Abstract
LncRNAs (long noncoding RNAs) have been shown to be potentially critical regulators in head and neck squamous cell carcinoma (HNSCC). LncRNA LINC00460 (long intergenic non-protein coding RNA 460), an “oncogene”, regulates progression of various tumors. However, the tumorigenic mechanism of LINC00460 on HNSCC is yet to be investigated. In the current study, we discovered that LINC00460 was relatively up-regulated in both HNSCC cancer tissues and cell lines, and predicted a poor prognosis in HNSCC patients. Gain- and loss-of functional studies established that over-expression of LINC00460 promoted cell proliferation, invasion and migration of HNSCC cells in vitro, while the promotion abilities were suppressed via knockdown of LINC00460. Our results identified miR-612 as a novel target of LINC00460, whose expression suggested a negative correlation with LINC00460 in HNSCC tissues and cell lines. LINC00460 increased the expression of serine/threonine kinase AKT2 via sponging miR-612. Rescue experiments indicated that LINC00460 could promote HNSCC progression partially through inhibition of miR-612. Subcutaneous xenotransplanted tumor model confirmed that interference of LINC00460 suppressed in vivo tumorigenic ability of HNSCC via down-regulation of AKT2. In conclusion, our findings clarified the biologic significance of LINC00460/miR-612/AKT2 axis in HNSCC progression and provided novel evidence that LINC00460 may be a new potential therapeutic target for HNSCC.
Keywords: LINC00460, miR-612, AKT2, HNSCC, progression
Introduction
Head and neck squamous cell carcinoma (HNSCC) ranks the sixth most common cancer worldwide [1]. By the means of TNM (tumor-node-metastasis) classification of malignant tumors, HNSCC is assigned with stage I, II, III and IV [2]. Although the therapeutical treatment for HNSCC has been progressed tremendously in the last several decades, the overall survival rate for HNSCC patients has not been improved due to local recurrence and distant metastasis [3]. Therefore, it is of great clinical significance to investigate the genetic and epigenetic molecular alterations in HNSCC to improve the survival rate of patients.
LncRNAs (long noncoding RNAs), with more than 200 nucleotides in length, have been considered as transcriptional or post-transcriptional regulators of gene expression [4] and participate in various physiological and pathological processes [5,6]. According to the critical regulation on cell proliferation, migration and invasion [7], lncRNAs are recently acquired great attention in cancer research. LINC00460 (long intergenic non-protein coding RNA 460), as an “oncogene”, has been reported to play an important role in nasopharyngeal cancer [8], esophageal squamous cell carcinoma [9], and lung cancer [10]. Recently, research has shown that LINC00460 might be a novel prognosis predictor of patients with HNSCC [11]. However, the mechanism of LINC00460 in the progression process of HNSCC is still under investigation.
Studies have investigated the application of miRNAs as diagnostic/prognostic biomarkers or potential therapeutic targets for improvement of HNSCC [12]. Moreover, considering that lncRNAs often act as a miRNA sponges or ceRNAs (competitive endogenous RNAs) to participate in various biological process [13], it is an urgent need to discover the target for LINC00460 in HNSCC. MiR-612 plays an important role in liver cancer [14], hepatocellular carcinoma [15] and melanoma [16] as a tumor suppressor. MiR-612 was reported to be involved in development and metastasis of esophageal squamous cell carcinoma [17]. In addition, several studies have shown that miR-612 could bind to AKT2 [18,19], which is involved in the promotion of the development of HNSCC [20]. Therefore, we evaluated the hypothesis that LINC00460, together with the downstream regulators miR-612/AKT2, may be involved in regulation of HNSCC progression.
We not only detect the impact of LINC00460 on tumor proliferation, migration and invasion of HNSCC, but also explore the underlying mechanism. The meaning results would provide a reference for development of new therapies for HNSCC.
Materials and methods
Patients and tumor tissues collection
60 HNSCC patients, diagnosed via pathology examination and confirmed with imaging modalities, were recruited in this study. The clinicopathological characteristics of the patients were shown in Table 1. The present study was approved by Tongde Hospital of Zhejiang Province, and all the patients signed written informed consent. The HNSCC tissues and adjacent normal tissues were collected from the patients.
Table 1.
Primer Sequence
| ID | Sequence (5’-3’) |
|---|---|
| GAPDH F | ACCACAGTCCATGCCATCAC |
| GAPDH R | TCCACCACCCTGTTGCTGTA |
| LINC00460 F | TTGAATAGAGTAGTTCCTTGCGCTG |
| LINC00460 R | GGCTACTTCACCCAGCTGTCTAG |
| miR-612 F | GCTGGGCAGGGCTTCT |
| miR-612 R | CAGTGCGTGTCGTGGAGT |
| AKT2 F | ACCACAGTCATCGAGAGGACC |
| AKT2 R | GGAGCCACACTTGTAGTCCA |
| Cyclin D F | CTGGCCATGAACTACCTGGA |
| Cyclin D R | GTCACACTTGATCACTCTGG |
| p21 F | CCTGTCACTGTCTTGTACCCT |
| p21 R | GCGTTTGGAGTGGTAGAAATCT |
| E-cadherin F | CCCGGGACAACGTTTATTAC |
| E-cadherin R | GCTGGCTCAAGTCAAAGTCC |
| N-cadherin F | GGTGGAGGAGAAGAAGACCAG |
| N-cadherin R | GGCATCAGGCTCCACAGT |
| U6 F | CTCGCTTCGGCAGCACA |
| U6 R | AACGCTTCACGAATTTGCGT |
In situ hybridization
5 μm sections made from HNSCC and adjacent normal tissues were firstly fixed with 10% formalin, and embedded in paraffin. Following by dewaxing and rehydrating, the tissue sections were then digested with 20 µg/mL proteinase K for 30 minutes. 4% paraformaldehyde-fixed samples were hybridized overnight with 8 ng/µL specific antisense oligonucleotide DNA probe 5’-GGCTGAGGCATTTCTAACAGGGCTGGAGGA-3’ synthesized by Invitrogen (Carlsbad, CA, USA) at 55°C. The samples were then incubated with HRP (horseradish peroxidase, Sigma Aldrich, St. Louis, MO, USA) at 4°C for 30 minutes. Hybridization signals were amplified with diaminobenzidine (DAB, Sigma Aldrich), and the images were taken by fluorescent microscope (DP12 SZX7, Olympus Inc., Japan).
Cell culture and transfection
HNSCC cell lines (HSC3, Fadu and SAS) and normal immortal keratinocyte cell line from adult human skin (HACA), purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA), were cultured in Dulbecco modified Eagle medium (DMEM; Lonza, Basel, Switzerland) supplemented with 10% fetal bovine serum (FBS, Gibco, MA, USA) with additional streptomycin (100 μg/ml) and penicillin (100 U/ml). 37°C constant temperature incubator with 5% CO2 was utilized to incubating cells.
For the over-expression of LINC00460, full-length of LINC00460 was amplified and cloned into pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). Fadu or SAS cells were seeded at 4×105 cells per well in 12-well plates and then transfected with pcDNA3.1-LINC00460 or the negative (pcDNA3.1-NC) via Lipofectamine 2000 (Invitrogen). For the knock down of LINC00460, shRNAs (1#: 5’-GCTAAGACCTAATAGCCAATA-3’ and 2#: 5’-GCCATCCACTTCAAAGTATTC-3’) were inserted into pLKO.1 (Biosettia, San Diego, CA, USA). 4×105/well HEK-293T cells were cotransfected with pLKO-1# LINC00460 (sh-LINC00460#1), pLKO-2# LINC00460 (sh-LINC00460#2, sh-LINC00460) or pLKO Scramble (sh-NC) with psPAX2 and pMD2.G via Lipofectamine 2000. Two days after transfection, lentiviruses containing released lentiviral vectors were harvested. Fadu or SAS cells were infected with sh-LINC004601#, sh-LINC004602# or sh-NC lentiviruses in the presence of ViraPower™ Packaging Mix (Thermo Fisher, Waltham, MA, USA) and 8 mg/mL polybrene. With 5 μg/mL puromycin (Sigma Aldrich, St. Louis, MO, USA) treatment for one week, the stable cell lines were obtained.
MiR-612 mimics, inhibitor and the negative controls (miR-NC, NC inh) were synthesized by GenePharma (Suzhou, China). Fadu or SAS cells were transfected with miR-612 mimics/inhibitor (40 nM) or their NC via Lipofectamine 2000.
Cell proliferation
1×103 Fadu or SAS cells per well were seeded in s in 96-well plates, and cultured as before for 5 days. Cells were washed with PBS after the medium was removed, and then incubated with 10 μL CCK8 (cell counting kit-8) reagent (Beyotime, Shanghai, China) at 37°C for 2 hours. The cell viabilities were calculated via detection of absorbance at 450 nm in 24, 48, 72 and 96 hours after the treatment. For the colony formation assay, 1×103 Fadu or SAS cells per well were plated in six-well plate, and cultured as before for 14 days. The colonies were stained by 1% crystal violet-2% ethanol suspended in PBS (Beyotime), counted and photographed under light microscope (Olympus, Tokyo, Japan).
Transwell assay
For cell invasion, 2×104 Fadu or SAS cells, suspending in 200 μL DMEM medium without FBS, were plated into the upper wells of chamber (BD Biosciences, Bedford, MA, USA) per well with the Matrigel-coated membrane (BD Biosciences). 500 μL DMEM medium supplemented with 10% FBS were added to the lower wells of chambers. 24 hours later, invasive cells at the bottom of chambers were fixed in 100% methanol for 30 minutes, washed with PBS and then stained with 0.1% crystal violet for 1 hour. Stained cells were imaged and counted under microscope. For cell migration experiment, the upper chambers were not precoated with Matrigel, and then subjected to the protocol as cell invasion assay.
Fluorescence in situ hybridization (FISH)
Fadu or SAS cells were firstly fixed in 4% formaldehyde solution for 15 minutes, and then incubated with 0.1% Triton X-100 for another 10 minutes. Fluorescence-conjugated LINC00460 probes (Invitrogen) were hybridized at 37°C with cells in the dark for 5 hours. The cells were then photographed via laser scanning confocal microscopy (Carl Zeiss, Jena, Germany).
Dual luciferase reporter assay
Sequences of wildtype or mutant 3’-UTR of LINC00460 or AKT2 were synthesized and then subcloned into pmirGLO luciferase reporter vector (Promega, Madison, Wisconsin, USA). 3×104 HEK-293T cells per well were seeded in 24-well plates and co-transfected miR-612 mimic or negative control (miR-NC) with pmirGLO-wt-LINC00460, pmirGLO-mut-LINC00460, pmirGLO-wt-AKT2 or pmirGLO-mut-AKT2 via Lipofectamine 2000. 48 hours after transfection, the luciferase activities were performed with the Lucifer Reporter Assay System (Promega) and normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP)
Fadu or SAS cells at 80-90% confluency were collected and lysate in RIP lysis buffer (EZ-Magna RIP kit; Millipore, Billerica, MA, USA), and then 100 μL samples were incubated with protein G Sepharose beads (GE Healthcare, Eindhoven, The Netherlands) coated with anti-AGO2 antibody (Abcam, Cambridge, MA, USA) at 4°C overnight, and anti-IgG antibody was used as the negative control for the RIP procedure, while anti-SNRNP70 was used as positive control. The samples were incubated with Proteinase K to digest the protein and then immunoprecipitated RNA was isolated for qRT-PCR as mentioned below to demonstrate the presence of the binding targets using respective primers.
qRT-PCR
Total RNAs from HNSCC tissues or cell lines were isolated with Trizol (Invitrogen), miRNAs were extracted via miRcute miRNA isolation kit (Tiangen, Beijing, China). RNAs were then reverse-transcribed by PrimeScript RT Reagent (Takara, Shiga, Japan). qRT-PCR was conducted with SYBR Green Master (Roche, Mannheim, Germany) on ViiA 7 (Applied Biosystems, Austin, TX, USA). GAPDH or U6 were used as endogenous control. The primer sequences were showed as follows.
Western blot
30 µg proteins extracted from cultured HNSCC cells were separated by SDS-PAGE, and then electro-transferred onto PVDF membrane. After blocking with 5% BSA, the membrane was incubated overnight with primary antibody: anti-Cyclin D1, p21 antibodies (1:1500, Abcam), E-cadherin, N-cadherin (1:2000, Abcam), AKT2, p-AKT2 (1:2500, Abcam), GAPDH (1:3000, Abcam) at 4°C. Following incubation with HRP labeled secondary antibody (1:5000; Abcam), the immunoreactivities were detected by enhanced chemiluminescence (KeyGen, Nanjin, China).
Mouse xenograft assay
All studies involving animals were approved by xxx and conducted in accordance with the guidelines set out by xxx. 12 six-week-old female BALB/c nude mice, obtained from ARS/Sprague Dawley Division (Madison, WI, USA), were randomly seperated into two groups and used for the tumor formation assay. 100 μL 1×108 Fadu cells in PBS with stable down-regulation of LINC00460 via pLKO-2# LINC00460 or the scrambled shRNA were subcutaneously injected into the position of hind flank of nude mice. Tumors were measured with digital calipers every three days and the tumor volume was calculated. 30 days later, the mice were sacrificed with 40 mg/kg sodium pentobarbital, and the tumor tissues were isolated and weighted, and RNAs and proteins were extracted for analysis.
Immunohistochemistry
HNSCC tissues were firstly fixed with 10% formalin, and embedded in paraffin. After dewaxing and rehydration, 4 µm thick sections were incubated in 3% H2O2 for blocking endogenous peroxidase. The sections were immersed in Tris-EDTA buffer containing 0.05% Tween 20, pH 9.0, in water bath at 95°C for 30 minutes for antigen retrieval. After washing with PBS, the sections were firstly incubated in 4% dry milk and 0.3% goat serum in PBS solution for 20 minutes to block non-specific binding, and then incubated overnight with anti-N-cadherin (ab18203) or Ki-67 (ab16667) antibody in the presence of 10% rabbit serum. After washing with PBS, the sections were then incubated with HRP goat anti-rabbit IgG secondary antibody. Slides were counterstained with hematoxylin to stain cell nuclei in order to identify cells, dehydrated, and examined under a light microscope (Olympus).
Statistical analysis
All results are expressed as mean ± SEM. By the means of GraphPad Prism software (GraphPad Prism Software Inc., San Diego, USA) and one-way analysis of variance (ANOVA), we determined the statistical analyses. Survival curves plotted via Kaplan-Meier method and log-rank test were used to analyze difference between patients with high or low levels of FEZF1-AS1 expression and overall survival. The differences among Multiple-group (more than two groups) are measured with the one-way analysis of variance (ANOVA), and the differences between two groups designs are measured with TurKey method or Student’s t-test. The overall analysis of the observation data at multiple time points was performed by repeated measures analysis of variance, and multiple comparisons between the two groups or two time points within groups were conducted by TurKey method or difference t test, respectively. For co-expressed pairs, Pearson correlation coefficients were calculated based on the expression value between every differentially expressed pairs. P < 0.05, P < 0.01 or P < 0.001 was considered as a mark of statistically significant.
Results
LncRNA LINC00460 was induced in both HNSCC tumor tissues and cell lines
We firstly detect the expression level of lncRNA LINC00460 in HNSCC to explore the effect of LINC00460 on HNSCC progression. qRT-PCR analysis demonstrated that LINC00460 was significantly upregulated in HNSCC tumor tissues (HNSCC) compared to the adjacent normal tissues (Normal) (P < 0.001) (Figure 1A). Kaplan-Meier survival analysis showed that higher expression of LINC00460 (fold change ≥ 2.5, N = 28) was related to short overall survival (OS) (P = 0.0386) (Figure 1B). Further refinement analysis of correlation between LINC00460 expression and clinic pathologic characteristics of HNSCC patients showed that among the 60 patients, higher expression of LINC00460 was more frequently occurred in patients with T3+T4 (primary tumor stage) (P = 0.0031), III+IV (clinical stage) (P < 0.001) and lymph node metastasis (P < 0.001) (Table 2). Moreover, no significant correlation was shown among the age (P = 0.8306), gender (P = 0.5501), smoking status (P = 0.0978) and histological grade (P = 0.3107) with LINC00460 expression. Theseresults revealed that up-regulation of LINC00460 was associated with poor prognosis of HNSCC. In line with the increase of LINC00460 in HNSCC tissues, LINC00460 was also up-regulated in HNSCC cell lines (HSC3, Fadu and SAS) compared with normal immortal keratinocyte cell line from adult human skin (HACA) (Figure 1C). In situ hybridization (ISH) furtherly confirmed the augment of LINC00460 in HNSCC tissues (Figure 1D).
Figure 1.
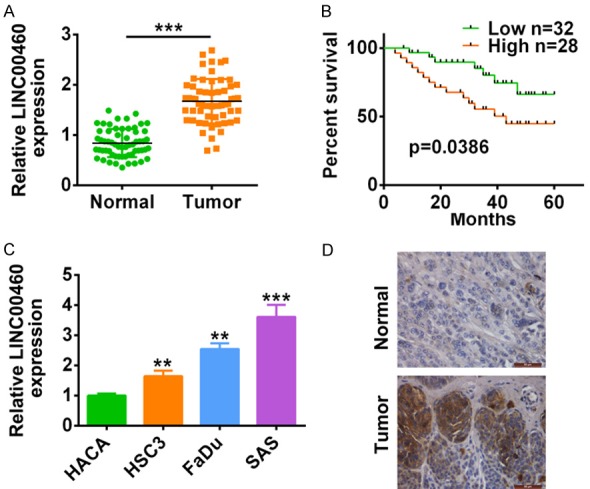
LncRNA LINC00460 was induced in both HNSCC tumor tissues and cell lines. A. The expression of lncRNA LINC00460 in HNSCC tissues and adjacent normal tissues was detected by qRT-PCR (N=60). ***represents tumor vs. adjacent normal tissues, P < 0.001. B. OS analysis was performed in HNSCC patients with high LINC00460 expression and low levels of LINC00460. C. The expression of LINC00460 in HNSCC cell lines (HSC3, Fadu and SAS) and normal immortal keratinocyte cell line from adult human skin (HACA) was detected by qRT-PCR. **, ***represents HNSCC cell lines vs. HACA, P < 0.01, P < 0.001. D. In situ hybridization (ISH) analysis was conducted to analyze the LINC00460 expression in HNSCC tissues.
Table 2.
Demographics and clinical characteristics of the cohort of HNSCC patients (N = 60)
| Parameters | No.of patients | PVT1 expression | p value |
|---|---|---|---|
| Age | |||
| < 59 | 28 | 1.66±0.437 | 0.8306 |
| ≥ 59 | 32 | 1.685±0.460 | |
| Gender | |||
| Female | 29 | 1.709±0.405 | 0.5501 |
| Male | 31 | 1.64±0.478 | |
| Smoking | |||
| Yes | 33 | 1.59±0.371 | 0.0978 |
| No | 27 | 1.78±0.503 | |
| Histological grade | |||
| G1+G2 | 24 | 1.602±0.405 | 0.3107 |
| G3 | 36 | 1.721±0.464 | |
| T classification | |||
| T1+T2 | 30 | 1.509±0.335 | 0.0031 |
| T3+T4 | 30 | 1.838±0.478 | |
| Clinical stage | |||
| I+II | 27 | 1.407±0.32 | 0.000 |
| III+IV | 33 | 1.891±0.42 | |
| Lymph node metastasis | |||
| N0 | 35 | 1.28±0.242 | 0.000 |
| N+ | 25 | 1.956±0.318 | |
LINC00460 promoted HNSCC cell proliferation, migration and invasion
The effect of LINC00460 on HNSCC was examined via gain-of functional assays. Fadu and SAS cell lines were transfected with pcDNA3.1-LINC00460 for the over-expression of LINC00460, which were confirmed by qRT-PCR in Figure 2A. ThenCCK8 (Figure 2B) and colony formation (Figure 2C) assays revealed the promotion ability of LINC00460 over-expression on cell proliferation of Fadu and SAS cells. Furthermore, transwell assay indicated the promotion ability of LINC00460 over-expression on cell migration and invasion of Fadu and SAS cells (Figure 2D). Expression of genes involved in cell progression was also detected via qRT-PCR and western blot analysis. The results showed that both mRNA (Figure 2E) and protein (Figure 2F) expression of Cyclin D and N-cadherin were increased by LINC00460 over-expression, whereas the expression of p21 and E-cadherin were decreased by LINC00460 over-expression (Figure 2E and 2F).
Figure 2.
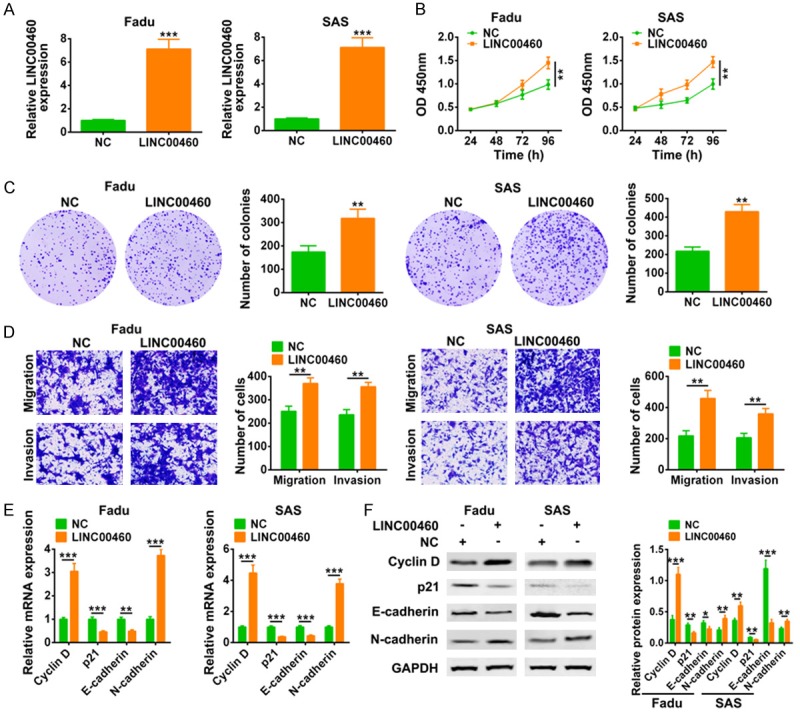
LINC00460 promoted HNSCC cell proliferation, migration and invasion. A. Transfection efficiency of pcDNA3.1-LINC00460 in Fadu and SAS cell lines was detected by qRT-PCR. ***Represents pcDNA3.1-LINC00460 vs. pcDNA3.1 (NC), P < 0.001. B. The influence of pcDNA3.1-LINC00460 on cell viability of Fadu and SAS cells was detected by CCK8. **Represents pcDNA3.1-LINC00460 vs. pcDNA3.1 (NC), P < 0.01. C. The influence of pcDNA3.1-LINC00460 on cell proliferation of Fadu and SAS cells was detected by colony formation assay. **Represents pcDNA3.1-LINC00460 vs. pcDNA3.1 (NC), P < 0.01. D. The influence of pcDNA3.1-LINC00460 on cell migration and invasion of Fadu and SAS cells was examined by transwell assay. **Represents pcDNA3.1-LINC00460 vs. pcDNA3.1 (NC), P < 0.01. E. The influence of pcDNA3.1-LINC00460 on mRNA expression of Cyclin D, p21, N-cadherin and E-cadherin in Fadu and SAS cells was detected by qRT-PCR. **, *** represents pcDNA3.1-LINC00460 vs. pcDNA3.1 (NC), P < 0.01, P < 0.001. F. The influence of pcDNA3.1-LINC00460 on protein expression of Cyclin D, p21, N-cadherin and E-cadherin in Fadu and SAS cells was detected by western blot. *, **, ***Represents pcDNA3.1-LINC00460 vs. pcDNA3.1 (NC), P < 0.05, P < 0.01, P < 0.001.
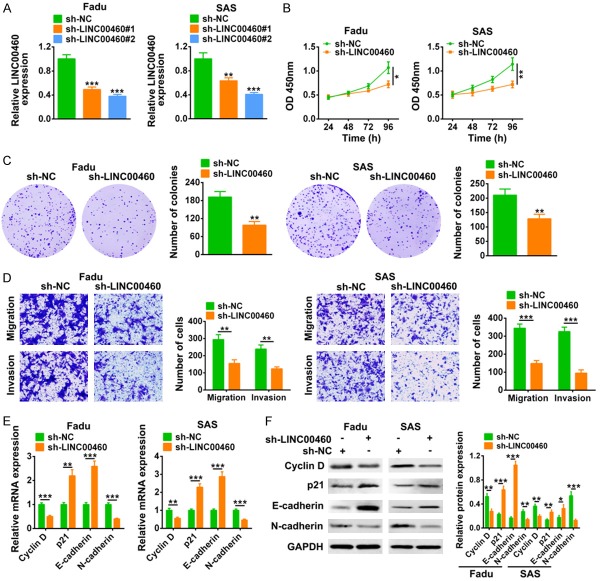
In addition, loss-of functional assays were also conducted. Similarly, Fadu and SAS cell lines transfected with sh-LINC00460#1 or sh-LINC00460#2 for the knock down of LINC00460, and the knockdown efficiency was confirmed by qRT-PCR (Figure 3A). sh-LINC00460#2 showed a better knockdown efficiency and was chosen for the following experiments, and named as sh-LINC00460. CCK8 (Figure 3B) and colony formation (Figure 3C) assays revealed the suppression ability of LINC00460 knockdown on the cell proliferation of Fadu and SAS cells. Transwell assay indicated the suppression ability of LINC00460 knockdown on cell migration and invasion of Fadu and SAS cells (Figure 3D). In contrary to the effect of LINC00460 over-expression on genes expression, both mRNA (Figure 3E) and protein (Figure 3F) expression of Cyclin D and N-cadherin were decreased by sh-LINC00460, while the expression of p21 and E-cadherin were increased by sh-LINC00460 (Figure 3E and 3F). All these results further suggested that LINC00460 may account for the malignant phenotypes of HNSCC.
Figure 3.
LINC00460 knockdown inhibited HNSCC cell proliferation, migration and invasion. A. Transfection efficiency of sh-LINC00460#1 or sh-LINC00460#2 in Fadu and SAS cell lines was detected by qRT-PCR. **, ***Represents sh-LINC00460#1 or sh-LINC00460#2 vs. sh-NC, P < 0.01, P < 0.001. B. The influence of sh-LINC00460 on cell viability of Fadu and SAS cells was detected by CCK8. *, **Represents sh-LINC00460 vs. sh-NC, P < 0.05, P < 0.01. C. The influence of sh-LINC00460 on cell proliferation of Fadu and SAS cells was detected by colony formation assay. **Represents sh-LINC00460 vs. sh-NC, P < 0.01. D. The influence of sh-LINC00460 on cell migration and invasion of Fadu and SAS cells was detected by transwell assay. **, *** represents sh-LINC00460 vs. sh-NC, P < 0.01, P < 0.001. E. The influence of sh-LINC00460 on mRNA expression of Cyclin D, p21, N-cadherin and E-cadherin in Fadu and SAS cells was detected by qRT-PCR. **, ***Represents sh-LINC00460 vs. sh-NC, P < 0.01, P < 0.001. F. The influence of sh-LINC00460 on protein expression of Cyclin D, p21, N-cadherin and E-cadherin in Fadu and SAS cells was detected by western blot. *, **, ***Represents sh-LINC00460 vs. sh-NC, P < 0.05, P < 0.01, P < 0.001.
LINC00460 directly bound to miR-612 and inhibited its expression
In order to detect the subcellular localization of LINC00460, nuclear/cytoplasmic extract isolation of Fadu and SAS cells were conducted. qRT-PCR analysis suggested that LINC00460 was mainly localized in cytoplasm (Figure 4A). Moreover, the results from fluorescent in situ hybridization (FISH) detected by microscope further indicated the cytoplasm location of LINC00460 (Figure 4B). LINC00460 was predicted to bind with miR-612 via miRDB (http://www.mirdb.org/) (Figure 4C), and dual luciferase reporter assay revealed that miR-612 mimics decreased the luciferase activity of pmirGLO-wt-LINC00460, while did not affect the luciferase activity of pmirGLO-mut-LINC00460 (Figure 4C). RNA immunoprecipitation (RIP) assay further confirmed the direct binding between LINC00460 and miR-612, as shown by an enrichment of LINC00460 and miR-612 with AGO2 antibody in both Fadu and SAS cells (Figure 4D). The expression of miR-612 was decreased by LINC00460 over-expression and increased by LINC00460 knockdown (Figure 4E). Collectively, these results indicated that LINC00460 could directly bind to miR-612 and inhibited its expression. A down-regulation of miR-612 was found in HNSCC tumor tissues (Figure 4F). Bivariate correlation analysis showed that expression of miR-612 was negatively correlated with LINC00460 in HNSCC (Figure 4G).
Figure 4.
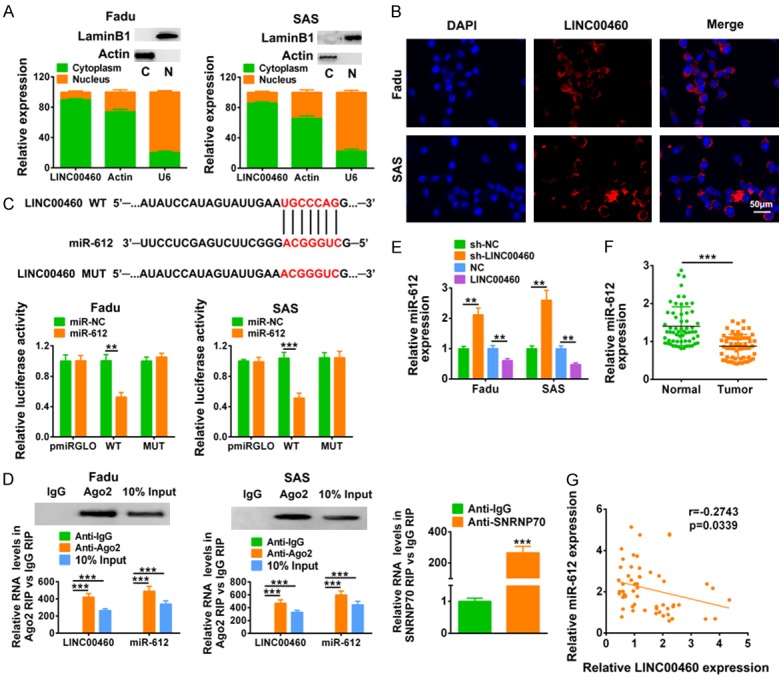
LINC00460 directly bound to miR-612 and inhibited its expression. A. LINC00460, actin and U6 expression levels were detected in nuclear/cytoplasmic extract isolation of Fadu and SAS cells via qRT-PCR. Cytoplasmic marker Actin and envelope marker Lamin B1 were detected to confirm nuclear/cytoplasmic extract isolation. B. Subcellular localization of LINC00460 in Fadu and SAS cells was examined by FISH. C. The predicted binding site of miR-612 in LINC00460, as well as the effect of miR-612 mimics on luciferase activity of reporter gene with wild-type or mutant LINC00460 were detected by luciferase reporter assay. **, ***Represents miR-612 mimics vs. miR NC, P < 0.01, P < 0.001. D. RNA immunoprecipitation (RIP) assay were performed using the AGO2, IgG and SNRNP70 antibody, and qRT-PCR was used to detect the enrichment of miR-612 and LINC00460 in Fadu and SAS cells were determined by Western-blot. ***Represents Anti-Ago2 or input vs. Anti-IgG, P < 0.001. E. The influence of LINC00460 over-expression or sh-LINC00460 on miR-612 expression in Fadu and SAS cells was detected by qRT-PCR. **Represents sh-LINC00460 vs. sh-NC or LINC00460 vs. NC, P < 0.01. F. The expression of miR-612 in HNSCC tissues and adjacent normal tissues was detected by qRT-PCR (N=60). ***Represents tumor vs. adjacent normal tissues, P < 0.001. G. Negative correlation between LINC00460 and miR-612 in HNSCC tissues was analyzed.
AKT2 was a direct target of miR-612
Similarly, AKT2 was predicted as miR-612 binding target via Targetscan, and confirmed by luciferase reporter assay as demonstrated in Figure 5A. miR-612 mimics decreased the luciferase activity of pmirGLO-wt-AKT2, while it had no effect on the luciferase activity of pmirGLO-mut-AKT2. Then Fadu and SAS cells were transfected with miR-612 mimics or inhibitor, and the transfection efficiency was confirmed via qRT-PCR (Figure 5B). The results showed that miR-612 mimics decreased mRNA (Figure 5B) and protein (Figure 5C) expression of AKT2, while miR-612 inhibitor increased mRNA (Figure 5B) and protein (Figure 5C) expression of AKT2. Interestingly, phosphorylation of AKT2 (p-AKT2) was also decreased by miR-612 mimics and increased by miR-612 inhibitor (Figure 5C), suggesting the inactivation of AKT2 signaling pathway by miR-612. Consistent with LINC00460 expression in HNSCC tissues, mRNA expression of AKT2 was also up-regulated in HNSCC tissues (Figure 5D). Bivariate correlation analysis showed that expression of miR-612 was negative correlated with AKT2 in HNSCC (Figure 5E). The up-regulation of AKT2 in HNSCC tissues was also confirmed by western blot analysis (Figure 5F). In summary, these results suggested that AKT2 was a direct target of miR-612.
Figure 5.
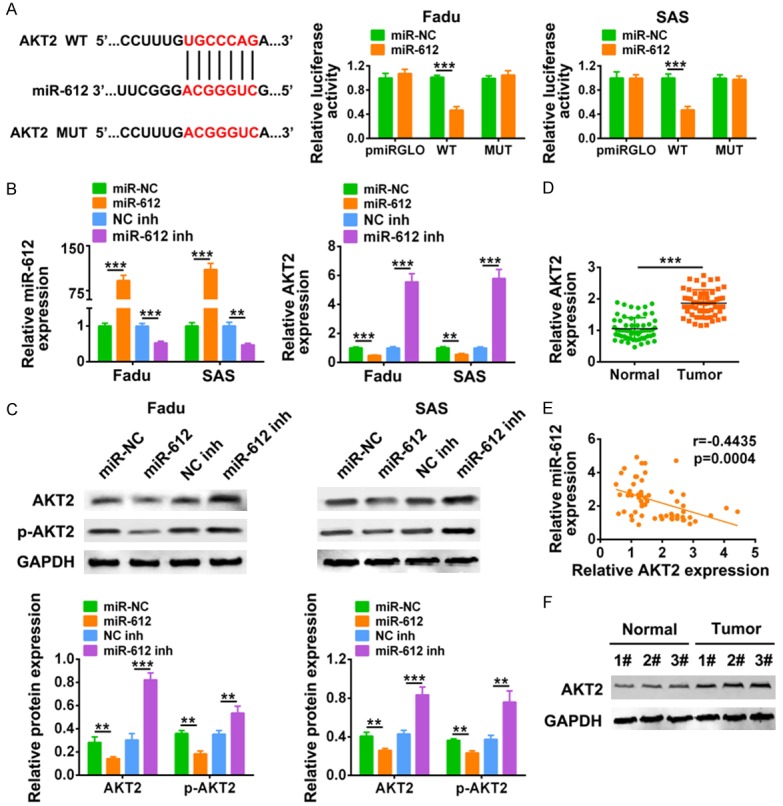
AKT2 was a direct target of miR-612. A. The predicted binding site of miR-612 in AKT2, as well as the effect of miR-612 mimics on luciferase activity of reporter gene with wild-type or mutant AKT2 were detected by luciferase reporter assay. ***Represents miR-612 mimics vs. miR NC, P < 0.001. B. Transfection efficiency of miR-612 mimics or inhibitor, as well as the influence of miR-612 mimics or inhibitor on mRNA expression of AKT2 in Fadu and SAS cells was detected by qRT-PCR. **, ***Represents miR-612 mimics vs. miR NC or miR-612 inh vs. NC inh, P < 0.01, P < 0.001. C. The influence of miR-612 mimics or inhibitor on protein expression of AKT2 and p-AKT2 in Fadu and SAS cells were detected by western blot. **, ***Represents miR-612 mimics vs. miR NC or miR-612 inh vs. NC inh, P < 0.01, P < 0.001. D. The mRNA expression of AKT2 in HNSCC tissues and adjacent normal tissues was detected by qRT-PCR (N = 60). ***Represents tumor vs. adjacent normal tissues, P < 0.001. E. Negative correlation between AKT2 and miR-612 in HNSCC tissues was analyzed. F. Protein expression of AKT2 in HNSCC tissues and adjacent normal tissues was detected by western blot. *Represents tumor vs. adjacent normal tissues, P < 0.05.
LINC00460 induced HNSCC progression via sponging miR-612
To test whether the promotion ability of LINC00460 on HNSCC progression was partially through binding with miR-612, Fadu cells were cotransfected with sh-LINC00406 and miR-612 inhibitor. CCK8 (Figure 6A) and colony formation (Figure 6B) assays showed that the inhibition of cell proliferation by sh-LINC00460 was promoted by additional transfection with miR-612 inhibitor. Repression of miR-612 via miR-612 inhibitor reversed the suppression abilities of sh-LINC00460 on cell migration and invasion of Fadu (Figure 6C). The decreased mRNA (Figure 6D) and protein (Figure 6E) expression of AKT2, Cyclin D and N-cadherin, as well as the increased mRNA (Figure 6D) and protein (Figure 6E) expression of p21 and E-cadherin, were reversed by additional transfection with miR-612 inhibitor. Taken together, these results indicated that LINC00460 functioned as miR-612 sponge to promote cell progression of HNSCC.
Figure 6.
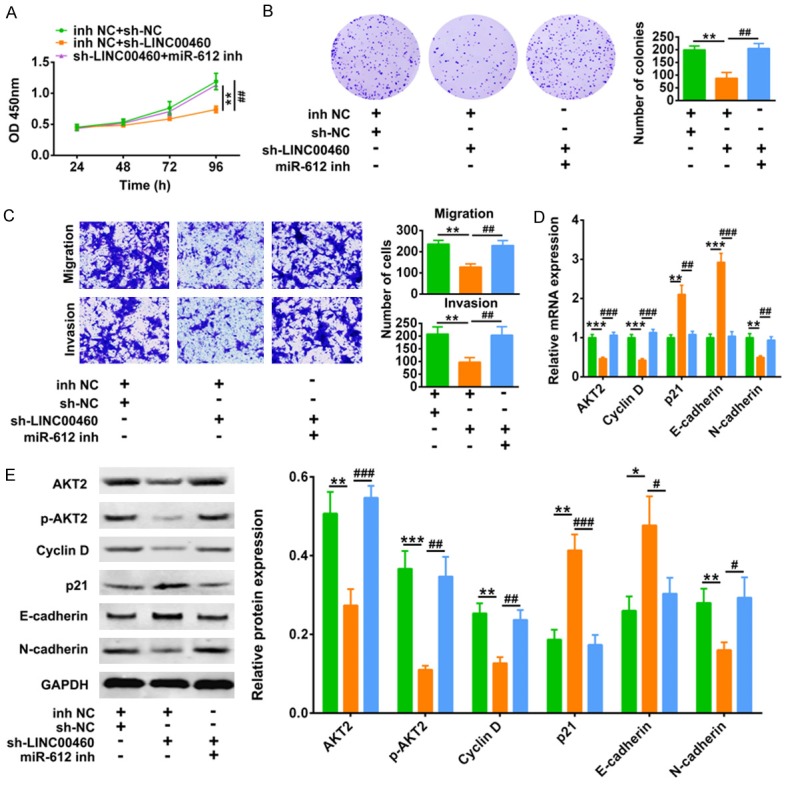
LINC00460 induced HNSCC progression via sponging miR-612. A. The influence of sh-LINC00406 and miR-612 inhibitor on cell viability of Fadu cells was detected by CCK8. **Represents inh NC +sh-NC vs. inh NC+ sh-LINC00406, P < 0.01. ##represents inh NC +sh-NC vs. miR-612 inh+ sh-LINC00406, P < 0.01. B. The influence of sh-LINC00406 and miR-612 inhibitor on cell proliferation of Fadu cells were detected by colony formation assay. **Represents inh NC +sh-NC vs. inh NC+ sh-LINC00406, P < 0.01. ##represents inh NC +sh-NC vs. miR-612 inh+ sh-LINC00406, P < 0.01. C. The influence of sh-LINC00406 and miR-612 inhibitor on cell migration and invasion of Fadu cells were detected by transwell assay. **Represents inh NC +sh-NC vs. inh NC+ sh-LINC00406, P < 0.01. ##Represents inh NC +sh-NC vs. miR-612 inh+ sh-LINC00406, P < 0.01. D. The influence of sh-LINC00406 and miR-612 inhibitor on mRNA expression of AKT2, Cyclin D, p21, E-cadherin and N-cadherin in Fadu cells were detected by qRT-PCR. **, ***Represents inh NC +sh-NC vs. inh NC+ sh-LINC00406, P < 0.01, P < 0.001. ##, ###represents inh NC +sh-NC vs. miR-612 inh+ sh-LINC00406, P < 0.01, P < 0.001. E. The influence of sh-LINC00406 and miR-612 inhibitor on protein expression of AKT2, p-AKT2, Cyclin D, p21, E-cadherin and N-cadherin in Fadu cells were detected by qRT-PCR. *, **, ***Represents inh NC +sh-NC vs. inh NC+ sh-LINC00406, P < 0.05, P < 0.01, P < 0.001. #, ##, ###Represents inh NC +sh-NC vs. miR-612 inh+ sh-LINC00406, P < 0.05, P < 0.01, P < 0.001.
LINC00460 knockdown inhibited in vivo HNSCC tumor growth
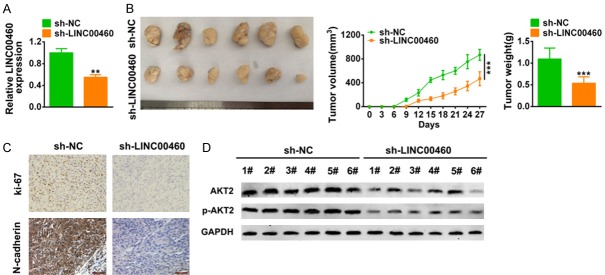
In order to explore the clinical application of LINC00460 on HNSCC, we inoculated Fadu cells transfected with sh-LINC00460 or the scrambled shRNA into nude mice. The downregulation efficiency of LINC00460 was confirmed by qRT-PCR in Figure 7A. Results showed that the intratumoral injection of lentiviral vector with LINC00460 knockdown inhibited tumor growth, wherein the tumor volume and weight were dramatically decreased compared to that of scramble (P < 0.001) (Figure 7B). Moreover, full specimen staining with immunohistochemistry revealed that the xenograft tumor tissues injected with sh-LINC00460 reduced the expression of N-cadherin and Ki67 (Figure 7C). The down-regulation of AKT2 and p-AKT2 (Figure 7D) in tumors via LINC00460 knockdown were confirmed by western blot. These results suggest that LINC00460 knockdown suppressed xenograft tumor growth through the regulation of AKT2.
Figure 7.
LINC00460 knockdown inhibited in vivo HNSCC tumor growth. A. Transfection efficiency of sh-LINC00460 in xenograft tumor mice was detected by qRT-PCR. **represents sh-LINC00460 vs. sh-NC, P < 0.01. B. The effect of sh-LINC00460 on tumor growth in xenograft tumor mice was shown. The tumor volume and weight were calculated. ***Represents sh-LINC00460 vs. sh-NC, P < 0.001. C. Immunohistochemistry staining was used to determine expression of N-cadherin and Ki-67 affected by sh-LINC00460. Black bar: 50 μm. D. The effect of sh-LINC00460 on protein expression of AKT2 and p-AKT2 in xenograft tumor mice was determined by western-blot. ***Represents sh-LINC00460 vs. sh-NC, P < 0.001.
Discussion
LncRNAs function as a promoter or inhibitor of cancer-critical genes of HNSCC, and involved in regulation of diverse cellular processes [21]. Increasing evidence reveals that LINC00460, located at chromosome 13q33.2 [22], plays a vital role in carcinogenesis and progression of various tumors. One of the major findings of the present study is that elevated expression of LINC00460, occurred in HNSCC tissues, was tightly associated with some clinical parameters and predicted a poor prognosis in HNSCC patients. In line with Cao et al [11], elevated expression of LINC00460 was associated with poor OS of HNSCC patients with more frequently occurred in patients with T3+T4 (primary tumor stage), III+IV (clinical stage) and lymph node metastasis, suggesting that LINC00460 might be a poor prognosis indicator for HNSCC. However, due to the small sample size of our current clinical analysis (N = 60), a larger patient cohort is needed to strengthen the clinical significance of LINC00460 in HNSCC patients in the future.
In line with the clinical results, in vitro gain- and loss- of function assays for the first time showed that over-expression of LINC00460 not only promoted cell proliferation, but also induced cell invasion and migration of HNSCC cells, while LINC00460 knockdown reversed the effect on HNSCC progression. Moreover, in vivo subcutaneous xenotransplanted tumor model revealed that interference of LINC00460 suppressed in vivo tumorigenic ability of HNSCC, indicating that LINC00460 is also a potential therapeutic target in HNSCC.
Since the “oncogene” function of LINC00460 on HNSCC has been found, the underlying mechanism remains to be clarified. To regulate cell progression, Cyclin D1, p21, E-cadherin and N-cadherin were found to be functional targets of LINC00460 in HNSCC. Generally, Cyclin D1 promoted G1-to-S phase transition during cell cycle [23], and over-expression of Cyclin D1 was significantly correlated with poor prognosis for HNSCC [24,25]. Down-regulating Cyclin D1 suppressed the malignant phenotype of HNSCC [26]. Moreover, as a cyclin-dependent kinases inhibitors, p21 suppressed cell growth as an antitumor agent [27], and the expression of p21 was significantly correlated with an increased prognosis of oral squamous cell carcinomas [28]. Down-regulation of Cyclin D1 and up-regulation of p21 were associated with inhibition of HNSCC cell proliferation, therefore inhibition of cell progression [29]. The results in the present study showed that LINC00460 increased the expression of Cyclin D1 and decreased the expression of p21, while LINC00460 knockdown decreased the expression of Cyclin D1 and increased the expression of p21. Moreover, in addition to the regulation of cell cycle, Cyclin D and p21 are also involved in regulation of cancer cell migration and invasion. TGF β-mediated Cyclin D1 and p21 results in increased cancer migration and invasion and inhibition of these two cell cycle regulators significantly reduces both tumor formation and local tumor invasion [30]. Therefore, Cyclin D1 and p21 may also be involved in regulation of HNSCC migration and invasion.
Epithelial to mesenchymal transition (EMT), essential for the development of metastasis, contributes to the unfavorable prognosis in cancer [10]. Decrease of E-cadherin accompanied by increase of N-cadherin is associated with positive lymph nodes and metastasis in HNSCC [31]. TNF receptor-associated factor 6 (TRAF6) knockdown elevated E-cadherin and down-regulated N-cadherin to inhibit the migration and invasion abilities of HNSCC [32]. The results in the present study showed that LINC00460 increased expression of N-cadherin and decreased expression of E-cadherin, while LINC00460 knockdown decreased N-cadherin and increased E-cadherin, showing the anti-migration and anti-invasion abilities of LINC00460 knockdown in HNSCC. Moreover, EMT also participates in mediation of cancer stemness. LncRNA PVT1 induced EMT and promoted stemness properties of HNSCC for the metastasis [10]. Considering that LINC00460 is associated with EMT in lung cancer [10] and non-small cell lung cancer [33], the effect of LINC00460 on stemness properties of HNSCC will be explored in the future studies.
As well known, biological function of lncRNAs dependents on the miRNAs and proteins they bind. There are no reported targets for LINC00460 in HNSCC before. The present study revealed a new miRNA target for LINC00460, miR-162, a common “oncogene” which was shown to be involved in the pathogenesis of various tumors. MiR-612/AKT2 axis negatively regulates progression of colorectal cancer [18] and glioblastoma [19]. The results of this study for the first time indicated that LINC00460 could promote HNSCC progression by sponging miR-612 to up-regulate AKT2. Serine/threonine kinase AKT plays an important role in the regulation of cell proliferation and survival, where AKT activation is related to tumor development and poor prognosis [34]. Decreased phosphorylation of AKT showed anti-tumor activity in HNSCC [35]. Moreover, AKT2 acts as a metastasis promoter, with its over-expression increasing the incidence of metastases [36] while its knockdown suppressing cell migration and EMT [37]. In vitro and in vivo loss- of function assays showed that LINC00460 knockdown decreased the expression of AKT2 and p-AKT2, thus suppressing cell progression of HNSCC. However, AKT/NF-κB signaling responsible for inflammatory regulation in tumors is yet to be investigated in the present study.
Conclusion
In summary, our results demonstrated that lncRNA LINC00460, as an “oncogene”, whose knockdown inhibited cell proliferation, migration and invasion of HNSCC cells via sponging miR-612 and targeting AKT2. This finding illuminated the relation between LINC00460/miR-612/AKT2 regulatory axis and HNSCC progression, suggesting potential application of LINC00460 in treatment for the disease.
Acknowledgements
All data generated or analyzed during this study are included in this published article. XXX and GYX conceived and designed the experiments, QLW analyzed and interpreted the results of the experiments, YPG and XYC performed the experiments. All procedures performed in studies involving human participants were in accordance with the standards upheld by the Ethics Committee of Tongde Hospital of Zhejiang Province and with those of the 1964 Helsinki Declaration and its later amendments for ethical research involving human subjects. (XMSC2018146). All animal experiments were approved by the Ethics Committee of Tongde Hospital of Zhejiang Province for the use of animals and conducted in accordance with the National Institutes of Health Laboratory Animal Care and Use Guidelines. (XMSC2019037).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Ling DC, Bakkenist CJ, Ferris RL, Clump DA. Role of immunotherapy in head and neck cancer. Semin Radiat Oncol. 2018;28:12–16. doi: 10.1016/j.semradonc.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Sawicki M, Szudy A, Szczyrek M, Krawczyk P, Klatka J. Molecularly targeted therapies in head and neck cancers. Otolaryngol Pol. 2012;66:307–312. doi: 10.1016/j.otpol.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 6.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han F, Wang C, Wang Y, Zhang L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am J Cancer Res. 2017;7:770–783. [PMC free article] [PubMed] [Google Scholar]
- 8.Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. doi: 10.1016/j.gene.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Liang Y, Wu Y, Chen X, Zhang S, Wang K, Guan X, Yang K, Li J, Bai Y. A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep. 2017;37 doi: 10.1042/BSR20171019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu C, Wang Y, Li G, She L, Zhang D, Chen X, Zhang X, Qin Z, Cao H, Liu Y. LncRNA PVT1 promotes malignant progression in squamous cell carcinoma of the head and neck. J Cancer. 2018;9:3593–3602. doi: 10.7150/jca.26465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji T, Chen WT, Zou X. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101. doi: 10.1016/j.oraloncology.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Masood Y, Kqueen CY, Rajadurai P. Role of miRNA in head and neck squamous cell carcinoma. Expert Rev Anticancer Ther. 2015;15:183–197. doi: 10.1586/14737140.2015.978294. [DOI] [PubMed] [Google Scholar]
- 13.Bossi L, Figueroa-Bossi N. Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria. Nat Rev Microbiol. 2016;14:775–784. doi: 10.1038/nrmicro.2016.129. [DOI] [PubMed] [Google Scholar]
- 14.Tang J, Tao ZH, Wen D, Wan JL, Liu DL, Zhang S, Cui JF, Sun HC, Wang L, Zhou J, Fan J, Wu WZ. MiR-612 suppresses the stemness of liver cancer via Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2014;447:210–215. doi: 10.1016/j.bbrc.2014.03.135. [DOI] [PubMed] [Google Scholar]
- 15.Tao ZH, Wan JL, Zeng LY, Xie L, Sun HC, Qin LX, Wang L, Zhou J, Ren ZG, Li YX, Fan J, Wu WZ. miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. 2013;210:789–803. doi: 10.1084/jem.20120153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, Zhang HL, Wang QY, Chen MJ, Liu LB. Overexpression of microRNA-612 restrains the growth, invasion, and tumorigenesis of melanoma cells by targeting Espin. Mol Cells. 2018;41:119–126. doi: 10.14348/molcells.2018.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou P, Dong H, He S, Fang L, Jiang N, Sun Q. miR612 is associated with esophageal squamous cell carcinoma development and metastasis, mediated through TP53. Mol Med Rep. 2017;16:1855–1863. doi: 10.3892/mmr.2017.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheng L, He P, Yang X, Zhou M, Feng Q. miR-612 negatively regulates colorectal cancer growth and metastasis by targeting AKT2. Cell Death Dis. 2015;6:e1808. doi: 10.1038/cddis.2015.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai J, Zhang J, Wu P, Yang W, Ye Q, Chen Q, Jiang C. Blocking LINC00152 suppresses glioblastoma malignancy by impairing mesenchymal phenotype through the miR-612/AKT2/NF-kappaB pathway. J Neurooncol. 2018;140:225–236. doi: 10.1007/s11060-018-2951-0. [DOI] [PubMed] [Google Scholar]
- 20.Pedrero JM, Carracedo DG, Pinto CM, Zapatero AH, Rodrigo JP, Nieto CS, Gonzalez MV. Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int J Cancer. 2005;114:242–248. doi: 10.1002/ijc.20711. [DOI] [PubMed] [Google Scholar]
- 21.Luo X, Qiu Y, Jiang Y, Chen F, Jiang L, Zhou Y, Dan H, Zeng X, Lei YL, Chen Q. Long non-coding RNA implicated in the invasion and metastasis of head and neck cancer: possible function and mechanisms. Mol Cancer. 2018;17:14. doi: 10.1186/s12943-018-0763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen L, Zhang X, Bian J, Han L, Huang H, He M, Wei M, Wang P. The long non-coding RNA LINC00460 predicts the prognosis and promotes the proliferation and migration of cells in bladder urothelial carcinoma. Oncol Lett. 2019;17:3874–3880. doi: 10.3892/ol.2019.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.E Hermosilla V, Salgado G, Riffo E, Escobar D, Hepp MI, Farkas C, Galindo M, Morin V, Garcia-Robles MA, Castro AF, Pincheira R. SALL2 represses cyclins D1 and E1 expression and restrains G1/S cell cycle transition and cancer-related phenotypes. Mol Oncol. 2018;12:1026–1046. doi: 10.1002/1878-0261.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gioacchini FM, Alicandri-Ciufelli M, Kaleci S, Magliulo G, Presutti L, Re M. The prognostic value of cyclin D1 expression in head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2016;273:801–809. doi: 10.1007/s00405-014-3426-3. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Pavelic ZP, Li YQ, Wang L, Gleich L, Radack K, Gluckman JL, Stambrook PJ. Amplification and overexpression of the cyclin D1 gene in head and neck squamous cell carcinoma. Clin Mol Pathol. 1995;48:M256–259. doi: 10.1136/mp.48.5.m256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanken H, Grobe A, Cachovan G, Smeets R, Simon R, Sauter G, Heiland M, Blessmann M. CCND1 amplification and cyclin D1 immunohistochemical expression in head and neck squamous cell carcinomas. Clin Oral Investig. 2014;18:269–276. doi: 10.1007/s00784-013-0967-6. [DOI] [PubMed] [Google Scholar]
- 27.Zhao YF, Wang CR, Wu YM, Ma SL, Ji Y, Lu YJ. P21 (waf1/cip1) is required for non-small cell lung cancer sensitive to Gefitinib treatment. Biomed Pharmacother. 2011;65:151–156. doi: 10.1016/j.biopha.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Fillies T, Woltering M, Brandt B, Van Diest JP, Werkmeister R, Joos U, Buerger H. Cell cycle regulating proteins p21 and p27 in prognosis of oral squamous cell carcinomas. Oncol Rep. 2007;17:355–359. [PubMed] [Google Scholar]
- 29.Barzegar M, Ma S, Zhang C, Chen X, Gu Y, Shang C, Jiang X, Yang J, Nathan CA, Yang S, Huang S. SKLB188 inhibits the growth of head and neck squamous cell carcinoma by suppressing EGFR signalling. Br J Cancer. 2017;117:1154–1163. doi: 10.1038/bjc.2017.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai M, Al-Odaini AA, Fils-Aime N, Villatoro MA, Guo J, Arakelian A, Rabbani SA, Ali S, Lebrun JJ. Cyclin D1 cooperates with p21 to regulate TGFbeta-mediated breast cancer cell migration and tumor local invasion. Breast Cancer Res. 2013;15:R49. doi: 10.1186/bcr3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pectasides E, Rampias T, Sasaki C, Perisanidis C, Kouloulias V, Burtness B, Zaramboukas T, Rimm D, Fountzilas G, Psyrri A. Markers of epithelial to mesenchymal transition in association with survival in head and neck squamous cell carcinoma (HNSCC) PLoS One. 2014;9:e94273. doi: 10.1371/journal.pone.0094273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Li YC, Wu L, Yu GT, Zhang WF, Huang CF, Sun ZJ. TRAF6 regulates tumour metastasis through EMT and CSC phenotypes in head and neck squamous cell carcinoma. J Cell Mol Med. 2018;22:1337–1349. doi: 10.1111/jcmm.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue QY, Zhang Y. Effects of Linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22:1003–1010. doi: 10.26355/eurrev_201802_14382. [DOI] [PubMed] [Google Scholar]
- 34.Nicholson KM, Anderson NG. The protein kinase B/Akt signalling pathway in human malignancy. Cell Signal. 2002;14:381–395. doi: 10.1016/s0898-6568(01)00271-6. [DOI] [PubMed] [Google Scholar]
- 35.Fritsch C, Huang A, Chatenay-Rivauday C, Schnell C, Reddy A, Liu M, Kauffmann A, Guthy D, Erdmann D, De Pover A, Furet P, Gao H, Ferretti S, Wang Y, Trappe J, Brachmann SM, Maira SM, Wilson C, Boehm M, Garcia-Echeverria C, Chene P, Wiesmann M, Cozens R, Lehar J, Schlegel R, Caravatti G, Hofmann F, Sellers WR. Characterization of the novel and specific PI3Kalpha inhibitor NVP-BYL719 and development of the patient stratification strategy for clinical trials. Mol Cancer Ther. 2014;13:1117–1129. doi: 10.1158/1535-7163.MCT-13-0865. [DOI] [PubMed] [Google Scholar]
- 36.Dillon RL, Marcotte R, Hennessy BT, Woodgett JR, Mills GB, Muller WJ. Akt1 and akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res. 2009;69:5057–5064. doi: 10.1158/0008-5472.CAN-08-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dillon RL, Muller WJ. Distinct biological roles for the akt family in mammary tumor progression. Cancer Res. 2010;70:4260–4264. doi: 10.1158/0008-5472.CAN-10-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]