Abstract
LncRNA is gradually considered as a vital regulator in various physiological and pathological processes. Recently, the role of LINC01234 in several cancers has been reported. However, the function and underlying regulatory mechanism of LINC01234 in multiple myeloma (MM) remain unclear. Our results indicated that LINC01234 was over-expressed in tumor tissues of MM patients, and LINC01234 down-regulation remarkably inhibited cell proliferation, cycle progression and promoted cell apoptosis in MM cells. Mechanistic studies revealed that LINC01234 could sponge miR-124-3p and decreased its expression, thereby up-regulated the protein level of miR-124-3p’s targets growth factor receptor-bound protein 2 (GRB2). Additionally, in vivo experiments revealed that LINC01234 shRNA could serve as a tumor suppressor through down-regulating GRB2 in MM. In this study, a novel established regulatory manner of LINC01234/miR-124-3p/GRB2 axis was systematically studied, which may hold promise as a promising target for MM treatment.
Keywords: Multiple myeloma, LINC01234, miR-124-3p, GRB2, cell proliferation, cell cycle
Introduction
As the second most prevalent hematologic malignancy, multiple myeloma (MM) is mainly caused by the disorder of tumorous plasma cells, which infiltrates into the bone marrow [1]. The survival time of MM patients ranges from several months to more than 10 years, with a 5-year survival rate of 40% [2]. The etiology and underlying molecular manner of MM are largely unknown, leading to the complexity of treatment [3]. Despite progress in diagnosis and treatment, the prognosis of MM patients is still poor [4]. Therefore, to supplement or replace existing therapeutic strategies, finding new molecular targets is very important.
Long non-coding RNA, whose length is greater than 200 nt, exerts key functions in cell proliferation, differentiation, apoptosis and other biological processes [5]. Some lncRNAs, such as MEG3, PDIA3P, lncRUNX2-AS1 and OIP5-AS1 have been implicated in MM [6-9]. MicroRNAs are key non coding RNA (ncRNAs) (18-22 nt), which mainly bind to gene’s 3’UTR and regulate their expression, serving as oncogenic miRNAs or tumour suppressor [10]. Studies have showed that lncRNAs could target and regulate miRNA, therefore affect target gene expressions [11]. For example, LncRNA OIP5-AS1 could negatively regulate miR-410, modulate KLF10/PTEN/AKT axis and influence cell proliferation in MM [7]. Long non-coding RNA CCAT1 accelerates MM development by targeting miR-181a-5p and regulating HOXA1 expression [12]. Both lncRNAs and miRNAs play key functions in MM, but the underlying mechanism remains to be illustrated.
Recent studies demonstrated that LINC01234 overexpression could promote cell proliferation in colon cancer by regulating miR642a-5p-SHMT2 axis [13]. Chen revealed that LINC01234 targets miR-204-5p and upregulates CBFB expression in gastric cancer [14]. Additionally, LINC01234 could affect esophageal cancer growth and metastasis [15]. However, the role and the underlying mechanism of LINC01234 in MM are still unclear. As an important miRNA, miR-124-3p has been found to be dysregulated and exerts vital roles in many cancers. A decrease in miR-124-3p level was demonstrated in hepatocellular carcinoma (HCC) tissues and patients with lower miR-124-3p level had a shorter overall survival and poor prognosis [16]. Besides, down-regulated miR-124-3p could promote breast cancer growth and metastasis by regulating N-acetylglucosaminyltransferase V (MGAT5) [17]. Although the functions of miR-124-3p in some cancers have been illustrated, its role in MM is still unclear.
Growth factor receptor-bound protein 2 (GRB2) is an adaptor protein in Ras signaling pathway, which has been recently reported to be involved in multiple tumor malignancies [18]. Including Src homology 2 (SH2) and Src homology 3 (SH3) domains, mature GRB2 mediate molecular signals from cell-surface activated receptors to downstream targets, therefore having important effects on cell proliferation, cycle progression and apoptosis [19,20]. Previous studies showed that IL-6 might promote tyrosine phosphorylation of Shc, recruit Grb2 and activate the Ras signaling pathway in MM cells [21]. However, potential miRNA targeting GRB2 and its function in MM progression are still unclear.
In this study, we determined the up-regulation of LINC01234 in tissue samples of MM patients and studied LINC01234’s role in MM. Through miRDB analysis, potential miRNA that may be targeted by LINC01234 were predicted and identified. In addition, as predicted by TargetScan, miRDB and starBase, miR-124-3p may target GRB2’s 3’UTR. Thus, we speculate that LINC01234 might play a role in MM development by affecting GRB2 expression through modulating the level of miR-124-3p. Here, the function and molecular mechanism of LINC01234/miR-124-3p/GRB2 in MM were systematically researched, which may exert new candidates for MM therapy.
Materials and methods
Clinical samples
Multiple myeloma tissues (MM) and healthy control samples (Control) were collected at The First Affiliated Hospital of Sun Yat-sen University from 2015 to 2018. Before section, the MM patients had received neither chemotherapy nor radiotherapy. The procedures were approved by the Institutional Review Board of The First Affiliated Hospital of Sun Yat-sen University [22]. Pathological diagnosis were performed by pathologists. Informed consent was acquired from all patients.
In situ hybridization (ISH)
After tissues were fixed and paraffin-embedded, sections (5 μm) were cut. For ISH, a peroxidase-labeled LINC01234 probe was obtained (Thermo Fisher Scientific). ISH assay was performed with LINC01234 probe using an ISH kit for lncRNA detection (RiboBio, Guangzhou, China). The staining intensity was quantified using Image-ProPlus 6.0 by scanning 10 nonoverlapping fields in each section. The mean staining intensity was calculated and served as an evaluation criterion for LINC01234 expression “Low and high”. Tissue was identified as “Low expression” if the LINC01234 intensity was under the mean value. Accordingly, tissue was identified as “High expression” if LINC01234 intensity was higher than the average value. An overall survival curve was determined using the Kaplan-Meier method based on the follow-up data.
Additionally, the RNA fluorescence in situ hybridization (RNA-FISH) detection for LINC01234 and U6 was performed as previously described [23]. Briefly, the deparaffinized 5 μm sections were firstly blocked in pre-hybridization buffer, then incubated with a FITC-conjugated LINC01234 probe or Cy5-conjugated U6 probe (Exiqon, Vedbaek, Denmark) in the hybridization buffer, followed by the DAPI (Sigma-Aldrich) staining.
Cell culture
Human normal plasma cells (nPCs) and four MM cell lines (RPMI-8226, U266, MM.1S and H929) were purchased from ATCC cell lines (Manassas, USA). Cells were incubated in RPMI-1640 Medium, supplemented with 10% FBS (Thermo Fisher Scientific) and cultured limpidly at 37°C in a humidified atmosphere (5% CO2).
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the MM tissues and cultured cells using the TRIzol reagent (Thermo Fisher Scientific). cDNA was synthesised from total RNA using a PrimeScript RT Reagent Kit (Takara, Shiga, Japan). SYBR Premix Ex Taq (Takara) was used to perform qRT-PCR assays. A SYBR PrimeScript miRNA RT-PCR Kit (Takara) was used to examine miRNA levels. LINC01234 and GRB2 mRNA expressions were normalized to GAPDH level, and U6 snRNA level was used for miRNA expression normalization. Primer sequences for qRT-PCR analysis were listed in Table 1.
Table 1.
The primers for qRT-PCR assay
| Primer Name | Primer Sequence (5’-3’) |
|---|---|
| GAPDH RT primer | random primer |
| GAPDH Reverse primer | AAATGAGCCCCAGCCTTC |
| GAPDH Forward primer | AATCCCATCACCATCTTCCAG |
| LINC01234 RT primer | random primer |
| LINC01234 Reverse primer | CCTTTCCTCTGATTCCACCTC |
| LINC01234 Forward primer | GACATCTCACCTTTCAAACGC |
| GRB2 RT primer | random primer |
| GRB2 Reverse primer | ACATGGATAAAATCTCCCCGG |
| GRB2 Forward primer | ACATAGAACAGGTGCCACAG |
| U6 RT primer | AACGCTTCACGAATTTGCGT |
| U6 Reverse primer | AACGCTTCACGAATTTGCGT |
| U6 Forward primer | CTCGCTTCGGCAGCACA |
| miR-124-3p RT primer | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTTACGGTT |
| miR-124-3p Reverse primer | TGGTGTCGTGGAGTCG |
| miR-124-3p Forward primer | ACACTCCAGCTGGGTAAGGCACGCGGTGAA |
Cell transfection
RPMI-8226 or U266 cells were transfected with siRNA for LINC01234 (si-LINC01234 1#, si-LINC01234 2#, si-LINC01234 3#), miR-124-3p mimics (miR-124-3p mimic), miR-124-3p inhibitors (miR-124-3p inh) or their corresponding controls (si-NC, NC-mimic or NC inh). The sequences for three siRNAs were listed in Table 2. si-LINC01234 2#, si-LINC01234 3# were selected for the following analysis. For cell transfection, a Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) was used.
Table 2.
The sequences of siRNAs for LINC01234
| siRNA Name | shRNA Sequence (5’-3’) |
|---|---|
| si-LINC01234 #1 | AAAGAGAGACAGCAGAGAACTGATT |
| si-LINC01234 #2 | AGAGAGACAGCAGAGAACTGATTCT |
| si-LINC01234 #3 | AGAGACAGCAGAGAACTGATTCTCT |
Cell proliferation assay
RPMI-8226 or U266 cells were respectively transfected with si-NC, si-LINC01234 2# or si-LINC01234 3#. These cells (6×103 cells/well) were then seeded onto 96-well plates. To detect the number of viable cells after 0, 24, 48, 72, 96 hours, a Cell Counting Kit-8 (CCK-8, Dojindo, Tokyo, Japan) was used. A microplate spectrometer (Thermo Fisher Scientific) was used to measure the absorbance at 450 nm.
For 5-ethynyl-2’-deoxyuridine (EDU) assay, to measure cells undergoing DNA replication, incorporation of the thymidine analog EDU into DNA during DNA replication was used. Briefly, after fixation and permeabilization, cells were incubated with EDU (50 µM, 3 h). The cell nuclei were stained with DAPI (1 µg/ml, 10 min, Sigma-Aldrich). The EDU positive cells were determined using fluorescence microscopy.
For colony formation assay, RPMI-8226 or U266 cells (1×103) were mixed into top agar (1.5 ml), and then added onto base agar. 3 weeks later, colonies were stained by Crystal Violet. And a dissection microscope (TE2000-U, Nikon, Japan) was used to count the colonies.
Cell cycle and cell apoptosis assay
At 48 hours post transfection, RPMI-8226 or U266 cells were collected. After fixed with 70% ethanol overnight, cells were incubated with propidium iodide (PI, BD Biosciences, San Jose, CA). PI and Annexin V-FITC (BD Biosciences) were used for cell apoptosis assay. Then, a FACSCalibur system (BD Biosciences) was used for DNA content and Annexin V analysis. The results of cell cycle and cell apoptosis were analyzed using the ModFit_LT software.
Protein extraction and western blotting
RIPA lysis buffer (Sigma-Aldrich) was used for protein extraction. For total protein content quantification, a BCA protein assay kit (Thermo Fisher Scientific) was used. included . The membranes were incubated with the primary antibodies, p21, CDK4, p27, Bcl-2, Bax, cleaved caspase3, GAPDH (an internal control), overnight at 4°C, and then incubated with a horseradish peroxidase-conjugated anti-mouse/anti-rabbit IgG (Thermo Fisher Scientific). Antibodies were listed in Table 3.
Table 3.
Antibodies used in this study
| Antibody name | Corporation name | Source | Poly/monoclonal | Dilution ratio |
|---|---|---|---|---|
| P21 | Cell Signaling Technology | rabbit | monoclonal | 1:1000 |
| CDK4 | Cell Signaling Technology | rabbit | monoclonal | 1:1000 |
| P27 | Cell Signaling Technology | rabbit | monoclonal | 1:1000 |
| Bcl-2 | Cell Signaling Technology | rabbit | monoclonal | 1:1000 |
| Bax | Cell Signaling Technology | rabbit | monoclonal | 1:1000 |
| Cleaved caspase3 | Cell Signaling Technology | rabbit | monoclonal | 1:1000 |
| GAPDH | Thermo Fisher Scientific | mouse | monoclonal | 1:2000 |
| Goat anti-rabbit IgG | Thermo Fisher Scientific | goat | monoclonal | 1:2000 |
| Goat anti-mouse IgG | Thermo Fisher Scientific | goat | monoclonal | 1:2000 |
Plasmid constructs
A human genomic DNA from U266 cells was used to amplify the sequence of LINC01234 using PCR. To insert the sequence of LINC01234 into the p-MIR-reporter plasmid (Promega, Madison, WI, USA), a One Step Cloning Kit ClonExpress II (Vazyme Biotech, Nanjing, P.R. China) was used. According to the above methods, p-MIR-reporter plasmid containing the 3’UTR of GRB2 was constructed. The binding-site mutant luciferase plasmid (binding site: GUGCCUU replaced by GUCGGAA) was also transfected as a control. The corresponding primer sequences for plasmid construct were listed in Table 4.
Table 4.
The primers for plasmid constructs
| Primer Name | Primer Sequence (5’-3’) |
|---|---|
| LINC01234 Reverse primer | GAAGCATGAATTCAAGGTACCTACTGCAGGGTGAAGAATTACTCAG |
| LINC01234 Forward primer | TAATAACTAAGATCTGGTACCTGTCCTCCATTGTAAGATAAAAAGAGC |
| GRB2 3’-UTR Reverse primer | GAAGCATGAATTCAAGGTACCTGAAGAATTCATTGTGTATTTATTATTCACA |
| GRB2 3’-UTR Forward primer | TAATAACTAAGATCTGGTACCGAGTCAAGAAGCAATTATTTAAAGAAAGT |
Luciferase reporter assay
When the cell confluence is at 70-80%, RPMI-8226 or U266 cells were co-transfected with the firefly luciferase reporter plasmid (1 μg), β-galactosidase expression vector (0.5 μg, Promega) and miR-124-3p-mimic or NC-mimic. The protein was extracted 24 hours after transfection. A Luciferase Reporter Assay System (Promega) was used to test the luciferase activities.
RNA binding protein immunoprecipitation (RIP) assay
RIP assay was performed as previously described [24]. Briefly, whole-cell lysates from RPMI-8226 or U266 cells were incubated with antibodies for Ago2, and the pull down complexes were subjected to RNA extraction and qRT-PCR analyses of LINC01234 and miR-124-3p.
RNA pull-down assay
RNA pull-down assay was performed as previously described [25]. Briefly, biotin-labeled LINC01234 containing miR-124-3p binding site was transcribed from LINC01234 expressing plasmid with the Biotin RNA Labeling Mix (Roche, Indianapolis, IN, USA) and T7 RNA polymerase (Promega). Then the RNA was treated with RNase-free Dnase I (Promega) and purified with a RNeasy Mini Kit (Qiagen, Valenia, CA). Biotinylated RNA was denatured at 95°C, put in ice, left at room temperature to allow secondary structure formation, and then incubated with whole-cell lysates from U266 cells at 25°C. The LINC01234-RNA complexes were isolated with Streptavidin agarose beads (Thermo Fisher Scientific) and the pull-down miR-124-3p was detected by qRT-PCR. According to the above methods, biotin-labeled GRB2 3’UTR was obtained. And the enriched miR-124-3p level of in the pull-down RNA with biotin-labeled GRB2 3’UTR was detected by qRT-PCR.
Establishment of a mouse MM xenograft model
Four-week-old thymic BALB/c nude mice were used to establish a mouse MM xenograft model. Animal care was performed according to the Guidelines for the Care and Use of Laboratory Animals, presented by the National Institutes of Health [26]. Firstly, using a recombinant lentivirus, U266 cells stably expressing shRNA for LINC01234 (LINC01234-shRNA) or the corresponding control cells (NC-shRNA) were constructed. Then, stable cells (1×106) were subcutaneously injected into the right flanks of mice (6 mice/group). The tumors were measured on day 3, 6, 9, 12, 15, 18 and 21 after injection. The tumor volume was calculated as 1/2LW2, where W and L represent the smallest and largest perpendicular tumor diameter. The mice were sacrificed, weighted and photographed at day 21 post-implantation. Additionally, the IHC analysis of Ki67 and GRB2, TUNEL analysis and the expression analysis of LINC01234 and miR-124-3p were performed.
Statistical analysis
Results are expressed as the mean ± standard error of the mean (SEM). Student’s t-test was used to analyze the differences between two groups. One-way ANOVA was used to analyze the differences among multiple groups. Each group contained three parallel lines and experiments were repeated at least three independent times. The correlations between LINC01234 and miR-124-3p, LINC01234 and GRB2 mRNA, miR-124-3p and GRB2 mRNA were respectively analyzed by Pearson correlation analysis. P < 0.05 was considered significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Results
LINC01234 level is up-regulated in both MM tissues and cells
As shown in Figure 1A, LINC01234 levels in tissue samples from MM patients (n=50) were determined using qRT-PCR, the results revealed that LINC01234 level was significantly increased in MM samples compared to Control samples. Additionally, the ISH data also indicated that LINC01234 up-regulation is a common event in MM (Figure 1C). In addition, using the Kaplan-Meier method, the relationship between the survival of MM patients and LINC01234 level was analyzed. As indicated in Figure 1B, MM patients with high LINC01234 expression have a lower survival rate. Additionally, LINC01234 levels in the four MM cell lines (RPMI-8226, U266, MM.1S and H929) were higher than that in human normal plasma cells, nPCs (Figure 1D). Among the MM cell lines, RPMI-8226 and U266 cells expressed higher levels of LINC01234, which were thus used in the following analysis. Additionally, the RNA-FISH results in Figure 1E demonstrated that LINC01234 was mainly distributed in cytoplasm, suggesting its post-transcriptional regulation manner. Together, these results revealed that LINC01234 is overexpressed in MM tissues and cells, which might play an oncogenic role in MM.
Figure 1.
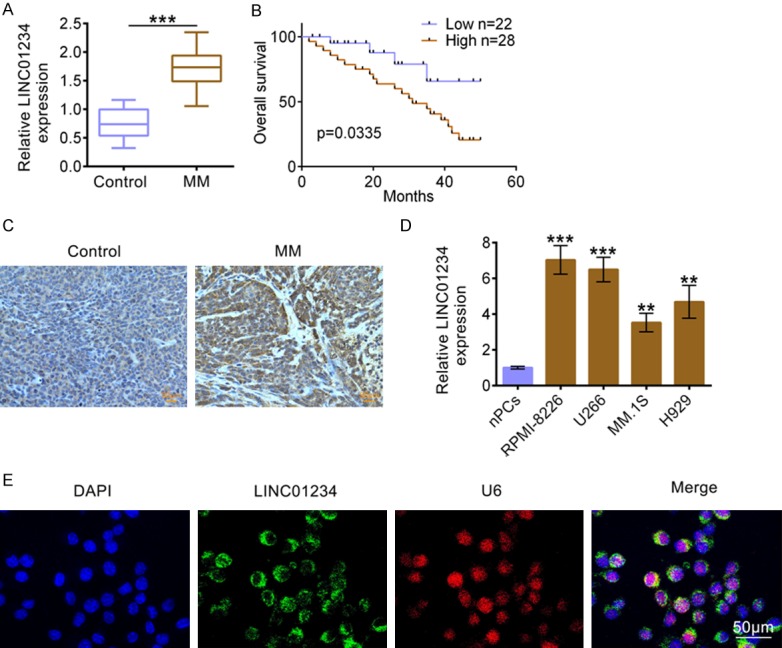
LINC01234 is up-regulated in MM tissues and cells. A. The relative LINC01234 expression levels in MM tissues (MM, n=50) and normal control tissues (Control, n=50), as determined using qRT-PCR (Student’s t-test). B. Kaplan-Meier curves of overall survival of 50 MM patients, stratified by LINC01234 expression. C. The LINC01234 expression levels in MM tissues (MM, n=50) and normal control tissues (Control, n=50), as determined using ISH. D. The relative LINC01234 expression levels in human normal plasma cells (nPCs) and MM cell lines (RPMI-8226, U266, MM.1S and H929), as determined using qRT-PCR (One-way ANOVA). E. Co-location detection of LINC01234 and U6 in U266 cells, as determined using ISH. The data are expressed as mean ± SEM, **P < 0.01, ***P < 0.001.
LINC01234 effects the proliferation, cycle progression and apoptosis of MM cells
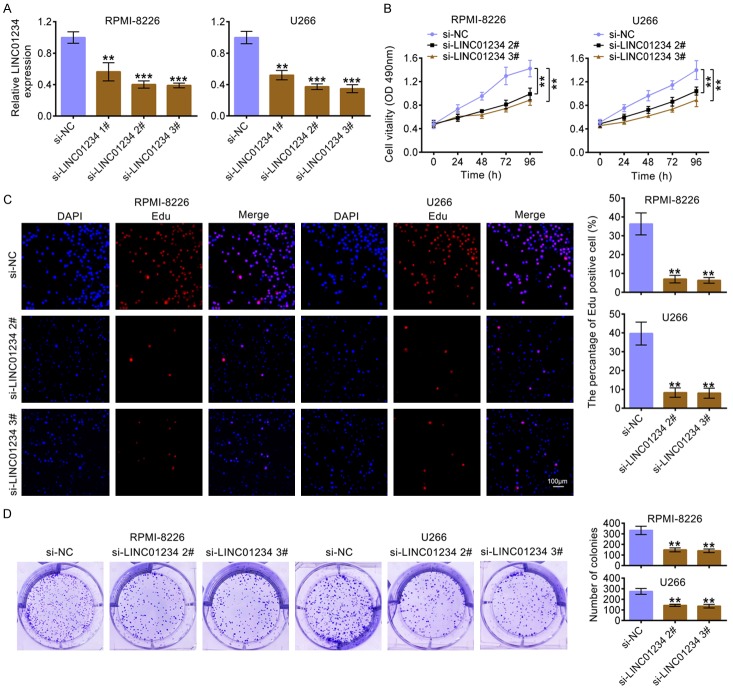
To determine the effects of LINC01234 on MM cells, LINC01234 was knocked down by three siRNAs. The results in Figure 2A demonstrated that LINC01234 level was markedly decreased after transfecting with LINC01234 siRNAs as compared to the control group. Because si-LINC01234 2# and si-LINC01234 3# had more effective interference efficiency, which were therefore selected for the following experiments. The results in Figure 2B revealed that the cell growth was inhibited in RPMI-8226 or U266 cells after LINC01234 was knockdown. Figure 2C indicated that the number of RPMI-8226 or U266 cells incorporating EDU in the si-LINC01234-treated group was significantly decreased as compared to the control group. Moreover, colony formation results demonstrated that LINC01234 down-expression significantly decreased colony numbers as compared to the control group (Figure 2D).
Figure 2.
LINC01234 affects the proliferation of MM cells. A. The relative expression level of LINC01234 in RPMI-8226 or U266 cells, after transfecting with siRNAs for LINC01234 (si-LINC01234 1#, si-LINC01234 2#, si-LINC01234 3#), or the corresponding control (si-NC) (One-way ANOVA). B. Growth curves of RPMI-8226 or U266 cells after transfecting with siRNAs for LINC01234 (si-LINC01234 2#, si-LINC01234 3#) or the corresponding control (si-NC) (One-way ANOVA). The cell growth rate was measured using a CCK-8 kit. C. Representative profiles of EDU cell growth in RPMI-8226 or U266 cells after transfecting with siRNAs for LINC01234 (si-LINC01234 2#, si-LINC01234 3#) or the corresponding control (si-NC). EDU positive cell numbers were quantified and shown as histograms (One-way ANOVA). D. Colony formation analysis of RPMI-8226 or U266 cells after transfecting with siRNAs for LINC01234 (si-LINC01234 2#, si-LINC01234 3#) or the corresponding control (si-NC). Colony numbers were quantified and shown as histograms (One-way ANOVA).
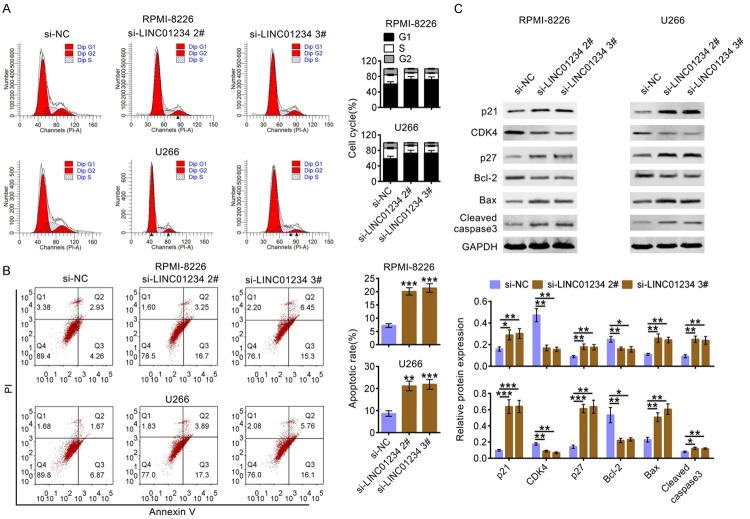
Additionally, as shown in Figure 3A, LINC01234 interference in RPMI-8226 or U266 cells led to an increase in the G1-phase cell population and a decrease in the S-phase cell population. Furthermore, the results of cell apoptosis assay in Figure 3B showed that LINC01234 down-expression significantly increased the percentage of apoptotic cells. Additionally, the western blot data in Figure 3C showed that the expression levels of cell cycle associated proteins (p21, CDK4, p27) and cell apoptosis related proteins (Bcl-2, Bax, Cleaved caspase3) were significantly changed after LINC01234 was knockdown. Among them, the protein levels of p21, p27, Bax and Cleaved caspase3 were increased, and the levels of CDK4 and Bcl-2 were decreased after LINC01234 was knocked down. These above results indicated that LINC01234 down-regulation could inhibit cell proliferation, cell cycle progression and promote cell apoptosis of RPMI-8226 or U266 cells.
Figure 3.
LINC01234 affects cycle progression and apoptosis of MM cells. A. Cell cycle analysis of RPMI-8226 or U266 cells after transfecting with siRNAs for si-LINC01234 (si-LINC01234 2#, si-LINC01234 3#) or the corresponding control (si-NC). Cell percentage in G1, S and G2 phages were quantified and shown as histograms (One-way ANOVA). B. Cell apoptosis analysis of RPMI-8226 or U266 cells after transfecting with siRNAs for LINC01234 (si-LINC01234 2#, si-LINC01234 3#) or the corresponding control (si-NC). Apoptosis rates were quantified and shown as histograms (One-way ANOVA). C. The protein levels of p21, CDK4, p27, Bcl-2, Bax and Cleaved caspase3 in RPMI-8226 or U266 cells after transfecting with siRNAs for LINC01234 (si-LINC01234 2#, si-LINC01234 3#) or the corresponding control (si-NC), using western blotting. Relatively quantitative results was determined by Image J and shown as histogram (One-way ANOVA). The data are expressed as mean ± SEM, *P < 0.05; **P < 0.01; ***P < 0.001.
LINC01234 directly binds to miR-124-3p and negatively regulates its expression
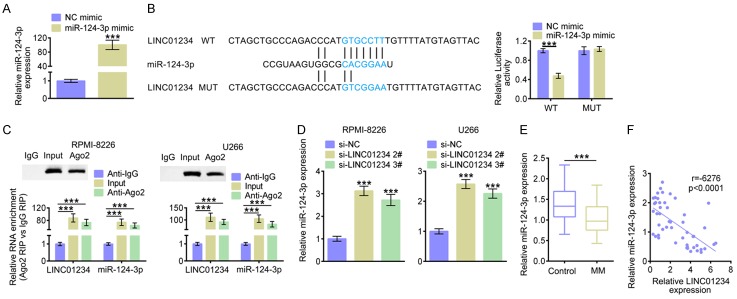
As indicated above, LINC01234 down-regulation inhibited cell proliferation, cell cycle progression and promoted cell apoptosis of MM cells. However, the underlying molecular mechanism remains unclear. Therefore, the potential target miRNA of LINC01234 was analyzed. By means of miRDB (http://http://mirdb.org/), LINC01234 was potentially identified to interact with miR-124-3p. As Figure 4A shown, the miR-124-3p level was significantly up-regulated after transfecting with miR-124-3p mimic. Figure 4B showed that there was potential base pairing sites between the binding seeds of miR-124-3p and LINC01234 (marked in blue). MiR-124-3p up-regulation inhibited the luciferase activity of the LINC01234 reporter plasmids, while had no significant effects after the miR-124-3p binding sites were mutated. Additionally, biotinylated LINC01234 harboring miR-124-3p binding site was synthesized in vitro and the biotin-avidin pull-down assay was performed. The qRT-PCR results demonstrated that miR-124-3p was significantly enriched in the RNA immunoprecipitation (RIP) pulled down by biotinylated LINC01234 (Figure 4C). As shown in Figure 4C, both LINC01234 and miR-124-3p were significantly enriched in the RNA, which were immunoprecipitated by the antibody for Ago2 protein, a core component of miRNA-mediated RISC protein complex. Accordingly, siRNA for LINC01234 caused significantly increased miR-124-3p levels in RPMI-8226 or U266 cells (Figure 4D). Additionally, the results in Figure 4E showed that miR-124-3p level was significantly decreased in MM tissue samples. The Pearson’s correlation analysis demonstrated that there was a negative relationship between miR-124-3p expression and LINC01234 levels in tissue samples from MM patients (Figure 4F). These findings together revealed that LINC01234 can negatively regulate miR-124-3p by targeting its binding seeds.
Figure 4.
LINC01234 directly binds to miR-124-3p and negatively regulates its expression. A. The relative expression levels of miR-124-3p in U266 cells, after transfecting with miR-124-3p mimics (miR-124-3p mimic) or the corresponding control (NC mimic) (Student’s t-test). B. StarBase identified the predicted seed-recognition sites between miR-124-3p and LINC01234, marked in blue. The relative luciferase activity of the LINC01234 reporter plasmid was detected in U266 cells after transfecting with miR-124-3p mimic or the NC mimic. The mutant LINC01234 reporter was also used as a control (Student’s t-test). C. RPMI-8226 or U266 cells were harvested and mixed with Ago2 antibodies to perform miRNA-based pull down. LINC01234 or miR-124-3p enrichments were tested by qRT-PCR and compared to Anti IgG and Input (One-way ANOVA). D. The relative miR-124-3p expression levels in RPMI-8226 or U266 cells after transfecting with siRNAs for LINC01234 (si-LINC01234 2#, si-LINC01234 3#) or the corresponding control (si-NC), as determined using qRT-PCR (One-way ANOVA). E. The relative miR-124-3p expression levels in MM tissues (MM, n=50) and normal control tissues (Control, n=50), as determined using qRT-PCR (Student’s t-test). F. The Pearson correlation analysis on miR-124-3p expression and LINC01234 level. The data are expressed as mean ± SEM, ***P < 0.001.
miR-124-3p negatively regulates GRB2 through targeting its 3’UTR
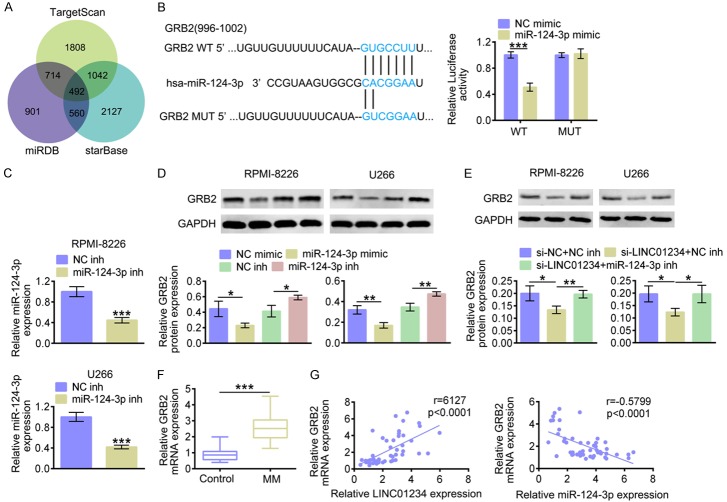
In MM cells, LINC01234 could directly bind to miR-124-3p. However, the role of miR-124-3p in MM remains largely unclear. Next, TargetScan, miRDB and starBase were then used to predicate the target genes of miR-124-3p (Figure 5A). We found that GRB2 are probably potential target of miR-124-3p. Accordingly, the seed binding sites between miR-124-3p and GRB2 were depicted in Figure 5B (marked in blue). As Figure 5B shown, miR-124-3p up-regulation decreased the luciferase activity of the GRB2 3’UTR reporter plasmids, while the luciferase activities were not significantly affected after the miR-124-3p binding sites were mutated. Additionally, biotinylated GRB2 3’UTR harboring miR-124-3p binding site was synthesized in vitro and the biotin-avidin pull-down assay was performed. The qRT-PCR results (Figure 5C) demonstrated that miR-124-3p level was markedly decreased after transfecting with miR-124-3p inhibitors (miR-124-3p inh). The western blot results (Figure 5D) showed that the GRB2 protein levels were markedly decreased after transfecting with miR-124-3p mimics, and miR-124-3p inhibitor caused reversed results. As Figure 5E shown, western blot results demonstrated that GRB2 protein level was markedly decreased after LINC01234 was knocked down. While, the simultaneous transfection of miR-124-3p inhibitors restored the protein expression of GRB2, which was inhibited by LINC01234 siRNA. The results in Figure 5F showed that GRB2 mRNA levels were markedly increased in MM tissue samples. According to the Pearson’s correlation analysis, miR-124-3p expression was negatively correlated with GRB2 mRNA level in tissue samples from MM patients (Figure 5F). These investigations suggested that LINC01234 might regulate GRB2 expression through sponging miR-124-3p.
Figure 5.
miR-124-3p negatively regulates GRB2 through targeting its 3’UTR. A. Analysis on miR-124-3p’s potential targets using TargetScan, miRDB and starBase. B. The predicted seed-recognition sites between 3’UTR of GRB2 and miR-124-3p are marked in blue. The relative luciferase activity of the GRB2 3’UTR reporter plasmid was detected in RPMI-8226 or U266 cells after transfecting with miR-124-3p mimics (miR-124-3p mimic) or the corresponding control (NC mimic). The mutant GRB2 3’UTR reporter was also used as a control (Student’s t-test). C. The relative miR-124-3p expression levels in RPMI-8226 or U266 cells after transfecting with miR-124-3p inhibitors (miR-124-3p inh) or the corresponding control (NC inh), as determined using qRT-PCR (Student’s t-test). D. The protein level of GRB2 in RPMI-8226 or U266 cells after transfecting with miR-124-3p mimics (miR-124-3p mimic), miR-124-3p inhibitors (miR-124-3p inh) or the corresponding controls (NC mimic or NC inh), as determined using western blotting. Relatively quantitative results were determined by Image J and shown as histogram (Student’s t-test). E. The protein level of GRB2 in RPMI-8226 or U266 cells after transfecting with si-LINC01234+NC inh, si-LINC01234+miR-124-3p inh or the corresponding controls (siNC+NC inh), as determined using western blotting. Relatively quantitative results were determined by Image J and shown as histogram (One-way ANOVA). F. The relative GRB2 mRNA expression level in MM tissues (MM, n=50) and normal control tissues (Control, n=50), as determined using qRT-PCR (Student’s t-test). G. The Pearson correlation analysis on GRB2 mRNA expression and LINC01234/miR-124-3p level. The data are expressed as mean ± SEM, *P < 0.05; **P < 0.01; ***P < 0.001.
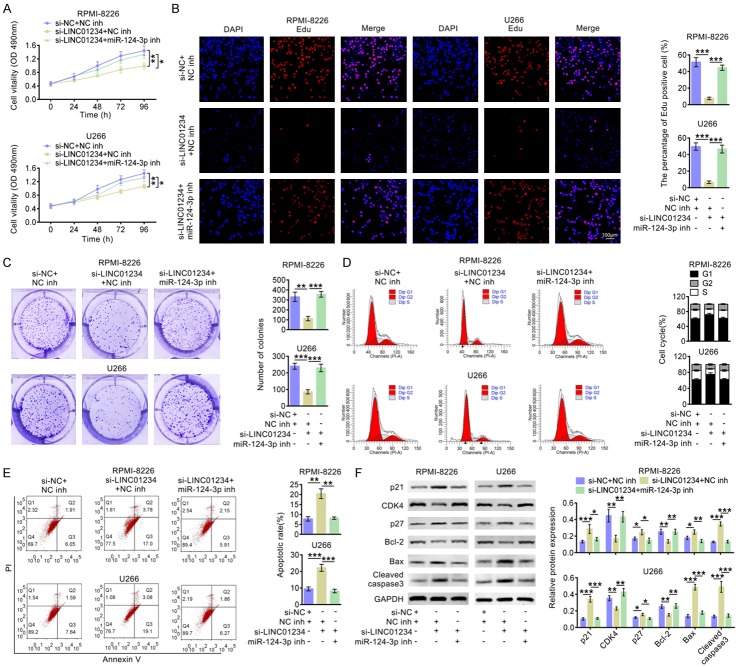
LINC01234 effects MM cell proliferation, cycle progression and apoptosis through inhibiting the miR-124-3p expression
The effects of LINC01234/miR-124-3p axis on cell proliferation, cycle progression and apoptosis were then investigated. As Figure 6A shown, the cell growth was inhibited after LINC01234 was down-regulated, and this effect was abolished after inhibiting LINC01234 and miR-124-3p simultaneously. Figure 6B indicated that LINC01234 down-regulation significantly decreased the number of RPMI-8226 or U266 cells incorporating EDU, and the number of EDU-positive cells was increased after inhibiting LINC01234 and miR-124-3p simultaneously. The colony formation results demonstrated that LINC01234 down-expression markedly decreased colony numbers, and si-LINC01234+miR-124-3p inh eliminated the suppression of colony formation (Figure 6C). Additionally, as shown in Figure 6D, si-LINC01234 resulted in a distinct increase in the G1-phase cell population and a decrease in the S-phase cell population in RPMI-8226 or U266 cells. While, after inhibiting LINC01234 and miR-124-3p simultaneously, the cell cycle arrest caused by LINC01234 down-regulation was abolished. As shown in Figure 6E, the percentage of apoptosis cells was significantly up-regulated after transfecting si-LINC01234, compared to that of si-NC+inh NC cells. While, after inhibiting LINC01234 and miR-124-3p simultaneously, the percentage of apoptosis cells was significantly decreased. In addition, the western blot data in Figure 6F showed that after LINC01234 was inhibited, the expression levels of p21, p27, Bax and Cleaved caspase3 were increased, and the levels of CDK4 and Bcl-2 were decreased. While, simultaneous miR-124-3p suppression abolished the expression changes of these proteins caused by si-LINC01234. These results above indicated that LINC01234 inhibition can suppress cell proliferation, cell cycle progression and promote cell apoptosis in RPMI-8226 or U266 cells, which largely depends on negatively regulating miR-124-3p.
Figure 6.
LINC01234/miR-124-3p affects the proliferation, cycle progression and apoptosis of MM cells. A. Growth curves of RPMI-8226 or U266 cells after transfecting with si-LINC01234+NC inh, si-LINC01234+miR-124-3p inh or the corresponding control (si-NC+NC inh) (One-way ANOVA). Cell growth rate was measured using a CCK-8 kit. B. Representative profiles of EDU cell growth in RPMI-8226 or U266 cells after transfecting with si-LINC01234+NC inh, si-LINC01234+miR-124-3p inh or the corresponding control (si-NC+NC inh). EDU positive cell numbers were quantified and shown as histograms (One-way ANOVA). C. Colony formation analysis of RPMI-8226 or U266 cells after transfecting with si-LINC01234+NC inh, si-LINC01234+miR-124-3p inh or the corresponding control (si-NC+NC inh). Colony numbers were quantified and shown as histograms (One-way ANOVA). D. Cell cycle analysis of RPMI-8226 or U266 cells after transfecting with si-LINC01234+NC inh, si-LINC01234+miR-124-3p inh or the corresponding control (si-NC+NC inh). Cell percentage in G1, S and G2 phages were quantified and shown as histograms (One-way ANOVA). E. Cell apoptosis analysis of RPMI-8226 or U266 cells after transfecting with si-LINC01234+NC inh, si-LINC01234+miR-124-3p inh or the corresponding control (si-NC+NC inh). Apoptosis rates were quantified and shown as histograms (One-way ANOVA). F. The protein levels of p21, CDK4, p27, Bcl-2, Bax and Cleaved caspase3 in RPMI-8226 or U266 cells after transfecting with si-LINC01234+NC inh, si-LINC01234+miR-124-3p inh or the corresponding control (si-NC+NC inh), as determined using western blotting. Relatively quantitative results was determined by Image J and shown as histogram (One-way ANOVA). The data are expressed as mean ± SEM, *P < 0.05; **P < 0.01; ***P < 0.001.
LINC01234 down-regulation inhibits tumor growth in a mouse xenograft model
The function of LINC01234 was finally analyzed in vivo. U266 cells stably down-expressing LINC01234 were were subcutaneously implanted into two groups of nude mice, the corresponding tumors were excised and Figure 7A showed the representative photograph. At day 21 post-implantation, the tumors in the LINC01234-shRNA group exhibited smaller sizes and less weight, compared with that of the NC-shRNA group. As Figure 7B shown, LINC01234 inhibition caused significant reduction in Ki67 positive cell number and decreased GRB2 expression in xenograft tumors. The TUNEL assay indicated higher percentage of apoptotic cells in LINC01234 shRNA group. Additionally, LINC01234 inhibition led to higher miR-124-3p levels in xenograft tumors (Figure 7C). These results indicated that LINC01234 down-expression could increase miR-124-3p expression, suppress the protein expression of GRB2, decrease cell proliferation and finally inhibit the MM growth in vivo.
Figure 7.
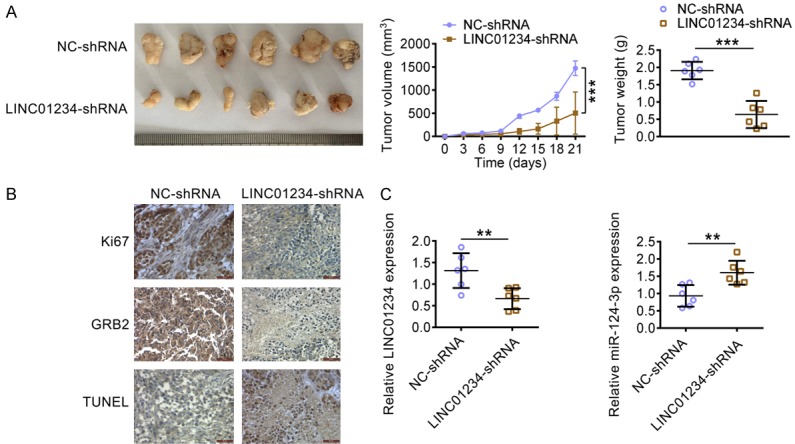
LINC01234 down-regulation inhibits tumor growth in a MM mouse xenograft model. A. A representative photograph of corresponding tumors dissected from mice at day 21 post-implantation. The volumes (left) and weights (right) of the xenograft tumors of nude mice derived from subcutaneous implantation of U266 cells stably expressing shRNA for LINC01234 (LINC01234-shRNA) or the control cells (NC-shRNA) (Student’s t-test). B. IHC staining of Ki67 and GRB2 and TUNEL assay in the xenograft tumors in nude mice derived from subcutaneous implantation of U266 cells stably expressing shRNA for LINC01234 (LINC01234-shRNA) or the control cells (NC-shRNA). C. The relative expression levels of LINC01234 and miR-124-3p in the xenograft tumors in nude mice derived from subcutaneous implantation of U266 cells stably expressing shRNA for LINC01234 (LINC01234-shRNA) or the control cells (NC-shRNA), as determined using qRT-PCR (Student’s t-test). The data are expressed as the mean ± SEM, **P < 0.01; ***P < 0.001.
Discussion
In the present study, the anti-proliferative, anti-cycle progression and pro-apoptotic functions of LINC01234 down-regulation was found, and theregulatory relationship between LINC01234 and miR-124-3p/GRB2 was also investigated. It was gradually demonstrated that lncRNAs were closely associated with MM progression. Recently, Lu found that Linc00515 could regulate miR-140-5p/ATG14 expression and enhance autophagy and chemoresistance of melphalan-resistant myeloma [17]. Li suggested that FEZF1-AS1 regulated miR-610/Akt3 axis and promoted MM cells proliferation [27]. These results demonstrated that lncRNAs exert their functional roles through sponging their target miRNAs. In our study, the results revealed that miR-124-3p could be sponged by LINC01234 in MM. Except for miR-124-3p, other miRNAs have been found to be regulated by LINC01234. These miRNAs mainly include miR-642a-5p in colon cancer [13] and miR-204-5p [14] in Gastric Cancer. So far, the expression signature of miR-642a-5p and miR-204-5p in MM has not been demonstrated. Thus, whether LINC01234 might also sponge these two miRNAs needs further study. Probably, LINC01234 may exert its function via affecting several miRNAs and regulating their target genes. Therefore, the potential target genes of miR-124-3p were next studied.
Using Targetscan, miRDB and starBase, potential seed binding sites between miR-124-3p and GRB2 were predicted. Other cancer-related genes have been reported to be targeted by miR-124-3p, mainly including MGAT5 in breast cancer [17], ITGA3 in bladder cancer [28], PIM1 in astrocytoma [29], IGF2BP1 in cervical cancer [30]. Based on the miRNA’s characteristics of multiple targets, whether miR-124-3p exerts its regulatory activity by negatively regulating these genes simultaneously in MM needs further study. On the other hand, miR-124-3p have been found to be sponged by other lncRNAs, which mainly including OGFRP1 in non-small cell lung cancer [31], SNHG16 in acute lymphoblastic leukemia [32] and LINC00511/HOXA11-AS in glioma [33,34]. The roles of these lncRNAs in MM have not yet been known. It might be hypothesized that in MM development, miR-124-3p act as an intermediary agent, which could be sponged by several lncRNAs and target multiple genes meanwhile.
Because GRB2 is enormously involved in the development of different cancers, which stands out as a candidate of anticancer drug target [19]. On account of the insufficient bioavailability of peptide-based inhibitors and the side effects of small molecule compounds, it may be of benefit for developing novel anticancer drugs, such as gene durgs based on ncRNAs. Our data showed that miR-124-3p inhibitors restored the effect of MM cell proliferation, cycle and apoptosis, which were caused by LINC01234 inhibition. Except for miR-124-3p, miR-329, miR-411-5p, miR-1258, miR-433-3p and miR-378a-3p have been reported to regulate GRB2 at post-transcriptional level in other cancers [35-39]. Although the roles of these miRNAs in MM remain unclear, they hold promise for targeting GRB2. Nevertheless, our data revealed that LINC01234 down-regulation also inhibited the expression of GRB2 by up-regulating miR-124-3p. Thus, a combined utilization of chemotherapy and GRB2 oligonucleotide inhibitors, such as LINC01234 inhibitors, miR-124-3p or other miRNAs, are helpfulp for MM treatment.
In summary, our findings revealed that LINC01234 down-expression could suppress cell proliferation, cell cycle progression and promote cell apoptosis in RPMI-8226 or U266 cells. LINC01234 could competitively sponge miR-124-3p and decrease its expression. Additionally, LINC01234/miR-124-3p play a crucial part in affecting GRB2 expression in MM through binding its 3’UTR. Furthermore, LINC01234 down-expression could inhibit the GRB2 expression and tumor growth in vivo. Based on the functions of anti-proliferation, anti-cell cycle progression and pro-apoptosis, the signaling axis of LINC01234/miR-124-3p/GRB2 might hold promise as promising candidates for MM treatment.
Disclosure of conflict of interest
None.
References
- 1.Rollig C, Knop S, Bornhauser M. Multiple myeloma. Lancet. 2015;385:2197–2208. doi: 10.1016/S0140-6736(14)60493-1. [DOI] [PubMed] [Google Scholar]
- 2.Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, Morgan G, Van Ness B, Chesi M, Minvielle S, Neri A, Barlogie B, Kuehl WM, Liebisch P, Davies F, Chen-Kiang S, Durie BG, Carrasco R, Sezer O, Reiman T, Pilarski L, Avet-Loiseau H International Myeloma Working Group. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corre J, Munshi N, Avet-Loiseau H. Genetics of multiple myeloma: another heterogeneity level? Blood. 2015;125:1870–1876. doi: 10.1182/blood-2014-10-567370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Licht JD, Shortt J, Johnstone R. From anecdote to targeted therapy: the curious case of thalidomide in multiple myeloma. Cancer Cell. 2014;25:9–11. doi: 10.1016/j.ccr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B, Xu H, Han H, Song S, Zhang X, Ouyang L, Qian C, Hong Y, Qiu Y, Zhou W, Huang M, Zhuang W. Exosome-mediated transfer of lncRUNX2-AS1 from multiple myeloma cells to MSCs contributes to osteogenesis. Oncogene. 2018;37:5508–5519. doi: 10.1038/s41388-018-0359-0. [DOI] [PubMed] [Google Scholar]
- 7.Yang N, Chen J, Zhang H, Wang X, Yao H, Peng Y, Zhang W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017;8:e2975. doi: 10.1038/cddis.2017.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Ye H, He M, Zhou X, Sun N, Guo W, Lin X, Huang H, Lin Y, Yao R, Wang H. LncRNA PDIA3P interacts with c-Myc to regulate cell proliferation via induction of pentose phosphate pathway in multiple myeloma. Biochem Biophys Res Commun. 2018;498:207–213. doi: 10.1016/j.bbrc.2018.02.211. [DOI] [PubMed] [Google Scholar]
- 9.Zhuang W, Ge X, Yang S, Huang M, Zhuang W, Chen P, Zhang X, Fu J, Qu J, Li B. Upregulation of lncRNA MEG3 promotes osteogenic differentiation of mesenchymal stem cells from multiple myeloma patients by targeting BMP4 transcription. Stem Cells. 2015;33:1985–1997. doi: 10.1002/stem.1989. [DOI] [PubMed] [Google Scholar]
- 10.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 11.Zhang G, Pian C, Chen Z, Zhang J, Xu M, Zhang L, Chen Y. Identification of cancer-related miRNA-lncRNA biomarkers using a basic miRNA-lncRNA network. PLoS One. 2018;13:e0196681. doi: 10.1371/journal.pone.0196681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Hu N, Wang C, Zhao H, Gu Y. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle. 2018;17:319–329. doi: 10.1080/15384101.2017.1407893. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Lin C, Zhang Y, Chen Y, Bai Y, Zhang Y. Long noncoding RNA LINC01234 promotes serine hydroxymethyltransferase 2 expression and proliferation by competitively binding miR-642a-5p in colon cancer. Cell Death Dis. 2019;10:137. doi: 10.1038/s41419-019-1352-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, Chen Q, Wei C, Fu H, Xu T, Ren S, Sun M, Wang Z. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin Cancer Res. 2018;24:2002–2014. doi: 10.1158/1078-0432.CCR-17-2376. [DOI] [PubMed] [Google Scholar]
- 15.Ghaffar M, Khodahemmati S, Li J, Shahzad M, Wang M, Wang Y, Li C, Chen S, Zeng Y. Long non-coding RNA LINC01234 regulates proliferation, invasion and apoptosis in esophageal cancer cells. J Cancer. 2018;9:4242–4249. doi: 10.7150/jca.26095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long HD, Ma YS, Yang HQ, Xue SB, Liu JB, Yu F, Lv ZW, Li JY, Xie RT, Chang ZY, Lu GX, Xie WT, Fu D, Pang LJ. Reduced hsa-miR-124-3p levels are associated with the poor survival of patients with hepatocellular carcinoma. Mol Biol Rep. 2018;45:2615–2623. doi: 10.1007/s11033-018-4431-1. [DOI] [PubMed] [Google Scholar]
- 17.Yan G, Li Y, Zhan L, Sun S, Yuan J, Wang T, Yin Y, Dai Z, Zhu Y, Jiang Z, Liu L, Fan Y, Yang F, Hu W. Decreased miR-124-3p promoted breast cancer proliferation and metastasis by targeting MGAT5. Am J Cancer Res. 2019;9:585–596. [PMC free article] [PubMed] [Google Scholar]
- 18.Buday L, Downward J. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell. 1993;73:611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 19.Ijaz M, Wang F, Shahbaz M, Jiang W, Fathy AH, Nesa EU. The role of Grb2 in cancer and peptides as Grb2 antagonists. Protein Pept Lett. 2018;24:1084–1095. doi: 10.2174/0929866525666171123213148. [DOI] [PubMed] [Google Scholar]
- 20.Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 21.Neumann C, Zehentmaier G, Danhauser-Riedl S, Emmerich B, Hallek M. Interleukin-6 induces tyrosine phosphorylation of the Ras activating protein Shc, and its complex formation with Grb2 in the human multiple myeloma cell line LP-1. Eur J Immunol. 1996;26:379–384. doi: 10.1002/eji.1830260217. [DOI] [PubMed] [Google Scholar]
- 22.World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 23.de Planell-Saguer M, Rodicio MC, Mourelatos Z. Rapid in situ codetection of noncoding RNAs and proteins in cells and formalin-fixed paraffin-embedded tissue sections without protease treatment. Nat Protoc. 2010;5:1061–1073. doi: 10.1038/nprot.2010.62. [DOI] [PubMed] [Google Scholar]
- 24.Bierhoff H. Analysis of lncRNA-protein interactions by RNA-protein pull-down assays and RNA immunoprecipitation (RIP) Methods Mol Biol. 2018;1686:241–250. doi: 10.1007/978-1-4939-7371-2_17. [DOI] [PubMed] [Google Scholar]
- 25.Ma G, Tang M, Wu Y, Xu X, Pan F, Xu R. LncRNAs and miRNAs: potential biomarkers and therapeutic targets for prostate cancer. Am J Transl Res. 2016;8:5141–5150. [PMC free article] [PubMed] [Google Scholar]
- 26.Guide for the Care and Use of Laboratory Animals. Washington (DC): 2011. [Google Scholar]
- 27.Li QY, Chen L, Hu N, Zhao H. Long non-coding RNA FEZF1-AS1 promotes cell growth in multiple myeloma via miR-610/Akt3 axis. Biomed Pharmacother. 2018;103:1727–1732. doi: 10.1016/j.biopha.2018.04.094. [DOI] [PubMed] [Google Scholar]
- 28.Wang JR, Liu B, Zhou L, Huang YX. MicroRNA-124-3p suppresses cell migration and invasion by targeting ITGA3 signaling in bladder cancer. Cancer Biomark. 2019;24:159–172. doi: 10.3233/CBM-182000. [DOI] [PubMed] [Google Scholar]
- 29.Deng D, Wang L, Chen Y, Li B, Xue L, Shao N, Wang Q, Xia X, Yang Y, Zhi F. MicroRNA-124-3p regulates cell proliferation, invasion, apoptosis, and bioenergetics by targeting PIM1 in astrocytoma. Cancer Sci. 2016;107:899–907. doi: 10.1111/cas.12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang P, Zhang L, Zhang J, Xu G. MicroRNA-124-3p inhibits cell growth and metastasis in cervical cancer by targeting IGF2BP1. Exp Ther Med. 2018;15:1385–1393. doi: 10.3892/etm.2017.5528. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Tang LX, Chen GH, Li H, He P, Zhang Y, Xu XW. Long non-coding RNA OGFRP1 regulates LYPD3 expression by sponging miR-124-3p and promotes non-small cell lung cancer progression. Biochem Biophys Res Commun. 2018;505:578–585. doi: 10.1016/j.bbrc.2018.09.146. [DOI] [PubMed] [Google Scholar]
- 32.Yang T, Jin X, Lan J, Wang W. Long non-coding RNA SNHG16 has Tumor suppressing effect in acute lymphoblastic leukemia by inverse interaction on hsa-miR-124-3p. IUBMB Life. 2019;71:134–142. doi: 10.1002/iub.1947. [DOI] [PubMed] [Google Scholar]
- 33.Yang JX, Liu B, Yang BY, Meng Q. Long non-coding RNA homeobox (HOX) A11-AS promotes malignant progression of glioma by targeting miR-124-3p. Neoplasma. 2018;65:505–514. doi: 10.4149/neo_2018_170705N462. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Liu H, Yang J, Yang J, Yang L, Wang Y, Yan Z, Sun Y, Sun X, Jiao B. Long noncoding RNA LINC00511 induced by SP1 accelerates the glioma progression through targeting miR-124-3p/CCND2 axis. J Cell Mol Med. 2019;23:4386–4394. doi: 10.1111/jcmm.14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W, Wei K, Pan C, Li H, Cao J, Han X, Tang Y, Zhu S, Yuan W, He Y, Xia Y, Chen L, Chen Y. MicroRNA-1258 suppresses tumour progression via GRB2/Ras/Erk pathway in non-small-cell lung cancer. Cell Prolif. 2018;51:e12502. doi: 10.1111/cpr.12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Lu X, Zhang T, Wen C, Shi M, Tang X, Chen H, Peng C, Li H, Fang Y, Deng X, Shen B. mir-329 restricts tumor growth by targeting grb2 in pancreatic cancer. Oncotarget. 2016;7:21441–21453. doi: 10.18632/oncotarget.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu ZH, Yao TZ, Liu W. miR-378a-3p sensitizes ovarian cancer cells to cisplatin through targeting MAPK1/GRB2. Biomed Pharmacother. 2018;107:1410–1417. doi: 10.1016/j.biopha.2018.08.132. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Xu G, Liu G, Ye Y, Zhang C, Fan C, Wang H, Cai H, Xiao R, Huang Z, Luo Q. miR-411-5p inhibits proliferation and metastasis of breast cancer cell via targeting GRB2. Biochem Biophys Res Commun. 2016;476:607–613. doi: 10.1016/j.bbrc.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Shi Q, Wang Y, Mu Y, Wang X, Fan Q. MiR-433-3p inhibits proliferation and invasion of esophageal squamous cell carcinoma by targeting GRB2. Cell Physiol Biochem. 2018;46:2187–2196. doi: 10.1159/000489548. [DOI] [PubMed] [Google Scholar]