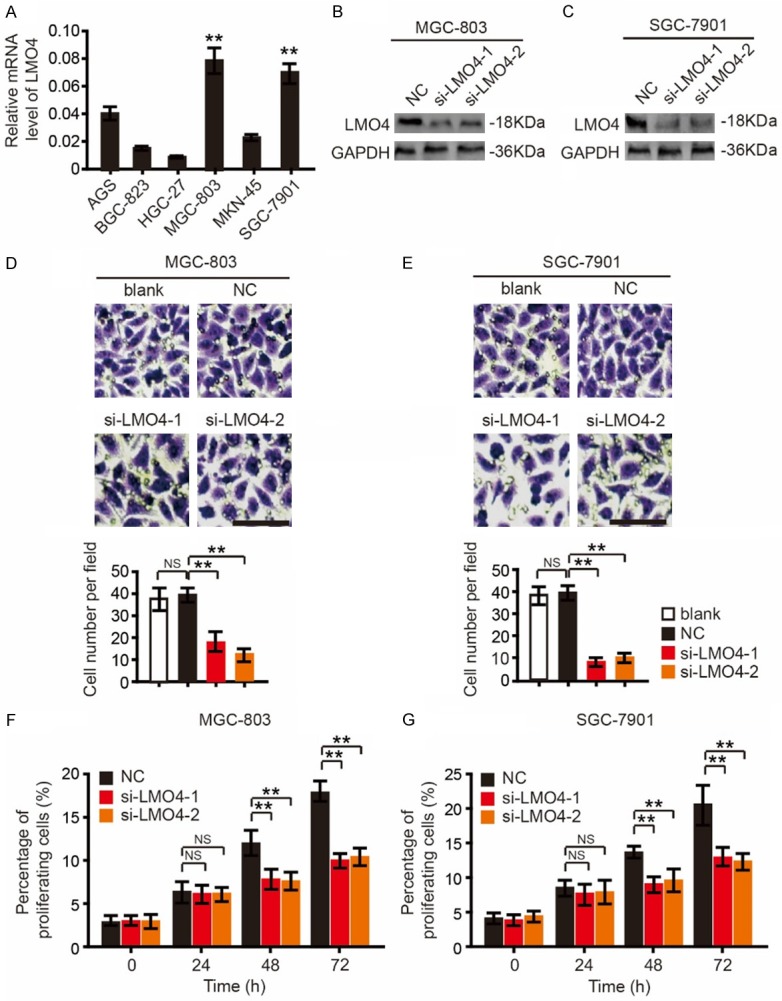
LMO4 knockdown suppresses the invasion and proliferation of gastric cancer cells. (A) Expression of LMO4 in the GC cell lines AGS, BGC-823, HGC-27, MGC-803, MKN-45 and SGC-7901. **P<0.01. (B and C) Protein expression levels of LMO4 in (B) MGC-803 and (C) SGC-7901 cells following transfection with siRNA targeting LMO4. (D) Representative photomicrographs of invaded MGC-803 cells transfected with siRNA targeting LMO4 (scale bar, 10 μm) and numbers of invaded MGC-803 cells in the three groups. (E) Representative photomicrographs of invaded SGC-7901 cells transfected with siRNA targeting LMO4 (scale bar, 10 μm) and numbers of invaded SGC-7901 cells in the three groups. (F and G) Cell Counting Kit-8 cell viability assay of (F) MGC-803 and (G) SGC-7901 cells transfected with siRNA targeting LMO4 at 0, 24, 48 and 72 h. **P<0.01.
The present study investigated the role of LMO4 in the invasion of GC cells. A Transwell Matrigel invasion assay demonstrated that knockdown of LMO4 suppressed the invasive capacity of MGC-803 (Figure 2D) and SGC-7901 (Figure 2E) cells after 48 h.
Furthermore, a CCK-8 cell viability assay was employed to investigate the role of LMO4 in the proliferation of GC cells. The viability of MGC-803 and SGC-7901 cells was significantly suppressed by knockdown of LMO4 at 48 and 72 h (Figure 2F and 2G).
Knockdown of LMO4 attenuates tumor growth in vivo and suppresses PI3K, Akt and mTOR signaling in GC cells
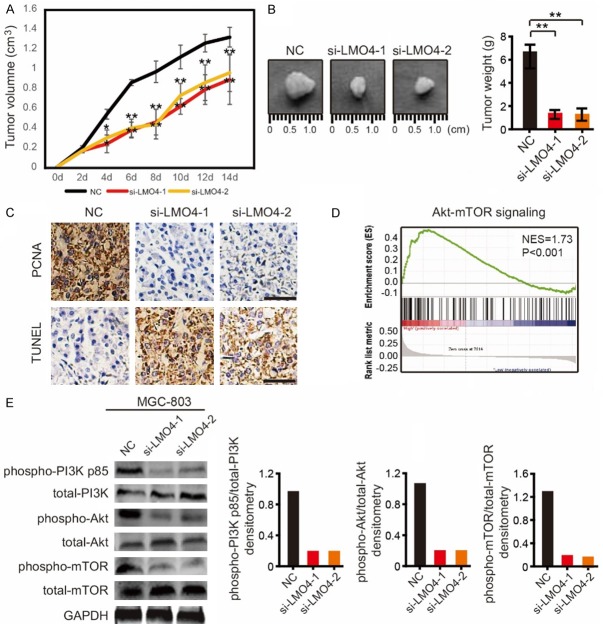
MGC-803 cells were subcutaneously injected into the lower back of male NU/NU mice. After 4 days, the tumor volumes in the si-LMO4-1 and si-LMO4-2 groups of mice were significantly smaller than those of the NC group (Figure 3A). After 2 weeks, all mice were sacrificed. The tumors in the si-LMO4-1 and si-LMO4-2 groups were smaller and weighed less than those of the NC group (Figure 3B). By IHC and TUNEL staining, it was found that the PCNA expression in the NC group was higher than in the si-LMO4-1 and si-LMO4-2 groups, while apoptosis in the NC group was lower than in the si-LMO4-1 and si-LMO4-2 groups (Figure 3C).
Figure 3.
LMO4 knockdown attenuates tumor growth in vivo and suppresses the phosphorylation of Akt, mTOR and GSK3β. A. The tumor volumes of mice in the NC, si-LMO4-1 and si-LMO4-2 groups over two weeks. **P<0.01. B. Mice in the si-LMO4-1 and si-LMO4-2 groups showed relatively larger tumors compared with the NC group after two weeks. The tumor weights of mice in the NC, si-LMO4-1 and si-LMO4-2 groups are shown at right. **P<0.01. C. The expression of PCNA and apoptosis in the tumors of NC, si-LMO4-1 and si-LMO4-2 group mice by IHC staining and TUNEL staining. D. LMO4 is closely related to Akt-mTOR signaling according to GSEA analysis. E. Western blot analysis of phospho-PI3K p85, total-PI3K, phospho-Akt, total-Akt, phospho-mTOR and total-mTOR in LMO4-knockdown and control MGC-803 cells and quantified phospho-PI3K p85/PI3K, phospho-Akt/total-Akt, phospho-mTOR/total-mTOR ratios.
To investigate the underlying mechanism of the association of LMO4 with GC, we performed GSEA analysis and found that LMO4 was closely related to Akt-mTOR signaling (Figure 3D). Then, the PI3K-Akt-mTOR signaling pathways were assessed in MGC-803 cells by western blotting analysis. Interestingly, LMO4 knockdown significantly suppressed the phosphorylation of PI3K p85 and Akt (Figure 3E). The phosphorylation of mTOR, a downstream signaling event of Akt, was also suppressed by silencing LMO4 (Figure 3E).
Taken together, the above results suggest that LMO4 promotes the invasion and proliferation of GC cells through PI3K-Akt-mTOR signaling.
LMO4 promotes GC cell invasion and proliferation through PI3K-Akt-mTOR signaling
The effects of LMO4 on GC cell invasion and proliferation were investigated in the presence of miltefosine (inhibitor of PI3K/Akt) or dactolisib (inhibitor of mTOR). rLMO4 protein was added to BGC-823 and HGC-27 cells, which had low LMO4 expression levels, and miltefosine and dactolisib were added 2 h later. rLMO4 protein treatment increased the phosphorylation of PI3K p85, Akt and mTOR (Figure 4A). Miltefosine and dactolisib treatment abrogated rLMO4-induced GC cell invasion (Figure 4B and 4C) and proliferation (Figure 4D and 4E).
Figure 4.
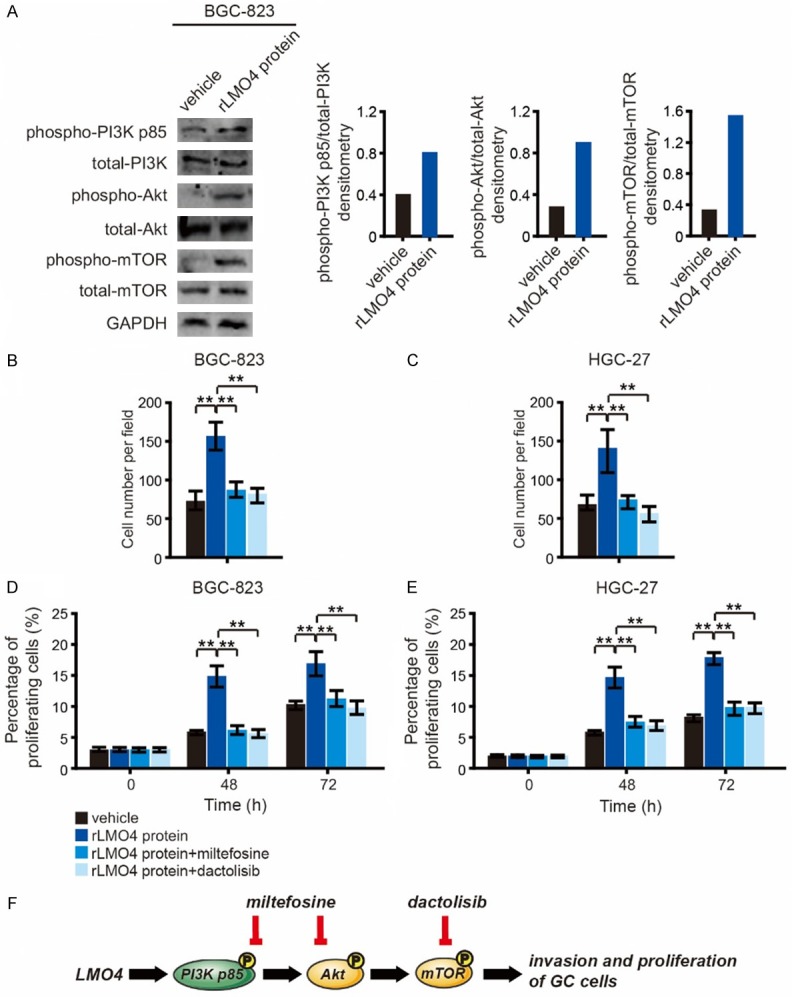
LMO4 promotes GC cell invasion and proliferation via PI3K-Akt-mTOR signaling. (A) Western blot analysis of phospho-PI3K p85, total PI3K, phospho-Akt, total Akt, phospho-mTOR and total mTOR in rLMO4-treated and vehicle-treated BGC-823 cells. (B and C) (B) BGC-823 and (C) HGC-27 cells were treated with 50 nM rLMO4 protein, 50 nM rLMO4 protein plus 50 nM miltefosine (an inhibitor of PI3K/Akt), or 50 nM rLMO4 protein plus 50 nM dactolisib (an inhibitor of mTOR). Cell invasion was analyzed after 48 h. (D and E) (D) BGC-823 and (E) HGC-27 cells were subjected to the above-mentioned treatments, and cell viability was detected with a Cell Counting Kit-8 at 0, 48 and 72 h. **P<0.01. (F) Schematic depicting the mechanism of LMO4-induced GC cell invasion and proliferation.
These results indicate that LMO4-induced GC cell invasion and proliferation are mainly dependent on PI3K-Akt-mTOR signaling. The signaling cascades are outlined in a schematic in Figure 4F.
Discussion
Numerous studies have assessed LMO4 in various cancer types. It has been reported that LMO4 is associated with the malignant phenotype of breast cancer, pancreatic cancer, non-small-cell lung cancer, head and neck cancer, and other tumors [21-26]. However, the detailed biological functions of LMO4 in GC and the underlying mechanisms have remained unclear. In this study, the exact roles of LMO4 in GC were investigated for the first time. By analyzing GC tissues and GC microarrays, it was found that the expression of LMO4 was closely associated with tumor size, differentiation, vascular embolism and TNM stage, as well as poor patient prognosis. These results suggest that LMO4 may have important roles in the development of GC.
We further revealed the biological functions of LMO4 in GC. The invasion ability and cell viability of GC cells were significantly suppressed by LMO4 knockdown, indicating that LMO4 is involved in the invasion and proliferation of GC cells. We also revealed that knockdown of LMO4 attenuated tumor growth in vivo. The expression of PCNA was decreased, and apoptosis was increased by LMO4 knockdown. All of these results suggest that LMO4 plays important roles in the invasion and proliferation of GC cells.
Invasion and metastasis are major concerns during the prognosis and progression of cancer. The PI3K-Akt-mTOR pathway is pivotal in modulating the invasion, migration and proliferation of tumor cells [28-31]. The present study indicated that knockdown of LMO4 decreases the phosphorylation of PI3K, Akt and mTOR. By using an inhibitor of PI3K/Akt, miltefosine, and an inhibitor of mTOR, dactolisib, it was further determined that LMO4-induced GC cell invasion and proliferation are dependent on PI3K-Akt-mTOR signaling.
In conclusion, the present study revealed that LMO4 has an important role in GC cell invasion and proliferation. LMO4 promotes GC cell invasion and proliferation through PI3K-Akt-mTOR signaling. LMO4 may be used as a potential prognostic or therapeutic target for GC in the future.
Acknowledgements
This research is supported by the National Natural Science Foundation of China (No. 81670735).
Disclosure of conflict of interest
None.
References
- 1.Hansson LE, Sparen P, Nyren O. Survival in stomach cancer is improving: results of a nationwide population-based Swedish study. Ann Surg. 1999;230:162–169. doi: 10.1097/00000658-199908000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun X, Mu R, Zhou Y, Dai X, Qiao Y, Zhang S, Huangfu X, Sun J, Li L, Lu F. [1990-1992 mortality of stomach cancer in China] . Zhonghua Zhong Liu Za Zhi. 2002;24:4–8. [PubMed] [Google Scholar]
- 3.Werner M, Becker KF, Keller G, Hofler H. Gastric adenocarcinoma: pathomorphology and molecular pathology. J Cancer Res Clin Oncol. 2001;127:207–216. doi: 10.1007/s004320000195. [DOI] [PubMed] [Google Scholar]
- 4.Becker KF, Keller G, Hoefler H. The use of molecular biology in diagnosis and prognosis of gastric cancer. Surg Oncol. 2000;9:5–11. doi: 10.1016/s0960-7404(00)00016-5. [DOI] [PubMed] [Google Scholar]
- 5.Wong SS, Kim KM, Ting JC, Yu K, Fu J, Liu S, Cristescu R, Nebozhyn M, Gong L, Yue YG, Wang J, Ronghua C, Loboda A, Hardwick J, Liu X, Dai H, Jin JG, Ye XS, Kang SY, Do IG, Park JO, Sohn TS, Reinhard C, Lee J, Kim S, Aggarwal A. Genomic landscape and genetic heterogeneity in gastric adenocarcinoma revealed by whole-genome sequencing. Nat Commun. 2014;5:5477. doi: 10.1038/ncomms6477. [DOI] [PubMed] [Google Scholar]
- 6.Kang G, Hwang WC, Do IG, Wang K, Kang SY, Lee J, Park SH, Park JO, Kang WK, Jang J, Choi MG, Lee JH, Sohn TS, Bae JM, Kim S, Kim MJ, Kim S, Park CK, Kim KM. Exome sequencing identifies early gastric carcinoma as an early stage of advanced gastric cancer. PLoS One. 2013;8:e82770. doi: 10.1371/journal.pone.0082770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu B, El Hajj N, Sittler S, Lammert N, Barnes R, Meloni-Ehrig A. Gastric cancer: classification, histology and application of molecular pathology. J Gastrointest Oncol. 2012;3:251–261. doi: 10.3978/j.issn.2078-6891.2012.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Y, Yang Q, Wang B, Ye G, Tong X. The correlation of MGMT promoter methylation and clinicopathological features in gastric cancer: a systematic review and meta-analysis. PLoS One. 2016;11:e0165509. doi: 10.1371/journal.pone.0165509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baroudi O, Benammar-Elgaaied A. Involvement of genetic factors and lifestyle on the occurrence of colorectal and gastric cancer. Crit Rev Oncol Hematol. 2016;107:72–81. doi: 10.1016/j.critrevonc.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Shen J, Seifer BJ, Jiang S, Wang J, Xiong H, Xie L, Wang L, Sui X. Approaches and genetic determinants in predicting response to neoadjuvant chemotherapy in locally advanced gastric cancer. Oncotarget. 2017;8:30477–30494. doi: 10.18632/oncotarget.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi J, Zhang P, Wang Y, Chen H, Li Y. Does total gastrectomy provide better outcomes than distal subtotal gastrectomy for distal gastric cancer? a systematic review and meta-analysis. PLoS One. 2016;11:e0165179. doi: 10.1371/journal.pone.0165179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 15.Fisch P, Boehm T, Lavenir I, Larson T, Arno J, Forster A, Rabbitts TH. T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene. 1992;7:2389–2397. [PubMed] [Google Scholar]
- 16.McGuire EA, Rintoul CE, Sclar GM, Korsmeyer SJ. Thymic overexpression of Ttg-1 in transgenic mice results in T-cell acute lymphoblastic leukemia/lymphoma. Mol Cell Biol. 1992;12:4186–4196. doi: 10.1128/mcb.12.9.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neale GA, Rehg JE, Goorha RM. Disruption of T-cell differentiation precedes T-cell tumor formation in LMO-2 (rhombotin-2) transgenic mice. Leukemia. 1997;11(Suppl 3):289–290. [PubMed] [Google Scholar]
- 18.Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, Tokita H, Ohira M, Nakagawara A. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65:4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- 19.Isogai E, Ohira M, Ozaki T, Oba S, Nakamura Y, Nakagawara A. Oncogenic LMO3 collaborates with HEN2 to enhance neuroblastoma cell growth through transactivation of Mash1. PLoS One. 2011;6:e19297. doi: 10.1371/journal.pone.0019297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grutz G, Forster A, Rabbitts TH. Identification of the LMO4 gene encoding an interaction partner of the LIM-binding protein LDB1/NLI1: a candidate for displacement by LMO proteins in T cell acute leukaemia. Oncogene. 1998;17:2799–2803. doi: 10.1038/sj.onc.1202502. [DOI] [PubMed] [Google Scholar]
- 21.Yu J, Ohuchida K, Nakata K, Mizumoto K, Cui L, Fujita H, Yamaguchi H, Egami T, Kitada H, Tanaka M. LIM only 4 is overexpressed in late stage pancreas cancer. Mol Cancer. 2008;7:93. doi: 10.1186/1476-4598-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Wu S, Guo M, He J. LMO4 is a prognostic marker involved in cell migration and invasion in non-small-cell lung cancer. J Thorac Dis. 2016;8:3682–3690. doi: 10.21037/jtd.2016.12.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simonik EA, Cai Y, Kimmelshue KN, Brantley-Sieders DM, Loomans HA, Andl CD, Westlake GM, Youngblood VM, Chen J, Yarbrough WG, Brown BT, Nagarajan L, Brandt SJ. LIM-only protein 4 (LMO4) and LIM domain binding protein 1 (LDB1) promote growth and metastasis of human head and neck cancer (LMO4 and LDB1 in head and neck cancer) PLoS One. 2016;11:e0164804. doi: 10.1371/journal.pone.0164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Visvader JE, Venter D, Hahm K, Santamaria M, Sum EY, O’Reilly L, White D, Williams R, Armes J, Lindeman GJ. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci U S A. 2001;98:14452–14457. doi: 10.1073/pnas.251547698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sum EY, Segara D, Duscio B, Bath ML, Field AS, Sutherland RL, Lindeman GJ, Visvader JE. Overexpression of LMO4 induces mammary hyperplasia, promotes cell invasion, and is a predictor of poor outcome in breast cancer. Proc Natl Acad Sci U S A. 2005;102:7659–7664. doi: 10.1073/pnas.0502990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montanez-Wiscovich ME, Seachrist DD, Landis MD, Visvader J, Andersen B, Keri RA. LMO4 is an essential mediator of ErbB2/HER2/Neu-induced breast cancer cell cycle progression. Oncogene. 2009;28:3608–3618. doi: 10.1038/onc.2009.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 28.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 29.Lin HP, Jiang SS, Chuu CP. Caffeic acid phenethyl ester causes p21 induction, Akt signaling reduction, and growth inhibition in PC-3 human prostate cancer cells. PLoS One. 2012;7:e31286. doi: 10.1371/journal.pone.0031286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Feng L, Tian C, Tang YL, Tang Y, Hu FQ. Long noncoding RNA-JPX predicts the poor prognosis of ovarian cancer patients and promotes tumor cell proliferation, invasion and migration by the PI3K/Akt/mTOR signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:8135–8144. doi: 10.26355/eurrev_201812_16505. [DOI] [PubMed] [Google Scholar]
- 31.Tian L, Cheng F, Wang L, Qin W, Zou K, Chen J. CLE-10 from Carpesium abrotanoides L. suppresses the growth of human breast cancer cells (MDA-MB-231) in vitro by inducing apoptosis and pro-death autophagy via the PI3K/Akt/mTOR signaling pathway. Molecules. 2019;24 doi: 10.3390/molecules24061091. [DOI] [PMC free article] [PubMed] [Google Scholar]