Abstract
The co-production of MCR and carbapenemase in Enterobacteriaceae has been previously reported. Here, we describe a clinical strain of Escherichia coli from Vietnam carrying both mcr-1 and blaNDM–1. Whole-genome sequencing showed that the genome of this strain consists of a 4,975,832-bp chromosome and four plasmids. The mcr-1 and blaNDM–1 genes are located on IncI2 and IncA/C2-type plasmids, respectively. Genetic analysis revealed the presence of a multidrug-resistant region with the structure of a novel complex class 1 integron including a class 1 integron region bearing two 5′ conserved segments and one 3′ conserved segment and two complete structures of ISCR1. The complex integron contains aminoglycoside resistance genes aadA2, aadB, strA, strB, and aphA6, quinolone resistance gene qnrA1, extended-spectrum β-lactamase gene blaOXA–4, and a Tn125-like transposon bearing blaNDM–1. In addition, the dfrA12-gcuF-aadA2-cmlA1-aadA1-qacH gene cassette array belonging to the sul3-type integron was also identified, but the region found downstream of the gene cassette array is the IS440-tet(M)-IS26 element instead of the sul3 gene. The results further support that Enterobacteriaceae isolates co-harboring mcr and blaNDM are widely being distributed. The structural characteristics of the complex integron reveal that ISCR1 elements play an important role in the mobilization of blaNDM–1 and the development of multidrug-resistant regions.
Keywords: mcr-1, blaNDM–1, colistin, carbapenemase, complex class 1 integron, sul3-type integron, Escherichia coli
Introduction
Infections caused by carbapenem-resistant Enterobacteriaceae (CRE) are a serious problem for global health. The main mechanism for carbapenem resistance in bacteria is the production of carbapenemases, out of which New Delhi metallo-lactamase (NDM) is one of the most common groups (Iovleva and Doi, 2017). The blaNDM–1 gene was mainly found on multiple plasmid types, including IncA/C, IncFIA, IncFIB, IncFII, and IncX3 (Wu et al., 2019). This indicates that the horizontal gene transfer mediated by plasmids plays a vital role in the widespread dissemination of blaNDM–1. Infections caused by NDM-1-producing Enterobacteriaceae are of major concern because those isolates tend to spread among patients, which poses a challenge to control the outbreak of CRE carrying blaNDM–1. In addition, NDM-1-producing plasmids also carry additional genes conferring resistance to other antibiotics, such as aminoglycosides (aacA29 and aadA16), chloramphenicol (catA1 and cmlA1), and tetracyclines (tetB) (Rojas et al., 2016), resulting in the limited choices of antibiotics in treatment. In several plasmids, the blaNDM–1 gene was flanked by two copies of insertion sequences (e.g., ISAba125, IS26, and IS3000), which generate the composite transposons carrying blaNDM–1 (Wu et al., 2019). Class 1 integrons were found linked to the complete structures of insertion sequence common region 1 (ISCR1) including a copy of ISCR1, a variable region (VR) bearing resistance genes, and a copy of the 3′ conserved segment (CS) of the class 1 integron to construct large genetic elements that are usually called “complex class 1 integrons” (Toleman et al., 2006). Some reports showed that blaNDM–1 was also inserted into the ISCR1-linked VR of complex class 1 integrons (Chen et al., 2014; Li et al., 2014), suggesting that the mobilization of blaNDM–1 is most likely associated with ISCR1 as well as the formation of composite transposons.
Colistin is one of the few antibiotics that are still effective against CRE. However, the mutation of chromosomal genes involved in lipopolysaccharide production have impacts on the susceptibility of bacteria to colistin (Olaitan et al., 2014). Moreover, colistin resistance has become more serious since plasmid-mediated colistin resistance gene mcr-1 was first reported in 2015 (Liu et al., 2016). Presently, eight additional mcr genes (mcr-2 to mcr-9) and their variants have been described in Enterobacteriaceae (Xavier et al., 2016; AbuOun et al., 2017; Borowiak et al., 2017; Carattoli et al., 2017; Yin et al., 2017; Wang et al., 2018c; Yang et al., 2018; Carroll et al., 2019). Although the presence of mcr was found on chromosomes (Zhou et al., 2017), the mcr genes were mainly reported to be carried on different plasmid types, such as IncI2, IncHI2, IncX4, IncP, and IncFII (Xavier et al., 2016; Matamoros et al., 2017; Wang et al., 2018c). IncX4, IncI2, and IncHI2 are the most common types of plasmids carrying mcr-1 (Matamoros et al., 2017). On the other hand, these plasmids also harbor additional resistance genes (e.g., blaCTX–M–14, fosA3, floR, and oqxAB) and can be transferred via conjugation (Wu et al., 2018). However, the ISApl1 composite transposon was considered as the main vehicle for the widespread distribution of mcr-1 (Wang et al., 2018a).
The mcr-1 gene was detected in colistin-resistant Enterobacteriaceae isolated from clinical samples (Du et al., 2016; Tada et al., 2017) and has been commonly found in colistin-resistant Escherichia coli recovered from farm animals and foods in China (Wang et al., 2018b; Liu et al., 2019) and Vietnam (Trung et al., 2017; Yamaguchi et al., 2018). More importantly, a recent study showed that the existence of mcr-1 is the predominant genotype of colistin-resistant E. coli isolated from healthy residents in Vietnam (Yamamoto et al., 2019). The prevalence of mcr-1 in multiple sources revealed that these sources might be potential reservoirs for the wide spread of colistin-resistant bacteria producing MCR-1 in livestock-producing countries such as Vietnam. Significantly, clinical mcr-1- and blaNDM-positive Enterobacteriaceae isolates, particularly E. coli and Klebsiella pneumoniae, were recently found in various regions (Du et al., 2016; Mediavilla et al., 2016; Zheng et al., 2016), which poses a threat to the effectiveness of carbapenems and colistin. In Vietnam, blaNDM is the most common genotype reported in carbapenemase-producing E. coli (Malchione et al., 2019), and mcr-1 has been frequently found in colistin-resistant E. coli isolated from different sources (Trung et al., 2017; Yamaguchi et al., 2018). Thus, we hypothesized that multidrug-resistant Enterobacteriaceae strains that simultaneously harbor mcr-1 and blaNDM are currently circulating in Vietnam. Recently, we have obtained a clinical E. coli strain resistant to multiple antibiotics, including carbapenems and colistin. The aim of this study was to elucidate the antibiotic resistance mechanisms of the isolate, especially to detect the coexistence of mcr-1 and blaNDM.
Materials and Methods
Bacterial Isolate
Escherichia coli EC17GD31 was recovered from the blood sample of a patient admitted to the intensive care unit of Nhan Dan Gia Dinh hospital (Ho Chi Minh City, Vietnam) in March 2017. The patient was diagnosed as having acute respiratory distress syndrome, pneumonia, and acute kidney injury and had a medical history of cerebral infarction. EC17GD31 was isolated in the routine diagnostic laboratory of the hospital and then sent to Pasteur Institute in Ho Chi Minh City for further analysis. The isolate was grown on brain heart infusion agar overnight at 37°C and species-identified by using VITEK 2 system (BioMérieux, United States).
This study received ethical approval from the Institutional Ethics Committee in Biomedical Research, Pasteur Institute in Ho Chi Minh City. Informed consent was not required for this study, since the strain was isolated as a part of the routine hospital laboratory procedures.
Antibiotic Susceptibility
The minimum inhibitory concentrations (MICs) of amikacin, gentamicin, ampicillin, amoxicillin-clavulanic acid, cefotaxime, ceftazidime, ceftriaxone, cefepime, ertapenem, imipenem, meropenem, cefoxitin, ciprofloxacin, chloramphenicol, tetracycline, and trimethoprim-sulfamethoxazole were determined by agar dilution method, although MICs of colistin and tigecycline were performed with broth microdilution. Colistin and tigecycline resistance was interpreted according to the breakpoints of the European Committee on Antimicrobial Susceptibility Testing (EUCAST1), whereas other antibiotics were identified according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2018).
Conjugation Assay
The transferability of the mcr-1-bearing plasmid was examined by conjugation experiment. E. coli J53, which is resistant to sodium azide, was used as the recipient. E. coli J53 transconjugants that harbor MCR-1-encoding plasmids were selected on brain heart infusion agar plates containing 100 μg/ml sodium azide and 2 μg/ml colistin. The presence of mcr-1 in the transconjugants was confirmed by PCR as previously described (Liu et al., 2016).
Whole-Genome Sequencing and Bioinformatics
The genomic DNA was extracted from EC17GD31 using a QIAamp DNA Mini Kit (Qiagen, Germany). Whole-genome sequencing was carried out using the MinION sequencer (Oxford Nanopore Technologies, United Kingdom). Genome assembly was performed by Canu 1.6 (Koren et al., 2017) and then corrected using Pilon 1.22 (Walker et al., 2014) with reads obtained from 250-bp paired-end Illumina MiSeq sequencing (Illumina Inc., United States). NCBI Prokaryotic Genome Annotation Pipeline2 was used to annotate the genomic sequences. Multilocus sequence typing of the isolate, plasmid replicon typing, and identification of resistance genes were done using tools from the Center for Genomic Epidemiology website3. Insertion sequences were identified based on the ISfinder database4. Sequence comparison was completed using BLASTn5. The circular image and linear comparison figures of sequences were generated by CGView (Stothard and Wishart, 2004) and Easyfig (Sullivan et al., 2011), respectively.
Phylogenetic Analysis of IncI2 Plasmids
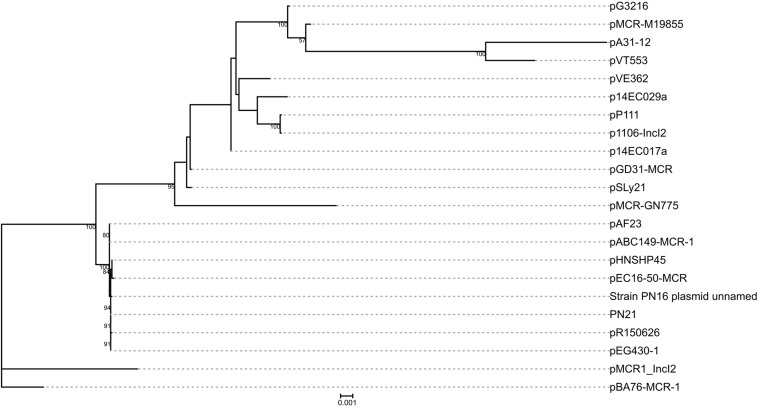
Complete sequences of IncI2 plasmids carrying mcr-1 (n = 21) were obtained from GenBank (Supplementary Table 1). A sum of 35 genes were identified as the core genome of the plasmids (including pGD31-MCR) using OrthoFinder version 2.3.3 (Emms and Kelly, 2015). The alleles of these genes were aligned as protein sequences using MAFFT version 7.427 (Katoh and Standley, 2013), and then the protein sequences of each plasmid were concatenated into a single sequence. RAxML version 8.2.12 (Stamatakis, 2014) was used to infer a phylogenetic tree with a 1000-bootstrap test. The phylogenetic tree was drawn using the iTOL web server (Letunic and Bork, 2019).
Nucleotide Sequence Accession Numbers
Complete sequences of the chromosome and plasmids of E. coli EC17GD31 have been deposited in GenBank under the accession numbers CP031293 to CP031297.
Results and Discussion
Genomic Characteristics of E. coli EC17GD31
The genome of E. coli EC17GD31 consists of a 4,975,832 bp chromosome with an average 50.46% GC content and four circular plasmids (Table 1). The isolate was resistant to all tested antibiotics, except for tigecycline (Table 2). It is notable that EC17GD31 was resistant to both carbapenems (MICs ≥ 8 μg/ml) and colistin (MIC = 4 μg/ml). Identification of 22 antibiotic resistance genes showed good correlation to the antibiotic susceptibility pattern of E. coli EC17GD31 (Table 1). The detection of blaTEM–1B, blaCTX–M–55, blaOXA–4, and blaNDM–1 explained the resistance to all tested β-lactam antibiotics, including extended-spectrum cephalosporins and carbapenems (Table 2). The existence of mcr-1 and efflux pump-encoding genes (cmlA1 and floR) was related to the resistance to colistin and chloramphenicol, respectively. In addition, the genome of EC17GD31 carries genes conferring the resistance to aminoglycosides (aadA1, aadA2, aadB, aph(3′)-Ia, aphA6, strA, and strB), quinolones (qnrA1), tetracyclines [tet(A) and tet(M)], sulfonamides (sul1, sul2, and sul3), and trimethoprim (dfrA12) (Table 1). The multidrug resistance transporter-encoding gene mdf(A) was also found in EC17GD31, which probably contributed to the resistance of this isolate to tetracycline, chloramphenicol, and ciprofloxacin (Edgar and Bibi, 1997). The blaCTX–M–55 encoding extended-spectrum beta-lactamase (ESBL) and mdf(A) genes are located on the chromosome, whereas other resistance genes are present on three plasmids. Moreover, three mutations in type II topoisomerases were identified, including substitution Ser83Leu and Asp87Tyr in GyrA and Ser80Ile in ParC. These mutations are the most common quinolone resistance-associated substitutions in E. coli (Ruiz, 2003), and their coexistence confers high-level fluoroquinolone resistance (Morgan-Linnell and Zechiedrich, 2007). Chromosomal genes (mgrB, phoP, phoQ, pmrB, and pmrA) involved in the susceptibility of bacteria to colistin were found to have no mutations, suggesting that colistin resistance of EC17GD31 is only related to the presence of mcr-1.
TABLE 1.
Genomic features of E. coli EC17GD31.
| Chromosome/plasmid | Type | Size (bp) | GC content (%) | Resistance gene (s) |
| EC17GD31 | ST457 | 4,975,832 | 50.46 | blaCTX–M–55, mdf(A) |
| pGD31-MCR | IncI2 | 60,038 | 42.28 | mcr-1 |
| pGD31-NDM | IncA/C2 | 188,230 | 51.62 | aadA2, aadB, aphA6, blaNDM–1, blaOXA–4, bleMBL, qacEΔ1, qnrA1, strA, strB, sul1 |
| pGD31-F25 | IncFIB (pLF82) | 110,686 | 45.70 | - |
| pGD31-F1928 | IncFIB (AP001918), IncFII, IncR | 245,305 | 51.37 | aadA1, aadA2, aph(3′)-Ia, blaTEM–1B, cmlA1, dfrA12, floR, Δmef(B), Δmig-14, qacH, sul2, sul3, tet(A), tet(M) |
TABLE 2.
Minimum inhibitory concentrations of tested antibiotic agents for E. coli EC17GD31, the transconjugant, and E. coli J53 recipient strain.
| Isolate |
MIC (μg/ml) |
|||||||||||||||||
| AMK | GEN | AMP | AMC | CTX | CAZ | CRO | FEP | ETP | IPM | MEM | FOX | CIP | CHL | CST | TET | TGC | SXT | |
| EC17GD31 | 128 | >256 | >256 | 32 | >256 | >256 | >256 | 24 | 32 | 8 | 128 | >256 | 128 | 256 | 4 | 256 | 0.06 | >32 |
| Transconjugant | 2 | 1.5 | 6 | 4 | 0.06 | 1 | 0.03 | 0.16 | 0.008 | 1 | 0.015 | 8 | 0.03 | 8 | 4 | 2 | 0.06 | 0.47 |
| E. coli J53 | 2 | 1.5 | 4 | 3 | 0.125 | 0.25 | 0.03 | 0.016 | 0.008 | 1 | 0.015 | 8 | 0.03 | 4 | 0.5 | 2 | 0.125 | 0.047 |
AMK, amikacin; GEN, gentamicin; AMP, ampicillin; AMC, amoxicillin-clavulanic acid; CTX, cefotaxime; CAZ, ceftazidime; CRO, ceftriaxone; FEP, cefepime; ETP, ertapenem; IPM, imipenem; MEM, meropenem; FOX, cefoxitin; CIP, ciprofloxacin; CHL, chloramphenicol; CST, colistin; TET, tetracycline; TGC, tigecycline; SXT, trimethoprim-sulfamethoxazole.
Multilocus sequence typing indicated that EC17GD31 belongs to ST457 according to the Warwick scheme. Several reports have described E. coli ST457 isolated from animals and hospitals also carries the mcr-1 gene (Suzuki et al., 2016; Tada et al., 2017). Furthermore, this clone and ST410 were the first identified clones of clinical E. coli from Vietnam harboring mcr-1 (Tada et al., 2017). Significantly, carbapenem resistance gene blaNDM–1 and colistin resistance gene mcr-1 coexist in EC17GD31. To the best of our knowledge, this is the first report of a clinical E. coli strain from Vietnam that co-harbors mcr-1 and blaNDM–1. NDM- and MCR-coproducing Enterobacteriaceae isolates, mainly E. coli, were reported in numerous other countries, e.g., China (Zheng et al., 2016), the United States (Mediavilla et al., 2016), the Arabian Peninsula (Sonnevend et al., 2016), Venezuela (Delgado-Blas et al., 2016), and Thailand (Paveenkittiporn et al., 2017). The NDM genes blaNDM–1 (Delgado-Blas et al., 2016; Zheng et al., 2016; Paveenkittiporn et al., 2017) and blaNDM–5 (Mediavilla et al., 2016; Yang et al., 2016; Zheng B. et al., 2018), followed by blaNDM–9 (Yao et al., 2016; Lai et al., 2017), were commonly detected in E. coli isolates co-carrying blaNDM and mcr-1. The emergence and dissemination of NDM- and MCR-coproducing Enterobacteriaceae has become an urgent threat to clinical treatment and infection control.
Genetic Features of Plasmid pGD31-MCR
pGD31-MCR is a 60,038-bp circular plasmid bearing the mcr-1 gene, with an average GC content of 42.28%. The plasmid belongs to the IncI2 group, which is the most prevalent MCR-1-producing plasmid group in Asia (Matamoros et al., 2017). pGD31-MCR only carries mcr-1, whereas some other IncI2-type plasmids harbor both the mcr-1 and CTX-M ESBL genes (Sun et al., 2016; Matamoros et al., 2017). Colistin-resistant transconjugant indicated that the plasmid carrying the mcr-1 gene was transferred to the E. coli J53 recipient strain (Table 2). This result showed that pGD31-MCR is a self-conjugative plasmid. Based on the core genome of IncI2 plasmids harboring mcr-1, the phylogenetic analysis revealed that pGD31-MCR shared the highest level of sequence similarity with pSLy21 (Genbank accession no. CP016405) in E. coli strain 210221272 isolated from a pig (Meinersmann et al., 2016; Figure 1). The sequence comparison of pGD31-MCR using BLASTn showed 99% identity and 97% coverage with pHNSHP45 (Genbank accession no. KP347127), the first identified plasmid harboring mcr-1 (Liu et al., 2016). Similar to pHNSHP45, the mcr-1 gene is located in a region with a structure of nikB-ISApl1-mcr-1-pap2. The ISApl1-mediated composite transposon was considered to be the primary genetic context of mcr-1 (Snesrud et al., 2016; Wang et al., 2018a). Illegitimate recombination may results in the loss of ISApl1 flanking elements, leading to the stable existence of mcr-1 in plasmids or chromosomes (Snesrud et al., 2016).
FIGURE 1.
Phylogenetic analysis on pGD31-MCR and 21 other IncI2 mcr-1-carrying plasmids obtained from GenBank. The accession numbers of the plasmids are shown in Supplementary Table 1. Bootstrap values are shown next to nodes.
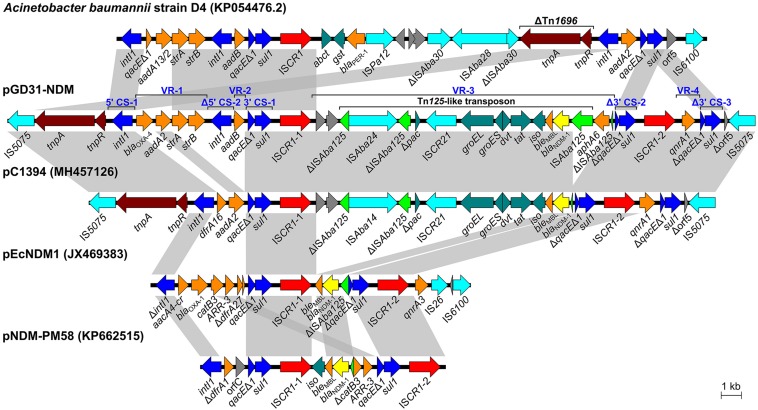
A Novel Complex Class 1 Integron in Plasmid pGD31-NDM
Plasmid pGD31-NDM is a circular molecule of 188,230 bp with an average GC content of 51.37% that belongs to the IncA/C2 group. The plasmid harbors numerous antibiotic resistance genes, which together provide resistance to multiple classes of antibiotics, including aminoglycosides, β-lactams, quinolones, and sulfonamides (Table 1). Interestingly, all resistance genes of the plasmid are located in a 30,659-bp multidrug-resistant region with the structure of a complex class 1 integron consisting of two 5′ CSs, three 3′ CSs, two ISCR1s, and four VRs bearing resistance genes (Figure 2). The class 1 integron region contains a complete copy and a partial copy of the 5′ CS and a copy of the 3′ CS, similar to that of Acinetobacter baumannii strain D4 (Genbank accession no. KP054476.2) (Hamidian et al., 2015). The cassette regions of the class 1 integron in pGD31-NDM carry five resistance genes (blaOXA–4, aadA2, strA, strB, and aadB), three of which were found in those of A. baumannii strain D4. The blaOXA–4 gene encoding an ESBL is usually found in Pseudomonas aeruginosa (Marumo et al., 1999; Kainuma et al., 2018).
FIGURE 2.
Schematic representation of the complex class 1 integron in pGD31-NDM and structural comparison with Acinetobacter baumannii strain D4, Vibrio alginolyticus plasmid pC1394, E. coli plasmid pEcNDM1, and Proteus mirabilis plasmid pNDM-PM58. Boxed arrows indicate the positions of open reading frames and their directions of transcription. Open reading frames encoding hypothetical proteins are portrayed by gray-shaded arrows. Gray-shaded areas denote more than 99% DNA identity between sequences. CS, conserved segment; VR, variable region.
The ISCR1-1-linked VR-3 in pGD31-NDM contains two genes of unknown function, aphA6 and a Tn125 transposon carrying the blaNDM–1 gene (Figure 2). However, ISAba125 lying downstream of blaNDM–1 was interrupted by the insertion of ISAba24. Interestingly, the ISCR1-1-associated VR in plasmid pC1394 (Genbank accession no. MH457126) is identical to that found in pGD31-NDM, except for the truncated ISAba125 lying downstream of blaNDM–1 and the lack of aphA6. ISCR1 was found linked to various VRs carrying blaNDM–1 in E. coli plasmid pEcNDM1 (Genbank accession no. JX469383) and Proteus mirabilis plasmid pNDM-PM58 (Genbank accession no. KP662515). The construction of complex class 1 integrons harboring blaNDM–1 indicated that ISCR1 is also an element responsible for the mobilization of the blaNDM–1 gene.
Similar to pECNDM1, pGD31-NDM harbors a second copy of ISCR1 followed by quinolone-resistant gene qnrA (Figure 2). In pGD31-NDM, however, a duplicate of the 3′ CS of the class 1 integron is located immediately downstream of the qnrA1 gene, revealing the presence of a complete structure of ISCR1 containing qnrA1. The ampR gene, which is involved in the induction of class C β-lactamase, is usually found adjacent to qnrA1 in ISCR1-linked regions (Rui et al., 2018), whereas it was missing in that of pGD31-NDM.
Unlike other plasmids, the complex class 1 integron in pGD31-NDM carries the class 1 integron region bearing two 5′ CSs and one 3′ CS and the two complete structures of ISCR1. The presence of these two complete structures of ISCR1 may be the result of successive integration events following the model of Toleman et al. (2006). Free circular intermediates carrying the orf-orf-Tn125-like transposon-aphA6-Δ3′ CS-ISCR1 element and qnrA1-Δ3′ CS-ISCR1 element were generated via aberrant rolling-circle replications. These circular intermediates might then be rescued by homologous recombination via the 3′ CS of the class 1 integron. Thus, it would be possible for the complex integron to expand structure. Surprisingly, the overall form of the complex class 1 integron in pGD31-NDM is identical to that of pC1394 in Vibrio alginolyticus strain Vb1394 isolated from shrimp (Zheng Z. et al., 2018), except for the structure of the class 1 integron, the truncation of ISAba125, and the existence of aphA6 (Figure 2). Two copies of IS5075 are adjacent to the boundaries of the complex integron, leading to putative genesis of an IS5075-mediated composite transposon, which may move the complex integron via transposition or homologous recombination. The formation of potential IS5075-mediated composite transposon raises a concern about the prevalence of this complex integron.
The linkage of the class 1 integron and ISCR1 results in the extension of multidrug-resistant regions by adding resistance genes. In addition, the mcr and blaNDM genes may be transferred to a same transconjugant, although they are located on various self-transmissible plasmids (Long et al., 2019). Altogether, ISCR1-mediated construction of complex class 1 integrons and co-transfer of mcr and blaNDM play a significant role in forming extensive drug-resistant and pandrug-resistant bacteria, thus causing great challenges in the fight against antibiotic resistance.
sul3-Type Integron in Plasmid pGD31-F1928
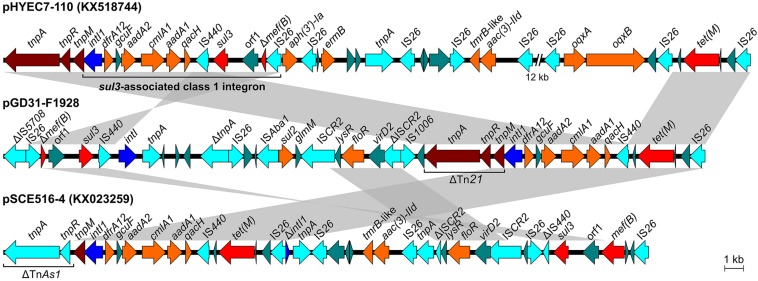
pGD31-F1928 is a 245,305-bp circular plasmid with an average GC content of 51.37% (Table 1). The plasmid harbors three replication systems (IncR, IncFIB, and IncFII), two conjugation regions, sitABCD operon, copper/silver resistance genes, Tn2 transposon, a class 1 integron, and some resistance genes bounded by various insertion sequences (Figure 3). Antibiotic resistance genes are associated with resistance to aminoglycosides, β-lactams (penicillins and first-generation cephalosporins), chloramphenicol, florfenicol, trimethoprim, sulfonamides, and tetracyclines. The genetic context of sulfonamide resistance gene sul2 and florfenicol resistance gene floR, sul2-glmM-ISCR2-lysR-floR-virD2-ΔISCR2, was identified (Figure 4). This genetic environment is identical to that of plasmid pHNSHP45-2 (Genbank accession no. KU341381) and Tn6450 transposon of Proteus mirabilis strain SNYG17 (Genbank accession no. MF805806) (Chen et al., 2018). The sul2 and floR genes are usually found associated with ISCR2, the amino acid sequence of which is 65% similar to that of ISCR1, but ISCR2 is not responsible for the formation of the complex class 1 integron (Toleman et al., 2006). In addition, pGD31-F1928 also carries a truncated mig-14 gene, which relates to antimicrobial peptide resistance in Salmonella enterica (Brodsky et al., 2002) and P. aeruginosa (Jochumsen et al., 2011).
FIGURE 3.
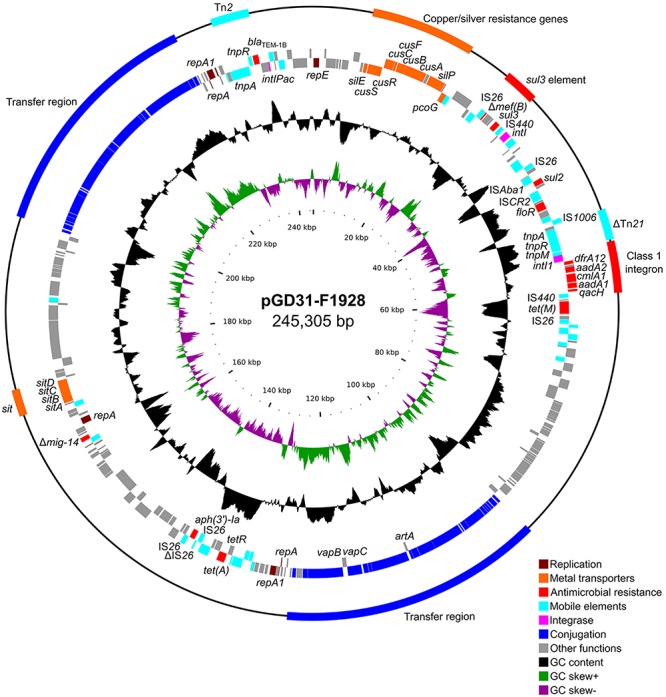
Circular representation of plasmid pGD31-F1928. The outermost circle indicates genetic elements, including resistance region, transposon, integron, and transfer region. The second and third circles show coding sequences on the forward and reverse strand of the plasmid, respectively. The fourth and fifth circles show the GC plot and GC skew graph, respectively. The innermost circle shows size in kb.
FIGURE 4.
Structural comparison of regions harboring class 1 integron and sul3 element of pGD31-F1928 and E. coli plasmid pSCE516-4 with sul3-associated class 1 integron of E. coli plasmid pHYEC7-110. Boxed arrows indicate the positions of open reading frames and their directions of transcription. Gray-shaded areas denote more than 99% DNA identity between sequences.
The aadA1, aadA2, cmlA1, dfrA12, and qacH genes are located in a gene cassette array of class 1 integron (Figure 4). The integron with a structure of intI1-dfrA12-gcuF-aadA2-cmlA1-aadA1-qacH-IS440-sul3 is known as sul3-associated integron type I (Antunes et al., 2007). Surprisingly, only IS440 was found downstream of the qacH gene, whereas sul3 and a duplicate of IS440 were located in another region of the plasmid (Figure 4). The IS440-sul3 element lay upstream of an untyped integrase-encoding intI gene and linked to the region harboring a putative oxidoreductase gene (orf1) and mef(B) truncated by an IS26 insertion. The linkage of sul3 with mef(B), a macrolide efflux gene, was first described in E. coli isolated from pigs (Liu et al., 2009). The intI1-dfrA12-gcuF-aadA2-cmlA1-aadA1-qacH-IS440 region is adjacent to the tetM-IS26 element, creating a new structure of a non-classic class 1 integron. This structure was also observed in plasmid pSCE516-4 (Genbank accession no. KX023259), but the integron is located upstream of the TnAs1 tnpA-tnpR element and Tn21 tnpM instead of the Tn21 tnpA-tnpR-tnpM element as in pGD31-F1928 and pHYEC7-110 (Genbank accession no. KX518744) (Figure 4). In addition, the genetic context of sul3 in pSCE516-4 bears a truncated IS440 and complete mef(B), whereas pGD31-F1928 contains a complete IS440 and truncated mef(B). Altogether, we propose a hypothesis that the original class 1 integron in pGD31-F1928 is similar to the sul3-associated integron in pHYEC7-110. It is likely that the region containing sul3, orf1, and Δmef(B) was flanked by IS440 and IS26, leading to a rearrangement of the plasmid sequence promoted by the insertion sequences. As a result, sul3 element, sul3-orf1-Δmef(B)-IS26, was separated out of the integron, thus breaking the linkage between sul3 and the class 1 integron. The connection of the class 1 integron and the tet(M)-IS26 element may construct a putative tet(M)-associated class 1 integron.
Conclusion
A multidrug-resistant E. coli strain EC17GD31 simultaneously harboring mcr-1 and blaNDM–1 is identified, which alerts health officials to the presence of mcr- and blaNDM-positive Enterobacteriaceae in Vietnam. The close monitoring of MCR- and NDM-coproducing Enterobacteriaceae isolates is essential in order to safeguard the effectiveness of last-resort antibiotics. Additionally, this study revealed that the blaNDM–1 gene is located in a novel complex class 1 integron containing a class 1 integron region bearing two VRs and two complete structures of ISCR1. The characterization of the complex integron further demonstrates that ISCR1 is a powerful genetic element that can mobilize a single gene as well as a composite transposon (e.g., Tn125-like transposon), therefore, the spread of resistance genes including blaNDM–1 can be anticipated. Although additional intermediates and progenitor plasmids were not found, the formation of the complex integron was most likely based on successive events of resistance gene mobilization. The findings of this study further support that complex interactions between antibiotic resistance genes and mobile elements play an important role in the creation and evolution of multidrug-resistant regions, thus facilitating bacteria to achieve either extensive drug resistance or pandrug resistance.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be shared upon request submitted to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Ethics Committee in Biomedical Research, Pasteur Institute in Ho Chi Minh City, Vietnam. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
VC and JJ conceived and designed the study. PT and LL performed the microbiological and molecular experiments. TI, SN, and DM performed the whole-genome sequencing experiments. H-NL-V, PT, and YM analyzed the data and prepared figures. H-NL-V wrote the manuscript with support from VC, PT, and LL. All authors contributed to manuscript revision, read, and approved the submitted version.
Disclaimer
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70-25.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Nhan Dan Gia Dinh Hospital in Ho Chi Minh City for providing the isolated bacterium for the research. We gratefully acknowledge the staffs of Molecular Biology Laboratory (Pasteur Institute in Ho Chi Minh City, Vietnam) for technical assistance.
Funding. This study was supported by the Armed Forces Research Institute of Medical Sciences (AFRIMS). Whole-genome sequencing of the isolate was supported by the Grant for Joint Research Project of the Research Institute for Microbial Diseases, Osaka University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02472/full#supplementary-material
References
- AbuOun M., Stubberfield E. J., Duggett N. A., Kirchner M., Dormer L., Nunez-Garcia J., et al. (2017). mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 72 2745–2749. 10.1093/jac/dkx286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes P., Machado J., Peixe L. (2007). Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3′ conserved sequence region among Salmonella isolates. Antimicrob. Agents Chemother. 51 1545–1548. 10.1128/AAC.01275-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiak M., Fischer J., Hammerl J. A., Hendriksen R. S., Szabo I., Malorny B. (2017). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 72 3317–3324. 10.1093/jac/dkx327 [DOI] [PubMed] [Google Scholar]
- Brodsky I. E., Ernst R. K., Miller S. I., Falkow S. (2002). mig-14 is a Salmonella gene that plays a role in bacterial resistance to antimicrobial peptides. J. Bacteriol. 184 3203–3213. 10.1128/JB.184.12.3203-3213.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Villa L., Feudi C., Curcio L., Orsini S., Luppi A., et al. (2017). Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 22:30589. 10.2807/1560-7917.ES.2017.22.31.30589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll L. M., Gaballa A., Guldimann C., Sullivan G., Henderson L. O., Wiedmann M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e853-19. 10.1128/mBio.00853-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-J., Wu T.-L., Lu P.-L., Chen Y.-T., Fung C.-P., Chuang Y.-C., et al. (2014). Closely related NDM-1-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. PLoS One 9:e104899. 10.1371/journal.pone.0104899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-P., Lei C.-W., Kong L.-H., Zeng J.-X., Zhang X.-Z., Liu B.-H., et al. (2018). Tn6450, a novel multidrug resistance transposon characterized in a Proteus mirabilis isolate from chicken in China. Antimicrob. Agents Chemother. 62:e2192-17. 10.1128/AAC.02192-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. (2018). Performance Standards For Antimicrobial Susceptibility Testing, 28th Edn, Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Delgado-Blas J. F., Ovejero C. M., Abadia-Patiño L., Gonzalez-Zorn B. (2016). Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob. Agents Chemother. 60 6356–6358. 10.1128/AAC.01319-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Chen L., Tang Y.-W., Kreiswirth B. N. (2016). Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect. Dis. 16 287–288. 10.1016/S1473-3099(16)00056-6 [DOI] [PubMed] [Google Scholar]
- Edgar R., Bibi E. (1997). MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 179 2274–2280. 10.1128/jb.179.7.2274-2280.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D. M., Kelly S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidian M., Holt K. E., Hall R. M. (2015). Genomic resistance island AGI1 carrying a complex class 1 integron in a multiply antibiotic-resistant ST25 Acinetobacter baumannii isolate. J. Antimicrob. Chemother. 70 2519–2523. 10.1093/jac/dkv137 [DOI] [PubMed] [Google Scholar]
- Iovleva A., Doi Y. (2017). Carbapenem-resistant Enterobacteriaceae. Clin. Lab. Med. 37 303–315. 10.1016/j.cll.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochumsen N., Liu Y., Molin S., Folkesson A. (2011). A Mig-14-like protein (PA5003) affects antimicrobial peptide recognition in Pseudomonas aeruginosa. Microbiology 157 2647–2657. 10.1099/mic.0.049445-0 [DOI] [PubMed] [Google Scholar]
- Kainuma A., Momiyama K., Kimura T., Akiyama K., Inoue K., Naito Y., et al. (2018). An outbreak of fluoroquinolone-resistant Pseudomonas aeruginosa ST357 harboring the exoU gene. J. Infect. Chemother. 24 615–622. 10.1016/j.jiac.2018.03.008 [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H., Phillippy A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Chuang Y.-C., Chen C.-C., Tang H.-J. (2017). Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int. J. Antimicrob. Agents 49 517–518. 10.1016/j.ijantimicag.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47 W256–W259. 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lan R., Xiong Y., Ye C., Yuan M., Liu X., et al. (2014). Sequential isolation in a patient of Raoultella planticola and Escherichia coli bearing a novel ISCR1 element carrying blaNDM-1. PLoS One 9:e89893. 10.1371/journal.pone.0089893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.-T., Li X., Zhang Q., Shan H., Zou M., Song F.-J. (2019). Colistin-resistant mcr-positive Enterobacteriaceae in fresh vegetables, an increasing infectious threat in China. Int. J. Antimicrob. Agents 54 89–94. 10.1016/j.ijantimicag.2019.04.013 [DOI] [PubMed] [Google Scholar]
- Liu J., Keelan P., Bennett P. M., Enne V. I. (2009). Characterization of a novel macrolide efflux gene, mef(B), found linked to sul3 in porcine Escherichia coli. J. Antimicrob. Chemother. 63 423–426. 10.1093/jac/dkn523 [DOI] [PubMed] [Google Scholar]
- Liu Y.-Y., Wang Y., Walsh T. R., Yi L.-X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Long H., Feng Y., Ma K., Liu L., McNally A., Zong Z. (2019). The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3. 5, the carbapenemase gene blaNDM-5 and the 16S methylase gene rmtB from Escherichia coli. Sci. Rep. 9:696. 10.1038/s41598-018-37125-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malchione M. D., Torres L. M., Hartley D. M., Koch M., Goodman J. (2019). Carbapenem and colistin resistance in Enterobacteriaceae in Southeast Asia: review and mapping of emerging and overlapping challenges. Int. J. Antimicrob. Agents 54 381–399. 10.1016/j.ijantimicag.2019.07.019 [DOI] [PubMed] [Google Scholar]
- Marumo K., Takeda A., Nakamura Y., Nakaya K. (1999). Detection of OXA-4 β-lactamase in Pseudomonas aeruginosa isolates by genetic methods. J. Antimicrob. Chemother. 43 187–193. 10.1093/jac/43.2.187 [DOI] [PubMed] [Google Scholar]
- Matamoros S., Hattem J. M., Arcilla M. S., Willemse N., Melles D. C., Penders J., et al. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:15364. 10.1038/s41598-017-15539-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mediavilla J. R., Patrawalla A., Chen L., Chavda K. D., Mathema B., Vinnard C., et al. (2016). Colistin-and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio 7:e1191-16. 10.1128/mBio.01191-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinersmann R. J., Ladely S. R., Bono J. L., Plumblee J. R., Hall M. C., Genzlinger L. L., et al. (2016). Complete genome sequence of a colistin resistance gene (mcr-1)-bearing isolate of Escherichia coli from the United States. Genome Announc. 4:e1283-16. 10.1128/genomeA.01283-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan-Linnell S. K., Zechiedrich L. (2007). Contributions of the combined effects of topoisomerase mutations toward fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 51 4205–4208. 10.1128/AAC.00647-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaitan A. O., Morand S., Rolain J.-M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paveenkittiporn W., Kerdsin A., Chokngam S., Bunthi C., Sangkitporn S., Gregory C. J. (2017). Emergence of plasmid-mediated colistin resistance and New Delhi metallo-β-lactamase genes in extensively drug-resistant Escherichia coli isolated from a patient in Thailand. Diagn. Microbiol. Infect. Dis. 87 157–159. 10.1016/j.diagmicrobio.2016.11.005 [DOI] [PubMed] [Google Scholar]
- Rojas L. J., Wright M. S., De La Cadena E., Motoa G., Hujer K. M., Villegas M. V., et al. (2016). Initial assessment of the molecular epidemiology of blaNDM-1 in Colombia. Antimicrob. Agents Chemother. 60 4346–4350. 10.1128/AAC.03072-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y., Lu W., Li S., Cheng C., Sun J., Yang Q. (2018). Integrons and insertion sequence common region 1 (ISCR1) of carbapenem-non-susceptible Gram-negative bacilli in fecal specimens from 5,000 patients in southern China. Int. J. Antimicrob. Agents 52 571–576. 10.1016/j.ijantimicag.2018.06.015 [DOI] [PubMed] [Google Scholar]
- Ruiz J. (2003). Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 51 1109–1117. 10.1093/jac/dkg222 [DOI] [PubMed] [Google Scholar]
- Snesrud E., He S., Chandler M., Dekker J. P., Hickman A. B., McGann P., et al. (2016). A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 60 6973–6976. 10.1128/AAC.01457-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnevend A., Ghazawi A., Alqahtani M., Shibl A., Jamal W., Hashmey R., et al. (2016). Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int. J. Antimicrob. Agents 50 85–90. 10.1016/j.ijid.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P., Wishart D. S. (2004). Circular genome visualization and exploration using CGView. Bioinformatics 21 537–539. 10.1093/bioinformatics/bti054 [DOI] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Li X.-P., Yang R.-S., Fang L.-X., Huo W., Li S.-M., et al. (2016). Complete nucleotide sequence of an IncI2 plasmid coharboring blaCTX-M-55 and mcr-1. Antimicrob. Agents Chemother. 60 5014–5017. 10.1128/AAC.00774-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Ohnishi M., Kawanishi M., Akiba M., Kuroda M. (2016). Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect. Dis. 16 284–285. 10.1016/S1473-3099(16)00008-6 [DOI] [PubMed] [Google Scholar]
- Tada T., Nhung P. H., Shimada K., Tsuchiya M., Phuong D. M., Anh N. Q., et al. (2017). Emergence of colistin-resistant Escherichia coli clinical isolates harboring mcr-1 in Vietnam. Int. J. Infect. Dis. 63 72–73. 10.1016/j.ijid.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Toleman M. A., Bennett P. M., Walsh T. R. (2006). ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70 296–316. 10.1128/MMBR.00048-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trung N. V., Matamoros S., Carrique-Mas J. J., Nghia N. H., Nhung N. T., Chieu T. T. B., et al. (2017). Zoonotic transmission of mcr-1 colistin resistance gene from small-scale poultry farms, Vietnam. Emerg. Infect. Dis. 23 529–532. 10.3201/eid2303.161553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Dorp L., Shaw L. P., Bradley P., Wang Q., Wang X., et al. (2018a). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. 10.1038/s41467-018-03205-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Liu Y., Zhang Q., Jin L., Wang Q., Zhang Y., et al. (2018b). The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: coexistence of mcr-1 and blaNDM with low fitness cost. Int. J. Antimicrob. Agents 51 739–744. 10.1016/j.ijantimicag.2018.01.023 [DOI] [PubMed] [Google Scholar]
- Wang X., Wang Y., Zhou Y., Li J., Yin W., Wang S., et al. (2018c). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7:122. 10.1038/s41426-018-0124-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Yi L.-X., Yu L.-F., Wang J., Liu Y., Chen X., et al. (2018). Fitness Advantage of mcr-1–bearing IncI2 and IncX4 plasmids in vitro. Front. Microbiol. 9:331. 10.3389/fmicb.2018.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Feng Y., Tang G., Qiao F., McNally A., Zong Z. (2019). NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32 e115–e118. 10.1128/CMR.00115-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier B. B., Lammens C., Ruhal R., Kumar-Singh S., Butaye P., Goossens H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 21:30280. 10.2807/1560-7917.ES.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Kawahara R., Harada K., Teruya S., Nakayama T., Motooka D., et al. (2018). The presence of colistin resistance gene mcr-1 and-3 in ESBL producing Escherichia coli isolated from food in Ho Chi Minh City, Vietnam. FEMS Microbiol. Lett. 365:fny100. 10.1093/femsle/fny100 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Kawahara R., Fujiya Y., Sasaki T., Hirai I., Khong D. T., et al. (2019). Wide dissemination of colistin-resistant Escherichia coli with the mobile resistance gene mcr in healthy residents in Vietnam. J. Antimicrob. Chemother. 74 523–524. 10.1093/jac/dky435 [DOI] [PubMed] [Google Scholar]
- Yang R.-S., Feng Y., Lv X.-Y., Duan J.-H., Chen J., Fang L.-X., et al. (2016). Emergence of NDM-5 and MCR-1-producing Escherichia coli clone ST648 and ST156 from a single muscovy duck (Cairina moschata). Antimicrob. Agents Chemother. 60 6899–6902. 10.1128/AAC.01365-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.-Q., Li Y.-X., Lei C.-W., Zhang A.-Y., Wang H.-N. (2018). Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 73 1791–1795. 10.1093/jac/dky111 [DOI] [PubMed] [Google Scholar]
- Yao X., Doi Y., Zeng L., Lv L., Liu J.-H. (2016). Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 16 288–289. 10.1016/S1473-3099(16)00057-8 [DOI] [PubMed] [Google Scholar]
- Yin W., Li H., Shen Y., Liu Z., Wang S., Shen Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e543-17. 10.1128/mBio.00543-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Dong H., Xu H., Lv J., Zhang J., Jiang X., et al. (2016). Coexistence of MCR-1 and NDM-1 in clinical Escherichia coli isolates. Clin. Infect. Dis. 63 1393–1395. 10.1093/cid/ciw553 [DOI] [PubMed] [Google Scholar]
- Zheng B., Lv T., Xu H., Yu X., Chen Y., Li J., et al. (2018). Discovery and characterisation of an Escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China. Int. J. Antimicrob. Agents 51 273–275. 10.1016/j.ijantimicag.2017.09.005 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Li R., Ye L., Chan E. W.-C., Chen S. (2018). Identification and characterization of IncA/C conjugative, blaNDM-1-bearing plasmid in Vibrio alginolyticus of food origin. Antimicrob. Agents Chemother. 62:e1897-18. 10.1128/AAC.01897-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-W., Zhang T., Ma J.-H., Fang Y., Wang H.-Y., Huang Z.-X., et al. (2017). Occurrence of plasmid-and chromosome-carried mcr-1 in waterborne Enterobacteriaceae in China. Antimicrob. Agents Chemother. 61:e00017-17. 10.1128/AAC.00017-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be shared upon request submitted to the corresponding author.