Abstract
Introduction
US claims data contain medical data on large heterogeneous populations and are excellent sources for medical research. Some claims data do not contain complete death records, limiting their use for mortality or mortality-related studies. A model to predict whether a patient died at the end of the follow-up time (referred to as the end of observation) is needed to enable mortality-related studies.
Objective
The objective of this study was to develop a patient-level model to predict whether the end of observation was due to death in US claims data.
Methods
We used a claims dataset with full death records, Optum© De-Identified Clinformatics® Data-Mart-Database—Date of Death mapped to the Observational Medical Outcome Partnership common data model, to develop a model that classifies the end of observations into death or non-death. A regularized logistic regression was trained using 88,514 predictors (recorded within the prior 365 or 30 days) and externally validated by applying the model to three US claims datasets.
Results
Approximately 25 in 1000 end of observations in Optum are due to death. The Discriminating End of observation into Alive and Dead (DEAD) model obtained an area under the receiver operating characteristic curve of 0.986. When defining death as a predicted risk of > 0.5, only 2% of the end of observations were predicted to be due to death and the model obtained a sensitivity of 62% and a positive predictive value of 74.8%. The external validation showed the model was transportable, with area under the receiver operating characteristic curves ranging between 0.951 and 0.995 across the US claims databases.
Conclusions
US claims data often lack complete death records. The DEAD model can be used to impute death at various sensitivity, specificity, or positive predictive values depending on the use of the model. The DEAD model can be readily applied to any observational healthcare database mapped to the Observational Medical Outcome Partnership common data model and is available from https://github.com/OHDSI/StudyProtocolSandbox/tree/master/DeadModel.
Electronic supplementary material
The online version of this article (10.1007/s40264-019-00827-0) contains supplementary material, which is available to authorized users.
Key Points
| Death can be incompletely recorded in US claims data and this can limit drug safety studies that use these datasets. |
| We present a model that can predict whether the end of observation was due to death in US claims data with a discriminative performance of 0.986 on the area under the receiver operating characteristic curve. |
| The model is available online and can be readily applied to any dataset in the Observational Medical Outcomes Partnership common data model. |
Introduction
Large observational healthcare datasets can be utilized by epidemiologists to learn new insights about disease and the effects of medical interventions in a real-world setting where patient populations are more heterogeneous than in randomized clinical trials [1]. They are essential for learning about rare or delayed outcomes [2]. Studies have shown that well-designed epidemiologic studies using observational data can yield similar results to randomized clinical trial data [3], the gold standard for epidemiological analysis. Administrative claims databases are particularly valuable for pharmacoepidemiologic research, as they commonly represent a closed population with a defined period of eligibility, during which time there is strong confidence in the capture of covered inpatient and outpatient medical services and outpatient pharmacy dispensing records, which allows for inferring associations between drug exposure and outcome incidence that can be defined by diagnostic or procedure codes. Unfortunately, one outcome of particular interest is not consistently available, which limits the utility of these data: mortality. End of observation, that is the time when a person is no longer followed in claims data can occur for multiple reasons, e.g., it was the cut-off calendar date for all patients in the database, the person changed insurance coverage, or the person died, but not all claims databases offer sufficient information to know the reason for each patient with complete confidence. The claims data used in this study do not provide reasons for the end of enrollment. When a patient is no longer observed, it could be because they discontinued/switched insurance or because he/she is deceased.
Many US insurance claims databases only have death at discharge recorded, thus people who die outside the hospital are unlikely to have their death recorded. If methods could be developed to impute death into claims data, then these large datasets could be used to generate additional evidence in mortality-related studies.
There are various comorbidity models that can be implemented for claims data, with the most commonly used being the Charlson Comorbidity Index [4] and the Elixhauser Comorbidity Index [5]. These models have been shown to be predictive of mortality [6], even across non-US and non-claims data [7]. However, these models are prognostic, thus they have not been developed to impute death, instead, they predict future mortality risk.
The Optum© De-Identified Clinformatics® Data Mart Database—Date of Death claims dataset has complete death records up to 2013. The objective of this study was to use this dataset to develop and validate a model to predict whether a patient’s end of observation is due to death. We then investigate whether the model can be used to impute death in other US claims datasets that lack complete death records.
Methods
We followed the Observational Healthcare and Data Science Informatics (OHDSI) Patient Level Prediction framework [8] to develop and validate the model. The framework is available as an open source R package and implements a process for developing patient-level prediction models while addressing existing best practices towards ensuring models are clinically useful and transparent. The datasets used for development and validation are described below.
All the datasets were mapped to the Observational Medical Outcome Partnership common data model [9], as having the datasets in a homogeneous data structure enables re-use of the code between model development and validation to ensure the model can be externally validated efficiently and reduces model reproducibility errors. The use of IBM MarketScan® and Optum databases was reviewed by the New England Institutional Review Board and was determined to be exempt from broad institutional review board approval. We share all the code required to perform the analysis in this article or apply the developed model on new data via online supplements or on github (https://github.com/OHDSI/StudyProtocolSandbox/tree/master/DeadModel).
Development Dataset
The Optum© De-Identified Clinformatics® Data Mart Database—Date of Death (Optum DOD)—Optum© Clinformatics® Extended Data Mart (Eden Prairie, MN, USA) is an adjudicated US administrative health claims database for members of private health insurance. The database contains insurance claims data (inpatient/outpatient medical conditions and drug dispensing plus some laboratory data) for US patients aged between 0 and 90 years. In this dataset, each state had a mandatory requirement to report patient deaths up until 2013, after 2013, it was no longer mandatory and therefore death may not be complete from 2013 onwards. The DOD table is sourced from the Death Master File maintained by the Social Security Office. The DeathMaster data provide year and month of death only. The dataset contains 73,969,539 individuals with data recorded between 1 May, 2000 and 31 March, 2016.
Validation Datasets
The IBM MarketScan® Medicare Supplemental Database (MDCR) represents health services of retirees in USA with primary or Medicare supplemental coverage through privately insured fee-for-service, point-of-service, or capitated health plans. These patients are aged 65 years or older and have additional private insurance and are thus likely to be more affluent than average. These data include adjudicated health insurance claims (e.g., inpatient, outpatient, and outpatient pharmacy). The database contains 9,559,877 individuals with data recorded between 1 January, 2000 and 30 April, 2016.
The IBM MarketScan® Multi-State Medicaid Database (MDCD) contains adjudicated US health insurance claims for Medicaid enrollees from multiple states and includes hospital discharge diagnoses, outpatient diagnoses and procedures, and outpatient pharmacy claims as well as ethnicity and Medicare eligibility. Patients in the MDCD have subsidized health insurance and are aged less than 65 years. The database contains 21,577,517 individuals with data recorded between 1 January, 2006 and 31 December, 2014.
The IBM MarketScan® Commercial Database (CCAE) represents data from individuals enrolled in US employer-sponsored insurance health plans. The data include adjudicated health insurance claims (e.g., inpatient, outpatient, and outpatient pharmacy) as well as enrollment data from large employers and health plans who provide private healthcare coverage to employees, their spouses, and dependents. The patients in the CCAE are aged 65 years or younger. The database contains 131,533,722 individuals with data recorded between 1 January, 2000 and 30 April, 2016.
The Optum DOD development dataset contains complete death records (death at discharge and all other deaths) up to 2013 and contains older and younger patients. The IBM datasets used for validation only have death at discharge (when a patient dies in hospital) recorded. The validation datasets also tend to have limited patient age ranges (e.g., older than 65 years or younger than 65 years). The databases capture all inpatient and outpatient medical services and outpatient pharmacy claims, which are submitted through the payer. This generally would include claims submitted and subsequently adjudicated to be reimbursed by a secondary payer as co-insurance. For example, the MDCR database includes claims from services reimbursed by national Medicare coverage in addition to the supplemental coverage.
Target Population
The target population for model development was defined as patients with an end of observation in Optum DOD data between 2011-01-01 or 2012-11-01 who had been in the database for at least 1 year prior to their end of the observation date. The target population index date was the end of the observation date (the date they left the database).
We chose to use the end of observation between 2011-01-01 or 2012-11-01 as we investigated death up to 61 days afterwards and death records were complete in the Optum DOD during this period. Since 2013, the death records have become incomplete.
Outcome
A patient in the target population was classified as ‘dead’ if they had a death record within 61 days of the end of observation. To review the SQL code used to create the datasets, see Electronic Supplementary Material (ESM) 1.
Predictors
The model variables were constructed using the records on or prior to the target population index date. The variables included were:
Age in 5-year groups (e.g., 0–5, 5–10).
Sex.
Month of target index.
Conditions (singular and grouped using a vocabulary hierarchy) in prior 365 days and prior 30 days;
Drugs (singular and grouped into ingredients) in prior 365 days and prior 30 days.
Procedures in prior 365 days and prior 30 days.
Measurements in prior 365 days and prior 30 days.
Observations in prior 365 days and prior 30 days.
Healthcare utility in prior 365 days and prior 30 days.
Note, discharge status (which indicates patients who died while in the hospital) was not included as a predictor in the model. We chose to only use the records that occurred in the prior 365 days to allow for the death risk model to be readily applied to any other dataset with at least 1 year of observation.
Statistical Analysis Methods
The development dataset was split into a training set (75% of the data) and a testing set (25% of the data) to perform an internal validation of the model. The chosen classifier was a regularized logistic regression with lasso regularization [10] and the hyper-parameter controlling the amount of regularization was acquired using an adaptive search and three-fold cross validation on the train set.
To internally evaluate the model, the model discrimination on the test set was assessed using the area under the receiver operating characteristic curve (AUC) and the model calibration was assessed by inspecting a calibration plot generated by binning the patients into ten groups based on their predicted death risk and comparing the observed fraction of the group with a death record observed around the end of observation vs. the mean predicted death risk for the group. To investigate whether the model is reproducible or transportable across US claims data, an external validation was implemented across three US claims datasets. However, in the other claims datasets, only death at discharge is recorded; therefore, to externally validate the model we applied it to a set of patients where the patients had a death at discharge recorded (only a subset of actual deaths) or had an end of observation followed by a future observation period (were definitively alive). If a patient had a death at discharge recorded they were defined as dead. For more details see ESM 2.
Results
Data Summary
A random sample of 1,000,000 patients in Optum DOD with an observation period ending within 2011-01-01–2012-11-01 was selected as the target population and for 24,531 of these patients, death was recorded within 61 days of the observation end date (2.45%). The median and mean time between the recorded death and the end of observation was 0 and 3 days, respectively, and 91% of deaths were at the end of the observation date. The characteristics of the sampled target population for the demographics and key predictors of death are presented in Table 1.
Table 1.
Characteristics of development target population and validation datasets
| Characteristic | Development dataset | Validation datasets | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OPTUM DOD test/train | OPTUM validation | CCAE | MDCD | MDCR | ||||||
| Non dead | Dead (all deaths) | Non dead | Dead (death at discharge) | Non dead | Dead (death at discharge) | Non dead | Dead (death at discharge) | Non dead | Dead (death at discharge) | |
| % (n = 975,469) | % (n = 24,531) | % (n = 389,080) | % (n = 996) | % (n = 409,238) | % (n = 1000) | % (n = 289,870) | % (n = 989) | % (n = 456,399) | % (n = 960) | |
| Sex: female | 50.6 | 49.8 | 50.1 | 42.3 | 51.7 | 45 | 60.5 | 63.8 | 51.9 | 47.2 |
| Acute respiratory disease | 26.2 | 48.0 | 19.6 | 40.8 | 21.6 | 58.3 | 25.3 | 50.2 | 15.2 | 57 |
| Chronic liver disease | 1 | 4.4 | 0.5 | 3 | 0.5 | 9.2 | 0.4 | 6.1 | 0.7 | 1.9 |
| Chronic obstructive lung disease | 2.1 | 36.5 | 1.1 | 29.8 | 0.6 | 22.8 | 1.6 | 38.4 | 7.2 | 38.2 |
| Dementia | 0.8 | 23.7 | 0.3 | 14.8 | 0.1 | 1.8 | 0.5 | 31.2 | 2.2 | 16.7 |
| Hypertensive disorder | 18.3 | 76.0 | 10 | 73.1 | 10 | 50.4 | 6.9 | 70.8 | 44.9 | 64.5 |
| Obesity | 4.1 | 7.6 | 2.2 | 11.4 | 2.2 | 9.9 | 2.9 | 10.9 | 2.6 | 4 |
| Osteoarthritis | 8.0 | 34.9 | 4.3 | 35.3 | 4.2 | 18 | 2.7 | 30.1 | 19.3 | 27.4 |
| Pneumonia | 2.0 | 39.2 | 1.2 | 25.6 | 1.1 | 36 | 2 | 38.7 | 3.3 | 44.7 |
| Renal impairment | 2.0 | 46.6 | 0.8 | 34.2 | 0.4 | 35.9 | 1 | 43.5 | 5.3 | 44.7 |
| Heart disease | 8.3 | 75.9 | 4.6 | 79.3 | 4 | 71.1 | 3.9 | 76.1 | 29.4 | 87.4 |
| Heart failure | 1.4 | 42.0 | 0.7 | 34.4 | 0.3 | 23.9 | 1 | 41.4 | 5.2 | 52 |
| Malignant neoplastic disease | 3.7 | 37.5 | 1.8 | 20.3 | 1.7 | 53.5 | 0.8 | 18.9 | 13.9 | 31.3 |
| Characteristic | Value | Value | Value | Value | Value | Value | Value | Value | Value | Value |
| Charlson Comorbidity Index | ||||||||||
| Mean | 0.9 | 7.5 | 0 | 6 | 0 | 7 | 0 | 6 | 2 | 7 |
| Age, years | ||||||||||
| Mean | 37.4 | 75.3 | 33 | 70 | 32 | 54 | 20 | 70 | 73 | 82 |
CCAE IBM MarketScan® Commercial Database, DOD date of death, MDCD IBM MarketScan® Multi-State Medicaid Database, MDCR IBM MarketScan® Medicare Supplemental Database, OPTUM Optum© De-Identified Clinformatics® Data Mart Database
Table 1 presents the baseline characteristics of the development datasets and validation datasets. Illnesses that are the most associated with death included cardiac diagnoses, disorders related to old age, and neoplasms. The Charlson Comorbidity Index was on average 7 for those with a death around the end of the observation whereas it was 1 or less for people with an end of observation but no death recorded, except for the MDCR where it was 2, as the patients are older and sicker in that database. An extended version of Table 1 with more variables is available, see ESM 4.
Model Specification
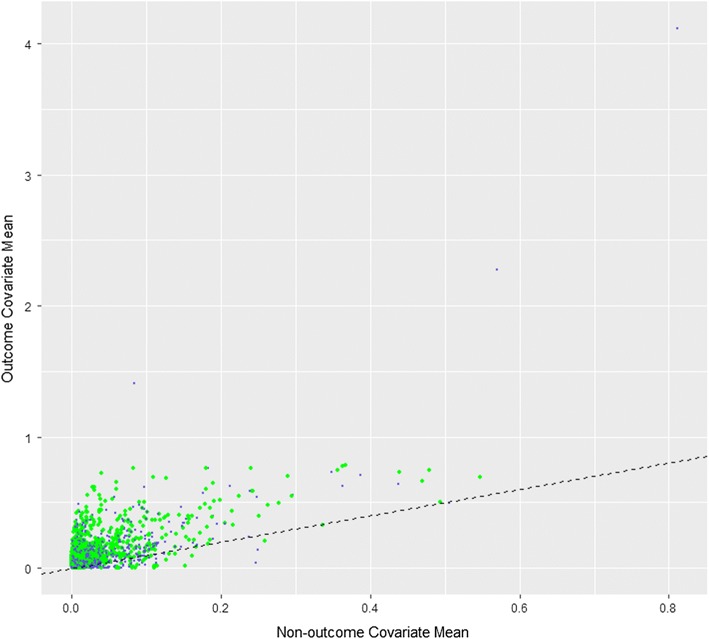
The Discriminating End of observation into Alive and Dead (DEAD) model developed in Optum DOD with a target size of 1,000,000 and an outcome count of 24,531 is described in ESM 5 and is available from https://github.com/OHDSI/StudyProtocolSandbox/tree/master/DeadModel. The model can be interactively explored at http://data.ohdsi.org/DeadImputation/. Of 88,514 candidate predictors, 2097 were selected into the final model. The trained model’s coefficient values are available, see ESM 5. Figure 1 is a visualization showing the differences between patients with an end of observation due to death and those without. It is a scatter plot with the mean value for each of the 88,514 variables within patients whose end of observation is not due to death (x axis) and patients whose end of observation is due to death (y axis). Variables that do not discriminate whether an end of observation is due to death or not fall on the diagonal. Those that fall above the diagonal are more common in patients whose end of observation was due to death. Those that fall below the diagonal are less common in patients whose end of observation was due to death.
Fig. 1.
Scatter plot of variable means for people with death recorded within 61 days of the end of observation (y axis) vs. people without death recorded within 61 days of the end of observation (x axis). Green points correspond to variables included in the trained model and blue dots are variables that were not included in the trained model
Model Performance
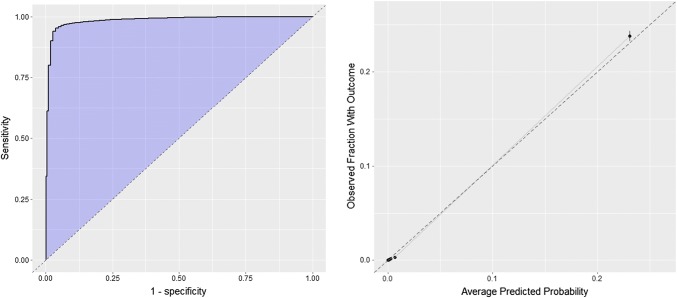
The internal validation of the DEAD model obtained an AUC of 0.986 on the test set (0.989 on the train set), the receiver operating characteristic curve plot is presented in Fig. 2. The calibration plot for the internal validation of the model is presented in Fig. 2.
Fig. 2.
Receiver operating characteristic curve and calibration plots for the internal validation
The DEAD model can be used to identify patients with an end of observation likely to be due to death or patients with an end of observation unlikely to be due to death. The required sensitivity, specificity, and positive predictive value (PPV) of a model is often context dependent, thus we present various predicted risk cut-offs in Tables 2 and 3. If a user wishes to find an end of observation due to death, then Table 2 should be used. If a study requires definitive deaths, the user may require a high specificity of 99.9%, thus he/she can go down the specificity of the death column to find the cell with 99.9 and then move to the prediction threshold column to find the cut-off value to use, 0.905 in this example. He/she would then select the end of observation with a risk of 0.905 or higher as being due to death, this would have a PPV of 87.2% but only have a sensitivity of 25.6%. If a user wished to find 50% of all deaths (50% sensitivity), then the prediction threshold corresponds to a cut-off of 0.66. This threshold suggests that classifying a risk ≥ 0.66 as being due to death would have a PPV of 79.5% and a specificity of 99.7%. Alternatively, a user may wish to identify the end of observations that are unlikely to be due to death, thus he/she would use Table 3. If he/she wanted to find the end of observations that are not due to death with a specificity of 99.9%, then he/she would use Table 3 to scroll down the specificity of the alive column to the cell with 99.9, this would indicate a prediction threshold of 0.00055 and then he/she could select the end of observations with a risk less than 0.00055 and class these as not being due to death. This would result in a PPV of 99.993% and a sensitivity of 34.6% (i.e., would identify 34.6% of all end of observations not due to death).
Table 2.
Results at various prediction threshold cut-offs that can be used to select the threshold used by any future epidemiology study when selecting an end of observations due to death
| Finding people who are dead … | |||||
|---|---|---|---|---|---|
| Prediction threshold | Sensitivity of death | Specificity of death | Positive predictive value of death | Proportion of target population | |
| If you choose by prediction threshold to be greater than … | |||||
| 0.9 | 26.170 | 99.901 | 86.898 | 0.007 | |
| 0.5 | 61.895 | 99.474 | 74.754 | 0.020 | |
| 0.1 | 90.282 | 98.149 | 55.095 | 0.040 | |
| If you choose by sensitivity … | |||||
| 0.666 | 50 | 99.676 | 79.549 | 0.015 | |
| 0.1046 | 90 | 98.184 | 55.489 | 0.040 | |
| 0.0019 | 99 | 69.321 | 7.510 | 0.324 | |
| If you choose by specificity… | |||||
| 0.253 | 78.608 | 99 | 66.515 | 0.029 | |
| 0.905 | 25.599 | 99.9 | 87.174 | 0.007 | |
| 0.990 | 7.207 | 99.99 | 95.054 | 0.002 | |
Table 3.
Results at various prediction threshold cut-offs that can be used to select the threshold used by any future epidemiology study when selecting the non-death end of observations
| Finding people who are still alive…. | |||||
|---|---|---|---|---|---|
| Prediction threshold | Sensitivity of alive | Specificity of alive | Positive predictive value of alive | Proportion of target population | |
| If you choose by prediction threshold less than … | |||||
| 0.5 | 99.474 | 61.895 | 99.046 | 0.980 | |
| 0.1 | 98.149 | 90.282 | 99.752 | 0.960 | |
| 0.01 | 91.351 | 97.196 | 99.923 | 0.892 | |
| If you choose by sensitivity … | |||||
| 0.00103 | 50 | 99.658 | 99.983 | 0.488 | |
| 0.0085 | 90 | 97.375 | 99.927 | 0.879 | |
| 0.252 | 99 | 78.624 | 99.460 | 0.971 | |
| If you choose by specificity … | |||||
| 0.104 | 98.177 | 90 | 99.747 | 0.960 | |
| 0.00196 | 70.264 | 99 | 99.964 | 0.686 | |
| 0.00055 | 34.636 | 99.9 | 99.993 | 0.338 | |
The external validation on CCAE, MDCR, and MDCD returned AUCs of 0.995, 0.951, and 0.977, respectively. Full details for the external validation models and results, see the ESM 2.
Differences between the patients the model incorrectly, but confidently, predicted were dead compared to patients the model incorrectly predicted were alive can be seen in ESM 3. As expected, if a patient did not have obvious health issues, then the model struggled to predict the patient as dead, whereas if a patient had many serious comorbidities the model was likely to predict an end of observation as due to death.
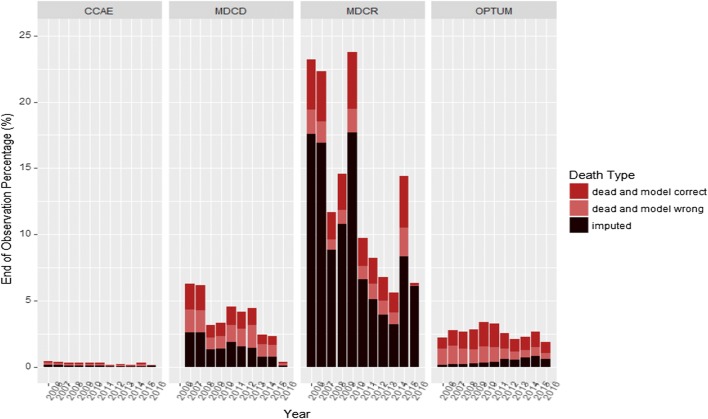
Figure 3 shows the recorded deaths (death identified by model, death not identified by model) and imputed death (where the DEAD model predicted the risk of death ≥ 0.5 but the patient did not have death recorded) for all end of observations where the patient is never seen again in the data. This is broken down by year between 2006 and 2016 in all databases.
Fig. 3.
Percentage of the final end of observation per year that are due to death or imputed as due to death by the DEAD model for each databases across the years 2006–16. CCAE IBM MarketScan® Commercial Database, MDCD IBM MarketScan® Multi-State Medicaid Database, MDCR IBM MarketScan® Medicare Supplemental Database, OPTUM Optum© De-Identified Clinformatics® Data Mart Database
The trends in Fig. 3 are as expected and provide face validity of the DEAD model. As CCAE is a database for employed patients aged 65 years or younger and their dependents, these patients are less likely to stop being observed in the database due to death, with more probable causes being changing insurance provider or job. Alternatively, the MDCR contains an older patient population, thus we would expect to see a higher percentage of patients leaving the database due to death. The imputation rate trend seen in Optum DOD (the imputation rate is initially low but increases until 2013 where it seems to stabilize) also makes sense as we know in 2013 onwards it was not mandatory for states to report deaths, thus not all deaths are likely to be recorded after 2013 and therefore the model will need to impute more. In CCAE/MDCD and MDCR, the imputation rate appears to be fairly stable over time, this is because these datasets only contain deaths at discharge, thus the deaths outside of the hospital will be identified by the DEAD model.
Discussion
Interpretation
The discriminative ability of the DEAD model was excellent in obtaining an internal AUC of 0.986 and external AUCs between 0.951 and 0.995, indicating the model can distinguish between end of observation due to death and end of observation due to non-death reasons. As the death records are incomplete in the other US claims databases, the external validation used death at discharge or non-death cases, but this may result in an optimistic AUC.
The results show the model is also well calibrated on the development dataset. It is difficult to access calibration on the external validation datasets because of these datasets being constructed of definitive death and non-deaths, thus extreme predictions (close to 1 or 0) may be more common. To determine whether the model needs to be recalibrated on new datasets, it will be useful to validate the model on another dataset that has death well recorded.
The most predictive variables were age, hospital discharge records (indicating an inpatient visit), and cardiovascular- or cancer-related diagnoses. These variables are known to be linked to death and are therefore expected. The month that the end of observation occurred (e.g., December vs. January) was also informative in predicting death. This may be oiwng to insurance policies often ending at the same time, thus a certain end of observation month may indicate non-death and therefore decrease the risk of the end of observation being due to death.
Implications
The results showed that developing a model using a claims dataset with complete death records for identifying which end of observations were due to death or non-death reasons resulted in a good discriminative ability and this model was validated across several datasets and showed a consistently high externally validated AUC. This suggests the model could be a useful tool to identify death in US claims data and this would be useful for epidemiology studies. The suitable risk threshold chosen to classify end of observations into death vs. non-death would depend on the study. Studies that want to be confident that the end of observation predicted to be death would truly be dead should use a higher threshold such as 0.9, whereas studies that want to capture as many deaths as possible and are less interested in the false-positive rate should use a lower threshold such as 0.10. We provided Table 2 that can guide the user into a suitable threshold to use. For example, if the user wanted a group of patients very likely to be dead then he/she can pick all patients with a DEAD risk score greater than 0.9, whereas if he/she wanted a group of patients very likely to be alive, then he/she could select all patients with a DEAD risk score less than 0.001.
The model can be used to impute death by applying it at the point in time when a patient stops being observed in a database (the end of observation date). If a patient is assigned a sufficiently high risk value by DEAD, then they can be considered to have died on the date their observation ended. If you want to identify the majority of true deaths, then a risk threshold of 0.1046 would identify 90% of all deaths (anyone who has a DEAD risk of 0.1046 or more on the date they leave the databases are considered to have died on the date they leave the database and otherwise they are considered to still be alive). If you want to identify a set of patients likely to be dead, then a risk threshold of 0.9 would identify approximately 26% of all deaths with a high specificity and PPV of 86.9%.
At prediction thresholds > 0.9 or specificity thresholds > 0.999, the performance of this death phenotype compares favorably with phenotypes developed for other conditions. For example, Rubbo et al. performed a systematic review of acute myocardial infarction phenotype validations, which estimated PPVs between 88 and 92% [11]. Kumamaru et al. estimated a 90% PPV for ischemic stroke [12]. Many published definitions only present PPV with sensitivity an unknown. In addition, our definition is a model rather than a set of codes, thus the user can pick the operating characteristic that best suits the application (e.g., high PPV and low sensitivity or high sensitivity and lower PPV).
Limitations
The main limitation of this study is that death is not well recorded in the datasets used to validate the model (the validation data only have death at discharge). This is a predicament, as the lack of complete death records in observational data prompted this research but also makes validation difficult. The results show the DEAD model appears to transport to other US claims datasets. However, it would be beneficial to validate it on more datasets, including as non-US and non-claims data, to gain additional confidence into the transportability. As the model has been developed on the Observational Medical Outcome Partnership common data model, it is possible in future work to design a network study to share the model with other researchers who may have complete death records.
Because the model training was performed on data prior to 2013 and no complete validation data were available in later years, we cannot assure the external validity, and changes in healthcare practice and data coding strategies (such as the International Classification of Diseases, Ninth Revision, Clinical Modification to the International Classification of Diseases, Tenth Revision, Clinical Modification transition) could impact its performance. Additionally, the requirement of looking for a death record within 61 days of an end of observation to classify it as being due to death may result in some misclassification where an end of observation is incorrectly labeled as not being due to death (e.g., the death record date was more than 61 days before/after the observation end). This was a trade-off between using a longer time period around an end of observation that would increase the chance of misclassification where an end of observation not due to death is incorrectly labeled as being due to death.
Conclusion
In this article, we developed the DEAD model that determines whether an end of observation in US claims data is due to death. The model was developed using a US claims database with complete death records, thus the target population was individuals with an end of observation and the outcome was an end of observation due to death. The model obtained a discrimination performance AUC of 0.986 on the internal validation and AUCs ranging from 0.951 to 0.995 on the external validation. The internal validation suggested the model was well calibrated but further application of the model to more datasets with complete death records is required to access the external calibration of the model. Mortality is often an outcome of interest in epidemiological studies but is often poorly recorded in US claims data. The model developed in this article can now be implemented to impute death in US claims data at various sensitivities or specificities. In the future, it would be useful to extend the external validation across the OHDSI network and outside the OHDSI network, so the performance can also be evaluated on non-US or non-claims data.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Gayle Murray for her editorial review.
Compliance with Ethical Standards
Funding
No sources of funding were used to assist in the preparation of this study.
Conflict of interest
Jenna Reps is an employee of Janssen Research & Development and a shareholder of Johnson & Johnson. Patrick Ryan is an employee of Janssen Research & Development and a shareholder of Johnson & Johnson. Peter Rijnbeek works for a research group who received unconditional research grants from Boehringer-Ingelheim, GSK, Janssen Research & Development, Novartis, Pfizer, Yamanouchi, and Servier. None of these grants result in a conflict of interest with the content of this paper.
References
- 1.Mariani AW, Pego-Fernandes PM. Observational studies: why are they so important? Sao Paulo Med J. 2014;132(1):1–2. doi: 10.1590/1516-3180.2014.1321784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black N. Why we need observational studies to evaluate the effectiveness of health care. BMJ. 1996;312(7040):1215–1218. doi: 10.1136/bmj.312.7040.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Concato J, Shah N, Horwitz RI. Randomized, controlled trials, observational studies, and the hierarchy of research designs. N Engl J Med. 2000;342(25):1887–1892. doi: 10.1056/NEJM200006223422507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlson ME, Pompei P, Ales KA, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 5.Elixhauser A, Steiner C, Harris R, Coffey R. Comorbidity measures for use with administrative data. Med Care. 1998;36:8–27. doi: 10.1097/00005650-199801000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Sharabiani M, Aylin P, Bottle A. Systematic review of comorbidity indices for administrative data. Med Care. 2012;50:1109–1118. doi: 10.1097/MLR.0b013e31825f64d0. [DOI] [PubMed] [Google Scholar]
- 7.Gutacker N, Bloor K, Cookson R. Comparing the performance of the Charlson/Deyo and Elixhauser comorbidity measures across five European countries and three conditions. Eur J Public Health. 2015;25:15–20. doi: 10.1093/eurpub/cku221. [DOI] [PubMed] [Google Scholar]
- 8.Reps JM, Schuemie MJ, Suchard MA, Ryan PB, Rijnbeek PR. Design and implementation of a standardized framework to generate and evaluate patient-level prediction models using observational healthcare data. J Am Med Inf Assoc. 2018;25(8):969–975. doi: 10.1093/jamia/ocy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overhage JM, Ryan PB, Reich CG, et al. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19(1):54–60. doi: 10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suchard MA, Simpson SE, Zorych I, Ryan P, Madigan D. Massive parallelization of serial inference algorithms for a complex generalized linear model. ACM Trans Model Comput Simul. 2013 doi: 10.1145/2414416.2414791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubbo B, Fitzpatrick NK, Denaxas S, Daskalopoulou M, Yu N, Patel RS, et al. Use of electronic health records to ascertain, validate and phenotype acute myocardial infarction: a systematic review and recommendations. Int J Cardiol. 2015;187:705–711. doi: 10.1016/j.ijcard.2015.03.075. [DOI] [PubMed] [Google Scholar]
- 12.Kumamaru H, Judd SE, Curtis JR, Ramachandran R, Hardy NC, Rhodes JD, et al. Validity of claims-based stroke algorithms in contemporary Medicare data: reasons for geographic and racial differences in stroke (REGARDS) study linked with Medicare claims. Circ Cardiovasc Qual Outcomes. 2014;7:611–619. doi: 10.1161/CIRCOUTCOMES.113.000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.