Abstract
Diffusion magnetic resonance images typically suffer from spatial distortions due to susceptibility induced off-resonance fields, which may affect the geometric fidelity of the reconstructed volume and cause mismatches with anatomical images. State-of-the art susceptibility correction (for example, FSL’s TOPUP algorithm) typically requires data acquired twice with reverse phase encoding directions, referred to as blip-up blip-down acquisitions, in order to estimate an undistorted volume. Unfortunately, not all imaging protocols include a blip-up blip-down acquisition, and cannot take advantage of the state-of-the art susceptibility and motion correction capabilities. In this study, we aim to enable TOPUP-like processing with historical and/or limited diffusion imaging data that include only a structural image and single blip diffusion image. We utilize deep learning to synthesize an undistorted non-diffusion weighted image from the structural image, and use the non-distorted synthetic image as an anatomical target for distortion correction. We evaluate the efficacy of this approach (named Synb0-DisCo) and show that our distortion correction process results in better matching of the geometry of undistorted anatomical images, reduces variation in diffusion modeling, and is practically equivalent to having both blip-up and blip-down non-diffusion weighted images.
Keywords: Image synthesis, Distortion correction, Diffusion MRI, Conditional generative network, Echo planar imaging
INTRODUCTION
Diffusion magnetic resonance imaging (dMRI) allows non-invasive mapping of the diffusion process of water molecules in biological tissue [1, 2], providing valuable information about tissue microstructure [3] and facilitating study of the anatomical connections of the brain in a process known as fiber tractography [4–6]. To date, a vast majority of dMRI is acquired using some variation of echo-planar imaging (EPI) [7], which allows collection of a large number of diffusion-weighted images (DWIs) in a very short time. However, a well-known problem with EPI is geometric and intensity distortions of DWIs caused by susceptibility induced field imperfections (B0 field inhomogeneities) in conjunction with a low bandwidth in the phase-encode (PE) direction, causing spatial distortion along the PE axis [8, 9]. These distortions may limit the accuracy of image analysis in affected regions and can lead to misalignment with alternative contrasts which provide structural information [10, 11].
Reducing or correcting EPI distortions generally requires sequence modification (i.e., parallel imaging, increased bandwidth) or acquisition of additional data. Strategies to correct distortions include B0 mapping [8, 12, 13], point spread function estimation [14, 15], image registration from the distorted diffusion images to an undistorted non-EPI anatomical target (typically a T1- or T2-weighted contrast, or a synthesized T2-contrast [16], using a non-rigid registration framework) [11, 17–20], or recently developed reverse gradient / “blip-up blip-down” acquisitions [9]. Each of these approaches suffers from practical imaging concerns that limit general applicability. Field mapping of the B0 inhomogeniety can be corrupted by motion, artifacts, or phase errors, and geometric correction with corrupt/limited B0 fields can actually degrade image quality. Similarly, point spread function estimation is dependent on empirical estimation of imaging properties and applying a forward model for correction. Although widely applied, registration-based correction suffers from difficulty in matching contrasts between scans. Diffusion images can have very different contrasts than the standard structural scans to which they are registered, and alternative optimization metrics or pre-processing techniques are required. Acquisition of reference images with contrast matched to the diffusion images consumes precious additional scan time.
The final distortion correction strategy is to acquire pairs of images with reverse phase encoding (PE) gradients, often referred to as a “blip-up blip-down” acquisition [9], yielding pairs of images with the same contrast, but distortions reversed in directions. The undistorted image can then be derived by warping the two images to the mid-way point between them. Initial work estimated the un-warped images independently along lines in the PE direction [21, 22] and normalizing by the geometric mean. However, these methods did not recognize that the underlying field should be continuous, and typically smooth, and due to their 1D nature (along PE lines), these methods are sensitive to noise, edge effects, and numerical instabilities. To improve upon this, Andersson et al. [23] proposed to model the warping field as a linear combination of smooth basis functions, which, in addition to regularizing the problem by operating on a 3D space instead of 1D lines, allows the inclusion of subject motion into the correction. This technique has been incorporated into the FSL software package [24, 25] under the name TOPUP as part of their standard dMRI preprocessing pipeline, and has since become a popular blip-up blip-down correction technique, as well as the tool of choice for the Human Connectome project [26]. Later modifications to this technique include hierarchical smoothing [27], integration of registration-based algorithms [28], alternative acquisitions [29, 30], and incorporation of field drift and structural image information [31]. An attractive feature of the blip-up blip-down acquisitions is that the pairs of images can be both used for distortion information and, after correction, contribute to the diffusion imaging microarchitecture estimation routines. Hence, time is not “wasted” on distortion correction. In summary, blip-up blip-down distortion correction methods significantly improve correction quality for EPI distortions with minimal penalty for overall scan time.
Unfortunately, not all imaging protocols include a blip-up blip-down, or similar, acquisition, and cannot take advantage of the state-of-the art susceptibility and motion correction capabilities of TOPUP. For example, many datasets of the past were not collected with this modification, and adoption in research and clinical settings may be hindered due to limited, familiarity with best-practices, or programming expertise. Additionally, it is not possible to implement blip-up blip-down acquisitions in many clinically deployed software releases. It would be beneficial to be able to perform advanced distortion corrections (using modern processing pipelines) relying only on the most widely available diffusion imaging data and clinically common sequences that accompany them (e.g., anatomical structural MRI along with diffusion imaging data with a single PE direction).
Here, we recognize that there has been extensive progress in computed tomography (CT) synthesis from MRI to support attenuation correction and reconstruction for PET/MRI scanners without the need for an independent CT scan. Initially these efforts were based on multi-atlas methods [32, 33] and non-local patches [34, 35]. Recently adapted to the computer vision field, deep learning with pix2pix conditional generative networks [36] and cycle generative adversarial networks [37] have proven especially effective at a variety of medical imaging synthesis tasks [38–40]. We note that the validity of synthetic images for medical interpretation of contrast “hallucination” remains highly controversial [41]. Yet, use of synthetic images as image processing intermediates is becoming accepted in much the same way as different matching criteria (e.g., mutual information, patch based similarity, structural similarity, etc. have become commonplace for image registration [42–45].
To enable TOPUP processing with historical and/or limited diffusion imaging data with only a structural image and single blip diffusion image, we propose to synthesize an undistorted EPI image from the structural image and use the non-distorted synthetic image as an anatomical target. Our approach only requires the (distorted) diffusion images and an undistorted anatomical image. To train the synthesis approach, we use a database of 568 pairs of structural images and multi-shot EPI images with contrast mapped to the diffusion imaging minimally weighted “b0” image. Briefly, we create three orthogonal 3 channel pix2pix Generative Adversarial Networks (GANs) at 1 mm resolution in MNI space to synthesize three-slice stacks of diffusion imaging data in the axial, coronal, and sagittal planes. Median filtering is used to combine the resulting model into a single estimate for each target voxel. We then enter both the “real” and “synthesized” b0 images as input into TOPUP, but inform the algorithm that the synthesized image has an infinite bandwidth in the PE direction (i.e., it is undistorted), which allows warping of only the real b0, forcing its geometry to match the undistorted synthesized image.
Herein, we evaluate the efficacy of this approach on data from a different site (different subject population, different scanners, different scanner software versions). We show that our distortion correction process results in better matching the geometry of undistorted T1, reduces variation in diffusion modeling, and is practically equivalent to having both blip-up and blip-down data.
METHODS
Acquisition – synthesis dataset
All human datasets were acquired after informed consent under supervision of the appropriate Institutional Review Board. This study accessed only de-identified patient information. The image synthesis GAN was conducted on 586 pairs of T1-weighted and diffusion b0 EPI brain images from healthy controls (ages 29–94 years old) as part of the Baltimore Longitudinal Study of Aging study, which is a study of aging operated by the National Institute of Aging [46]. For detailed demographics and scanner protocols, we refer the reader to [47]. Briefly, data were acquired on a two 3 Tesla Philips Achieva scanners using an 8-channel head coil. A T1-weighted contrast and diffusion acquisition was acquired for each participant.
T1-weighted images were acquired using an MPRAGE sequence (TE=3.1ms, TR=6.8 ms, slice thickness=1.2mm, number of Slices=170, flip angle=8 deg, FOV=256×240mm, acquisition matrix=256×40, reconstruction matrix=256×256, reconstructed voxel size=1×1mm). Diffusion acquisition was acquired using a single-shot EPI sequence, and consisted of a single b-value (b=700 s/mm2), with 33 volumes (1 b0 + 32 DWIs) acquired axially (TE=75ms, TR=6801ms, slice thickness=2.2mm, number of slices=65, flip angle=90 degrees, FOV=212*212, acquisition matrix=96*95, reconstruction matrix=256*256, reconstructed voxel size=0.83*0.83mm). Importantly, to obtain a non-distorted image with b0 contrast, for 586 sessions, a high resolution, readout-segmented (multi-shot) EPI image was acquired [48] (same acquisition settings as diffusion data, with no diffusion gradients applied). This technique significantly reduces susceptibility artifacts in the PE direction and allows alignment directly with T1 through rigid body registration. This (ideally undistorted) b0 image is used for image synthesis to learn the mapping from T1 to b0 contrast.
b0 Synthesis
For 586 subjects, the multi-shot EPI b0 scans were rigid body registered (6 degree of freedom with FSL flirt [24]) to the paired T1-weighted MPRAGE scan. The MPRAGE scans for each subject were affine registered (12 degree of freedom using FSL flirt) to the MNI-152 1 mm isotropic T1-weighted atlas included with FSL [24]. The paired multi-shot EPI scans were transformed to MNI space using cubic interpolation. Pre-aligning to MNI space reduces the variation in the data and reduces the need to learn pose invariance.
To perform image synthesis, we train three separate GAN networks, each learning the mapping from T1-weighted contrast to a b0 contrast using stacks of 2D slices, with one network learning mapping of axial 2D slices, the second learning coronal, and the third sagittal. The three different orientations allow pseudo 3-D information to be used. Some image features may be clearer in one plane over another. Additionally, we can utilize standard 2-dimensional GAN networks (pix2pix) and (although the underlying data remains the same) this approach increases the paired data 3-fold (i.e., 3 orientations).
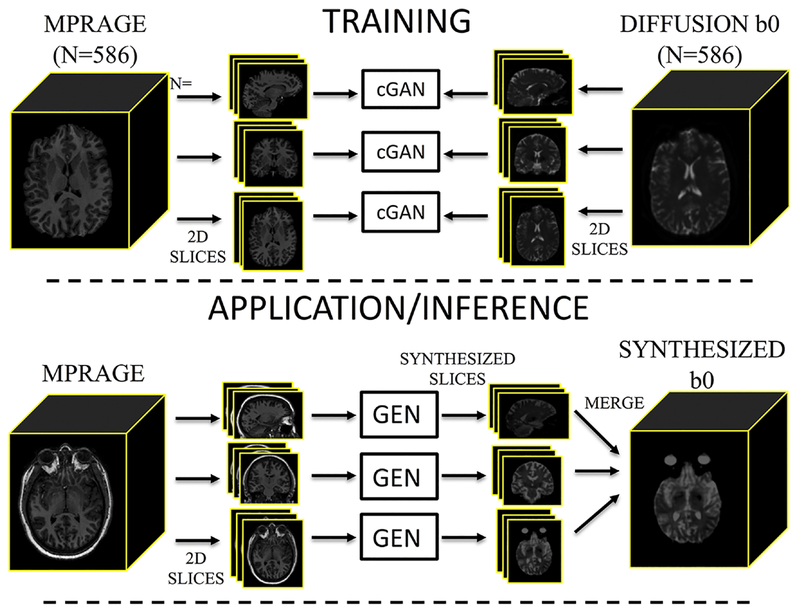
The training pipeline is shown in Figure 1 (top). We created three orthogonal pix2pix networks with an input resolution of 1 mm x 1mm (in plane) and 3 consecutive slices in the three channels of the network. All overlapping sets of three contiguous slices for all subjects were created for all three orthogonal planes in a paired manner for corresponding MPRAGE and multi-shot EPI datasets for a total of 113098, 134194, and 113098 training pairs (axial, coronal, sagittal, respectively) that were split 80/20 % for training and validation. The models were independently trained on an NVIDIA GTX 1080i GPU using pix2pix [36] implementation in pytorch (https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix). All networks were trained from scratch with a learning rate of 0.002. We keep the same learning rate for the first 100 epochs and linearly decay the rate to zero over the next 100 epochs.
Figure 1.
Image synthesis training (top) and application of the network (bottom). Paired MPRAGE and diffusion b0 images are sliced into groups of orthogonal slices to act as input to three independent conditional GANs (cGAN). Upon application of the network, an MPRAGE is used to synthesize an undistorted b0 contrast. The MPRAGE is sliced into orthogonal stacks, input into the trained networks (the generator – GEN) which synthesize orthogonal slices, and merged into the synthesized b0.
Application, or inference, of the network is shown in Figure 1 (bottom). To apply the trained networks, an MPRAGE dataset without a paired multi-shot EPI scan is affine registered (12 degree of freedom using FSL flirt [24]) to the MNI-152 1 mm isotropic T1-weighted atlas. All overlapping sets of three contiguous slices for the scan are written out (193, 229, and 193 stacks of 2D images per subject, axial, coronal, sagittal, respectively). Then, the corresponding generative models are applied to each stack independently. All stacks are then loaded to reconstruct 3 estimated volumes per subject by averaging the 3 output channels for each reconstruction (e.g., +1 mm, same slice, −1 mm) and repeated for each orientation. Finally, the three different orientations are combined by median filtering, resulting in a synthesized b0 image. It is important to emphasize that, for application of this network, only a single T1-weighted volume is required for image synthesis (along with the pre-trained network weights). We have made this image synthesis network freely available with python code at (https://github.com/MASILab/Synb0-DISCO) and in a dockerized version (https://hub.docker.com/r/vuiiscci/synb0-disco).
Acquisition – validation datasets
Two additional datasets were acquired for validating the fidelity of resulting distortion corrections. The first dataset was intended for comparisons with state of the art TOPUP corrections. This data was acquired on a healthy subject scanned at Vanderbilt University, consisting of a T1-weighted MPRAGE and diffusion-weighted EPI (b = 1500 s/mm2, 5 interspersed b0’s + 32 DWIs, 2.5mm isotropic resolution) with the PE direction anterior to posterior (PE-P). This protocol was then repeated with PE direction posterior to anterior (PE-A).
The second set of data was intended for validating the proposed distortion correction when no reverse PE scans are available. This dataset contained 25 healthy subjects scanned at Vanderbilt University, consisting of a T1-weighted MPRAGE and diffusion-weighted EPI (b = 1000 s/mm2, 1 b0 + 32 DWIs, 2.5mm isotropic resolution). Importantly, these datasets do not have reverse PE scans for full blip-up/blip-down corrections.
Distortion correction
Distortion correction was performed by first applying the trained network to a given MPRAGE dataset, resulting in the synthesized b0 image. The synthesized b0 was rigidly registered to the real b0 image (note that these will not, and do not have to, align perfectly as both motion and susceptibility distortions are estimated with TOPUP), and concatenated, which formed the input to TOPUP. TOPUP was then run on the merged b0 volume. Importantly, when setting up the acquisition parameters, the readout time (i.e., time between the center of the first echo and center of last echo) for the synthesized image is set to 0 (while the real b0 retains the correct readout time). This tells the algorithm that the synthesized b0 has an infinite bandwidth in the PE direction, with no distortions, thus fixing its geometry when estimating the susceptibility field. After this, conventional processing is performed, including eddy current correction (FSL eddy) and non-linear least squares diffusion tensor fitting and fractional anisotropy (FA) calculation.
Quality Assessment
Distortion correction quality is assessed both qualitatively and quantitatively. Qualitatively, we overlaid the corrected and/or uncorrected EPI images and calculated FA maps on the T1 image, which allows observations and comparisons of boundaries, edges, and contrasts before and after correction. Quantitative measures of distortion correction includes calculating mutual information (MI) between pairs of image [49]. For example, calculating the MI between PE-A and PE-P images, PE-A and PE-P image corrected using TOPUP, and those corrected using the proposed methods. Additionally, the mean-square-difference (MSD) of FA values between pairs of images is calculated, where a lower FA MSD would indicate more agreement (or alignment) between calculated DTI metrics.
RESULTS
Qualitative Results
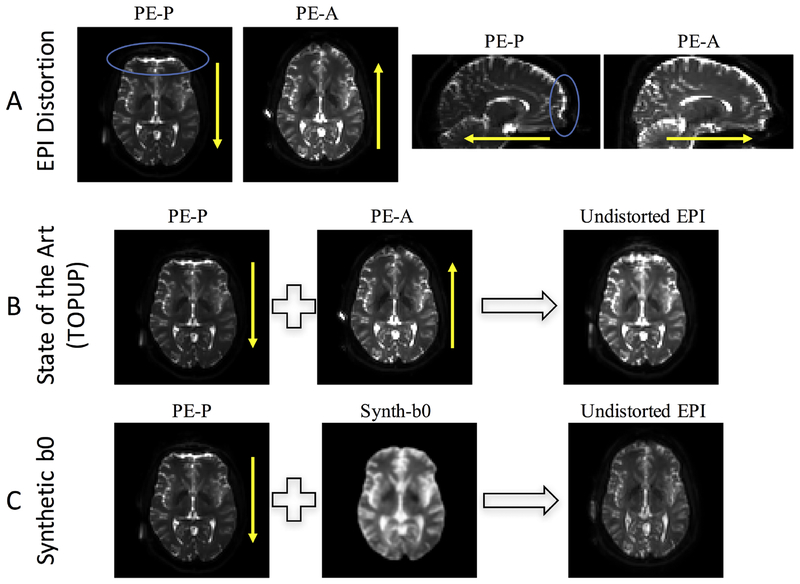
Figure 2 shows typical susceptibility artifacts associated with EPI scans (Figure 2, A), distortion correction results from state-of-the art blip-up/blip-down methods (i.e., TOPUP) (Figure 2, B), as well as the proposed distortion correction (Figure 2, C). Distortions in EPI images typically manifest as stretching or compression of geometry in the PE direction, accompanied by signal “pile-up” resulting in low and high intensity regions. In this case, b0 images with a PE-P acquisition demonstrate compression and signal pileup in the anterior portions of the brain, with opposite effects apparent in the PE-P acquisition (Figure 2, A). TOPUP correction results in an undistorted EPI that has a geometry largely intermediate between the two original images (Figure 2, B). Finally, the proposed correction estimates an anatomically faithful synthetic b0 (Figure 2, C) with contrast similar to that from the EPI scans, and the full pipeline yields a corrected EPI with similar geometry to the synthesized image, demonstrating the feasibility of the proposed method.
Figure 2.
Distortion and correction in diffusion MRI. (A) EPI susceptibility distortion occurs along the phase encode direction, with phase encoding in the posterior (PE-P) direction leading to displacements of identical distance but opposite direction of that in the anterior (PE-A) direction. (B) State of the art distortion correction (topup) typically uses distortions in two opposite directions to iteratively estimate the undistorted image. (C) The proposed method uses an undistorted T1-weighted image to synthesize an undistorted volume with b0 contrast, which can be used to correct the distorted (in this case, PE-P) image without requiring an additional phase encoding acquisition. The blue circles highlight areas of observable signal distortion. Yellow arrows indicate phase encode direction.
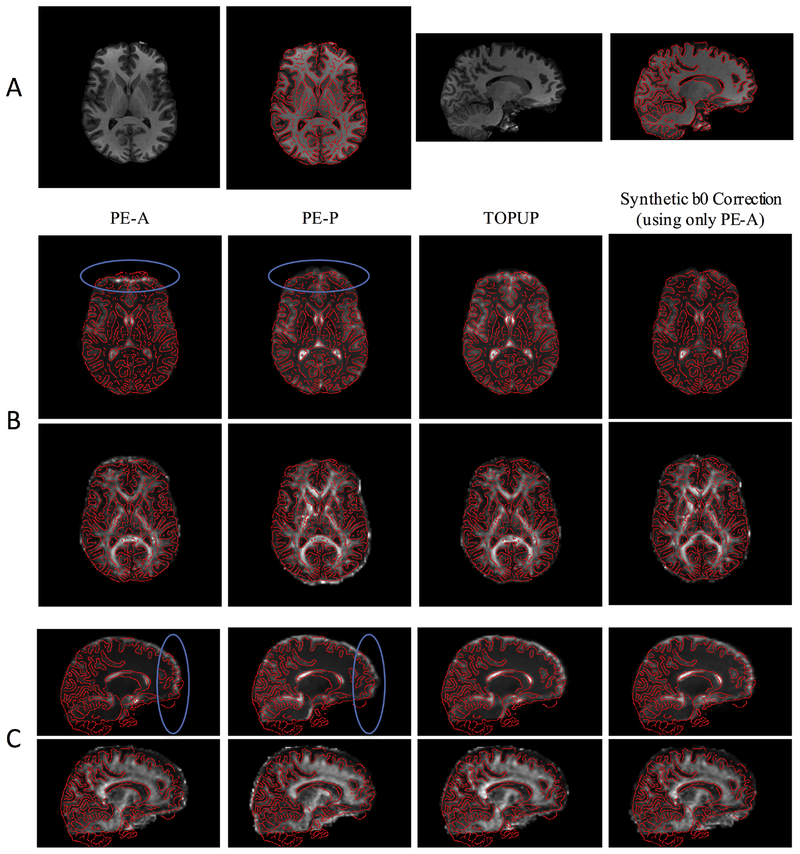
The distorted and undistorted images, and calculated FA maps, are shown overlaid on the anatomically-correct T1-weighted image (Figure 3, A), in axial (Figure 3, B) and sagittal (Figure 3, C) planes. Again, signal pile-up or signal stretching is apparent in uncorrected PE-A and PE-P images, respectively, along with clear anatomical mismatch in the FA maps. For example, misalignment of FA with T1 is noticeable in several white matter regions, as well as in high-intensity outlines at the front or back of the brain. Both TOPUP and Synb0-DiCso results in reasonable distortion corrections.
Figure 3.
Synthetic b0 correction results in reasonable distortion corrections, qualitatively similar to state of the art (topup). (A) Axial and sagittal slices of the T1-weighted image are shown with edges highlighted. (B) Axial slices of the volumes with PE in the anterior (PE-A) and posterior (PE-P) directions, the topup corrected image, and that corrected using the synthetic b0 image are shown for both b0 image and FA maps, shown in T1 space. (C) Sagittal slices of the volumes with PE in the anterior (PE-A) and posterior (PE-P) directions, the topup corrected image, and that corrected using the synthetic b0 image are shown for both b0 image and FA maps, shown in T1 space. The blue circles highlight areas of observable signal distortion, which are qualitatively corrected in the topup-corrected and synthetic b0-corrected images.
Quantitative Comparison to State-of-the art
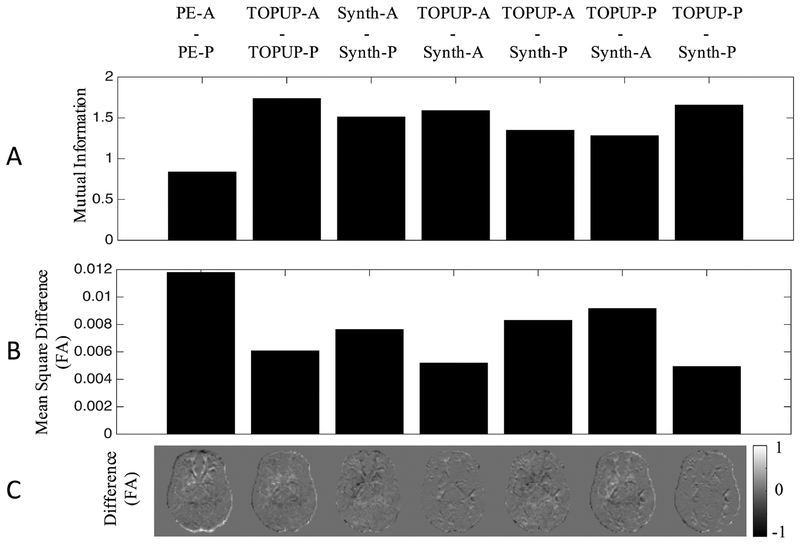
We next quantified the MI in pairs of images (Figure 4, A) to quantitatively assess correction quality. As expected, PE-A and PE-P has a relatively lower MI compared to the topup-corrected PE-A scan (TOPUP-A) and topup-corrected PE-A scan (TOPUP-P), which has the highest MI, indicating the correction results in images with more similar contrast. Similarly, the Synb0-DiCso-correct PE-A (Synth-A) and Synb0-DiCso-corrected PE-P scan (Synth-B) show an increased MI, although smaller than TOPUP corrections (resulting in >80% of the topup improvement). Additionally, the PE-A after correction with TOPUP shows high MI with PE-A after correction with Synb0-DiCso, with similar results for the PE-P volume.
Figure 4.
Synthetic b0 correction performs nearly as well as state of the art (topup) at correcting the shape of the b0 and reducing variation in FA caused by distortions. The mutual information (A), mean square difference in FA (B) and difference in FA (C) are shown for pairs of image volumes. PE-A: phase encoding in anterior direction; PE-P: phase encoding in posterior direction; TOPUP-A: topup-corrected PE-A; TOPUP-P: topup-corrected PE-P; Synth-A: synthetic b0-corrected PE-A; Synth-P: synthetic b0-corrected PE-P.
Quantifying (Figure 4, B), and visualizing (Figure 4, C) FA differences between the same pairs of images further reaffirms that (1) uncorrected scans show the greatest variation in FA, (2) TOPUP results in the highest similarity after correction, and (3) Synb0-DiCso shows nearly as much improvement as TOPUP correction. The greatest FA differences appear in the most anterior and most posterior portions of the brain (as expected), which are largely corrected using both methods. Again, the PE-A (and PE-P) images corrected with both methods indicate high similarities, suggesting that they result in similar distortion correction. However, neither method is perfect (residual differences still exist in both TOPUP and Synb0-DisCo corrections), although the corrections for both methods result in very similar output corrected images and quantification of metrics (i.e. TOPUP-A is similar to Synth-A, TOPUP-P is similar to Synth-P). Thus, Synb0-DiCso performs nearly as well as TOPUP at correcting the geometry and reducing variation in DTI-derived measures.
Distortion correction with no reverse PE
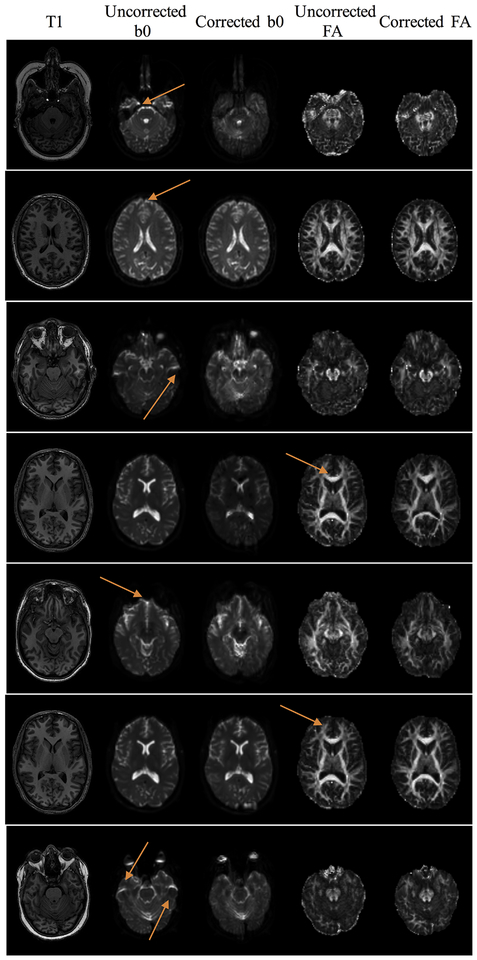
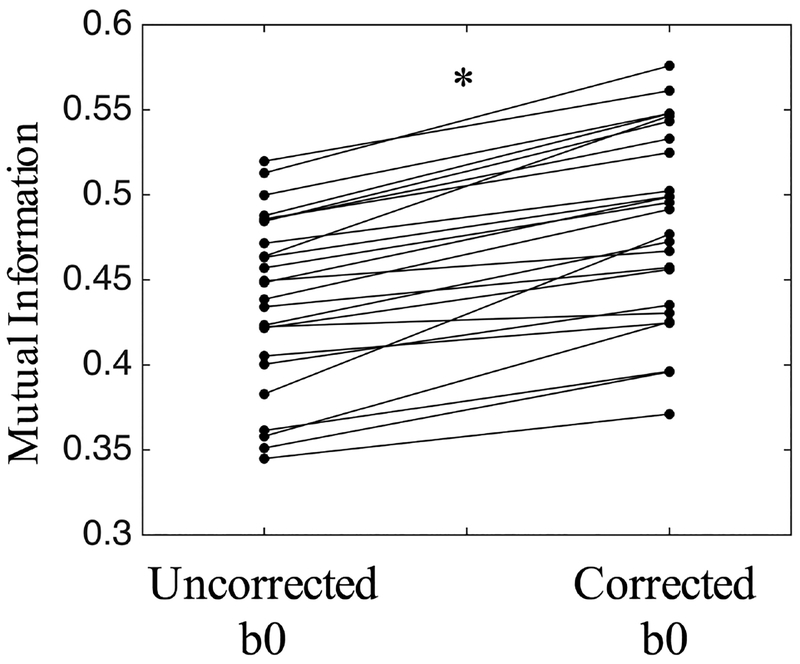
We further evaluate Synb0-DiCso when no reverse PE direction is available using dataset #2 (Figure 5). Distortions, as expected, are apparent in the frontal lobe due to the presence of the sinus. Qualitatively, the b0 better matches the T1 geometry after correction, both near the sinus (as seen on corrected b0) and in several white matter regions (more discernable on corrected FA maps). Thus, Synb0-DiCso qualitatively appears to be an adequate correction strategy in the absence of additional PE acquisitions. Finally, quantifying the similarity of the uncorrected and corrected b0 volumes with the T1 image (Figure 6) indicates a statistically significant increase in MI with the T1 after Synb0-DiCso correction (paired t-test, p<.001), with all corrected volumes increasing MI, indicating a more anatomically faithful geometry.
Figure 5.
Synthetic b0 distortion correction is able to consistently correct distortions in scans without additional phase encodings. Example slices of the T1 image, uncorrected and corrected b0, and uncorrected and corrected FA are shown for 7 different subjects, with arrows highlighting locations that are un-warped after correction, noticeable as reduced distortion, mitigated signal pile-up, or more clearly delineated FA maps.
Figure 6.
Mutual information with T1 increases after synthetic b0 correction, indicating more anatomically faithful shape. Asterisk indicates significant increase (p<0.001) in mutual information using paired t-test.
DISCUSSION
In this study, we have proposed a distortion correction technique, Synb0-DiCso, that can be utilized in the absence of additional reverse PE acquisitions, and only requires a standard T1-weighted acquisition (Figure 2). We have shown qualitatively that Synb0-DiCso results in reasonable distortion corrections (Figure 3), largely similar to state-of-the art methods that require blip-up blip-down acquisitions. In agreement with this, quantitative comparisons suggest that Synb0-DiCso performs nearly as well as TOPUP at both correcting image geometry and at reducing variation in DTI-derived parameters (Figure 4). This methodology was applied to 25 diffusion-weighted EPI scans without blip-up blip-down acquisitions, consistently correcting observable distortions (Figure 5), and resulting in brain shape more similar to the anatomical images than pre-correction (Figure 6).
There are a number of distortion correction methods that utilize additional data in the form of field maps or additional data and subsequent signal and image processing. We have chosen TOPUP as the standard for comparison, as it is well integrated into excellent image processing packages, has arguably seen the most widespread use, and has become the tool of choice for one of the larger neuroimaging initiatives. Another popular package, DR-BUDDI [31], is worth mentioning, as it is integrated into the TORTOISE software package, and utilizes a deformation model capable of dealing with large distortions and allows compensation for B0 field changes between acquisitions. Importantly, DR-BUDDI can also include structural images as a priori undistorted information that further constrains the deformation fields [31]. It would be of interest to the field of diffusion MRI to perform a comprehensive evaluation of the large number of existing distortion correction algorithms, in addition to eddy-currents, movement, and other artifacts associated with EPI acquisitions. Towards this end, a standardized dataset and evaluation method, utilizing similar measures of intensity similarities and metric variabilities, would provide valuable benchmarking for algorithms and acquisitions.
There are several potential limitations to using image synthesis to create the standard undistorted space. First, the contrast is not perfect, and median filtering of sagittal, axial, and coronal slices creates a slight blurring of fine details and structures (see synthesized image in Figure 1, C). Second, the learned network structure may not be appropriate with dramatic differences in acquisition settings, including varying resolution (for example, HCP datasets) or echo times. However, the data from training and testing acquisitions were not exactly matched (and in fact came from different scanners), and distortion correction was successful. We urge caution when applying to datasets that may be dramatically different either in T1 or b0 contrast. Future training should incorporate a range of contrasts, or alternatively, a different network could include the b0 itself as a second input when learning and applying this technique in order to learn a histogram match and force perfect contrast-matching to the b0. Finally, we do not expect the network to be able to predict the appropriate b0 contrast in certain regions of non-healthy populations (for example, tumors), and this should be an area of future research. We note that the contrast between the real and synthetic b0 is not perfect (see Figure 2 for a comparison) and that the synthesized b0 is smoothed due to the merging operations of the three networks outputs. However, relevant structures are still apparent, with the similar contrast magnitude, and the TOPUP algorithms begins with an image normalization between volumes (which is traditionally to account for varying gain factors across scans).
Deep learning methods have been successfully applied to numerous medical imaging tasks due to the superior performance. The recent advances in GAN based deep learning methods enabled the high quality medical image synthesis in a model-free manner between two different medical imaging modalities such as, but not limit to, MRI to CT [50], CT to X-ray [51], ultrasound to CT [52], T1 MRI to T2 MRI [53] etc. In this study, we proposed a new application by synthesizing undistorted b0 diffusion MRI from T1 MRI using image-to-image GAN. However, other imaging modals and modalities could also be used as the source images to synthesize undistorted b0 diffusion MRI (e.g., T2 MRI), or we can even directly learn the undistorted b0 diffusion MRI from k-space images.
In this study, paired image-to-image synthesis was performed between two imaging modalities. In the future, more training data could be incorporated into the training by combining the unpaired image-to-image training using CycleGAN [37]. Another direction is to expand the training from using two modalities into three or more modalities like the recent proposed StarGAN [54]. Meanwhile, the recent proposed BigGAN [55] can be used to further improve the synthesis performance for the high-resolution scenarios.
The qualitative and quantitative evaluation on “how real are the synthesized images?” is still an open question. Therefore, beyond pursuing high realistic image-to-image synthesis performance, recent studies have utilized the synthesized images in advancing other medical imaging tasks (e.g., segmentation) in multi-task learning manner [50, 51, 56]. Thus, an interesting future direction is to use the synthesized undistorted scans for the following diffusion weighted image analyses (e.g., tractography). Moreover, the proposed method can be generalized to other imaging modalities (e.g, T2 MRI, fMRI, CT etc.) and applications (e.g., chest, abdomen, pelvis, etc.).
CONCLUSIONS
In this work, we have implemented a diffusion susceptibility distortion correction method that utilized an undistorted non-diffusion weighted image synthesized from an anatomical scan. This pipeline integrates well with standard diffusion pre-processing packages. Importantly, this distortion correction can be utilized without additional blip-up blip-down acquisitions. This method is implemented in open sourced Python code (https://github.com/MASILab/Synb0-DISCO) in addition to example TOPUP processing scripts, and a Docker image for system-agnostic processing (https://hub.docker.com/r/vuiiscci/synb0-disco).
ACKNOWLEDGEMENTS
This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. This work was supported by the National Institutes of Health under award numbers R01EB017230, and T32EB001628, and in part by ViSE/VICTR VR3029 and the National Center for Research Resources, Grant UL1 RR024975–01. This research was conducted with the support from Intramural Research Program, National Institute on Aging, NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161(2):401–7. doi: 10.1148/radiology.161.2.3763909. [DOI] [PubMed] [Google Scholar]
- 2.Taylor DG, Bushell MC. The spatial mapping of translational diffusion coefficients by the NMR imaging technique. Phys Med Biol. 1985;30(4):345–9. [DOI] [PubMed] [Google Scholar]
- 3.Novikov DS, Fieremans E, Jespersen SN, Kiselev VG. Quantifying brain microstructure with diffusion MRI: Theory and parameter estimation. eprint arXiv. 2016. doi: 2016arXiv161202059N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori S, van Zijl PCM. Fiber tracking: principles and strategies – a technical review. NMR in biomedicine. 2002;15(7–8):468–80. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- 5.Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annals of neurology. 1999;45(2):265–9. [DOI] [PubMed] [Google Scholar]
- 6.Xue R, van Zijl PC, Crain BJ, Solaiyappan M, Mori S. In vivo three-dimensional reconstruction of rat brain axonal projections by diffusion tensor imaging. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1999;42(6):1123–7. [DOI] [PubMed] [Google Scholar]
- 7.Ordidge R The development of echo-planar imaging (EPI): 1977–1982. MAGMA. 1999;9(3):117–21. [PubMed] [Google Scholar]
- 8.Jezzard P, Balaban RS. Correction for geometric distortion in echo planar images from B0 field variations. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1995;34(1):65–73. [DOI] [PubMed] [Google Scholar]
- 9.Andersson JLR, Skare S. Image Distortion and Its Correction in Diffusion MRI In: Jones DK, editor. Diffusion MRI: Theory, Methods, and Applications: Oxford University Press; 2010. [Google Scholar]
- 10.Irfanoglu MO, Walker L, Sarlls J, Marenco S, Pierpaoli C. Effects of image distortions originating from susceptibility variations and concomitant fields on diffusion MRI tractography results. NeuroImage. 2012;61(1):275–88. doi: 10.1016/j.neuroimage.2012.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu M, Chang LC, Walker L, Lemaitre H, Barnett AS, Marenco S, et al. Comparison of EPI distortion correction methods in diffusion tensor MRI using a novel framework. Med Image Comput Comput Assist Interv. 2008;11(Pt 2):321–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen NK, Wyrwicz AM. Correction for EPI distortions using multi-echo gradient-echo imaging. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1999;41(6):1206–13. [DOI] [PubMed] [Google Scholar]
- 13.Techavipoo U, Okai AF, Lackey J, Shi J, Dresner MA, Leist TP, et al. Toward a practical protocol for human optic nerve DTI with EPI geometric distortion correction. Journal of magnetic resonance imaging : JMRI. 2009;30(4):699–707. doi: 10.1002/jmri.21836. [DOI] [PubMed] [Google Scholar]
- 14.Zeng H, Constable RT. Image distortion correction in EPI: comparison of field mapping with point spread function mapping. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2002;48(1):137–46. doi: 10.1002/mrm.10200. [DOI] [PubMed] [Google Scholar]
- 15.Zaitsev M, Hennig J, Speck O. Point spread function mapping with parallel imaging techniques and high acceleration factors: fast, robust, and flexible method for echo-planar imaging distortion correction. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2004;52(5):1156–66. doi: 10.1002/mrm.20261. [DOI] [PubMed] [Google Scholar]
- 16.Roy S, Chou YY, Jog A, Butman JA, Pham DL. Patch Based Synthesis of Whole Head MR Images: Application to EPI Distortion Correction. Simul Synth Med Imaging (2016). 2016;9968:146–56. doi: 10.1007/978-3-319-46630-9_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kybic J, Thevenaz P, Nirkko A, Unser M. Unwarping of unidirectionally distorted EPI images. IEEE transactions on medical imaging. 2000;19(2):80–93. doi: 10.1109/42.836368. [DOI] [PubMed] [Google Scholar]
- 18.Huang H, Ceritoglu C, Li X, Qiu A, Miller MI, van Zijl PC, et al. Correction of B0 susceptibility induced distortion in diffusion-weighted images using large-deformation diffeomorphic metric mapping. Magnetic resonance imaging. 2008;26(9):1294–302. doi: 10.1016/j.mri.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao R, Fletcher PT, Gerber S, Whitaker RT. A variational image-based approach to the correction of susceptibility artifacts in the alignment of diffusion weighted and structural MRI. Inf Process Med Imaging. 2009;21:664–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irfanoglu MO, Walker L, Sammet S, Pierpaoli C, Machiraju R. Susceptibility distortion correction for echo planar images with non-uniform B-spline grid sampling: a diffusion tensor image study. Med Image Comput Comput Assist Interv. 2011;14(Pt 2):174–81. [DOI] [PubMed] [Google Scholar]
- 21.Bowtell RW, McIntyre DJO, Commandre MJ, Glover PM, Mansfield P, editors. Correction of geometric distortion in echo planar images. Proceedings of 2nd Annual Meeting of the SMR; 1994; San Francisco. [Google Scholar]
- 22.Morgan PS, Bowtell RW, McIntyre DJO, Worthington BS. Correction of spatial distortion in EPI due to inhomogeneous static magnetic fields using the reversed gradient method. Journal of Magnetic Resonance Imaging. 2004;19(4):499–507. doi: doi: 10.1002/jmri.20032. [DOI] [PubMed] [Google Scholar]
- 23.Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage. 2003;20(2):870–88. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 24.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage. 2012;62(2):782–90. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23 Suppl 1:S208–19. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 26.Sotiropoulos SN, Jbabdi S, Xu J, Andersson JL, Moeller S, Auerbach EJ, et al. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. NeuroImage. 2013;80:125–43. doi: 10.1016/j.neuroimage.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holland D, Kuperman JM, Dale AM. Efficient correction of inhomogeneous static magnetic field-induced distortion in Echo Planar Imaging. NeuroImage. 2010;50(1):175–83. doi: 10.1016/j.neuroimage.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olesch J, Ruthotto L, Kugel H, Skare, Fischer B, Wolters CH, editors. A variational approach for the correction of field-inhomogeneities in EPI sequences. SPIE Medical Imaging; 2010: SPIE. [Google Scholar]
- 29.Gallichan D, Andersson JL, Jenkinson M, Robson MD, Miller KL. Reducing distortions in diffusion-weighted echo planar imaging with a dual-echo blip-reversed sequence. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2010;64(2):382–90. doi: 10.1002/mrm.22318. [DOI] [PubMed] [Google Scholar]
- 30.Chang HC, Chuang TC, Lin YR, Wang FN, Huang TY, Chung HW. Correction of geometric distortion in Propeller echo planar imaging using a modified reversed gradient approach. Quant Imaging Med Surg. 2013;3(2):73–81. doi: 10.3978/j.issn.2223-4292.2013.03.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irfanoglu MO, Modi P, Nayak A, Hutchinson EB, Sarlls J, Pierpaoli C. DR-BUDDI (Diffeomorphic Registration for Blip-Up blip-Down Diffusion Imaging) method for correcting echo planar imaging distortions. NeuroImage. 2015;106:284–99. doi: 10.1016/j.neuroimage.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgos N, Cardoso MJ, Modat M, Punwani S, Atkinson D, Arridge SR, et al. CT synthesis in the head & neck region for PET/MR attenuation correction: an iterative multi-atlas approach. EJNMMI Phys. 2015;2(Suppl 1):A31. doi: 10.1186/2197-7364-2-S1-A31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgos N, Cardoso MJ, Thielemans K, Modat M, Pedemonte S, Dickson J, et al. Attenuation correction synthesis for hybrid PET-MR scanners: application to brain studies. IEEE transactions on medical imaging. 2014;33(12):2332–41. doi: 10.1109/TMI.2014.2340135. [DOI] [PubMed] [Google Scholar]
- 34.Lee J, Carass A, Jog A, Zhao C, Prince JL. Multi-atlas-based CT synthesis from conventional MRI with patch-based refinement for MRI-based radiotherapy planning. Proc SPIE Int Soc Opt Eng. 2017;10133. doi: 10.1117/12.2254571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jog A, Roy S, Carass A, Prince JL. Magnetic Resonance Image Synthesis through Patch Regression. Proc IEEE Int Symp Biomed Imaging. 2013;2013:350–3. doi: 10.1109/ISBI.2013.6556484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isola P, Zhu J-Y, Zhou T, Efros AA. Image-to-Image Translation with Conditional Adversarial Networks. ArXiv e-prints [Internet]. 2016. November 01, 2016 Available from: https://ui.adsabs.harvard.edu/-abs/2016arXiv161107004I. [Google Scholar]
- 37.Zhu J-Y, Park T, Isola P, Efros AA. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. ArXiv e-prints [Internet]. 2017. March 01, 2017 Available from: https://ui.adsabs.harvard.edu/-abs/2017arXiv170310593Z. [Google Scholar]
- 38.Wolterink JM, Dinkla AM, Savenije MHF, Seevinck PR, van den Berg CAT, Isgum I. Deep MR to CT Synthesis using Unpaired Data. ArXiv e-prints [Internet]. 2017. August 01, 2017 Available from: https://ui.adsabs.harvard.edu/-abs/2017arXiv170801155W. [Google Scholar]
- 39.Welander P, Karlsson S, Eklund A. Generative Adversarial Networks for Image-to-Image Translation on Multi-Contrast MR Images - A Comparison of CycleGAN and UNIT. ArXiv e-prints [Internet]. 2018. June 01, 2018 Available from: https://ui.adsabs.harvard.edu/-abs/2018arXiv180607777W. [Google Scholar]
- 40.Huo Y, Xu Z, Bao S, Assad A, Abramson RG, Landman BA. Adversarial Synthesis Learning Enables Segmentation Without Target Modality Ground Truth. ArXiv e-prints [Internet]. 2017. December 01, 2017 Available from: https://ui.adsabs.harvard.edu/-abs/2017arXiv171207695H. [Google Scholar]
- 41.Cohen JP, Luck M, Honari S. Distribution Matching Losses Can Hallucinate Features in Medical Image Translation. ArXiv e-prints [Internet]. 2018. May 01, 2018 Available from: https://ui.adsabs.harvard.edu/-abs/2018arXiv180508841C. [Google Scholar]
- 42.Roy S, Carass A, Jog A, Prince JL, Lee J. MR to CT Registration of Brains using Image Synthesis. Proc SPIE Int Soc Opt Eng. 2014;9034. doi: 10.1117/12.2043954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen M, Carass A, Jog A, Lee J, Roy S, Prince JL. Cross contrast multi-channel image registration using image synthesis for MR brain images. Med Image Anal. 2017;36:2–14. doi: 10.1016/j.media.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao X, Yang J, Gao Y, Guo Y, Wu G, Shen D. Dual-core steered non-rigid registration for multi-modal images via bi-directional image synthesis. Med Image Anal. 2017;41:18–31. doi: 10.1016/j.media.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao X, Yang J, Gao Y, Wang Q, Shen D. Region-adaptive Deformable Registration of CT/MRI Pelvic Images via Learning-based Image Synthesis. IEEE Trans Image Process. 2018. doi: 10.1109/TIP.2018.2820424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrucci L The Baltimore Longitudinal Study of Aging (BLSA): a 50-year-long journey and plans for the future. J Gerontol A Biol Sci Med Sci. 2008;63(12):1416–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkatraman VK, Gonzalez CE, Landman B, Goh J, Reiter DA, An Y, et al. Region of interest correction factors improve reliability of diffusion imaging measures within and across scanners and field strengths. NeuroImage. 2015;119:406–16. doi: 10.1016/j.neuroimage.2015.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porter DA, Heidemann RM. High resolution diffusion-weighted imaging using readout-segmented echo-planar imaging, parallel imaging and a two-dimensional navigator-based reacquisition. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2009;62(2):468–75. doi: 10.1002/mrm.22024. [DOI] [PubMed] [Google Scholar]
- 49.Pluim JP, Maintz JB, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE transactions on medical imaging. 2003;22(8):986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 50.Huo Y, Xu Z, Moon H, Bao S, Assad A, Moyo TK, et al. SynSeg-Net: Synthetic Segmentation Without Target Modality Ground Truth. IEEE transactions on medical imaging. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Miao S, Mansi T, Liao R. Task Driven Generative Modeling for Unsupervised Domain Adaptation: Application to X-ray Image Segmentation. arXiv preprint arXiv:180607201. 2018. [Google Scholar]
- 52.Liao H, Tang Y, Funka-Lea G, Luo J, Zhou SK, editors. More Knowledge Is Better: Cross-Modality Volume Completion and 3D+ 2D Segmentation for Intracardiac Echocardiography Contouring International Conference on Medical Image Computing and Computer-Assisted Intervention; 2018: Springer. [Google Scholar]
- 53.Dar SUH, Yurt M, Karacan L, Erdem A, Erdem E, Çukur T. Image Synthesis in Multi-Contrast MRI with Conditional Generative Adversarial Networks. arXiv preprint arXiv:180201221. 2018. [DOI] [PubMed] [Google Scholar]
- 54.Choi Y, Choi M, Kim M, Ha J-W, Kim S, Choo J. Stargan: Unified generative adversarial networks for multi-domain image-to-image translation. arXiv preprint. 2017;1711. [Google Scholar]
- 55.Brock A, Donahue J, Simonyan K. Large scale gan training for high fidelity natural image synthesis. arXiv preprint arXiv:180911096. 2018. [Google Scholar]
- 56.Zhang Z, Yang L, Zheng Y, editors. Translating and segmenting multimodal medical volumes with cycle-and shapeconsistency generative adversarial network.