Abstract
INTRODUCTION:
Downregulation of brain-derived neurotrophic factor (BDNF) and its cognate neurotrophin receptor, TrkB, were observed during the progression of dementia, but whether the Alzheimer’s disease (AD) pathological lesions diffuse plaques, (DPs), neuritic plaques (NPs), and neurofibrillary tangles (NFTs) are related to this alteration remains to be clarified.
METHODS:
Negative binomial (NB) regressions were performed using gene expression data accrued from a single population of CA1 pyramidal neurons and regional hippocampal dissections obtained from participants in the Rush Religious Orders Study (RROS).
RESULTS:
Downregulation of Bdnf is independently associated with increased entorhinal cortex NPs. Downregulation of TrkB is independently associated with increased entorhinal cortex NFTs and CA1 NPs during the progression of AD.
DISCUSSION:
Results indicate that BDNF and TrkB dysregulation contribute to AD neuropathology, most notably hippocampal NPs and NFTs. These data suggest attenuating BDNF/TrkB signaling deficits either at the level of BDNF, TrkB, or downstream of TrkB signaling may abrogate NPs and/or NFTs.
Keywords: Alzheimer’s disease, brain-derived neurotrophic factor, microarray, mild cognitive impairment, neuritic plaques, neurofibrillary tangles, TrkB, negative binomial model
Introduction
The neurotrophin brain-derived neurotrophic factor (BDNF) and its cognate neurotrophin receptor, TrkB, regulate many key brain functions involved in survival, neurogenesis, neuroplasticity, and learning and memory, among others (Cowansage et al., 2010; Leal et al., 2015; Naito et al., 2017; Tapia-Arancibia et al., 2008; Yoshii and Constantine-Paton, 2010). Defects in the expression or activity of BDNF and TrkB are found in a number of neurodegenerative disorders (Autry and Monteggia, 2012; Belrose et al., 2014; Michalski et al., 2015; Thompson Ray et al., 2011), including mild cognitive impairment (MCI) (Forlenza et al., 2015; Ginsberg et al., 2006, 2010; Mufson et al., 2007a, 2012; Peng et al., 2005), Alzheimer’s disease (AD) (Allen et al., 2011; Erickson et al., 2010; Ginsberg et al., 2019; Tanila, 2017), and Huntington’s disease (Autry and Monteggia, 2012;Zuccato and Cattaneo, 2009).
Downregulation of Bdnf occurs in the hippocampus in end-stage AD (Alvarez et al., 2014; Garzon et al., 2002; Holsinger et al., 2000; Michalski et al., 2015; Murray et al., 1994; Phillips et al., 1991; Sen et al, 2017), and those who died with a clinical diagnosis of MCI (Ginsberg et al., 2019). In contrast, TrkB, both the extracellular domain (ECD) and tyrosine kinase (TK) forms, display significant downregulation in MCI and AD compared to those with no cognitive impairment (NCI) in CA1 pyramidal neurons and regional hippocampal dissections (Ginsberg et al., 2019). Moreover, downregulation of TrkB ECD and TrkB TK significantly correlate with the abundance of neuritic plaques (NPs) and neurofibrillary tangles (NFTs), but not diffuse plaques (DPs) (Ginsberg et al., 2019). Specifically, decreased TrkB levels are associated with increased hippocampal CA1 NP load, entorhinal cortex NFTs, and hippocampal CA1 NFTs (Ginsberg et al., 2019). These correlative findings indicate Bdnf and/or TrkB dysfunction may associate with AD lesions during the progression of dementia, suggesting boosting BDNF-TrkB signaling as a potential therapeutic approach. This association remains an under investigated area in the AD field.
To determine whether Bdnf and TrkB expression levels are associated with AD neuropathology during the progression of AD, we applied negative binomial (NB) regressions to expression profile data accrued from a homogeneous population of CA1 pyramidal neurons and regional hippocampal dissections. Postmortem samples were obtained from subjects enrolled in the Rush Religious Orders Study (RROS), a longitudinal clinical pathological study of aging and dementia in retired Catholic clergy (Bennett and Launer, 2012; Bennett et al., 2002, 2012; Malek-Ahmadi et al., 2018; Mufson et al., 1999, 2012, 2016a). Each RROS participant received an annual detailed premortem clinical and postmortem neuropathological evaluation, enabling an assessment of Bdnf and TrkB levels in relation to entorhinal cortex and hippocampal NPs, NFTs, and DPs in the same cases.
Materials and Methods
Clinical evaluation
All subjects came to autopsy with no coexisting clinical conditions contributing to cognitive impairment as judged by the examining neurologist and were not receiving anticholinesterase drug therapy. A complete description of the RROS cohort has been published previously (Bennett and Launer, 2012; Bennett et al., 2002; Mufson et al., 2000, 2002b, 2016a, 2016b). Premortem cognitive assessments were performed each year prior to death. Memory tests consisted of the East Boston Memory immediate and delayed recall, Logical Memory immediate and delayed story recall, Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) Word List Memory immediate and delayed recall, and CERAD Word List Recognition. A board-certified neurologist with expertise in the evaluation of the elderly, and blinded to age, sex, race/ethnicity, and clinical data other than occupation, education, and information about sensory or motor deficits, used these results to summarize impairments in each of the five cognitive domains. At consensus conferences, neurologists and neuropsychologists reviewed clinical and medical records and family interviews before assigning a final clinical diagnosis. Subjects were clinically categorized as NCI, MCI insufficient to meet criteria for dementia, and AD according to the criteria that our group and others have reported in a large number of clinical pathology studies that used tissue from the RROS (Bennett and Launer, 2012; Bennett et al., 2002; Mufson et al., 2000, 2002b, 2016a, 2016b). Within the RROS cohort the MCI designation continues to be attributed to subjects with impaired cognitive testing who did not display frank dementia (Abner et al., 2017; Han et al., 2012, 2015). Case selection for this study was performed in association with the Neuropathology Core of the RROS (Oveisgharan et al., 2018; Yu et al., 2019), similar to our previously published studies (Ginsberg et al., 2019; Malek-Ahmadi et al., 2016, 2018; Mufson et al., 2016a, 2016b).
Neuropathological evaluation
For microarray and neuropathological analyses, hippocampal tissue blocks were immersion-fixed in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.2, paraffin embedded, and cut at 6 μm thickness (Ginsberg et al., 2010, 2019). A series of tissue sections for each case were prepared for neuropathological evaluation including visualization and quantitation of NPs, DPs, and NFTs using a modified Bielschowsky silver stain, Thioflavine-S, and antibodies directed against amyloid-β peptide (Aβ; 4G8, monoclonal, BioLegend, San Diego, CA) and tau (PHF1, monoclonal, a gift of Dr. Peter Davies, Hofstra Northwell School of Medicine) (Ginsberg et al., 2010; Malek-Ahmadi et al., 2016; Mufson et al., 2000, 2002a). Amyloid plaque load and NFT burden were determined by counting the total number of NPs, DPs, and NFTs per subject using a modified Bielschowsky silver stain in one square mm area (100x magnification) from the entorhinal cortex and hippocampal CA1 region (Ginsberg et al., 2019; Malek-Ahmadi et al., 2016; Mufson et al., 2016b).
All cases were evaluated by a board-certified neuropathologist blinded to clinical diagnosis. Designation of “normal”, “possible AD”, “probable AD”, or “definite AD” was based on estimation of NP density, as established by CERAD (Mirra et al., 1991). Each case also received a Braak score using NFT pathology staging (Braak and Braak, 1991). RROS cases also received NIA-Reagan diagnosis of Alzheimer’s disease and NINCDS-ADRDA designations (Bennett et al., 2012; Dubois et al., 2007; Hyman and Trojanowski, 1997). Exclusion criteria include hippocampal sclerosis, Lewy body disease, Parkinson’s disease, and identified strokes. Apolipoprotein E (ApoE) allele status was determined as described previously (Counts et al., 2007; Mufson et al., 2000, 2016b).
Bdnf and TrkB gene expression in CA1 pyramidal neurons and regional hippocampal dissections
The procedures for single population and regional hippocampal microarray analysis on human postmortem tissue have been described in detail previously (Ginsberg et al., 2006, 2010, 2012, 2019; Tieman et al., 2016). Briefly, individual CA1 pyramidal neurons were microaspirated via laser capture microdissection (Arcturus PixCell IIe, ThermoFisher Scientific, South San Francisco, CA). Fifty CA1 pyramidal neurons were captured for population analysis. After accruing CA1 neurons, the remaining tissue containing the entorhinal cortex, CA1, CA2, and CA3 sectors, dentate gyrus, and subicular complex medial to the rhinal sulcus was microaspirated in total as described previously (Ginsberg et al., 2012, 2019). RNAs were amplified by the terminal continuation (TC) RNA amplification method developed as previously described (Alldred et al., 2008, 2009; Che and Ginsberg, 2004; Ginsberg 2005, 2008, 2014). The TC RNA amplification protocol is available at http://cdr.rfmh.org/pages/ginsberglabpage.html. Microaspirated samples were homogenized in 100 μl of Trizol (ThermoFisher Scientific, Waltham, MA), extracted with chloroform, and precipitated utilizing isopropanol (Alldred et al., 2009). RNAs were reverse transcribed in the presence of poly d(T) primer (100 ng/μl) and TC primer (100 ng/μl) in 1X first strand buffer (ThermoFisher Scientific), 2 μg of linear acrylamide (Applied Biosystems, Foster City, CA), 10 mM dNTPs, 100 μM dithiothreitol (DTT), 20 U of SuperRNase Inhibitor (Applied Biosystems) and 200 U of reverse transcriptase (Superscript III, ThermoFisher Scientific). Single stranded cDNAs were digested and placed in a thermal cycler in a solution consisting of 10 mM Tris (pH 8.3), 50 mM KC1, 1.5 mM MgCl2, and 10 U RNase H (ThermoFisher Scientific) in a final volume of 100 μl. The thermal cycler program ran as follows: RNase H digestion step at 37 °C, 30 minutes; denaturation step 95 °C, 3 minutes; primer re-annealing step 60 °C, 5 minutes (Alldred et al., 2009; Che and Ginsberg 2004). Samples were purified by column filtration (MilliporeSigma, Billerica, MA). Hybridization probes were synthesized by in vitro transcription using 33P incorporation in 40 mM Tris (pH 7.5), 6 mM MgCl2, 10 mM NaCl, 2 mM spermidine, 10 mM DTT, 2.5 mM ATP, GTP and CTP, 100 μM of cold UTP, 20 U of RNase inhibitor, 2 KU of T7 RNA polymerase (LGC, Middleton, WI), and 120 μCi of 33P -UTP (Perkin-Elmer, Boston, MA) (Alldred et al., 2009; Ginsberg 2008). The reaction was performed at 37 °C for 4 hours. Radiolabeled TC RNA probes were hybridized to custom-designed cDNA arrays without further purification.
Custom-designed cDNA array platforms and hybridization
Arrays were prehybridized (2 hours) and hybridized (12 hours) in a solution consisting of 6X saline-sodium phosphate-ethylenediaminetetraacetic acid (SSPE), 5X Denhardt’s solution, 50% formamide, 0.1% sodium dodecyl sulfate (SDS), and denatured salmon sperm DNA (200 μg/ml) at 42 °C in a rotisserie oven (Che and Ginsberg 2004; Ginsberg 2008). Following hybridization, arrays were washed sequentially in 2X SSC/0.1% SDS, 1X SSC/0.1% SDS and 0.5X SSC/0.1% SDS for 15 min each at 37 °C. Arrays were placed in a phosphor screen for 24 hours and developed on a phosphor imager (GE Healthcare, Piscataway, NJ). All array phosphor images were adjusted to the same brightness and contrast levels for data acquisition (Alldred et al., 2012, 2015, 2018).
Hybridization signal intensity was determined utilizing ImageQuant software (GE Healthcare). Each array was compared to negative control arrays without any starting RNA. Bdnf and TrkB expression was expressed as a ratio of the total hybridization signal intensity of the array (Ginsberg et al., 2000, 2006, 2010, 2012, 2019).
Statistical Analysis
For demographic and neuropathological variables, the Chi-square test was used to analyze categorical variables. The Kruskall-Wallis test was used to analyze group differences for continuous and count variables. The Conover-Inman test was used to test groupwise comparisons.
Expression levels of Bdnf and TrkB with AD neuropathology associations were examined using NB regression models, employing Bdnf or TrkB as predictor variables with NPs, DPs, and NFTs (entorhinal cortex for Model 1 and hippocampal CA1 for Model 2) as the outcome variable. An interaction term was included for Bdnf/TrkB and APOE ε4 carrier status. Age, gender, and APOE ε4 carrier status were included as covariates in all models.
NB regression models were used due to significantly skewed distributions for entorhinal cortex and hippocampal CA1 NPs, DPs, and NFTs in CA1 pyramidal neurons and regional hippocampal dissections (Figure 1), which were analogous to NB distributions (Berk and MacDonald, 2008). Additional justification for the use of NB regression models (Kim and Kriebel, 2009) is presented, as the variance values for each of the pathology measures were substantially greater than their means. For CA1 pyramidal neurons, mean entorhinal cortex NFT counts were 22.03 with a variance of 359.48 and mean hippocampal CA1 sector NFT counts were 23.08 with a variance of 615.54. For regional hippocampal dissections, mean entorhinal cortex NFT counts were 16.55 with a variance of 275.23 and mean hippocampal CA1 sector NFT counts were 22.82 with a variance of 606.14. Accordingly, entorhinal cortex and hippocampal CA1 NP, DP, and NFT data were tested to determine whether they followed a NB distribution (p>0.05). NB model fits were assessed by testing residual deviance values and their associated degrees of freedom in the chi-square distribution where (p>0.05) indicated good model fit (Tjur, 1998). Statistical analyses were performed using the ‘glm.nb’ function in R 3.4.0 using the “MASS” package while model fit was determined by using the ‘pchisq’ function. NB distribution testing for NP, DP, and NFT data was performed using the ‘goodfit’ function in the “vcd” package {The R Project for Statistical Computing, www.r-project.org; (Vose, 2008)}. Statistical significance was set at (p≤0.05).
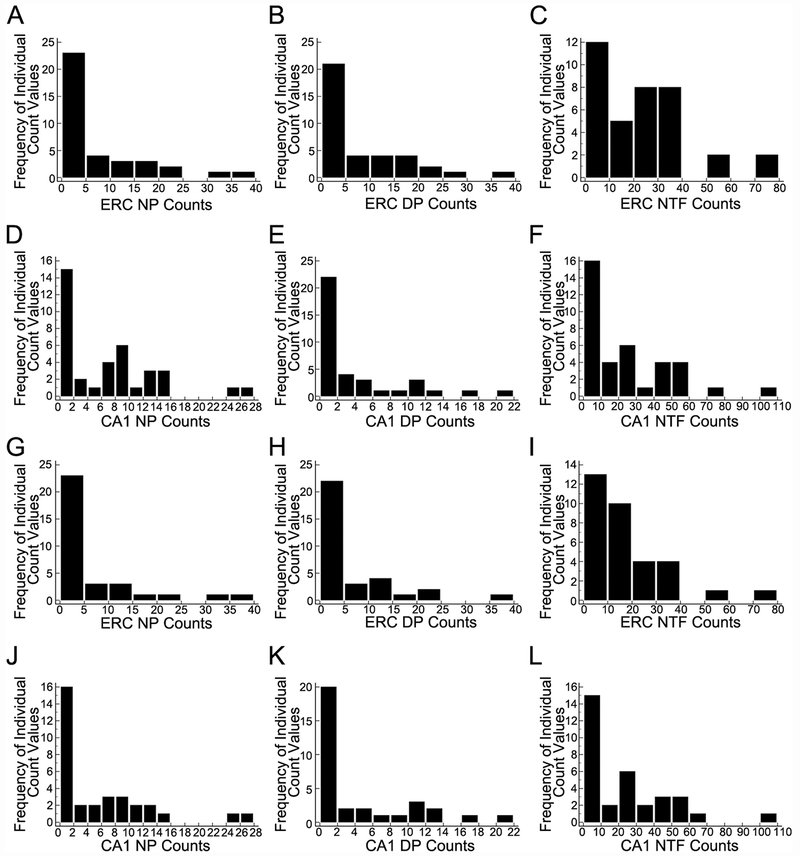
Figure 1.
Frequency distributions of entorhinal cortex (ERC) and CA1 neuritic plaques (NPs), diffuse plaques (DPs), and neurofibrillary tangles (NFTs) for homogeneous CA1 pyramidal neurons (A-F) and regional hippocampal dissections (G-L). Individual histograms indicate the distributions for NP, DP, and NFT counts are heavily skewed toward lower values. Since these count data are the outcome variables, negative binomial (NB) regression models were used in order to obtain valid estimates of Bdnf and TrkB associations with NP, DP, and NFT counts.
Results
Demographic and neuropathological characteristics for dissections of CA1 pyramidal neurons
Demographic and neuropathological characteristics for each group are shown in Table 1. Age at death and years of education were not significantly different (p = 0.68, p = 0.19, respectively). No significant difference in the proportions of males and females was found (p = 0.33). The APOE ε4 allele was more prevalent in AD relative to NCI and MCI (p<0.001). Postmortem interval (PMI) and brain weight at autopsy were not significantly different (p = 0.45, p = 0.23, respectively).
Table 1.
Demographic and neuropathological characteristics of cases used for homogeneous CA1 pyramidal neuron microarray analysis.
| NCI | MCI | AD | p-value | Groupwise Comparisons | |
|---|---|---|---|---|---|
| N | 13 | 15 | 9 | na | na |
| Gender (M/F) | 7/6 | 6/9 | 2/7 | 0.33 | na |
| APOE ε4 allele | 1/12 | 7/8 | 8/1 | <0.001 | na |
| Age at Death (years) | 83.0 ± 7.7 | 85.3 ± 4.5 | 86.8 ± 6.6 | 0.68 | na |
| Education (years) | 17.5 ± 4.1 | 19.1 ± 2.2 | 17.6 ± 1.7 | 0.19 | na |
| PMI (hours) | 7.5 ± 8.2 | 6.9 ± 4.0 | 7.6 ± 3.6 | 0.45 | na |
| Brain Weight (grams) | 1245.8 ± 170.4 | 1239.7 ± 212.0 | 1123.8 ± 152.6 | 0.23 | na |
| Entorhinal Cortex NP Counts | 0 {0 - 10} | 4 {0 - 30} | 13 {1 - 38} | <0.001 | NCI<MCI, NCI<AD |
| Entorhinal Cortex DP Counts | 0 {0 – 23} | 10 {0 – 38} | 3 {0 – 17} | 0.02 | NCI<MCI |
| Entorhinal Cortex NFT Counts | 5 {1 - 30} | 20 {1 - 74} | 34 {2 0 74} | <0.001 | NCI<MCI, NCI<AD |
| Hippocampus CA1 NP Counts | 0 {0 - 7} | 9 {0 - 27} | 12 {6 - 25} | <0.001 | NCI<MCI<AD |
| Hippocampus CA1 DP Counts | 0 {0 - 13} | 1 {0 –−20} | 3 {0 - 11} | 0.01 | NCI<MCI, NCI<AD |
| Hippocampus CA1 NET Counts | 2 {0 - 57} | 20 {0 - 58} | 40 {3 - 104} | <0.001 | NCI<AD, MCI<AD |
| Bdnf (relative expression) | 6.0 ± 3.8 | 2.5 ± 2.3 | 4.3 ± 3.4 | 0.09 | na |
| TrkB (relative expression) | 1.9 ± 0.6 | 1.1 ± 0.5 | 0.9 ± 0.3 | <0.001 | NCI>MCI, NCI>AD |
| Braak Stage | 0.06 | na | |||
| 0 – II | 5 | 2 | 1 | ||
| III – IV | 8 | 7 | 3 | ||
| V | 0 | 6 | 5 | ||
| CERAD Diagnosis | 0.02 | na | |||
| No AD | 7 | 1 | 0 | ||
| Possible AD | 2 | 2 | 0 | ||
| Probable AD | 2 | 5 | 3 | ||
| Definite AD | 2 | 7 | 6 | ||
| NIA Reagan Diagnosis | 0.01 | na | |||
| Not AD | 0 | 0 | 0 | ||
| Low Likelihood | 9 | 4 | 1 | ||
| Intermediate Likelihood | 4 | 8 | 3 | ||
| High Likelihood | 0 | 3 | 5 | ||
Results are presented as the mean ± standard deviation. Median neuropathological counts presented as {minimum - maximum}, na = not applicable.
In entorhinal cortex, NCI displayed significantly lower NP counts relative to MCI (p = 0.01) and AD (p<0.001), whereas MCI and AD were not significantly different (p =0.11). MCI had significantly higher DP counts than NCI (p<0.001). However, differences between NCI and AD as well MCI and AD were not statistically significant (p = 0.10, p = 0.30, respectively). NCI had significantly lower entorhinal cortex NFTs relative to MCI (p = 0.03) and AD (p<0.001). MCI and AD displayed similar levels of NFT pathology (p = 0.08).
Hippocampal CA1 region NP counts were significantly different between all three clinical groups (NCK<MCI, p<0.001; NCI<AD, p = 0.001; MCI<AD, p = 0.04). For CA1 region DPs, NCI displayed significantly lower counts compared to MCI (p = 0.01) and AD (p<0.001). MCI and AD were not significantly different (p = 0.28). For CA1 NFTs, AD had significantly greater counts than NCI (p<0.001) and MCI (p=0.02). NCI and MCI were not significantly different (p = 0.08).
Braak stage was not significantly different between the clinical groups (p = 0.06). Stage I: NCI = 23%, MCI = 13%, AD = 0%; Stage II: NCI = 15%, MCI = 0%, AD = 11%; Stage III: NCI= 23%, MCI = 13%, AD = 0%; Stage IV: NCI = 39%, MCI = 40%, AD = 22%; Stage V: NCI= 0%, MCI = 33%, AD = 67%. The CERAD criteria Definite AD classification was more prevalent in MCI and AD relative to NCI (p = 0.05). The NIA-Reagan High Likelihood classification was most prevalent in AD (p<0.001) while the Intermediate Likelihood classification was most prevalent in MCI.
Relation of single population CA1 pyramidal neuron Bdnf and TrkB expression levels with AD pathology
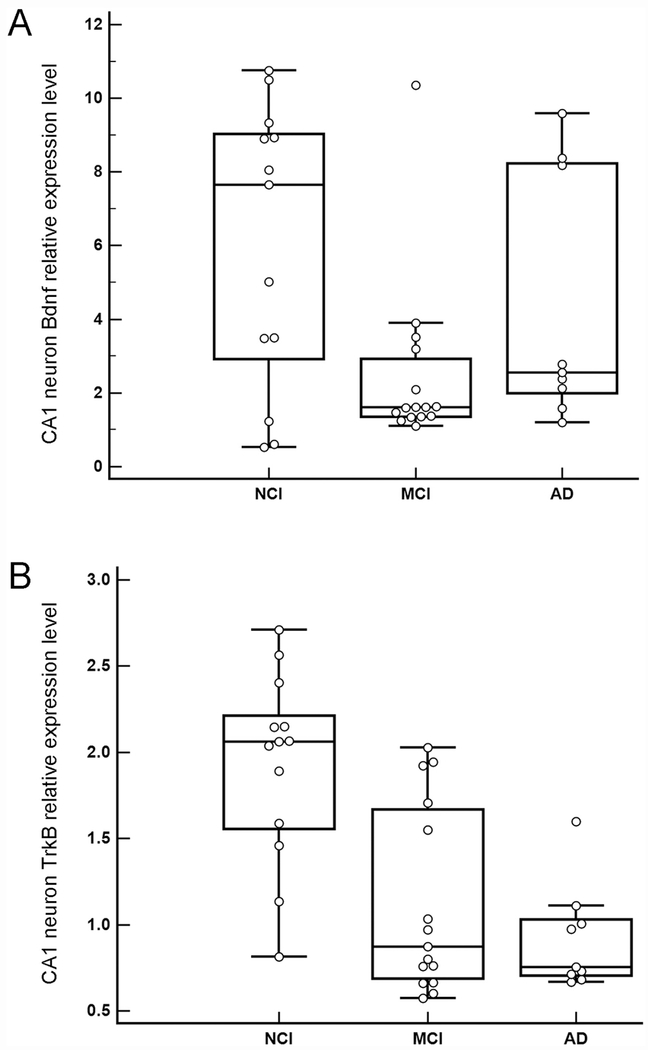
Bdnf expression within a homogeneous population of CA1 pyramidal neurons was not significantly different between clinical groups (p = 0.09; Figure 2A). A separate analysis of NCI and MCI demonstrated significant downregulation (p = 0.03). TrkB expression showed significant downregulation with disease progression (NCI>MCI, p<0.001; NCI>AD, p<0.001; MCI=AD, p = 0.50; Figure 2B). Both Bdnf and TrkB were not significantly associated with PMI (β = −0.06, SE = 0.10, p = 0.57) and (β = 0.00, SE = 0.02, p = 0.95), respectively.
Figure 2.
A. Boxplots illustrating clinical group differences within homogeneous CA1 pyramidal neurons for Bdnf. Bdnf expression was not significantly different between the clinical groups (p = 0.09). A separate analysis yielded a significant downregulation in MCI compared to NCI (p = 0.03). B. Boxplots illustrating downregulation of TrkB during the progression of dementia within homogeneous CA1 pyramidal neurons. TrkB expression showed significant group differences where expression decreased with disease progression (NCI>MCI, p<0.001; NCI>AD, p<0.001; MCI=AD, p = 0.50).
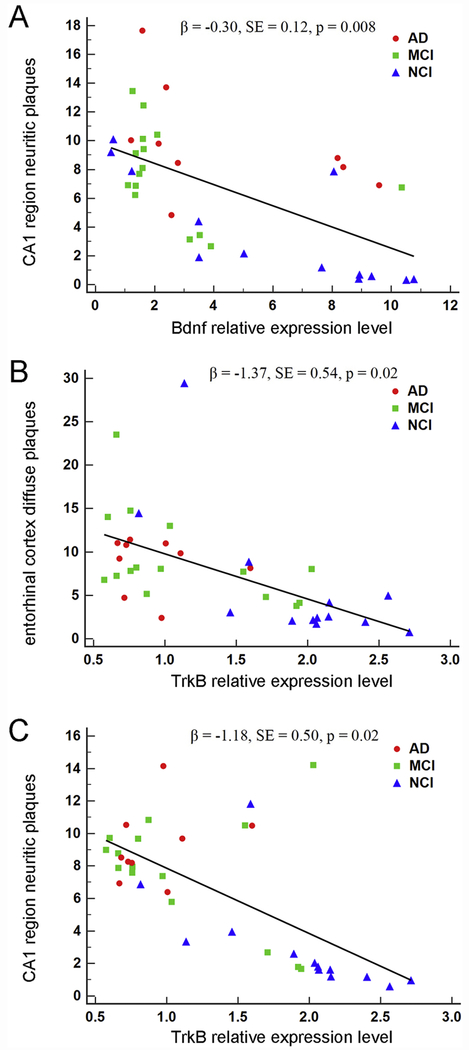
Single population CA1 pyramidal neuron Bdnf expression was not significantly associated with entorhinal cortex NPs (β = 0.03, SE = 0.11, p = 0.79), DPs (β = −0.06, SE = 0.11, p = 0.62), or NFTs (β = −0.05, SE = 0.08, p = 0.31) (Table 2). Interactions for APOE ε4 status and Bdnf were not statistically significant for NPs (β = −0.10, SE = 0.15, p = 0.53), DPs (β = 0.07, SE = 0.16, p = 0.65), or NFTs (β = −0.08, SE = 0.08, p = 0.31). Model fit was deemed to be good for the NP (p = 0.14), DP (p = 0.13), and NFT (p = 0.13) models. Bdnf was significantly associated with CA1 NPs (β = −0.30, SE = 0.12, p = 0.008; Figure 3A), but not with DPs (β = −0.07, SE = 0.11, p = 0.53) or NFTs (β = 0.04, SE = 0.07, p = 0.54) (Table 2). Interactions for APOE ε4 status and Bdnf for CA1 region pathology were not statistically significant for NPs (β = 0.27, SE = 0.16, p = 0.09), DPs (β = −0.23, SE = 0.16, p = 0.15), or NFTs (β = −0.16, SE = 0.10, p = 0.11). Model fit was deemed to be good for all models (NPs: p = 0.14, DPs: p = 0.23, NFTs: p = 0.09).
Table 2.
CA1 pyramidal neuron negative binomial regression models using Bdnf as a predictor of entorhinal cortex and hippocampal AD pathology.
| Entorhinal Cortex | ||||
| Coefficient | SE | p-value | ||
| Outcome = NPs | ||||
| Bdnf | 0.03 | 0.11 | 0.79 | |
| APOE ε4 | 1.40 | 0.80 | 0.08 | |
| APOE ε4 x Bdnf | −0.10 | 0.15 | 0.53 | |
| Outcome = DPs | ||||
| Bdnf | −0.06 | 0.11 | 0.62 | |
| APOE ε4 | 0.17 | 0.87 | 0.84 | |
| APOE ε4 x Bdnf | 0.07 | 0.16 | 0.65 | |
| Outcome = NFTs | ||||
| Bdnf | −0.05 | 0.05 | 0.32 | |
| APOE ε4 | 1.01 | 0.39 | 0.01* | |
| APOE ε4 x Bdnf | −0.08 | 0.08 | 0.31 | |
| Hippocampal CA1 | ||||
| Coefficient | SE | p-value | ||
| Outcome = NPs | ||||
| Bdnf | −0.30 | 0.12 | 0.008* | |
| APOE ε4 | 0.05 | 0.76 | 0.95 | |
| APOE ε4 x Bdnf | 0.27 | 0.16 | 0.09 | |
| Outcome = DPs | ||||
| Bdnf | −0.07 | 0.11 | 0.53 | |
| APOE ε4 | 2.47 | 0.81 | 0.002* | |
| APOE ε4 x Bdnf | −0.23 | 0.16 | 0.15 | |
| Outcome = NFTs | ||||
| Bdnf | 0.04 | 0.07 | 0.54 | |
| APOE ε4 | 1.26 | 0.53 | 0.02* | |
| APOE ε4 x Bdnf | −0.16 | 0.10 | 0.11 | |
Models included age at death, gender, and APOE ε4 carrier status. SE = standard error.
= significant.
Figure 3.
Association of Bdnf and TrkB expression in homogeneous CA1 pyramidal neurons with hippocampal CA1 region neuritic plaques and entorhinal cortex diffuse plaques. A. A significant inverse association was found between Bdnf expression and hippocampal CA1 region neuritic plaques by clinical group (β = −0.30, SE = 0.12, p = 0.008). B. A significant inverse association was found between TrkB expression and entorhinal cortex diffuse plaques by clinical group (β = −1.37, SE = 0.54, p = 0.02). C. A significant inverse association was found between TrkB expression and hippocampal CA1 region neuritic plaques by clinical group (β = −1.18, SE = 0.50, p = 0.02).
Single population CA1 neuron TrkB expression was not associated with entorhinal cortex NPs (β = −0.43, SE = 0.27, p = 0.11) or NFTs (β = −1.02, SE = 0.52, p = 0.10), but showed a significant association with DPs (β = −1.37, SE = 0.54, p = 0.02) (Figure 3B and Table 3). Model fit was deemed to be good for all models (NPs: p = 0.13, DPs: p = 0.14, NFTs: p = 0.14). For the CA1 analyses, TrkB was significantly associated with NPs (β = −1.18, SE = 0.50, p = 0.02; Figure 3C) but not DPs (β = −1.10, SE = 0.63, p = 0.08) or NFTs (β = −0.13, SE = 0.35, p = 0.71) (Table 3). The CA1 models all demonstrated good fit (NPs: p = 0.11, DPs: p = 0.30, NFTs: p = 0.09).
Table 3.
CA1 pyramidal neuron NB regression models using TrkB as a predictor of entorhinal cortex and hippocampal AD pathology.
| Entorhinal Cortex | ||||
| Coefficient | SE | p-value | ||
| Outcome = NPs | ||||
| TrkB | −1.02 | 0.52 | 0.10 | |
| APOE ε4 | −0.70 | 1.26 | 0.58 | |
| APOE ε4 x TrkB | 1.18 | 0.98 | 0.23 | |
| Outcome = DPs | ||||
| TrkB | −1.37 | 0.54 | 0.02* | |
| APOE ε4 | −1.16 | 1.34 | 0.39 | |
| APOE ε4 x TrkB | 0.99 | 1.04 | 0.34 | |
| Outcome = NFTs | ||||
| TrkB | −0.43 | 0.27 | 0.11 | |
| APOE ε4 | 0.16 | 0.67 | 0.81 | |
| APOE ε4 x TrkB | 0.37 | 0.52 | 0.48 | |
| Hippocampal CA1 | ||||
| Coefficient | SE | p-value | ||
| Outcome = NPs | ||||
| TrkB | −1.18 | 0.50 | 0.02* | |
| APOE ε4 | −1.25 | 1.21 | 0.30 | |
| APOE ε4 x TrkB | 1.64 | 0.94 | 0.08 | |
| Outcome = DPs | ||||
| TrkB | −1.10 | 0.63 | 0.08 | |
| APOE ε4 | −0.16 | 1.41 | 0.91 | |
| APOE ε4 x TrkB | 1.46 | 1.09 | 0.18 | |
| Outcome = NFTs | ||||
| TrkB | −0.13 | 0.35 | 0.71 | |
| APOE ε4 | 0.34 | 0.90 | 0.70 | |
| APOE ε4 x TrkB | 0.22 | 0.69 | 0.75 | |
Models included age at death, gender, and APOE ε4 carrier status. SE = standard error.
= significant.
Demographic and neuropathological characteristics for regional hippocampal dissections
Demographic and neuropathological characteristics for each group are shown in Table 4. Age at death and years of education were not significantly different across groups examined (p = 0.18, p = 0.27, respectively). The proportions of male and female subjects was not statistically different (p = 0.74). The APOE ε4 allele was more prevalent in MCI and AD relative to NCI (p = 0.03). PMI and brain weight at autopsy were not significantly different (p = 0.54, p = 0.14, respectively). Braak stage was significantly different between clinical groups (p = 0.02). NCI cases had lower Braak stages relative to MCI and AD. Stage I: NCI = 23%, MCI = 10%, AD = 0%; Stage II: NCI = 31%, MCI = 0%, AD = 10%; Stage III: NCI= 15%, MCI = 10%, AD = 0%; Stage IV: NCI= 31%, MCI = 60%, AD = 30%; Stage V: NCI = 0%, MCI = 20%, AD = 60%). A CERAD diagnosis of Definite AD classification was more prevalent in MCI and AD relative to NCI (p = 0.05). The NIA-Reagan High Likelihood classification was most prevalent in AD (p<0.001).
Table 4.
Demographic and neuropathological characteristics of cases for regional hippocampal data.
| NCI | MCI | AD | p-value | Groupwise Comparisons | |
|---|---|---|---|---|---|
| N | 13 | 10 | 10 | na | na |
| Gender (M/F) | 7/6 | 4/6 | 4/6 | 0.74 | na |
| APOE ε4 allele | 2/11 | 4/6 | 7/3 | 0.03 | na |
| Age at Death (years) | 80.3 ± 9.1 | 83.0 ± 4.6 | 85.8 ± 4.6 | 0.18 | na |
| Education (years) | 17.8 ± 4.5 | 18.9 ± 2.4 | 16.3 ± 3.9 | 0.27 | na |
| PMI (hours) | 11.1 ± 10.6 | 7.7 ± 4.8 | 6.0 ± 3.4 | 0.54 | na |
| Brain Weight (grams) | 1291.2 ± 161.4 | 1255.7 ± 201.6 | 1155.6 ± 135.6 | 0.14 | na |
| Entorhinal Cortex NP Counts | 0 {0 - 10} | 2.5 {0 - 30} | 5 {1 - 38} | <0.001 | NCI<AD, MCI<AD |
| Entorhinal Cortex DP Counts | 0 {0 - 23} | 7.5 {0 - 38} | 2.5 {0 - 13} | 0.05 | NCI<MCI |
| Entorhinal Cortex NFT Counts | 3 {0 - 22} | 15 {1 - 36} | 28 {2 - 74} | <0.001 | NCI<MCI, NCI<AD |
| Hippocampus CA1 NP Counts | 0 {0 - 7} | 4.5 {0 - 27} | 9 {0 - 25} | <0.001 | NCI<MCI, NCI<AD |
| Hippocampus CA1 DP Counts | 0 {0 - 13} | 2.5 {0 - 20} | 2.50 {0 - 10} | 0.18 | na |
| Hippocampus CA1 NET Counts | 2 {0 - 57} | 22.5 {1 - 66} | 38 {3 - 104} | <0.001 | NCI<MCI, NCI<AD |
| Bdnf (relative expression) | 7.3 ± 1.8 | 3.9 ± 1.6 | 5.7 ±2.0 | <0.001 | NCI<MCI, MCI<AD |
| TrkB (relative expression) | 5.6 ± 0.7 | 2.6 ± 0.6 | 2.3 ± 1.0 | <0.001 | NCI>MCI, NCI>AD |
| Braak Stage | 0.02 | na | |||
| 0 – II | 7 | 1 | 1 | ||
| III – IV | 6 | 7 | 3 | ||
| V | 0 | 2 | 6 | ||
| CERAD Diagnosis | 0.05 | na | |||
| No AD | 6 | 2 | 0 | ||
| Possible AD | 2 | 2 | 0 | ||
| Probable AD | 4 | 2 | 4 | ||
| Definite AD | 1 | 4 | 6 | ||
| NIA Reagan Diagnosis | <0.001 | na | |||
| Not AD | 0 | 0 | 0 | ||
| Low Likelihood | 9 | 4 | 1 | ||
| Intermediate Likelihood | 4 | 6 | 4 | ||
| High Likelihood | 0 | 0 | 5 | ||
Results are presented as the mean ± standard deviation. Median neuropathological counts presented as {minimum - maximum}. na = not applicable.
Significantly greater entorhinal cortex NP pathology was observed in AD relative to both NCI (p<0.001) and MCI (p = 0.02). NCI and MCI were not significantly different (p = 0.17). Entorhinal cortex DPs were significantly greater in MCI compared to NCI(p = 0.01). No significant difference in DPs were found between NCI and AD (p = 0.14) as well as MCI and AD (p = 0.29). Entorhinal cortex NFTs were significantly lower in NCI compared to MCI (p = 0.03) and AD (p<0.001). NFT pathology was similar between MCI and AD (p = 0.08). Both Bdnf and TrkB were not significantly associated with PMI (β = 0.05, SE = 0.05, p = 0.31) and (β = 0.06, SE = 0.04, p = 0.12), respectively.
CA1 region NPs were significantly greater in AD than NCI (p<0.001) while MCI had significantly greater CA1 regional NP neuropathology compared to NCI (p = 0.02). MCI and AD were not significantly different (p = 0.14). CA1 regional DPs were not significantly different between groups (p = 0.18). CA1 NFTs showed group differences that were similar to the entorhinal cortex (NCI<MCI, p = 0.01; NCI<AD, p<0.001; MCI=AD, p = 0.15).
Regional hippocampal dissection Bdnf and TrkB expression with AD pathology
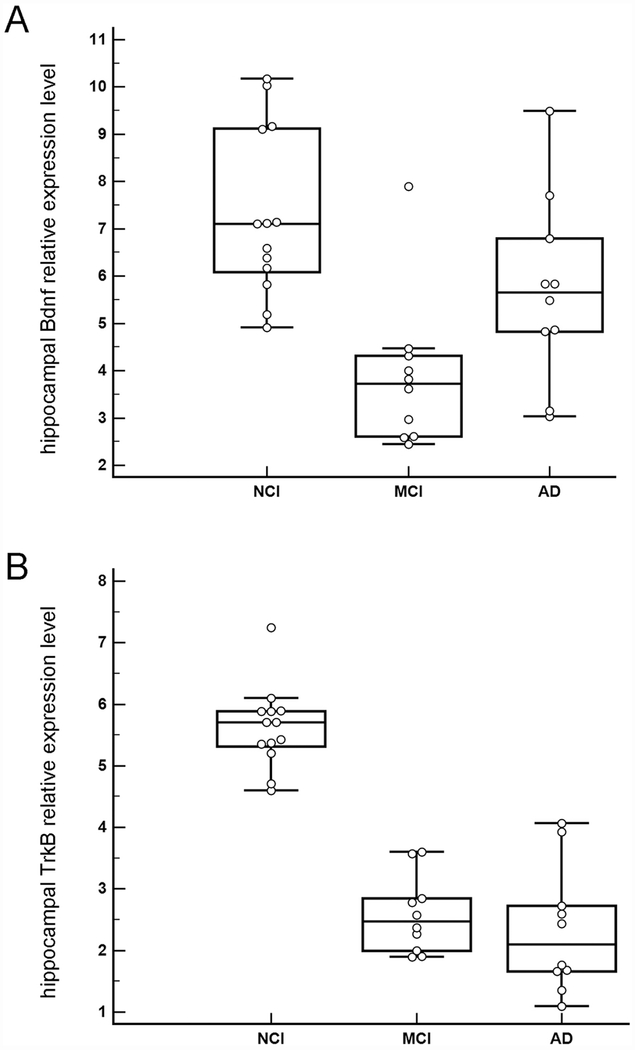
Downregulation of hippocampal Bdnf was found between NCI and MCI (p<0.001) while upregulation was observed between MCI and AD groups (p = 0.01) (Figure 4A). Downregulation of hippocampal TrkB was found between NCI and MCI (p<0.001) and NCI and AD (p<0.001) (Figure 4B). No significant difference in TrkB expression was found between MCI and AD (p = 0.27).
Figure 4.
A. Boxplots illustrating clinical group differences within regional hippocampal dissections for Bdnf. Significant downregulation of Bdnf was found between NCI and MCI (p<0.001) while significant upregulation was observed between MCI and AD (p = 0.01). B. Boxplots illustrating clinical group differences within regional hippocampal dissections for TrkB. Significant downregulation was found for TrkB during the progression of dementia as MCI (p<0.001) and AD (p<0.001) displayed significantly less expression than NCI. No significant difference in TrkB expression was found between MCI and AD (p = 0.27).
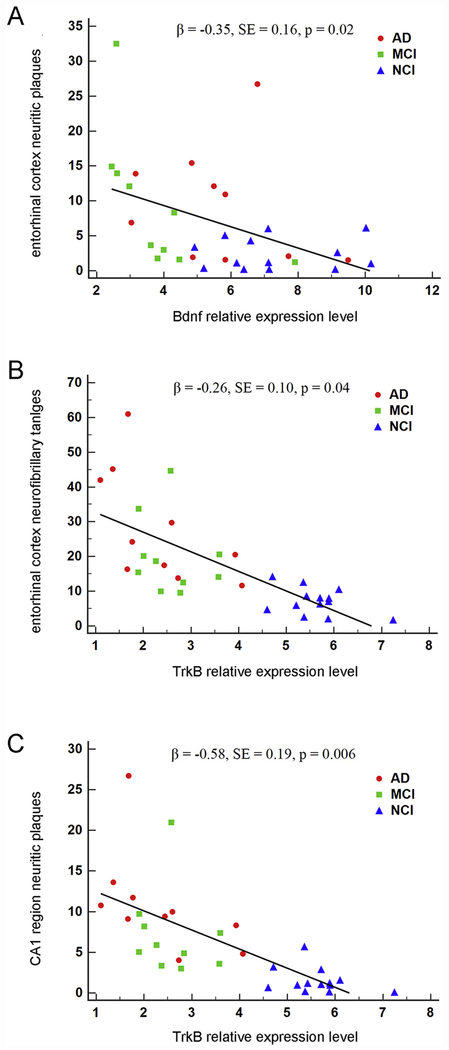
Among entorhinal cortex pathology measures, hippocampal Bdnf was a significant predictor of NPs (β = −0.35, SE = 0.16, p = 0.02; Figure 5A), but not DPs (β = −0.21, SE = 0.17, p = 0.38) or NFTs (β = −0.04, SE = 0.09, p = 0.70) (Table 5). Bdnf interactions with APOE ε4 carrier status were not significant for NPs (β = 0.27, SE = 0.24, p = 0.25), DPs (β = 0.23, SE = 0.27, p = 0.38), or NFTs (β = −0.09, SE = 0.14, p = 0.54). Entorhinal cortex pathology models for Bdnf all demonstrated good fit (NPs: p = 0.19, DPs: p = 0.15, NFTs: p = 0.11). For CA1 region pathology measures, Bdnf was not a significant predictor of NPs (β = 0.09, SE = 0.18, p = 0.64), DPs (β = −0.16, SE = 0.22, p = 0.45) or NFTs (β = −0.10, SE = 0.11, p = 0.35) (Table 5). Bdnf interactions with APOE ε4 carrier status were not significant for NPs (β = −0.18, SE = 0.28, p = 0.51), DPs (β = 0.28, SE = 0.32, p = 0.37), or NFTs (β = −0.08, SE = 0.17, p = 0.64). Each CA1 pathology model demonstrated good fit (NPs: p = 0.18, DPs: p = 0.37, NFTs: p = 0.10).
Figure 5.
Association of Bdnf and TrkB expression in regional hippocampal dissections with entorhinal cortex neuritic plaques, entorhinal cortex neurofibrillary tangles, and hippocampal CA1 region neuritic plaques. A. A significant inverse association was found between Bdnf expression and entorhinal cortex neuritic plaques by clinical group (β = −0.35, SE = 0.16, p = 0.02). B. A significant inverse association was found between TrkB expression and entorhinal cortex neurofibrillary tangles by clinical group (β = −0.26, SE = 0.10, p = 0.04). C. A significant inverse association was found between TrkB expression and hippocampal CA1 neuritic plaques by clinical group (β = −0.58, SE = 0.19, p = 0.006).
Table 5.
Regional hippocampal negative binomial regression models using Bdnf as a predictor of entorhinal cortex and hippocampal CA1 AD pathology.
| Entorhinal Cortex | ||||
| Coefficient | SE | p-value | ||
| Outcome = NPs | ||||
| Bdnf | −0.35 | 0.16 | 0.02* | |
| APOE ε4 | −0.79 | 1.45 | 0.58 | |
| APOE ε4 x Bdnf | 0.27 | 0.24 | 0.25 | |
| Outcome = DPs | ||||
| Bdnf | −0.21 | 0.17 | 0.23 | |
| APOE ε4 | −1.05 | 1.66 | 0.53 | |
| APOE ε4 x Bdnf | 0.23 | 0.27 | 0.38 | |
| Outcome = NFTs | ||||
| Bdnf | −0.04 | 0.09 | 0.70 | |
| APOE ε4 | 1.04 | 0.88 | 0.24 | |
| APOE ε4 x Bdnf | −0.09 | 0.14 | 0.54 | |
| Hippocampal CA1 | ||||
| Coefficient | SE | p-value | ||
| Outcome = NPs | ||||
| Bdnf | 0.09 | 0.18 | 0.64 | |
| APOE ε4 | 1.85 | 1.75 | 0.29 | |
| APOE ε4 x Bdnf | −0.18 | 0.28 | 0.51 | |
| Outcome = DPs | ||||
| Bdnf | −0.16 | 0.22 | 0.45 | |
| APOE ε4 | 0.22 | 1.97 | 0.91 | |
| APOE ε4 x Bdnf | 0.28 | 0.32 | 0.37 | |
| Outcome = NFTs | ||||
| Bdnf | −0.10 | 0.11 | 0.35 | |
| APOE ε4 | 0.78 | 1.06 | 0.46 | |
| APOE ε4 x Bdnf | −0.08 | 0.17 | 0.64 | |
Models included age at death, gender, and APOE ε4 carrier status. SE = standard error.
= significant.
Using hippocampal TrkB as a predictor of entorhinal cortex pathology, a significant association was found for NFTs (β = −0.26, SE = 0.10, p = 0.04; Figure 5B), but not DPs (β = −0.26, SE = 0.21, p = 0.22) or NPs (β = −0.22, SE = 0.21, p = 0.29) (Table 6). TrkB and APOE ε4 carrier status associations were not significant for NPs (β = 0.05, SE = 0.37, p = 0.89), DPs (β = 0.05, SE = 0.37, p = 0.88), or NFTs (β = 0.00, SE = 0.19, p = 0.99). Entorhinal cortex pathology models for TrkB demonstrated good fit (NPs: p = 0.20, DPs: p = 0.15, NFTs: p = 0.12). For CA1 region pathology measures, TrkB was significantly associated with NPs (β = −0.58, SE = 0.19, p = 0.006; Figure 5C), but not DPs (β = −0.31, SE = 0.27, p = 0.26) or NFTs (β = −0.26, SE = 0.12, p = 0.08) (Table 6). TrkB interactions with APOE ε4 carrier status were not significant for NPs (β = 0.48, SE = 0.39, p = 0.22), DPs (β = 0.08, SE = 0.42, p = 0.85), or NFTs (β = −0.28, SE = 0.22, p = 0.21). CA1 region pathology models for TrkB all demonstrated good fit (NPs: p = 0.21, DPs: p = 0.35, NFTs: p = 0.11).
Table 6.
Hippocampal dissection negative binomial regression models using TrkB as a predictor of entorhinal cortex and hippocampal CA1 AD pathology.
| Entorhinal Cortex | ||||
| Coefficient | SE | p-value | ||
| Outcome = NPs | ||||
| TrkB | −0.22 | 0.21 | 0.29 | |
| APOE ε4 | 0.50 | 1.34 | 0.71 | |
| APOE ε4 x TrkB | 0.05 | 0.37 | 0.89 | |
| Outcome = DPs | ||||
| TrkB | −0.26 | 0.21 | 0.22 | |
| APOE ε4 | −0.12 | 1.37 | 0.93 | |
| APOE ε4 x TrkB | 0.05 | 0.37 | 0.88 | |
| Outcome = NFTs | ||||
| TrkB | −0.26 | 0.10 | 0.04* | |
| APOE ε4 | 0.45 | 0.68 | 0.51 | |
| APOE ε4 x TrkB | 0.00 | 0.19 | 0.99 | |
| Hippocampal CA1 | ||||
| Coefficient | SE | p-value | ||
| Outcome = NPs | ||||
| TrkB | −0.58 | 0.19 | 0.006* | |
| APOE ε4 | −0.88 | 1.38 | 0.52 | |
| APOE ε4 x TrkB | 0.48 | 0.39 | 0.22 | |
| Outcome = DPs | ||||
| TrkB | −0.31 | 0.27 | 0.26 | |
| APOE ε4 | 1.68 | 1.56 | 0.28 | |
| APOE ε4 x TrkB | 0.08 | 0.42 | 0.85 | |
| Outcome = NFTs | ||||
| TrkB | −0.26 | 0.12 | 0.08 | |
| APOE ε4 | 0.98 | 0.81 | 0.22 | |
| APOE ε4 x TrkB | −0.28 | 0.22 | 0.21 | |
Models included age at death, gender, and APOE ε4 carrier status. SE = standard error.
= significant.
Discussion
The present investigation assessed associations between Bdnf and TrkB expression and NPs, DPs, and NFTs within a homogeneous population of CA1 pyramidal neurons as well as regional hippocampal dissections using postmortem tissues. The NB model using regional hippocampal entorhinal cortex NPs as an outcome measure demonstrates that downregulation of Bdnf is independently associated with increased regional entorhinal cortex NPs. A parallel observation was found with TrkB and entorhinal cortex NFTs. Specifically, downregulation of TrkB is independently associated with increased entorhinal cortex NFTs. These novel results suggest dysfunctional hippocampal neurotrophin signaling, namely BDNF and its cognate receptor TrkB, is associated with, and may possibly be upstream of entorhinal cortex NPs and NFTs, early sites of AD pathology. Perhaps ameliorating BDNF/TrkB signaling deficits, especially within the entorhinal cortex and hippocampus, may potentially abrogate the formation of NP and NFT pathology. Although this hypothesis is conjectural, the power of this timely study is that it is an attempt at teasing apart a spatio-temporal association between neuronal transcript expression and AD pathology that can be validated or rejected in representative model systems. In this scenario, findings derived from human disease dictates what to assess in model systems, and not vice versa.
Significance of the possible temporal sequence of BDNF/TrkB deficits preceding neuropathology lies in its therapeutic potential. Several research laboratories, including our group, hypothesize deficits in BDNF signaling leads to selective vulnerability of specific neuronal populations, including hippocampal CA1 pyramidal neurons and cholinergic basal forebrain neurons of the nucleus basalis of Meynert (Erickson et al., 2010; Ginsberg et al., 2006, 2010, 2011, 2012, 2019; Mufson et al., 2007a, 2007b; Peng et al., 2005; Tanila, 2017). Restoration of BDNF/TrkB signaling, either through neurotrophin delivery (Mitra et al., 2019; Nagahara et al., 2009, 2013), transactivation of the TrkB receptor, or activation of downstream pathways (Mitre et al., 2017; Rajagopal et al., 2004; Skaper, 2008) may provide neuroprotection by targeting mechanisms of neuroplasticity as well as preventing and/or attenuating amyloid and tau pathology (Arancibia et al., 2008; Mufson et al., 2007a, 2007b, 2015; Nagahara et al., 2009).
The association(s) between BDNF signaling and the cascades that lead to plaque and tangle formation are likely not unidirectional. Rather, a growing number of studies using relevant AD animal and cellular models suggests amyloid-β precursor protein (APP) and its metabolites, including amyloid-β peptide (Aβ) downregulates BDNF expression and BDNF-TrkB retrograde trafficking (Garzon and Fahnestock, 2007; Peng et al., 2009; Poon et al., 2011, 2013), possibly through cyclic adenosine monophosphate response element binding protein transcriptional downregulation (Rosa and Fahnestock, 2015). Alternatively, loss of BDNF leads to changes in synaptic protein levels (Mariga et al., 2015), cleavage and metabolism of APP (Matrone et al., 2008) and tau (Corsetti et al., 2008). Since exogenous delivery of BDNF ameliorates synaptic as well as learning and memory deficits in primate and rodent AD models (Arancibia et al., 2008; Iwasaki et al., 2012; Nagahara et al., 2009, 2013), extensive development has gone into devising BDNF brain delivery methods (Jiang et al., 2018; Mitra et al., 2019). Moreover, increased tau pathology has been related to a depletion of BDNF signaling through the glucocorticoid receptor pathway (Arango-Lievano et al., 2016). Tau overexpression decreases BDNF signaling in cellular and mouse models of AD and in tauopathy (Mazzaro et al., 2016; Rosa et al., 2016). A defective BDNF response, likely through NMDA receptor signaling, has also been observed in a mouse model of tauopathy (Burnouf et al., 2013). The present postmortem human tissue findings suggest multiple levels of regulation exist between neurotrophin signaling and amyloid and tau pathology that merit further investigation in both well-characterized clinical pathological cohorts of human subjects during the progression of dementia as well as in animal and cellular models that enable temporal sequences and mechanistic interactions to be elucidated.
Similar to evaluations of NCI subjects with low- and high-AD pathology (Malek-Ahmadi et al., 2016, 2018, 2019) that failed to reveal correlations between DPs and cognitive decline, we found Bdnf and TrkB were not associated with DP load. However, CA1 TrkB showed a significant inverse association with entorhinal cortex DPs. The present dataset also lends support to the contention that NPs are the salient plaque species related to the pathogenesis of dementia, while DPs are more likely to be bystanders in disease progression (Dickson, 2009; Ginsberg et al., 2019; Malek-Ahmadi et al., 2018, 2019; Mufson et al., 2016a).
This study provides proof-of-concept that individual transcripts within a functional gene ontology group (e.g., neurotrophins and neurotrophin receptors) can be used to predict (or to be an outcome measure) associated with AD neuropathology. Future studies evaluating other potential drivers of NPs and NFTs, such as endosomal-lysosomal and autophagy markers, or potential drivers of synaptic pathology, such as synaptic-related markers, glutamatergic neurotransmission, and GABAergic neurotransmission genes in relation to NPs and NFTs is warranted. Moreover, present results highlight the utility of nontraditional informatics/biostatistics on human postmortem data to drive mechanistic assessments of AD pathogenesis. Future studies using microarray and/or RNA-sequencing derived data from individual populations and discrete vulnerable brain regions is likely to shed light on nodal interactions between gene/encoded protein dysregulation, AD pathological lesions, and cognitive decline during the progression of dementia. Machine learning or related systems biology modeling approaches (Donnelly-Kehoe et al., 2018; Grassi et al., 2018; Lins et al., 2017; Signaevsky et al., 2019) appear to be feasible methods to pair neurotrophin dysregulation with AD pathology deposition/accumulation.
There are limitations to consider including a lack of direct causality between neurotrophin/neurotrophin receptor mRNA levels and AD neuropathological lesions in postmortem human tissue based clinical pathological investigations. Future mechanistic studies using in vitro and in vivo models (Arango-Lievano et al., 2016; Mariga et al., 2015; Mitra et al., 2019; Nagahara et al., 2009, 2013; Rosa et al., 2016) are warranted based on our findings derived from bona fide human disease. Another caveat is the number of subjects relative to the number of predictors in the NB regression analyses. All of the NB models we employed contained five predictor variables (four main effect and one interaction), which by some standards, is too many for the sample sizes used in this study. Although the statistical convention that ten subjects per independent variable is ideal to provide valid estimates, studies have revealed that as few as two subjects per covariate can provide adequate estimates for regression models (Austin and Steyerberg, 2015). Nonetheless, careful consideration was given to the selection of covariates and interaction terms used in the models to account for biologically relevant factors. In addition, goodness of fit assessments for each model indicated our data met the underlying assumptions of NB distribution. Therefore, we contend these models accurately support relationships between Bdnf and TrkB expression and NP/NFT pathology.
In summary, NB regression models were employed to infer Bdnf and TrkB within homogenous CA1 pyramidal neurons and heterogeneous hippocampal complex dissections in relation to NP, NFT, and DP pathology. Based upon our findings, we propose that BDNF signaling defects may be upstream of classic AD lesions. This does not preclude the possibility that amyloid and tau cascades impinge upon BDNF and TrkB regulation as part of a reciprocal regulatory loop exacerbating neurodegenerative pathology and serve as a node or hub for therapeutic intervention.
Highlights.
Negative binomial models associate Bdnf & TrkB levels with Alzheimer neuropathology.
CA1 neuron and hippocampal Bdnf & TrkB levels are similar in Alzheimer’s disease.
Hippocampal Bdnf inversely correlates with entorhinal cortex neuritic plaques.
Hippocampal TrkB inversely correlates with entorhinal cortex neurofibrillary tangles.
Hippocampal TrkB inversely correlates with hippocampal CA1 neuritic plaques.
Acknowledgments
Supported by grants PO1 AG014449, RO1 AG043375, PO1 AG107617, R01 NS21072, R01 AG025970, P30 AG010161, and P30 AG053769 from the National Institutes of Health, the Alzheimer’s Association, and Barrow and Beyond at the Barrow Neurological Institute. We are indebted to the Catholic nuns, priests, and lay brothers who participated in the RROS. We thank Shaoli Che, M.D., PhD. Irina Elarova, M.S., and Arthur Saltzman, M.S. for expert technical assistance.
Abbreviations
- Aβ
amyloid-β peptide
- AD
Alzheimer’s disease
- ApoE
apolipoprotein E
- BDNF
brain-derived neurotrophic factor
- DTT
dithiothreitol
- DPs
diffuse plaques
- ECD
extracellular domain
- MCI
mild cognitive impairment
- NB
negative binomial
- NCI
no cognitive impairment
- NFTs
neurofibrillary tangles
- NPs
neuritic plaques
- PMI
postmortem interval
- RROS
Rush Religious Orders Study
- SDS
sodium dodecyl sulfate
- SSPE
6X saline–sodium phosphate–ethylenediaminetetraacetic acid
- TC
terminal continuation
- TK
tyrosine kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosure
Stephen D. Ginsberg, Michael H. Malek-Ahmadi, Melissa J. Alldred, Yinghua Chen, Kewei Chen, Moses V Chao, Scott E. Counts have no financial disclosures to declare. Elliott J. Mufson consults for Rexgen and RegenxBio.
References
- Abner EL, et al. , 2017. Outcomes after diagnosis of mild cognitive impairment in a large autopsy series. Ann Neurol. 81, 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, et al. , 2018. CA1 pyramidal neuron gene expression mosaics in the Ts65Dn murine model of Down syndrome and Alzheimer’s disease following maternal choline supplementation. Hippocampus. 28, 251–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, et al. , 2008. Terminal continuation (TC) RNA amplification enables expression profiling using minute RNA input obtained from mouse brain. Int J Mol Sci. 9, 2091–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, et al. , 2009. Terminal continuation (TC) RNA amplification without second strand synthesis. J Neurosci Methods. 177, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, et al. , 2012. Microarray analysis ofCA1 pyramidal neurons in a mouse model of tauopathy reveals progressive synaptic dysfonction. Neurobiol Dis. 45, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, et al. , 2015. Expression profile analysis of hippocampal CA1 pyramidal neurons in aged Ts65Dn mice, a model of Down syndrome (DS) and Alzheimer’s disease (AD). Brain Struct Funct. 220, 2983–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen SJ, et al. , 2011. The neurotrophins and their role in Alzheimer’s disease. Curr Neuropharmacol. 9, 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez A, et al. , 2014. Apathy and APOE4 are associated with reduced BDNF levels in Alzheimer’s disease. J Alzheimers Dis. 42, 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arancibia S, et al. , 2008. Protective effect of BDNF against beta-amyloid induced neurotoxicity in vitro and in vivo in rats. Neurobiol Dis. 31, 316–326. [DOI] [PubMed] [Google Scholar]
- Arango-Lievano M, et al. , 2016. Deletion of neurotrophin signaling through the glucocorticoid receptor pathway causes tau neuropathology. Sci Rep. 6, 37231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin PC, Steyerberg EW, 2015. The number of subjects per variable required in linear regression analyses. J Clin Epidemiol. 68, 627–636. [DOI] [PubMed] [Google Scholar]
- Autry AE, Monteggia LM, 2012. Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacol Rev. 64, 238–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belrose JC, et al. , 2014. Increased pro-nerve growth factor and decreased brain-derived neurotrophic factor in non-Alzheimer’s disease tauopathies. Neurobiol Aging. 35, 926–933. [DOI] [PubMed] [Google Scholar]
- Bennett DA, Launer LJ, 2012. Longitudinal epidemiologic clinical-pathologic studies of aging and Alzheimer’s disease. Curr Alzheimer Res. 9, 617–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, et al. , 2012. Overview and findings from the religious orders study. Curr Alzheimer Res. 9, 628–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, et al. , 2002. Natural history of mild cognitive impairment in older persons. Neurology. 59, 198–205. [DOI] [PubMed] [Google Scholar]
- Berk R, MacDonald J, 2008. Overdispersion and Poisson regression. J Quant Criminol. 24, 269–284. [Google Scholar]
- Burnouf S, et al. , 2013. NMDA receptor dysfunction contributes to impaired brain-derived neurotrophic factor-induced facilitation of hippocampal synaptic transmission in a Tau transgenic model. Aging Cell. 12,11–23. [DOI] [PubMed] [Google Scholar]
- Che S, Ginsberg SD, 2004. Amplification of transcripts using terminal continuation. Lab Invest. 84, 131–137. [DOI] [PubMed] [Google Scholar]
- Corsetti V, et al. , 2008. Identification of a caspase-derived N-terminal tau fragment in cellular and animal Alzheimer’s disease models. Mol Cell Neurosci. 38, 381–392. [DOI] [PubMed] [Google Scholar]
- Counts SE, et al. , 2007. {alpha}7 Nicotinic receptor up-regulation in cholinergic basal forebrain neurons in Alzheimer disease. Arch Neurol. 64, 1771–1776. [DOI] [PubMed] [Google Scholar]
- Cowansage KK, et al. , 2010. Brain-derived neurotrophic factor: a dynamic gatekeeper of neural plasticity. Curr Mol Pharmacol. 3, 12–29. [DOI] [PubMed] [Google Scholar]
- Dickson DW, 2009. Neuropathology of non-Alzheimer degenerative disorders. Int J Clin Exp Pathol. 3, 1–23. [PMC free article] [PubMed] [Google Scholar]
- Donnelly-Kehoe PA, et al. , 2018. Looking for Alzheimer’s disease morphometric signatures using machine learning techniques. J Neurosci Methods. 302, 24–34. [DOI] [PubMed] [Google Scholar]
- Erickson KI, et al. , 2010. Brain-derived neurotrophic factor is associated with age-related decline in hippocampal volume. J Neurosci. 30, 5368–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlenza OV, et al. , 2015. Lower cerebrospinal fluid concentration of brain-derived neurotrophic factor predicts progression from mild cognitive impairment to Alzheimer’s disease. Neuromolecular Med. 17, 326–332. [DOI] [PubMed] [Google Scholar]
- Garzon D, et al. , 2002. A new brain-derived neurotrophic factor transcript and decrease in brain-derived neurotrophic factor transcripts 1, 2 and 3 in Alzheimer’s disease parietal cortex. J Neurochem. 82, 1058–1064. [DOI] [PubMed] [Google Scholar]
- Garzon DJ, Fahnestock M, 2007. Oligomeric amyloid decreases basal levels of brain-derived neurotrophic factor (BDNF) mRNA via specific downregulation of BDNF transcripts IV and V in differentiated human neuroblastoma cells. J Neurosci. 27, 2628–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, 2005. RNA amplification strategies for small sample populations. Methods. 37, 229–237. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, 2008. Transcriptional profiling of small samples in the central nervous system. Methods Mol Biol. 439, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, 2014. Considerations in the use of microarrays for analysis of the CNS. Reference Module in Biomedical Research. 1–7. [Google Scholar]
- Ginsberg SD, et al. , 2012. Gene expression levels assessed by CA1 pyramidal neuron and regional hippocampal dissections in Alzheimer’s disease. Neurobiol Dis. 45, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, et al. , 2010. Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer’s disease progression. Biol Psychiatry. 68, 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, et al. , 2006. Down regulation of trk but not p75NTR gene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer’s disease. J Neurochem. 97, 475–487. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, et al. , 2000. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Ann Neurol. 48, 77–87. [PubMed] [Google Scholar]
- Ginsberg SD, et al. , 2019. Selective decline of neurotrophin and neurotrophin receptor genes within CA1 pyramidal neurons and hippocampus proper: Correlation with cognitive performance and neuropathology in mild cognitive impairment and Alzheimer’s disease. Hippocampus. 29, 422–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, et al. , 2011. Upregulation of select rab GTPases in cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer’s disease. J Chem Neuroanat. 42, 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi M, et al. , 2018. A clinically-translatable machine learning algorithm for the prediction of Alzheimer’s disease conversion in individuals with mild and premild cognitive impairment. J Alzheimers Dis. 61, 1555–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SD, et al. , 2012. Functional connectivity variations in mild cognitive impairment: associations with cognitive function. J Int Neuropsychol Soc. 18, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SD, et al. , 2015. Mild cognitive impairment is associated with poorer decision-making in community-based older persons. J Am Geriatr Soc. 63, 676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger RM, et al. , 2000. Quantitation of BDNF mRNA in human parietal cortex by competitive reverse transcription-polymerase chain reaction: decreased levels in Alzheimer’s disease. Brain Res Mol Brain Res. 76, 347–354. [DOI] [PubMed] [Google Scholar]
- Iwasaki Y, et al. , 2012. Sendai virus vector-mediated brain-derived neurotrophic factor expression ameliorates memory deficits and synaptic degeneration in a transgenic mouse model of Alzheimer’s disease. J Neurosci Res. 90, 981–989. [DOI] [PubMed] [Google Scholar]
- Jiang Y, et al. , 2018. Nanoformulation of brain-derived neurotrophic factor with target receptor-triggered-release in the central nervous system. Adv Funct Mater. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kriebel D, 2009. Regression models for public health surveillance data: a simulation study. Occup Environ Med. 66, 733–739. [DOI] [PubMed] [Google Scholar]
- Leal G, et al. , 2015. Regulation of hippocampal synaptic plasticity by BDNF. Brain Res. 1621, 82–101. [DOI] [PubMed] [Google Scholar]
- Lins A, et al. , 2017. Using artificial neural networks to select the parameters for the prognostic of mild cognitive impairment and dementia in elderly individuals. Comput Methods Programs Biomed. 152, 93–104. [DOI] [PubMed] [Google Scholar]
- Malek-Ahmadi M, et al. , 2018. Cognitive composite score association with Alzheimer’s disease plaque and tangle pathology. Alzheimers Res Ther. 10, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek-Ahmadi M, et al. , 2019. Cerebral amyloid angiopathy and neuritic plaque pathology correlate with cognitive decline in elderly non-demented individuals. J Alzheimers Dis. 67, 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek-Ahmadi M, et al. , 2016. Neuritic and diffuse plaque associations with memory in non-cognitively impaired elderly. J Alzheimers Dis. 53, 1641–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariga A, et al. , 2015. Withdrawal of BDNF from hippocampal cultures leads to changes in genes involved in synaptic function. Dev Neurobiol. 75, 173–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrone C, et al. , 2008. NGF and BDNF signaling control amyloidogenic route and Abeta production in hippocampal neurons. Proc Natl Acad Sci USA. 105, 13139–13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzaro N, et al. , 2016. Tau-driven neuronal and neurotrophic dysfunction in a mouse model of early tauopathy. J Neurosci. 36, 2086–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski B, et al. , 2015. Brain-derived neurotrophic factor and TrkB expression in the “oldest-old,” the 90+ Study: correlation with cognitive status and levels of soluble amyloid-beta. Neurobiol Aging. 36, 3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, et al. , 2019. Innovative therapy for Alzheimer’s disease-with focus on biodelivery of NGF. Front Neurosci. 13, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitre M, et al. , 2017. Neurotrophin signalling: novel insights into mechanisms and pathophysiology. Clin Sci (Lond). 131, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufson EJ, et al. , 2012. Mild cognitive impairment: pathology and mechanisms. Acta Neuropathol. 123, 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufson EJ, et al. , 1999. Entorhinal cortex beta-amyloid load in individuals with mild cognitive impairment. Exp. Neurol. 158, 469–490. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, et al. , 2007a. Cholinotrophic molecular substrates of mild cognitive impairment in the elderly. Curr Alzheimer Res. 4, 340–350. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, et al. , NGF family of neurotrophins and their receptors: early involvement in the progression of Alzheimer’s disease In: Dawbarn D, Allen SJ, (Eds.), Neurobiology of Alzheimer’s Disease, Third Edition Oxford University Press, Oxford, 2007b, pp. 283–321. [Google Scholar]
- Mufson EJ, et al. , 2002a. Gene expression profiles of cholinergic nucleus basalis neurons in Alzheimer’s disease. Neurochem Res. 27, 1035–1048. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, et al. , 2016a. Molecular and cellular pathophysiology of preclinical Alzheimer’s disease. Behav Brain Res. 311, 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufson EJ, et al. , 2000. Loss of nucleus basalis neurons containing trkA immunoreactivity in individuals with mild cognitive impairment and early Alzheimer’s disease. J Comp Neurol. 427, 19–30. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, et al. , 2002b. Loss of basal forebrain p75(NTR) immunoreactivity in subjects with mild cognitive impairment and Alzheimer’s disease. J Comp Neurol. 443, 136–153. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, et al. , 2015. Hippocampal plasticity during the progression of Alzheimer’s disease. Neuroscience. 309, 51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufson EJ, et al. , 2016b. Braak staging, plaque pathology, and APOE status in elderly persons without cognitive impairment. Neurobiol Aging. 37, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray KD, et al. , 1994. Differential regulation of brain-derived neurotrophic factor and type II calcium/calmodulin-dependent protein kinase messenger RNA expression in Alzheimer’s disease. Neuroscience. 60, 37–48. [DOI] [PubMed] [Google Scholar]
- Nagahara AH, et al. , 2013. Early BDNF treatment ameliorates cell loss in the entorhinal cortex of APP transgenic mice. J Neurosci. 33, 15596–15602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahara AH, et al. , 2009. Neuroprotective effects of brain-derived neurotrophic factor in rodent and primate models of Alzheimer’s disease. Nat Med. 15, 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, et al. , 2017. Emerging roles of the neurotrophin receptor TrkC in synapse organization. Neurosci Res. 116, 10–17. [DOI] [PubMed] [Google Scholar]
- Oveisgharan S, et al. , 2018. Sex differences in Alzheimer’s disease and common neuropathologies of aging. Acta Neuropathol. 136, 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, et al. , 2009. Decreased brain-derived neurotrophic factor depends on amyloid aggregation state in transgenic mouse models of Alzheimer’s disease. J Neurosci. 29, 9321–9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, et al. , 2005. Precursor form of brain-derived neurotrophic factor and mature brain-derived neurotrophic factor are decreased in the pre-clinical stages of Alzheimer’s disease. J Neurochem. 93, 1412–21. [DOI] [PubMed] [Google Scholar]
- Phillips HS, et al. , 1991. BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer’s disease. Neuron. 7, 695–702. [DOI] [PubMed] [Google Scholar]
- Poon WW, et al. , 2011. beta-Amyloid impairs axonal BDNF retrograde trafficking. Neurobiol Aging. 32, 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon WW, et al. , 2013. beta-Amyloid (Abeta) oligomers impair brain-derived neurotrophic factor retrograde trafficking by down-regulating ubiquitin C-terminal hydrolase, UCH-L1. J Biol Chem. 288, 16937–16948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal R, et al. , 2004. Transactivation of Trk neurotrophin receptors by G-protein-coupled receptor ligands occurs on intracellular membranes. J Neurosci. 24, 6650–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa E, Fahnestock M, 2015. CREB expression mediates amyloid beta-induced basal BDNF downregulation. Neurobiol Aging. 36, 2406–2413. [DOI] [PubMed] [Google Scholar]
- Rosa E, et al. , 2016. Tau downregulates BDNF expression in animal and cellular models of Alzheimer’s disease. Neurobiol Aging. 48, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, et al. , 2017. ApoE isoforms differentially regulates cleavage and secretion of BDNF. Mol Brain. 10, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signaevsky M, et al. , 2019. Artificial intelligence in neuropathology: deep learning-based assessment of tauopathy. Lab Invest, 99, 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaper SD, 2008. The biology of neurotrophins, signalling pathways, and functional peptide mimetics of neurotrophins and their receptors. CNS Neurol Disord Drug Targets. 7, 46–62. [DOI] [PubMed] [Google Scholar]
- Tanila H, 2017. The role of BDNF in Alzheimer’s disease. Neurobiol Dis. 97, 114–118. [DOI] [PubMed] [Google Scholar]
- Tapia-Arancibia L, et al. , 2008. New insights into brain BDNF function in normal aging and Alzheimer disease. Brain Res Rev. 59, 201–220. [DOI] [PubMed] [Google Scholar]
- Thompson Ray M, et al. , 2011. Decreased BDNF, TrkB-TK+ and GAD67 mRNA expression in them hippocampus of individuals with schizophrenia and mood disorders. J Psychiatry Neurosci. 36, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiernan CT, et al. , 2016. Protein homeostasis gene dysregulation in pretangle-bearing nucleus basalis neurons during the progression of Alzheimer’s disease. Neurobiol Aging. 42, 80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjur T, 1998. Nonlinear regression, quasi likelihood, and overdispersion in generalized linear models. Am Stat. 52, 222–227. [Google Scholar]
- Vose D, 2008. Risk Analysis: A Quantitative Guide, 3rd Edition John Wiley & Sons, Chichester. [Google Scholar]
- Yoshii A, Constantine-Paton M, 2010. Postsynaptic BDNF-TrkB signaling in synapse maturation, plasticity, and disease. Dev Neurobiol. 70, 304–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, et al. , 2019. Association of cortical beta-amyloid protein in the absence of insoluble deposits with Alzheimer disease. JAMA Neurol. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E, 2009. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 5, 311–322. [DOI] [PubMed] [Google Scholar]