Abstract
The potential for genetic variation to cause adult unconjugated hyperbilirubinemia is increasingly being recognized. However, the cumulative effects of genetic variants have not been fully illuminated. The current study aimed to investigate the effects of uridine diphospho-glucuronosyl transferase 1A1 (UGT1A1) and/or solute carrier organic anion transporter family member 1B (SLCO1B) polymorphic variants and their combined effects on mild unconjugated hyperbilirubinemia in Chinese adults. Fourteen genetic variants in the UGT1A1 or SLCO1B gene were genotyped through sequencing in 148 adults with unconjugated hyperbilirubinemia and 158 healthy controls. Variants c.-3275T > G, (TA)6>(TA)7, c.211G > A or c.1091C > T within the UGT1A1 gene as well as c.521T > C within the SLCO1B1 gene appear to be genetic risk factors for inherited unconjugated hyperbilirubinemia. After adjusting for covariates, the results of multivariate logistic regressions revealed that odds ratios (ORs) [(with 95% confidence interval (CI)] of these five variants were 2.35 (95% CI: 1.37–4.01, p = 0.002), 2.38 (95% CI: 1.35–4.20, p = 0.003), 2.99 (95% CI: 1.71–5.21, p < 0.001), 7.60 (95% CI: 1.99–28.96, p = 0.003), and 2.54 (95% CI: 1.27–5.11, p = 0.009), respectively. The OR for unconjugated hyperbilirubinemia is positively correlated with the cumulative number of these five variants in adults. And the greater the number of genetic variations, the higher the total bilirubin level. Adults carrying diplotype 3/4 (homozygous c.-3275T > G and heterozygous (TA)6>(TA)7) had higher bilirubin levels than those with diplotypes 1/3 (heterozygous c.-3275T > G and (TA)6>(TA)7)) or 1/4 (heterozygous c.-3275T > G) (P < 0.05). Similarly, bilirubin levels in individuals with diplotype 2/4 (heterozygous c.-3275T > G and c.211G > A) were higher than adults carrying diplotypes 1/2 (heterozygous c.211G > A) or 1/4 (P < 0.001). For subjects with heterozygous or homozygous variant c.211G> A, as the number of c.521T > C alleles variation increased, the incidence of unconjugated hyperbilirubinemia increased, but it was not statistically significant. Our results indicate that variants of UGT1A1 and/or SLCO1B1 have combined effects on Chinese adult mild unconjugated hyperbilirubinemia.
Keywords: unconjugated hyperbilirubinemia, uridine diphospho-glucuronosyl transferase 1A1, solute carrier organic anion transporter family member 1B, variant, combined effect
Introduction
Bilirubin is produced mainly by the turnover of red blood cells and then transported to the liver via the solute carrier organic anion transporter family member 1B (SLCO1B) (Tiribelli and Ostrow, 1993). In the liver, Uridine diphospho-glucuronosyl transferase 1A1 (UGT1A1) catalyzes the binding of unconjugated bilirubin and glucuronic acid molecules into hydrophilic form that is excreted into bile (Clarke et al., 1997).
Several studies have demonstrated the potential contribution of genetic factors to human serum unconjugated bilirubin levels (Johnson et al., 2009; Dai et al., 2013). Among the genes involved in bilirubin metabolism, UGT1A1 gene is considered to be a major factor controlling serum bilirubin levels (Johnson et al., 2009). Polymorphisms in the promoter and coding region of the UGT1A1 gene often underline Gilbert Syndrome (GS), a mild unconjugated hyperbilirubinemia condition (Chiddarwar et al., 2017; Wagner et al., 2018). In Caucasian and African populations, the most common polymorphism of UGT1A1 is an additional TA repeat in the TATA box region of the promoter, i.e., A(TA)7TAA (35.7–41.5%) (Beutler et al., 1998; Huang et al., 2004; Erlinger et al., 2014), while the predominant variation in Asians is a missense mutation, c.211G > A (p.G71R) (11–33.2%) (Chen et al., 2014; Chiddarwar et al., 2017; Yanagi et al., 2017). An exome-wide association study demonstrated that UGT1A1 intronic variations may also reduce UGT1A1 enzyme activity and cause unconjugated hyperbilirubinemia (Oussalah et al., 2015). Moreover, the compound heterozygous UGT1A1 variant has also been reported in GS (Skierka et al., 2013), suggesting that UGT1A1 variant may have a cumulative effect on unconjugated hyperbilirubinemia. However, the evidence for this issue is limited.
Another liver-expressing gene, SLCO1B, which is closely related to the transportation of bilirubin and organic anions into hepatocytes, has also been found to play a role in bilirubin homeostasis (D'Silva et al., 2014). Some genome-wide association studies (GWAS) suggested that polymorphisms in SLCO1B1 and SLCO1B3 reached genome-wide significance associated with bilirubin levels (Johnson et al., 2009; Sanna et al., 2009; Kang et al., 2010; Dai et al., 2013). Further, SLCO1B1 variations c.388G > A (p.N130D, rs2306283) and c.521T > C (p.V174A, rs4149056) were also found to be risk factors for neonatal unconjugated hyperbilirubinemia (Huang et al., 2004; D'Silva et al., 2014). But the relationship between these SLCO1B polymorphisms and unconjugated hyperbilirubinemia in adults is unclear. Furthermore, the genetic basis, especially the combined effects of UGT1A1 and SLCO1B genes for unconjugated hyperbilirubinemia in Chinese descendants has not been fully illuminated. This study aims to determine the potential effects of liver-expressing UGT1A1 and SLCO1B polymorphic variants and their combined effects on mild non-hemolytic unconjugated hyperbilirubinemia.
Patients and Methods
Study Subjects
This is a retrospective case-control study. Adult patients who attended routine health checkups at the second affiliated hospital of Luohe Medical College from June to July in 2017, and with the bilirubin detection results of predominantly unconjugated hyperbilirubinemia (total bilirubin ranged from 17.1 to 85 µmol/l), were enrolled in this study. The 158 control subjects were enrolled from the same health examination people with bilirubin values of less than 17.1 µmol/l. Blood samples were taken in 10–12 h fasting state in the morning and the subjects were kept from strenuous exercise. And clinical records including the gender, age as well as biochemical test results were collected. Exclusion criteria were: (1) a history of chronic liver disease (viral hepatitis, autoimmune liver disease, or cirrhosis and so on); (2) abnormal liver functions (alanine aminotransferase (ALT) or aspartate aminotransferase (AST) > 50 U/L); (3) hemolysis signs (hemoglobin <100 g/L, positive Coomb's test); (4) evidence of infection or biliary obstruction or pregnancy; (5) a history of drug use within 6 months.
This Study Was Approved by the Ethics Committee of Beijing You an Hospital, Capital Medical University and a Written Informed Consent Form Was Obtained From All Participants.
Molecular Analysis
Genomic DNA was extracted from whole blood using a QIAamp DNA Blood Mini Kit 51106 (QIAGEN, Germany) according to the manufacturer's recommended protocol. The promoter, all 5 exons, exon–intron boundaries, and a region in the distal promoter (the phenobarbital response enhancer module, PBREM) of UGT1A1 were PCR amplified and sequenced, as reported before (Huang et al., 2004). Similarly, the four known SLCO1B polymorphisms (388A > G and 521T > C within the SLCO1B1 gene, IVS8+2087T > C and g.21074122C > T within the SLCO1B3 gene) were investigated. The PCR products were purified from agarose gel and sequenced via an ABI3730XL sequencer (Applied Biosystems, Foster City, CA, USA).
Selection of SNPs was done based on their association with serum bilirubin levels as well as their prevalence in Asia population (Huang et al., 2005; Kang et al., 2010; Zhang et al., 2012; Dai et al., 2013; Chiddarwar et al., 2017). Finally, a total of 14 SNPs (10 of UGT1A1 and 4 of SLCO1B) were selected. Analysis of linkage disequilibrium (LD) and haplotype were performed by using SNPStats website (https://www.snpstats.net/start.htm). Strong LD was defined as both |D′| and r2 > 0.8.
Statistical Analysis
Continuous variables were expressed as mean and standard deviation (SD) and then statistically evaluated by using Student's t-test. Categorical variables were compared by using the chi-squared (χ2) test. Logistic regression models were performed to evaluate the effects of SNPs on hyperbilirubinemia. The odds ratios (ORs) with corresponding 95% confidence interval (CI) were estimated between the case and the control groups by using binary logistic regression models after adjusting for clinical factors (including sex, age, hemoglobin, platelet, and albumin). The relation between log-transformed total bilirubin (TB) and different genotypes was also assessed. All statistical analyses were performed by using SPSS version 23.0 (SPSS, Chicago, IL, USA). A P-value < 0.05 was considered to be statistically significant.
Results
Clinical Characteristics
A total of 146 adults with hyperbilirubinemia were enrolled in the case group and 158 adults with bilirubin values of less than 17.1µmol/L in the control group. The clinical features of the case and control groups are shown in Table 1. The average total bilirubin of the case group and the control group were 24.93 ± 9.71 µmol/l and 11.83 ± 3.04 µmol/L, respectively. Compared with control subjects, case group had a higher level of hemoglobin (HB) (142.19 ± 14.44 g/L vs 137.24 ± 16.47 g/L, P = 0.006) and albumin (ALB) (44.98 ± 3.65 g/L vs 44.12 ± 2.27 g/L, P = 0.016), but lower platelet (PLT) values (218.53 ± 54.10×109/L vs 231.38 ± 52.13 × 109/L, P = 0.026). Besides, there were no obvious differences between the case and the control groups concerning age, gender, blood routine, and other plasma biochemical results.
Table 1.
Clinical characteristics of case group and control group.
| Parameter | Cases (N = 146) | Controls (N = 158) | P |
|---|---|---|---|
| Gender, female (%) | 66 (45.21%) | 77 (48.73%) | 0.538 |
| Age (years) | 40.86 ± 12.32 | 40.00 ± 6.98 | 0.462 |
| WBC(×109/L) | 6.01 ± 1.38 | 6.33 ± 1.51 | 0.052 |
| HB (g/L) | 142.19 ± 14.44 | 137.24 ± 16.47 | 0.006 |
| PLT (×109/L) | 218.53 ± 54.10 | 231.38 ± 52.13 | 0.026 |
| ALT (U/L) | 20.61 ± 10.28 | 20.06 ± 9.22 | 0.624 |
| AST (U/L) | 19.25 ± 5.05 | 18.56 ± 4.66 | 0.217 |
| γ-GT (U/L) | 21.87 ± 19.13 | 20.97 ± 11.88 | 0.62 |
| ALP (U/L) | 69.95 ± 24.82 | 66.92 ± 17.55 | 0.218 |
| TB (μmol/L) | 24.93 ± 9.71 | 11.83 ± 3.04 | <0.001 |
| DB (μmol/L) | 8.95 ± 4.54 | 4.41 ± 1.20 | <0.001 |
| IB (μmol/L) | 15.97 ± 6.82 | 7.46 ± 2.00 | <0.001 |
| TP (g/L) | 76.07 ± 6.73 | 75.54 ± 4.02 | 0.414 |
| ALB (g/L) | 44.98 ± 3.65 | 44.12 ± 2.27 | 0.016 |
| GLB(g/L) | 5.51 ± 3.66 | 6.10 ± 2.66 | 0.121 |
| FPG (mmol/L) | 4.99 ± 0.80 | 4.86 ± 0.51 | 0.106 |
| TC (mmol/L) | 4.40 ± 0.84 | 4.41 ± 0.71 | 0.896 |
| TG (mmol/L) | 1.40 ± 0.78 | 1.53 ± 1.02 | 0.211 |
| HDL (mmol/L) | 1.32 ± 0.29 | 1.27 ± 0.28 | 0.096 |
| LDL (mmol/L) | 2.64 ± 0.69 | 2.65 ± 0.63 | 0.907 |
| BUN (mmol/L) | 4.72 ± 1.03 | 4.85 ± 1.13 | 0.319 |
| CR (mmol/L) | 67.63 ± 11.58 | 67.27 ± 12.39 | 0.803 |
| UA (μmol/L) | 324.59 ± 92.30 | 321.18 ± 87.54 | 0.824 |
P-values were calculated between groups by using t-test for continuous variables and the χ2 test for categorical variables.
WBC, white blood cell count; HB, hemoglobin; PLT, platelet; ALT, alanine aminotransferase; AST, aspartate aminotransferase; γ-GT, glutamyltransferase; ALP, alkaline phosphatase; TB, total bilirubin; DB, direct bilirubin; IB, indirect bilirubin; TP, total protein; ALB, albumin; GLB, globulin; FPG, fasting plasma glucose; TC, total cholesterol; TG, triglyceride; HDL, high-density lipoprotein; LDL, low-density lipoprotein; BUN, blood urea nitrogen; CR, creatinine; UA, uric acid.
UGT1A1 and SLCO1B Gene Polymorphisms
The frequencies of UGT1A1 variations are presented in Table S1. The incidence of the two most common variants (either heterozygous or homozygous variants) in the case group was significantly higher than that of the control group [(TA)6>(TA)7: 41.78% vs 22.78%, P < 0.001; c.211G > A: 50.68% vs 29.11%, P < 0.001]. In addition, 8.22% of cases featured homozygous (TA)7 variant, while no controls carried this homozygous variation. The incidence of other six UGT1A1 variations in the case group was also significantly higher for c.-3275T > G (65.07% vs 43.04%, P < 0.001), c.-3152G > A (43.84% vs 23.42%, P < 0.001), c.686C > A (6.85% vs 1.90%, P = 0.033), IVS1+2842G > T (43.15% vs 23.42%, P < 0.001), IVS1+2925T > G (43.15% vs 23.42%, P < 0.001), and c.1091C > T (11.49% vs 1.90%, P = 0.001), but lower for IVS2+15T > C (5.48% vs 13.29%, P = 0.021). The proportion of case subjects with c.1456T > G variant was not statistically different from that of control subjects (3.42% vs 0.63%; P = 0.182). Moreover, the three variants c.686C > A, c.1091C > T, and c.1456T > G presented only heterozygous forms or wild-type in cases and controls. The LD pattern across the multiple SNPs of UGT1A1 is shown in Table S2. Strong pairwise LD was observed among the four SNPs within UGT1A1 [(TA)n repeat, c.-3152G > A, IVS1+2842G > T and IVS1+2925T > G], where each |D′| > 0.8 and r2 > 0.8. Moderate LD was present between (TA)n repeat polymorphism and c.-3275T > G (|D′| > 0.8 and r2= 0.4320).
The frequencies of different genotypes of the SLCO1B gene observed among the cases and controls are also shown in Table S1. The four known polymorphisms (388G > A and 521T > C within SLCO1B1, IVS8+2087T > C and g.21074122C > T within SLCO1B3) were investigated in this study. However, there was only a statistical difference of the frequency of 521T > C variation between the case and the control groups (24.66% vs 13.92%, P = 0.017). A low pairwise LD was found between the four polymorphisms of SLCO1B (r2 < 0.3) (Table S3).
Effects of UGT1A1 or SLCO1B Gene Variants on Unconjugated Hyperbilirubinemia
Initially, the OR with 95% CI of single suspected variant for hyperbilirubinemia was assessed by using separate binary logistic regression (Table 2). Further, the results adjusted for gender, age, hemoglobin, PLT, and albumin were also calculated by multivariate logistic regression models (Table 2). Four variants of UGT1A1 and one variant of SLCO1B1 revealed statistical significance. Among the variations that significantly increased the risk of hyperbilirubinemia, the first priority in descending order was c.1091C > T (OR = 7.596, 95% CI: 1.992–28.956, P = 0.003). There were also strong associations between other common variations in UGT1A1 and hyperbilirubinemia, including c.-3275T > G (OR = 2.345, 95% CI: 1.371–4.012, P = 0.002), (TA)6>(TA)7 (OR = 2.383, 95% CI: 1.351-4.203, P = 0.003), and c.211G > A (OR = 2.985, 95% CI: 1.71–5.211, P < 0.001). Variant IVS2+15T > C appeared to reduce the risk of hyperbilirubinemia (OR = 0.278, 95% CI: 0.095–0.82, P = 0.02). Specifically, variant c.521T > C within SLCO1B1 was found to have a significantly increased risk of hyperbilirubinemia (OR = 2.544, 95% CI: 1.267–5.11, P = 0.009). The log10-transferred TB of different variants is shown in Figure S1. After log10-transferred, the upper limit of the normal TB level is 1.23. In summary, subjects with most homozygous or heterozygous variants of UGT1A1 or variant c.521T > C of SLCO1B1 have higher bilirubin values than those with wild type. However, in the cases of variants IVS2+15T > C or IVS7+2087T > C, the opposite is true.
Table 2.
ORs and 95% CIs for unconjugated hyperbilirubinemia associated with genetic variation.
| SNP | OR | 95% CI | P | OR* | 95% CI* | P* |
|---|---|---|---|---|---|---|
| Variant within UGT1A1 gene | ||||||
| c.-3275T > G | 2.47 | 1.55–3.92 | <0.001 | 2.35 | 1.37–4.01 | 0.002 |
| (TA)6>(TA)7 | 2.43 | 1.48–4.00 | <0.001 | 2.38 | 1.35–4.20 | 0.003 |
| c.211G > A | 2.50 | 1.56–4.01 | <0.001 | 2.99 | 1.71–5.21 | <0.001 |
| c.686C > A | 3.80 | 1.02–14.09 | 0.046 | 2.19 | 0.48–10.02 | 0.312 |
| IVS2 + 15T > C | 0.38 | 0.16–0.88 | 0.025 | 0.28 | 0.09–0.82 | 0.02 |
| c.1091C > T | 6.81 | 1.95–23.75 | 0.003 | 7.60 | 1.99–28.96 | 0.003 |
| c.1456T > G | 5.57 | 0.64–48.23 | 0.119 | 4.28 | 0.45–40.59 | 0.205 |
| Variant within SLCO1B gene | ||||||
| c.388G > A | 1.25 | 0.51–3.06 | 0.62 | 1.11 | 0.40–3.10 | 0.84 |
| c.521T > C | 2.02 | 1.13–3.64 | 0.02 | 2.54 | 1.27–5.11 | 0.009 |
| IVS7 + 2087T > C | 0.74 | 0.19–2.80 | 0.65 | 1.20 | 0.22–6.56 | 0.834 |
| g.21074122C > T | 0.68 | 0.38–1.204 | 0.186 | 0.72 | 0.36–1.42 | 0.34 |
*Adjusted with gender, age, HB, PLT, ALB, WBC, FPG, and HDL. SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
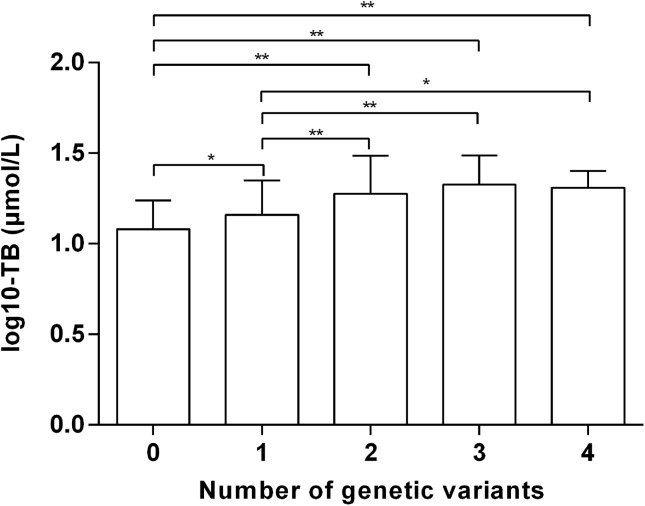
To assess the impact of co-expression of UGT1A1 or SLCO1B variants, the relationship between the number of genetic variants and the risk of unconjugated hyperbilirubinemia was investigated. The OR is positively correlated with the cumulative number of five variants in individuals (Table 3). Adjust ORs in adults with one, two, three, and four risk variants were 3.98 (95% CI: 1.48–10.69, P = 0.006), 10.92 (95% CI: 3.91–30.53, P < 0.001), 18.43 (95% CI: 5.95–57.04, P < 0.001) and 34.38 (95% CI: 3.05–387.32, P = 0.004), respectively. Besides, the greater the number of genetic variations, the higher the total bilirubin level (Figure 1). These data indicated that different variants in UGT1A1 or SLCO1B have combined effects on unconjugated hyperbilirubinemia.
Table 3.
Adjusted ORs and 95% CI of unconjugated hyperbilirubinemia associated with the number of genetic variation.
| Number of genetic variantsa | Cases (N = 146) | Controls (N = 158) | OR | 95% CI | P |
|---|---|---|---|---|---|
| 0 | 10 | 45 | 1.00 | – | – |
| 1 | 38 | 68 | 3.98 | 1.48–10.69 | 0.006 |
| 2 | 54 | 30 | 10.92 | 3.91–30.53 | <0.001 |
| 3 | 39 | 13 | 18.43 | 5.95–57.04 | <0.001 |
| 4 | 5 | 2 | 34.38 | 3.05–387.32 | 0.004 |
All results were adjusted with gender, age, HB, PLT, ALB, WBC, FPG, and HDL.
Variants including c.-3275T > G, (TA)6>(TA)7, c.211G > A, and c.1091C > T in UGT1A1 as well as variant c.521T > C in SLCO1B1.
OR, odds ratio; CI, confidence interval.
Figure 1.
Total bilirubin values (log10-transformed) amongst different numbers of genetic variants. TB, total bilirubin. *P < 0.05 **P < 0.01.
Combined Effects of UGT1A1 Variants on Unconjugated Hyperbilirubinemia
To further analyze the combined effects of UGT1A1 variations on unconjugated hyperbilirubinemia, haplotype and diplotype analyses were performed. Based on multivariate logistic regression results, four SNPs of UGT1A1, which increased the risk of hyperbilirubinemia, formed 5 common haplotypes with frequencies > 1% (Table 4). Among them, haplotype 1 [–3275T–(TA)6–211G–1091C] is the most common (43.52%), representing wild type. Compared with haplotype 1, haplotypes 2 [–3275T–(TA)6–211A–1091C], 3 [–3275G–(TA)7–211G–1091C], 4 [–3275G–(TA)6–211G–1091C] and 5 [–3275G–(TA)6–211G–1091A] all increased the risk of unconjugated hyperbilirubinemia (P < 0.05).
Table 4.
ORs and 95% CIs for unconjugated hyperbilirubinemia associated with UGT1A1 haplotypes.
| Haplotype | Variant | OR (95%CI) | P | OR (95%CI)* | P* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| c.-3275T > G | (TA)6> (TA)7 |
c.211G > A | c.1091C > T | Cases, frequency | Controls, frequency | Total, frequency | |||||
| 1 | T | (TA)6 | G | C | 26.15% | 59.49% | 43.52% | 1.00 | – | 1.00 | – |
| 2 | T | (TA)6 | A | C | 30.70% | 15.82% | 22.93% | 4.51 (2.77–7.35) | <0.001 | 5.72 (3.19–10.26) | <0.001 |
| 3 | G | (TA)7 | G | C | 25.00% | 11.39% | 17.93% | 5.29 (3.07–9.11) | <0.001 | 6.24 (3.29–11.86) | <0.001 |
| 4 | G | (TA)6 | G | C | 11.86% | 12.34% | 12.07% | 2.07 (1.16–3.68) | 0.014 | 2.27 (1.15–4.47) | 0.019 |
| 5 | G | (TA)6 | G | T | 5.82% | 0.95% | 3.29% | 12.48 (3.36–46.38) | <0.001 | 16.64 (3.89–71.12) | <0.001 |
*Adjusted with gender, age, HB, PLT, ALB, WBC, FPG, and HDL. OR, odds ratio; CI, confidence interval.
All subjects were divided into 14 diplotypes, covering a total of 303 (99.7%) adults. All bilirubin values (log10-transformed) among UGT1A1 diplotypes are shown in Figure 2A. Except for 4/4, subjects with homozygous variants or compound heterozygous variants (2/2, 2/3, 2/4, 2/5, 3/3, 3/4, 3/5, 4/5) had the highest levels of total bilirubin, followed by those with heterozygous variants (1/2, 1/3, 1/4, 1/5), while wild type (1/1) the lowest. Of all UGT1A1 diplotypes, 3/3 (homozygous variation of c.-3275T > G and (TA)6>(TA)7) featured the highest levels of bilirubin. The diplotypes 1/4 and 4/4 represent heterozygous or homozygous c.-3275T > G variant, respectively, and their bilirubin levels are lower than normal. Moreover, adults carrying diplotype 3/4 (homozygous c.-3275T > G and heterozygous (TA)6>(TA)7) had higher bilirubin levels than those with diplotypes 1/3 (heterozygous c.-3275T > G and (TA)6>(TA)7)) or 1/4 (heterozygous c.-3275T > G) (P < 0.05) (Figure 2B). Similarly, bilirubin levels in individuals with diplotype 2/4 (heterozygous c.-3275T > G and c.211G > A) were higher than those of adults carrying diplotypes 1/2 (heterozygous c.211G > A) or 1/4 (P < 0.001). However, there was no significant difference in bilirubin levels among adults with diplotype 1/4, 1/5 or 4/5. These data demonstrated an additive effect of variants c.-3275T > G and (TA)6>(TA)7 or c.211G > A on unconjugated hyperbilirubinemia.
Figure 2.
Total bilirubin values (log10-transformed) amongst adults with various UGT1A1 genotypes. a: –3275T–(TA)6–211G–1091C; b: –3275T–(TA)6–211A–1091C; c: –3275G–(TA)7–211G–1091C; d: –3275G–(TA)6–211G–1091C; e: –3275G–(TA)6–211G–1091A. *: P < 0.05 **: P < 0.01.
Combined Effects of UGT1A1 and SLCO1B Variants on Unconjugated Hyperbilirubinemia
The association between the two common variations (TA)6/(TA)7 and c.211G > A in UGT1A1 and variation c.521T > C in SLCO1B was further explored. In UGT1A1-normal subjects, homozygous variation c.521T > C increased total bilirubin level and the incidence of unconjugated hyperbilirubinemia compared to wild-type or heterozygous forms (Table S4). When carried heterozygous variation c.211G > A, the total bilirubin level of heterozygous c.521T > C adults was higher than that of wild type (22.92µmol/L vs 17.21µmol/L, P = 0.046); as the number of variation c.521T > C alleles increased, the incidence of unconjugated hyperbilirubinemia increased, but it was not statistically significant (P = 0.186). When carried homozygous variation c.211G > A, the incidence of hyperbilirubinemia for individuals featured c.521T > C was higher, but it did not reach statistically significant (100% vs 75%, P = 0.214). In addition, among the 21 adults with the homozygous c.211G > A, all 5 subjected with the heterozygous variation c.521T > C suffered from hyperbilirubinemia. But for (TA)6>(TA)7, we did not observe similar results. These data indicated a cumulative effect of variants c.521T > C and c.211G > A on unconjugated hyperbilirubinemia.
Discussion
GS, a benign condition that occurs in 5–10% of the population (Wagner et al., 2018), is characterized by mild unconjugated hyperbilirubinemia, in the absence of liver disease or hemolysis (Strassburg, 2010). Although it has long been considered a benign condition and does not require treatment, recent data suggest that affected individuals may be prone to liver damage following various drugs and xenobiotic treatments (Ehmer et al., 2012). Molecular defects in genes involved in the bilirubin metabolic pathway account for hyperbilirubinemia. In the present study, we evaluated the effects of UGT1A1 and SLCO1B on mild unconjugated hyperbilirubinemia in adults. Our results indicated that variations of UGT1A1 and SLCO1B genes had a substantial impact on unconjugated bilirubin levels in Chinese adults. Notably, variant c.521T > C in the SLCO1B may be independent genetic risk factors for inherited unconjugated hyperbilirubinemia. Furthermore, the risk of unconjugated hyperbilirubinemia is positively correlated with the cumulative number of variants in adults. To our knowledge, this is the first report that showed the combined effect of UGT1A1 and SLCO1B variants on mild unconjugated hyperbilirubinemia in adults.
Two common variants (TA)6>(TA)7 and c.211G > A of UGT1A1 both reduce UGT1A1 enzyme activity and increase unconjugated bilirubin level (Sato et al., 1996; Beutler et al., 1998; Maruo et al., 2016). Our results confirmed the strong association of variants (TA)6>(TA)7 and c.211G > A with the incidence of unconjugated hyperbilirubinemia. Additionally, the frequencies of variants (TA)6>(TA)7 and c.211G > A in the case group were significantly higher than those in the control group. And subjects with these two heterozygous or homozygous variants had higher mean total serum bilirubin levels than adults carrying the wild type.
Other mutations occurring in the coding region of UGT1A1 c.686C > A, c.1091C > T and c.1456T > G can also lead to Gilbert syndrome amongst the Asian population (Huang et al., 2005; Yang et al., 2016). In this study, the frequencies of variants c.686C > A and c.1091C > T in the UGT1A1 gene in cases and controls were statistically different, but only the OR of c.1091C > T was statistically significant when adjusting the covariates. The role of variants c.686C > A and c.1456T > G needs to be explored through large-scale investigations. Besides, we found that variant IVS2+15T > C appears to reduce the risk of hyperbilirubinemia, and is associated with lower bilirubin levels. However, there is no relevant evidence and further research is needed.
Earlier studies suggested the decrease in transcription caused by both (TA)6>(TA)7 and c.-3275T > G mutations together may be essential to the Gilbert syndrome (Maruo et al., 2004). In the present study, variation c.-3275T > G was found to be moderately linked with TA repeats. The results of multivariate logistic regression and haplotype analysis demonstrated that variant c.-3275T > G was an independent risk factor of unconjugated hyperbilirubinemia. Interestingly, subjects carrying homozygous or heterozygous variant c.-3275T > G (4/4 or 1/4 diplotype) featured normal mean bilirubin levels. This result is consistent with other researches, suggesting that variant c.-3275T > G was not associated with increased bilirubin levels (Chen et al., 2014; Pasternak et al., 2017). In addition, diplotype analyses revealed that subjects with co-inheriting heterozygous variants c.-3275T > G as well as (TA)6>(TA)7 or c.211G > A had higher bilirubin levels than those carrying any of the above heterozygous variants. These data indicate a combined effect on unconjugated hyperbilirubinemia for different variants in UGT1A1.
Some GS cases have been reported in which plasma clearance of cholephilic dyes such as sulfobromophthalein (BSP) and indocyanine green (ICG) was markedly impaired, indicating a role of impaired hepatic bilirubin uptake (Metreau et al., 1977; Gentile et al., 1990). Genetic analyses suggest that such cases of unconjugated hyperbilirubinemia could be linked to polymorphisms in SLCO1B1 and SCLO1B3 (Sanna et al., 2009; Watchko, 2013; Erlinger et al., 2014). In the present research, four polymorphisms of SLCO1B identified by previous GWAS were detected (Johnson et al., 2009; Sanna et al., 2009; Kang et al., 2010; Dai et al., 2013). A novel founding is that variant c.521T > C in SLCO1B1 is closely related to bilirubin levels and significantly increases the risk of unconjugated hyperbilirubinemia (OR = 2.54, 95% CI: 1.27–5.11, P = 0.009). Besides, the variant c.521T > C showed an additive effect on UGT1A1 gene variants, particularly the variant c.211G > A. These results indicated that the change of amino acid (valine to alanine, encoded by nucleotides 520–522) at codon 174 of SLCO1B1 may reduce the function in unconjugated bilirubin transportation. However, the function of the variants should be validated in cell and animal model experiments. Interestingly, simultaneous homozygous mutations in SLCO1B1 and SLCO1B3, resulting in disruption of hepatic reuptake of bilirubin, may account for the predominantly conjugated hyperbilirubinemia of Rotor syndrome (van de Steeg et al., 2012). These findings suggested that the function of solute carrier organic anion transporter is more complex than transporting unconjugated bilirubin, and thus mutations in these coding genes result in different hyperbilirubinemia propensities.
Besides, previous researches have shown that genes expressed outside the liver also affect bilirubin homeostasis, including glucose-6-phosphate dehydrogenase (G6PD), heme oxygenase (HMOX), biliverdin reductase A (BLVRA), nucleoporin 153 (NUP153) and UGT1A6 (Huang et al., 2002; Huang et al., 2005; Datta et al., 2012; Oussalah et al., 2015; Chiddarwar et al., 2017). However, our study mainly discussed the effects of intrahepatic genetic variations on non-hemolytic unconjugated hyperbilirubinemia. More studies are encouraged to further explore the effect of genetic variation of extrahepatic expressed genes on Chinese adult unconjugated hyperbilirubinemia.
This study has some limitation. First, the level of bilirubin in GS is fluctuating, especially in particular conditions such as prolonged fasting, fever, and strenuous exercise (Vitek et al., 2019). Although we standardized blood collection and adjusted clinical factors as covariables for all association tests, there are still uncontrollable factors affecting bilirubin levels, such as mood and season. Second, the study is a descriptive study. Based on previous GWAS, our study investigated the UGT1A1 and SLCO1B genetic variation of Chinese adults with hyperbilirubinemia and emphatically explored the combined effects of multiple variants. Consistent with the GWAS, our study confirmed the role of the single genetic variation. Moreover, we found that the variants of UGT1A1 and SLCO1B1 have combined effects on the level of bilirubin. The function of the variants should be validated in cell and animal model experiments.
In conclusion, this study demonstrates the impact of genetic factors on non- hemolytic unconjugated hyperbilirubinemia in Chinese population. Variations c.-3275T > G, (TA)6>(TA)7, c.211G > A, and c.1091C > T within UGT1A1 as well as c.521T > C within SLCO1B1 are independent risk factors of mild unconjugated hyperbilirubinemia. Our results also reveal that there is a combined effect on unconjugated hyperbilirubinemia for c.-3275T > G as well as (TA)6>(TA)7 or c.211G > A in UGT1A1. Further, co-expression of c.211G > A of UGT1A1 and c.521T > C of SLCO1B1 has a cumulative effect on unconjugated hyperbilirubinemia.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics committee of Beijing Youan Hospital, Capital Medical University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
JB, LL, SZ, and ZD: Conception and design. LL: Patient recruitment. JB and CL: Collection of data. SL and LB: Analysing data. JB: Manuscript draft. JB, LL, YC, SZ and ZD: Editing and revision. All authors read and approved the final manuscript.
Funding
This study was supported by Beijing Municipal Science and Technology Project (No.Z171100002217070), National Key R&D Program of China (No.2017YFA0103000), National Science and Technology Key Project on “Major Infectious Diseases such as HIV/AIDS, Viral Hepatitis Prevention and Treatment” (NO. 2012ZX10002004-006, No.2017ZX10203201-005, 2017ZX10201201, No.2017ZX10202203-006-001 and No.2017ZX10302201-004-002), “Beijing Municipal Administration of Hospitals” Ascent Plan (No. DFL20151601), Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (No.ZYLX201806), the Digestive Medical Coordinated Development Center of Beijing Municipal Administration of Hospitals (No.XXZ0503), the You An fund for liver diseases and AIDS (YNKTTS201801189), and the Basic-Clinical Cooperation Project of Capital Medical University (17JL47). Science and technology innovation service capacity building - high-precision discipline construction project (11920703).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01073/full#supplementary-material
References
- Beutler E., Gelbart T., Demina A. (1998). Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc. Natl. Acad. Sci. U. S. A. 95 (14), 8170–8174. 10.1073/pnas.95.14.8170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Su D., Ai L., Jiang X., Wu C., Xu Q., et al. (2014). UGT1A1 sequence variants associated with risk of adult hyperbilirubinemia: a quantitative analysis. Gene 552 (1), 32–38. 10.1016/j.gene.2014.09.009 [DOI] [PubMed] [Google Scholar]
- Chiddarwar A. S., D'Silva S. Z., Colah R. B., Ghosh K., Mukherjee M. B. (2017). Genetic Variations in Bilirubin Metabolism Genes and Their Association with Unconjugated Hyperbilirubinemia in Adults. Ann. Hum. Genet. 81 (1), 11–19. 10.1111/ahg.12179 [DOI] [PubMed] [Google Scholar]
- Clarke D. J., Moghrabi N., Monaghan G., Cassidy A., Boxer M., Hume R., et al. (1997). Genetic defects of the UDP-glucuronosyltransferase-1 (UGT1) gene that cause familial non-haemolytic unconjugated hyperbilirubinaemias. Clin. Chim. Acta. 266 (1), 63–74. 10.1016/s0009-8981(97)00167-8 [DOI] [PubMed] [Google Scholar]
- D'Silva S., Colah R. B., Ghosh K., Mukherjee M. B. (2014). Combined effects of the UGT1A1 and OATP2 gene polymorphisms as major risk factor for unconjugated hyperbilirubinemia in Indian neonates. Gene 547 (1), 18–22. 10.1016/j.gene.2014.05.047 [DOI] [PubMed] [Google Scholar]
- Dai X., Wu C., He Y., Gui L., Zhou L., Guo H., et al. (2013). A genome-wide association study for serum bilirubin levels and gene-environment interaction in a Chinese population. Genet. Epidemiol. 37 (3), 293–300. 10.1002/gepi.21711 [DOI] [PubMed] [Google Scholar]
- Datta S., Chowdhury A., Ghosh M., Das K., Jha P., Colah R., et al. (2012). A genome-wide search for non-UGT1A1 markers associated with unconjugated bilirubin level reveals significant association with a polymorphic marker near a gene of the nucleoporin family. Ann. Hum. Genet. 76 (1), 33–41. 10.1111/j.1469-1809.2011.00688.x [DOI] [PubMed] [Google Scholar]
- Ehmer U., Kalthoff S., Fakundiny B., Pabst B., Freiberg N., Naumann R., et al. (2012). Gilbert syndrome redefined: a complex genetic haplotype influences the regulation of glucuronidation. Hepatology 55 (6), 1912–1921. 10.1002/hep.25561 [DOI] [PubMed] [Google Scholar]
- Erlinger S., Arias I. M., Dhumeaux D. (2014). Inherited disorders of bilirubin transport and conjugation: new insights into molecular mechanisms and consequences. Gastroenterology 146 (7), 1625–1638. 10.1053/j.gastro.2014.03.047 [DOI] [PubMed] [Google Scholar]
- Gentile S., Persico M., Tiribelli C. (1990). Abnormal hepatic uptake of low doses of sulfobromophthalein in Gilbert's syndrome: the role of reduced affinity of the plasma membrane carrier of organic anions. Hepatology 12 (2), 213–217. 10.1002/hep.1840120206 [DOI] [PubMed] [Google Scholar]
- Huang C. S., Chang P. F., Huang M. J., Chen E. S., Chen W. C. (2002). Glucose-6-phosphate dehydrogenase deficiency, the UDP-glucuronosyl transferase 1A1 gene, and neonatal hyperbilirubinemia. Gastroenterology 123 (1), 127–133. 10.1053/gast.2002.34173 [DOI] [PubMed] [Google Scholar]
- Huang C. S., Huang M. J., Lin M. S., Yang S. S., Teng H. C., Tang K. S. (2005). Genetic factors related to unconjugated hyperbilirubinemia amongst adults. Pharmacogenet. Genomics 15 (1), 43–50. 10.1097/01213011-200501000-00007 [DOI] [PubMed] [Google Scholar]
- Huang M. J., Kua K. E., Teng H. C., Tang K. S., Weng H. W., Huang C. S. (2004). Risk factors for severe hyperbilirubinemia in neonates. Pediatr. Res. 56 (5), 682–689. 10.1203/01.pdr.0000141846.37253.af [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Kavousi M., Smith A. V., Chen M. H., Dehghan A., Aspelund T., et al. (2009). Genome-wide association meta-analysis for total serum bilirubin levels. Hum. Mol. Genet. 18 (14), 2700–2710. 10.1093/hmg/ddp202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T. W., Kim H. J., Ju H., Kim J. H., Jeon Y. J., Lee H. C., et al. (2010). Genome-wide association of serum bilirubin levels in Korean population. Hum. Mol. Genet. 19 (18), 3672–3678. 10.1093/hmg/ddq281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo Y., D'Addario C., Mori A., Iwai M., Takahashi H., Sato H., et al. (2004). Two linked polymorphic mutations (A(TA)7TAA and T-3279G) of UGT1A1 as the principal cause of Gilbert syndrome. Hum. Genet. 115 (6), 525–526. 10.1007/s00439-004-1183-x [DOI] [PubMed] [Google Scholar]
- Maruo Y., Nakahara S., Yanagi T., Nomura A., Mimura Y., Matsui K., et al. (2016). Genotype of UGT1A1 and phenotype correlation between Crigler-Najjar syndrome type II and Gilbert syndrome. J. Gastroenterol. Hepatol. 31 (2), 403–408. 10.1111/jgh.13071 [DOI] [PubMed] [Google Scholar]
- Metreau J. M., Dhumeaux D., Gisselbrecht C., Preaux A. M., Berthelot P. (1977). Constitutional unconjugated hyperbilirubinaemia. Lancet 1 (8025), 1319. 10.1016/s0140-6736(77)91359-9 [DOI] [PubMed] [Google Scholar]
- Oussalah A., Bosco P., Anello G., Spada R., Gueant-Rodriguez R. M., Chery C., et al. (2015). Exome-wide association study identifies new low-frequency and rare UGT1A1 coding variants and UGT1A6 coding variants influencing serum bilirubin in elderly subjects: a strobe compliant article. Med. (Baltimore) 94 (22), e925. 10.1097/md.0000000000000925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A. L., Crews K. R., Caudle K. E., Smith C., Pei D., Cheng C., et al. (2017). The impact of the UGT1A1*60 allele on bilirubin serum concentrations. Pharmacogenomics 18 (1), 5–16. 10.2217/pgs-2016-0135 [DOI] [PubMed] [Google Scholar]
- Sanna S., Busonero F., Maschio A., McArdle P. F., Usala G., Dei M., et al. (2009). Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum. Mol. Genet. 18 (14), 2711–2718. 10.1093/hmg/ddp203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Adachi Y., Koiwai O. (1996). The genetic basis of Gilbert's syndrome. Lancet 347 (9001), 557–558. 10.1016/S0140-6736(96)91266-0 [DOI] [PubMed] [Google Scholar]
- Skierka J. M., Kotzer K. E., Lagerstedt S. A., O'Kane D. J., Baudhuin L. M. (2013). UGT1A1 genetic analysis as a diagnostic aid for individuals with unconjugated hyperbilirubinemia. J. Pediatr. 162 (6), 1146–11521152. e1141–1142. 10.1016/j.jpeds.2012.11.042 [DOI] [PubMed] [Google Scholar]
- Strassburg C. P. (2010). Hyperbilirubinemia syndromes (Gilbert-Meulengracht, Crigler-Najjar, Dubin-Johnson, and Rotor syndrome). Best. Pract. Res. Clin. Gastroenterol. 24 (5), 555–571. 10.1016/j.bpg.2010.07.007 [DOI] [PubMed] [Google Scholar]
- Tiribelli C., Ostrow J. D. (1993). New concepts in bilirubin chemistry, transport and metabolism: report of the Second International Bilirubin Workshop, April 9-11, 1992, Trieste, Italy. Hepatology 17 (4), 715–736. 10.1002/hep.1840170428 [DOI] [PubMed] [Google Scholar]
- van de Steeg E., Stranecky V., Hartmannova H., Noskova L., Hrebicek M., Wagenaar E., et al. (2012). Complete OATP1B1 and OATP1B3 deficiency causes human Rotor syndrome by interrupting conjugated bilirubin reuptake into the liver. J. Clin. Invest 122 (2), 519–528. 10.1172/jci59526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek L., Bellarosa C., Tiribelli C. (2019). Induction of mild hyperbilirubinemia: hype or real therapeutic opportunity? Clin. Pharmacol. Ther. 106 (3), 568–575. 10.1002/cpt.1341 [DOI] [PubMed] [Google Scholar]
- Wagner K. H., Shiels R. G., Lang C. A., Seyed Khoei N., Bulmer A. C. (2018). Diagnostic criteria and contributors to Gilbert's syndrome. Crit. Rev. Clin. Lab. Sci. 55 (2), 129–139. 10.1080/10408363.2018.1428526 [DOI] [PubMed] [Google Scholar]
- Watchko J. F. (2013). Genetics and pediatric unconjugated hyperbilirubinemia. J. Pediatr. 162 (6), 1092–1094. 10.1016/j.jpeds.2013.01.044 [DOI] [PubMed] [Google Scholar]
- Yanagi T., Nakahara S., Maruo Y. (2017). Bilirubin uridine diphosphate-glucuronosyltransferase polymorphism as a risk factor for prolonged hyperbilirubinemia in japanese preterm infants. J. Pediatr. 190, 159–162. e151. 10.1016/j.jpeds.2017.07.014 [DOI] [PubMed] [Google Scholar]
- Yang H., Wang Q., Zheng L., Zheng X. B., Lin M., Zhan X. F., et al. (2016). Clinical significance of UGT1A1 genetic analysis in chinese neonates with severe hyperbilirubinemia. Pediatr. Neonatol. 57 (4), 310–317. 10.1016/j.pedneo.2015.08.008 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ao G., Wang Y., Yan W., Wang M., Chen E., et al. (2012). Genetic variants and haplotypes of the UGT1A9, 1A7 and 1A1 genes in Chinese Han. Genet. Mol. Biol. 35 (2), 428–434. 10.1590/s1415-47572012005000036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.