Abstract
Rearing systems play an important role in animal welfare, health and the composition of the gut microbiome. Therefore, the purpose of this study was to investigate the effects of different rearing systems on the composition and function of cecal microbiota in chickens. The 120-day-old Lohmann hens of cage rearing systems (CRS) and free-range systems (FRS) were studied. The cecal bacterial populations of hens were surveyed by high-throughput sequencing (HTS) of the bacterial 16S rRNA hypervariable region V3–V4 combined with metagenomic sequencing analysis. The 16S rRNA sequencing analysis showed that the cecal microbiota differed between the FRS and CRS. The three most abundant bacteria phyla in the two systems were the Bacteroidetes (> 48%), Firmicutes (> 37%), and Proteobacteria (> 6%), the Deferribacteres (> 2.4%) were found in FRS and almost absent in CRS (< 0.01%). The three most abundant genera were the Bacteroides, Rikenellaceae_RC9, and Faecalibacterium, and we found relative abundance of the Parabacteroides (P < 0.05), Prevotellaceae_Ga6A1 (P < 0.01), unclassified Proteobacteria (P < 0.05), and unclassified Spirochaetaceae (P < 0.01) was greater in FRS, whereas abundance of Faecalibacterium, Ruminococcaceae, and Helicobacter was greater in CRS (P < 0.05). Functional gene classification of metagenomic sequencing suggested that energy production and conversion, carbohydrate transport and metabolism, as well as amino acid transport and metabolism were significantly more abundant in FRS, and we identified a range of antibiotic resistance categories in gut microbes of hens reared under both systems. We confirmed differences in microbe gut composition and function in hens reared using two contrasting systems, and ARGs were also identified in the microbiota of these hens. This work has produced new data for laying hens in different production systems and increased the understanding of intestinal microorganisms in laying hens.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1970-7) contains supplementary material, which is available to authorized users.
Keywords: 16Sr RNA sequencing, Metagenomic sequencing, Cecal microbiota, Hen rearing systems, Microbe composition and function
Introduction
It is well known that the trend of future development is healthy and rational rearing that has become a hot spot for livestock husbandry. The rearing systems of hens is closely related to the health (Siegerstetter et al. 2017), physiological state (Best et al. 2017), and animal welfare (Wang et al. 2016a). Rearing systems affect the microbe composition of the host gut (Stanley et al. 2013a; Waite and Taylor 2015) that plays an important role in the host’s health (Clavijo and Vives Florez 2018) There are more than 100 bacterial species in the intestinal tract, which are termed intestinal microbiota (Ubeda et al. 2017), where their community composition tends to be stable in a healthy host. The intestinal tract is a metabolic organ of the host that is involved in feed conversion (Cui et al. 2017), absorption of nutrients (Pan and Yu 2014), and development of the immune system (Yeoman et al. 2012) including prevention of colonization by pathogens (Clavijo and Vives Florez 2018). Disruption of the dynamic balance of intestinal microbiota in livestock and poultry leads to impaired digestion and immunity, and an increase in susceptibility to pathogens that result in diarrhea and reduced growth performance (Cao et al. 2013).
Chyme residues, comprising gastric juice and partly digested food, reach the cecum and are converted to other substances (Adil and Magray 2012). Avian intestinal flora in the cecum comprise high proportions of anaerobic bacteria (Barnes 1972), and the cecum is important for the survival and activity of the intestinal microbiota in chickens. Previous studies have shown that the intestinal microbes of chickens are closely related to the health status of the host and level of production intensity. For example, composition and function of intestinal microbiota of chickens differed between commercial large-scale and semi-wild production systems, and compared with microbiomes in broiler chickens, that of free-range chickens contained more genes involved in the acetate production pathway (Mancabelli et al. 2016), and a previous study found variations in microbe composition and diversity among populations of chicken cecal bacteria from five locations in Tibet (Lhasa, Ganzi, Aba, Qinghai, and Diqing) (Zhou et al. 2016).The intestinal microbiota of broiler chickens are affected by litter management (Wang et al. 2016a).
While there is a wealth of research about intestinal microbiota in broiler chickens that are bred purely for meat production, little is known about intestinal microorganisms (Han et al. 2016) or effects of rearing systems on their composition and function in hens that are used for egg production. Therefore, the aim of this study was to investigate the effects of different rearing systems on gut microbes in hens to improve rearing information for hen breeders and farmers.
Materials and methods
Animals and sample collection
We compared the intestinal microbiome in Lohmann hens reared within the same geographical location (in Hefei, Anhui Province, China) using two rearing systems: CRS rearing is characterized by high bird density that limits movement, the feeding density is 2 chickens per cage, 1 layer has 100 cages, 1 row has 6 layers. While FRS rearing comprises free-ranging birds in a natural environment, the breeding density is 120 chickens per 660 square meters. All chickens have no history of intestinal infectious disease. We randomly selected ten individuals with similar body weight (1500–1550 g) at 120-day old Lohmann hens from the farms that were immediately dissected using sterile scissors to aseptically remove the intestines from the abdominal cavity; contents of cecal were gently squeezed out, and the cecal contents were collected aseptically, rapidly injected into liquid nitrogen, and then stored at − 80 °C prior to analysis.
DNA extraction and sequencing
Microbe DNA was extracted using a Qiagen QIAamp Fast DNA Stool Mini Kit, according to the manufacturer’s protocols. Final DNA concentration and purification were determined using a NanoDrop 2000 UV–Vis spectrophotometer (Thermo Scientific, Wilmington, USA). The DNA quality was tested by 1.0% agarose gel electrophoresis. The V3–V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) on a thermocycler PCR system (GeneAmp 9700, ABI, USA), and then PCR products were extracted from a 2.0% agarose gel, further purified using a AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA), and quantified using QuantiFluor-ST (Promega, USA), according to the manufacturer’s protocol.
Equimolar amounts of the purified amplicons were pooled and paired-end sequenced (2 × 300) on the Illumina MiSeq platform (Illumina, San Diego, CA, USA), with standard protocols (Majorbio Bio-Pharm Technology Co., Ltd, Shanghai, China). The raw FASTQ files were demultiplexed and quality-filtered with Trimmomatic, and then merged using FLASH, with the following criteria: (1) reads that were truncated at any site, with an average quality score of < 20 over a 50-bp sliding window; (2) primers were exactly matched, two nucleotide mismatches were allowed, and reads containing ambiguous bases were removed; and (3) sequences overlaps > 10 bp were merged according to the overlap sequence. Sequences with ≥ 97% similarity were clustered into operational taxonomic units (OTUs) with UPARSE (version 7.1; http://drive5.com/uparse/), and chimeric sequences were identified and removed with UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed with the RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU123) 16S rRNA database, using a confidence threshold of 70%.
We used DNA that was fragmented to 300 bp with a Covaris M220 focused ultrasonicator (Gene Company Limited, China) to construct the paired-end library, which was prepared with the TruSeq™ DNA Sample Prep Kit (Illumina). Adapters containing the full complement of sequencing primer hybridization sites were ligated to the blunt-ended fragments, and paired-end sequencing was performed on the Illumina HiSeq 4000 platform (Illumina Inc.) at Majorbio Bio-Pharm Technology Co., Ltd (Shanghai, China) with the HiSeq 3000/4000 PE Cluster Kit and HiSeq 3000/4000 SBS Kits, according to the manufacturer’s instructions (http://www.illumina.com). The 3′ and 5′ ends were then stripped with SeqPrep (https://github.com/jstjohn/SeqPrep), and the low-quality reads (length < 50 bp, quality value < 20, or N bases) were removed with Sickle (https://github.com/najoshi/sickle). The reads were aligned to the chicken genome (https://www.ncbi.nlm.nih.gov/genome/?term=chicken) with BWA (http://bio-bwa.sourceforge.net), and any hit that associated with a read with a corresponding reads was removed. We used a de-Bruijn-graph-based assembler (SOAPdenovo, http://soap.genomics.org.cn, version 1.06) for the short reads (Li et al. 2008), and the K-mers, varying from 1/3 to 2/3 of the read length, were tested for each sample. We evaluated the quality and quantity of the scaffolds generated by each assembly and selected the best K-mer, i.e. the K-mer that yielded the minimum scaffold number and maximum values for N50 and N90. The scaffolds > 500 bp in length were then extracted and broken into contigs, without gaps, for gene prediction and annotation.
Sequencing data analysis
The 16S rRNA data and their richness were investigated with the Quantitative Insights Into Microbial Ecology 1.9.0 software (QIIME; http://qiime.org). The Illumina adapters and primers were removed from the raw sequences. The trimmed forward and reverse sequences were combined (Eren et al. 2013). These sequences were clustered into OTUs (97% similarity) with UCLUST (Edgar 2010). The reference OTU sequences were taxonomically assigned with the UCLUST Consensus TaxonAssigner (DeSantis et al. 2006) against the Greengenes database (McDonald et al. 2012), with a 0.5 confidence threshold, and identified to the species level. Rarefied OTUs were used to measure the bacterial richness from the total lengths of the phylogenetic branches (Faith and Baker 2006) and the relative proportions of rare sequences (Chao1) (Neufeld and Mohn 2005). The unweighted UniFrac distances (Chang et al. 2011) were used to compare the bacterial communities depending on the chicken breed. Based on the sample information, a redundancy analysis (RDA) with clustered OTUs was used to compare the chicken breed with the bacterial community structures using the R statistical software version 3.3.0 (Stanley et al. 2013b). To assess whether both chicken-breed-specific microbiomes were significantly distinguished, we used a nonparametric statistical test, analysis of similarity (ANOSIM). The significance of differences between groups was determined with permutations (n = 999) using the vegan package in the R statistical software. Using mothur software, co-occurrence analysis among genera was investigated by calculating C-scores, and Spearman’s rank correlations of the 50 most abundant genera were calculated. Network analysis using the genera with rho > 0.6 and p < 0.01 was visualized using Cytoscape (version 3.4.0).
The open reading frames (ORFs) in each metagenomic sample were predicted with MetaGene (http://metagene.cb.k.u-tokyo.ac.jp/) (Noguchi et al. 2006), and the predicted ORFs with lengths > 100 bp were extracted and translated to amino acid sequences using the National Center for Biotechnology Information (NCBI) translation table (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=tgencodes#SG). The sequences from gene sets with 95% sequence identity (90% coverage) (Fu et al. 2012) were clustered as a nonredundant gene catalogue with CD-HIT (http://www.bioinformatics.org/cd-hit/). After quality control, the reads were mapped to the representative genes with 95% identity using SOAPaligner (http://soap.genomics.org.cn/), and the gene abundance in each sample was evaluated.
We used BLASTP (version 2.2.28+; http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al. 1997) for taxonomic annotation, by aligning the nonredundant gene catalogues against the NCBI nonredundant (NR) database, with an e-value cutoff of 1e−5. An analysis of the ORF annotations (Clusters of Orthologous Groups [COG]) was performed with BLASTP against the eggNOG database (v4.5) (Jensen et al. 2008), with an e-value cutoff of 1e−5. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation was performed with a BLASTP search (version 2.2.28+) against the KEGG database (http://www.genome.jp/kegg/) (Xie et al. 2011), with an e-value cutoff of 1e−5. Carbohydrate-active enzyme annotations were made with hmmscan (http://hmmer.janelia.org/search/hmmscan) against the CAZy database v5.0 (http://www.cazy.org/), with an e-value cutoff of 1e−5.
Statistical analyses
Differences between two groups were analyzed using Student’s t test in the SAS statistical software package version 9.3 (SAS Institute Inc., Cary, NC, United States). and we used the statistical analysis of metagenomic profiles (STAMP) probability model to identify biologically relevant differences between metagenomic communities.
Data deposition
The raw 16S rRNA gene sequences are accessible through Sequence Read Archive (SRA) study accession number SRP139155, and the raw shotgun metagenomic data are accessible through SRA study accession number SRP158884.
Results
16S rRNA profiling of FRS and CRS
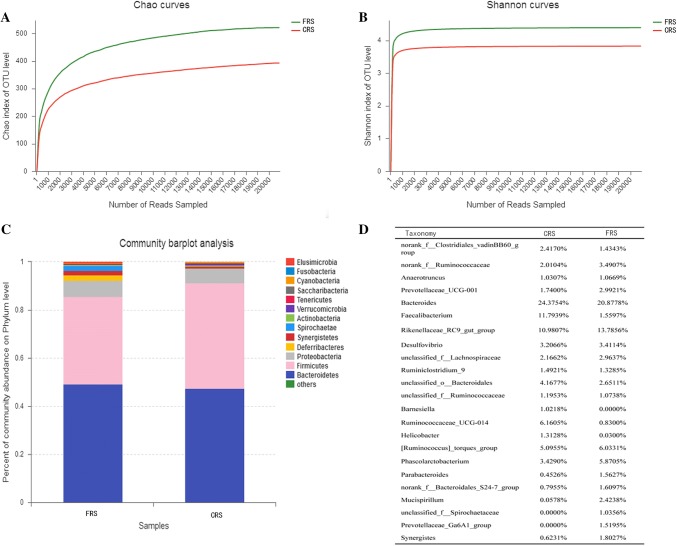
We obtained a total of 528,592 sequenced reads after quality control that averaged 52,859 reads per sample, and average sequence length was 438 bp (Table S1). We used Shannon curve and Chao index to evaluate the amount of sequence data for all samples: there was an increase in the number of bands obtained by sequencing, and the plateaus in the curves indicated that the sequenced data represented the majority of the diversity of microorganisms in the samples (Fig. 1a, b, Table S2). Average rarefaction curves showed a different cecal microbes between two rearing systems (Fig. 1), where there was greater diversity of bacteria phyla in FRS (Fig. 1c).
Fig. 1.
16S rRNA profiling of gut microbes from hens rearing under CRS and FRS. a Average rarefaction curve of the Chao diversity index; b average rarefaction curve of the Shannon diversity index; and, c average taxonomic composition of phyla. The legend shows the average of relative abundance of each gene in all samples
16S rRNA profiling analysis of gut microbiota composition between FRS and CRS
We found the dominant microbe phyla in the two systems comprised the Bacteroidetes, Firmicutes, and Proteobacteria (Fig. 1c), where the Bacteroidetes was most dominant phylum, accounting for more than 47%, followed by the Firmicutes (36.49% in FRS and 43.84% in CRS; Table S3). Relative abundance of the Proteobacteria was low in each system (FRS: 6.08%; CRS: 6.63%), and the Deferribacteres represented 2.42% relative abundance in FRS, but were undetected in CRS (Table S3).
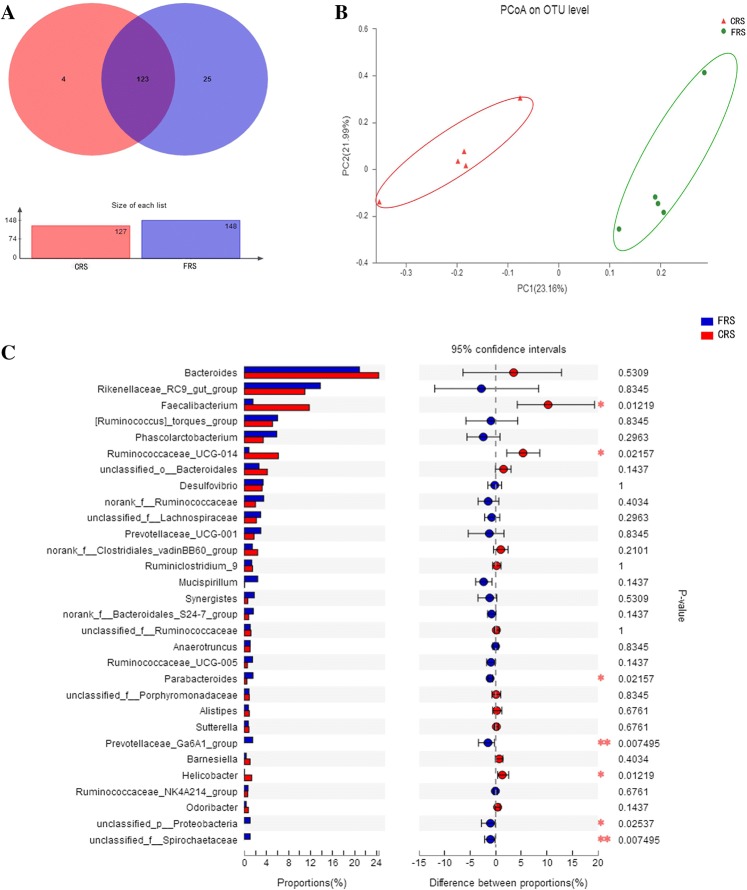
The most dominant genera were the Bacteroides (FRS: 20.88%; CRS: 24.38%), Rikenellaceae_RC9_group (FRS: 13.79%; CRS: 10.98%), and Faecalibacterium (FRS: 1.56%; CRS: 11.79%; Fig. 1d). We identified 152 genera, of which 123 were present in all samples, while 4 and 25 genera were unique to FRS and CRS, respectively (Fig. 2a).
Fig. 2.
Diversity of gut microbes in hens reared under FRS and CRS. a Venn diagram of total, unique, and shared number of predicted genera in FRS and CRS; b phylogenetic differences in cecum microbe OTUs between FRS and CRS using principal coordinate analysis; and c extended error bar plot showing differences between the 30 most abundant genera in FRS and CRS. Positive differences indicate genus overrepresented in FRS, while negative differences indicate greater abundance in CRS. The Wilcoxon rank-sum test within STAMP was used to test for differences in genera abundance between the two rearing systems groups (confidence interval method); the abscissa indicates abundance of a species and the ordinate indicates the species name; and colors indicate different groups. *P < 0.05, **P < 0.01
Analysis of the relationships among the bacterial communities in hen cecum
To measure the degree of similarity between microbial communities, we have analyzed the beta-diversity based on the ordination of the distance matrix generated using Bray–Curtis complementary algorithm, the clear demarcation between bacterial assemblages from FRS and CRS were apparent along principal coordinate axis 1 (PCoA1) of the PCoA plot (Fig. 2b), and ANOSIM analysis showed there were overall differences in the communities (P = 0.002, R = 0.225; Table S4). We compared the composition and average relative abundance of the 30 most abundant genera and found that there was a greater abundance of Parabacteroides (P < 0.05), Prevotellaceae_Ga6A1_group (P < 0.01), Unclassified_p__Proteobacteria (P < 0.05), and unclassified_f__Spirochaetaceae (P < 0.01) in hens reared under the FRS system, while there was a greater abundance of Faecalibacterium (P < 0.05), Ruminococcaceae (P < 0.05), and Helicobacter (P < 0.05) in hens reared under the CRS system (Fig. 2c).
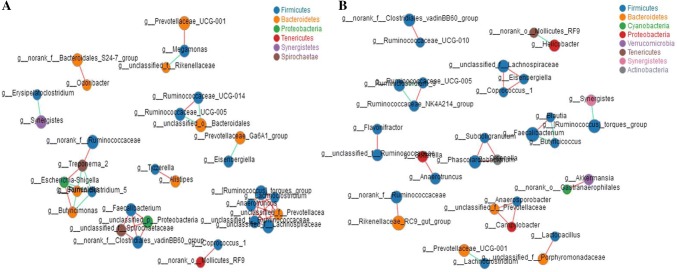
Based on the analysis of beta-diversity, there was a difference in the structure of the cecum intestinal microbiota between FRS and CRS. At the genus level, the results of the correlation network analysis showed that the cecum microbiota network of FRS and CRS hens were different in different rearing systems, and we found that the relationships within the cecal microbe community of FRS was more closely than that of CRS, especially between the Firmicutes and Bacteroidetes (Fig. 3a), whereas there was a relatively simple relationship in CRS, mainly driven by the Firmicutes (Fig. 3b).
Fig. 3.
Network analysis of cecal microbes in hens reared under FRS and CRS. Network correlation of the 50 most abundant species in a FRS and b CRS. Node size indicates relative abundance of a species; different colors indicate different species; red line indicates positive correlation and green indicates negative correlation; line thickness indicates magnitude of the Pearson correlation coefficient, where thickness increases with magnitude
Summary of the metagenomic datasets
Metagenomic sequencing of representative samples from the two rearing systems produced a total of 97,602,410 raw and 96,661,039 filtered reads, and comparison of the predicted taxonomic distribution from the 16S rRNA profiling analysis (Fig. 1c) with that from the metagenomic data (Table S5) showed that the sequencing results of the two methods are consistent.
Comparison of functionality of the FRS and CRS hens gut metagenomes
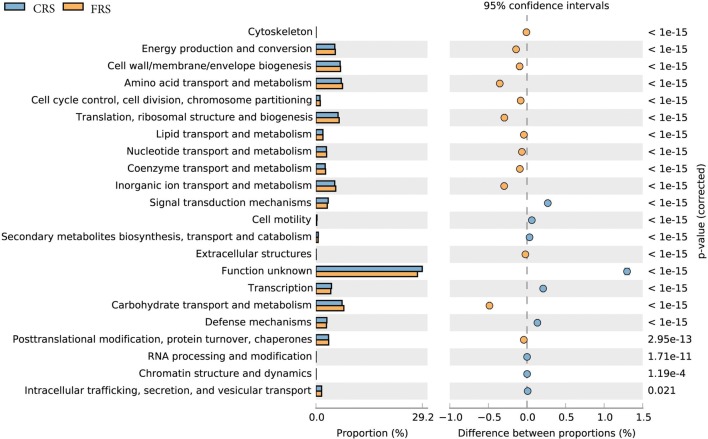
Functional classification of open reading frames based on the Cluster of Orthologous Genes (COG) obtained from assembled metagenomic datasets allowed detection of significant differences in relative abundance of COG functional categories between the two datasets. The analysis results showed that the abundance of annotation in the FRS dataset is significantly higher than that of CRS, mainly including energy production and conversion, carbohydrate transport and metabolism, as well as amino acid transport and metabolism were shown to be the most overrepresented in both datasets (P < 0.05; Fig. 4).
Fig. 4.
COG functional classification and difference in COG abundance
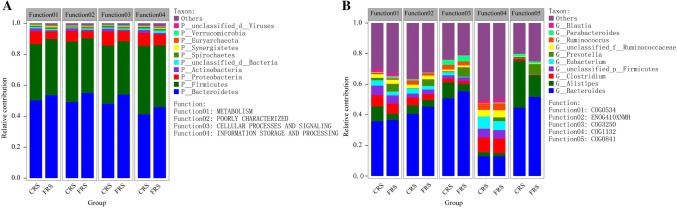
Based on the corresponding relationship between species and functions in the sample, the correlation analysis between species and functional contribution relative abundances is performed. At the Category function level, we found that the main functional contributions of Bacteroidetes (more than the contribution 41%), Firmicutes (more than the contribution 36%) and Proteobacteria (more than the contribution 2%) in FRS were metabolism, cellular processes and signals, and information storage and processing functions, while in CRS, these functions enriched in the Bacteroidetes (more than the contribution 41%), Firmicutes (more than the contribution 36%), Proteobacteria (more than the contribution 2%), and Actinobacteria (more than the contribution 1%) (Fig. 5a). In addition, in the analysis of species and COG contribution, We annotated functions of the Bacteroides and Prevotella species in FRS as COG0841 (acriflavin resistance protein, inorganic ion transport and metabolism, METABOLISM), COG0534 (Mate efflux family protein, defense FRShanisms, CELLULAR PROCESSES AND SIGNALING), COG0325 (alanine racemase domain protein, function unknown, POORLY CHARACTERIZED), COG1132 ((ABC) transporter, defense FRShanisms, CELLULAR PROCESSES AND SIGNALING), and ENOG410XNMH (Histidine kinase, signal transduction FRShanisms, CELLULAR PROCESSES AND SIGNALING), and found they had a higher contribution than in CRS; however, contribution of the Alistipes and Clostridium spp. was lower in FRS (Fig. 5b).
Fig. 5.
Species and functional contribution analysis. Contributions of a species contribution relationship of category functions at the phylum level. Contributions of b species contribution relationship of NOG functions at the gene level
Antibiotic resistance profiles of chicken intestinal microbiome
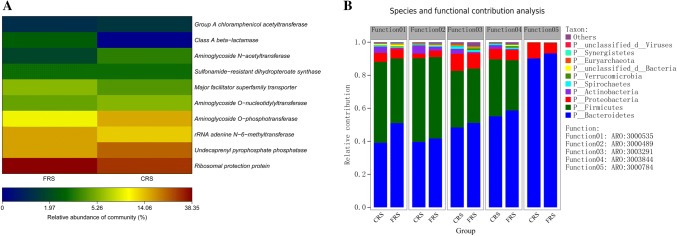
To investigate the antibiotic resistance genes (ARGs) present in the gut microbiota of free-range system (FRS) and cage rearing systems (CRS), the metagenome data were screened for antibiotic resistance factors using the Antibiotic Resistance Genes Database (ARBD) and the Comprehensive Antibiotic Resistance Database (CARD). At the class level, we chose the relative abundance of antibiotic resistance genes as greater than 1% for annotation. The result of the annotation is that the Ribosomal protection protein encoding genes were the most abundant (CRS: 8.96%), represented by undecaprenyl pyrophosphate phosphatase, aminoglycoside O-phosphotransferase, and aminoglycoside N-acetyltransferase-encoding genes (6.84, 6.55, and 1.65%, respectively). In FRS, we observed abundance of genes encoding for rRNA adenine N-6-methyltransferase resistance was 3.16% (Fig. 6a). The top five most abundant antibiotic resistance ontology (ARO) genes based on CARD are shown in Fig. 6b, and the range of identified AROs (Table 1) included ARO:3000535, ARO:3000489, ARO:3003291, ARO:3003844, and ARO:3000784. Based on Species and functional contribution analysis in the sample, the correlation analysis between the microbial origin and the relative abundance of these AROs were further analyzed. In FRS, functional species contributions of ARO were highly enriched in the Bacteroidetes (41%), Firmicutes (30%), and Proteobacteria (3%) (Fig. 6b), while in CRS, functional species contributions were found enriched in the Bacteroidetes (39%), Firmicutes (34%), Proteobacteria (2%), and Actinobacteria (2%). Bacteroidetes (90%) and Proteobacteria (6%) were detected almost to the exclusion of other phyla in ARO:3000784 (Fig. 6b).
Fig. 6.
a Relative abundance of predictive enzymes involved in transmitting antibiotic resistance present in the FRS and CRS metagenomic datasets. The names of the coding genes are listed on the right, and the names of the sample groups used are listed at the bottom, and b relationship between contribution of species and antibiotic resistance genes contribution analysis
Table 1.
Antibiotic resistance gene annotation
| ARO_accession | ARO_name | ARO_description | CRS | FRS | Total |
|---|---|---|---|---|---|
| ARO:3000535 | macB | Efflux pump conferring antibiotic resistance | 56,658 | 31,964 | 88,622 |
| ARO:3000489 | sav1866 | Efflux pump conferring antibiotic resistance | 48,522 | 25,396 | 73,918 |
| ARO:3003291 | Staphylococcus aureus rpoC conferring resistance to daptomycin | Antibiotic resistant gene variant or mutant, lipopeptide antibiotic resistance gene | 41,090 | 31,636 | 72,726 |
| ARO:3003844 | mfd | Antibiotic target protection protein, fluoroquinolone resistance gene | 42,674 | 27,964 | 70,638 |
| ARO:3000784 | cmeB | Efflux pump conferring antibiotic resistance | 34,602 | 18,908 | 53,510 |
Discussion
Gut microbiota are known to be influenced by many factors, such as diet, age, and feeding regimen (Li et al. 2012). The study (Mancabelli et al. 2016) suggested that large-scale commercial production and semi-wild rearing systems influenced intestinal microbial composition and diversity in broiler chickens. However, intestinal microbiota in hens has previously been unclear. In this study, we observed that composition of gut microbes in hens differed between the two rearing systems by analysis of the 16S rRNA profiling. At the phylum level, the intestinal flora of laying hens was mainly composed of Bacteroidetes and Firmicutes, and abundance of the Proteobacteria was relatively low. These results support other studies (Wei et al. 2013) that have shown chicken cecal microbes are dominated by the Firmicutes and Bacteroidetes that are also commonly observed in the gut environments of many birds (Waite and Taylor 2014; Wang et al. 2016b). The role of members of these two phyla in food digestion has been frequently studied. For example, members of Firmicutes are involved in the degradation of insoluble fibers (Berry 2016), and Proteobacteria members are associated with cellulose activity (Reid et al. 2011). Interestingly, we found high levels of abundance of Deferribacteres in FRS, but low levels in CRS (Fig. 1d). This difference may be due to the difference between the two rearing systems, and further research is needed.
In this study, there were 25 unique genera in hens reared under the FRS system, and 4 unique genera in CRS hens (Fig. 2a). Diversity of cecal microbiota was greater in FRS than in CRS (Fig. 1a, b, d), and we found that abundance of Faecalibacterium prausnitzii and Ruminococcaceae spp. differed between hens reared under the FRS and CRS systems (P < 0.05; Fig. 2c). The Firmicutes mainly comprised Faecalibacterium and Clostridium (Hold et al. 2002). Some intestinal clostridial microorganisms produce short-chain fatty acids, such as Clostridium IX, Faecalibacterium, and Clostridium, that play a role in resistance to colonization by pathogenic microorganisms (Van den Abbeele et al. 2010). Ruminococcaceae, which is related to growth performance in broilers (Torok et al. 2011), is highly abundant in efficient feed conversion in birds (Stanley et al. 2016) and it can be an indicator of feed efficiency in cecal digesta (Siegerstetter et al. 2017). The differences in cecal microbes between the rearing systems are likely to be a result of differences in breeding environment, environmental pressure, and space.
The gut microecosystem performs numerous functions that alter microbe composition (Zhang et al. 2018). In this study, dominant microbes found in hens gut, such as the phyla Bacteroidetes, Firmicutes, and Deferribacteres, as well as the genus Deferribacter, may be involved in host nutrient metabolism, especially of carbohydrates, amino acids, and peptides. We found that abundance of functional genes (carbohydrate transport and metabolism, energy production and conversion, amino acid transport and metabolism, and genes related to replication, recombination, and repair; Fig. 4) in hens reared under the FRS system was higher than in CRS. Numerous functional genes are involved in metabolic pathways, such as metabolism of carbohydrates, amino acids, lipids, and energy, and it is possible that hens reared under the FRS system require more energy to provide enhanced muscle development and synthesis of body protein to support the greater amounts of physical movement associated with this system.
An analysis of the overall functional profiles in the present study indicated that the gut microbes associated with both rearing systems exhibited high metabolic activities. These results were consistent with earlier studies on the gut metagenomes of chicken (Mancabelli et al. 2016). This high metabolic rate may be related to the energy consumption required to fulfill the demands of outdoor sport, FRS hens exceed CRS in outdoor sport. Avian metabolism was reported to be approximately 60% higher than that of most mammals (Scanes and Braun 2013). The comparison of the functional profiles of the datasets from FRS and CRS revealed many differences, which suggested that not only the bacterial compositions but their functionalities were important. For example, functions related to carbohydrate metabolism, lipid metabolism, amino acid metabolism, and glycan biosynthesis and metabolism were significantly more abundant in FRS than that in CRS. Thus, it is reasonable to hypothesize that the microbiota of the FRS is probably specialized to degrade more diverse types of foods than that of the CRS, which eats a more homogenous type of food.
In this study, ARGs were identified in the microbiota of hens in both rearing systems, and the ARG spectrum was related to the composition of the microbe community, where most of the ARG species contributed to the dominant phyla Firmicutes (> 30%) and Bacteroidetes (> 39%). Previous work has shown that the phyla of these bacteria, commonly known as antibiotic producing bacteria, were also present in the gut microbes of four bird species (Wright 2007), therefore, we suggest potential environmental pollution risks associated with the widespread and persistent use of antibiotics and associated resistance should be reduced.
Conclusions
Bacteroidetes, Firmicutes, and Proteobacteria were the main cecal bacterial phyla in FRS and CRS and Deferribacteres were found in FRS and almost absent in CRS. Parabacteroides, Prevotellaceae_Ga6A1, Unclassified Proteobacteria, and unclassified Spirochaetaceae were more abundant in FRS, and Faecalibacterium, Ruminococcaceae, and Helicobacter were more abundant in CRS. The main functional annotations of species were replication, recombination and repair, energy production and transformation, cell wall/membrane/envelope biogenesis, and amino acid transport and metabolism-related functions. These results indicate that in hens of the different rearing systems can cause changes in the abundance and structural composition of the gut microbiota, although the underlying mechanisms are unclear. And ARGs were also identified in the microbiota of these hens. This work has produced new data for laying hens in different production systems and increased the understanding of intestinal microorganisms in laying hens.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the National Science Foundation of China (Grant no. 31772707) for supporting the high-throughput sequencing. The collection of the experimental samples was supported by the Integration and Demonstration of Quality and Safety Control Technology for Green Ecological Livestock and Poultry Products Industry Chain (Grant no. 1604a0702033) and the Animal Food Quality and Safety Control, Anhui Province 115 Industry Innovation Team.
Authors’ contributions
KQ and SL conceived and designed the experiments; SS, ZQ, and BG performed the experiments; SS, BC, YS, and XS analyzed the data; SS and JT contributed reagents/materials/Únalysis tools; SS, ZQ, and BG wrote the paper. All authors critically read and contributed to the manuscript and approved the final version. All authors critically read and contributed to the manuscript and approved the final version.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval and consent to participate
This study was performed in accordance with the Chinese Laboratory Animal Administration Act of 1988. Before the experiments, the research protocol was reviewed and approved by the Research Ethics Committee of Anhui Agricultural University. Permission was obtained from all managers of the chicken farms studied before the samples were collected.
Footnotes
Shuiqin Shi and Zhao Qi contributed equally to this work.
Contributor Information
Shuiqin Shi, Phone: 15755101655, Email: 879237477@qq.com.
Zhao Qi, Phone: 15755101655, Email: 403069355@qq.com.
Bintao Gu, Phone: 15755101655, Email: 289003407@qq.com.
Baoyan Cheng, Phone: 15755101655, Email: 1776547064@qq.com.
Jian Tu, Phone: 15755101655, Email: 157518784@qq.com.
Xiangjun Song, Phone: 15755101655, Email: 593258787@qq.com.
Yin Shao, Phone: 15755101655, Email: 1611623019@qq.com.
Hongmei Liu, Phone: 15755101655, Email: liu2844707@sina.com.
Kezong Qi, Phone: 15755101655, Email: qkz@ahau.edu.cn.
Shaowen Li, Phone: 15755101655, Email: shwli@ahau.edu.cn.
References
- Adil S, Magray SN. Impact and manipulation of gut microflora in poultry: a review. J Anim Vet Adv. 2012;11(6):873–877. doi: 10.3923/javaa.2012.873.877. [DOI] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes EM. The avian intestinal flora with particular reference to the possible ecological significance of the cecal anaerobic bacteria. Am J Clin Nutr. 1972;25(12):1475–1479. doi: 10.1093/ajcn/25.12.1475. [DOI] [PubMed] [Google Scholar]
- Berry D. The emerging view of Firmicutes as key fibre degraders in the human gut. Environ Microbiol. 2016;18(7):2081–2083. doi: 10.1111/1462-2920.13225. [DOI] [PubMed] [Google Scholar]
- Best AA, Porter AL, Fraley SM, Fraley GS. Characterization of gut microbiome dynamics in developing pekin ducks and impact of management system. Front Microbiol. 2017 doi: 10.3389/fmicb.2016.02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao GT, Zeng XF, Chen AG, Zhou L, Zhang L, Xiao YP, Yang CM. Effects of a probiotic, Enterococcus faecium, on growth performance, intestinal morphology, immune response, and cecal microflora in broiler chickens challenged with Escherichia coli K88. Poult Sci. 2013;92(11):2949–2955. doi: 10.3382/ps.2013-03366. [DOI] [PubMed] [Google Scholar]
- Chang Q, Luan Y, Sun F. Variance adjusted weighted UniFrac: a powerful beta diversity measure for comparing communities based on phylogeny. BMC Bioinform. 2011 doi: 10.1186/1471-2105-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo V, Vives Florez MJ. Non-invited review the gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: a review. Poult Sci. 2018;97(3):1006–1021. doi: 10.3382/ps/pex359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Wang Q, Liu S, Sun R, Zhou Y, Li Y. Age-Related variations in intestinal microflora of free-range and caged hens. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/aem.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Eren AM, Vineis JH, Morrison HG, Sogin ML. a filtering method to generate high quality short reads using illumina paired-end technology. PLoS One. 2013 doi: 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform. 2006;2:121–128. [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GG, Kim EB, Lee J, Lee J-Y, Jin G, Park J, Huh C-S, Kwon I-K, Kil DY, Choi Y-J, Kong C. Relationship between the microbiota in different sections of the gastrointestinal tract, and the body weight of broiler chickens. Springerplus. 2016 doi: 10.1186/s40064-016-2604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hold GL, Pryde SE, Russell VJ, Furrie E, Flint HJ. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol. 2002;39(1):33–39. doi: 10.1111/j.1574-6941.2002.tb00904.x. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Li M, Zhou M, Adamowicz E, Basarab JA, Guan LL. Characterization of bovine ruminal epithelial bacterial communities using 16S rRNA sequencing, PCR-DGGE, and qRT-PCR analysis. Vet Microbiol. 2012;155(1):72–80. doi: 10.1016/j.vetmic.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Mancabelli L, Ferrario C, Milani C, Mangifesta M, Turroni F, Duranti S, Lugli GA, Viappiani A, Ossiprandi MC, van Sinderen D, Ventura M. Insights into the biodiversity of the gut microbiota of broiler chickens. Environ Microbiol. 2016;18(12):4727–4738. doi: 10.1111/1462-2920.13363. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Green genes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld JD, Mohn WW. Unexpectedly high bacterial diversity in arctic tundra relative to boreal forest soils, revealed by serial analysis of ribosomal sequence tags. Appl Environ Microbiol. 2005;71(10):5710–5718. doi: 10.1128/aem.71.10.5710-5718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34(19):5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Yu Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes. 2014;5(1):108–119. doi: 10.4161/gmic.26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid NM, Addison SL, Macdonald LJ, Lloyd-Jones G. Biodiversity of active and inactive bacteria in the gut flora of wood-feeding huhu beetle larvae (Prionoplus reticularis) Appl Environ Microbiol. 2011;77(19):7000–7006. doi: 10.1128/aem.05609-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanes CG, Braun Avian metabolism: its control and evolution. Front Biol. 2013;8(2):134–159. doi: 10.1007/s11515-012-1206-2. [DOI] [Google Scholar]
- Siegerstetter S-C, Schmitz-Esser S, Magowan E, Wetzels SU, Zebeli Q, Lawlor PG, O’Connell NE, Metzler-Zebeli BU. Intestinal microbiota profiles associated with low and high residual feed intake in chickens across two geographical locations. PLoS One. 2017 doi: 10.1371/journal.pone.0187766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D, Geier MS, Denman SE, Haring VR, Crowley TM, Hughes RJ, Moore RJ. Identification of chicken intestinal microbiota correlated with the efficiency of energy extraction from feed. Vet Microbiol. 2013;164(1–2):85–92. doi: 10.1016/j.vetmic.2013.01.030. [DOI] [PubMed] [Google Scholar]
- Stanley D, Geier MS, Hughes RJ, Denman SE, Moore RJ. Highly variable microbiota development in the chicken gastrointestinal tract. PLoS One. 2013 doi: 10.1371/journal.pone.0084290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D, Hughes RJ, Geier MS, Moore RJ. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol. 2016 doi: 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok VA, Hughes RJ, Mikkelsen LL, Perez-Maldonado R, Balding K, MacAlpine R, Percy NJ, Ophel-Keller K. Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl Environ Microbiol. 2011;77(17):5868–5878. doi: 10.1128/aem.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Djukovic A, Isaac S. Roles of the intestinal microbiota in pathogen protection. Clin Transl Immunol. 2017 doi: 10.1038/cti.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abbeele P, Grootaert C, Marzorati M, Possemiers S, Verstraete W, Gerard P, Rabot S, Bruneau A, El Aidy S, Derrien M, Zoetendal E, Kleerebezem M, Smidt H, Van de Wiele T. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for bacteroidetes and clostridium cluster IX. Appl Environ Microbiol. 2010;76(15):5237–5246. doi: 10.1128/aem.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite DW, Taylor MW. Characterizing the avian gut microbiota: membership, driving influences, and potential function. Front Microbiol. 2014 doi: 10.3389/fmicb.2014.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite DW, Taylor MW. Exploring the avian gut microbiota: current trends and future directions. Front Microbiol. 2015 doi: 10.3389/fmicb.2015.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lilburn M, Yu Z. Intestinal microbiota of broiler chickens as affected by litter management regimens. Front Microbiol. 2016 doi: 10.3389/fmicb.2016.00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cao J, Yang F, Wang X, Zheng S, Sharshov K, Li L. High-throughput sequencing reveals the core gut microbiome of bar-headed goose (Anser indicus) in different wintering areas in tibet. Microbiol Open. 2016;5(2):287–295. doi: 10.1002/mbo3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Morrison M, Yu Z. Bacterial census of poultry intestinal microbiome. Poult Sci. 2013;92(3):671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5(3):175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C-Y, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoman CJ, Chia N, Jeraldo P, Sipos M, Goldenfeld ND, White BA. The microbiome of the chicken gastrointestinal tract. Anim Health Res Rev. 2012;13(1):89–99. doi: 10.1017/s1466252312000138. [DOI] [PubMed] [Google Scholar]
- Zhang B, Lv Z, Li Z, Wang W, Li G, Guo Y. Dietary l-arginine supplementation alleviates the intestinal injury and modulates the gut microbiota in broiler chickens challenged by Clostridium perfringens. Front Microbiol. 2018 doi: 10.3389/fmicb.2018.01716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jiang X, Yang C, Ma B, Lei C, Xu C, Zhang A, Yang X, Xiong Q, Zhang P, Men S, Xiang R, Wang H. Cecal microbiota of tibetan chickens from five geographic regions were determined by 16S rRNA sequencing. Microbiol Open. 2016;5(5):753–762. doi: 10.1002/mbo3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.