Abstract
Identification of T cell targets to improve immunotherapies is of prime interest. To facilitate large-scale CRISPR screens directly in T cells in vivo, here, we developed a hybrid genetic screening system with adeno associated virus (AAV) and the Sleeping Beauty (SB) transposon, where the transposon is nested in the viral vector. The approach enables efficient gene editing in primary murine T cells and genomic integration of the sgRNA cassette for screen readout. We performed focused in vivo AAV-SB-CRISPR screens in CD8+ T cells in mouse models of glioblastoma (GBM) and identified membrane protein targets. Adoptive transfer of CD8+ T cells with Pdia3, Mgat5, Emp1, or Lag3 gene editing enhance the survival of GBM-bearing mice in both syngeneic and TCR-transgenic models. Transcriptome profiling, single cell sequencing, cytokine assays, and T cell signaling analysis showed that Pdia3 editing in T cells enhances effector functions. Engineered PDIA3 mutant EGFRvIII chimeric antigen T cells are more potent in antigen-specific killing of human GBM cells.
Introduction
Recently, immunotherapy has offered transformative clinical benefits for diverse cancer types1. Checkpoint blockade enhances the antitumor response by neutralizing cytotoxic T cell lymphocyte antigen 4 (CTLA-4), programmed cell death protein 1 (PD-1), or its ligand PD-L12–5. However, for patients with glioblastoma (GBM), the most common and deadliest primary malignant brain tumor in adults, checkpoint blockade efficacy is minimal. A recent clinical trial showed that PD-1 blockade does not prolong survival of GBM patients (). Combination of anti-PD-1 and anti-CTLA-4 in GBM patients failed to provide clinical benefits and engendered serious adverse effects6. EGFR-vIII chimeric antigen receptor (CAR)-T therapy evaluated in GBM clinical trials showed little overall survival benefit7. These clinical failures underscores the need to identify targets that enhance anti-tumor activity of CD8+ T cells in GBM. Although genetic screens on human primary T cells are feasible, given the complexity of the GBM tumor microenvironment, it is important to carry out screens in an in vivo setting in hosts with intact immune systems. However, mouse primary T cells are difficult to edit, hindering in vivo genetic screens directly on T cells in immunologically relevant animal models.
To overcome these challenges, we explored viral vectors and found that adeno associated virus (AAV) is a potent carrier of CRISPR-Cas9 gene editing components in primary murine immune cells (Fig. S1a). However, unlike lentivirus commonly used for CRISPR screens, AAV is a non-integrating virus, making readout of single guide RNA (sgRNA) library impractical, except a direct readout on target sites with targeted capture sequencing8. The limitation of capture sequencing is that the number of capturable targets limits the number of genes in a “screenable” library, usually to the order of dozens to a few hundred8, 9. We thus set out to develop more efficient tools for large-scale in vivo target discovery in otherwise difficult-to-edit murine primary T cells. Here, we harness AAV-CRISPR and genomic integration of the transposon system to develop a hybrid genetic screening platform where CRISPR libraries are embedded in the Sleeping Beauty transposon carried between the inverted terminal repeats (ITRs/IRs) of AAV. This enables efficient gene editing in primary murine T cells and genomic integration of the sgRNA cassette for screen readout. We focused our screen on membrane targets for enhancement of CD8+ T cell activity, because membrane-bound proteins are amenable to monoclonal antibody (mAb) based therapies, and their encoding genes can also be targeted by direct T cell gene editing.
Results
Generation of AAV-SB-CRISPR hybrid vector and surface proteome knockout library
We generated a hybrid AAV vector for CRISPR perturbation of primary T cells that additionally utilizes a hyperactive Sleeping Beauty (SB) transposon system, SB100x10. By AAV transduction, the transgene can integrate into the genome thereby allowing direct sgRNA library readout, while simultaneously expresses sgRNA to allow gene editing, enhancing high-throughput screenability (Fig. 1a). We named this vector as AAV-SB100x, and the hybrid system as AAV-SB-CRISPR, thereafter. We generated AAV, transduced mouse primary naïve CD8+ T cells, and tested the genomic integration of AAV-SB100x using splinkerette PCR (Methods) (Fig. S1b). Electrophoresis of the splinkerette PCR amplification products from AAV-SB100x infected T cells, but not from uninfected T cells, showed multiple bands of varying intensity, indicating random genomic integration (Methods) (Fig. S1c). We sequenced the splinkerette PCR products and revealed that they indeed mapped to the mouse genome with junctions to the SB transposon inverted repeats (IRs) (Fig. S1d). The genomic reads span across 18 out of 19 autosomes and both sex chromosomes (X and Y) in the mouse genome (Fig. S1e) (Dataset S1). Most of the integration sites mapped to intergenic regions and intronic regions, as compared to promoters, coding regions, or exonic untranslated regions (UTRs) (Fig. S1f) (Dataset S1), suggesting that these random integrations rarely disrupt essential coding or key functional elements.
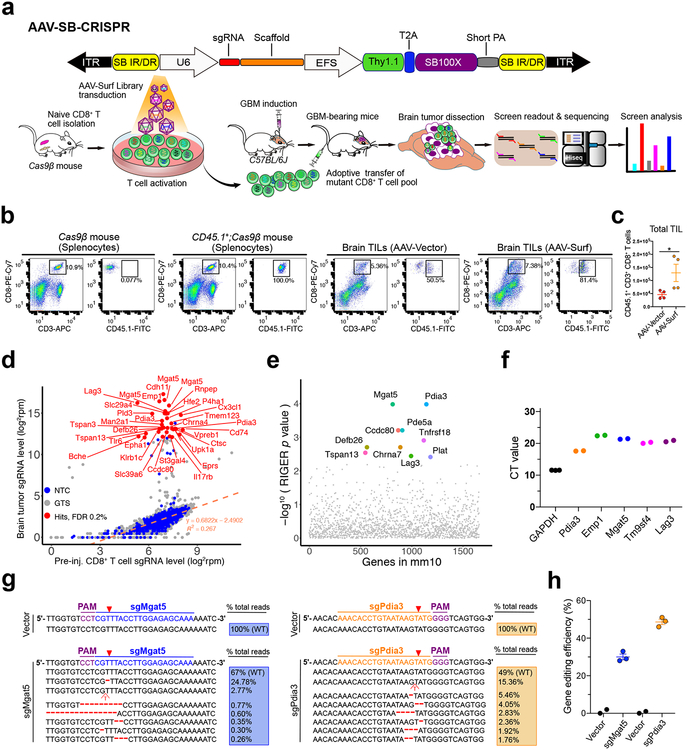
Figure 1. In vivo AAV-CRISPR CD8+ T cell screen of membrane bound proteome knockouts in GBM.
(a) (Top) Schematics of the hybrid AAV-SB-CRIPSR vector;
(Bottom) Schematics of in vivo AAV-SB-CRIPSR screen in a syngeneic mouse model of GBM. Schematics of naïve CD8+ T cell isolation, AAV library transduction, GBM cell transplantation, adoptive cell transfer (ACT), organs isolation, sgRNA readout and deep sequencing. 5 × 105 GL261 cancer cells were injected in to the brain, and 3 × 106 Cas9β CD8+ T cells were intravenously injected after 10 days of tumor engraftment. Brain tumors were dissected at the endpoint of survival.
(b) Flow cytometry analysis of TILs in the GBM bearing brain. 5e5 GL261-FLuc cancer cells were injected per mouse, at day 12 after tumor injection, luciferase imaging was performed to reasonably group mice based on luminescence intensity, then 4e6 CD45.1+;Cas9β CD8+ T cells were i.v. injected. Mice were euthanized at day 6 after T cell injection, brains (without olfactory and hindbrain) were dissected for TIL isolation. The i.v. injected CD45.1+;CD3+;CD8+ T cells were quantified and sorted for TCR-seq. Cas9β mouse and CD45.1+;Cas9β mouse splenocytes were used as gating controls. Data was collected from one experiment.
(c) Quantification of TIL number after transduction with AAV-Vector and AAV-Surf virus. Data was collected from two mice per group, two independent stainings were performed for each mouse. Data shown are mean ± s.e.m.. * p < 0.05, Mann Whitney test, two-tailed.
(d) Bulk analysis for brain tumor vs. cell sgRNA library representation of an AAV-Surf GBM CD8+ T cell screen experiment. A list of most significantly enriched sgRNAs in brain tumors are highlighted as red dots (FDR <= 0.2%). Custom methods by comparing sgRNAs to NTCs were used to estimate enriched sgRNAs (one-sided). FDR was calculated based on the ranks of sgRNAs relative to NTCs.
(e) RIGER analysis for brain tumor vs. cell gene level significance of AAV-Surf screen experiment, taken the metrics from multiple sgRNAs. The top 10 most enriched genes (by RIGER p-value, second-best sgRNA method) in brain tumors are highlighted.
(f) CD8+ T cell mRNA levels of several top hits from the AAV-Surf GBM screen. The mRNA levels of all candidates were measured with RT-qPCR using gene-specific probes, indicating that all genes tested are expressed in mouse primary CD8+ T cells. (n = 3 for Gapdh, n = 2 for other genes).
(g-h) Nextera indel analysis for Mgat5 and Pdia3 knock-out in mouse CD8+ T cells. (g) Representative mutations were shown around predicted sgRNA target sites. (h) Quantification of total indel frequency for each gene were shown, demonstrating that AAV-mediated primary mouse CD8+ T cell gene editing was efficient. (n = 2 for Vector group, n = 3 for sgMgat5 and sgPdia3 groups). Data are shown as mean ± s.e.m.., plus individual data points on the bar graph.
We designed a focused sgRNA library (mouse surface and membrane protein encoding gene library, Surf), targeting 1,658 genes genes associated with the term “cell surface” with 6,628 sgRNAs and 1,000 non-targeting controls (NTCs) (Methods) (Table S1). We cloned Surf into AAV-SB100x vector (AAV-Surf) and verified successful cloning by sgRNA readout using Illumina sequencing (Methods). We pool-packaged AAV-Surf plasmid library into a viral library at a titer of approximately 1.4e12 viral genome copy per milliliter (1.4e12 vg/mL). While AAV titer estimated by gc is often high, functional transduction can be multiple orders of magnitudes lower due to empty viral particles, defective particles, non-infectious particles, non-productive infections, and clearance by host cells11, 12. Therefore, we performed analysis of functional multiplicity of infection (MOI) via single cell sgRNA qPCR of T cells, which were transduced with AAV-Surf library for 5 days. Single cells with functional sgRNA expression is estimated at 48%, or a functional MOI of 0.65 (Fig. S2).
In vivo AAV-Surf T cell GBM screen identified robust hits
Considering the delicacy of the brain tumor microenvironment, we first performed primary T cell screening in GBM using fully immunocompetent syngeneic models. We set up syngeneic mouse models with native or firefly luciferase-expressing GL261 cell lines (GL261 or GL261-Fluc/GL261-Luc) orthotopically transplanted into the lateral ventricle (LV) of C57BL/6J mice via intracranial injection using a stereotaxic instrument (Fig. 1a). With intracranial injection of GL261 or its derivatives, the penetrance of brain tumor induction in untreated mice is at or near 100% (Fig. S3a–b). We isolated Cas9+ naïve CD8+ T cells from constitutive Cas9 mice (Rosa26-Cas9, or Cas9β, generated by crossing Rosa26-LSL-Cas9 to β-actin Cre driver13). We activated Cas9+CD8+ T cells with anti-CD3ε and anti-CD28, and transduced them with AAV-Surf library to mutagenize the membrane proteome. CD8+ T cells 5 days in culture showed no difference between AAV-Surf and AAV-Vector groups in PD-1, Lag3, or Tim-3 (Fig. S4a). Using CD45.1 transgenic mice, we were able to distinguish the donor T cells from those in the host (Fig. 1b). With CD8+ T cells isolated from Cas9β;CD45.1 mice and transduced with AAV, we measured the number of infiltrated donor-derived CD8+ T cells (Fig. 1c). It is interesting that mice from the AAV-Surf group have increased number of infiltrated T cells, potentially linked to enhanced trafficking and/or survival of certain mutant T cells, or more complicated cell-cell interactions with a complex mutant pool. These TILs also have no difference in surface PD-1 level (Fig. S4b–c). We performed TCR-seq on the pre-injection T cells and post-injection TILs, and observed a large number of different TCR clonotypes in pre-injection T cells as well as reduction of clonality in post-injection T cells, potentially due to limited number of TILs in the brain (Fig. S4d–f; Dataset S2).
We then performed adoptive transfer of the AAV-Surf pool mutant CD8+ T cells into GBM engrafted mice via tail vein injection (Fig. 1a). In parallel, we performed two independent screens using CD8+ T cells from T cell receptor (TCR) transgenic mice (OT-I)14 bred to Cas9β (OT-I;Cas9β) (Fig. S5a). We monitored injected mice for brain tumor development by observation of macrocephaly and by in vivo luciferase imaging where GL261-FLuc cells were used (Fig. S5b–c). Results showed that adoptive transfer of CD8+ T cells increased overall survival (Fig. S5d). Brain tumors were found in most mice at the endpoint (Fig. S5e), except 3 mice in the AAV-Surf group that were luciferase-negative after T cell treatment.
Using barcoded primers (Table S2) specific for AAV-SB100x, we performed sgRNA library readout (Tables S3–S5). The majority of the 1,000 NTC sgRNAs follow a linear regression line between brain and cell pellet (Fig. 1d) representing a null joint distribution without selection; whereas a fraction of sgRNAs are highly enriched in the brain, suggesting expansion of these specific mutant T cells (Fig. 1d). At a stringent false-discovery rate (FDR) of 0.2%, we identified 33 significantly enriched sgRNAs targeting various membrane proteins, which include Mgat5, Cdh11, Emp1, Lag3, Slc29a4, Rnpep, Hef2, P4ha1, Man2a1, and Pdia3 (Fig. 1d). We performed RIGER analysis for gene level significance (Methods), which showed Mgat5, Pdia3, Pde5a, Ccdc80, Tnfrsf18, Defb26, Chrna7, Tspan13, Plat, and Lag3 as the top 10 hits (Fig. 1e). All top 10 hits have 2 or more independent sgRNAs targeting different regions of the same genes ranking at the top 200 out of 7,628 sgRNAs (Table S6, n = 7, RIGER “second-best guide” algorithm), which makes it unlikely that such enrichment is simply mediated by off-target effects.
Lag3 is a well-known immune checkpoint regulator expressed on T cells15–18 and a prime target for immunotherapy5, currently with anti-LAG-3 mAb clinical trials for GBM (, ). In two independent screens (one shorter term and one longer term) using OT-I;Cas9β TCR transgenic system, we also identified highly similar lists of hits (Fig. S5f–g). Mouse-to-mouse variation exist, which is even more challenging in the brain setting where the number of TILs per brain is limited. Therefore, we considered reproducibility among multiple animal replicates, multiple sgRNAs, and multiple screens when choosing hits for further investigation. Notably, Pdia3, Mgat5, and Emp1 were among the top hits supported by multiple independent animals and/or multiple sgRNAs, across all three screens. We performed RT-qPCR and confirmed that these hits are abundantly expressed in mouse CD8+ T cells (Fig. 1f). We hypothesized that these genes may modulate T cells’ anti-tumor activity against GBM.
Pre-clinical efficacy testing of top hits by direct T cell editing and adoptive transfer
Single-gene targeting sgRNAs showed that AAV-SB-CRISPR generated high-efficiency gene editing of Pdia3 and Mgat5 in mouse primary CD8+ T cells (Fig. 1g–h). T7EI assay and RT-qPCR confirmed on-target gene editing and mRNA downregulation before T cell adoptive transfer (Fig. S6b, d). By surveyor assay, we found no editing in any of the top four predicted off-target sites by sgPdia3 (Fig. S6c). In a syngeneic orthotopic GBM model with GL261 intracranial implantation in C57BL/6J mice, survival analysis of GBM engrafted mice showed that the individual knockouts of each of the three genes (Lag3, Mgat5, and Pdia3) in the adoptively transferred CD8+ T cells prolonged overall survival when compared to AAV-Vector control (Fig. S6e–f). We then used an antigen-specific OT-I;Cas9β CD8+ T cells with transgenic TCR that recognizes GL261 brain tumors expressing a model antigen, chicken ovalbumin (cOVA). We established single cell derived clonal GL261-FLuc-mCh-cOVA cell lines (Fig. S6a). We transplanted GL261-FLuc-mCh-cOVA#1 cells into LV Rag1−/− mouse to induce GBM, then adoptively transferred single gene edited or control OT-I;Cas9β CD8+ T cells intravenously into GBM-bearing recipients (Fig. 2a). Survival analysis showed that AAV-CRISPR perturbation of Mgat5, Pdia3, or Emp1 each significantly improved overall survival of GL261-cOVA GBM bearing mice when compared to AAV-Vector control (Fig. 2b). Furthermore, flow cytometry analysis of infiltrating CD45.2+;CD8+ immune cells revealed higher abundance of Mgat5 and Pdia3 knockout CD8+ T cells following adoptive transfer (Fig. 2c). All animals developed brain tumors with the pathology of GBM (Fig. S6g–h). These data suggested that single gene AAV-CRISPR perturbations of Mgat5, Pdia3, Lag3, or Emp1 enhanced the efficacy of adoptive T cell transfer against GBM in mice with both immunocompetent and antigen-specific transgenic TCR models.
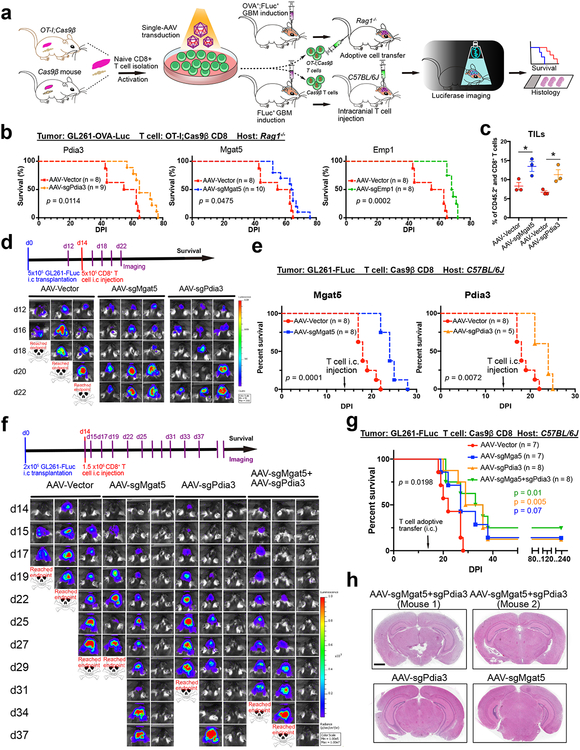
Figure 2. In vivo validation and efficacy testing of top candidates by adoptive transfer of mutant CD8+ T cells in mouse models of GBM.
(a) Schematic of the pre-clinical therapeutic efficacy testing strategy for top candidates from the AAV- Surf screens using an independent model of GBM immunotherapy, where cancer cells express a cognate cOVA model tumor antigen recognized by CD8+ T cells from TCR transgenic OT-I mice. And a syngeneic mouse model of GBM was used to evaluate therapeutic efficacy by intracranially (i.c) delivering T cells.
(b) Survival plots of adoptive transfer top candidate validations in Rag1−/− mice. Mgat5, Pdia3, and Emp1, were chosen for gene editing in CD8+ T cells for therapeutic efficacy testing. All mice were engrafted with 1 × 105 GL261-FLuc-mCh-cOVA cells, and adoptive transfer was performed after 10 days of tumor engraftment by intravenous injection of 1 × 106 OT-I;Cas9β CD8+ T cells infected with AAV-Vector (n = 8), AAV-sgMgat5 (n = 10), AAV-sgPdia3 (n = 9), and AAV- sgEmp1 (n = 8). Vector control from the same group and each gene was plotted against Vector separately for visibility. Survival significance was assessed by a log-rank Mantel-Cox test.
(c) Barplot of quantitative results for CD45.2+ and CD8+ CD8+ T cell infiltration in GBM bearing mice (TILs) with or without Mgat5 or Pdia3 knockout (n = 3 for each group). Unpaired t test was used for assess significance. * p < 0.05. Data are shown as mean ± s.e.m., plus individual data points on the bar graph.
(d) Representative IVIS images. In vivo imaging illustrate that all mouse brains had a growing tumor at day 12. The luciferase imaging was performed every 2 days using an IVIS system. The tumor growth rate significantly slowed down after injecting T cells infected with AAV- sgMgat5 or AAV-sgPdia3 virus compared with the AAV-Vector group. Data was collected from one independent experiment, each group included 5–8 mice.
(e) Survival plots of mice treated with T cells. Survival significance was assessed by a log-rank Mantel-Cox test. DPI, days post tumor implantation.
(f) (Top) A time line for tumor induction, T cell i.c injection, and imaging for therapeutic efficacy testing of AAV-SB-CRISPR targeting Pdia3, Mgat5, and combination in CD8+ T cells in a syngeneic mouse model of GBM. C57BL/6J mice were implanted intracranially with 2 × 105 GL261-FLuc cancer cells on day 0. In vivo imaging was performed at day 14 before T cell injection for randomization with tumor-burden matched subgrouping. 1.5 × 106 T cells were injected intracranially at the same coordinate as tumor injection. The luciferase imaging was performed every 2–3 days. (Bottom) Representative IVIS images of brain tumor growth in GL261-FLuc cancer cell injected mice receiving i.c. injection of T cells infected with AAV-Vector, AAV-sgMgat5 and AAV-sgPdia3 virus groups. Data was collected from one independent experiment, each group with 7–8 mice.
(g) Survival plot of mice treated with T cells. Overall survival significance was assessed by a log-rank Mantel-Cox test between Vector and mutant groups. Comparison between groups, Log-Rank test. DPI, days post tumor implantation.
(h) Whole brain section H&E staining of four long-term survivor mice. Scale bar, 2 mm for whole brain sections. Data was collected from one independent experiment, survivor mice were from the same experiment as in f-g.
The p-values and number of mice used in each group are indicated in the plots and/or in a supplemental excel table.
Validating efficacy of Pdia3 and Mgat5 perturbation in CD8+ T cells using independent models
While i.v. delivery offers simplicity of infusion, it encounters hurdles such as lack of local concentration, or systemic side effect. Intracranial infusion is a local delivery that avoids potential systemic toxicity, although naturally along with different complications or caveat. Because GBM therapy naturally involves surgical procedures, and intracranial T cell therapy has entered clinical trials for recurrent GBM (). Bearing in mind its advantages and caveats, we tested intracranial adoptive transfer of the CRISPR-perturbed T cells. We injected 5e5 GL261-FLuc cells via intracranial procedure to induce tumor (Fig. 2a). Because of the complete penetrance of this model (Fig. S3a–b), it is expected that all mice will develop brain tumors with this amount of cells injected. 14 days post brain tumor challenge, we performed intracranial injection of AAV-sgMgat5 or AAV-sgPdia3 virus infected CD8+ T cells at 1:1 initial seeding ratio (cancer cell : T cell) into the LV of tumor bearing mouse brains (Fig. 2a). Luciferase imaging showed disease progression (Fig. 2d; Fig. S7a), and that mice receiving Mgat5 or Pdia3 knockout CD8+ T cells had significantly improved overall survival (Fig. 2e). We repeated the intracranial adoptive T cell transfer experiment with a lower cancer cell : T cell initial seeding ratio (1:7.5) (Fig. 2f). Of note, because of the variable kinetics of the number of animals deemed for euthanasia, we noted that the quantifications are not comparable between groups at later time points when a number of mice were euthanized (Fig. S7b). All mice receiving AAV-Vector infected CD8+ T cells quickly reached survival endpoints due to rapid GBM progression (Fig. 2f–g), whereas all three AAV-CRISPR CD8+ T cell perturbation groups had significantly prolonged survival (AAV-sgMgat5, AAV-sgPdia3, or AAV-sgMgat5+AAV-sgPdia3), with a fraction of mice becoming tumor-free and having long-term survival (Fig. 2f–g). We examined the brain of long-term survivor mice by histology at 8 months (approximately 240 days) post injection and found that their brains were indeed tumor-free (Fig. 2h). These data suggested that single gene AAV-CRISPR perturbation of Mgat5, Pdia3, or their combination enhanced the efficacy of intracranial adoptive T cell transfer against GBM.
Granzymes and interferon gamma (Ifnγ) as effector molecules for the enhanced anti-tumor activity of Pdia3 mutant CD8 T cells
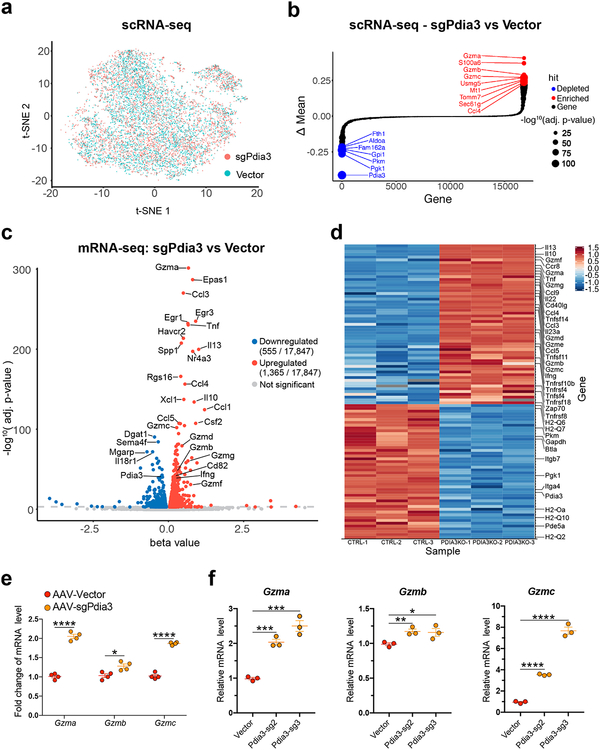
We performed single cell RNA sequencing (scRNA-seq) and profiled the transcriptomes of a total of 9,193 single CD8 T cells (Fig. 3a; Fig. S8a) (Dataset S3). Comparing AAV-sgPdia3 to AAV-Vector treated groups showed that Pdia3 was dramatically and significantly downregulated (Fig. 3b; Fig. S8b), indicating a clear on-target effect. To our surprise, multiple effector cytokines are significantly upregulated after Pdia3 knockout. The top 5 upregulated genes are Granzyme a (Gzma), S100a6, Gzmb, Gzmc, and Usmg5 (Fig. 3b; Fig. S8b), which implied that granzyme family upregulation may account for the Pdia3 mutant CD8+ T cells’ augmented ability to kill tumor cells.
Figure 3. Single-cell RNA-seq and bulk mRNA-seq analysis of Pdia3 knockout in CD8+ T cells.
(a) t-SNE plot of sample distribution based on the transcriptome of 9,193 single cells from AAV-sgPdia3 and AAV-Vector treated CD8+ T cells.
(b) Bubble-rank plot of differential gene expression of scRNA-seq. Delta-mean is the difference of mean expression value between AAV-sgPdia3 and AAV-Vector treated single CD8+ T cells (n = 3 each group). Differential expression: Two-sided Wilcoxon signed-rank test by gene, with p-values adjusted by Benjamini & Hochberg. Statistical significance is scaled by –log10, p-value as shown in the size key.
(c) A volcano plot of all differentially expressed genes between AAV-Vector and AAV-sgPdia3 transduced mouse primary CD8+ T cells (n = 3 biological replicates). Differential gene expression was performed with Sleuth using Wald test, the FDR adjusted q-value was used for the plot.
(d) Heatmap of representative immune-related differentially expressed genes between AAV-Vector and AAV-sgPdia3 transduced mouse primary CD8+ T cells (n = 3 biological replicates).
(e) RT-qPCR validation of the scRNA-seq and bulk mRNA-seq results confirmed the upregulation of granzyme genes upon AAV-sgPdia3 perturbation (n = 4). Unpaired t test, two-tailed. * p < 0.05, **** p < 0.0001.
(f) RT-qPCR validation of scRNA-seq and bulk mRNA-seq results using two independent Pdia3 sgRNAs (n =3). Unpaired t test, two-tailed. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
The p-values and number of mice used in each group are indicated in the plots and/or in a supplemental excel table.
To show the bulk differences while minimizing other heterogeneous effects, we also performed transcriptome profiling of AAV-sgPdia3 and AAV-Vector treated CD8+ T cells by bulk mRNAseq (Methods) (Dataset S4). Differential expression analysis revealed a striking set of differentially expressed genes between Pdia3 knockout and control CD8+ T cells, with 1,365 genes upregulated and 555 genes downregulated at FDR-adjusted q value < 0.001 (Fig. 3c). Gene set and pathway analysis revealed a strong signature of T cell effector and pro-inflamatory immune gene upregulation upon Pdia3 knockout in CD8+ T cells (Fig. 3c–d; Dataset S4). These include genes encoding T cell effector cytokines (Gzma / b / c / e / f / g, Perforin, Tnf, Ifng), inflammatory cytokines and their receptors (IL-10 / 13 / 22 / 23a, Ccl-1 / 3 / 5 / 9, Ccr8), as well as costimulatory or activation receptors and their ligands (CD40L, Tnfsf4/OX40L, Tnfsf14/Light, Tnfrsf4/OX40R, Tnfrsf18/GITR, Tnfrsf10b/TRAIL) (Fig. 4d). These transcriptome profiles showed that Pdia3 knockout enhanced effector phenotypes in CD8+ T cells.
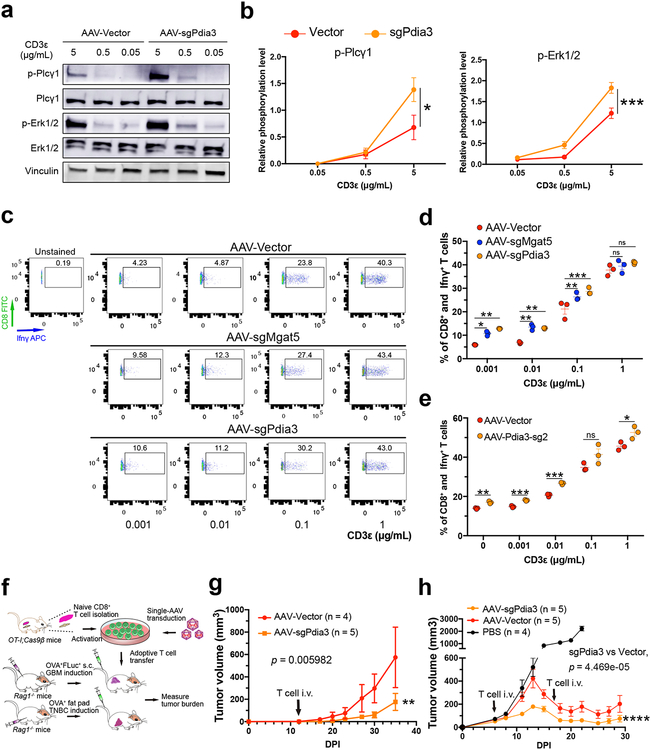
Figure 4. Mechanistic analysis and pre-clinical efficacy testing of Pdia3 knockout in CD8+ T cells.
(a-b) Dose-dependent TCR signaling experiment for Pdia3 KO showing upregulation of the phosphorylation level of Plcγ and Erk1/2. (a) Original western blot gel of a representative experiment among the three independent replicate experiments. (b) Quantification of relative phosphorylation level of Plcγ and Erk1/2, (n = 3). Data are shown as mean ± s.e.m.. Two-way ANOVA, sgPdia3 vs vector, * p < 0.05, *** p < 0.001.
(c-e) Intracellular flow cytometry was performed to detect the expression levels of Ifnγ. The OT-I;Cas9β CD8+ T cells were infected with AAV-Vector and AAV-sgMgat5 or AAV-sgPdia3 after isolation. Before Ifnγ detection assay, T cells were rested for 12 h, then reactivated with different concentration of anti-CD3ε for 4 h. (c) Flow cytometry results suggested that Mgat5 or Pdia3 knockout significantly improved T cell sensitivity to the low concentration anti-CD3ε and secreted more Ifnγ. (d) The quantification result of (c). (e) Ifnγ intracellular staining after Pdia3 KO using a different sgRNA. Two-sided multiple t test was used to assess the significance, Holm-Sidak method was used for multiple comparisons correction. * p < 0.05, ** p < 0.01, *** p < 0.001, ns, not significant.
(f) Schematic of the therapeutic efficacy testing strategy for Pdia3 knockout T cell using a subcutaneous model of GBM and a syngeneic triple-negative breast cancer (TNBC) model.
(g) Tumor growth curves of GL261-FLuc-mCh-cOVA tumor bearing mice receiving T cells infected with AAV-Vector (n = 4) or AAV-sgPdia3 (n = 5). Wilcoxon rank sum test with continuity correction, two sided, p = 0.005982. DPI, days post tumor implantation.
(h) Tumor growth curves of E0771-mCh-cOVA TNBC bearing mice receiving CD8+ T cell therapy. Wilcox test, two sided, using only data points on or after T cell adoptive transfer: AAV-Vector vs. AAV-sgPdia3, p < 0.001. DPI, days post tumor implantation.
The p-values and number of mice used in each group are indicated in the plots and/or in a supplemental excel table.
We performed RT-qPCR to validate the RNA-seq results, which confirmed the upregulation of granzyme genes upon AAV-sgPdia3 perturbation (Fig. 3e). To further exclude off-target factors, we validated the result using two independent sgRNAs targeting different regions of the Pdia3 gene (Fig. 3f). It was shown that Mgat5-deficient T cells augment phosphorylation of Plcγ and Erk, critical molecules required for T cell activation19–21. We investigated T cell signaling pathways upon Pdia3 perturbation. Quantification of immunoblots showed that the phosphorylation of Plcγ and Erk1/2 was significantly upregulated across a dose-dependent anti-CD3ε stimulation (Fig. 4a–b). In concordance with the more sensitive TCR signaling pathway, intracellular flow cytometry experiments revealed that Ifnγ production was upregulated in AAV-sgPdia3 infected CD8 T cells, which secreted more Ifnγ with low anti-CD3ε stimulation (Fig. 4c, d). This was again validated using an independent sgRNA targeting Pdia3 (Fig. 4f). Collectively, these data suggested that inhibition of Pdia3 led to upregulation of granzyme gene expression and a more sensitive induction threshold for TCR signaling and Ifnγ production.
Pre-clinical anti-tumor efficacy testing of Pdia3 editing in CD8+ T cell using independent tumor models of immunotherapy
Because granzymes and Ifnγ are T cell intrinsic properties, we speculated that Pdia3 editing could also have anti-tumor effects in other models. We performed experiments using two independent models of antigen-specific orthotopic tumor immunotherapy. First, we induced GL261 tumors with subcutaneous injection, and treated the tumor-bearing mice by adoptive transfer of mutant CD8+ T cells via intravenous injection (Fig. 4f). Knocking out Pdia3 using AAV-sgPdia3 in CD8+ T cells significantly enhanced the anti-tumor effect (Fig. 4g). Second, we used a different cancer cell line, E077122, to induce syngeneic orthotopic triple-negative breast cancer (TNBC) via mammary fat pad injection, and treated the tumor-bearing mice by T cell adoptive transfer (Fig. 4f, h). PBS-treated TNBC grew aggressively to >2000 cubic-mm in 3 weeks, Vector T cell adoptive transfer controlled the tumor growth, and Pdia3 knockout in T cells further enhanced the efficacy (Fig. 4h). Together, these data suggest that the enhanced anti-tumor activity of Pdia3 perturbation encompasses, at least in part, T cell intrinsic phenotypes.
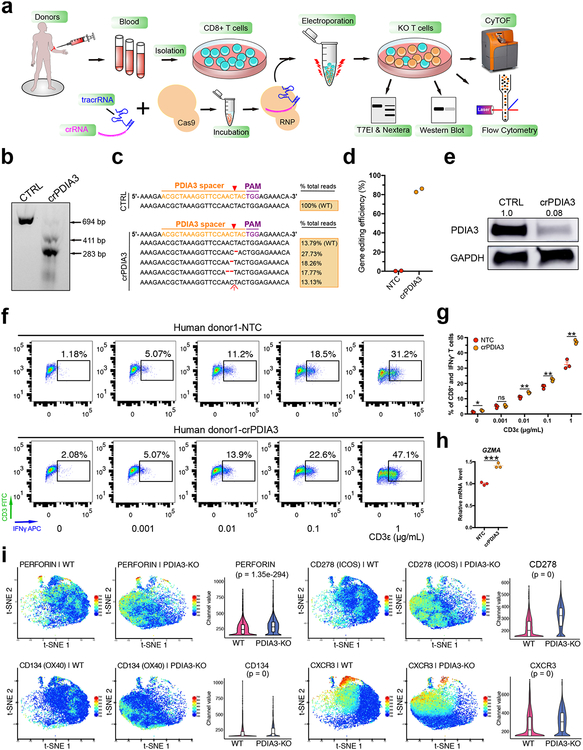
Mass Cytometry (CyTOF) analysis of PDIA3 knockout in human CD8+ T cell
To investigate PDIA3 in human CD8+ T cells, we generated high-efficiency editing of PDIA3 in human CD8+ T cells using Cas9 ribonucleoprotein (RNP) (Fig. 5a–e). DNA level analyses by surveyor and Nextera-NGS demonstrated PDIA3 knockout was highly efficient (86%) (Fig. 5b–d). Western blot using an antibody specific to human PDIA3 showed that 92% protein level knockdown in crPDIA3 treated T cells (Fig. 5e). We performed anti-CD3 dose-dependent analysis of IFNγ production and found that PDIA3 edited CD8+ T cells had significantly higher IFNγ (Fig. 5f–g). RT-qPCR analysis of human GZMA also showed upregulation upon PDIA3 loss (Fig. 5h).
Figure 5. Human CD8+ T cells PDIA3 knockout and effector function analysis.
(a) Schematics of human CD8+ T cell isolation, culture, RNP electroporation, T7EI assay, Nextera sequencing, Flow cytometry and CyTOF analysis.
(b) T7EI assay showed human PDIA3 knockout with a high efficiency compared with control. Arrows pointed to pre- and post- cleavage products of predicted sizes. Data shown are representative of three independent experiments.
(c) Nextera data quantification of gene editing efficiency of (b).
(d) Quantification of Nextera data (n = 2 each).
(e) Western blot for PDIA3 change in protein level upon CRISPR knockout. Data from one experiment.
(f) IFNγ intracellular staining after PDIA3 KO.
(g) Quantification of (f). Two-sided multiple t test was used to assess the significance. Holm-Sidak method was used for multiple comparisons correction. * p < 0.05, ** p < 0.01, ns, not significant. The p-values and number of mice used in each group are indicated in the plots and/or in a supplemental excel table.
(h) qPCR validation of GZMA expression. Unpaired t test, two-tailed. *** p < 0.001.
(i) t-SNE plots of representative markers detected by the CyTOF. Perforin, two co-stimulatory molecules (CD134/OX40 and CD278/ICOS) and CXCR3 were found to be significantly upregulated at the single cell level upon PDIA3 KO (n = 3 replicates each, sampled 7,000 cells per replicate for comparison). Violin plots were used for visualizing marker levels quantitatively in single cells. Violins show kernel probability density on side, and boxplot is standard, i.e. middle band is median, hinges/ends of box are interquartile range (25% and 75% quantiles), lower whisker = smallest observation greater than or equal to lower hinge - 1.5 * IQR, upper whisker = largest observation less than or equal to upper hinge + 1.5 * IQR. Wilcoxon test, two-sided, p value adjusted by Benjamini & Hochberg method. KO vs WT, PERFORIN, p = 1.35e-294; CD278, p = 0 (below algorithm detection limit); CD134, p = 0; CXCR3, p = 0.
Next, we performed mass cytometry (CyTOF) and profiled multiple immune markers in a total of 227,848 single cells (Fig. S9a–b), which mapped the high-dimensional landscapes of multiple immune checkpoints and other functional molecules in PDIA3 knockout and wildtype human CD8+ T cells. Clustering analysis showed that the 3 PDIA3-KO samples clustered together and were distinct from the 3 wildtype samples (Fig. S9a–b), revealing the consistency of datasets. Perforin, co-stimulation markers OX40/CD134 and ICOS/CD278, as well as CXCR3, were significantly upregulated in PDIA3 knockout T cells (Fig. 5i; Fig. S9c). 4–1BB/CD137 and IL7R/CD127 were moderately altered, but not Fas/CD95 (Fig. S9c). Interestingly, TIM3 was significantly upregulated in PDIA3 knockout, consistent with mRNA-seq (Fig. S9c). Together, these data showed PDIA3 CRISPR editing influenced the surface expression of multiple of immune regulators and effectors in human CD8+ T cells.
Analysis of for PDIA3’s cytotoxic T lymphocyte signature with human clinical data
To investigate whether PDIA3 expression was clinically relevant, we performed patient data analysis using Tumor Immune Dysfunction and Exclusion (TIDE) algorithm (Methods). PDIA3 expression has strong signatures in cytotoxic T lymphocyte (CTL) dysfunction, where PDIA3-low patient groups have CTL-associated overall survival benefits across multiple cancer types including GBM, TNBC, and lung adenocarcinoma, while high-levels of PDIA3 abolishes or weakens the overall survival benefit of CTL-high patients (Fig. S10a–c). In melanoma patients treated with immune checkpoint blockade antibodies, PDIA3-high patients had significantly poorer survival (Fig. S10d), although checkpoint antibodies are not yet commonly used for treating GBM. These data pointed to the significance of clinical association of PDIA3 with T cell dysfunction in human cancer.
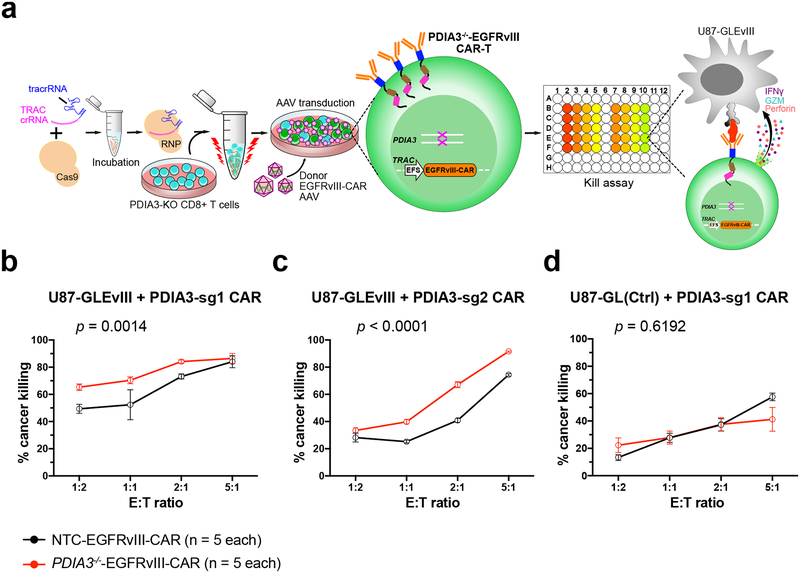
PDIA3 engineering enhanced CAR-T killing of human EGFRvIII+ GBM cells
To further establish PDIA3 as an immunotherapy target of T cell engineering, especially against GBM, we established PDIA3 mutant and control human EGFRvIII CAR-T cells by Cas9-RNP mediated gene editing of primary CD8+ T cells, along with AAV donor mediated knockin of an EGFRvIII CAR-T cassette into the TCR Alpha Constant chain (TRAC) locus (Methods) (Fig. 6a). We also generated an EGFRvIII-antigen expressing U87 GBM cell line (U87-Luc-EGFRvIII). We then performed CAR-T cell : cancer cell co-culture assays to test the cytolytic (killing) activity. PDIA3 knockout compared to wildtype EGFRvIII CAR-T cells had significantly higher killing ability against the cognate U87-Luc-EGFRvIII cells (Fig. 6b). This was confirmed using an independent sgRNA targeting PDIA3 (Fig. 6c), further minimizing the probability of off-target. The killing ability of CAR-T is not different towards the parental U87 cells without EGFRvIII antigen (Fig. 6d), supporting CAR-T’s antigen-specificity. These data together demonstrate that the major effect of PDIA3 knockout in EGFRvIII CAR-T cells is dependent on CAR-antigen recognition, and minimally due to TCR off target or gene editing off-target effects.
Figure 6. Human PDIA3−/−-EGFRvIII CAR-T cell establishment and GBM cell killing.
(a) Schematics of human PDIA3−/−-EGFRvIII CAR-T cell generation. CD8 T cells were electroporated with crPDIA3:tracRNA:Cas9 first, then PDIA3−/− T cells were knock-in (KI) with an EGFRvIII-CAR construct which consists of TRAC locus homology-directed repair (HDR) 5’ and 3’ arms, an EFS promoter, an EGFRvIII-CAR expression cassette, and a short polyA. The donor KI constructs were packaged into AAV6, then introduced into T cells by viral transduction after TRAC first-exon targeting RNP electroporation. U87-GFP-Luc-EGFRvIII (U87-GLEvIII) and PDIA3−/−-EGFRvIII CAR-T cell co-culture assay was set up after CAR-T cells were established to test PDIA3−/−-EGFRvIII CAR-T cell killing ability.
(b-d) Kill assay of NTC-EGFRvIII-CAR and PDIA3−/−-EGFRvIII-CAR T cells with U87-GLEvIII and U87-GL (parental line control) human GBM cells, with a titration series of Effector : Target (E:T) ratios at 24h post co-culture:
(b) Kill assay with PDIA3-sg1, on U87-GLEvIII cells;
(c) Kill assay with PDIA3-sg2, on U87-GLEvIII cells;
(d) Kill assay with PDIA3-sg1, on U87-GL parental control cells;
Data are shown as mean ± s.e.m., plus individual data points, n = 5 biological replicates. Two-way ANOVA test was used to evaluated significance. The p-values and number of mice used in each group are indicated in the plots and/or in a supplemental excel table.
Discussion
There are only 4 approved drugs for GBM to date, none involve immunotherapy. The brain is highly immune-privileged, with the exception of the meningeal lymphatic system23. Recent studies demonstrated active adaptive immune cell trafficking in the brain in the arachnoid meninges and dura, leptomeninges, cerebrospinal fluid (CSF), and the CNS parenchyma24. The active brain surveillance by the adaptive immune system may provide a new window to identify targets that modulate T cell function against GBM. The U.S. Food and Drug Administration (FDA) approved checkpoint antibodies against PD-1/PD-L1 and CTLA-4 for various cancer types1, 5, 25, 26. Although clinical trials with checkpoint inhibitors have entered the GBM clinic both as mono-therapy and as combinations27, the efficacy and survival benefits are limited, except in a recent trial using anti-PD1 in a neoadjuvant setting28. GBM typically has low mutational load and T cell infiltration, possibly making it less responsive to checkpoint blockade29. Anti-PD-1 antibody (Nivolumab) did not improve overall survival compared with bevacizumab in patients with recurrent GBM in a recent trial30. Combination of Nivolumab and Ipilimumab (anti-CTLA-4) treatment resulted in severe toxicity in 50% of GBM patients6, leading to discontinuation. Clinical trials with LAG-3 mAbs either alone or in combinations are ongoing (, ). Several preclinical studies showed promises with various other forms of immunotherapies in GBM, such as GM-CSF vaccination and CTLA-4 blockade31,32, local chemotherapy with anti-PD-133, a triple combination of anti-CTLA-4, anti-PD-1, and oHSV G47Δ expressing murine IL-12 (G47Δ-mIL12)34, as well as dual PD-1 and TIM-3 blockade with radiation35. In summary, limited clinical efficacy and high toxicity remain unresolved problems for combinatorial checkpoint blockade immunotherapy in GBM.
CAR-T cell therapy is recently approved by FDA for refractory pre-B cell acute lymphoblastic leukemia and diffuse large B cell lymphoma. However, in solid tumours, currently available CAR-Ts alone most often showed insufficient single agent activity36. EGFR-vIII CAR-T has completed several clinical trials, yet have shown little overall survival benefit for GBM patients7. Intracranial infusion of CAR-T cells targeting IL-13Rα2 against GBM initially showed tumor regression although with ultimate recurrence37. A ten patient trial with recurrent EGFRvIII-positive GBM receiving a single intravenous infusion of autologous anti-EGFRvIII CAR-T cells did not improve overall survival38. Recent pre-clinical studies developed a number of promising GBM CARs, such as those targeting chondroitin sulfate proteoglycan (CSPG4)39, the disialoganglioside GD240. Therefore, improvement of CAR-T efficacy in GBM and solid tumors are critically needed. AAV-SB-CRISPR screens may facilitate rapid identification of novel factors modulating T cell function provide direct targets to enhance CAR-T efficacy. Our co-culture model showed that PDIA3 editing can enhance the killing ability of human CAR-T against EGFRvIII+ GBM cells, while in vivo efficacy has yet to be determined.
Functional genetic screens can discover diagnostic markers and therapeutic targets41. A study using an immortalized T cell line screened for regulators of PD-1 expression42. In vitro screens were reported in primary human T cells43, 44. Genetic screens of human T cells in tumor models require immunodeficient mice. Screening in primary murine T cells offers the advantage of unbiased discovery of genes that modulate immune function in immunocompetent settings using syngeneic tumor models. RNAi T cell screens identified genes for T cell infiltration and cytokine production45, 46. CRISPR-based approaches demonstrated superior performance in head-to-head comparisons47 such as higher consistency, lower off-target rate, and fewer false-negatives. We developed a new hybrid AAV-SB-CRISPR system, which combines the power of AAV for transduction and transposon for genomic integration, enabling efficient genome editing of murine T cells and opens up large-scale knockout screens in vivo. We screened a focused membrane proteome in CD8+ T cells in syngeneic models of GBM in immunocompetent mice, identified and validated previously uncharacterized targets across several GBM models. Among top hits, Lag3 encodes an important cell surface immune checkpoint. Mgat5 has been shown to negatively regulate of T-cell activation48 and Mgat5 knockout mice are tumor resistant49. Scoring these genes benchmarked the success of the screens. We validated the pre-clinical efficacy of Lag3 and Mgat5 in this study in a GBM model, together with Pdia3 and Emp1, which have not been previously investigated in T cells or GBM. We performed studies with both immunocompetent B6 and antigen-specific OT-I / Rag1−/− settings, carried out adoptive transfer via i.v. as well as intracranial approaches, although these were not in head-to-head comparisons. Nevertheless, these experiments demonstrate in a convergent manner that genetic perturbation of these targets enhanced anti-GBM efficacy. Our characterization experiments provided data on the previously unknown roles of PDIA3 in immune regulation and immunooncology relevant phenotypes in CD8 T cells.
Finally, AAV-SB-CRISPR-mediated in vivo screens of membrane protein coding genes is advantageous because the hits can serve as immunooncology targets either via mAbs, or via direct T cell engineering. While therapeutic-grade mAbs may require multiple years of effort, direct T cell engineering can enhance the efficacy of cell therapies such as CAR-T, TCR-T and TIL adoptive transfer. Albeit with limitations, targeting PDIA3 and other genes via genome-engineering of T cells also avoid toxicity issues due to target expression in other cell types. CAR-T cell therapy faces various challenges including persistence in vivo, resistance to immunosuppression, and exhaustion, all of which are critical for in vivo efficacy and potential clinical success. Genetic screens in CD4 T cells or other immune cell types may be performed similarly in the future. Direct editing or pharmacological perturbation of PDIA3 or other targets identified from these screens may provide different routes to improve T cell based immunotherapy for GBM and potentially more broadly for other difficult-to-treat cancer types.
Online Methods
Institutional Approval
This study has received institutional regulatory approval. All recombinant DNA work was performed under the guidelines of Yale Environment, Health and Safety (EHS) Committee with an approved protocol (Chen-rDNA-15–45). All animal work was performed under the guidelines of Yale University Institutional Animal Care and Use Committee (IACUC) with approved protocols (Chen-2015–20068 and Chen-2018–20068). All human sample work was performed under the guidelines of Yale University Institutional Review Board (IRB) with an approved protocol (HIC#2000020784). This study uses existing de-identified human samples and data under NIH Exemption 4.
Mice
Rosa26-Cas9–2A-EGFP constitutive expressed mice (Cas9β mice), OT-I TCR transgenic mice14, Rag1−/− and C57BL/6J mice were used in this study. For OT-I;Cas9β mice, which were generated by breeding OT-I and Cas9β mice. OT-I;Cas9β and Cas9β mice, both female and male, aged 8–12 weeks were used for naïve CD8+ T cell isolation. For the lateral ventricle (LV) injection, 8 week-old Female C57BL/6J or 7–9 week-old Rag1−/− mice were used. Mice were randomly classified into different groups.
Design and synthesis of membrane bound protein CRISPR knockout library
The GO term GO0009986 was chosen to focus on a set of genes associated with the term “cell surface”. A total of 1,657 membrane bound protein coding genes were selected. Four sgRNAs were chosen per gene similar to the mBrie library design50, given a total of 6,628 sgRNAs. A total of 1,000 non-targeting controls (NTCs) were spiked into the library, making the Surf library a total of 7,628 sgRNAs in size (Table S1). This membrane bound protein gene targeting single-strand RNA (sgRNA) library was named Surf. The membrane bound protein library was synthesized by massively parallel oligo array synthesis and pooled (CustomArray).
Generation of AAV-CRISPR vector and AAV-Surf library for primary T cell editing and screening
A hybrid AAV-SB-CRISPR vector for targeting primary mouse T cells (AAV-SB100x) was constructed by gBlock fragments (IDT) followed by Gibson assembly (NEB). Synthesized library was firstly PCR amplified, then cloned sgRNAs into double Bbs I sites of AAV-CRISPR vector by the Gibson assembly (NEB). The Gibson assembly products were transformed into high efficiency competent cells (Endura) by electroporation methods. An estimated library coverage of ≥ 60 x was observed after electroporation. The cloned library was PCR amplified using barcoded primers to ensure proper representation. The cloned library was named AAV-Surf.
AAV production
AAV-SB100x plasmid cloned with library or single sgRNA was packaged similarly to our previously described approach8. Details in supplementary notes.
Cell culture for cell lines and primary T cells
HEK293FT, U87, GL261, and E0771 cell lines were cultured in D10 medium. Mouse naïve CD8+ T cells were cultured in RPMI-1640 (Gibico) medium supplemented with 10% FBS, 2 mM L-Glutamine, 200 U/mL penicillin-streptomycin (Gibico), and 49 μM β-mercaptoethanol (Sigma). Details in supplementary notes.
Generation of stable cell lines
Stable cell lines GL261-FLuc-mCh-cOVA, U87-GFP-Luc-EGFRvIII (U87-GLEvIII) and U87-GFP-Luc (U87-GL) were generated by a combination of procedures including viral transduction, antibiotics selection, FACS sorting, and/or single cell cloning. Flow cytometry was performed again after stable cell lines were established to ensure purity. Details in supplementary notes.
Splinkerette PCR
Sleeping beauty transposon integration was detected by splinkerette PCR51. Splink 1 and SB-Right1 primers (Table S10) were used for 1st round PCR, Splink 2 and SB-Right 2 primers (Table S10) were used for 2nd round PCR. PCR products were prepared using a Nextera kit (Illumina) and sequenced, then analyzed by custom codes. Details in supplementary notes.
GBM induction by intracranial surgery and cancer cell transplantation
Same gender mice were used in each batch of experiments. Mice were anesthetized by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg). They were also administered carprofen (5 mg/kg) intraperitoneally as a pre-emptive analgesic. Once mice were in deep anesthesia, they were immobilized in a stereotaxic apparatus (Kopf or Stoelting) using intra-aural positioning studs and a tooth bar to immobilize the skull, similar to a previous method8. The lateral ventricle (LV) was targeted according to the mouse brain stereotaxic coordinates, approximately at 0.6–1.0 mm caudal/posterior to bregma, 0.8–1.5 mm right-side lateral to bregma and 2.0–3.0 mm deep from the pial surface for injection (coordinates: A/P −0.6 to −1.0, M/L 0.8 to 1.5, D/V −2.0 to −3.0). A ~1 mm hole was drilled on the skull surface, via which 1 × 105 to 1.2 × 106 cancer cells were injected into LV with a volume of 4–8 μL, dependent on specific experiments. The injection rate was controlled at 2 μL/min by an UltraMicroPump 3 (World Precision Instruments). After injection, the incision was closed with tissue adhesive (3 M Vetbond) and subcutaneously injected 500 μL lactated Ringer’s solution. Mice were placed under the heat lamp until they recovered. Of note, in certain cases, injection of cells into LV may cause leptomeningeal growth instead of glioblastoma. In this study, all GBMs that grew from these experiments that were examined by histology represented a single tumor in each mouse that expanded on the site of injection in a relatively even manner, which also pathologically resembles malignant glioma.
AAV-Surf CD8+ T cell screen in a syngeneic mouse model of GBM
Naïve CD8+ T cells were isolated from the spleen and lymph modes of Cas9+ mice. A total of 2 × 107 Naïve OT-I;Cas9β or Cas9β CD8+ T cells were transduced with 1011 AAV-Surf virus. Syngeneic mouse models of GBM were setup with intracranial injection of native or luciferase-expressing GL261 cells (GL261 and GL261-Luc, respectively) transplanted into the lateral ventricle (LV) of C57BL/6J mice. AAV-Surf infected CD8+ T cells were adoptively cell transfer of into GBM engrafted mice via intravenous (tail vein) injection. Three screens were performed. The one with native GL261 GBM (injected with OT-I;Cas9β T cells) reached endpoint sooner (all mice euthanized by 20 dpi, “shorter term screen”); The one with GL261-Luc GBM (injected with OT-I;Cas9β T cells) reached endpoint later (all mice euthanized by 92 dpi, “longer term screen”); The third screen with GL261 GBM (injected with Cas9β T cells) reached endpoints with a duration between the first two (all mice euthanized by 26 dpi, “medium term screen”). With transduction at this level, single cells with functional sgRNA expression is estimated at 48%, which gives a functional MOI estimate to be 0.65, In the screen, we euthanize the mice at a synchronized time point, or at the endpoint when the body condition score < 2 if the animal does not survive to the synchronized endpoint (per Yale IACUC protocol). We performed 3 independent screens, “medium-term” used 7 mice, “short-term” used 3 mice, and “long-term” used 9 mice for library readout. The number of TILs recovered per brain averages at 5e4 from AAV-Vector mouse and 1.25e5 from AAV-Surf mouse.
Adoptive cell transfer (ACT)
Naïve CD8+ T cells were infected with virus after isolation, and cultured for 3–5 days before intravenous injection. For the shorter term AAV-Surf screen, 1.8 × 106 OT-I;Cas9β CD8+ T cells were injected. For the longer term screen, 4 × 106 OT-I;Cas9β CD8+ T cells were injected. In the Cas9β CD8+ T cell screen, 3 × 106 Cas9β CD8+ T cells were intravenously injected after 10 days of tumor engraftment. For the validation experiments, OT-I;Cas9β or Cas9β CD8+ T cells were injected, where cell number of cancer cells and T cells injected were illustrated in figures and legends. Adoptive transfer was also performed using an intracranial approach in validation experiments. Noted that these two methods of T cell infusion were not performed head-to-head. After T cell injection, mice were monitored every day. All animals are deemed for euthanasia when they developed macrocephaly, poor body condition or other euthanasia criteria according to the approved animal protocol. Brains were isolated and stored at −80 °C for genomic DNA extraction and readout, or fixed in 4% PFA for hematoxylin and eosin (H&E) staining.
SgRNA readout and deep sequencing
Two rounds of PCR reactions were used for the sgRNA library readout. PCR primers were provided (Table S1). Details in supplementary notes.
AAV-SB-CRISPR screen data processing
Raw single-end fastq read files were filtered and demultiplexed using Cutadapt52. To remove extra sequences downstream (i.e. 3’ end) of the sgRNA spacer sequences, the following settings were used: cutadapt --discard-untrimmed -a GTTTTAGAGCTAGAAATGGC. As the forward PCR primers used to readout sgRNA representation were designed to have a variety of barcodes to facilitate multiplexed sequencing, these filtered reads were then demultiplexed with the following settings: cutadapt -g file:fbc.fasta --no-trim, where fbc.fasta contained the 12 possible barcode sequences within the forward primers. Finally, to remove extra sequences upstream (i.e. 5’ end) of the sgRNA spacers, the following settings were used: cutadapt --discard-untrimmed –g GTGGAAAGGACGAAACACCG. Through this procedure, the raw fastq read files could be pared down to the 20 bp sgRNA spacer sequences. The 20 bp sgRNA spacer sequences from each demulitplexed sample were then mapped the designed sgRNA spacers in the Surface library (Table S1). A bowtie index of the sgRNA library was generated using the bowtie-build command in Bowtie 1.1.253. The filtered fastq read files were mapped to the index using the following settings: bowtie -v 1 --suppress 4,5,6,7 --chunkmbs 2000 –best. Using the resultant mapping output, the number of reads that had mapped to each sgRNA within the library was quantified.
Analysis of CRISPR screens using RIGER
For RIGER analysis of CRISPR screens, read count tables were used to calculate log fold changes for tumor versus cell samples in order to score and rank sgRNAs, with ties in rank broken by random order. This data was then used as input to a Java-based implementation of RIGER (https://github.com/broadinstitute/rigerj) in order to generate p-values and gene rankings based on consistent enrichment across multiple sgRNAs for identification of candidate genes47. Both the second highest-ranking sgRNA and the weighted sum scoring methods were used for computation of gene rankings, and compared to ensure consistency between methods.
T cell adoptive transfer with a subcutaneous tumor models
GL261-FLuc-mCh-rOVA cells and E0771-mCh-rOVA cells were subcutaneously injected into male Rag1−/− mice for modeling GBM and TNBC, respectively. Adoptive transfer of T cells were performed similarly as above. Details in supplementary notes.
Mouse brain tumor studies
Mouse brain tumor monitoring, IVIS imaging, brain dissection and histology were performed using standard tumor study procedures. Details in supplementary notes.
Flow cytometry
Flow cytometry were performed using standard immunology methods. Details in supplementary notes.
Standard molecular biology
Experiments such as DNA / RNA prep, T7 endonuclease I assay (T7EI), RT-qPCR and electrophoresis were performed following standard molecular biology procotols. Primers for T7E1 were provided in (Table S7). Additional details in supplementary notes.
Detection of AAV-mediated mutagenesis by Nextera
The PCR products were used for Nextera library preparation following manufacturer protocols (Illumina). Reads were mapped to the amplicon sequences using BWA-MEM54 at default settings. Indel variants were first processed with Samtools55 with the settings samtools mpileup -d 1000000, then piped into VarScan v2.4.156 with the settings pileup2indel --min-coverage 2 --min-reads2 2 --min-var-freq 0.00001.
Human primary CD8+ T cell endogenous gene knockout
Human primary CD8+ T cells were isolated from health donors, stimulated with anti-CD3 / CD28 beads (Invitrogen) and cultured in X-VIVO™ 15 media (Lonza) supplied with 5 % human serum and IL-2. Gene knockout was performed by Cas9 RNP electroporation. Details in supplementary notes.
PDIA3−/−-EGFRvIII-CAR-T cell establishment
NTC (non-targeting control crRNA electroporated T cells) and PDIA3−/− primary CD8+ T cells were targeted with TRAC locus RNP complex, a total of ~ 6e9 viral genome copy of AAV6 HDR donor (LHA-EFS-EGFRvIII-CAR-RHA) was added into each electroporated T cell reaction (3e6 T cell / reaction) within 1 h after electroporation.
Human PDIA3−/−-EGFRvIII-CAR-T cell co-culture (kill) assay
To sensitively detect PDIA3−/−-EGFRvIII-CAR-T cell killing efficacy, U87-GL and U87-GLEvIII cell lines were established. 2e4 U87-GL or U87-GLEvIII cells were seeded in a 96-well white polystyrene plate, then different T cell : cancer cell ratio (E : T ratio) co-cultures were set up. Cancer cell killing was measured after 24 h of co-culture by adding 150 μg / mL D-Luciferin (PerkinElmer) using a multichannel pipette. Luciferase intensity was measured by a Plate Reader (PerkinElmer).
Mass cytometry (CyTOF)
High targeting efficiency of PDIA3 was confirmed by surveyor assay and Nextera sequencing. Human CD8+ T cells were collected and washed with PBS, resuspended cell to 1 × 107 / mL in PBS and add Cell-ID Cisplatin (Fluidigm) to a final concentration of 5 μM. Cells were incubated at room temperature for 5 min, then washed with Maxpar Cell Staining Buffer (Fluidigm). Each replicate was aliquoted with 2 × 106 cells in a volume of 50 μL, adding 50 μL staining buffer with surface marker antibody cocktail (Table S8) (Fluidigm or provided by the Yale CyTOF core) in each tube. The tube was gently mixed with pipette and incubated at room temperature for 30 min. Following the incubation, cells were washed with Maxpar Cell Staining Buffer two times. Cells were fixed by adding 500 μL Maxpar Fix I Buffer (Fluidigm) to each tube, and incubated for 15 min at room temperature. Cells were then washed with Maxpar Perm-S Buffer (Fluidigm) for two times. 50 μL staining buffer with cytoplasmic / secreted antibody cocktail (Table S8) was added into fixed cells which was resuspended in 50 μL Maxpar Cell Staining Buffer. Cells were incubated at room temperature for 30 min. After incubation, cells were washed with Maxpar Cell Staining Buffer for two times. Finally, cells were incubated in intercalation solution (Fluidigm) in a final concentration of 125 nM, then incubated overnight at 4 °C. Before running on a CyTOF machine, cells were washed with Maxpar Cell Staining Buffer and adjusted cell concentration to 5–7 × 105 / mL with water. All data were collected on a CyTOF Helios instrument (Fluidigm).
CyTOF data processing
CyTOF quality control pre-filtering performed by gating in FlowJo (live-dead, CD3, CD8). Channel signal values were exported as CSV and analyzed using custom scripts in R. Dimensionality reduction was performed by t-SNE (Rtsne package), followed by k-means and hierarchical clustering.
Immunoblot and TCR signaling
Immunoblots were performed with standard molecular biology methods. TCR signaling was performed using standard western with phospho-specific antibodies. Details in supplementary notes.
Single-cell RNA sequencing (scRNA-seq)
Naïve CD8+ T cells were isolated from OT-I;Cas9 mice, T cells were stimulated with anti-CD3ε and anti-CD28 as previously described. CD8+ T cells were infected with AAV6-sgPdia3 and AAV6-Vector after being activated. At day 5 after AAV infection, T cells were collected and dead cells were removed using Dead Cell Removal Kit (Miltenyi Biotec). T cells were resuspended in PBS in a concentration of 1 × 106 / mL. 10,000 CD8+ T cells per samples were used for scRNA-seq by following the protocol as 10x Genomics provided. Data processing details in supplementary notes.
scRNA-seq data processing
Read count matrices from single cell RNA sequencing samples were obtained by mapping using native 10x Cell Ranger output. Samples were pooled together into a single CSV and analyzed using custom scripts in R. Reads were pre-filtered by ribosomal and mitochondrial genes, normalized by cell per 10000 reads, then log transformed. For cell percentage quantifications, cells were first pre-filtered, in order, by Ptprc+, Cd3e+, Cd8a+, and Cd4− expression. Marker expression status on high-confidence Cd8 cells was then quantified individually for each marker of interest. Expression status for a given gene was thresholded at 0.1 normalized read value. Differential expression between sgPdia3 and AAV-vector control was performed by two-sided Wilcoxon signed-rank test by gene, with p-values adjusted by Benjamini & Hochberg. Significance was compared to differences in mean expression between populations. Dimensionality reduction was performed by t-SNE (Rtsne package), followed by k-means and hierarchical clustering. Heatmap.2 function used to show normalized gene expression for most variable genes.
Bulk mRNA-seq
Activated OT-I;Cas9β CD8+ T cells were transduced with AAV-sgPdia3 or AAV-vector virus. At day 4 after transduction, a small portion of T cells was collected for surveyor assay to ensure Pdia3 knockout before doing the RNA-seq library preparations. mRNA library preparation was performed at day 5 after virus transduction using a NEBNext® Ultra™ RNA Library Prep Kit for Illumina. Samples were multiplexed using barcoded primers provided by NEBNext® Multiplex Oligos for Illumina® (Index Primers Set 1). Libraries were sequenced with HiSeq 4000 or Novaseq systems (Illumina).
Metadata of mRNA-seq can be found in Table S9.
mRNA-seq data processing
Raw FASTQ files from mRNA-seq were analyzed for transcript quantification using Kalliso quant algorithm57 with the setting -b 100, and differential gene expression was performed with Sleuth58. Transcriptome references were obtained from Ensembl. Differentially upregulated and downregulated genes with cutoff of q-value 1e-3 were used for DAVID analysis59, and genes belonging to select enriched gene ontology terms were used for generating heatmaps. Z-scores for heatmaps were calculated on log2 normalized gene counts scaled by genes. Visualization of differentially expressed genes including volcano plot and heatmaps were performed using standard R packages including ggplot2 and VennDiagram.
Large-scale patient T cell immune signature data analysis using TIDE
The gene signatures of T cell dysfunction and prediction of cancer immunotherapy response on cancer patient data was performed using the TIDE algorithm as previously described60. Gene expression level of PDIA3 was associated to CTL-mediated patient survival with or without checkpoint blockade treatment.
Standard statistical analysis
All statistical methods are described in figure legends and/or supplementary excel tables. The p values and statistically significance were estimated for all analyses. Prism (GraphPad Software Inc.) and RStudio were used for these analyses.
Data and resource availability
Source data and statistics for non-NGS experiments such as tumor studies, flow cytometry, T7E1, qPCR, protein experiments, and co-culture assays are provided in an excel table as Supplementary Statistics “original data_v8.xlsx”. Genome sequencing data are deposited to SRA with an accession number PRJNA553676. Other data, reagents, methods, computational codes and materials that support the findings of this research are available from the corresponding author upon reasonable request.
Code availability
Custom codes used to support the findings of this research are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank Drs. Charles Fuchs, David Hafler, Ruth Montgomery, David Rimm, and Murat Gunel for discussion. We thank Stanley Lam, Li Shen, Zhigang Bai, Hanghui Ye, Ray Kim and other members in the Chen laboratory, as well as various colleagues in Department of Genetics, Systems Biology Institute, Cancer Systems Biology Center, MCGD Program, Immunobiology Program, BBS Program, Cancer Center and Stem Cell Center at Yale for assistance and/or discussion. We thank the Center for Genome Analysis, Center for Molecular Discovery, Pathology Tissue Services, Histology Services, High Performance Computing Center, West Campus Analytical Chemistry Core and West Campus Imaging Core, Mass Cytometry Core, and Keck Biotechnology Resource Laboratory at Yale, for technical support.
S.C. is supported by Yale SBI/Genetics Startup Fund, NIH/NCI (1DP2CA238295–01, 1R01CA231112–01, 1U54CA209992–8697, 1R33CA225498–01A1, 5P50CA196530-A10805, 4P50CA121974-A08306), Damon Runyon Dale Frey Award (DFS-13–15), Melanoma Research Alliance (412806, 16–003524), St-Baldrick’s Foundation (426685), Breast Cancer Alliance, Cancer Research Institute (CLIP), AACR (499395, 17–20-01-CHEN), The Mary Kay Foundation (017–81), The V Foundation (V2017–022), Ludwig Family Foundation, DoD (W81XWH-17–1-0235), Sontag Foundation, and Chenevert Family Foundation. GW is supported by CRI Irvington and RJ Anderson Fellowships. MBD, RDC and JJP are supported by the Yale MSTP training grant from NIH (T32GM007205).
Footnotes
Competing interests declaration
A patent application has been filed related to the data in this manuscript.
References
- 1.Chen DS & Mellman I Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Tumeh PC et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568-+ (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kvistborg P et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8(+) T cell response. Science Translational Medicine 6 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Ribas A Tumor Immunotherapy Directed at PD-1. New Engl J Med 366, 2517–2519 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Pardoll DM The blockade of immune checkpoints in cancer immunotherapy. Nature reviews. Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omuro A et al. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neuro-Oncology 20, 674–686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Rourke DM et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Science Translational Medicine 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow RD et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G et al. Mapping a functional cancer genome atlas of tumor suppressors in mouse liver using AAV-CRISPR-mediated direct in vivo screening. Sci Adv 4, eaao5508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mates L et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41, 753–761 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Colella P, Ronzitti G & Mingozzi F Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol Ther Methods Clin Dev 8, 87–104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D, Tai PWL & Gao GP Adeno-associated virus vector as a platform for gene therapy delivery. Nature Reviews Drug Discovery 18, 358–378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platt RJ et al. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogquist KA et al. T-Cell Receptor Antagonist Peptides Induce Positive Selection. Cell 76, 17–27 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Workman CJ, Dugger KJ & Vignali DA Cutting edge: molecular analysis of the negative regulatory function of lymphocyte activation gene-3. J Immunol 169, 5392–5395 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Workman CJ et al. Lymphocyte activation gene-3 (CD223) regulates the size of the expanding T cell population following antigen activation in vivo. J Immunol 172, 5450–5455 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Anderson AC, Joller N & Kuchroo VK Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 44, 989–1004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang CT et al. Role of LAG-3 in regulatory T cells. Immunity 21, 503–513 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Smith LK et al. Interleukin-10 Directly Inhibits CD8(+) T Cell Function by Enhancing N-Glycan Branching to Decrease Antigen Sensitivity. Immunity 48, 299–312 e295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibui A et al. Alteration of immune responses by N-acetylglucosaminyltransferase V during allergic airway inflammation. Allergol Int 60, 345–354 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Morgan R et al. N-acetylglucosaminyltransferase V (Mgat5)-mediated N-glycosylation negatively regulates Th1 cytokine production by T cells. J Immunol 173, 7200–7208 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Sugiura K & Stock CC Studies in a tumor spectrum. II. The effect of 2,4,6-triethylenimino-s-triazine on the growth of a variety of mouse and rat tumors. Cancer 5, 979–991 (1952). [DOI] [PubMed] [Google Scholar]
- 23.Louveau A et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlager C et al. Effector T-cell trafficking between the leptomeninges and the cerebrospinal fluid. Nature 530, 349-+ (2016). [DOI] [PubMed] [Google Scholar]
- 25.Khalil DN, Smith EL, Brentjens RJ & Wolchok JD The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat Rev Clin Oncol 13, 273–290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribas A et al. Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (KEYNOTE-002): a randomised, controlled, phase 2 trial. Lancet Oncol 16, 908–918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preusser M, Lim M, Hafler DA, Reardon DA & Sampson JH Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat Rev Neurol 11, 504–514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cloughesy TF et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med 25, 477–486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 17, 174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reardona DA et al. Randomized Phase 3 Study Evaluating the Efficacy and Safety of Nivolumab Vs Bevacizumab in Patients with Recurrent Glioblastoma: Checkmate 143. Neuro-Oncology 19, 21–21 (2017). [Google Scholar]
- 31.Agarwalla P, Barnard Z, Fecci P, Dranoff G & Curry WT Sequential Immunotherapy by Vaccination With GM-CSF-expressing Glioma Cells and CTLA-4 Blockade Effectively Treats Established Murine Intracranial Tumors. J Immunother 35, 385–389 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fecci PE et al. Systemic CTLA-4 blockade ameliorates glioma-induced changes to the CD4(+) T cell compartment without affecting regulatory T-cell function. Clinical Cancer Research 13, 2158–2167 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Mathios D et al. Anti-PD-1 antitumor immunity is enhanced by local and abrogated by systemic chemotherapy in GBM. Science Translational Medicine 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saha D, Martuza RL & Rabkin SD Macrophage Polarization Contributes to Glioblastoma Eradication by Combination Immunovirotherapy and Immune Checkpoint Blockade. Cancer Cell 32, 253–267 e255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JE et al. Combination Therapy with Anti-PD-1, Anti-TIM-3, and Focal Radiation Results in Regression of Murine Gliomas. Clinical Cancer Research 23, 124–136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai H, Wang Y, Lu X & Han W Chimeric Antigen Receptors Modified T-Cells for Cancer Therapy. J Natl Cancer Inst 108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown CE et al. Regression of Glioblastoma after Chimeric Antigen Receptor T-Cell Therapy. N Engl J Med 375, 2561–2569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Rourke DM et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pellegatta S et al. Constitutive and TNF alpha-inducible expression of chondroitin sulfate proteoglycan 4 in glioblastoma and neurospheres: Implications for CAR-T cell therapy (vol 10, eaao2731, 2018). Science Translational Medicine 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mount CW et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M(+) diffuse midline gliomas. Nat Med 24, 572–579 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernards R Finding effective cancer therapies through loss of function genetic screens. Current Opinion in Genetics & Development 24, 23–29 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Okada M et al. Blockage of Core Fucosylation Reduces Cell-Surface Expression of PD-1 and Promotes Anti-tumor Immune Responses of T Cells. Cell Rep 20, 1017–1028 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Shifrut E et al. Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ting PY et al. Guide Swap enables genome-scale pooled CRISPR-Cas9 screening in human primary cells. Nat Methods 15, 941–946 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Zhou P et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 506, 52–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen R et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity 41, 325–338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demetriou M, Granovsky M, Quaggin S & Dennis JW Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature 409, 733–739 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Granovsky M et al. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat Med 6, 306–312 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Doench JG et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32, 1262–1267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uren AG et al. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat Protoc 4, 789–798 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 53.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koboldt DC et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Pimentel H, Bray NL, Puente S, Melsted P & Pachter L Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Jiang P et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 24, 1550–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.