Abstract
Iron (Fe) is an essential micronutrient that is frequently inaccessible to plants. Rice (Oryza sativa L.) plants employ the Combined Strategy for Fe uptake, which is composed by all features of Strategy II, common to all Poaceae species, and some features of Strategy I, common to non-Poaceae species. To understand the evolution of Fe uptake mechanisms, we analyzed the root transcriptomic response to Fe deficiency in O. sativa and its wild progenitor O. rufipogon. We identified 622 and 2,017 differentially expressed genes in O. sativa and O. rufipogon, respectively. Among the genes up-regulated in both species, we found Fe transporters associated with Strategy I, such as IRT1, IRT2 and NRAMP1; and genes associated with Strategy II, such as YSL15 and IRO2. In order to evaluate the conservation of these Strategies among other Poaceae, we identified the orthologs of these genes in nine species from the Oryza genus, maize and sorghum, and evaluated their expression profile in response to low Fe condition. Our results indicate that the Combined Strategy is not specific to O. sativa as previously proposed, but also present in species of the Oryza genus closely related to domesticated rice, and originated around the same time the AA genome lineage within Oryza diversified. Therefore, adaptation to Fe2+ acquisition via IRT1 in flooded soils precedes O. sativa domestication.
Subject terms: Plant domestication, Plant stress responses, Plant molecular biology
Introduction
Iron (Fe) is an essential micronutrient for virtually all organisms. In humans, anemia prevalence was one third of world population in 2010, and Fe deficiency anemia (IDA) is correlated with decreased cognitive performance, low weight at birth, and child and maternal mortality1. In plants, the capacity to change redox states from Fe2+ (ferrous) to Fe3+ (ferric) allows Fe to participate in electron transfer reactions in both photosynthesis, respiration and chlorophyll biosynthesis2,3. Higher plants cultivated under Fe deficiency experience severe chlorosis, reduction on biomass, yield and nutritional value of their grains4. In calcareous soils, which cover approximately one third of the Earth’s surface4,5, Fe is less bioavailable to plants, which can lead to Fe deficiency. In contrast, Fe can become harmful when present at elevated concentrations in plant tissues, since it can react with oxygen and catalyze the formation of reactive oxygen species through the Fenton reaction6. Thus, plants must tightly regulate internal Fe concentration to avoid both Fe toxicity and Fe deficiency2.
To cope with Fe deficiency, plants possess two different ways to maintain adequate levels of Fe7. Strategy I, or reduction strategy, found in all plants except those from the Poaceae family, consists in: (1) lowering soil pH by extrusion of H+ to increase Fe3+ solubility, which relies on P-type ATPases such as AtAHA2 in Arabidopsis8; (2) reduction of Fe3+ to Fe2+ at the root surface by a plasma membrane-bound ferric-chelate reductase, named AtFRO2 (Ferric Reductase Oxidase)9; and uptake of Fe2+ into root cells by the Fe high-affinity transporter AtIRT1 (Iron-Regulated Transporter)10. All proteins involved in this strategy increase their expression level under Fe deficiency. On the other hand, plants from the Poaceae family rely on Strategy II, or chelation strategy, to absorb Fe. This strategy employs the release of phytosiderophores into the rhizosphere. Phytosiderophores are derivatives of mugineic acid (MAs) family of modified amino acids11. MAs synthesis involves the trimerization of S-adenosyl Met to form nicotianamine (NA), catalyzed by nicotianamine synthase (NAS)12; the conversion of NA in 3-keto intermediate by the transfer of an amino group by nicotianamine aminotransferase (NAAT)13, and DMA synthesis by deoxymugineic acid synthase (DMAS)13. The first MA synthesized in the pathway is DMA, but different grass species may secrete other forms of MAs. Phytosiderophore secretion is performed by OsZIFL4/TOM1 (transporter of mugineic acid family phytosiderophores 1) in rice14,15. In the rhizosphere, phytosiderophores chelate Fe3+ and the formed complex Fe(III)-MA is transported into root cells through specific transmembrane proteins of the Yellow Stripe (YS) family, such as Yellow Stripe 1 (YS1) in maize16 and its ortholog Yellow Stripe-like 15 (OsYSL15) in rice17.
In spite of being a grass and relying on Strategy II, O. sativa was shown to induce the Fe2+ transporters OsIRT1 and OsIRT2 under Fe deficiency18,19. It was proposed that O. sativa uses a Combined Strategy (CS) to absorb Fe from the rhizosphere, using all features of Strategy II plants and partial features of Strategy I, such as IRT-type transporters20. The capability to absorb Fe2+ would have evolved in O. sativa as an adaptation to flooded paddies, since cultivated rice is well adapted for growth under submerged conditions, in which Fe2+ is frequently more abundant than Fe3+, unlike most graminaceous crops11,18,21. Thus, O. sativa is the only plant described as using the CS for Fe uptake to date22.
The Oryza genus is composed by two domesticated species, O. sativa and O. glaberrima, and 25 wild species which diverged from their wild progenitors 9,000 to 8,000 years ago23. The Oryza genus comprehends 11 genome types, 6 diploid (n = 12: AA, BB, CC, EE, FF, and GG) and 5 polyploid (n = 24: BBCC, CCDD, HHJJ, HHKK, and KKLL)24, and a genome variation size of 3.6 times25. O. rufipogon, an Asian wild grass, is the species most closely related to O. sativa23.
In the present study, a transcriptomic analysis was performed to compare the regulation of genes involved in Fe deficiency response in O. sativa and its wild progenitor O. rufipogon. We also analyzed the expression of the orthologs of OsYSL15, OsIRT1, OsNRAMP1 and OsIRO2 in other wild Oryza species, as well as in sorghum (Sorghum bicolor) and maize (Zea mays). Our results indicate that the CS observed in rice, based on OsYL15 and OsIRT1 as Fe3+ -phytosiderophore and Fe2+ transporters, is not an evolutionary novelty restricted to O. sativa, but has an origin that precedes the split of most AA genome Oryza species. This suggests a common origin for the CS in these species, and indicates that adaptation to Fe2+ acquisition in flooded soils precedes rice domestication.
Results
Plants of O. sativa and O. rufipogon show similar Fe deficiency physiological responses
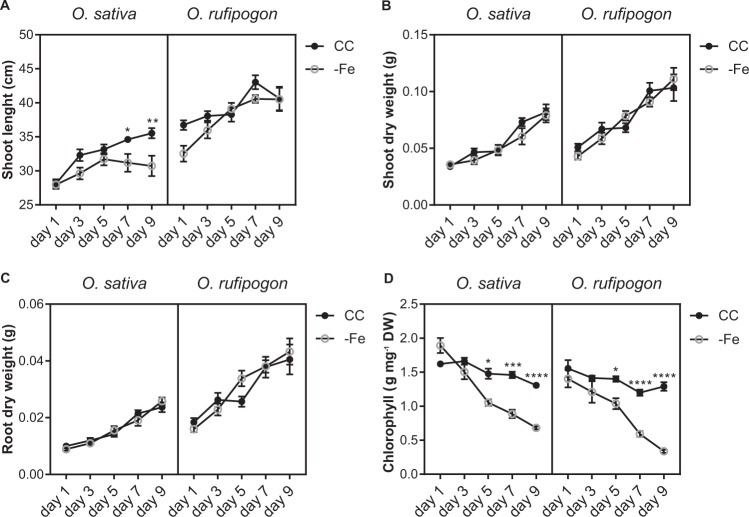
Plants of O. sativa and O. rufipogon were cultivated under control condition (CC, containing 100 μM Fe+3-EDTA) and Fe deficiency (−Fe) in non-aerated hydroponics for nine days. Overall, both species responded similarly to low Fe conditions. Shoot length decreased significantly in O. sativa plants compared with plants in CC after seven days of −Fe, while no change was observed in O. rufipogon plants (Fig. 1A). Shoot and root dry weight showed no significant differences in both species when comparing plants under CC or −Fe conditions (Fig. 1B,C). Chlorophyll concentration decreased clearly in both species after five days of −Fe treatment (Fig. 1D). These data indicate that both O. sativa and O. rufipogon are at the early stage of the Fe deficiency response at five days of treatment.
Figure 1.
Phenotypic analysis of Oryza sativa and Oryza rufipogon plants grown under control (CC) and Fe deficiency (−Fe) conditions. Four week-old plants were grown with or without Fe for up to 9 days. (A) Shoot length (cm) (n = 10). (B) Shoot dry weight (g) (n = 10). (C) Root dry weight (g) (n = 10). (D) Chlorophyll concentration (µg mg−1 DW) (n = 4, 3 plants each). The x-axis represents days after onset of −Fe treatment. Values are the averages ± SE. Asterisks indicate statistical difference between plants grown under CC and −Fe conditions (Student t-test, *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001, ****P-value < 0.0001). DW = dry weight.
O. sativa and O. rufipogon transcriptomic changes under Fe deficiency
In order to compare the Fe deficiency gene expression responses in O. sativa and O. rufipogon, transcriptomic analyses of roots from plants submitted to CC and −Fe treatments were performed. Deep sequencing generated 33,253,905 reads from O. sativa CC libraries, 36,581,515 from O. sativa −Fe libraries; 34,973,923 from O. rufipogon CC libraries, and 36,365,135 from O. rufipogon −Fe libraries. Transcripts were considered differentially expressed when FDR < 0.05 (Supplementary Fig. 1). Comparing −Fe to CC, O. sativa showed a total of 622 differentially expressed genes (340 up- and 282 down-regulated), which represent 1.77% of all annotated genes in the genome of this species (Supplementary Table 1). O. rufipogon showed a total of 2,017 differentially expressed genes (1,433 up- and 584 down-regulated), which represent 5.43% of all annotated genes in the genome of this species (Supplementary Table 2).
Functional annotation of differentially expressed transcripts associated with Fe deficiency response in O. sativa and O. rufipogon shows similar processes
Aiming to compare the transcriptomic changes in roots under −Fe from O. sativa and O. rufipogon, we performed Gene Ontology (GO) categories enrichment analysis. From a total of 622 and 2,017 differentially expressed genes in O. sativa and O. rufipogon, respectively, 485 and 1,516 genes could be assigned a GO term and were considered in the enrichment analysis. We found 216 terms enriched in the set of differentially expressed genes of O. sativa, with 120 terms up- and 96 terms down-regulated by −Fe, and 280 terms enriched in the set of differentially expressed genes of O. rufipogon, with 138 terms up- and 142 terms down-regulated. When comparing O. sativa and O. rufipogon, we found processes regulated in both: 26 processes were up- and 29 were down-regulated in the two Oryza species. Among the up-regulated processes in both species (Supplementary Fig. 2A), we found “iron ion transport”, “zinc II ion transport”, “organic acid biosynthetic process”, processes related to “amino acid salvage” including “L-methionine salvage” and “L-methionine salvage from methylthioadenosine” as well as several amino acid biosynthetic processes, mainly related to “methionine biosynthetic process”, “S-adenosylmethionine biosynthetic process” and “L-methionine biosynthetic process”. Among the commonly down-regulated processes (Supplementary Fig. 2B), we found terms associated to “cellular response to nutrient levels”, “cellular responses to starvation”, “responses to extracellular stimulus”. Four similar categories, including metabolic and biosynthetic processes related to glycolipid, glycosilceramide, glucosylceramide and glycosphingolipid; and three categories and processes related to sulfur transport, such as “sulfate transport”, “sulfur compound transport” and “sulfate transmembrane transport” were also identified.
GO terms exclusively up-regulated in O. sativa include “formate metabolic process”, “ureide catabolic process”, “ureide metabolic process”, “divalent metal ion transport” and “divalent inorganic cation transport”, whereas down-regulated terms exclusively in O. sativa, which include “chemical homeostasis”, “response to toxic substance”, “cellular oxidant detoxification” and “response to oxidative stress”, showed high level of significance (Supplementary Fig. 3A). O. rufipogon up-regulated GO terms include “nitrogen cycle metabolic process”, “nitrate assimilation”, “ammonium transport”, and “response to oxidative stress”, and down-regulated terms include “cellular nitrogen compound catabolic process”, “divalent metal ion transport” and “cellular response to phosphate starvation” (Supplementary Fig. 3B).
The Fe deficiency regulon genes are responsive in both O. sativa and O. rufipogon
Although the number of differentially expressed genes was different in roots of O. sativa and O. rufipogon under −Fe, we found orthologous genes regulated in both species. Fifty-seven genes up-regulated by −Fe are orthologous between the two species, whereas 45 orthologous genes were down-regulated in both species after −Fe treatment (Table 1). Interestingly, a number of genes encoding proteins involved in Strategy I for Fe2+ uptake from the rhizosphere were up-regulated under −Fe in both species, such as IRT1, IRT2 and NRAMP1. We also found genes related to Strategy II for Fe uptake which were up-regulated in both species, including enzymes involved in biosynthesis and secretion of mugineic acids (MAs), such as S-adenosylmethionine synthase 2 (SAM2), deoxymugineic acid synthase (DMAS) and nicotianamine synthase 1 (NAS1), and genes involved in the biosynthesis of methionine, as MTK1 (Methylthioribose kinase 1), a precursor for the MAs biosynthetic pathway13. Known regulators of Strategy II, such as the iron-regulated bHLH transcription factor IRO2, which plays a role in transcriptional regulation of genes that participate in Fe acquisition26; IRO3, which plays an important role as a negative regulator of the Fe deficiency response in O. sativa; and HRZ1 and HRZ2, which are iron-binding sensors that negatively regulate iron acquisition under conditions of Fe sufficiency27, were also up-regulated in roots of both species. Finally, the genes encoding the main Fe(III)-deoxymugineic acid transporter in roots (YSL15, which is essential for Fe uptake17), were also up-regulated in both O. sativa and O. rufipogon. These results strongly indicate that these two species utilize a common mechanism, the Combined Strategy, for Fe uptake.
Table 1.
List of differentially expressed genes identified by RNAseq in response to Fe-deficiency in roots of Oryza sativa and Oryza rufipogon.
| O. sativa | O. rufipogon | Gene name | Regulation | Description |
|---|---|---|---|---|
| Phytosiderophore biosynthesis | ||||
| OS01G0323600/LOC_Os01g22010 | ORUFI01G14960 | SAM2 | up | S-adenosylmethionine synthase [Source:UniProtKB/TrEMBL;Acc:A0A0R7VIP9] |
| OS02G0306401/LOC_Os02g20360 | ORUFI02G13810 | NAAT1 | up | Os02g0306401 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0VI36] |
| OS03G0237100/LOC_Os03g13390 | ORUFI03G09980 | DMAS | up | NADH-dependent oxidoreductase 1, putative, expressed [Source:UniProtKB/TrEMBL;Acc:Q10PE7] |
| OS03G0307300/LOC_Os03g19427 | ORUFI03G15260 | NAS1 | up | Nicotianamine synthase 1 [Source:UniProtKB/TrEMBL;Acc:H9BE58] |
| OS04G0669800/LOC_Os04g57400 | ORUFI04G30700 | MTK1 | up | Methylthioribose kinase 1 [Source:UniProtKB/Swiss-Prot;Acc:Q7XR61] |
| OS06G0112200/LOC_Os06g02220 | ORUFI06G00790 | up | Methylthioadenosine/S-adenosyl homocysteine nucleosidase [Source:UniProtKB/TrEMBL;Acc:Q9LHZ0] | |
| Iron homeostasis | ||||
| OS01G0689451/LOC_Os01g49470 | ORUFI01G30140 | HRZ1 | up | Hemerythrin motif-containing really interesting new gene (RING)-and zinc-finger protein 1 [Source:UniProtKB/TrEMBL;Acc:V9G2Z0] |
| OS05G0551000/LOC_Os05g47780 | ORUFI05G26690 | HRZ2 | up | Os05g0551000 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0WPZ3] |
| OS03G0751100/LOC_Os03g54000 | ORUFI03G35800 | OPT7 | up | Oligopeptide transporter 3, putative, expressed [Source:UniProtKB/TrEMBL;Acc:Q75LM0] |
| OS11G0151500/LOC_Os11g05390 | ORUFI11G03210 | ENA1 | up | Os11g0151500 protein [Source:UniProtKB/TrEMBL;Acc:Q0IUK3] |
| OS12G0568500/LOC_Os12g38051 | ORUFI12G18020 | up | Metallothionein-like protein 1, putative, expressed [Source:UniProtKB/TrEMBL;Acc:Q2QNE5] | |
| OS12G0570700/LOC_Os12g38270 | ORUFI12G18180 | MT4A | up | Metallothionein-like protein [Source:UniProtKB/TrEMBL;Acc:A1L4T7] |
| Iron transport | ||||
| OS02G0650300/LOC_Os02g43410 | ORUFI02G27480 | YSL15 | up | Iron-phytosiderophore transporter YSL15 [Source:UniProtKB/Swiss-Prot;Acc:Q6H3Z3] |
| OS03G0667300/LOC_Os03g46454 | ORUFI03G29890 | IRT2 | up | Fe(2+) transport protein 2 [Source:UniProtKB/Swiss-Prot;Acc:Q6L8G1] |
| OS03G0667500/LOC_Os03g46470 | ORUFI03G29910 | IRT1 | up | Fe(2+) transport protein 1 [Source:UniProtKB/Swiss-Prot;Acc:Q75HB1] |
| OS04G0542200/LOC_Os04g45860 | ORUFI04G21940 | YSL9 | up | Os04g0542200 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0WDA4] |
| OS07G0258400/LOC_Os07g15460 | ORUFI07G09420 | NRAMP1 | up | Metal transporter Nramp1 [Source:UniProtKB/Swiss-Prot;Acc:Q0D7E4] |
| OS09G0396900/LOC_Os09g23300 | ORUFI09G10010 | VIT2 | down | Os09g0396900 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0XMS4] |
| Transcription factors | ||||
| OS01G0952800/LOC_Os01g72370 | ORUFI01G47560 | IRO2 | up | Os01g0952800 protein [Source:UniProtKB/TrEMBL;Acc:Q0JFZ0] |
| OS03G0379300/LOC_Os03g26210 | ORUFI03G20000 | IRO3 | up | Helix-loop-helix DNA-binding domain containing protein, expressed [Source:UniProtKB/TrEMBL;Acc:Q10KL8] |
| OS07G0573300/LOC_Os07g38580 | ORUFI07G20280 | zinc finger family protein | down | Os07g0573300 protein [Source:UniProtKB/TrEMBL;Acc:Q6ZL20] |
| Other transporters | ||||
| OS08G0207500/LOC_Os08g10630 | ORUFI08G06860 | ZIP4 | up | Zinc transporter 4 [Source:UniProtKB/Swiss-Prot;Acc:Q6ZJ91] |
| OS08G0369000/LOC_Os08g28170 | ORUFI08G14240 | nucleobase-ascorbate transporter | up | Os08g0369000 protein [Source:UniProtKB/TrEMBL;Acc:Q0J648] |
| OS09G0440700/LOC_Os09g26900 | ORUFI09G12590 | COPT5.1 | up | Copper transporter 5.1 [Source:UniProtKB/Swiss-Prot;Acc:Q69P80] |
| OS11G0235200/LOC_Os11g12740 | ORUFI11G07890 | up | Nitrate transporter NTL1, putative, expressed [Source:UniProtKB/TrEMBL;Acc:Q53JI5] | |
| OS06G0554800/LOC_Os06g36090 | ORUFI06G19420 | ABC-2 type transporter | down | |
| OS08G0156600/LOC_Os08g06010 | ORUFI08G03760 | transporter, major facilitator family | down | Os08g0156600 protein [Source:UniProtKB/TrEMBL;Acc:Q0J7X7] |
| OS12G0581600/LOC_Os12g39180 | ORUFI12G18860 | NRAMP7 | down | Metal transporter Nramp6 [Source:UniProtKB/Swiss-Prot;Acc:Q2QN30] |
| Other genes | ||||
| OS01G0495701 | ORUFI01G40710 | up | Os01g0495701 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0V2W9] | |
| OS01G0605100/LOC_Os01g42030 | ORUFI01G24790 | BCS1 | up | BCS1 protein-like [Source:UniProtKB/TrEMBL;Acc:Q5ZDA1] |
| OS01G0608101 | ORUFI01G25040 | up | Os01g0608101 protein [Source:UniProtKB/TrEMBL;Acc:C7IWA4] | |
| OS01G0655500/LOC_Os01g46720 | ORUFI01G27970 | up | Probable plastid-lipid-associated protein 14, chloroplastic [Source:Projected from Arabidopsis thaliana (AT5G53450) UniProtKB/Swiss-Prot;Acc:Q9LV04] | |
| OS01G0775400/LOC_Os01g56810 | ORUFI01G36000 | CKX5 | up | Cytokinin dehydrogenase 5 [Source:UniProtKB/Swiss-Prot;Acc:Q5ZAY9] |
| OS01G0878700/LOC_Os01g65670 | ORUFI01G42590 | up | Os01g0878700 Amino acid transporter, transmembrane domain containing protein [Source:UniProtKB/TrEMBL;Acc:Q5N9H2] | |
| OS01G0952900/LOC_Os01g72360 | ORUFI01G47550 | up | Os01g0952900 protein [Source:UniProtKB/TrEMBL;Acc:Q0JFZ1] | |
| OS02G0509500/LOC_Os02g30600 | ORUFI02G18770 | up | Os02g0509500 protein [Source:UniProtKB/TrEMBL;Acc:Q0E0Z7] | |
| OS02G0579800/LOC_Os02g36940 | ORUFI02G22960 | up | Os02g0579800 protein [Source:UniProtKB/TrEMBL;Acc:Q6EP48] | |
| OS02G0714600/LOC_Os02g48390 | ORUFI02G31750 | up | Ribose-phosphate pyrophosphokinase 4 [Source:UniProtKB/Swiss-Prot;Acc:Q6ZFT5] | |
| OS02G0731900/LOC_Os02g49920 | ORUFI02G33040 | up | 3-ketoacyl-CoA synthase [Source:UniProtKB/TrEMBL;Acc:A0A0N7KG16] | |
| OS02G0744700/LOC_Os02g51070 | ORUFI02G33990 | SSII-2 | up | Starch synthase, chloroplastic/amyloplastic [Source:UniProtKB/TrEMBL;Acc:Q0DXM0] |
| OS02G0791300/LOC_Os02g54870 | ORUFI02G36990 | up | Os02g0791300 protein [Source:UniProtKB/TrEMBL;Acc:Q6KAE4] | |
| OS03G0161800/LOC_Os03g06620 | ORUFI03G04320 | ARD2 | up | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Source:UniProtKB/Swiss-Prot;Acc:Q10RE5] |
| OS03G0327600/LOC_Os03g21040 | ORUFI03G16490 | R40C1 | up | Ricin B-like lectin R40C1 [Source:UniProtKB/Swiss-Prot;Acc:Q10M12] |
| OS03G0401100/LOC_Os03g28300 | ORUFI03G21290 | up | Os03g0401100 protein [Source:UniProtKB/TrEMBL;Acc:Q10K10] | |
| OS03G0431800/LOC_Os03g31730 | ORUFI03G23070 | up | Os03g0431800 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0VZP7] | |
| OS03G0736900/LOC_Os03g52680 | ORUFI03G34750 | up | Os03g0736900 protein [Source:UniProtKB/TrEMBL;Acc:Q0DNS7] | |
| OS01G0776600/LOC_Os01g56880 | ORUFI01G36050 | down | Purple acid phosphatase 10 [Source:Projected from Arabidopsis thaliana (AT2G16430) UniProtKB/Swiss-Prot;Acc:Q9SIV9] | |
| OS01G0801600/LOC_Os01g58740 | ORUFI01G37440 | down | Probable glycerol-3-phosphate dehydrogenase [NAD(+)] 2, cytosolic [Source:UniProtKB/Swiss-Prot;Acc:Q8S2G5] | |
| OS01G0814400/LOC_Os01g59900 | ORUFI01G38230 | down | lisH domain-containing protein C1711.05 [Source:UniProtKB/TrEMBL;Acc:A0A0P0V9N0] | |
| OS01G0906000/LOC_Os01g67870 | ORUFI01G44340 | down | Os01g0906000 protein [Source:UniProtKB/TrEMBL;Acc:Q0JGT3] | |
| OS01G0908200/LOC_Os01g68020 | ORUFI01G44480 | BTBZ2 - Bric-a-Brac, Tramtrack, and Broad Complex BTB domain with TAZ zinc finger and Calmodulin-binding domains | down | Os01g0908200 protein [Source:UniProtKB/TrEMBL;Acc:Q8L3R7] |
| OS01G0941800/LOC_Os01g71420 | ORUFI01G47010 | down | Probable inactive purple acid phosphatase 16 [Source:Projected from Arabidopsis thaliana (AT3G10150) UniProtKB/Swiss-Prot;Acc:Q9SR79] | |
| OS02G0202200/LOC_Os02g10780 | ORUFI03G26300 | SPX2 | down | SPX domain-containing protein 2 [Source:UniProtKB/Swiss-Prot;Acc:Q6Z784] |
| OS02G0327000/LOC_Os02g22130 | ORUFI02G15000 | GAP1 | down | GTPase activating protein 1 [Source:UniProtKB/Swiss-Prot;Acc:Q6YWF1] |
| OS02G0514500/LOC_Os02g31030 | ORUFI02G19040 | glycerophosphoryl diester phosphodiesterase family protein | down | Os02g0514500 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0VJM6] |
| OS02G0542400/LOC_Os02g33770 | ORUFI02G20760 | homeodomain | down | Os02g0542400 protein [Source:UniProtKB/TrEMBL;Acc:Q6ESZ1] |
| OS04G0675000/LOC_Os04g57870 | ORUFI04G31160 | up | Os04g0675000 protein [Source:UniProtKB/TrEMBL;Acc:B9FDA0] | |
| OS05G0482400/LOC_Os05g40384 | ORUFI05G21860 | CYP714D1 | up | Cytochrome P450 714D1 [Source:UniProtKB/Swiss-Prot;Acc:Q5KQH7] |
| OS06G0114000/LOC_Os06g02380 | ORUFI06G00900 | up | Os06g0114000 protein [Source:UniProtKB/TrEMBL;Acc:Q9LWT6] | |
| OS06G0486800/LOC_Os06g29180 | ORUFI06G16260 | up | Formate dehydrogenase, mitochondrial [Source:UniProtKB/TrEMBL;Acc:Q0DC43] | |
| OS06G0549600/LOC_Os06g35650 | ORUFI06G19050 | up | Os06g0549600 protein [Source:UniProtKB/TrEMBL;Acc:Q5Z957] | |
| OS06G0628032/LOC_Os06g42280 | ORUFI06G23230 | up | Os06g0628032 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0WZ43] | |
| OS06G0702700/LOC_Os06g48960 | ORUFI06G28520 | up | Os06g0702700 protein [Source:UniProtKB/TrEMBL;Acc:Q5Z825] | |
| OS08G0108700/LOC_Os08g01710 | ORUFI08G00660 | up | Os08g0108700 protein [Source:UniProtKB/TrEMBL;Acc:Q6ZC75] | |
| OS08G0557200/LOC_Os08g44300 | ORUFI08G25860 | RETROTRANSPOSON | up | Calcineurin-like phosphoesterase family-like [Source:UniProtKB/TrEMBL;Acc:Q6ZJ14] |
| OS09G0129600/LOC_Os09g04339 | ORUFI09G01420 | up | Os09g0129600 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0XKA3] | |
| OS09G0453800/LOC_Os09g28050 | ORUFI09G13500 | up | Aminotransferase [Source:UniProtKB/TrEMBL;Acc:Q67UZ0] | |
| OS10G0195250/LOC_Os10g11889 | ORUFI12G07700 | up | Os10g0195250 protein [Source:UniProtKB/TrEMBL;Acc:B9G7W2] | |
| OS11G0484000/LOC_Os11g29370 | ORUFI11G14510 | up | Probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 [Source:UniProtKB/Swiss-Prot;Acc:Q2R483] | |
| OS12G0260500/LOC_Os12g16010 | ORUFI12G08470 | sex determination protein tasselseed-2 | up | Os12g0260500 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0Y8Z9] |
| OS12G0589100/LOC_Os12g39860 | ORUFI12G19290 | up | Adenine phosphoribosyltransferase 1, putative, expressed [Source:UniProtKB/TrEMBL;Acc:Q2QMV8] | |
| OS01G0142300/LOC_Os01g04920 | ORUFI01G02770 | glycosyl transferase | down | Os01g0142300 protein [Source:UniProtKB/TrEMBL;Acc:Q5ZBM2] |
| OS01G0227100/LOC_Os01g12710 | ORUFI01G08790 | NYC1 | down | Probable chlorophyll(ide) b reductase NYC1, chloroplastic [Source:UniProtKB/Swiss-Prot;Acc:Q5N800] |
| OS01G0741900/LOC_Os01g53880 | ORUFI01G33490 | IAA6 | down | Auxin-responsive protein IAA6 [Source:UniProtKB/Swiss-Prot;Acc:Q8LQ74] |
| OS02G0695600/LOC_Os02g46830 | ORUFI02G30390 | down | Os02g0695600 protein [Source:UniProtKB/TrEMBL;Acc:Q6YUQ1] | |
| OS02G0704900/LOC_Os02g47600 | ORUFI02G31040 | IPP | down | Soluble inorganic pyrophosphatase [Source:UniProtKB/TrEMBL;Acc:B7E5N1] |
| OS02G0802700/LOC_Os02g55910 | ORUFI02G37800 | MGD3 | down | Os02g0802700 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0VQS5] |
| OS03G0130400/LOC_Os03g03820 | ORUFI03G02170 | down | Probable adenylate kinase 1, chloroplastic [Source:UniProtKB/Swiss-Prot;Acc:Q10S93] | |
| OS03G0214400/LOC_Os03g11560 | ORUFI03G08360 | down | Digalactosyldiacylglycerol synthase 2, putative, expressed [Source:UniProtKB/TrEMBL;Acc:Q10Q06] | |
| OS03G0238600/LOC_Os03g13540 | ORUFI03G10060 | down | Purple acid phosphatase [Source:UniProtKB/TrEMBL;Acc:Q10PD0] | |
| OS04G0110600/LOC_Os04g02000 | ORUFI04G00600 | down | Os04g0110600 protein [Source:UniProtKB/TrEMBL;Acc:Q0JFE3] | |
| OS04G0438600/LOC_Os04g35790 | ORUFI04G14680 | GLTP domain containing protein | down | Os04g0438600 protein [Source:UniProtKB/TrEMBL;Acc:Q0JD08] |
| OS04G0480900/LOC_Os04g40490 | ORUFI04G17670 | down | OSJNBb0011N17.5 protein [Source:UniProtKB/TrEMBL;Acc:Q7XUQ7] | |
| OS04G0508200/LOC_Os04g42920 | ORUFI04G19660 | down | Isocitrate dehydrogenase [NADP] [Source:UniProtKB/TrEMBL;Acc:Q0JBV4] | |
| OS04G0627300/LOC_Os04g53560 | ORUFI04G27820 | down | Os04g0627300 protein [Source:UniProtKB/TrEMBL;Acc:B9FCM8] | |
| OS04G0640600/LOC_Os04g54800 | ORUFI04G28560 | shikimate kinase, | down | Os04g0640600 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0WFI9] |
| OS04G0652700/LOC_Os04g55850 | ORUFI04G29370 | nuclease PA3, | down | Os04g0652700 protein [Source:UniProtKB/TrEMBL;Acc:B9FCW0] |
| OS05G0178300/LOC_Os05g08554 | ORUFI05G05490 | OsCDT5 | down | Similar to Cadmium tolerant 1. (Os05t0178300-01) [Source: https://rapdb.dna.affrc.go.jp/tools/search/run?id=on&attr=desc&keyword=Os05g0178300] |
| OS05G0566400/LOC_Os05g49140 | ORUFI05G27900 | down | Mitogen-activated protein kinase [Source:UniProtKB/TrEMBL;Acc:Q0DFW7] | |
| OS06G0115800/LOC_Os06g02540 | ORUFI06G01060 | down | Os06g0115800 protein [Source:UniProtKB/TrEMBL;Acc:Q0DF43] | |
| OS06G0603600/LOC_Os06g40120 | ORUFI06G21760 | SPX1 | down | SPX domain-containing protein 1 [Source:UniProtKB/Swiss-Prot;Acc:Q69XJ0] |
| OS07G0187700/LOC_Os07g09000 | ORUFI07G05590 | WD domain, G-beta repeat domain containing protein | down | Os07g0187700 protein [Source:UniProtKB/TrEMBL;Acc:Q6Z4F3] |
| OS07G0598200/LOC_Os07g40710 | ORUFI07G22020 | circadian clock coupling factor-related | down | Os07g0598200 protein [Source:UniProtKB/TrEMBL;Acc:Q6ZJE5] |
| OS07G0668700/LOC_Os07g47250 | ORUFI07G26640 | lipase precursor, | down | Os07g0668700 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0X9 × 0] |
| OS08G0118900/LOC_Os08g02540 | ORUFI08G01360 | down | Probable adenylate kinase 7, mitochondrial [Source:UniProtKB/Swiss-Prot;Acc:Q6ZJ48] | |
| OS08G0245200/LOC_Os08g14760 | ORUFI08G08590 | 4CL1 | down | Probable 4-coumarate–CoA ligase 1 [Source:UniProtKB/Swiss-Prot;Acc:P17814] |
| OS08G0536000/LOC_Os08g42410 | ORUFI08G24240 | down | Pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial [Source:UniProtKB/Swiss-Prot;Acc:Q6Z1G7] | |
| OS09G0356000/LOC_Os09g19140 | ORUFI09G07640 | senescence-induced receptor-like serine/threonine-protein kinase precursor | down | Os09g0356000 protein [Source:UniProtKB/TrEMBL;Acc:A0A0P0XML9] |
| OS09G0569800/LOC_Os09g39620 | ORUFI09G21810 | protein kinase family protein | down | Os09g0569800 protein [Source:UniProtKB/TrEMBL;Acc:Q0IZH1] |
| OS10G0116800/LOC_Os10g02750 | ORUFI10G00780 | OsPAP3b | down | Purple acid phosphatase [Source:UniProtKB/TrEMBL;Acc:Q7XH73] |
| OS11G0126800/LOC_Os11g03290 | ORUFI11G01490 | nucleoside-triphosphatase | down | Os11g0126800 protein [Source:UniProtKB/TrEMBL;Acc:Q0IUZ4] |
| OS04G0523600/LOC_Os04g44240 | ORUFI04G20630 | Glycosyltransferase | up | Glycosyltransferase [Source:UniProtKB/TrEMBL;Acc:A0A0P0WCX8] |
Genes were up- and down-regulated in common in both species.
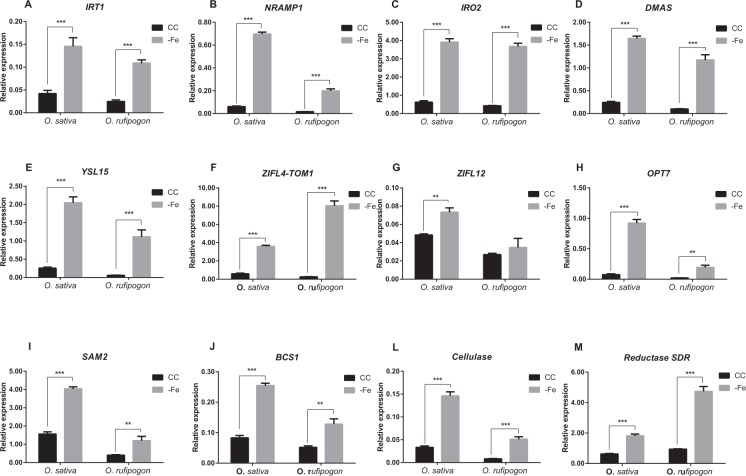
In order to confirm the results obtained by RNAseq, we analyzed the expression level of two genes associated with Strategy I (IRT1 and NRAMP1), five genes associated with Strategy II (IRO2, DMAS, YSL15, ZIFL4-TOM1, and OPT7), and ZIFL12, SAM2, mitochondrial chaperone BCS1, cellulase and one gene annotated as Reductase SDR Family (Supplementary Table 3). We selected genes that were either commonly regulated in both species, or were up-regulated only in O. sativa under −Fe condition. Differential expression was confirmed for all eight tested genes which were regulated similarly on both species according to the RNAseq data (IRT1, NRAMP1, IRO2, DMAS, YSL15, ZIFL4-TOM1, OPT7, SAM2, and mitochondrial chaperone BCS1; Fig. 2A–E,H–J). In addition, the genes ZIFL4/TOM1, cellulase and reductase SDR family, which were up-regulated in roots of O. sativa based on our RNAseq data, were shown to be also up-regulated in O. rufipogon (Fig. 2F,L,M). ZIFL12 was up-regulated only in roots of O. sativa (Fig. 2G). Thus, we confirmed that genes associated with Fe acquisition from the soil in O. sativa are also up-regulated in O. rufipogon under Fe deficiency. These data further indicate that the two species use similar mechanisms for Fe uptake.
Figure 2.
Expression analysis of selected genes ((A) IRT1; (B) NRAMP1; (C) IRO2; (D) DMAS; (E) YSL15; (F) ZIFL4-TOM1; (G) ZIFL12; (H) OPT7; (I) SAM2; (J) BCS1; (L) Cellulase; (M) Reductase SDR) in cultivated (Oryza sativa) and wild (Oryza rufipogon) rice. Relative expression levels (RT-qPCR, relative to OsUBQ5 expression) of genes identified by RNAseq, in roots of plants submitted to control (CC) or Fe deficiency (−Fe) conditions for five days. Roots were collected from rice plants grown in non-aerated nutrient solution, at three-leaf stage on both conditions at the time of RNA extraction. Values are the averages of four samples (3 plants each) ± SE. Asterisks indicate statistical difference between plants grown under CC and −Fe conditions (Student t-test, **P-value < 0.01, ***P-value < 0.001).
Analysis of synteny and collinearity allow the inference of OsIRT1, OsYSL15, OsIRO2, and OsNRAMP1 orthologs between Oryza species, maize and sorghum
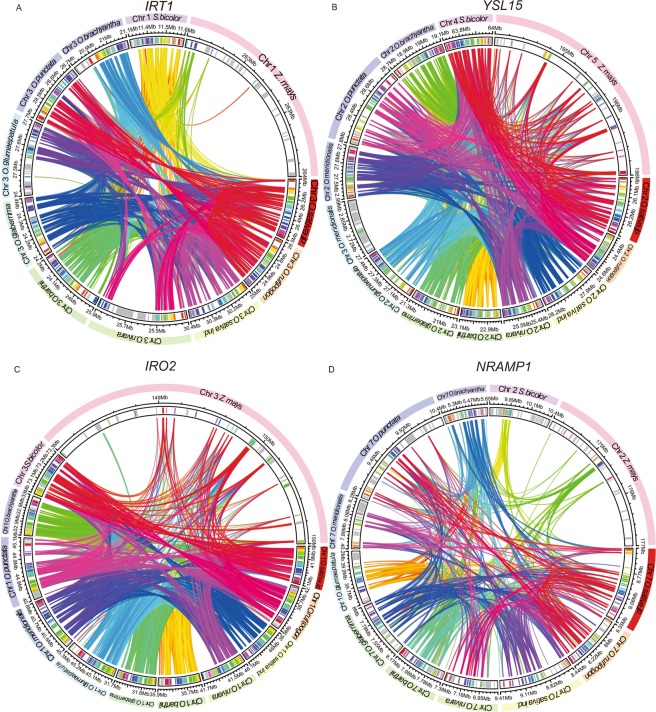
Based on the previous results, we hypothesized that wild rice species may also use the CS for Fe uptake. Since an expected characteristic of such strategy would be concomitant up-regulation of OsYSL15 and OsIRT1, we decided to analyze the expression of these genes in other species of the Oryza genus and also in other grasses. We also included IRO2, a transcription factor involved in regulating the Strategy II genes in rice, and NRAMP1, an Fe transporter that is also up-regulated under Fe deficiency in cultivated rice, but has no clear function in Fe uptake yet. To identify the orthologous genes in nine species of the Oryza genus, maize and sorghum, we analyzed the syntenic relationships between their genomes using O. sativa ssp. japonica as a reference genome. We identified synteny in the IRT1 locus for most species, with maize, O. nivara and O. glumaepatula having the least synteny compared to O. sativa (Fig. 3A). We found no IRT1 ortholog in O. meridionalis. Scanning the O. meridionalis genome sequence, we found that the genome region that contains IRT1 is likely missing from the current draft (Supplementary Fig. 4). We also identified syntenic genomic blocks for YSL15 and IRO2 (Fig. 3C) sequences in all genomes considered, except for YSL15 in O. meridionalis (Fig. 3B). In this species, YSL15 was found in chromosome 3, and not in chromosome 2, as in all other Oryza species. OsYSL15 and OsYSL2 are found in tandem in chromosome 2, an organization that is conserved in most Oryza species, but not in O. meridionalis (Fig. 3B). For NRAMP1, we identified synteny for most species considered (Fig. 3D).
Figure 3.
Synteny analysis of Fe deficiency-related genes in Oryza spp., maize and sorghum. Circular graphs displaying the results from the synteny analysis, performed through the MCSCanX tool, across the Oryza genus, Zea mays, and Sorghum bicolor gene orthologs. The analyses were performed on genomic segments containing 41 genes. Same-color ribbons connect syntenic genes from the same pair of genomic segments. Colored bars represent genes inside the genomic segments. The rainbow spectrum color bands inside the genomic segments are in reference to the 41 genes in the O. sativa’s chromosome segment containing the target gene (YSL15, IRO2, NRAMP1 or IRT1) plus 20 genes up and downstream of it. A color-coded scheme is used to represent the genes inside the genomic segments as such: if a given gene is a potential homolog of any of the 41 afore mentioned set of reference genes, then it receives the same color of its homolog; otherwise, it is colored grey. (A) Synteny analysis for IRT1. (B) Synteny analysis for YSL15. (C) Synteny analysis for IRO2. (D) Synteny analysis for NRAMP1.
Collinearity analyses showed the occurrence of inversions in the region containing IRT1 when comparing O. sativa ssp. japonica with O. nivara and O. glumaepatula (Supplementary Fig. 5A). The segment containing the YSL15 gene, located on chromosome 2 in O. sativa, tends to have genes in the same order and orientation as in O. sativa in most of the evaluated genomes, except in O. meridionalis and sorghum. When we consider O. meridionalis, the genomic block which is syntenic with O. sativa does not contain YSL15 (Supplementary Fig. 5B). In this species, YSL15 is located on chromosome 3. In relation to the transcription factor IRO2, the occurrence of collinearity with the O. sativa genome was observed in most of the analyzed species (Supplementary Fig. 5C). There was collinearity of the NRAMP1 chromosomal region from O. sativa ssp. japonica with sorghum and maize only in the up and downstream regions, respectively (Supplementary Fig. 5D). These analyses allowed us to identify the most likely orthologs for IRT1, YSL15, IRO2, and NRAMP1 in Oryza species, maize and sorghum.
Oryza species with AA genome up-regulate genes used in the Combined Strategy under Fe deficiency conditions
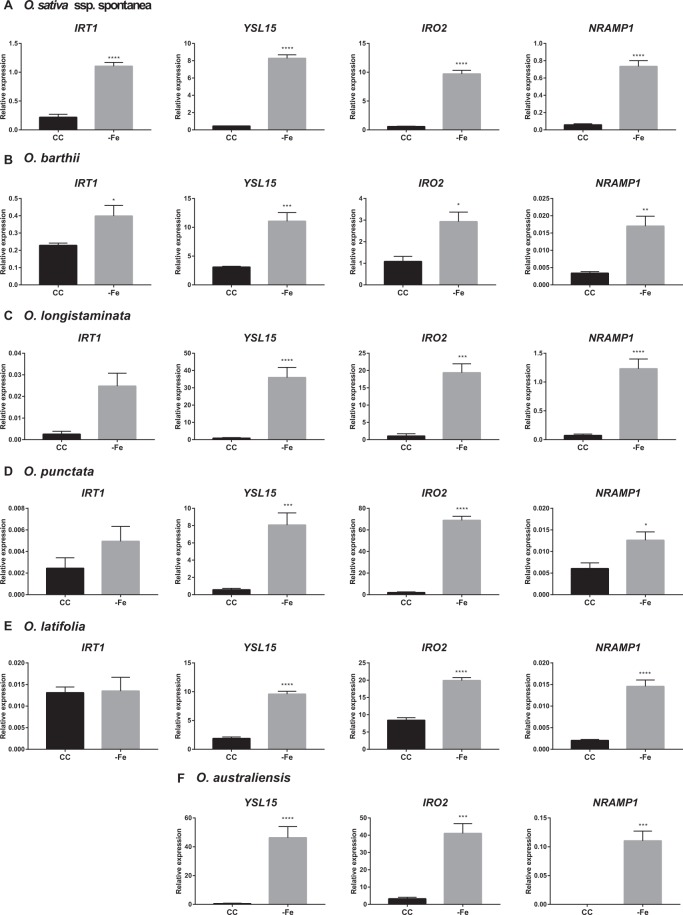
To test whether the rice orthologous genes involved in the CS are up-regulated in other Oryza species, plants of O. sativa ssp. spontanea (AA genome type), O. barthii (AA), O. longistaminata (AA), O. punctata (BB), O. latifolia (CCDD) and O. australiensis (EE) were submitted to control or −Fe conditions for five days, and gene expression of IRT1, YSL15, IRO2, and NRAMP1 was evaluated in roots. In O. sativa ssp. spontanea and O. barthii, expression of all four genes was up-regulated by −Fe (Fig. 4A,B). In O. longistaminata, IRT1 expression seemed to be induced under −Fe, although not statistically significant (p = 0.067). However, the expression levels of IRO2, YSL15, and NRAMP1 were up-regulated by −Fe in the same species (Fig. 4C). In O. punctata and O. latifolia roots, expression of IRT1 was not significantly induced upon −Fe, while the transcript levels of YSL15, IRO2, and NRAMP1 were up-regulated (Fig. 4D,E). O. australiensis also showed up-regulation of YSL15, IRO2, and NRAMP1 (in this last, the expression was not detected in CC) in roots under −Fe, while IRT1 expression was not detected, even under −Fe (Fig. 4F). These results suggest that it is possible that distinct species of Oryza genus evolved independently to Strategy II-exclusive or CS20, and indicate that the CS is common to other AA genome species besides O. sativa.
Figure 4.
Expression analysis of Fe deficiency-related genes in (A) Oryza sativa ssp. spontanea; (B) O. barthii; (C) O. longistaminata; (D) O. punctata; (E) O. latifolia; (F) O. australiensis. Relative expression levels (RT-qPCR, relative to OsUBQ5 expression) of selected genes (IRT1, YSL15, IRO2, and NRAMP1) in roots of plants submitted to control (CC) or Fe deficiency (−Fe) conditions for five days. Roots were collected from plants grown in non-aerated nutrient solution, at three-leaf stage on both conditions at the time of RNA extraction. Values are the averages of four samples (3 plants each) ± SE. Asterisks indicate statistical difference between plants grown under CC and −Fe conditions (Student t-test, *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001, ****P-value < 0.0001).
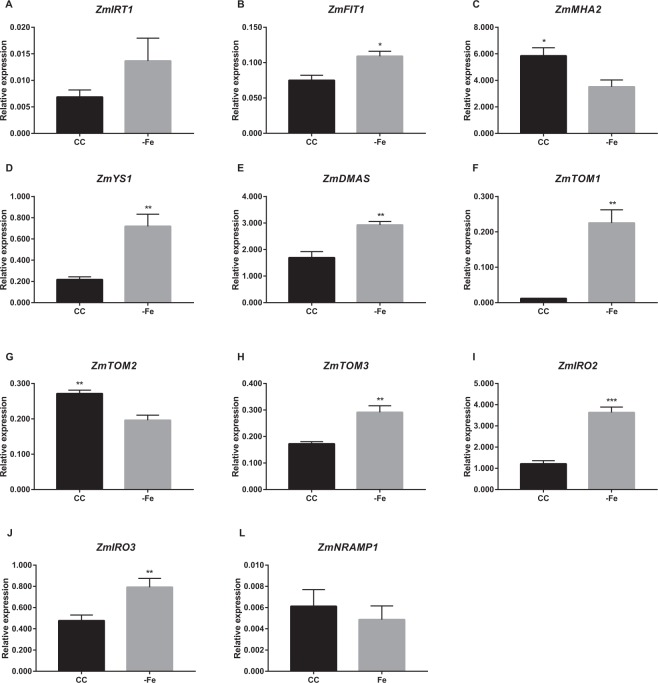
We also evaluated the expression of the same genes in both maize and sorghum roots under CC and −Fe conditions (Figs. 5 and 6). Chlorophyll quantification showed that maize plants are also at the early phase of Fe deficiency response after five days (Supplementary Fig. 6). Expression profiles of genes related to Strategy I (ZmIRT128, ZmFIT129 and ZmMHA229) were evaluated. ZmFIT1, orthologous to the transcription factor AtFIT, which regulates the expression of FRO2 and IRT1 in Arabidopsis30, was up-regulated in roots under −Fe (Fig. 5B). ZmMHA2, similar to plasma membrane H+ATPase from Arabidopsis (MHA2)29, was down-regulated (Fig. 5C). ZmIRT1 expression was not significantly affected by Fe deficiency (Fig. 5A). Genes related to Strategy II (ZmYS1, ZmDMAS, ZmTOM1, ZmTOM2, ZmTOM3, ZmIRO2, and ZmIRO3) were up-regulated in roots grown under −Fe condition compared to those cultivated in CC, except for ZmTOM2, as described31 (Fig. 5D–J). ZmNRAMP1 expression was not regulated by Fe deficiency (Fig. 5L).
Figure 5.
Expression analysis of Fe deficiency-related genes in Zea mays. Relative expression levels (RT-qPCR, relative to ZmUBQ expression) of selected genes ((A) IRT1; (B) FIT1; (C) MHA2; (D) YS1; (E) DMAS; (F) TOM1; (G) TOM2; (H) TOM3; (I) IRO2; (J) IRO3; (L) NRAMP1), in roots of plants submitted to control (CC) or Fe deficiency (−Fe) conditions for five days. Roots were collected from maize plants grown in aerated nutrient solution, at three-leaf stage on both conditions at the time of RNA extraction. Values are the averages of four samples (3 plants each) ± SE. Asterisks indicate statistical difference between plants grown under CC and −Fe conditions (Student t-test, *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001).
Figure 6.
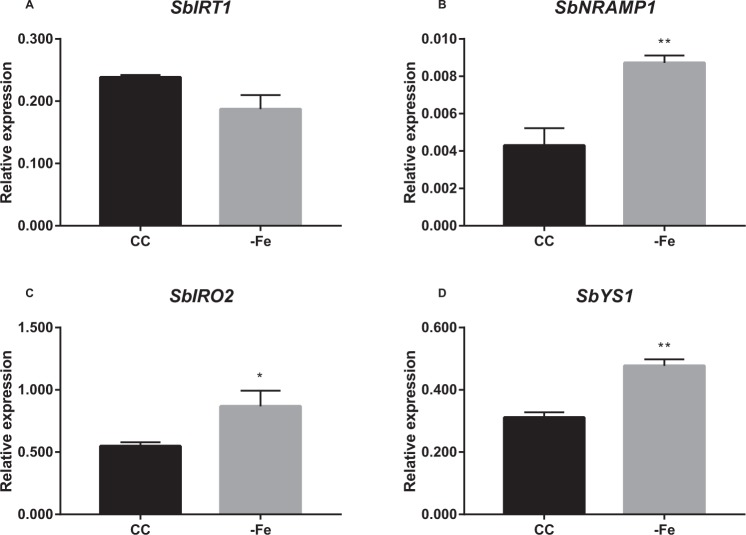
Expression analysis of Fe deficiency-related genes in Sorghum bicolor. Relative expression levels (RT-qPCR, relative to SbACT expression) of selected genes ((A) IRT1; (B) NRAMP1; (C) IRO2; (D) YS1), in roots of plants submitted to control (CC) or Fe-deficiency (−Fe) conditions for three days. Roots were collected from sorghum plants grown in aerated nutrient solution, at three-leaf stage on both conditions at the time of RNA extraction. Values are the averages of four samples (3 plants each) ± SE. Asterisks indicate statistical difference between plants grown under CC and −Fe conditions (Student t-test, *P-value < 0.05, **P-value < 0.01).
Sorghum plants showed decreased chlorophyll concentration after two days of treatment (Supplementary Fig. 7). SbIRT1 was not regulated by Fe deficiency in roots, while the transcript levels of SbNRAMP1, SbIRO2, and SbYS1 were up-regulated under −Fe compared to CC (Fig. 6A–D). These results suggest that both maize and sorghum do not induce IRT1 expression under −Fe conditions, and thus may be Strategy II-exclusive plants.
Discussion
Currently, there is little experimental evidence to support models of Fe deficiency response evolution in plants20. Evidence from Chlamydomonas reinhardtii and Marchantia polymorpha support the hypothesis that reduction-based Strategy I is ancestral in the plant lineage32,33. In this work, to add information to the evolution of Fe uptake strategies in Poaceae, we compared the root transcriptomic responses of O. sativa and O. rufipogon under −Fe. We choose O. rufipogon because this species is the wild progenitor of O. sativa ssp. japonica34. This is the first report of such comparison in the literature, and the first characterization of O. rufipogon Fe deficiency response. It is also the first RNAseq dataset for cultivated rice under −Fe, which allows us to confirm other studies using microarrays35,36. Our data also allowed us to propose a model for −Fe response evolution (see next sections).
We used physiological characterization to identify the early phase of −Fe response, which was established at five days for both O. sativa and O. rufipogon (Fig. 1). Transcriptomic data showed that both species have significant overlapping sets of regulated genes in roots upon −Fe treatment (Table 1 and Supplementary Fig. 2), although a large group of genes are exclusively regulated in one species (Supplementary Fig. 3). Among those regulated in both, we found genes linked to Fe homeostasis. HRZ1 and HRZ2, two hemerythrin domain-containing ubiquitin ligases that negatively regulate −Fe response through protein degradation27, are up-regulated in both species. HRZ1 and HRZ2 are involved in degradation of some components of the Fe uptake machinery, similar to the role of their Arabidopsis homolog BRUTUS37. Two bHLH transcription factors, IRO2 and IRO3, were also up-regulated in roots of both species under −Fe. IRO2 is a positive regulator of Fe uptake genes, whereas IRO3 is a negative regulator, orthologous to Arabidopsis POPEYE38.
We found transporters that are associated with Fe homeostasis in cultivated rice also regulated in O. rufipogon. OsYSL9 was described as an Fe-chelate transporter involved in Fe distribution in developing grains, which is up-regulated by −Fe in roots39 (Table 1). OsOPT7, also described as an up-regulated gene in roots upon −Fe40, but with unknown transport substrate, was also up-regulated in O. rufipogon (Table 1). ENA1, described as a NA efflux transporter in rice, was also up-regulated in both species in our experiments14. Interestingly, we also found genes related to homeostasis of other metals, such as zinc (Zn, ZIP441), copper (Cu; COPT5.142) and metallothioneins (Table 1). Transporters such as IRT1 are capable of transporting a variety of cations other than Fe, including cadmium (Cd), cobalt (Co), manganese (Mn) and Zn10, which can lead to accumulation of heavy metals such as Cd43 and Zn44 in plants when IRT1 is highly expressed. Thus, up-regulation of transporters of other metals might be a consequence of the broad substrate specificity of IRT1-like transporters45 and accumulation of toxic levels of heavy metals. Moreover, we found the vacuolar iron transporter 2 (VIT2), which is up-regulated by Fe excess and down-regulated by −Fe, among the genes down-regulated in both species, corroborating previous data46,47. Finally, the uncharacterized NRAMP7 transporter was also down-regulated in both species, suggesting a role for this transporter in Fe homeostasis.
Our GO analyses showed that Fe and methionine/S-adenosyl methionine-related processes are among the terms enriched in both species (Supplementary Fig. 2). Under −Fe, the methionine cycle feeds the biosynthesis of phytosiderophores in plants that rely on Strategy II to absorb Fe from the rhizosphere19,35. MAs are synthetized in a conserved pathway from S-adenosyl-L-methionine13, which is biosynthesized during the reaction of methionine with ATP by S-adenosyl-methionine synthase (SAMS)48,49. The pathway for biosynthesis of MA includes enzymatic reactions meditated by nicotianamine synthase (NAS1) which catalyzes the formation of NA. Next, nicotianamine transferase (NAAT1)21 converts NA into a 3′-keto intermediate, which is then reduced to deoxymugineic acid (DMA) by deoxymugineic acid synthase (DMAS)13,40. The synchronous expression of the methionine cycle-related genes was observed during the first 36 hours of −Fe in roots of rice50. Between the transcripts up-regulated only in O. sativa, we identified an aspartate aminotransferase (OsIDI4) which has been predicted to be the enzyme that catalyzes the conversion of 2-keto-4-methylthiobutyric acid to methionine by transamination, and is a candidate enzyme for the final step of the methionine cycle51–53. Up-regulation of genes involved in the biosynthesis of methionine and MA, like SAM2, NAS1, and DMAS were observed in O. sativa and O. rufipogon (Table 1). These results are indicative that both species responded to −Fe by increasing precursors and phytosiderophore synthesis for Fe uptake54.
We also found ZIFL4/TOM1 up-regulated in roots of both species when exposed to −Fe. In rice, OsZIFL4/TOM1 was characterized as the transporter involved in DMA secretion to the rhizosphere, and thus a crucial player in Strategy II. DMA binds to Fe(III), generating the Fe(III)-phytosiderophore complex. Barley plants were also shown to use the ortholog HvTOM1 to perform this key step in phytosiderophore release14, while maize ZmTOM1 was shown to be up-regulated by −Fe31. Thus, up-regulation of ZIFL4/TOM1 in O. rufipogon fits well with what is expected in a plant acquiring Fe using Strategy II/chelation strategy. In agreement with that, we also found both OsYSL15 and OrYSL15 genes up-regulated in roots of Fe deficient plants. As YSL15 transporters are necessary for Fe(III)-phytosiderophore uptake into root cells, it is clear that a full Strategy II is being induced in both O. sativa and O. rufipogon under −Fe conditions.
Among the genes up-regulated by −Fe in both species, we found genes classically characterized as part of the Strategy I, similar to IRT110 and IRT255. AtIRT1 encodes a high affinity Fe2+ transporter10,18 up-regulated in epidermis and exodermis of roots under −Fe. In rice, the most similar gene, OsIRT1, is induced by low Fe, and was able to complement yeast mutants defective in Fe transport18. It was shown that OsIRT1 is part of the rice Fe regulon, being regulated along with Strategy II genes36. OsIRT1 over-expression results in Fe, Zn, and Cd accumulation in rice tissues56. Based on these evidences, it was proposed that OsIRT1 is involved in Fe uptake in cultivated rice, allowing rice roots to acquire Fe2+ from the soil. This would be an adaptation to paddy fields where rice is commonly cultivated, and where Fe2+ is much more abundant than Fe3+ 18.
Another gene found up-regulated in O. sativa and O. rufipogon was NRAMP1. In cultivated rice, OsNRAMP1 was characterized as an Fe2+, Mn2+, and Cd2+ transporter up-regulated by −Fe57. High OsNRAMP1 expression was also linked to Cd2+ accumulation in rice cultivars57. A highly similar gene, OsNRAMP5, is also involved in Cd2+ accumulation, and is able to transport Fe2+ and Zn2+ 58. In Arabidopsis, the high-affinity metal transporter NRAMP1, which is closely related to OsNRAMP1/OsNRAMP5, is induced by −Fe, and was shown to be important for Fe transport under Fe sufficiency conditions, cooperating with IRT1 to absorb Fe+2 from the rhizosphere59. However, to date, the physiological function of the rice ortholog OsNRAMP1 is not clear. Thus, it is possible that rice plants use three transporters for primary Fe uptake from the soil under −Fe conditions (i.e., YSL15, IRT1, and NRAMP1), with OsNRAMP1 (or OsNRAMP5) having a role for Fe2+ uptake under Fe sufficient conditions. These hypotheses, however, need to be supported by additional evidence. Taken together, our data indicate that O. rufipogon also uses Strategy I-related genes such as IRT1 and NRAMP1 for Fe uptake, as observed in O. sativa.
After the first work showing that O. sativa is able to transport Fe2+ in addition to the ability to transport Fe(III)-phytosiderophore18, the current model for the evolution of Fe deficiency response was established, in which rice uses the CS as a recent adaptation to waterlogged soils18,20. Recently, we hypothesized that the underlying assumption that Fe2+ uptake by IRT1, considered as a new adaptation of cultivated rice, might not be true, and other Poaceae species might also use IRT1 for Fe uptake, which would mean the IRT1-based CS has a common, more ancient origin within the family or the Oryza genus. In order to test this hypothesis in wild Oryza species, maize and sorghum, we analyzed the expression of the orthologous genes of YSL15 and IRT1, which control Fe3+ and Fe2+ uptake, respectively, in plants under −Fe. We also included IRO2, a positive regulator of the Strategy II genes in rice; and NRAMP1, which is part of the Fe regulon in rice and may also be involved in Fe uptake. Expression of the orthologous genes (Fig. 3 and Supplementary Fig. 5) encoding each of these proteins in all species was analyzed. We assumed that concerted up-regulation of both YSL15 and IRT1 was indicative of CS use in a given species. NRAMP1 up-regulation was considered as further indication of a conserved role for this gene in −Fe response. Importantly, our approach aims at identifying a common origin of the CS in Poaceae, based on the IRT1 orthologs in these species. It is possible that independent CS strategies have evolved in this family, which could be based on other transporters that perform the same activity (i.e., NRAMP1 or IRT2).
O. sativa ssp. spontanea (AA genome) is the weedy rice variety, and is cultivated in flooded soil along with cultivated rice, being widely distributed in South and South-east Asia, South and North America, and southern Europe60. O. barthii and O. longistaminata (both AA genomes) are species cultivated in seasonally dry habitats. These three species are representative of the O. sativa complex, which include AA genome species, the closest ones to cultivated rice in the Oryza genus61,62. In these three species, we observed increased expression of both IRT1 and YSL15. NRAMP1 and IRO2 were also up-regulated (Fig. 4A–C). We also tested three species outside the O. sativa complex: O. punctata (BB genome), O. latifolia (CCDD genome), and O. australiensis (EE genome), all from the O. officinalis complex. These species are cultivated in seasonally dry habitats61. Interestingly, we observed up-regulation of YSL15, NRAMP1, and IRO2, but not of IRT1 (Fig. 4D–F). These results suggest that species from the O. officinalis complex do not use IRT1 for Fe uptake under low Fe conditions, while species from the O. sativa complex up-regulate IRT1 under the same conditions.
We also evaluated the expression of the same genes in maize and sorghum, two Poaceae species outside the Oryza genus that serve as out groups to test our hypothesis for a common origin of an IRT1-based CS in Poaceae. Even though the Fe uptake mechanism has not been as well characterized in maize and sorghum as in O. sativa, there has been some discussion whether maize can use a partial Strategy I29,31,63. We observed that ZmIRT1 expression was not significantly induced in maize roots exposed to −Fe, although there seems to be a trend for higher expression under iron deficiency (Fig. 5A). Nozoye et al.31 and Li et al.28 found ZmIRT1 up-regulated under similar conditions. ZmYS1, as expected, was clearly up-regulated (Fig. 5D). We also tested other candidate genes likely involved in −Fe regulation in maize already described in the literature to confirm that maize plants are responding properly to −Fe conditions29 (Fig. 5). When we tested the likely orthologs of our candidate genes in sorghum, we observed that SbIRT1 was not up-regulated, while SbYS1 was (Fig. 6D). Interestingly, ZmNRAMP1 was not regulated by −Fe, whereas SbNRAMP1 was. These results suggest that sorghum plants do not use the CS, whereas maize may use ZmIRT1 as an Fe2+ transporter upon Fe deficiency, but without a strong, consistent up-regulation. Alternatively, since we used a different genotype than Nozoye et al., ZmIRT1 up-regulation might be genotype specific, as observed for barley HvIRT164. Thus, as already suggested20, the use of CS might vary in Poaceae species outside the Oryza genus or even in genotypes within the genus, which may be linked to local adaptation to specific environments where each genotype is found. Considering the possibility that maize also uses the CS based on the IRT1 ortholog, our data suggest that maize CS is not homologous to what is observed in the AA Oryza group, which would indicate an independent/convergent evolution in the use of IRT1, together with the shared chelation strategy. Altogether, our data suggest a restricted up-regulation of IRT1 and YSL15 within the AA genome group of the Oryza genus, resembling the O. sativa CS. Moreover, NRAMP1 does not seem to have a conserved function in all Poaceae species analyzed.
Conclusion
In conclusion, our data suggest that Strategy II for Fe uptake, represented by YSL15 up-regulation under −Fe, is conserved in the Poaceae family, being an evolutionary novelty after the split between Poaceae last common ancestor and other monocots lineages (Fig. 7). Partial Strategy I for Fe uptake based on up-regulation of IRT1 orthologs under −Fe is conserved only within the O. sativa complex containing the AA genome group. Therefore, the species with AA genome share a common origin for the IRT1 up-regulation, which is an evolutionary novelty that arose after the split of the AA lineage from the last common ancestor with the BB genome lineage (Fig. 7). We also propose that the CS is not exclusive of cultivated rice, but rather common in wild species closely related to O. sativa, and likely an adaptation to flooded soils that preceded speciation within the O. sativa complex. This improves our knowledge about the evolution of Fe uptake mechanisms in plants, and especially in the Poaceae family.
Figure 7.
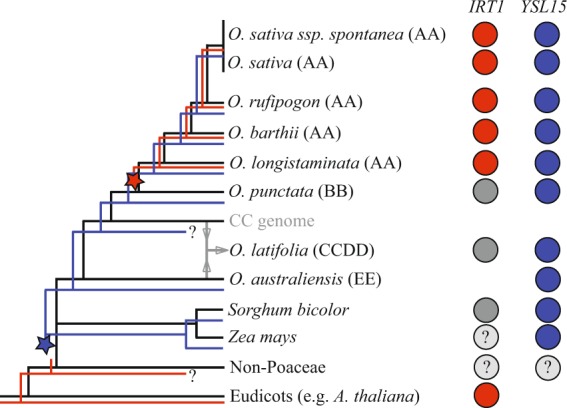
Model for the evolution of rice Combined Strategy (CS) of Fe uptake in the Poaceae family. Red and blue lines represent Oryza species using the CS for Fe uptake, showed by IRT1 up-regulation (partial Strategy I) and YSL15 up-regulation (Strategy II) under Fe deficiency. Only red or blue lines represent plants exclusively using Strategy I or Strategy II, respectively. Red and blue circles represent species in which IRT1 and YSL15 are expressed in roots under −Fe, respectively. Dark grey circles represent species in which IRT1 expression is not induced under −Fe. Light grey circles represent species in which regulation of key genes is unclear or not yet known. The red star represents the split of O. sativa complex within the Oryza genus, in which IRT1 is up-regulated under −Fe, and the CS is conserved. The blue star represents the split between Poaceae last common ancestor and other monocots lineages.
Methods
Plant material and treatments
Seeds of Oryza sativa L. (Nipponbare cultivar) and O. rufipogon (BRA 00004909-8 accession from EMBRAPA Rice & Beans) were germinated for four days in Petri dishes with filter paper soaked in distilled water at 28 °C (two days in the dark and two days in the light, 40 µmol.m−2.s−1). After germination, plants were transferred to vermiculite and cultivated for fifteen days in distilled water. Then, plants were transferred to plastic pots with 0.5 L of nutrient solution (five plants per pot) containing 700 μM K2SO4, 100 μM KCl, 100 μM KH2PO4, 2 mM Ca(NO3)2, 500 μM MgSO4, 10 μM H3BO3, 0.5 μM MnSO4, 0.5 μM ZnSO4, 0.2 μM CuSO4, 0.01 μM (NH4)6Mo7O24, and 100 μM Fe+3-EDTA. After seven days of acclimation, plants were transferred to control condition (CC; containing 100 μM Fe+3-EDTA) or to Fe deficiency treatment (−Fe; no iron added) for one, three, five, seven and nine days. To evaluate the response to Fe deficiency in the Oryza genus, seeds from the species O. australiensis (IRGC 86526), O. barthii (IRGC 86524), O. latifolia (IRGC 103808), O. longistaminata (IRGC 101254), O. punctata (IRGC 88825), and O. sativa ssp. spontanea (IRGC 86719) were cultivated. Before germination, seeds were submitted to 50 °C for seven days to break dormancy, according to instructions provided by the International Rice Research Institute (IRRI). After, seeds were germinated and seedlings were cultivated without aeration as described above.
Maize (genotype B73)65 and sorghum (genotype BTx623)66 were germinated in vermiculite and nutrient solution (as described above) for fifteen days, and transferred to pots with 6 L of hydroponic solution (30 plants per pot) aerated by air pumps. Plants were then transferred to control conditions (CC, 100 μM Fe+3-EDTA) or to Fe deficiency (−Fe; no iron added) treatment. The pH of the nutrient solutions was adjusted to 5.4. Plants were grown in a growth room at 26 °C ± 1 °C under photoperiod of 16 h/8 h light/dark (150 μmol.m−2.s−1). Solutions were replaced twice a week.
Chlorophyll quantification
Samples from the two youngest fully expanded leaves (about 100 mg) from plants submitted to CC or −Fe conditions were collected (n = 4, each sample consisting of three pooled plants), frozen in liquid nitrogen and stored at −80 °C. Chlorophyll extraction was performed in 85% (v/v) acetone. Chlorophyll a and b were quantified by measuring absorbance at 643 and 663 nm, and the concentrations calculated according to Ross67. Measurements of relative leaf chlorophyll level were performed with Soil Plant Analysis Development (SPAD-502, Minolta, Japan; n = 10).
RNA extraction and transcriptome analyses by RNAseq
Rice roots samples were harvested from plants grown in CC and from plants submitted to −Fe treatment for one, three, five, seven and nine days. Total RNA was extracted using Concert Plant RNA Reagent (Invitrogen®, Carlsbad, USA), according to the manufacturer’s instructions, quantified by Nanodrop and treated with DNAse I (Invitrogen®, Carlsbad, USA). Approximately 20 µg of total RNA was used to high-throughput cDNA sequencing by Illumina HiSeq. 2000 technology (Fasteris SA, Chemin du Pont-du-Centenaire, Switzerland – http://www.fasteris.com/). RNAs derived from three biological replicates were pooled to generate each library. A total of eight libraries were constructed, from samples harvested five days after the onset of treatments: two libraries from roots of each O. sativa and O. rufipogon grown in CC and two libraries from roots grown in −Fe for each species. The cDNA libraries were prepared according to Illumina’s protocols, as described68. After sequencing, read quality was checked by FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and all low quality reads (PHRED value < 30) were removed. Adapter sequences were trimmed using Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and the first twelve nucleotides were removed from the 5′ end. The abundance of each transcript was estimated using Kallisto69. Kallisto is a pseudoalignment RNAseq quantification method, in which the reads are pseudoaligned to a reference transcriptome, producing a list of transcripts that are compatible with each read. Kallisto does not assign each read to a physical position on a reference genome. Instead, it measures how likely the relative abundance of a transcript given a certain library dataset is. For transcripts proportions quantification, Kallisto uses an expectation-maximization algorithm to optimize a likelihood function. This optimization process will output a set of parameters that quantify the proportion of each transcript. Transcripts from different species or from different treatments were considered differentially expressed when the False Discovery Rate (FDR) < 0.05, according to the methodology proposed by Pimentel et al.70. Differentially expressed transcript annotations were downloaded from Ensembl plants database using the R package biomaRt71. The data is publicly available through the GEO database with accession number GSE131238.
Gene ontology (GO) terms enrichment analysis
Comparison of differentially expressed genes in CC or −Fe conditions for O. sativa and O. rufipogon datasets was performed to find enriched Gene Ontology (GO) terms. The enrichment analysis was performed using topGO72. We used Fisher’s Exact Test and the GO terms with p < 0.05 were considered enriched.
Gene expression analysis by RT-qPCR
Total RNA was extracted from roots of plants from eight species of the Oryza genus, maize and sorghum, submitted to CC or −Fe conditions for five days. After extraction with Concert Plant RNA Reagent (Invitrogen®, Carlsbad, USA), according to the manufacturer’s instructions, quantification was performed using Nanodrop® (Thermo Fisher Scientific, Waltham, USA). Total RNA was treated with DNAse I (Invitrogen®, Carlsbad, USA), and first-strand cDNA synthesis was performed with OligodT and reverse transcriptase M-MLV (Invitrogen®, Carlsbad, USA) using 1 µg of DNAse-treated RNA. RT-qPCRs were carried out in a StepOne Real-Time Cycler (Applied Biosystems, Foster City, USA). For all species from the Oryza genus, genomic and coding sequences of selected genes were aligned using DiAlign Local multiple alignment (http://www.genomatix.de/cgi-bin/dialign/dialign.pl) to find conserved regions. Primers that could amplify the same region of each gene in all species with available genomes were designed. All primers (listed in Supplementary Table 3) were designed to amplify 100–200 bp and to have similar Tm values (60 °C ± 1 °C). Reactions settings were composed of an initial denaturation step of 5 min at 94 °C, followed by 40 cycles of 10 s at 94 °C, 15 s at 60 °C, 15 s at 72 °C and 40 s at 60 °C (fluorescence data collection). Samples were held for 2 min at 40 °C for annealing of the amplified products and then heated from 55 to 99 °C with a ramp of 0.1 °C/s to produce the denaturation curve of the amplified products. RT-qPCRs were carried out in 20 µL final volume composed of 10 µL of cDNA sample diluted 100 times, 2 µL of 10 × PCR buffer, 1.2 µL of 50 mM MgCl2, 0.2 µL of 10 mM dNTPs, 0.4 µL of each primer pair (10 µM), 3.82 µL of water, 2 µL of SYBR green (1:10,000 Molecular Probe), and 0.05 µL of Platinum Taq DNA Polymerase (5 U µL−1, Invitrogen, Carlsbad, CA, USA). Data were analyzed using the Pfaffl method73. The PCR efficiency from the exponential phase was calculated for each individual amplification plot using the LinReg software74. Since the method using LinReg allows for assumption-free calculation of PCR efficiency for each PCR reaction (all technical and biological replicates), we used efficiency averages in each PCR run for calculation. PCR efficiencies are shown in Supplementary Table 3. We used four biological replicates composed by roots from three plants each, and three technical replicates.
Search for synteny and analysis of regions with similarities and/or differences between the genomes of species from the Oryza genus, maize, and sorghum
Whole genome and proteome sequences of nine species from the Oryza genus (O. meridionalis, O. brachyantha, O. barthii, O. punctata, O. glumaepatula, O. nivara, O. glaberrima, O. sativa ssp. indica, O. sativa ssp. japonica, and O. rufipogon), maize (genotype B73) and sorghum (genotype BTx623) were retrieved from Ensembl (http://ensembl.gramene.org/). The protein amino acid sequences of OsNRAMP1, OsIRT1, OsIRO2, and OsYSL15 were used as queries on a batch BLASTp75 search against the complete sets of annotated proteins for all species. For any queried protein X on each targeted species Y, the hit displaying the lowest E-value was selected as the best candidate for being the X homologous on species Y. This resulted in four groups composed by different sets of homologous proteins across the considered species. For each of these groups, synteny analysis was carried out separately as follows. For each gene that encodes a protein in the homologous set, we took a total of 20 genes upstream and 20 genes downstream from its locus. Next, in order to infer synteny degree among these genomic blocks, we used the McScanX software, following the methodology proposed on the original work76. The pairs of genes displaying an E-value < 10e−10 for the expected number of collinear blocks were considered to be in syntenic positions. Circular graphs displaying synteny relationships among the genomic blocks were built with the R package circlize77.
Statistical analysis
Mean values were compared by the Student’s t test (p ≤ 0.05) using the GraphPad Prism 7 (GraphPad Software) for Windows.
Supplementary information
Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001, and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), which granted fellowships to A.W., B.H.N.O., G.L.D., E.B.A., R.L.P. CNPq also helped with a research grant to R.A.S. (Grant Number 476450/2013-6).
Author contributions
A.W., E.B.A., G.L.D. and L.R.P. performed the experiments and collected the data. A.W., B.H.N.O., R.A.S. and F.K.R. analyzed the data. R.A.S., F.K.R. and J.P.F. conceptualized the work. A.W., R.A.S., F.K.R. and J.P.F. wrote the manuscript. All authors read and approved the manuscript.
Data availability
Authors declare that all data and materials used in this work are available to others upon request.
Competing interests
All authors declare to have no competing interests, both financial and non-financial.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Felipe Klein Ricachenevsky, Email: felipecruzalta@gmail.com.
Janette Palma Fett, Email: jpfett@cbiot.ufrgs.br.
Supplementary information
is available for this paper at 10.1038/s41598-019-52502-0.
References
- 1.Kassebaum, N. J. et al. A systematic analysis of global anemia burden from 1990 to 2010. 123, 615–624 (2014). [DOI] [PMC free article] [PubMed]
- 2.Curie C, Briat J-F. Iron transport and signaling in pants. Annu. Rev. Plant Biol. 2003;54:183–206. doi: 10.1146/annurev.arplant.54.031902.135018. [DOI] [PubMed] [Google Scholar]
- 3.Sperotto R, Ricachenevsky FK, Stein RJ, Waldow V, Fett JP. Iron stress in plants: dealing with deprivation and overload. Plant Stress. 2010;4:57–69. [Google Scholar]
- 4.Guerinot ML, Yi Y. Iron: nutritious, noxious, and not readily available. Plant Physiol. 1994;104:815–820. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerkeb, L. & Connolly, E. L. Iron transport and metabolism in plants. in Genetic Engineering (ed. J. K., S.) 119–140, 10.1007/0-387-25856-6_8 (Springer, 2006). [DOI] [PubMed]
- 6.Ravet K, Pilon M. Copper and iron homeostasis in plants: the challenges of oxidative stress. Antioxid. Redox Signal. 2013;19:919–932. doi: 10.1089/ars.2012.5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marschner H, Römheld V, Kissel M. Different strategies in higher plants on mobilizing and uptake of iron. J. Plant Nutr. 1986;9:695–713. doi: 10.1080/01904168609363475. [DOI] [Google Scholar]
- 8.Santi S, Schmidt W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009;183:1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x. [DOI] [PubMed] [Google Scholar]
- 9.Robinson NJ, Procter CM, Connolly EL, Guerinot M. Lou. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- 10.Eide D, Broderius M, Fett JP, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperotto RA, Ricachenevsky FK, Waldow VdeA, Fett JP. Iron biofortification in rice: It’s a long way to the top. Plant Sci. 2012;190:24–39. doi: 10.1016/j.plantsci.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi K, et al. Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 1999;119:471–479. doi: 10.1104/pp.119.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashir K, et al. Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J. Biol. Chem. 2006;281:32395–32402. doi: 10.1074/jbc.M604133200. [DOI] [PubMed] [Google Scholar]
- 14.Nozoye T, et al. Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 2011;286:5446–5454. doi: 10.1074/jbc.M110.180026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ricachenevsky FK, et al. ZINC-INDUCED FACILITATOR-LIKE family in plants: lineage-specific expansion in monocotyledons and conserved genomic and expression features among rice (Oryza sativa) paralogs. BMC Plant Biol. 2011;11:11–20. doi: 10.1186/1471-2229-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curie C, et al. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- 17.Inoue H, et al. Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 2009;284:3470–3479. doi: 10.1074/jbc.M806042200. [DOI] [PubMed] [Google Scholar]
- 18.Ishimaru Y, et al. Rice plants take up iron as an Fe3+ - phytosiderophore and as Fe2+ Plant J. 2006;45:335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- 19.Walker EL, Connolly EL. Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 2008;11:530–535. doi: 10.1016/j.pbi.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Ricachenevsky FK, Sperotto RA. There and back again, or always there? The evolution of rice combined strategy for Fe uptake. Front. Plant Sci. 2014;5:1–5. doi: 10.3389/fpls.2014.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng L, et al. Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol. 2007;145:1647–1657. doi: 10.1104/pp.107.107912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grillet Louis, Schmidt Wolfgang. Iron acquisition strategies in land plants: not so different after all. New Phytologist. 2019;224(1):11–18. doi: 10.1111/nph.16005. [DOI] [PubMed] [Google Scholar]
- 23.Callaway E. The birth of rice. Nature. 2014;514:S58–59. doi: 10.1038/514S58a. [DOI] [PubMed] [Google Scholar]
- 24.Stein JC, et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 2018;50:285–296. doi: 10.1038/s41588-018-0040-0. [DOI] [PubMed] [Google Scholar]
- 25.Jacquemin J, Bhatia D, Singh K, Wing RA. The International Oryza Map Alignment Project: development of a genus-wide comparative genomics platform to help solve the 9 billion-people question. Curr. Opin. Plant Biol. 2013;16:147–156. doi: 10.1016/j.pbi.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Ogo Y, et al. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J. Exp. Bot. 2006;57:2867–2878. doi: 10.1093/jxb/erl054. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi T, et al. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 2013;4:1–12. doi: 10.1038/ncomms3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, et al. Identification and characterization of the zinc-regulated transporters, iron-regulated transporter-like protein (ZIP) gene family in maize. BMC Plant Biol. 2013;13:1–14. doi: 10.1186/1471-2229-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Zhou X, Chen J, Chen R. Is there a strategy I iron uptake mechanism in maize? Plant Signal. Behav. 2018;13:e1161877. doi: 10.1080/15592324.2016.1161877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colangelo EP, Guerinot ML. The essential Basic Helix-Loop-Helix Protein FIT1 is required for the iron deficiency response. Plant Cell. 2004;16:3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nozoye T, Nakanishi H, Nishizawa NK. Characterizing the crucial components of iron homeostasis in the maize mutants ys1 and ys3. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0062567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urzica EI, et al. Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell. 2012;24:3921–3948. doi: 10.1105/tpc.112.102491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo J, et al. Evolutionary analysis of iron (Fe) acquisition system in Marchantia polymorpha. New Phytol. 2016;211:569–583. doi: 10.1111/nph.13922. [DOI] [PubMed] [Google Scholar]
- 34.Choi JY, et al. The rice paradox: Multiple origins but single domestication in Asian Rice. Mol. Biol. Evol. 2017;34:969–979. doi: 10.1093/molbev/msx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng L, et al. Physiological and transcriptome analysis of iron and phosphorus interaction in rice deedlings. Plant Physiol. 2009;151:262–274. doi: 10.1104/pp.109.141051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bashir K, et al. Transcriptomic analysis of rice in response to iron deficiency and excess. Rice. 2014;7:1–15. doi: 10.1186/s12284-014-0018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hindt MN, et al. BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics. 2017;9:876–890. doi: 10.1039/C7MT00152E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long TA, et al. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell. 2010;22:2219–2236. doi: 10.1105/tpc.110.074096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Senoura T, et al. The iron-chelate transporter OsYSL9 plays a role in iron distribution in developing rice grains. Plant Mol. Biol. 2017;95:375–387. doi: 10.1007/s11103-017-0656-y. [DOI] [PubMed] [Google Scholar]
- 40.Bashir K, et al. Iron deficiency regulated OsOPT7 is essential for iron homeostasis in rice. Plant Mol. Biol. 2015;88:165–176. doi: 10.1007/s11103-015-0315-0. [DOI] [PubMed] [Google Scholar]
- 41.Ishimaru Y, et al. OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 2005;56:3207–3214. doi: 10.1093/jxb/eri317. [DOI] [PubMed] [Google Scholar]
- 42.Klaumann S, et al. The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol. 2011;192:393–404. doi: 10.1111/j.1469-8137.2011.03798.x. [DOI] [PubMed] [Google Scholar]
- 43.Nakanishi H, Ogawa I, Ishimaru Y, Mori S, Nishizawa NK. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 2006;52:464–469. doi: 10.1111/j.1747-0765.2006.00055.x. [DOI] [Google Scholar]
- 44.Korshunova Y, Eide D, Gregg Clark W, Lou Guerinot M, Pakrasi H. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 1999;40:37–44. doi: 10.1023/A:1026438615520. [DOI] [PubMed] [Google Scholar]
- 45.Ricachenevsky FK, de Araújo AT, Jr., Fett JP, Sperotto RA. You shall not pass: root vacuoles as a symplastic checkpoint for metal translocation to shoots and possible application to grain nutritional quality. Front. Plant Sci. 2018;9:1–6. doi: 10.3389/fpls.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Xu YH, Yi HY, Gong JM. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012;72:400–410. doi: 10.1111/j.1365-313X.2012.05088.x. [DOI] [PubMed] [Google Scholar]
- 47.Liu T, et al. Evolutionary expansion and functional diversification of oligopeptide transporter gene family in rice. Rice. 2012;5:1–14. doi: 10.1186/1939-8433-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishikawa S, et al. Ion-bead irradiation, gene identification, and marker-assisted breeding in the development of low-cadmiun rice. Proc. Natl. Acad. Sci. 2012;109:19166–19171. doi: 10.1073/pnas.1211132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Markham GD, Hafner EW, Tabor CW, Tabor H. S-Adenosylmethionine synthetase from Escherichia coli. J Biol Chem. 1980;255:9082–9092. [PubMed] [Google Scholar]
- 50.Itai RN, Ogo Y, Kobayashi T, Nakanishi H, Nishizawa NK. Rice genes involved in phytosiderophore biosynthesis are synchronously regulated during the early stages of iron deficiency in roots. Rice. 2013;6:1–13. doi: 10.1186/1939-8433-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heilbronn J, Wilson J, Berger BJ. Tyrosine aminotransferase catalyzes the final step of methionine recycling in Klebsiella pneumoniae. J. Bacteriol. 1999;181:1739–1747. doi: 10.1128/jb.181.6.1739-1747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berger BJ, English S, Chan G, Knodel MH. Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis. J. Bacteriol. 2003;185:2418–2431. doi: 10.1128/JB.185.8.2418-2431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki M, et al. Biosynthesis and secretion of mugineic acid family phytosiderophores in zinc-deficient barley. Plant J. 2006;48:85–97. doi: 10.1111/j.1365-313X.2006.02853.x. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi T, Nishizawa NK. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012;63:131–152. doi: 10.1146/annurev-arplant-042811-105522. [DOI] [PubMed] [Google Scholar]
- 55.Vert G, Briat JF, Curie C. Arabidopsis IRT2 gene encodes a root-periphery iron transporter. Plant J. 2001;26:181–189. doi: 10.1046/j.1365-313x.2001.01018.x. [DOI] [PubMed] [Google Scholar]
- 56.Lee S, An G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant, Cell Environ. 2009;32:408–416. doi: 10.1111/j.1365-3040.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi R, et al. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 2011;62:4843–4850. doi: 10.1093/jxb/err136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishimaru Y, et al. Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Sci. Rep. 2012;2:1–8. doi: 10.1038/srep00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Castaings L, Caquot A, Loubet S, Curie C. The high-affinity metal transporters NRAMP1 and IRT1 team up to take up iron under sufficient metal provision. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep37222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao Q, et al. Genetic diversity and origin of weedy rice (Oryza sativa f. spontanea) populations found in North-eastern China revealed by simple sequence repeat (SSR) markers. Ann. Bot. 2006;98:1241–1252. doi: 10.1093/aob/mcl210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaughan DA, Morishima H, Kadowaki K. Diversity in the Oryza genus. Curr. Opin. Plant Biol. 2003;6:139–146. doi: 10.1016/S1369-5266(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 62.Menguer PK, Sperotto RA, Ricachenevsky FK. A walk on the wild side: Oryza species as source for rice abiotic stress tolerance. Genet. Mol. Biol. 2017;40:238–252. doi: 10.1590/1678-4685-gmb-2016-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li S, et al. Overexpression of ZmIRT1 and ZmZIP3 enhances iron and zinc accumulation in transgenic Arabidopsis. PLoS One. 2015;10:1–21. doi: 10.1371/journal.pone.0136647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pedas P, et al. Manganese efficiency in barley: identification and characterization of the metal ion transporter HvIRT1. Plant Physiol. 2008;148:455–466. doi: 10.1104/pp.108.118851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science (80-.). 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 66.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 67.Ross, C. W. Plant physiology laboratory manual. (Wadsworth, 1974).
- 68.Dametto A, et al. Cold tolerance in rice germinating seeds revealed by deep RNAseq analysis of contrasting indica genotypes. Plant Sci. 2015;238:1–12. doi: 10.1016/j.plantsci.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 69.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 70.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 71.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alexa, A. & Rahnenführer, J. Gene set enrichment analysis with topGO. In http://www.mpi-sb.mpg.de/alexa (2019).
- 73.Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT – PCR. 29, 16–21 (2001). [DOI] [PMC free article] [PubMed]
- 74.Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 75.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:1–14. doi: 10.1093/nar/gkr648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Authors declare that all data and materials used in this work are available to others upon request.